Abstract
In the last 300 thousand years, the genus Chlorocebus expanded from equatorial Africa into the southernmost latitudes of the continent, where colder climate was a probable driver of natural selection. We investigated population-level genetic variation in the mitochondrial uncoupling protein 1 (UCP1) gene region—implicated in non-shivering thermogenesis (NST)—in 73 wild savannah monkeys from three taxa representing this southern expansion (Chlorocebus pygerythrus hilgerti, Chlorocebus cynosuros and Chlorocebus pygerythrus pygerythrus) ranging from Kenya to South Africa. We found 17 single nucleotide polymorphisms with extended haplotype homozygosity consistent with positive selective sweeps, 10 of which show no significant linkage disequilibrium with each other. Phylogenetic generalized least-squares modelling with ecological covariates suggest that most derived allele frequencies are significantly associated with solar irradiance and winter precipitation, rather than overall low temperatures. This selection and association with irradiance is demonstrated by a relatively isolated population in the southern coastal belt of South Africa. We suggest that sunbathing behaviours common to savannah monkeys, in combination with the strength of solar irradiance, may mediate adaptations to thermal stress via NST among savannah monkeys. The variants we discovered all lie in non-coding regions, some with previously documented regulatory functions, calling for further validation and research.
Keywords: Chlorocebus, uncoupling protein 1, non-shivering thermogenesis, cold adaptation, vervet monkey
1. Background
Cold environments pose a physiological challenge for homeothermic endotherms, which must find ways to conserve or produce heat to maintain bodily functions. Severe or long-term exposure to cold temperatures can result in hypothermia or death, but there are also less obvious costs to warm-blooded species inhabiting cold environments. Even while surviving, energy allocated for successfully maintaining body temperature may diminish the energy available for other physiological needs, like reproduction [1]. Long-term cold exposure may also slow the rate of biochemical reactions such as the production of hormones and enzymes [2], which mediate many aspects such as reproduction and pregnancy, growth and development, even behaviour, potentially indirectly affecting fitness. Among endotherms, and mammals in particular, extreme cold has been an evolutionary problem that has resulted in several adaptive solutions. These range from heat- and energy-conserving phenomena like hibernation and torpor [3,4], shifts in circulation [5], and shifts in body size and proportions (e.g. Bergmann's and Allen's rules) [6,7], to physiological phenomena that generate heat to offset losses in body temperature such as shivering [8] and non-shivering thermogenesis (NST).
NST is a short-term, heat generating phenotype mediated by brown adipose tissue (BAT). Brown adipocytes are metabolically active fat cells, the metabolism of which is driven by the expression of the uncoupling protein 1 (UCP1) gene (which codes for the protein UCP1, or thermogenin) [9]. Thermogenin, a membrane protein, disrupts the electron transport chain at the inner mitochondrial membrane, effectively inhibiting adenosine triphosphate (ATP) synthesis and resulting in the production of heat [9]. More specifically, UCP1 proteins embedded in the mitochondrial membrane interact with free fatty-acids (FA) in the intermembrane space, resulting in an increase in the ability of UCP1 to transport hydrogen ions across the mitochondrial membrane. The presence of FA in the intermembrane space disrupts the activity of ATP synthase directing more ions to UCP1 proteins in order to reach the mitochondrial matrix. Heat is produced as hydrogen ions pass through the uncoupling protein into the matrix, primarily as a by-product of the metabolic breakdown of FAs. Although other genes may play a role in regulating NST in muscle tissue [10,11], UCP1 has been shown to be critical for NST in BAT, highlighting its adaptive significance [12].
Cold-induced heat production via UCP1-mediated NST is believed to have played a critical role in the evolution and expansion of eutherian mammals [13]. The phylogenetic history of the mammalian UCP1 gene shows that variants increasing NST function were evolutionarily favoured among small-bodied mammals and in larger mammals these variants may protect neonates, but NST is far from a universal solution to offsetting the cold, and several mammalian lineages have thrived while having lost the trait [14]. Even among different mammals that rely on BAT for NST, the ways in which they are used to physiologically offset cold may differ—in sympatric populations of bank voles (Clethrionomys glareolus) and wood mice (Apodemus flavicollis, Apodemus sylvaticus) in Germany, for example, each species showed distinctive changes in body mass and BAT depot mass even though increases in NST-based thermogenic capacity were similar, suggesting that BAT activation may be used as just one part of taxonomically distinct physiological strategies to counter cold [15]. These differing strategies are certainly, in part, evolutionary in nature, but may also be owing to changes in gene expression mediated by developmental/epigenetic effects from cold exposure during critical periods during ontogeny [16,17]. Among primates—most notably in humans—NST is hypothesized to have been a critically important adaptation to climatic variation. Adaptations to cold climates, including NST, probably contributed to the successful migration out of Africa by early humans [18] and the eventual colonization of more extreme cold environments in circumpolar regions [17,19]. Among humans, genotype variants of the UCP1 gene have a significant impact on the degree of NST in the BAT of healthy subjects [20]. A recent human study showed that young adults from an indigenous Siberian population had greater BAT thermogenic activity compared to participants from the midwestern USA [21], suggesting the potential for regional evolution leading to human population differences in BAT functionality. Unfortunately, outside of rodent and human studies little research has been done comparing BAT physiology or its underlying genetic variation. Although the evolution of UCP1 thermoregulatory function in mammals and beyond has been the focus of previous studies [2,13,20,22,23], we have a limited understanding of the selective pressures that have shaped this gene in the primate order.
Savannah monkeys (Chlorocebus spp.) are an excellent wild primate model in which to study thermoregulatory adaptations. The genus probably originated in equatorial central Africa [24] and, with the exception of the rainforest dwelling dryas monkey (Chlorocebus dryas) [25] and Bale monkey (Chlorocebus djamdjamensis) [26], are largely restricted to savannah environments. Genomic evidence suggests that, in the past approximately 200–500 thousand years (ky), savannah monkeys expanded from eastern equatorial Africa into the Southern Hemisphere [24,25,27]. Vervet monkeys (Chlorocebus pygerythrus, sensu lato) [28] represent this southern expansion, with extant populations ranging from Ethiopia through South Africa along the Indian Ocean coast. South African vervets (Chlorocebus pygerythrus pygerythrus), in particular, have presented an interesting case study for cold adaptation as they regularly experience subzero temperatures at the southern extremes of their range [29,30]. Within this range, vervets also occupy habitats at a variety of elevations, ranging from sea level to altitudes over 2000 m a.s.l. [31], with the higher elevations further exacerbating cold temperatures. Like our own species, vervet monkeys have successfully balanced adaptations to cold with the thermoregulatory demands of living in open savannahs, which can also reach very high temperatures [32]. Furthermore, their wide geographical and ecological range make vervets a compelling model system for studying the evolutionary significance of NST in humans and primates more broadly.
To test this idea, we investigated variation in the UCP1 gene region for signs of selection in three closely related taxa of savannah monkey, at various times all subsumed into the species Chlorocebus pygerythrus, representing what we refer to as the ‘southern expansion’ of the genus into temperate latitudes. We hypothesized that as vervet monkeys expanded their geographical range into parts of southern Africa, exposure to colder temperatures would have favoured UCP1 variants that promote NST. Given their wide geographical distribution, we predicted generally strong differentiation within the gene region between populations sampled from areas geographically distant from one another, consistent with isolation by distance. However, we also predicted evidence of positive selection in restricted, functionally relevant regions of the UCP1 gene region—including, potentially, in cis-acting regulatory regions—of populations sampled from higher latitudes more generally, and in geographical regions where climatic conditions are coldest, either generally (owing to high elevations) or during particularly cold seasons in more temperate latitudes.
2. Methods
(a) . Study populations
Data for this project were generated by the International Vervet Research Consortium (IVRC) from a sample of 163 savannah monkeys (Chlorocebus spp.) from six closely related taxa, captured as part of a larger sampling effort in 11 countries across Africa and the Caribbean, with particularly extensive sampling in South Africa [27,28,33]. We focused on 73 individuals from three vervet taxa: Chlorocebus pygerythrus hilgerti, Chlorocebus cynosuros and Chlorocebus pygerythrus pygerythrus (figure 1a; electronic supplementary material, table S1). In this paper, we adhere roughly to taxonomic distinctions made by Groves [34], although recent nuclear and mitochondrial genomic evidence both suggest a relatively deep division suggesting a species-level distinction between C. p. hilgerti and C. p. pygerythrus [27,35], while evidence of gene flow between these two taxa and C. cynosuros suggest that the latter is nested within the larger C. pygerythrus clade (as first proposed by Dandelot [36]). A taxonomic revision may be in order for the genus [37], but is not within the scope of this paper. Each taxon was further subdivided into distinct populations, when possible, based on inferences from the whole-genome phylogeny (see below).
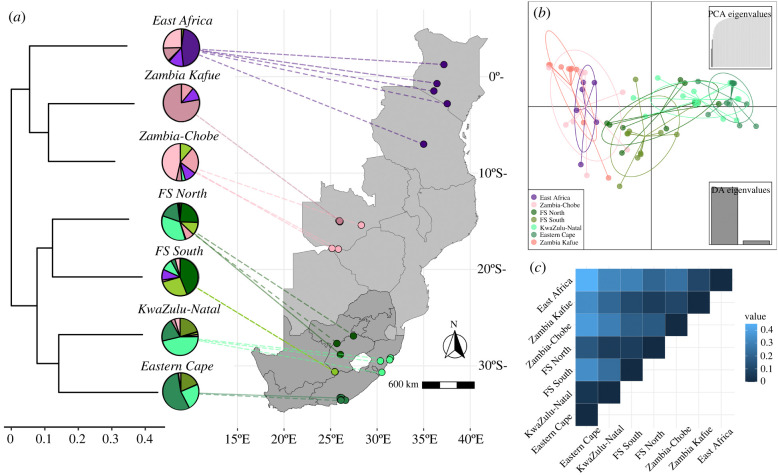
Figure 1.
Population differentiation in the southern expansion of savannah monkeys in the UCP1 gene region, including (a) population relationships based on the whole-genome phylogeny of savannah monkeys, with pie charts indicating substructuring in the UCP1 gene region in which purple clusters are primarily found in C. p. hilgerti, pink in C. cynosuros (darker pink = Zambia-Kafue; lighter pink = Zambia-Chobe) and shades of green in C. p. pygerythrus (dark green = Free State North (FS North); olive green = Free State South (FS South); light sea green = KwaZulu-Natal; dark sea green = Eastern Cape), with the map showing collection locations from southern and eastern Africa (South Africa, the more darkly shaded region, denotes regional boundaries, all others are national boundaries); (b) Discriminant analyses of principal components results for the UCP1 gene region; and (c) FST heatmap for the UCP1 gene region comparing defined populations in the southern expansion. (Online version in colour.)
(b) . Sequence data
Whole-genome sequences with median coverage of 4.4×, ranging from 1.8× to 44.8×, of the genus Chlorocebus were previously generated by collaborators at the McDonnell Genome Institute at Washington University in St Louis [27], aligned to the Chlorocebus sabaeus reference genome (ChlSab1.1) [24], and analysed in this study using a publicly available variant call format (VCF) file generated at the Gregor Mendel Institute [27]. We used tabix [38] in HTSlib v. 1.10.2 and vcftools [39] to isolate an approximately 28 kb gene region around UCP1 (ChlSab1.1 positions 7 : 87 492 195–87 502 665 on the reverse strand), including 10 kb upstream and downstream of the coding region itself to capture potential cis-regulatory regions. With the exception of the whole-genome phylogeny, all subsequent analyses in this study focus on this approximately 28 kb gene region around UCP1.
(c) . Whole-genome phylogeny
We used SNPRelate [40] v. 1.24.0 to generate FASTA alignments from the VCF file, and phangorn [41] v. 2.5.5, phytools [42] v. 0.6-99 and geiger [43] v. 2.0.6.4 to construct a neighbour-joining tree with a Jukes-Cantor mutation model representing whole-genome phylogenetic relationships for all 163 individuals in our original sample. We pruned this phylogeny to drop tips not represented in the study population sample.
(d) . Population structure in the uncoupling protein 1 gene region
We used discriminant analyses of principal components (DAPC) in adegenet [44,45] v. 2.1.2 and calculated the Fixation Index (FST) using hierfstat [46] v. 0.04-22 to model population structure for the approximately 28 kb region around UCP1. To statistically validate the patterns noted, we ran analyses of molecular variance (AMOVA) in poppr [47] v. 2.8.6. In both DAPC and FST analyses we used sample population as a grouping variable, while in AMOVA we used population nested within taxon. In the FST analysis, we used the Nei87 setting [48], to assess genetic distance between populations. We also ran principal component and admixture analysis using LEA [49] v. 3.0.0, using the snmf function with standard settings to visualize entropy criterion values to define K levels of population differentiation. To model isolation by distance in the UCP1 gene region, we assessed the correlation between genetic and geographical distance matrices in our sample using Mantel tests implemented in vegan [50] v. 2.5-6, using the Pearson method. We constructed our genetic distance matrix in poppr, also using Nei's [51] method, and our geographical distance matrix from GPS points associated with trapping locations (electronic supplementary material, table S1) using raster [52] v. 3.4-5.
(e) . Assessing selection in the uncoupling protein 1 gene region
We calculated Hardy-Weinberg equilibrium (HWE) values in the UCP1 gene region with pegas [53] v. 0.12 to identify candidate loci potentially experiencing selective forces, both within the whole southern expansion and in local populations. We assessed linkage disequilibrium (LD) in the gene region with gpart [54] v. 1.6.0, using standard BigLD settings and the r2 method (to avoid the ‘ceiling effect’ common to using D’ in small samples [55]). Single nucleotide polymorphisms (SNPs) considered to be in LD with each other (r2 > 0.85) were subsumed into a single representative locus for downstream analyses. We calculated site-frequency spectrum statistics for the whole-gene region including Tajima's D, and Fu and Li's D* and F* (PopGenome [56] v. 2.7.5; pegas v. 0.12), as well as a sliding window Tajima's D in vcftools with a window size of 500 bp, for both the southern expansion and each constituent subpopulation in South Africa. We compared UCP1 regional values of Tajima's D and Fu and Li's D* and F* to those from a sample of 1000 random, non-overlapping regions of equivalent size from vervet chromosome 7 to assess relative significance.
To calculate integrated haplotype scores (iHS) and assess extended haplotype homozygosity (EHH) both across the UCP1 gene region and for selected UCP1 loci, respectively, we inferred the ancestral allele sequence for our sample population using the program est-sfs [57] v. 2.03 using Kimura's mutation model. We chose the rhesus macaque reference (Macaca mulatta; BCM Mmul_8.0.1/rheMac8) as our single outgroup, which we downloaded using biomaRt [58,59] v. 2.44.4. We aligned it to the vervet reference genome (Chlorocebus sabaeus; Chlorocebus_sabeus 1.1/chlSab2) using rMSA [60] v. 0.99.0 and MAFFT [61] v. 7.467 using standard settings, and trimmed the macaque reference to the vervet extent visually in JalView [62] v. 2.11.1.3. Ancestral alleles were assigned to the major vervet allele when the probability assigned by est-sfs was above 0.70, and to the minor allele if below 0.30. When the probability was between these benchmarks, we assigned the ancestral allele to the allele shared by the Macaca and Chlorocebus references if it matched the population allele; when there was no concordance between the two outgroups (n = 7), we chose the Chlorocebus reference allele. We used rehh [63] v. 3.0.1 to estimate iHS and EHH using standard settings, but with a frequency bin of 0.15 to calculate iHS. We used a significance threshold of 2 for absolute iHS values [64] with a window size of 3000 bp and an overlap of 300 bp. We used a significance threshold value of 1.3 for considering individual SNPs for inclusion in further analyses.
(f) . Ecological covariates
We used GPS coordinates recorded at each trapping location to download altitude, annual mean temperature, mean temperature of the coldest month, winter precipitation levels, and mean temperature of the wet season, among other ecological covariates, for each population for the 30 year period from 1970 to 2000 from the WorldClim2 [65] online database. We also collected data on mean annual solar irradiance (measured in MJ m−2 d−1) for these points from the 10 year period from 2005 to 2015, originally generated by the NASA Langley Research Center (LaRC) POWER Project funded through the NASA Earth Science/Applied Science Program, using nasapower [66] v. 3.0.1. We standardized all covariates using z-scores, and then used the package Perfomance Analytics [67] v. 2.0.4 to reduce strongly correlated covariates. Correlations of standardized covariates, and the distributions of the final ecological covariate chosen for each population can be seen in the electronic supplementary material, figure S1.
(g) . Modelling allele frequency by ecological covariates
We used phylogenetic generalized least-squares (PGLS) regression, implemented in the package nlme [68] v. 3.1-150, to model variation in derived allele frequency of each target locus by geoclimatic variables including insolation/solar irradiation, annual mean temperature, minimum temperature of the coldest month, winter precipitation, and mean winter temperature. We incorporated our phylogenomic tree into our models using a Brownian correlation structure to account for average genetic distance across populations. We then used an information theoretic approach [69] to assess ecological covariate inclusion for each locus putatively experiencing selection, using the lowest Akaike information criterion modified for small sample sizes to select the appropriate model.
3. Results
(a) . Whole-genome phylogeny and isolation by distance
The maximum-likelihood phylogeny we constructed accords with previously published assessments of these taxa ([24,27]; electronic supplementary material, figure S2), with C. p. hilgerti and C. cynosuros clustering separately from C. p. pygerythrus, presumably reflecting the more recent and relatively larger amount of gene flow estimated to have occurred between these two taxa [27]. The whole-genome phylogeny further suggests that the monkeys sampled in South Africa are best represented by two geographically and genetically distinct populations, which can each be further divided into two clusters: the Free State (represented by Free State North and Free State South), and the southern coastal belt, represented by vervets from KwaZulu-Natal (KZN) and the Eastern Cape (figure 1a). One individual sampled at a rehabilitation centre in Limpopo (VSAJ2008) appears to have been transplanted from the Free State North population, and so was assigned to that population for subsequent analyses. Another rehabilitant (VSAI3005) did not fit easily into any of these populations, instead appearing basal to the KZN/Eastern Cape cluster, and so was excluded from further analyses. The two individuals sampled in Botswana (VBOA1003 and VBOA1005) and taxonomically assigned to C. p. pygerythrus appear to be genomically nested within C. cynosuros, and so we assigned them to that taxon for these analyses. We also divided C. cynosuros into two populations based on geographical and genetic distance. One clade from Kafue National Park in Zambia (Zambia Kafue) is nested within a larger clade comprised of both the Zambian Lusaka samples and Botswana samples from Chobe; we included the latter groups with those sampled in Livingstone as a population (Zambia-Chobe) owing to similarities in climate and to retain sufficient sample size for downstream modelling (figure 1a; electronic supplementary material, figure S2).
(b) . Population structure
DAPC indicates clear genetic differentiation in the UCP1 gene region among our study populations. Again in concordance with previous work on the whole genome [24,27], we see relatively high genetic similarity in the UCP1 gene region between C. cynosuros and C. p. hilgerti (presumably owing to more recent or extensive introgression), with C. p. pygerythrus showing a relative lack of shared variation with the other taxa in this gene region. Chlorocebus pygerythrus pygerythrus also showed the widest range of variation in the gene region (figure 1b), to the extent that the South African southern coastal belt populations (Eastern Cape and KZN) appear distinct from C. p. hilgerti/cynosuros. AMOVA results confirm this pattern, showing significant variation between taxa (13.8%, d.f. = 2, p < 0.05) and also between populations within taxa (12.6%, d.f. = 4, p < 0.01) (electronic supplementary material, table S2 and figure S3), with C. p. pygerythrus appearing to drive the former result and the latter driven by the separation between the Free State and southern coastal belt populations within South Africa. FST results are identical to those from DAPC, showing relatively high FST between the southern coastal belt population and all others, with the exception of relatively high similarity between KZN and Free State North (FST = 0.064) (figure 1c; electronic supplementary material, table S3).
Entropy criterion values from the principal component and admixture analyses strongly suggest isolation by distance across the southern expansion, with a population differentiation of K = 10 clusters (electronic supplementary material, figure S4). The gradual geographical pattern of genetic differentiation and generally low taxon- or population-specific substructure in genetic clusters also suggest isolation by distance, although there are a few patterns of note in keeping with the analyses above, including distinct clusters associated with the South African southern coastal belt that are shared with Free State North, and large overlap in shared clusters between C. p. hilgerti and C. cynosuros (figure 1a; electronic supplementary material, figure S5). Mantel tests of Nei's genetic distance between individual vervets compared to geographical distances between sample collection points show a strong positive correlation between genetic and geographical distances, again suggesting isolation by distance (Mantel's r = 0.44, p = 0.001).
(c) . Linkage disequilibrium
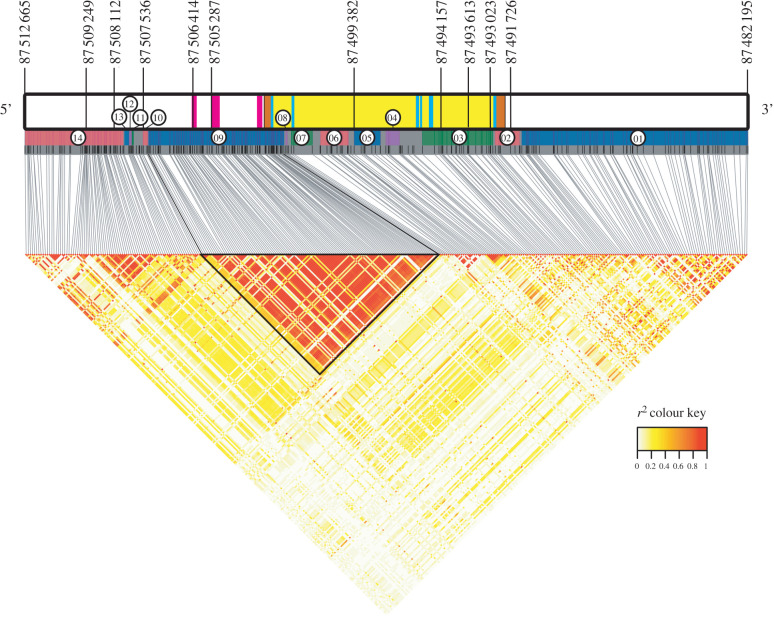
We identified 14 linkage blocks in the UCP1 gene region (figure 2; electronic supplementary material, figure S6 and table S4), including a large block (09) encompassing both known 5′ enhancer regions, the basal promoter region, the associated CpG island, the 5′ untranslated region (UTR), exon 1 and most of intron 1–2 of UCP1. We also found clear linkage between several loci of interest (see below), and used LD to reduce the number of loci assessed for selection to 10 unlinked loci of interest (electronic supplementary material, figure S7).
Figure 2.
UCP1 gene region highlighting candidate SNPs undergoing putative positive selective sweeps and LD blocks. Gene region includes UCP1 (positions 87 502 665 to 87 492 195, on the reverse strand) and 10 kilobases upstream and downstream. Thin black lines indicate SNPs located in the gene region, and those labelled above the gene diagram are SNPs undergoing putative positive selective sweeps (table 1 for details). In the gene diagram, magenta rectangles represent 5′ upstream enhancers and the basal promoter region, orange rectangles represent UTRs, blue rectangles represent exons and yellow rectangles represent introns. Coloured rectangles below the gene diagram represent LD blocks (positions available in the electronic supplementary material, table S4), with associated block numbers noted in circles. The LD heatmap below highlights the large LD block (09) encompassing the enhancers, promoter, 3′ UTR and exon 1. (Online version in colour.)
(d) . Assessing selection
Site-frequency spectrum statistics indicate no significant deviations from neutrality in the UCP1 gene region, as a whole, in the southern expansion. Tajima's D was overall positive (DTajima = 0.70), although in C. p. pygerythrus alone it was more strongly if not significantly positive (DTajima = 1.23). The only local population showing even a slightly negative, albeit insignificant Tajima's D was the Eastern Cape (DTajima = −0.021). Assessments of each statistic for 1000 random, non-overlapping regions of vervet chromosome 7 suggest that Fu and Li's D* and F* are both within the normal range of values across the chromosome. Tajima's D, however, is markedly positive compared to the rest of the chromosome (p = 0.060), again suggesting an excess of polymorphisms in the UCP1 region. A sliding window assessment of Tajima's D in the southern expansion indicates several regions with a strong excess of polymorphisms, including a 1000 bp window (7 : 87 497 500–87 498 500, DTajima = 2.04/2.33) in the 3′ UTR, a 500 bp window (7 : 87 503 000–87 503 500, DTajima = 2.04) in intron 2–3 and a 500 bp sequence in the 5′ upstream region (7 : 87 508 500–87 509 000, DTajima = 2.84) (electronic supplementary material, table S6a). Given we are discussing more localized selection processes, we also assessed individual populations in South Africa and found the same marked excess of polymorphisms, particularly upstream of the enhancer regions in Free State North (7 : 87 507 000–87 509 500; electronic supplementary material, table S6b), and including the promotor and enhancer regions in Free State South (7 : 87 501 500–87 509 500; electronic supplementary material, table S6c). This was less marked in the southern coastal belt populations (electronic supplementary material, table S6d-e), particularly in Eastern Cape where the entire gene region showed negative Tajima's D (electronic supplementary material, table S6d), although only one 500 bp segment in the 5′ upstream region neared the significance threshold (7 : 87 511 000–87 511 500, DTajima = −2.00).
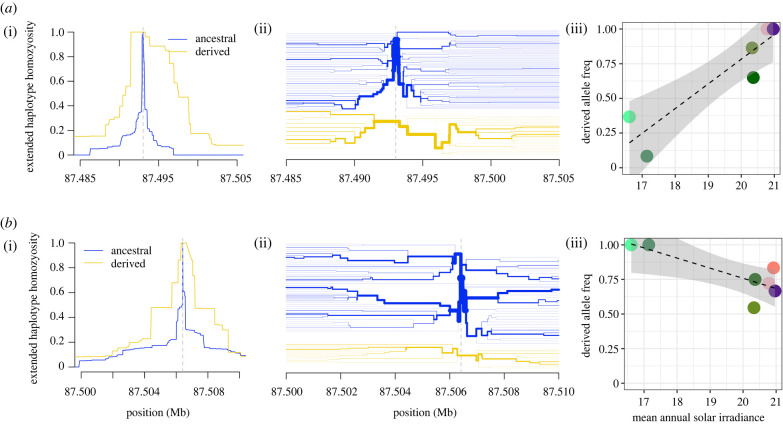
Seventeen SNPs met our threshold for consideration (piHS > 1.3) based on iHS in the southern expansion (electronic supplementary material, table S7), with the most compelling evidence for selection appearing in the upstream region of the gene (electronic supplementary material, figure S8). Of these loci, several showed compelling evidence for positive selective sweeps based on visual inspections of EHH (figure 3; electronic supplementary material, figure S9). Several of these SNPs also failed to meet the expectations of HWE in the larger southern expansion or among local populations (p < 0.05) (table 1).
Figure 3.
Examples of extended haplotype homozygosity (EHH) and ecological correlations with derived allele frequencies for (a) 7 : 87 493 023 and (b) 7 : 87 506 414 showing: (i) extended haplotype homozygosity around the candidate SNP, (ii) bifurcation patterns around the candidate SNP, and (iii) association of mean annual solar irradiation with derived allele frequencies among savannah monkey populations. In EHH diagrams, haplotypes around the ancestral allele are indicated in blue, and around the derived allele in yellow. For the plots of association between irradiance and population derived allele frequencies, population colours are the same as indicated in figure 1. Similar figures for the remaining candidate SNPs are available in the electronic supplementary material, figures S9 and S10. (Online version in colour.)
Table 1.
Targeted SNPs in the UCP1 gene region in the southern expansion and their associated indicators of selection. (Tajima's D values are from a 500 bp sliding window analysis conducted on either the South African (ZA) or southern expansion (SE) populations. Significance levels for Hardy–Weinberg equilibrium (HWE) are indicated with asterisks (* p < 0.05; ** p < 0.01; *** p < 0.001). Further details for each analysis/result are available in the electronic supplementary material.)
| SNP | position | polymorphism | location | block | Tajiima's D |
HWE |
iHS | best model | significant ecological covariates |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZA | SE | ZA | SE | sig. covariate(s) | β | p-value | R | p-value | outliers | |||||||
| 1 | 7 : 87 491 726 | *469C > T | 3′ downstream | 02 | 0.57 | 0.78 | — | * | 1.28 | 1 + winter precip. + irradiance | winter precipitation | 0.521 | 0.018 | 0.934 | 0.003 | EC/KZN |
| irradiance | 0.607 | 0.011 | ||||||||||||||
| 2 | 7 : 87 493 023 | 6-304G > A | intron 5–6 | 02 | 1.79 | 1.00 | * | *** | 1.29 | 1 + irradiance | irradiance | 0.310 | 0.011 | 0.920 | 0.003 | EC/KZN |
| 3 | 7 : 87 493 613 | 6-895C > T | intron 5–6 | 03 | 0.45 | −0.51 | — | — | 1.34 | 1 + winter precip. | winter precipitation | −0.262 | 0.024 | 0.831 | 0.021 | |
| 4 | 7 : 87 494 157 | 5 + 743C > T | intron 5–6 | 03 | 1.25 | −0.24 | — | — | 1.85 | null | — | — | — | — | — | |
| 5 | 7 : 87 499 382 | 2 + 2137T > C | intron 2–3 | 06 | −1.41 | −1.37 | — | — | 1.57 | null | — | — | — | — | — | |
| 6 | 7 : 87 505 287 | −2622C > A | 5' enhancer | 09 | 1.47 | 0.95 | — | — | 1.72 | 1 + winter precip. | winter precipitation | 0.162 | 0.073 | 0.796 | 0.032 | |
| 7 | 7 : 87 506 414 | −3749A > T | 5' enhancer | 09 | 1.39 | 0.91 | — | — | 2.84 | 1 + irradiance | irradiance | −0.150 | 0.031 | 0.812 | 0.026 | EC/KZN |
| 8 | 7 : 87 507 536 | −4871T > C | 5' upstream | 10 | −0.07 | 1.06 | — | *** | 1.84 | null | — | — | — | — | — | |
| 9 | 7 : 87 508 112 | −5447C > G | 5′ upstream | 11 | 1.40 | 1.56 | — | — | 2.00 | 1 + min. temp. coldest month | min. temp. coldest month | 0.125 | 0.000 | 0.731 | 0.062 | |
| 10 | 7 : 87 509 248 | −6583C > A | 5′ upstream | 14 | 2.66 | 1.02 | — | — | 2.72 | 1 + irradiance | irradiance | −0.094 | 0.000 | 0.982 | 0.000 | EC/KZN |
(e) . Modelling genotype by ecological covariates
We targeted 10 SNPs for PGLS analysis based on iHS/EHH and cumulative evidence of selection (table 1). Upon inspection of correlations between potential ecological covariates, we reduced them to include only normalized values for annual mean temperature (bio1), minimum temperature of the coldest month (bio6), mean temperature of the wet season (bio8), winter precipitation levels (bio19) and mean annual solar irradiance (insol). Using our model reduction approach, we found several environmental variables to be significantly associated with derived alleles at UCP1 loci showing potential for having undergone recent selection, including mean annual solar irradiance, winter precipitation levels and the minimum temperature of the coldest month (table 1 and figure 3; electronic supplementary material, figure S10).
4. Discussion
We hypothesized that the UCP1 gene would have undergone selection in relation to the expansion of Chlorocebus into more temperate geographical areas where temperatures regularly fall below freezing in the winter months, reflecting selection on NST as a way of offsetting these colder temperatures. This gene region generally shows differentiation consistent with both phylogenetic and geographical distance, and much of the variation therein is consistent with isolation by distance; however, there is clear divergence in excess of isolation by distance in South African C. p. pygerythrus populations when compared with the rest of the genus (figure 1).
This divergence in South Africa is particularly marked in the coastal belt south and east of the Drakensberg mountains, represented in this study by populations sampled in KZN and the Eastern Cape. These populations show high FST relative to all other populations, share relatively unique haplotypes to the exclusion of most other populations, and show similar patterns of variation relative to ecogeographic variables (figures 1 and 3), while also showing the strongest evidence for positive selective sweeps (see below). Strong differentiation of these populations from the rest of the South African vervets has been noted previously in subspecies taxonomy (which differentiated them as Chlorocebus pygerythrus cloeti [34]), studies of simian immunodeficiency virus unique to savannah monkeys (SIVagm) differentiation within South African vervets [70], and studies of mitochondrial DNA haplotypes [71]. Turner et al. [71] suggest that the Tugela River in the northeast and limited dispersal abilities across the Nama Karoo in the southwest have resulted in this coastal belt being a population isolate, potentially subject to strong drift. Our results here further suggest that unique patterns of selection could also be a factor in the differentiation of UCP1 in these populations, given that the southern coastal belt is as cold as the rest of our savannah monkey sample while also showing relatively low levels of solar irradiance (because of being at comparatively low elevations). The extent to which migration is constrained between the southern coastal belt populations and the rest of C. p. pygerythrus in South Africa is important, particularly given theoretical work demonstrating that isolated populations with constrained migration are more likely to see locally beneficial alleles increase in frequency in response to ecological variation [72]. The apparently high amount of shared variation between the Free State North and KZN populations in this gene region (figure 1) suggest recent/frequent migration, and further calls for greater sampling in this area to discover the relative contributions of drift and selection to these results.
Both haplotype- and site frequency spectrum-based analyses suggest that several SNPs in the UCP1 region may have undergone recent selection among savannah monkeys. Many of the SNPs identified in this study appear to be in a large linkage block (09; figure 2; electronic supplementary material, figure S6 and table S4) encompassing the UCP1 basal promoter region and associated CpG island, suggesting that these variants may have undergone positive selection related to some regulatory effect influencing UCP1 expression. Indeed, SNPs in this upstream region in humans have previously been associated with increased UCP1 expression, resulting in greater body heat generation when exposed to cold (in particular, the A-3826G variant, among others [20,73]). Positive Tajima's D results, primarily in the 5′ upstream region among local populations in the Free State, suggest either balancing selection or decreases in population size. The only populations where we observe the negative values of Tajima's D associated with positive selective sweeps are in the southern coastal belt, and particularly in the Eastern Cape.
It is important to note that these statistics indicating selection can be influenced by demographic parameters that we are unable to account for in this study. For example, site frequency spectrum-based statistics like Tajima's D and Fu & Li's D* and F* can be strongly influenced by demographic patterns, with the genomic signature of population expansions, in particular, mirroring that of positive selective sweeps [74]. Although we generally did not find significant results using these statistics in the UCP1 gene region (which is not unusual for recent selection [75]), inferred recent increases in effective population sizes in C. p. pygerythrus, and smaller increases in C. cynosuros, could interfere with these results [24]. To discover the difference between positive selection and background selection using these site frequency spectrum-based methods would require a better understanding of, for example, demographic history and recombination rate differences between these populations, which is not currently feasible [76]. For the linkage-based methods used here, strong bottlenecks and high migration between populations may reduce power to detect positive selective sweeps [74]. Even with the high admixture previously reported among these populations [27], we were able to detect positive selective sweeps. These methods are also generally not optimal for detecting soft selective sweeps [74], although it is difficult to infer how much standing ancestral variation in UCP1 may have been available in savannah monkeys prior to the southern expansion given the general dearth of annotated whole genomes in closely related cercopithecine primates [77]. Finally, linkage-based methods, in particular, perform poorly when populations are hierarchical in structure [74,78], which is a concern given the clear hierarchical structure and inferred localized migration and introgression patterns in this sample [27].
PGLS models indicate that the derived allele frequencies of many of these variants are associated with geoclimatic variables that indicate selection in relation to cold. Some associations, like increases in derived allele frequencies with the minimum temperature of the coldest month (7 : 87 508 112) and increases (7 : 87 491 726, 7 : 87 505 287) or decreases (7 : 87 493 613) with winter precipitation, suggest that these UCP1 variants may be under selection by modifying NST to offset the cold directly (with cold wet fur, as in winter rain conditions, being a particularly difficult thermoregulatory challenge [79]). A surprisingly common association was with the strength of mean annual solar irradiance, with derived allele frequencies strongly associated with weaker irradiance (i.e. 7 : 87 506 414, 7 : 87 509 248). Given that sunbathing has previously been identified as an important behaviour for offsetting cold temperatures in vervet monkeys [29] (figure 4), we propose that vervet populations exposed to relatively low levels of solar radiation—such as those in the low-elevation South African southern coastal belt—receive little thermoregulatory benefit from sunbathing behaviours, making adaptive changes in NST (via variation in UCP1 function/expression) necessary as a means of increasing thermoregulatory resistance to the cold. Loci 7 : 87 506 414 (along with linked locus 7 : 87 503 019) and 7 : 87 505 287 are particularly compelling as they reside within or near upstream regulatory regions: 7 : 87 503 019 is less than 100 bp upstream of the basal promoter region [80], 7 : 87 506 414 is in the enhancer region identified by Gaudry & Campbell [81] between the relatively conserved CRE-3 and RARE-1 motifs, while 7 : 87 505 287 lies 5 bp upstream of the conserved enhancer region identified by Shore et al. [82]. These three loci may be especially good candidates for functional validation of expression-based differences in UCP1-associated phenotypes in savannah monkeys.
Figure 4.
Sunbathing vervet monkeys (Chlorocebus pygerythrus pygerythrus) in Soetdoring Nature Reserve, Free State, South Africa. This photo was previously published by Danzy et al. [29] and is reproduced with permission from African Primates. (Online version in colour.)
In contrast to the negative association between mean annual solar irradiance and derived allele frequencies in the 5′ upstream region of UCP1 (i.e. 7 : 87 506 414, figure 3biii; 7 : 87 509 248, electronic supplementary material, figure S10g), we note that loci 7 : 87 491 756 (electronic supplementary material, figure S10a) and 7 : 87 493 023 (figure 3aiii) show strongly positive associations with irradiance, with each of these loci showing the southern coastal belt populations as outliers. In these cases, our hypothesized pattern for the derived allele appears to be inverted in that the signs of selection strongly point towards greater EHH and putative selection in what are probably warmer conditions. It may simply be the case that these alleles are simply linked with the actual allele of effect, making the direction of association arbitrary. In the case of 7 : 87 491 756, the evidence for EHH appears relatively weak (electronic supplementary material, figure S9a), and the relationship between the derived allele and irradiance seems to be exclusively driven by the southern coastal belt (electronic supplementary material, figure S10a); elimination of these outliers would eliminate the association with irradiance, but would also strengthen the relationship of derived allele frequencies with winter precipitation, creating a more significantly positive association suggesting selection in C. p. hilgerti populations. In the case of 7 : 87 493 023, the derived allele appears to be fixed in populations of C. cynosuros and C. p. hilgerti, where it shows strong EHH (electronic supplementary material, figures S9b and S10b); given that it eliminates one of the only CpG sites in intron 5–6, it is possible that the selection is somehow related to methylation potential at this locus and its effects on UCP1 expression. It is also possible this is an artefact of selection on other sequence variants in the larger linkage block (02) in this region of UCP1, which includes the functionally relevant 3′ UTR [83]. Aside from these potential functional explanations, it is unclear why this pattern is being observed.
Although the evidence of selection at certain loci in the UCP1 gene region and the apparent relationship between their allele frequencies and geoclimatic variables are compelling, there are many ways in which this study could be strengthened to better inform this question. Our sample sizes should be adequately informative given past studies on wild non-model organisms [84,85]; however, increased sampling, particularly across geoclimatic variables and within each population (particularly C. cynosuros and C. p. hilgerti) would strengthen our confidence in these results. Additionally, although we did not include elevation as a covariate in our PGLS models (owing to a 95% correlation with irradiance, which we considered to be more biologically meaningful for this study than elevation alone; electronic supplementary material, figure S1b), work in mice has suggested that hypoxia may also independently influence NST by significantly reducing thermogenic capacity [86]. Given that the southern coastal belt populations are the only populations in our sample near sea-level, increased variation in elevation in our sample at different latitudes might be necessary to disentangle a potential relationship between elevation, irradiance, and NST. Given the archived data available from the IVRC [28,33], this could be attainable with the resequencing of already-sampled individuals not included in this dataset. Further sampling in areas to increase geoclimatic diversity and in populations that bridge taxonomic and geographical gaps in our dataset—such as C. p. pygerythrus in Mozambique and Malawi, C. p. rufoviridis in Malawi and Tanzania, and C. p. pygerythrus in central and southern Botswana—could also provide greater information on the relative strength of selection and drift in the southern expansion.
To test explicitly in the wild whether behaviours like sunbathing may be implicated in selection for NST-related variants associated with irradiance would require an investigation of such behaviours in tandem with thermoregulatory physiology—including an assessment of BAT depots—across relevant genotypes and ecologies. BAT physiology and functionality may be complicated by competing thermoregulatory demands, such as those associated with reproductive condition and growth [87,88]. Given previous research, it is also likely that developmental exposures [16] and other behavioural factors beyond sunbathing also mediate the adaptive value of such variants (i.e. diet/feeding, dominance status and social integration [30,89]; sex, body size, residency status, social networks [90]; body position and microhabitat selection [91]; clustering and embracing behaviours [92]). Incorporating genotype data at key loci into already-existing biophysical modelling systems of thermoregulation in wild savannah monkeys [93] could be a relatively simple and effective means of testing these relationships.
One important question raised by this study is whether the variants we identified actually have a functional effect on the expression of UCP1 and NST. As has been shown in human and mouse models, upstream variants of UCP1 may yield significant differences in the thermogenic potential of brown fat, making those loci found in upstream enhancer and promotor regions (i.e. 7 : 87 503 019, 7 : 87 505 287, 7 : 87 506 414) particularly compelling candidates for validation. However, we presently know very little regarding how the thermoregulatory function of BAT is regulated in non-human primates. Quantifying changes in vervet UCP1 expression associated with the variants identified here, either through adipose biopsy in captive and wild individuals or through assessing relative UCP1 expression in biobanked samples that have the variants in question (such as those collected by the IVRC [28,33]), would help us to better characterize and study this critically important adaptive mammalian feature. Such studies of localized adaptive change in UCP1 expression may inform how our hominin ancestors managed to rapidly expand their geographical range beyond equatorial Africa, as well as how contemporary non-human primates may endure the thermoregulatory challenges of our changing global climate.
Acknowledgements
The authors gratefully acknowledge the associate editor and two anonymous reviewers for recommendations that greatly improved this manuscript. They further thank the many people with whom they have worked over the years to obtain and process these data. Lisa Nevell provided the original spark for this idea while teaching about human UCP1 at BU. Many students in the Sensory Morphology and Anthropological Genomics Laboratory at Boston University helped to refine this genomics pipeline at various times during its development, including especially Becca DeCamp, Gianna Grob, Erica Sun and Melissa Zarate, as well as Mansa Asiedu, Ishrat Chowdhury, Tennyson Cooper, Jane Hilsenrath, Alexia Lancea, Kimberly Louisor, Diem Maxwell, Elizabeth Shelton and Akshata Shukla (some with funding by the BU Undergraduate Research Opportunities Program). Original data collection included assistance in the field by Fred Brett in Ethiopia; Nicholas Dracopoli, James Else and The Institute for Primate Research in Kenya; Oliver ‘Pess’ Morton and Yoon Jung at UCLA; numerous field assistants in South Africa including especially Riel Coetzer, Magali Jacquier, Helene DeNys, JD Pampush, Elzet Answegen, Tegan Gaetano, Dewald DuPlessis, Micah Beller, David Beller and Pess Morton, with support by the University of Limpopo and the University of the Free State. Sampling was conducted under permits and permissions issued by the following: the Botswana Ministry of Environment & Wildlife and Tourism, the Ethiopian Wildlife Conservation Authority; Ministry of Tourism and Wildlife, Kenya; in South Africa the Department of Economic Development and Environmental Affairs, Eastern Cape; Department of Tourism, Environmental and Economic Affairs, Free State Province; Ezemvelo KZN Wildlife; Department of Economic Development, Environment and Tourism, Limpopo; the Department of Agriculture, Conservation and Environment, Mpumalanga; the Department of Environment and Nature Conservation, Northern Cape; the South African National Department of Environmental Affairs; and the Zambia Wildlife Authority. We thank J. Brenchley, K. Reimann (R24OD010976), and J. Baulu and the Barbados Primate Research Center and Wildlife Reserve for providing samples of Tanzanian origin. The authors also acknowledge their appreciation to all the veterinarians who worked with them to safely obtain samples, especially Lizanne Meiring and Murray Stokoe.
Ethics
This study was conducted using established in silico datasets. Original data collections adhered to the American Society of Primatologists' Principles for the Ethical Treatment of Non-Human Primates, and all techniques for trapping, sedation and sampling were approved by the Institutional Animal Care and Use Committee at the University of Wisconsin-Milwaukee, the Animal Research Committee at the University of California Los Angeles, and the Interfaculty Animal Ethics Committee of the University of the Free State.
Data accessibility
Raw sequence data are publicly available through the NCBI Sequence Read Archive (SRA) under BioProject nos. PRJNA168521, PRJNA168472, PRJNA168520, PRJNA168527 and PRJNA168522. The VCF file used in this study is available from the European Variation Archive (EVA) under accession PRJEB22988. The data used in this analysis and the associated analytical pipeline are publicly available via the Dryad Digital Repository (https://doi.org/10.5061/dryad.k3j9kd59z) [94], including instructions for downloading and processing all relevant online datasets and code.
Data are provided in the electronic supplementary material [95].
Authors' contributions
C.M.G.: formal analysis, methodology, software, visualization, writing—original draft, writing—review and editing; H.S.: data curation, resources, writing—review and editing; A.J.J.: data curation, resources, writing—review and editing; J.D.C.: visualization, writing—review and editing; N.B.F.: funding acquisition, resources; J.P.G.: funding acquisition, resources, writing—review and editing; T.R.T.: funding acquisition, resources, writing—review and editing; C.A.S.: conceptualization, formal analysis, methodology, project administration, software, supervision, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was funded by Boston University, and the original collections work funded by The Fulbright Foundation, NSF (SOC 74-24166, BNS 770-3322 and BCS 0938969), NIH (R01RR0163009), the University of Wisconsin- Milwaukee, The University of the Free State, the University of Limpopo and the Coriell Institute.
References
- 1.Bronson FH. 1985. Mammalian reproduction: an ecological perspective. Biol. Reprod. 32, 1-26. ( 10.1095/biolreprod32.1.1) [DOI] [PubMed] [Google Scholar]
- 2.D'Amico S, Claverie P, Collins T, Georlette D, Gratia E, Hoyoux A, Meuwis M-A, Feller G, Gerday C. 2002. Molecular basis of cold adaptation. Phil. Trans. R. Soc. B 357, 917-925. ( 10.1098/rstb.2002.1105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochachka PW, Guppy M. 1987. Metabolic arrest and the control of biological time, 227 p. Cambridge, MA: Harvard University Press. [Google Scholar]
- 4.Davenport J. 1992. Animal life at low temperature, 246 p. London, UK: Chapman & Hall. [Google Scholar]
- 5.Solonin YG, Katsyuba EA. 2003. Thermoregulation and blood circulation in adults during short-term exposure to extreme temperatures. Hum. Physiol. 29, 188-194. ( 10.1023/A:1022950728495) [DOI] [PubMed] [Google Scholar]
- 6.Meiri S, Dayan T. 2003. On the validity of Bergmann's rule. J. Biogeogr. 30, 331-351. ( 10.1046/j.1365-2699.2003.00837.x) [DOI] [Google Scholar]
- 7.Tilkens MJ, Wall-Scheffler C, Weaver TD, Steudel-Numbers K. 2007. The effects of body proportions on thermoregulation: an experimental assessment of Allen's rule. J. Hum. Evol. 53, 286-291. ( 10.1016/j.jhevol.2007.04.005) [DOI] [PubMed] [Google Scholar]
- 8.Hohtola E. 2004. Shivering thermogenesis in birds and mammals. In Life in the cold: evolution, mechanisms, adaptation, and application. 12th Int. Hibernation Symp. (eds Barnes BM, Carey H), pp. 241-252. Fairbanks, AK: Institute of Arctic Biology. [Google Scholar]
- 9.Cannon B, Nedergaard JAN. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277-359. ( 10.1152/physrev.00015.2003) [DOI] [PubMed] [Google Scholar]
- 10.Nowack J, Giroud S, Arnold W, Ruf T. 2017. Muscle non-shivering thermogenesis and its role in the evolution of endothermy. Front. Physiol. 8, 889. ( 10.3389/fphys.2017.00889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowack J, et al. 2019. Muscle nonshivering thermogenesis in a feral mammal. Sci. Rep. 9, 1-10. ( 10.1038/s41598-019-42756-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golozoubova V, Cannon B, Nedergaard J. 2006. UCP1 is essential for adaptive adrenergic nonshivering thermogenesis. Am. J. Physiol. Endocrinol. Metab. 291, E350-E357. ( 10.1152/ajpendo.00387.2005) [DOI] [PubMed] [Google Scholar]
- 13.Hughes DA, Jastroch M, Stoneking M, Klingenspor M. 2009. Molecular evolution of UCP1 and the evolutionary history of mammalian non-shivering thermogenesis. BMC Evol. Biol. 9, 4. ( 10.1186/1471-2148-9-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudry MJ, Campbell KL, Jastroch M. 2018. Evolution of UCP1. In Brown adipose tissue. Handbook of experimental pharmacology, vol. 251 (eds Pfeifer A, Klingenspor M, Herzig S), pp. 127-141. Cham, Switzerland: Springer. (Cited 22 August 2022.) See https://link.springer.com/chapter/10.1007/164_2018_116. [DOI] [PubMed] [Google Scholar]
- 15.Klaus S, Heldmaier G, Ricquier D. 1988. Seasonal acclimation of bank voles and wood mice: nonshivering thermogenesis and thermogenic properties of brown adipose tissue mitochondria. J. Comp. Physiol. B 158, 157-164. ( 10.1007/BF01075829) [DOI] [PubMed] [Google Scholar]
- 16.Robertson CE, McClelland GB. 2021. Ancestral and developmental cold alter brown adipose tissue function and adult thermal acclimation in Peromyscus. J. Comp. Physiol. B 191, 589-601. ( 10.1007/s00360-021-01355-z) [DOI] [PubMed] [Google Scholar]
- 17.Levy SB, Leonard WR. 2021. The evolutionary significance of human brown adipose tissue: Integrating the timescales of adaptation. Evol. Anthropol. 31, 75-91. ( 10.1002/evan.21930) [DOI] [PubMed] [Google Scholar]
- 18.Sazzini M, Schiavo G, De Fanti S, Martelli PL, Casadio R, Luiselli D. 2014. Searching for signatures of cold adaptations in modern and archaic humans: hints from the brown adipose tissue genes. Heredity (Edinb.) 113, 259-267. ( 10.1038/hdy.2014.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJJ. 2009. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500-1508. ( 10.1056/NEJMoa0808718) [DOI] [PubMed] [Google Scholar]
- 20.Nishimura T, Katsumura T, Motoi M, Oota H, Watanuki S. 2017. Experimental evidence reveals the UCP1 genotype changes the oxygen consumption attributed to non-shivering thermogenesis in humans. Sci. Rep. 7, 5570. ( 10.1038/s41598-017-05766-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy SB, et al. 2022. Brown adipose tissue thermogenesis among young adults in northeastern Siberia and Midwest United States and its relationship with other biological adaptations to cold climates. Am. J. Hum. Biol. 34, e23723. ( 10.1002/ajhb.23723) [DOI] [PubMed] [Google Scholar]
- 22.Klingenspor M, Fromme T, Hughes DA, Manzke L, Polymeropoulos E, Riemann T, Trzcionka M, Hirschberg V, Jastroch M. 2008. An ancient look at UCP1. Biochim. Biophys. Acta Bioenerg. 1777, 637-641. ( 10.1016/j.bbabio.2008.03.006) [DOI] [PubMed] [Google Scholar]
- 23.Mozo J, Emre Y, Bouillaud F, Ricquier D, Criscuolo F. 2005. Thermoregulation: what role for UCPs in mammals and birds? Biosci. Rep. 25, 227-249. ( 10.1007/s10540-005-2887-4) [DOI] [PubMed] [Google Scholar]
- 24.Warren WC, et al. 2015. The genome of the vervet (Chlorocebus aethiops sabaeus). Genome Res. 25, 1921-1933. ( 10.1101/gr.192922.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Valk T, Gonda CM, Silegowa H, Almanza S, Sifuentes-Romero I, Hart TB, Hart JA, Detwiler KM, Yoder A. 2020. The genome of the endangered dryas monkey provides new insights into the evolutionary history of the vervets. Mol. Biol. Evol. 37, 183-194. ( 10.1093/molbev/msz213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mekonnen A, et al. 2018. Population genetic structure and evolutionary history of Bale monkeys (Chlorocebus djamdjamensis) in the southern Ethiopian Highlands. BMC Evol. Biol. 18, 1-15. ( 10.1186/s12862-018-1217-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svardal H, et al. 2017. Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat. Genet. 49, 1705-1713. ( 10.1038/ng.3980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner TR, Schmitt CA, Cramer JD. 2019. Savanna monkeys: the genus chlorocebus. 342 p. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 29.Danzy J, Grobler JP, Freimer N, Turner TR. 2012. Sunbathing: a behavioral response to seasonal climatic change among South African vervet monkeys (Chlorocebus aethiops). Afr. Primates 7, 230-237. [Google Scholar]
- 30.McFarland R, Barrett L, Boner R, Freeman NJ, Henzi SP. 2014. Behavioral flexibility of vervet monkeys in response to climatic and social variability. Am. J. Phys. Anthropol. 154, 357-364. ( 10.1002/ajpa.22518) [DOI] [PubMed] [Google Scholar]
- 31.Turner TR, Schmitt CA, Cramer JD, Lorenz J, Grobler JP, Jolly CJ, Freimer NB. 2018. Morphological variation in the genus Chlorocebus: Ecogeographic and anthropogenically mediated variation in body mass, postcranial morphology, and growth. Am. J. Phys. Anthropol. 166, 682-707. ( 10.1002/ajpa.23459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lubbe A, Hetem RS, McFarland R, Barrett L, Henzi PS, Mitchell D, Meyer LCR, Maloney SK, Fuller A. 2014. Thermoregulatory plasticity in free-ranging vervet monkeys, Chlorocebus pygerythrus. J. Comp. Physiol. B 184, 799-809. ( 10.1007/s00360-014-0835-y) [DOI] [PubMed] [Google Scholar]
- 33.Jasinska AJ, et al. 2013. Systems biology of the vervet monkey. ILAR J. 54, 122-143. ( 10.1093/ilar/ilt049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groves CP. 2001. Primate taxonomy, 350 p. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 35.Dolotovskaya S, Torroba Bordallo J, Haus T, Noll A, Hofreiter M, Zinner D, Roos C. 2017. Comparing mitogenomic timetrees for two African savannah primate genera (Chlorocebus and Papio). Zool. J. Linn. Soc. 181, 471-483. ( 10.1093/zoolinnean/zlx001) [DOI] [Google Scholar]
- 36.Dandelot P. 1959. Note sur la classification des Cercopitheques du groupe aethiops. Mammalia 23, 357-368. ( 10.1515/mamm.1959.23.3.357) [DOI] [Google Scholar]
- 37.Svardal H, et al. 2018. Population genomics disentangles taxonomic relationships and identifies ancient hybridization in the genus Chlorocebus. Am. J. Phys. Anthropol. 162, 50. [Google Scholar]
- 38.Li H. 2011. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics 27, 718-719. ( 10.1093/bioinformatics/btq671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156-2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X, Levine D, Shen J, Gogarten S, Laurie C, Weir B. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326-3328. ( 10.1093/bioinformatics/bts606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27, 592-593. ( 10.1093/bioinformatics/btq706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Revell LJ. 2012. phytools: phylogenetic tools for comparative biology (and other things). Methods Ecol. Evol. 3, 217-223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 43.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. 2014. geiger v2. 0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216-2218. ( 10.1093/bioinformatics/btu181) [DOI] [PubMed] [Google Scholar]
- 44.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403-1405. ( 10.1093/bioinformatics/btn129) [DOI] [PubMed] [Google Scholar]
- 45.Jombart T, Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070-3071. ( 10.1093/bioinformatics/btr521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goudet J. 2005. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184-186. ( 10.1111/j.1471-8286.2004.00828.x) [DOI] [Google Scholar]
- 47.Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281. ( 10.7717/peerj.281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M. 1987. Molecular evolutionary genetics, 514 p. New York, NY: Columbia University Press. [Google Scholar]
- 49.Frichot E, François O. 2015. LEA: an R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925-929. ( 10.1111/2041-210X.12382) [DOI] [Google Scholar]
- 50.Oksanen J, et al. 2022. vegan: community ecology package version 2.5-6. See https://CRAN.R-project.org/package=vegan.
- 51.Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583-590. ( 10.1093/genetics/89.3.583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hijmans RJ, van Etten J. 2022. raster: geographic analysis and modeling with raster data. R package version 2.0-12. See https://CRAN.R-project.org/package=raster.
- 53.Paradis E. 2010. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419-420. ( 10.1093/bioinformatics/btp696) [DOI] [PubMed] [Google Scholar]
- 54.Kim S, Yoo Y. 2021. gpart: human genome partitioning of dense sequencing data by identifying haplotype blocks. R package version 1.0.3.
- 55.Marroni F, Pinosio S, Zaina G, Fogolari F, Felice N, Cattonaro F, Morgante M. 2011. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genet. Genomes 7, 1011-1023. ( 10.1007/s11295-011-0391-5) [DOI] [Google Scholar]
- 56.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929-1936. ( 10.1093/molbev/msu136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keightley PD, Jackson BC. 2018. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics 209, 897-906. ( 10.1534/genetics.118.301120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durinck S, Spellman PT, Birney E, Huber W. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184. ( 10.1038/nprot.2009.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439-3440. ( 10.1093/bioinformatics/bti525) [DOI] [PubMed] [Google Scholar]
- 60.Hahsler M, Manguy J. 2020. rMSA: interface for popular multiple sequence alignment tools. R package version 0.99.0. See https://rdrr.io/github/mhahsler/rMSA/.
- 61.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189-1191. ( 10.1093/bioinformatics/btp033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gautier M, Klassmann A, Vitalis R. 2017. rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol. Ecol. Resour. 17, 78-90. ( 10.1111/1755-0998.12634) [DOI] [PubMed] [Google Scholar]
- 64.Voight BF, Kuduravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4, e72. ( 10.1371/journal.pbio.0040072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fick SE, Hijmans RJ. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302-4315. ( 10.1002/joc.5086) [DOI] [Google Scholar]
- 66.Sparks A. 2018. nasapower: A NASA POWER global meteorology, surface solar energy and climatology data client for R. J. Open Source Softw. 3, 1035. ( 10.21105/joss.01035) [DOI] [Google Scholar]
- 67.Peterson BG, et al. 2020. PerformanceAnalytics: econometric tools for performance and risk analysis. R package version 2.0.4. See https://rdrr.io/cran/PerformanceAnalytics/.
- 68.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2021. nlme: linear and nonlinear mixed effects models. R package version 3.1-152. See https://rdrr.io/cran/nlme/.
- 69.Burnham KP, Anderson DR. 2002. Model selection and multimodal inference: a practical information-theoretic approach, 488 p. New York, NY: Springer. [Google Scholar]
- 70.Ma D, et al. 2013. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 9, 1-18. ( 10.1371/journal.ppat.1003011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turner TR, Coetzer WG, Schmitt CA, Lorenz JG, Freimer NB, Grobler JP. 2016. Localized population divergence of vervet monkeys (Chlorocebus spp.) in South Africa: evidence from mtDNA. Am. J. Phys. Anthropol. 159, 17-30. ( 10.1002/ajpa.22825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeaman S, Otto SP. 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 67, 2123-2129. ( 10.1111/j.1558-5646.2011.01277.x) [DOI] [PubMed] [Google Scholar]
- 73.Rose G, Crocco P, D'Aquila P, Montesanto A, Bellizi D, Passarino G. 2011. Two variants in the upstream enhancer region of human UCP1 gene affect gene expression and are correlated with human longevity. Exp. Gerontol. 46, 897-904. ( 10.1016/j.exger.2011.07.011) [DOI] [PubMed] [Google Scholar]
- 74.Weigand H, Leese F. 2018. Detecting signatures of positive selection in non-model species using genomic data. Zool. J. Linn. Soc. 184, 528-583. ( 10.1093/zoolinnean/zly007) [DOI] [Google Scholar]
- 75.Sabeti P, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832-837. ( 10.1038/nature01140) [DOI] [PubMed] [Google Scholar]
- 76.Stephan W. 2010. Genetic hitchhiking versus background selection: the controversy and its implications. Phil. Trans. R. Soc. B 365, 1245-1253. ( 10.1098/rstb.2009.0278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernandez M, Shenk MK, Perry GH. 2020. Factors influencing taxonomic unevenness in scientific research: a mixed-methods case study of non-human primate genomic sequence data generation. R. Soc. Open Sci. 7, 201206. ( 10.1098/rsos.201206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vatsiou AI, Bazin E, Gaggiotti OE. 2016. Detection of selective sweeps in structured populations: a comparison of recent methods. Mol. Ecol. 25, 89-103. ( 10.1111/mec.13360) [DOI] [PubMed] [Google Scholar]
- 79.Cristóbal-Azkarate J, Maréchal L, Semple S, Majolo B, MacLarnon A. 2017. Metabolic strategies in wild male Barbary macaques: evidence from faecal measurement of thyroid hormone. Biol. Lett. 12, 20160168. ( 10.1098/rsbl.2016.0168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shore A, Karamitri A, Kemp P, Speakman JR, Lomax MA. 2010. Role of UCP1 enhancer methylation and chromatin remodeling in the control of UCP1 expression in murine adipose tissue. Diabetologia 51, 1164-1173. ( 10.1007/s00125-010-1701-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaudry MJ, Campbell KL. 2017. Evolution of UCP1 transcriptional regulatory elements across the mammalian phylogeny. Front. Physiol. 8, 670. ( 10.3389/fphys.2017.00670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shore A, Emes RD, Wessely F, Kemp P, Cillo C, D'Armiento M, Hoggard N, Lomax MA. 2013. A comparative approach to understanding tissue-specific expression of uncoupling protein 1 expression in adipose tissue. Front. Genet. 3, 304. ( 10.3389/fgene.2012.00304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mayr C. 2019. What are 3′ UTRs doing? Cold Spring Harb. Perspect. Biol. 11, a034728. ( 10.1101/cshperspect.a034728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nazareno AG, Bemmels JB, Dick CW, Lohmann LG. 2017. Minimum sample sizes for population genomics: an empirical study from an Amazonian plant species. Mol. Ecol. Resour. 17, 1136-1147. ( 10.1111/1755-0998.12654) [DOI] [PubMed] [Google Scholar]
- 85.Trask JA, Malhi RS, Kanthaswamy S, Johnson J, Garnica WT, Malladi VS, Smith DG. 2011. The effect of SNP discovery method and sample size on estimation of population genetic data for Chinese and Indian rhesus macaques (Macaca mulatta). Primates 52, 129-138. ( 10.1007/s10329-010-0232-4) [DOI] [PubMed] [Google Scholar]
- 86.Beaudry JL, McClelland GB. 2010. Thermogenesis in CD-1 mice after combined chronic hypoxia and cold acclimation. Comp. Biochem. Physiol. B 157, 301-309. ( 10.1016/j.cbpb.2010.07.004) [DOI] [PubMed] [Google Scholar]
- 87.Baker FC, Siboza F, Fuller A. 2020. Temperature regulation in women: effects of the menstrual cycle. Temperature 7, 226-262. ( 10.1080/23328940.2020.1735927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maloney SK, Marsh MK, McLeod SR, Fuller A. 2017. Heterothermy is associated with reduced fitness in wild rabbits. Biol. Lett. 13, 20170521. ( 10.1098/rsbl.2017.0521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McFarland R, Fuller A, Hetem RS, Mitchell D, Maloney SK, Henzi SP, Barrett L. 2015. Social integration confers thermal benefits in a gregarious primate. J. Anim. Ecol. 84, 871-878. ( 10.1111/1365-2656.12329) [DOI] [PubMed] [Google Scholar]
- 90.Henzi SP, Hetem R, Fuller A, Maloney S, Young C, Mitchell D, Barrett L, Mcfarland R. 2017. Consequences of sex-specific sociability for thermoregulation in male vervet monkeys during winter. J. Zool. 302, 193-200. ( 10.1111/jzo.12448) [DOI] [Google Scholar]
- 91.Gestich CC, Caselli CB, Setz EZF. 2014. Behavioural thermoregulation in a small Neotropical primate. Ethology 120, 331-339. ( 10.1111/eth.12203) [DOI] [Google Scholar]
- 92.Gartlan JS, Brain CK. 1968. Ecology and social variability in Cercopithecus aethiops and C. mitis. In Primates. Studies in adaptation and variability (ed. Jay PC ), pp. 253-292, New York, NY: Rinehart and Winston. [Google Scholar]
- 93.Mathewson P, Porter WP, Barrett L, Fuller A, Henzi SP, Hetem RS, Young C, Mcfarland R. 2020. Field data confirm the ability of a biophysical model to predict wild primate body temperature. J. Therm. Biol 94, 102754. ( 10.1016/j.jtherbio.2020.102754) [DOI] [PubMed] [Google Scholar]
- 94.Schmitt CA, Gagnon CM, Svardal H, Jasinska AJ, Danzy Cramer J, Freimer NBet al. 2022. Savannah monkey (Chlorocebus spp.) population genetics/genomics pipeline. Dryad Digital Repository. (doi:20.5062/dryad.k3j9kd59z) [DOI] [PMC free article] [PubMed]
- 95.Gagnon CM, Svardal H, Jasinska AJ, Danzy Cramer J, Freimer NB, Paul Grobler J, Turner TR, Schmitt CA. 2022. Data from: Evidence of selection in the uncoupling protein 1 gene region suggests local adaptation to solar irradiance in savannah monkeys (Chlorocebus spp.). Figshare. ( 10.6084/m9.figshare.c.6169761) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Schmitt CA, Gagnon CM, Svardal H, Jasinska AJ, Danzy Cramer J, Freimer NBet al. 2022. Savannah monkey (Chlorocebus spp.) population genetics/genomics pipeline. Dryad Digital Repository. (doi:20.5062/dryad.k3j9kd59z) [DOI] [PMC free article] [PubMed]
- Gagnon CM, Svardal H, Jasinska AJ, Danzy Cramer J, Freimer NB, Paul Grobler J, Turner TR, Schmitt CA. 2022. Data from: Evidence of selection in the uncoupling protein 1 gene region suggests local adaptation to solar irradiance in savannah monkeys (Chlorocebus spp.). Figshare. ( 10.6084/m9.figshare.c.6169761) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Raw sequence data are publicly available through the NCBI Sequence Read Archive (SRA) under BioProject nos. PRJNA168521, PRJNA168472, PRJNA168520, PRJNA168527 and PRJNA168522. The VCF file used in this study is available from the European Variation Archive (EVA) under accession PRJEB22988. The data used in this analysis and the associated analytical pipeline are publicly available via the Dryad Digital Repository (https://doi.org/10.5061/dryad.k3j9kd59z) [94], including instructions for downloading and processing all relevant online datasets and code.
Data are provided in the electronic supplementary material [95].