Abstract
Plant genetic engineering emerged as a methodology to introduce only few transgenes into the plant genome. Following fast-paced developments of the past few decades, engineering of much larger numbers of transgenes became a reality, allowing to introduce full metabolic pathways from other organisms into plants and generate transgenics with startling new traits. From the advent of the classical plant genetic engineering, the transgenes were introduced into the nuclear genome of the plant cell, and this strategy still is quite successful when applied to few transgenes. However, for introducing large number of transgenes, we advocate that the chloroplast genome is a superior choice, especially for engineering of new complete metabolic pathways into plants. The ability to genetically engineer plants with complex and fully functional metabolic pathways from other organisms bears a substantial promise in generation of pharmaceuticals, i.e., biopharming, and new agricultural crops with that traits never existed before, leading to enhancement in quality of human life.
Introduction
Plant genetic engineering started with the ability to introduce one or two new genes into the plant genome, and for decades it has been optimized on that basis (Twyman et al., 2002; Gelvin, 1998). For the most part, the transgenes included a resistance marker, used to select the transformed cells and further regenerate them into a mature plant, and the gene of interest, frequently a plant gene the function of which was under investigation. Later on, this technique was further perfected, leading to production of agricultural crops carrying herbicide (Block et al., 1987) and pesticide resistance genes (van Wordragen et al., 1993), as well as other types of transgenes bestowing beneficial properties on the transgenic crops.
With time, from the biotechnology perspective, it has become increasingly important to introduce a larger number of transgenes into the same plant to allow reconstitution of full metabolic pathways, opening the door to generating a wide spectrum of new compounds, from recombinant proteins and drugs to plastics, within the transgenic plant (Daniell, 2006; Arai et al., 2004). This methodology promises significant benefits for many areas of biotechnology and research. For instance, many recombinant proteins are produced in immortalized mammalian cell lines or bacteria, requiring lengthy and costly purification procedures to ensure that no pathogens or other harmful agents are contained in the final product. Plants and humans do not share the same pathogens, and in-planta production of pharmaceuticals would have significant safety and cost benefits (Daniell, 2006). Also, the ability to produce plastics in plants is becoming increasingly important due to rising costs and adverse environmental impacts of petroleum (Arai et al., 2004; Bohmert-Tatarev et al., 2011). Furthermore, the potential for genetic transplantation of entire metabolic pathways from other organisms into plants is literarily unlimited. One example of an unorthodox application of this technology is introduction of a complete and fully functional luciferase pathway from bacteria into a plant cell (Krichevsky et al., 2010). The resulting plants emit light visible to the naked human eye - just as plants of Pandora forests in James Cameron’s “Avatar” - which in the future may lead to generation of glowing ornamental plants aimed to revitalize floriculture.
Multigene engineering of the plant cell remains a technologically challenging process (Dafny-Yelin and Tzfira, 2007; Halpin, 2005; Capell and Christou, 2004). The genes coding for all the necessary components of a metabolic pathway to be introduced into a plant cell can be placed in different cellular compartments, such as nucleus, plastids or mitochondria, each carrying its own genome. Plant genetic engineering via the nuclear genome has been and still remains the prevalent approach, and in the past few years improved technologies for multigene delivery into the nucleus have been developed (Goderis et al., 2002; Tzfira et al., 2005). We, however, advocate genetic engineering via the chloroplast genome as a particularly effective approach for introduction of multigene pathways into plants.
Creating plants expressing multiple transgenes
There are three fundamental approaches to introduce and “stack” multiple genes into a plant: (i) introducing all of the transgenes on a single vector (Krichevsky et al., 2010; Arai et al., 2004; Goderis et al., 2002; Tzfira et al., 2005), (ii) introducing transgenes on different vectors, either simultaneously by a co-transformation or in sequential transformation steps (Naqvi et al., 2010; Chen et al., 1998; Ye et al., 2000), and (iii) genetically intercrossing a number of various transgenic lines, each containing a different transgene (Ma et al., 1995; Datta et al., 2002). The latter two options have significant drawbacks. Introducing transgenes on a number of separate vectors usually requires each plasmid to bear a distinct selection marker, which substantially limits the number of vectors that can be used - and consequently the number of introduced transgenes - due to relatively few selection markers available for use in plants. Transgene stacking via genetic crosses of different transgene-bearing lines is an extremely lengthy process. Moreover, both of these techniques typically result in integration of the transgenes in different and often distant loci, which may lead to their segregation in the progeny and thus genetic instability. In contrast, introduction of all the transgenes required for reconstitution of a complete metabolic pathway on a single vector circumvents all these difficulties, making it an approach of choice.
In the past, introduction of multiple transgenes encoding a given metabolic pathway on a single vector was limited by the technical difficulty of gene cloning into large vectors. However, recent years saw a significant progress made in this area. For example, introduction of large, ca. 150 kb, DNA fragments into plant cells has been reported (Mullen et al., 1998; Wada et al., 2009). Another advance came with the use of homing endonucleases, rather than conventional restriction endonucleases. Most conventional restriction enzymes have an average length of their DNA recognition site of 6–8 bp whereas the homing endonucleases typically recognize rather long, 14–40-bp, sequences (Stoddard, 2005). For instance, the homing endonuclease I-SceI recognizes an 18-bp sequence, which statistically occurs once in every 7×1010 bp in a random DNA sequence (Jasin, 1996), whereas a conventional restriction enzyme paradigm EcoRI has a 6-bp recognition sequence, which appears on average every 4×103 bp (Gardiner, 1990). This makes homing endonucleases approximately 20×106 times less likely to cleave a given DNA sequence than a conventional restriction enzyme, which circumvents the increasing likelihood of the presence of the sequences used for cloning within one of the multiple transgenes. An elegant approach to employ homing endonucleases for plant multigene transformation was demonstrated in the pSAT (Tzfira et al., 2005) and similar modular vector systems (Goderis et al., 2002). Each pSAT vector can carry an expression cassette comprising the transgene of interest equipped with the desired promoter/terminator sequences and, if required, fused to a fluorescent (e.g., green fluorescent protein, GFP) or enzymatic tag (e.g., β-glucuronidase). This expression cassette within pSAT is flanked by homing endonucleases recognition sites, and up to 7 independent expression cassettes from different pSAT vectors can be mounted - using the corresponding homing endonucleases - into a single binary vector with a multiple cloning site that contains recognition sequences of all these homing endonucleases (Tzfira et al., 2005; Goderis et al., 2002); the resulting construct can be used for production of transgenic plants via Agrobacterium-mediated transformation (Tzfira et al., 2005).
Other approaches to generate high capacity plasmids include Agrobacterium-mediated transformation of an artificial chromosome of 150 kb, termed BIBAC (Hamilton et al., 1996). The transferred DNA fragment became stably inherited, and it can potentially serve as a carrier for multiple transgenes of interest. Similarly, minichromosomes, which allow transfer of even larger DNA fragments, have been employed to transform plants (Houben et al., 2008; Houben and Schubert, 2007). However, as compared to transgenes integrated into the native plant genet elements, minichromosomes, However, relatively to techniques where the transgenes are integrated into the host genome, the use of minichromosomes has significant stability issues since, in plants, unpaired or monosomic chromosomes are frequently lost during meiosis, leading to low transmission frequency for the transgenes (Houben et al., 2008; Houben and Schubert, 2007). For example, the maize B chromosome, specialized to function as a monosome and to be transmitted at high frequencies (Carlson and Roseman, 1992), exhibited a 95% transmission reduction rate when its centromeric region was reduced from approximately 700 kb to about 110 kb (Jin et al., 2005) to make it more manageable size-wise. This and other technical challenges must be overcome before minichromosomes can become a routine tool for introduction of multiple transgenes into plants.
Once a vector carrying the full set of transgenes required to reconstitute a complete metabolic pathway is constructed, it is used to transform the plant cell and then to regenerate the mature transgenic plant. The transgenes can be integrated into one of the cellular genomes, i.e., nuclear, plastidal or mitochondrial, or maintained as minichromosomes. While integration into the mitochondrial genome (Remacle et al., 2006) or maintenance as minichromosomes (Houben et al., 2008; Houben and Schubert, 2007) are relatively rarely used, nuclear and plastidal - typically chloroplast - genome transformations are widely employed, and they represent the focus of this paper.
A number of techniques are known to transform nuclear and chloroplast genomes, including particle bombardment (Kikkert et al., 2005), electroporation (Lazzeri, 1995), and polyethylene glycol (PEG)-mediated (Lazzeri, 1995; Radchuk et al., 2002) and Agrobacterium-mediated transformations (Lee and Gelvin, 2008; Lacroix et al., 2006). Among these, Agrobacterium-mediated transformation is the most efficient and widely used method for plant nuclear transformation. The routinely used binary vector systems are composed of two vectors, both carried by an Agrobacterium cell capable to transform stably a susceptible plant (Lee and Gelvin, 2008). Specifically, the helper vector represents an engineered Agrobacterium Ti plasmid carrying the bacterial virulence (vir) genes that encode the T-DNA transfer machinery, but lacking the T-DNA element itself, which is contained within the second vector. The T-DNA element is defined by 25-bp border sequences between which the transgenes of interest, as well as selection markers, are cloned (Lee and Gelvin, 2008). The T-DNA is transferred from Agrobacterium cell into the host plant cell, culminating in its integration into the plant nuclear genome (reviewed by Lacroix et al., 2006). An example of a sophisticated binary vector system is the pSAT vector system where multiple transgene expression cassettes derived from the pSAT vectors are inserted into the T-DNA region of a binary vector (Tzfira et al., 2005).
A standard tool for nuclear plant DNA transformation, Agrobacterium is ineffective for transformation of other organelles, notwithstanding the early reports of T-DNA transfer into chloroplasts (Block et al., 1985; Venkateswarlu and Nazar, 1991); this makes biological sense because in nature Agrobacterium transforms only nuclear genomes of its hosts. Thus, at present, biolistic gene delivery is the most efficient and broadly used method for chloroplast genetic modification (Lutz et al., 2006), (Verma et al., 2008). In this technique, the transforming DNA is adsorbed on gold or tungsten projectiles and bombarded using a high-pressure device, such as the PDS-1000/He gene gun, into plant tissue. DNA molecules that reach the chloroplast stroma are incorporated into the plastidal genome by the homologous recombination mechanism (Cerutti et al., 1995; Maliga et al., 1994) maintained by the chloroplasts from their bacterial evolutionary ancestry (Gould et al., 2008). While other transformation techniques are sometimes employed, Agrobacterium-mediated transformation and biolistic delivery remain by far the most popular and routinely applied methodologies for generation of transgenic plants worldwide.
Plastids: the gateway for engineering of multigene pathways
Plastids, and chloroplasts in particular, represent perhaps the best choice for transgenic expression of complete metabolic pathways. Plastids have retained bacterial gene expression properties rooted in their prokaryotic ancestry (Gould et al., 2008). Thus, plastids are not only able to express groups of transgenes in operons (Maliga, 2003; Quesada-Vargas et al., 2005; Zerges, 2000) and support high expression levels of the transgenes, but they also lack the gene silencing and other transcriptional suppression mechanisms present in the nucleus; these properties make plastids the organelle of choice for advanced metabolic engineering. Proteins expressed in the chloroplast typically reach levels as high as 20–25% of the total soluble protein (TSP) of the cell, or even up to 70% of TSP (Oey et al., 2009), while proteins expressed from nuclear transgenes normally do not exceed 1–2% of TSP. For instance, a comparative transgene expression study showed that human interferon gamma (hINF-γ) expressed from the chloroplast genome accumulated to levels 100 times higher than hINF-γ expressed from the nuclear genome (Leelavathi and Reddy, 2003). Moreover, a large number of plant cell biosynthetic pathways are located in chloroplast, thus providing a wealth of biomolecules potentially required as precursors for a newly introduced metabolic pathways. Finally, maternal inheritance of plastidal DNA precludes the spread of transgenes through pollen, eliminating the risk of cross-contamination with wild-type plants (Hagemann, 2004; Heifetz, 2000).
To date, the most complex transgenic metabolic pathway, containing the largest number of transgenes expressed in concert, was engineered into Nicotiana tabacum chloroplasts. Specifically, the complete and fully functional bacterial luciferase pathway from Photobacterium leiognathi was reconstituted in plant chloroplasts (Krichevsky et al., 2010) by integrating into their genome six genes of the bacterial lux operon (Meighen, 1993; Meighen, 1991). The resulting transgenic plants were autoluminescent, actively emitting light visible by a naked human eye (Figure 1) (Krichevsky et al., 2010). The light emission continued throughout the life of the plant and was stable from generation to generation. Interestingly, introduction of this bacterial metabolic pathway did not have any notable side effects on tobacco plant development or morphogenesis, suggesting that plants might be receptive to transplantation of other energetically costly metabolic pathways. In addition to demonstrating that a large number of in-concert acting transgenes can be expressed from plastidal genome, this particular innovation has a number of practical applications aimed to enhance the quality of human life, such as generation of “glow in the dark” ornamental plants for floriculture, as well as biosensor plants that react with light emission to pollutants or stress.
Figure 1.
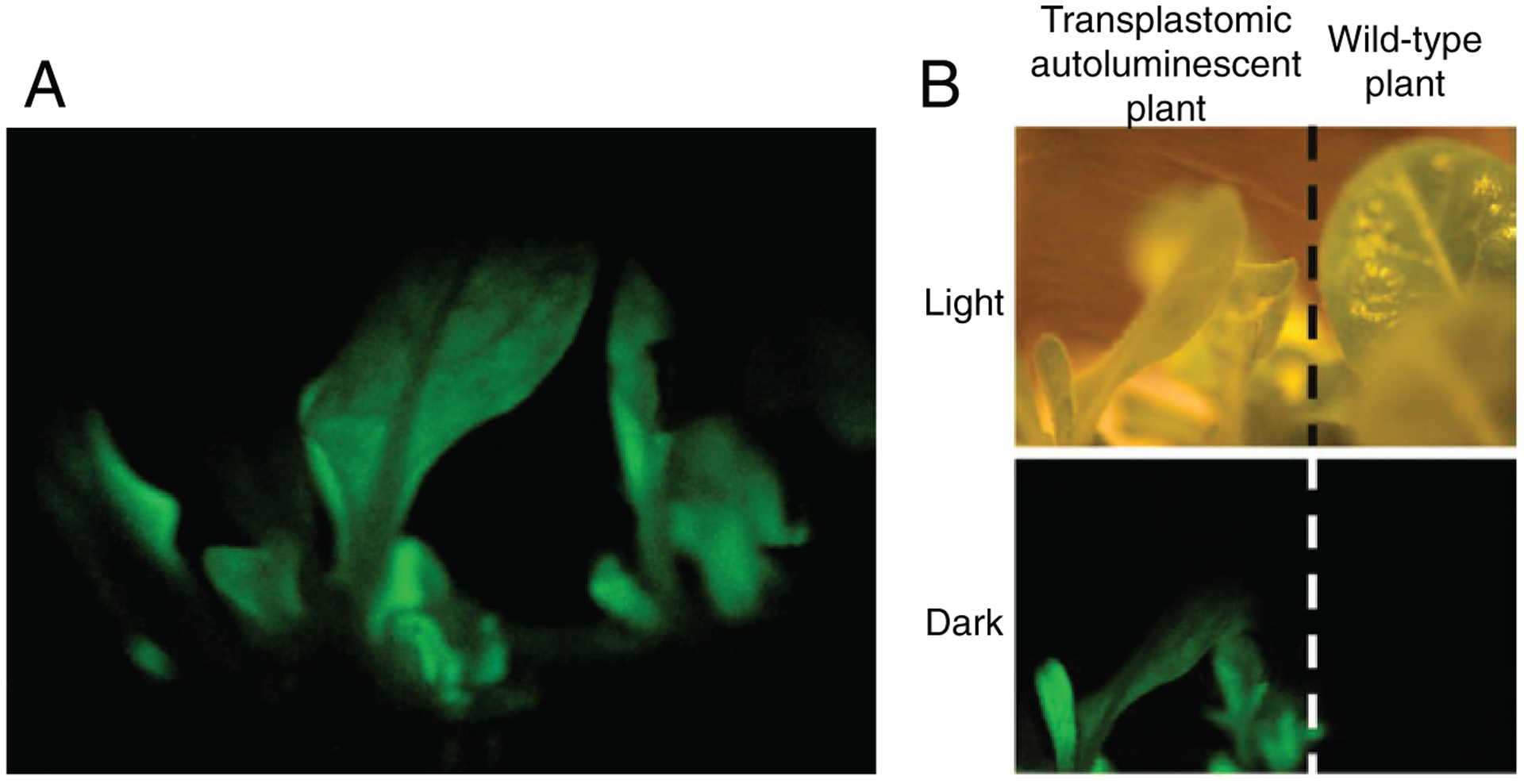
Expression of the fully functional luciferase pathway in transplastomic tobacco plants results in autoluminescence. (A) Transplastomic plants emit light clearly visible to the human eye. (B) Transplastomic autoluminescent and wild-type plants in the dark and light. Images were acquired with a standard hand-held consumer-quality camera. The images are adapted from (Krichevsky et al., 2010).
Additional examples of groups of transgenes used for concerted action of the encoded proteins include the expression of the phb operon, consisting of three bacterial genes for the biosynthesis of the biodegradable polyester, poly[(R)-3-hydroxybutyrate] (PHB), in tobacco chloroplasts (Arai et al., 2004). The phbC, phbA and phbB genes of Ralstonia eutropha with modified and enhanced regulatory sequences were integrated into the rbcL/accD locus of the tobacco chloroplast genome, resulting in production of PHB from acetyl-CoA in the leaves of the transgenic plants. PHB belongs to a class of biodegradable polyesters produced by a variety of bacteria as storage material. Polymers of this group have advantageous properties of biodegradability, thermoplasticity and elasticity (Doi, 1990), making them an environmentally friendly manufacturing material and reducing dependence on fossil sources of carbon. Interestingly, the technique of PHB production in transplastomic plants was further refined by the use of a synthetic phb operon, generating transgenic plants that produce up to 17.3% dry weight PHB in leaves and 8.8% dry weight PHB in the total biomass of the plant (Bohmert-Tatarev et al., 2011).
In yet another example, three genes of the Bacillus thuringiensis (Bt) Cry2Aa2 operon were introduced into the chloroplast genome of tobacco plants (De Cosa et al., 2001). In addition to the Cry toxin encoded by this operon and bestowing pesticide qualities onto the transgenic plant, this operon also encodes a putative chaperonin that facilitates the correct folding of the Cry toxin to form proteolytically stable cuboidal crystals, thus making this approach significantly more effective than expression the toxin sequence alone. In the resulting transplastomic plants, the Cry2Aa2 operon-encoded proteins represented up to 46% of TSP, an unprecedented expression levels, which are significantly higher than those ever achieved for plants with nucleus-encoded Cry toxin. Consequently, a 100% mortality rate of typically hard-to-control pests, such as cotton bollworm and beet armyworm, have been recorded for the Cry2Aa2 operon-expressing plants.
Whereas introduction of entire metabolic pathways into the plastidal genome is still in its infancy, these examples demonstrate the immense potential of this approach for multigene plant metabolic engineering and genetic transplantation of fully functional metabolic pathways into plants. Transgene expression from the chloroplast genome has the advantage of simplicity as compared to expression of multiple transgenes from the nuclear genome, which is burdened by the complexity of regulatory sequences and silencing mechanisms.
Perils of multigene expression from the nuclear genome
The cell nucleus lacks the relative simplicity of gene expression of the chloroplasts due to its numerous regulatory mechanisms, which complicate genetic engineering of large sets of transgenes. First, for nuclear genome transformation, each transgene has to contain its own regulatory sequences, e.g. promoter and terminator, due to the monocistronic nature of the nuclear transcriptional machinery. In contrast, for chloroplast genome transformation - and perhaps for future mitochondrial transformation - all transgenes can be introduced as a single operon controlled by one set of regulatory elements, due to the ability of plastids to support polycistronic gene expression (Meyers et al., 2010). Further, integration of transgenes into the nuclear genome occurs mainly by non-homologous recombination and therefore is random, leading to unpredicted patterns of expression due to position effects (e.g., Eike et al., 2005; Fischer et al., 2008), such as transgene silencing following integration into heterochromatic regions. In addition, even when a transgene is integrated into an active chromatin, it may become silenced by the RNA silencing machinery of the eukaryotic cell (Vaucheret, 2006). These mechanisms may affect different genes within the same transgenic pathway differently (Day et al., 2000), undermining the functionality of the entire metabolic pathway. Typically, due to position effects and gene silencing, screening of large numbers of transgenic plants is required to identify lines that steadily and consistently express even one or two transgenes; the use of expression cassettes of higher complexity, with multiple transgenes, makes this task exceedingly difficult and often prohibitive. Conversely, transgenes integrate into the chloroplast genome via homologous recombination and not randomly, decreasing the potential for adverse position effects. Furthermore, chloroplasts do not appear to possess RNA silencing mechanism (Hegeman et al., 2005), making them the preferred target for multigene engineering.
One interesting case demonstrating these points is engineering of the metabolic pathway for PHB biosynthesis through modification of the nuclear genome (Somleva et al., 2008), which can be compared to a strategy described in the previous section for generating PHB through manipulation of the chloroplast genome (Arai et al., 2004; Bohmert-Tatarev et al., 2011). For nuclear transformation, the phb operon genes were introduced into switchgrass using a binary vector that carried each of the three phb operon genes with separate regulatory elements, requiring multiple cloning steps during construction of the this vector (Somleva et al., 2008). In contrast, for plastid transformation, the phb genes were introduced as a single operon (Arai et al., 2004), making the vector fairly simple. Following nuclear transformation, several hundreds of transgenic lines needed to be analyzed to identify consistent phb expressers, with most of the lines exhibiting high variability in PHB production (Somleva et al., 2008), likely stemming from the position effects and silencing mechanisms. Conversely, only a small number of transplastomic lines needed to be assessed to identify high producers of PHB (Arai et al., 2004). Most importantly, the high-expressers of the phb transgenes integrated into the nuclear genome produced only up to 3.72% dry weight PHB in leaf tissues and 1.23% dry weight PHB in whole plants, which is 4–8 fold less than PHB-producing plants with the phb transgenes integrated into the chloroplast genome, which yielded up to 17.3% dry weight PHB in leaves and 8.8% dry weight PHB in whole plants (Somleva et al., 2008; Bohmert-Tatarev et al., 2011). This, quite typical, example shows that generation of transplastomic plants containing multigene pathways is less complex, owing to straightforward vector construction and screening of only few resulting lines, yet it yields much higher amounts of the desired product as compared to transgenic plants expressing the multigene pathway from their nuclear DNA.
Notwithstanding the advantages of transplastomic expression, obviously, nuclear transformations have produced several spectacular successes in commercial expression of, albeit relatively short, multigene pathways. One such example is “golden rice”, enriched in β-carotene and designed to overcome the lack of this essential nutrient in this crop, which causes serious public health issues in Asia, Africa, and Latin America; in this case, three genes of the β-carotene biosynthetic pathway were introduced on two separate vectors (Ye et al., 2000). In another example, genes coding for Δ9 elongase of Isochrysis galbana, Δ8 desaturase of Euglena gracilis and Δ5 desaturase of Mortierella alpina were introduced into Arabidopsis on a triple-cassette T-DNA to increase synthesis of long-chain polyunsaturated fatty acids (PUFA) (Qi et al., 2004), which in the future may allow development of PUFA-producing agricultural crops. Overall, although a few cases of introduction and expression of numerous transgenes, i.e., seven genes for ketocarotenoid biosynthesis in Brassica (Fujisawa et al., 2009), in the plant cell nucleus are known, most multigene-encoded pathways successfully expressed from the plant nuclear genome typically comprise only 3–4 transgenes. This suggests that whereas nuclear transgene expression is suitable for reconstruction of relatively short metabolic pathways, more complex pathways, with larger number of individual components, can be engineered with higher efficiency in the chloroplast genome. Finally, unlike the transgenes integrated into the maternally-inherited plastid genome, transgenes inserted into the nuclear genome can be transmitted by to other plants via pollen, which requires additional biosecurity measures, such as co-engineering of male sterility (Cho et al., 2001), to prevent the spread of the transgene to wild-type plant populations. This can further complicate and increase the cost of translation of fundamental research achievements in plant multigene metabolic engineering to biotechnology products if done through the nuclear route.
Concluding remarks
The ability of early plant genetic engineers to incorporate one or only few transgenes into the plant nuclear genome was sufficient for fundamental research purposes of the day as well as for generation of first genetically modified agricultural products. As plant genetic engineering became more sophisticated, a need emerged to integrate and express increased numbers of transgenes in the same plant for reconstitution of complete and complex metabolic pathways. This, in turn, requires a change from the existing paradigm of targeting the transgenes to the nuclear genome to genetic manipulation of plastidal genomes, which represent a better target for integration and optimal coexpression of numerous transgenes.
Several challenges still remain, however, in making chloroplast transformation a standard for multigene genetic engineering. The most significant is the shortage of optimized transformation protocols, which currently exist for only a handful of species. However, this technical hurdle stems from the fact that relatively few laboratories worldwide work in the field of chloroplast genetic engineering, and broadening research efforts will undoubtedly lead to closing the gap between the number of transformation techniques available for plastids as compared to those for nuclear transformations. Interestingly, mitochondrial genomes share with chloroplasts a number of traits, such as polycistronic gene expression, that are conducive to genetic engineering. Currently, only a few examples of mitochondrial transformation are known for photosynthetic organisms, with perhaps the most prominent being Chlamydomonas reinhardtii (Remacle et al., 2006). Development of mitochondrial transformation techniques for higher plants, including agriculturally important crops, will pave the way to engineering of multigene pathways into the mitochondrial genome and open a new range of possibilities for plant genetic engineering. Ultimately, however, all three types of transformation, nuclear, plastid and mitochondrial, should be available to researchers to be used depending on the specific requirements of the experiment. Figure 2 summarizes major considerations for and against each of these transformation approaches as applied to genetic engineering of multigene pathways into plants. The ability to reconstruct in transgenic plants such fully functional metabolic pathways derived not only from plants, but also from other organisms, holds great promise for research, agriculture, and biopharming.
Figure 2.
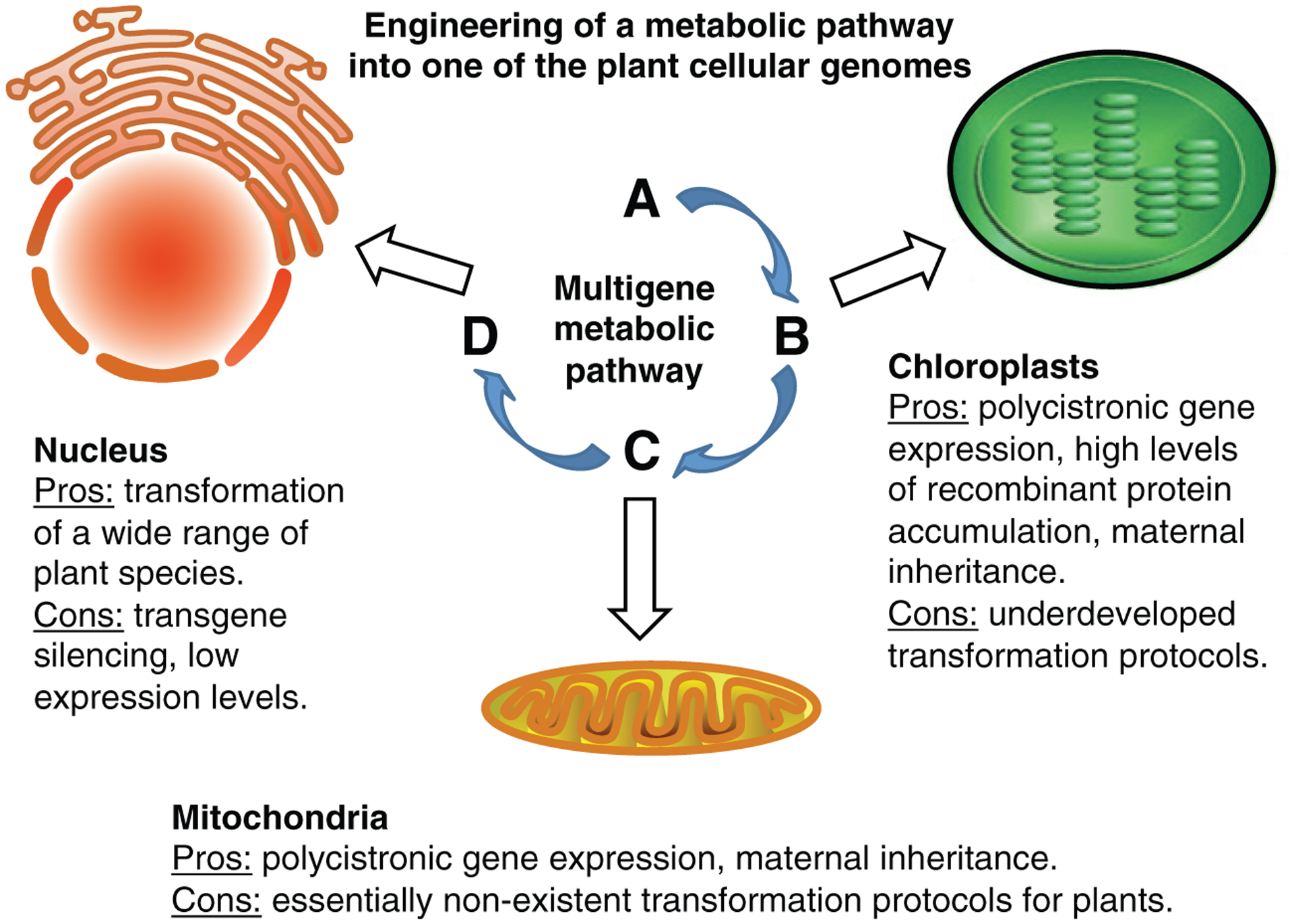
Major considerations for and against nuclear, plastid and mitochondrial transformations for genetic engineering of multigene pathways into plants.
References
- Arai Y, Shikanai T, Doi Y, Yoshida S, Yamaguchi I, and Nakashita H (2004). Production of polyhydroxybutyrate by polycistronic expression of bacterial genes in tobacco plastid. Plant Cell Physiol. 45, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Block MD, Botterman J, Vandewiele M, Dockx J, Thoen C, Gosselé V, Movva NR, Thompson C, Montagu MV, and Leemans J (1987). Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 6, 2513–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MD, Schell J, and Montagu MV (1985). Chloroplast transformation by Agrobacterium tumefaciens. EMBO J. 4, 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert-Tatarev K, McAvoy S, Daughtry S, Peoples OP, and Snell KD (2011). High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol. 155, 1690–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell T, and Christou P (2004). Progress in plant metabolic engineering. Curr. Opin. Biotechnol 15, 148–154. [DOI] [PubMed] [Google Scholar]
- Carlson WR, and Roseman RR (1992). A new property of the maize B chromosome. Genetics 131, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Johnson AM, Boynton JE, and Gillham NW (1995). Inhibition of chloroplast DNA recombination and repair by dominant negative mutants of Escherichia coli RecA. Mol. Cell. Biol 15, 3003–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Marmey P, Taylor NJ, Brizard JP, Espinoza C, D’Cruz P, Huet H, Zhang S, de Kochko A, Beachy RN, and Fauquet CM (1998). Expression and inheritance of multiple transgenes in rice plants. Nat. Biotechnol 16, 1060–1064. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Kim S, Kim M, and Kim BD (2001). Production of transgenic male sterile tobacco plants with the cDNA encoding a ribosome inactivating protein in Dianthus sinensis L. Mol. Cells 11, 326–333. [PubMed] [Google Scholar]
- Dafny-Yelin M, and Tzfira T (2007). Delivery of multiple transgenes to plant cells. Plant Physiol. 145, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H (2006). Production of biopharmaceuticals and vaccines in plants via the chloroplast genome. Biotechnol. J 1, 1071–1079. [DOI] [PubMed] [Google Scholar]
- Datta K, Baisakh N, Thet KM, Tu J, and Datta SK (2002). Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet 106, 1–8. [DOI] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi J, Holappa LD, Albert H, and Ow DW (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes. Dev 14, 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, and Daniell H (2001). Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol 19, 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y (1990). Microbial Polyesters. VCH Publishers, NY. [Google Scholar]
- Eike MC, Mercy IS, and Aalen RB (2005). Transgene silencing may be mediated by aberrant sense promoter sequence transcripts generated from cryptic promoters. Cell. Mol. Life Sci 62, 3080–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Kuhlmann M, Pecinka A, Schmidt R, and Mette MF (2008). Local DNA features affect RNA-directed transcriptional gene silencing and DNA Expression of complete metabolic pathways in transgenic plants methylation. Plant J. 53, 1–10. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Takita E, Harada H, Sakurai N, Suzuki H, Ohyama K, Shibata D, and Misawa N (2009). Pathway engineering of Brassica napus seeds using multiple key enzyme genes involved in ketocarotenoid formation. J. Exp. Bot 60, 1319–1332. [DOI] [PubMed] [Google Scholar]
- Gardiner K (1990). Pulsed field gel electrophoresis and investigations into mammalian genome organization. J. Cell Sci 96, 5–8. [DOI] [PubMed] [Google Scholar]
- Gelvin SB (1998). The introduction and expression of transgenes in plants. Curr. Opin. Biotechnol 9, 227–232. [DOI] [PubMed] [Google Scholar]
- Goderis IJ, De Bolle MF, Francois IE, Wouters PF, Broekaert WF, and Cammue BP (2002). A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol. Biol 50, 17–27. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, and McFadden GI (2008). Plastid evolution. Annu. Rev. Plant Biol 59, 491–517. [DOI] [PubMed] [Google Scholar]
- Hagemann R (2004). The sexual inheritance of plant organelles. In Molecular Biology and Biotechnology of Plant Organelles (Daniell H, and Chase CD, eds.), pp. 93–113, Springer, Dordrecht, The Netherlands. [Google Scholar]
- Halpin C (2005). Gene stacking in transgenic plants - the challenge for 21st century plant biotechnology. Plant Biotechnol. J 3, 141–155. [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, and Tanksley SD (1996). Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. USA 93, 9975–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman CE, Halter CP, Owens TG, and Hanson MR (2005). Expression of complementary RNA from chloroplast transgenes affects editing efficiency of transgene and endogenous chloroplast transcripts. Nucl. Acids Res 33, 1454–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz PB (2000). Genetic engineering of the chloroplast. Biochimie 82, 655–666. [DOI] [PubMed] [Google Scholar]
- Houben A, Dawe RK, Jiang J, and Schubert I (2008). Engineered plant minichromosomes: a bottom-up success? Plant Cell 20, 8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, and Schubert I (2007). Engineered plant minichromosomes: a resurrection of B chromosomes? Plant Cell 19, 2323–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M (1996). Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet 12, 224–228. [DOI] [PubMed] [Google Scholar]
- Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, and Jiang J (2005). Molecular and functional dissection of the maize B chromosome centromere. Plant Cell 17, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert JR, Vidal JR, and Reisch BI (2005). Stable transformation of plant cells by particle bombardment/biolistics. Methods Mol. Biol 286, 61–78. [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Meyers B, Vainstein A, Maliga P, and Citovsky V (2010). Autoluminescent plants. PLoS ONE 5, e15461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Li J, Tzfira T, and Citovsky V (2006). Will you let me use your nucleus? How Agrobacterium gets its T-DNA expressed in the host plant cell. Can. J. Physiol. Pharmacol 84, 333–345. [DOI] [PubMed] [Google Scholar]
- Lazzeri PA (1995). Stable transformation of barley via direct DNA uptake. Electroporation- and PEG-mediated protoplast transformation. Methods Mol. Biol 49, 95–106. [DOI] [PubMed] [Google Scholar]
- Lee LY, and Gelvin SB (2008). T-DNA binary vectors and systems. Plant Physiol. 146, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelavathi S, and Reddy VS (2003). Chloroplast expression of His-tagged GUS-fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol. Breed 11, 49–58. [Google Scholar]
- Lutz KA, Bosacchi MH, and Maliga P (2006). Plastid marker-gene excision by transiently expressed CRE recombinase. Plant J. 45, 447–456. [DOI] [PubMed] [Google Scholar]
- Ma JK, Hiatt A, Hein M, Vine ND, Wang F, Stabila P, van Dolleweerd C, Mostov K, and Lehner T (1995). Generation and assembly of secretory antibodies in plants. Science 268, 716–719. [DOI] [PubMed] [Google Scholar]
- Maliga P (2003). Progress towards commercialization of plastid transformation technology. Trends Biotechnol. 21, 20–28. [DOI] [PubMed] [Google Scholar]
- Maliga P, Staub J, Carrer H, Kanevski I, and Svab Z (1994). Homologous recombination and integration of foreign DNA in plastids of higher plants. In Homologous Recombination and Gene Silencing in Plants (Paszkowski J, ed.), pp. 83–93, Kluwer Academic, Amsterdam. [Google Scholar]
- Meighen EA (1991). Molecular biology of bacterial bioluminescence. Microbiol. Rev 55, 123–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighen EA (1993). Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7, 1016–1022. [DOI] [PubMed] [Google Scholar]
- Meyers B, Zaltsman A, Lacroix B, Kozlovsky SV, and Krichevsky A (2010). Nuclear and plastid genetic engineering of plants: comparison of opportunities and challenges. Biotechnol. Adv 28, 747–756. [DOI] [PubMed] [Google Scholar]
- Mullen J, Adam G, Blowers A, and Earle E (1998). Biolistic transfer of large DNA fragments to tobacco cells using YACs retrofitted for plant transformation. Mol. Breed 4, 449–457. [Google Scholar]
- Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, and Christou P (2010). When more is better: multigene engineering in plants. Trends Plant Sci. 15, 48–56. [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, and Bock R (2009). Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445. [DOI] [PubMed] [Google Scholar]
- Qi B, Fraser T, Mugford S, Dobson G, Sayanova O, Butler J, Napier JA, Stobart AK, and Lazarus CM (2004). Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat. Biotechnol 22, 739–745. [DOI] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, and Daniell H (2005). Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, and translation. Plant Physiol. 138, 1746–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk VV, Ryschka U, Schumann G, and Klocke E (2002). Genetic transformation of cauliflower (Brassica oleracea var. botrytis) by direct DNA uptake into mesophyll protoplasts. Physiol. Plant 114, 429–438. [DOI] [PubMed] [Google Scholar]
- Remacle C, Cardol P, Coosemans N, Gaisne M, and Bonnefoy N (2006). Highefficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. proc. Natl. Acad. Sci. USA 103, 4771–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somleva MN, Snell KD, Beaulieu JJ, Peoples OP, Garrison BR, and Patterson NA (2008). Production of polyhydroxybutyrate in switchgrass, a valueadded co-product in an important lignocellulosic biomass crop. Plant Biotechnol. J 6, 663–678. [DOI] [PubMed] [Google Scholar]
- Stoddard BL (2005). Homing endonuclease structure and function. Q. Rev. Biophys 38, 49–95. [DOI] [PubMed] [Google Scholar]
- Twyman RM, Christou P, and Stoger E (2002). Genetic transformation of plants and their cells. In Plant Biotechnology and Transgenic Plants (Oksman-Caldentey KM, and Barz W, eds.), pp. 111–141, Marcel Dekker Inc. [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, and Citovsky V (2005). pSAT vectors: a modular series of plasmids for fluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol 57, 503–516. [DOI] [PubMed] [Google Scholar]
- van Wordragen MF, Honée G, and Dons HJ (1993). Insect-resistant chrysanthemum calluses by introduction of a Bacillus thuringiensis crystal protein gene. Transgenic Res. 2, 170–180. [DOI] [PubMed] [Google Scholar]
- Vaucheret H (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, and Nazar RN (1991). Evidence for T-DNA mediated gene targeting to tobacco chloroplasts. Biotechnology 9, 1103–1105. [DOI] [PubMed] [Google Scholar]
- Verma D, Samson NP, Koya V, and Daniell H (2008). A protocol for expression of foreign genes in chloroplasts. Nat. Protoc 3, 739–758. [DOI] [PubMed] [Google Scholar]
- Wada N, Kajiyama S, Akiyama Y, Kawakami S, No D, Uchiyama S, Otani M, Shimada T, Nose N, Suzuki G, Mukai Y, and Fukui K (2009). Bioactive beads-mediated transformation of rice with large DNA fragments containing Aegilops tauschii genes. Plant Cell Rep. 28, 759–68. [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, and Potrykus I (2000). Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287, 303–305. [DOI] [PubMed] [Google Scholar]
- Zerges W (2000). Translation in chloroplasts. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]


