Abstract
In Arabidopsis thaliana , the Sigma factor B regulator RsbQ‐like family of α/β hydrolases contains the strigolactone (SL) receptor DWARF14 (AtD14), the karrikin receptor KARRIKIN INSENSITIVE2 (AtKAI2), and DWARF14‐LIKE2 (AtDLK2), a protein of unknown function. Despite very similar protein folds, AtD14 and AtKAI2 differ in size and architecture of their ligand binding pockets, influencing their substrate specificity. We present the 1.5 Å crystal structure of AtDLK2, revealing the smallest ligand binding pocket in the protein family, bordered by two unique glycine residues. We identified a gatekeeper residue in the protein's lid domain and present a pyrrolo‐quinoline‐dione compound that inhibits AtDLK2's enzymatic activity.
1. INTRODUCTION
Strigolactones (SLs) are a class of butenolide‐bearing phytohormones that have roles as germination stimulants for parasitic plants (Cook et al., 1966), as rhizospheric signals to arbuscular mycorrhizal fungi (Akiyama et al., 2005), and as branching inhibitors inside the plant body (Gomez‐Roldan et al., 2008; Umehara et al., 2008). SLs are perceived by the protein DWARF14 (D14), an enzyme with low or single substrate turnover (Machin et al., 2019). D14 and its homologs in other species are dual receptor hydrolases that bind and hydrolyze SLs (de Saint Germain et al., 2016; Hamiaux et al., 2012; Yao et al., 2016). In Arabidopsis thaliana, D14 has two paralogs, KARRIKIN INSENSITIVE2 (KAI2) and DWARF14‐LIKE2 (DLK2) (Waters et al., 2012). KAI2 is involved in many developmental processes, and it perceives karrikins, a class of butenolide‐containing compounds that are enriched in the soil after plant combustion (Flematti et al., 2004; Nelson et al., 2010). Besides acting as karrikin receptor, there is evidence that KAI2 perceives an unknown ligand, usually referred to as KAI2 ligand (KL) (Conn & Nelson, 2015). KAI2 gets also activated by binding of the nonnatural stereoisomer of the synthetic SL analog GR24 (Scaffidi et al., 2014), which somewhat works as a chemical mimic of the KL. DLK2's function is mostly unknown, but it has been associated with regulating photomorphogenesis in Arabidopsis seedlings (Végh et al., 2017) and being involved in symbiosis with arbuscular mycorrhizal fungi in tomato (Ho‐Plagaro et al., 2020) and in rice (Sisaphaithong et al., 2021). DLK2 might be involved in other processes that are not easily identifiable due to the aphenotypic nature of the dlk2 mutant (Végh et al., 2017; Waters et al., 2012). In addition, no DLK2 ligand has been identified. D14 and KAI2 both fold into an α/β hydrolase architecture containing a core domain and a four‐helix lid that covers the entrance to the ligand binding pocket harboring at its bottom a conserved catalytic triad of serine/histidine/aspartate. Whereas the overall protein folds are very similar (AtD14 has an RMSD of 1.23 Å over 257 amino acids when superimposed on AtKAI2), the ligand binding pockets differ in shape and size, with D14's pocket being wider and having a larger volume than in KAI2, which is the case in both Arabidopsis and rice (Kagiyama et al., 2013; Zhao et al., 2013). The differences between these pockets are important factors, as a combination of pocket shape and size seems to determine ligand specificity (Machin et al., 2019). Here, we report the crystal structure of A. thaliana DLK2, revealing that despite having a protein fold very similar to AtD14 and AtKAI2, it features a much narrower and smaller substrate binding pocket. In addition, we identified a specific γ‐lactam containing pyrrolo‐quinoline‐dione inhibitor for AtDLK2, which might help to elucidate the protein's function.
2. MATERIAL AND METHODS
2.1. Molecular cloning
For protein production in Escherichia coli, all genes (AtDLK2 wt and mutants, A. thaliana SUPPRESSOR OF AVRBST‐ELICITED RESISTANCE 1 [AtSOBER1], AtTIPSY1, AtD14, and AtKAI2) were synthesized codon‐optimized for E. coli and cloned into a pGEX 4T1 expression vector. Genes were designed to encode an N‐terminal site for HRV3 protease, leaving two amino acids Gly‐Pro as N‐terminal cloning artifact.
2.2. Protein expression and purification
E. coli BL21 (DE3) cells were transformed, grown overnight in Lysogenic Broth (LB) medium, and the next day, used to start a new culture in Terrific Broth (TB) medium with a 1:100 dilution. Cells were grown at 23°C until an OD600 of 0.6 and induced with 0.1 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) at 18°C overnight. Cells were harvested and lysed using sonication, cell debris was removed by centrifugation at 75,000 g for 45 min, and the supernatant was loaded onto a glutathione affinity column. The column was washed with 50 mM TRIS‐HCl, 150 mM NaCl, 5% glycerol, 1 mM TCEP, final pH 7.7 until no protein flow‐through was found by UV detection. HRV3 protease was added on the column overnight. The cleaved target protein was eluted using the same buffer and further purified to homogeneity by size exclusion chromatography using a GE Healthcare HiLoad 16/600 Superdex 75 pg column in 20 mM TRIS‐HCl, 30 mM NaCl, 1 mM TCEP‐HCl, final pH 7.7. Proteins were concentrated to at least 10 mg/ml and frozen in liquid nitrogen. For the chemical array screening of the Natural Products Depository (NPDepo), no HRV3 protease was added, but instead the intact GST‐fusion protein was eluted using 50 mM TRIS‐HCl, 150 mM NaCl, 5% glycerol, 1 mM TCEP, 20 mM glutathione, final pH 7.7, and further purified to homogeneity by size exclusion chromatography using a GE Healthcare HiLoad 16/600 Superdex 200 pg column in 20 mM TRIS‐HCl, 30 mM NaCl, 1 mM TCEP‐HCl, final pH 7.7.
2.3. Protein crystallization and structure solution
AtDLK2 crystals were grown under the following conditions in 2 μl hanging drops using a 1:1 protein:reservoir ratio: 100 mM TRIS, 1.9 M ammonium sulfate. 1.2 M Na malonate was used as cryoprotectant. X‐ray data were collected at the Advanced Light Source at Lawrence Berkeley National Laboratory at beamline 8.2.1. X‐ray data were processed with XDS (Kabsch, 2010). The AtDLK2 structure was solved by molecular replacement using chain A of PDB structure 4IH1 (AtKAI2). 5% of the data were flagged for R‐free, and initial models were build using AutoBuild (Terwilliger et al., 2008) as part of Phenix (Adams et al., 2010), manually corrected and finalized with Coot (Emsley et al., 2010), refined with phenix.refine (Afonine et al., 2012) and validated with MolProbity (Chen et al., 2010).
2.4. Structure visualization
Structures were visualized with CCP4mg (McNicholas et al., 2011) or UCSF Chimera (Pettersen et al., 2004). Ligand pocket volumes were calculated using the CASTp server (Tian et al., 2018).
2.5. Chemical array screening
The slides of the RIKEN NPDepo (Osada, 2010) were prepared and analyzed as previously published (Bürger et al., 2012; Hagiwara et al., 2010; Kanoh et al., 2006; Kondoh et al., 2015; Miyazaki et al., 2008; Zimmermann et al., 2013). Chemical array screening was performed in 20 mM HEPES, 150 mM NaCl, final pH 7.53–7.55. For protein and antibody incubation, the array slide was covered by a gap cover glass (24 × 60 mm) from Matsunami Glass Ind., Ltd and incubated using 50 μl of GST‐AtDLK2, GST‐AtSOBER1, or GST‐AtTIPSY1 solution (1 μM) in the above buffer containing 1% skim milk at 30°C for 1 h. After washing, array slides were incubated with anti‐GST antibody (rabbit IgG fraction, Invitrogen, 30 μg/ml) in the same buffer containing 1% skim milk at 30°C for 1 h. This incubation was followed by another wash step and incubation with a second antibody (Millipore, goat anti‐rabbit IgG, Cy5 conjugate, 50 μg/ml) at 30°C for 1 h. After the final wash step, slides were scanned at 635 nm on a GenePix 4300A microarray scanner (Molecular Devices).
2.6. Enzymatic analyses
Enzymatic activity was measured using colorimetric para‐nitrophenyl acetate, and the release of yellow para‐nitrophenol was monitored by recording the absorbance at 410 nm at room temperature using a Tecan Safire II microplate reader. Reactions were measured as triplicates in 20 mM HEPES, 150 mM NaCl, pH 7.52–7.55, .01% (v/v) Triton X‐100. Michaelis–Menten parameters and pharmacological curve fitting to determine IC50 values were carried out with Origin (OriginLab).
2.7. Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) experiments were performed in a CFX Opus 384 system (Bio‐Rad). Sypro Orange (Life Technologies) was used as reporter. Ten micrograms of protein was heat‐denatured using a linear 25–95°C gradient at a rate of 1°C per minute. The denaturation curve and its derivative were obtained using the CFX manager software. Final reaction mixtures were prepared in 20 μl volumes in triplicates in 384‐well white microplates. Reactions were carried out in 20 mM TRIS‐HCl, 30 mM NaCl, 1 mM TCEP‐HCl, final pH 7.7. A final 3× concentration of Sypro Orange was used.
2.8. Molecular docking
Files for ligand docking of compound 1 into AtDLK2 were prepared with AutoDockTools (Morris et al., 2009), and docking was performed with AutoDock Vina (Trott & Olson, 2010) using a 20 × 20 × 20 Å box covering the ligand binding site. The calculated affinity between compound 1 and AtDLK2 was −7.0 kcal/mol.
2.9. Data availability
The structural coordinates and diffraction data of AtDLK2 have been deposited in the Protein Data Bank under accession code 7TVW.
3. RESULTS
3.1. AtDLK2 has a smaller ligand binding pocket than AtD14 and AtKAI2
To investigate the similarities and differences of AtDLK2 to its paralogs AtD14 and AtKAI2, we solved the crystal structure of AtDLK2 at a resolution of 1.48 Å (Table 1). As expected, the overall fold of AtDLK2 turned out to be very similar to AtD14 and AtKAI2, with RMSDs of 1.14 Å and 1.10 Å over 259 amino acids, respectively. However, AtDLK2's ligand binding pocket volume was 321 Å3, smaller than the pockets in AtD14 (699 Å3) or in AtKAI2 (595 Å3) (Figure 1a–c). The structural reasons for the smaller pocket seem to derive from a shift of alpha helix αT1 in the lid domain of the protein, caused by a glycine residue at the end of the helix (G154), which corresponds to a glutamate and arginine in AtD14 and AtKAI2, respectively (Figure 1d). The shift causes AtDLK2 residues I149 and I153 to restrain the size of the pocket (Figure 1e–g). Another glycine residue in AtDLK2, G201 (corresponding to N196 in AtD14 and Q195 in AtKAI2), causes termination of the last alpha helix of the protein lid, αT4 (Figure 1d), producing a slightly shorter helix compared with AtD14 and AtKAI2. As a consequence, AtDLK2 residue F200 is moved further into the ligand binding pocket, restricting its volume (Figure 1h–j).
TABLE 1.
AtDLK2 X‐ray data collection and refinement statistics
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 42.3–1.48 (1.533–1.48) |
| Space group | C 2 2 21 |
| Unit cell | 79.56, 84.59, 76.88, 90.00, 90.00, 90.00 |
| Total reflections | 325,559 (26,013) |
| Unique reflections | 43,371 (4,281) |
| Multiplicity | 7.5 (6.1) |
| Completeness (%) | 99.70 (99.35) |
| Mean I/sigma(I) | 17.42 (3.17) |
| Wilson B‐factor | 14.65 |
| R‐merge | 0.07077 (0.7296) |
| R‐meas | 0.07602 |
| CC1/2 | 0.999 (0.881) |
| CC* | 1 (0.968) |
| R‐work | 0.1637 (0.2683) |
| R‐free | 0.1884 (0.3011) |
| Number of non‐hydrogen atoms | 2411 |
| Macromolecules | 2090 |
| Water | 321 |
| Protein residues | 265 |
| RMS (bonds) | 0.007 |
| RMS (angles) | 1.10 |
| Ramachandran favored (%) | 98 |
| Ramachandran allowed (%) | 2 |
| Ramachandran outliers (%) | 0 |
| Clashscore | 3.80 |
| Average B‐factor | 20.90 |
| Macromolecules | 19.00 |
| Solvent | 33.10 |
Note: Statistics for the highest‐resolution shell are shown in parentheses.
FIGURE 1.
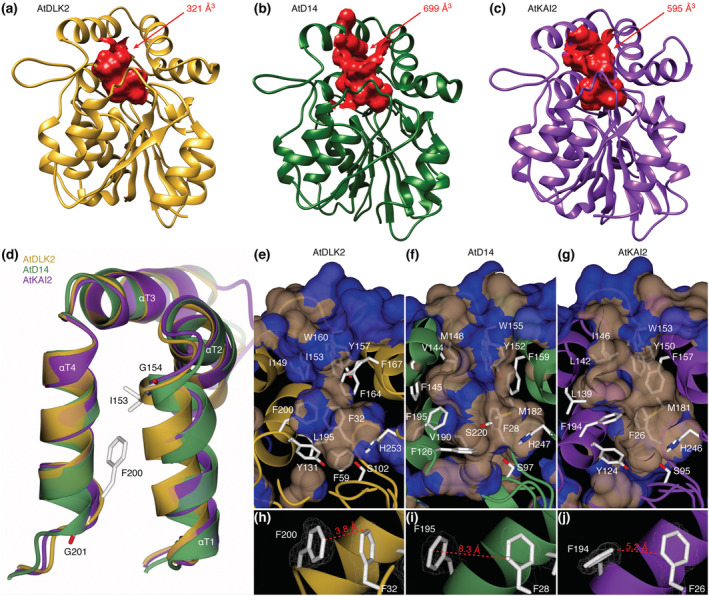
Two glycine residues border a smaller substrate binding pocket in AtDLK2. (a–c) AtDLK2 has a smaller ligand binding pocket than AtD14 or AtKAI2. (d) Superimposition of the lid domains of AtDLK2, AtD14, and AtKAI2, showing that the glycine residues G154 and G201 in AtDLK2 lead to shifts of helices αT1 and αT4, pushing residues I153 and F200 into the substrate binding pocket, restraining its size. (e–g) Visualization of the substrate binding pockets of AtDLK2, AtD14, and AtKAI2, respectively. Surface is colored by hydrophobicity, with blue as hydrophilic and gray as hydrophobic. (h–j) Distances between pocket wall forming phenylalanines, demonstrating that the shift of F200 in AtDLK2 leads to a narrower pocket diameter compared with AtD14 and AtKAI2.
3.2. AtDLK2 is an active hydrolase on para‐nitrophenyl acetate
Like AtD14 and AtKAI2, AtDLK2 has a conserved catalytic triad composed of serine, histidine, and aspartic acid. AtD14 is a poor hydrolase on the chemical SL analog rac‐GR24 with a turnover rate of 1 molecule/3 min (Zhao et al., 2013), and AtDLK2 exhibits moderate hydrolytic activity against the nonnatural SL stereoisomer (−)‐5‐deoxystrigol (Végh et al., 2017). To obtain a readout for ligand binding studies and to compare enzymatic activities within the RsbQ‐like family of α/β hydrolases in A. thaliana, we examined the hydrolytic activities of AtD14, AtKAI2, and AtDLK2 using the simple hydrolase substrate para‐nitrophenyl acetate (pNP acetate). In our assay, AtD14 displayed a catalytic efficiency (kcat/KM) of 1.6 × 102 s−1 M−1 against pNP acetate. In comparison, AtKAI2 had a catalytic efficiency of 1.5 × 104 s−1 M−1, 94 times higher than AtD14. When we tested AtDLK2, we measured its catalytic efficiency to be 1.0 × 105 s−1 M−1, 625 times higher than AtD14 and 6.7 times higher than AtKAI2 (Table 2), thus concluding that AtDLK2 is the most active enzyme against pNP acetate in the RsbQ‐like family of α/β hydrolases in A. thaliana.
TABLE 2.
Kinetic parameters of AtDLK2 proteins on para‐nitrophenyl acetate
| pNP acetate | AtDLK2 wt | AtDLK2 F167L | AtDLK2 F167A | AtD14 | AtKAI2 |
|---|---|---|---|---|---|
| KM [M] | 2.58 ± 0.10 × 10−4 | 2.53 ± 0.16 × 10−4 | 2.78 ± 0.08 × 10−4 | 2.00 ± 0.16 × 10−2 | 1.56 ± 0.06 × 10−3 |
| kcat [s−1] | 2.66 ± 0.07 × 101 | 6.98 ± 0.15 × 101 | 6.69 ± 0.06 × 101 | 3.20 ± 0.06 × 100 | 2.36 ± 0.06 × 101 |
| kcat/KM [s−1 M−1] | 1.03 ± 0.04 × 105 | 2.76 ± 0.17 × 105 | 2.41 ± 0.07 × 105 | 1.60 ± 0.12 × 102 | 1.51 ± 0.06 × 104 |
Notes: Enzymatic activity was measured in triplicates as absorbance of para‐nitrophenol at 410 nm as result of enzymatic activity against pNP acetate. ± symbols represent standard deviation.
3.3. Phe167 acts as a gatekeeper residue at the substrate pocket entrance of AtDLK2
In the AtDLK2 crystal structure, the entrance to the substrate binding pocket is partially blocked by the side chain of amino acid Phe167 (Figure 2). To test whether this residue affects the enzymatic properties of AtDLK2, we designed protein versions in which we replaced the phenylalanine with the smaller hydrophobic amino acid leucine or with an alanine (AtDLK2 F167L and AtDLK2 F167A, respectively). We found that although these substitutions only marginally changed the Km values to pNP acetate, the turnover numbers (kcat) increased from 27 molecules per second in AtDLK2 wt to 70 in AtDLK2 F167L and to 67 in AtDLK2 F167A (Table 2). These results suggest that Phe167 serves as a gatekeeper residue restricting the turnover rate in the AtDLK2 enzyme.
FIGURE 2.
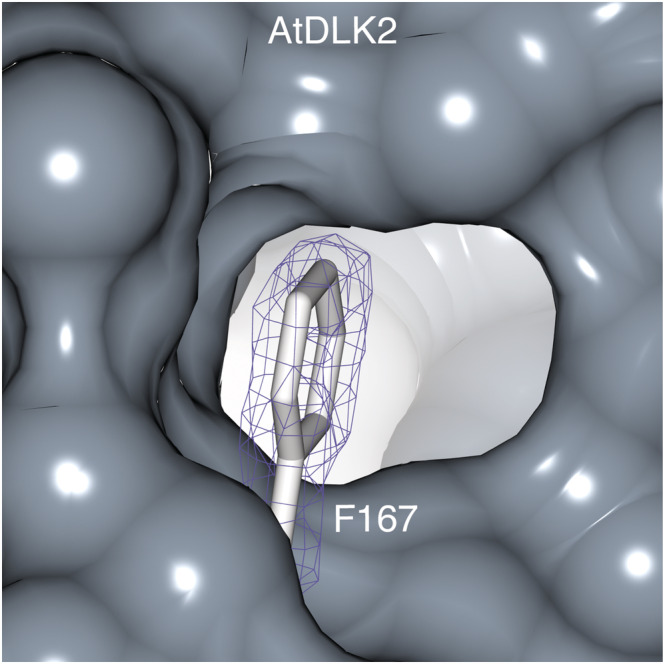
Phe167 partially blocks the entrance into the substrate binding pocket of AtDLK2. The 2mF o − DF c electron density map of the Phe167 side chain is contoured at 1 σ and shown in blue.
3.4. Phe167 is not the reason why AtDLK2 does not perceive SLs
To test whether the gatekeeper residue Phe167 restricts AtDLK2 from perceiving SLs, we used DSF to monitor protein destabilization in the presence of the chemical SL analog rac‐GR24. Consistent with a previous report in which the SL stereoisomers (+)‐5‐deoxystrigol and (−)‐5‐deoxystrigol were used (Végh et al., 2017), we observed no or marginal rac‐GR24‐induced destabilization of wild‐type AtDLK2 in the DSF assay (Figure 3a). Crucially, this was also the case when we used either AtDLK2 F167L (Figure 3b) or AtDLK2 F167A (Figure 3c), whereas rac‐GR24 did clearly destabilize the SL receptor AtD14 (Figure 3d).
FIGURE 3.
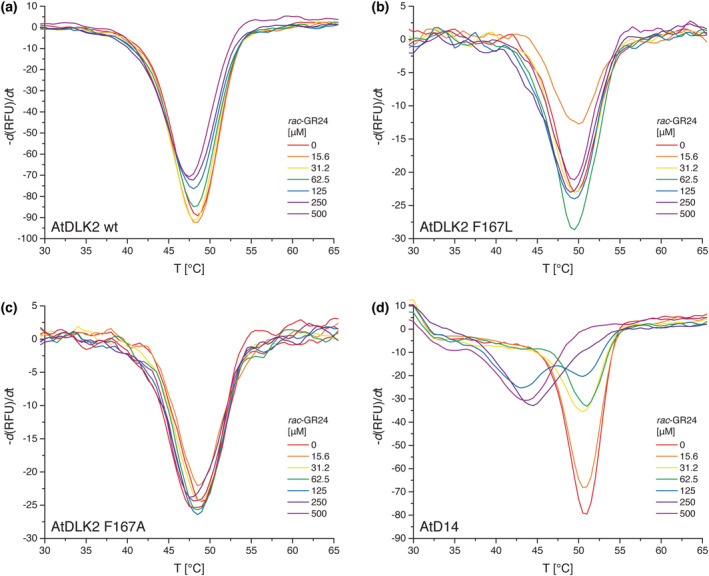
Thermal stability of (a) AtDLK2 wt, (b) AtDLK2 F167L, (c) AtDLK2 F167A, and (d) AtD14 in the presence of rac‐GR24. Proteins were heat‐denatured in triplicates in the presence of Sypro Orange dye using a linear 25–95°C gradient at a rate of 1°C per minute.
3.5. The experimental AtDLK2 structure differs from the AlphaFold prediction
Recently, AlphaFold, a novel protein structure prediction tool, was published (Jumper et al., 2021). AlphaFold outperforms other protein prediction methods and demonstrates accuracy that is competitive with experimental structures in many cases. In general, the α/β hydrolase fold is relatively easy to predict; however, the architecture is very flexible, and minor changes can alter protein function (Ollis et al., 1992). To assess how reliable AlphaFold is in this context, especially relating to pocket size and shape within the RsbQ‐like family, we compared the experimental AtDLK2 structure with the AlphaFold prediction. As expected, the AlphaFold structure had a very high backbone accuracy of 0.53 Å over 254 amino acids. When we analyzed the substrate binding pockets in both proteins, we found that the pocket in the AlphaFold structure was only 155 Å3 and that it was disconnected from the protein surface. In comparison, we found an open 321 Å3 pocket in the experimental AtDLK2 structure (Figure S1a,b). While AlphaFold correctly predicted the position and side‐chain orientation of the gatekeeper residue Phe167, the smaller pocket seemed to originate from an inaccurate prediction of the side‐chain orientation of residue Phe200, which was oriented away from the pocket in the experimental AtDLK2 structure (Figure S1c,d).
3.6. Small‐molecule screens for α/β hydrolases identify a specific AtDLK2 inhibitor
To identify new ligands for α/β hydrolases, we screened AtDLK2, A. thaliana SUPPRESSOR OF AVRBST‐ELICITED RESISTANCE 1 (AtSOBER1) (which has a shallow ligand binding site), and AtTIPSY1 (which has a tunnel‐like hydrophobic ligand binding site) (Bürger et al., 2017) against a large library of small molecules from the RIKEN NPDepo by chemical array, a protein‐small‐molecule binding assay. We then verified 639 binders from the three screens in a secondary screen using the above‐described enzymatic activity of AtDLK2 toward pNP acetate. We obtained 4,4,6‐trimethyl‐6‐phenyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione (1) (NPD14805), a small‐molecule hit for AtSOBER1, as strongest inhibitor for AtDLK2's enzymatic activity, with an IC50 of 3.1 μM (Figure 4a).
FIGURE 4.
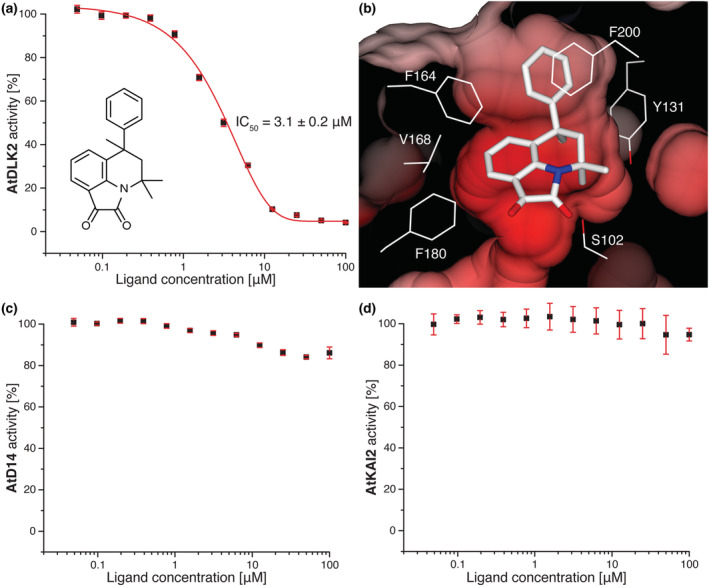
4,4,6‐Trimethyl‐6‐phenyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione (1) inhibits AtDLK2 activity. (a) Enzymatic activity was measured in triplicates as absorbance of para‐nitrophenol at 410 nm as result of enzymatic activity against pNP acetate. All values have been corrected for spontaneous pNP acetate hydrolysis. Error bars represent standard deviation. (b) Molecular docking of the inhibitor into the ligand binding pocket of AtDLK2. The protein surface is shown as electrostatic potential contoured from −12.8 kT e−1 (red) to +12.8kT e−1 (blue). (c–d) Compound 1 only marginally inhibits the enzymatic activity of AtD14 or AtKAI2.
Unfortunately, our attempts to co‐crystallize this compound with AtDLK2 or soak it into AtDLK2 protein crystals were not successful, possibly due to the low solubility of the chemical under the crystallization condition. We therefore used molecular docking to assess its binding to AtDLK2. Our docking results suggest that the inhibitor fits well into the substrate binding site of AtDLK2. Although most of the molecule was coordinated by the hydrophobic residues at the sides of the binding pocket, the more hydrophilic γ‐lactam unit was oriented towards the active site serine S102 (Figure 4b). We tested chemical derivatives of compound 1 and found that the addition of a chlorine moiety to the phenol ring (compound 2) led to an increase of the IC50 to about 25 μM and that either the addition of a fourth methyl group (compound 3) or removal of the phenol ring (compound 4) increased the IC50 to greater than 100 μM (Table 3 and Figure S2).
TABLE 3.
Inhibition of AtDLK2 by different pyrrolo‐quinoline‐dione compounds
| Compound | Chemical structure | Name | IC50 to AtDLK2 |
|---|---|---|---|
| 1 |
|
4,4,6‐Trimethyl‐6‐phenyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione | 3.1 ± .2 μM |
| 2 |
|
6‐(4‐Chlorophenyl)‐4,4,6‐trimethyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione | >25 μM |
| 3 |
|
4,4,6,9‐Tetramethyl‐6‐phenyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione | >100 μM |
| 4 |
|
4,4,6‐Trimethyl‐5,6‐dihydro‐4H‐pyrrolo[3,2,1‐ij]quinoline‐1,2‐dione | >100 μM |
Notes: Enzymatic activity was measured in triplicates as absorbance of para‐nitrophenol at 410 nm as result of enzymatic activity against pNP acetate. All values have been corrected for spontaneous pNP acetate hydrolysis. ± symbols represent standard deviation.
To assess specificity of compound 1 within the A. thaliana RsbQ‐like family, we tested the chemical in the same enzymatic assay using pNP acetate as a substrate and with AtD14 or AtKAI2 as proteins. We observed a slight decrease of AtD14's enzymatic activity when 1 was used at concentrations of 12.5 μM or higher and a marginal decrease of AtKAI2's activity when 1 was used at concentrations of 50 μM or higher (Figure 4c,d). Even at a 100 μM concentration of 1, AtD14 and AtKAI2 still retained activities of 84% and 95%, respectively. We thus conclude that compound 1 is a specific AtDLK2 inhibitor.
4. DISCUSSION
Here, we present the crystal structure of A. thaliana DLK2, which, as a major difference to its paralogs AtD14 and AtKAI2, features a significantly smaller substrate binding pocket. Previous structures of strigolactone receptors or KAI2 clade proteins have shown that not only the substitution of amino acids inside the substrate binding pocket determines its size or ligand specificity (Guercio et al., 2022; Toh et al., 2015) but that also interactions between residues located in different secondary structure elements can influence the volume and shape of the pocket. This has been observed in a hydrogen bond between helices αD1 and αD3 in hyposensitive to light proteins from Striga hermonthica (ShHTL) (Xu et al., 2018) and in a loop region between helices αE and αF in KAI2‐like proteins from Physcomitrium patens (Bürger et al., 2019). Here, two glycine residues in helices αT1 and αT4 possibly lead to a narrower and smaller substrate binding pocket in AtDLK2. The presence of a conserved catalytic triad in the DLK2 clade of proteins and the fact that AtDLK2 is an active hydrolase suggest that DLK2's real substrate is likely to undergo hydrolysis. This might be analogous to what is expected from the yet to be discovered KL, because the catalytic serine is required for KAI2's enzymatic activity and for the rescue of kai2 mutants (Waters et al., 2014; Waters, Scaffidi, Flematti, & Smith, 2015; Waters, Scaffidi, Moulin, et al., 2015). Moreover, there is increasing evidence that karrikins, despite being able to bind to KAI2 distant from the active site (Guo et al., 2013), need to get metabolized before they become the bioactive signal (Sepulveda et al., 2022). Although it has been shown that AtDLK2 has poor but detectable hydrolytic activity against the nonnatural SL isomer (−)‐5‐deoxystrigol and is able to react with it at high concentrations in DSF experiments (Végh et al., 2017), the protein's small pocket likely disfavors molecules from the SL class from getting fully accommodated. Unlike in AtD14 and AtKAI2, where the pocket entrance is open, it is partially blocked in AtDLK2 by the side chain of Phe167. Although the removal or replacement of Phe167 with a smaller side chain has increased the turnover number of the hydrolase substrate pNP acetate, these alterations have not changed the fact that AtDLK2 did not react to the SL analog GR24 in DSF assays. This might corroborate the assumption that the substrate binding pocket architecture of AtDLK2 disfavors SLs. The concept of gatekeeper regions in the protein family is not new. In a previous study, conformational differences of helix αD1 have been observed in S. hermonthica KAI2iB structures, suggesting that this helix could act as a gatekeeper for ligand entry and exit (Xu et al., 2016). In addition, a recent study has suggested that the conserved Phe28 residue in D14/KAI2 proteins may act as a gatekeeper and could be a possible explanation for the single turnover kinetics observed with some SL analogs (Lopez‐Obando et al., 2021). The recent interest of the scientific community in AlphaFold has prompted us to compare the experimental AtDLK2 structure with the in silico structure generated by the AlphaFold prediction tool. Despite the very high overall match with the experimental structure and the correct prediction of most side‐chain orientations, the orientation of the Phe200 side chain differed between prediction and experiment, resulting in an inaccurate substrate binding pocket size and shape in the AlphaFold‐predicted structure. This demonstrates that in silico prediction of mechanistic details in α/β hydrolases might remain challenging, despite the major improvement that AlphaFold represents.
We have identified a pyrrolo‐quinoline‐dione compound that targets AtDLK2's activity. Our docking simulation suggested that there would be little space for modifications at the pyrrolo‐quinoline‐dione core structure due to the narrow fit of this part of the molecule inside AtDLK2's binding pocket. This seems to be corroborated by the increase of the experimentally observed IC50 value from 3 to 100 μM upon the addition of a fourth methyl group to the inhibitor. Likewise, the docking study suggested that the phenyl ring is coordinated by hydrophobic residues in the binding pocket, which is in agreement with an increased IC50 to 25 μM after addition of a hydrophilic chlorine group to the phenol ring. The IC50 of 3 μM between compound 1 and AtDLK2 is in a comparable range to the affinities of SLs to their receptors, for example, 0.9 μM between (+)‐5‐deoxystrigol and ShHTL7 (Wang et al., 2021) and 17 μM between rac‐GR24 and P. patens KAI2‐like E (Bürger et al., 2019), and to the affinity between the karrikin KAR1 and AtKAI2, which is 9 μM (Guo et al., 2013). However, due to the non‐hydrolyzable nature of the inhibitor, we think that it would not be appropriate to draw conclusions from this compound about DLK2's natural ligand. In this study, compound 1 acted as specific inhibitor for AtDLK2, only marginally decreasing the activity of AtD14 or AtKAI2. However, compound 1 bears some similarities to a quinazolinone derivative that was identified as an inhibitor of DECREASED APICAL DOMINANCE 2 (DAD2) (Hamiaux et al., 2019), the Petunia × hybrida homolog of AtD14. In this case, the inhibitor showed an unexpected orientation inside the structure of DAD2, with the hydroxyl group of the compound's Y ring making contact to the protein's catalytic serine. Although in silico studies about ligand–protein interaction often deviate from experimental structural data, the γ‐lactam unit in compound 1 constitutes its only hydrophilic part. Therefore, it appears unlikely that it would orient away from AtDLK2's catalytic site. Because the biological role of DLK2 is unclear, we hope that the inhibitor presented here will contribute to elucidating this protein's function. It would also allow to conduct dose‐dependent studies and investigate DLK2's enzymatic function separately from other roles that the protein might have, for example scaffolding, which would be eliminated in dlk2 mutant lines. Finally, we hope that our results aid in the further development of inhibitors against different members of the RsbQ‐like protein family in plants by understanding the structural requirements for binding to its different paralogs.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Figure S1: Comparison of the AtDLK2 structure predicted by AlphaFold and the experimentally obtained AtDLK2 structure. (a‐b) The predicted AtDLK2 structure has a ligand binding pocket that is disconnected from the protein surface and has a smaller volume, compared to the experimental X‐ray structure of AtDLK2. (c‐d) The positions of the gatekeeper residue Phe167 match well between the predicted and the experimental structures, however, the side chain of Phe200 closes the pocket in the AlphaFold‐predicted structure. The 2mFo − DFc electron density map of the Phe167 and Phe200 side chains in the experimental structure is contoured at 1 σ and shown in blue.
Figure S2: Inhibition of AtDLK2 by pyrrolo‐quinoline‐dione compounds (a) 2, (b) 3, and (c) 4. Enzymatic activity was measured in triplicates as absorbance of para‐nitrophenol at 410 nm as result of enzymatic activity against paranitrophenyl acetate. All values have been corrected for spontaneous para‐nitrophenyl acetate hydrolysis. Error bars represent standard deviation.
ACKNOWLEDGMENTS
We thank members of RIKEN NPDepo for providing chemical compounds. We thank the staff at Advanced Light Source at the Berkeley Center for Structural Biology for assistance with X‐ray data collection. The Berkeley Center for Structural Biology is supported in part by the NIH, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the U.S. Department of Energy (contract DE‐AC0205CH11231). This study has been supported by the National Institutes of Health (NIH) grants (R01 GM52413, R01 GM094428, and R35 GM122604). J.C. is an investigator of the Howard Hughes Medical Institute.
Bürger, M. , Honda, K. , Kondoh, Y. , Hong, S. , Watanabe, N. , Osada, H. , & Chory, J. (2022). Crystal structure of Arabidopsis DWARF14‐LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture. Plant Direct, 6(9), e446. 10.1002/pld3.446
REFERENCES
- Adams, P. D. , Afonine, P. V. , Bunkoczi, G. , Chen, V. B. , Davis, I. W. , Echols, N. , Headd, J. J. , Hung, L. W. , Kapral, G. J. , Grosse‐Kunstleve, R. W. , McCoy, A. J. , Moriarty, N. W. , Oeffner, R. , Read, R. J. , Richardson, D. C. , Richardson, J. S. , Terwilliger, T. C. , & Zwart, P. H. (2010). PHENIX: A comprehensive Python‐based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography, 66, 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine, P. V. , Grosse‐Kunstleve, R. W. , Echols, N. , Headd, J. J. , Moriarty, N. W. , Mustyakimov, M. , Terwilliger, T. C. , Urzhumtsev, A. , Zwart, P. H. , & Adams, P. D. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica. Section D, Biological Crystallography, 68, 352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K. , & Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Bürger, M. , Mashiguchi, K. , Lee, H. J. , Nakano, M. , Takemoto, K. , Seto, Y. , Yamaguchi, S. , & Chory, J. (2019). Structural basis of karrikin and non‐natural strigolactone perception in Physcomitrella patens . Cell Reports, 26, 855–865. 10.1016/j.celrep.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger, M. , Willige, B. C. , & Chory, J. (2017). A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nature Communications, 8, 2201. 10.1038/s41467-017-02347-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger, M. , Zimmermann, T. J. , Kondoh, Y. , Stege, P. , Watanabe, N. , Osada, H. , Waldmann, H. , & Vetter, I. R. (2012). Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases. Journal of Lipid Research, 53, 43–50. 10.1194/jlr.M019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, V. B. , Arendall, W. B. III , Headd, J. J. , Keedy, D. A. , Immormino, R. M. , Kapral, G. J. , Murray, L. W. , Richardson, J. S. , & Richardson, D. C. (2010). MolProbity: All‐atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography, 66, 12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn, C. E. , & Nelson, D. C. (2015). Evidence that KARRIKIN‐INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Frontiers in Plant Science, 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, C. E. , Whichard, L. P. , Turner, B. , Wall, M. E. , & Egley, G. H. (1966). Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science, 154, 1189–1190. 10.1126/science.154.3753.1189 [DOI] [PubMed] [Google Scholar]
- de Saint Germain, A. , Clave, G. , Badet‐Denisot, M. A. , Pillot, J. P. , Cornu, D. , Le Caer, J. P. , Burger, M. , Pelissier, F. , Retailleau, P. , Turnbull, C. , Bonhomme, S. , Chory, J. , Rameau, C. , & Boyer, F. D. (2016). An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology, 12, 787–794. 10.1038/nchembio.2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W. G. , & Cowtan, K. (2010). Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography, 66, 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti, G. R. , Ghisalberti, E. L. , Dixon, K. W. , & Trengove, R. D. (2004). A compound from smoke that promotes seed germination. Science, 305, 977. 10.1126/science.1099944 [DOI] [PubMed] [Google Scholar]
- Gomez‐Roldan, V. , Fermas, S. , Brewer, P. B. , Puech‐Pages, V. , Dun, E. A. , Pillot, J. P. , Letisse, F. , Matusova, R. , Danoun, S. , Portais, J. C. , Bouwmeester, H. , Becard, G. , Beveridge, C. A. , Rameau, C. , & Rochange, S. F. (2008). Strigolactone inhibition of shoot branching. Nature, 455, 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Guercio, A. M. , Torabi, S. , Cornu, D. , Dalmais, M. , Bendahmane, A. , Le Signor, C. , Pillot, J.‐P. , Le Bris, P. , Boyer, F.‐D. , Rameau, C. , Gutjahr, C. , de Saint, G. A. , & Shabek, N. (2022). Structural and functional analyses explain pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception. Communications Biology, 5, 126. 10.1038/s42003-022-03085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Zheng, Z. , La Clair, J. J. , Chory, J. , & Noel, J. P. (2013). Smoke‐derived karrikin perception by the alpha/beta‐hydrolase KAI2 from Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 110, 8284–8289. 10.1073/pnas.1306265110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara, K. , Kondoh, Y. , Ueda, A. , Yamada, K. , Goto, H. , Watanabe, T. , Nakata, T. , Osada, H. , & Aida, Y. (2010). Discovery of novel antiviral agents directed against the influenza A virus nucleoprotein using photo‐cross‐linked chemical arrays. Biochemical and Biophysical Research Communications, 394, 721–727. 10.1016/j.bbrc.2010.03.058 [DOI] [PubMed] [Google Scholar]
- Hamiaux, C. , Drummond, R. S. , Janssen, B. J. , Ledger, S. E. , Cooney, J. M. , Newcomb, R. D. , & Snowden, K. C. (2012). DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology, 22, 2032–2036. 10.1016/j.cub.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Hamiaux, C. , Larsen, L. , Lee, H. W. , Luo, Z. , Sharma, P. , Hawkins, B. C. , Perry, N. B. , & Snowden, K. C. (2019). Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode. The Biochemical Journal, 476, 1843–1856. 10.1042/BCJ20190288 [DOI] [PubMed] [Google Scholar]
- Ho‐Plagaro, T. , Morcillo, R. J. L. , Tamayo‐Navarrete, M. I. , Huertas, R. , Molinero‐Rosales, N. , Lopez‐Raez, J. A. , Macho, A. P. , & Garcia‐Garrido, J. M. (2020). DLK2 regulates arbuscule hyphal branching during arbuscular mycorrhizal symbiosis. The New Phytologist, 229, 548–562. 10.1111/nph.16938 [DOI] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. , Tunyasuvunakool, K. , Bates, R. , Zidek, A. , Potapenko, A. , Bridgland, A. , Meyer, C. , Kohl, S. A. A. , Ballard, A. J. , Cowie, A. , Romera‐Paredes, B. , Nikolov, S. , Jain, R. , Adler, J. , … Hassabis, D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596, 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. (2010). XDS. Acta Crystallographica. Section D, Biological Crystallography, 66, 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama, M. , Hirano, Y. , Mori, T. , Kim, S. Y. , Kyozuka, J. , Seto, Y. , Yamaguchi, S. , & Hakoshima, T. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells, 18, 147–160. 10.1111/gtc.12025 [DOI] [PubMed] [Google Scholar]
- Kanoh, N. , Asami, A. , Kawatani, M. , Honda, K. , Kumashiro, S. , Takayama, H. , Simizu, S. , Amemiya, T. , Kondoh, Y. , Hatakeyama, S. , Tsuganezawa, K. , Utata, R. , Tanaka, A. , Yokoyama, S. , Tashiro, H. , & Osada, H. (2006). Photo‐cross‐linked small‐molecule microarrays as chemical genomic tools for dissecting protein‐ligand interactions. Chemistry, an Asian Journal, 1, 789–797. 10.1002/asia.200600208 [DOI] [PubMed] [Google Scholar]
- Kondoh, Y. , Honda, K. , & Osada, H. (2015). Construction and application of a photo‐cross‐linked chemical array. Methods in Molecular Biology, 1263, 29–41. 10.1007/978-1-4939-2269-7_3 [DOI] [PubMed] [Google Scholar]
- Lopez‐Obando, M. , Guillory, A. , Boyer, F. D. , Cornu, D. , Hoffmann, B. , Le Bris, P. , Pouvreau, J. B. , Delavault, P. , Rameau, C. , de Saint Germain, A. , & Bonhomme, S. (2021). The Physcomitrium (Physcomitrella) patens PpKAI2L receptors for strigolactones and related compounds function via MAX2‐dependent and independent pathways. Plant Cell, 33, 3487–3512. 10.1093/plcell/koab217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin, D. C. , Hamon‐Josse, M. , & Bennett, T. (2019). Fellowship of the rings: A saga of strigolactones and other small signals. The New Phytologist, 225, 621–636. 10.1111/nph.16135 [DOI] [PubMed] [Google Scholar]
- McNicholas, S. , Potterton, E. , Wilson, K. S. , & Noble, M. E. (2011). Presenting your structures: The CCP4mg molecular‐graphics software. Acta Crystallographica. Section D, Biological Crystallography, 67, 386–394. 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, I. , Simizu, S. , Ichimiya, H. , Kawatani, M. , & Osada, H. (2008). Robust and systematic drug screening method using chemical arrays and the protein library: Identification of novel inhibitors of carbonic anhydrase II. Bioscience, Biotechnology, and Biochemistry, 72, 2739–2749. 10.1271/bbb.80383 [DOI] [PubMed] [Google Scholar]
- Morris, G. M. , Huey, R. , Lindstrom, W. , Sanner, M. F. , Belew, R. K. , Goodsell, D. S. , & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30, 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C. , Flematti, G. R. , Riseborough, J. A. , Ghisalberti, E. L. , Dixon, K. W. , & Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 107, 7095–7100. 10.1073/pnas.0911635107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis, D. L. , Cheah, E. , Cygler, M. , Dijkstra, B. , Frolow, F. , Franken, S. M. , Harel, M. , Remington, S. J. , Silman, I. , Schrag, J. , Sussman, J. L. , Verschueren, K. H. G. , & Goldman, A. (1992). The alpha/beta hydrolase fold. Protein Engineering, 5, 197–211. 10.1093/protein/5.3.197 [DOI] [PubMed] [Google Scholar]
- Osada, H. (2010). Introduction of new tools for chemical biology research on microbial metabolites. Bioscience, Biotechnology, and Biochemistry, 74, 1135–1140. 10.1271/bbb.100061 [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F. , Goddard, T. D. , Huang, C. C. , Couch, G. S. , Greenblatt, D. M. , Meng, E. C. , & Ferrin, T. E. (2004). UCSF chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Scaffidi, A. , Waters, M. T. , Sun, Y. K. , Skelton, B. W. , Dixon, K. W. , Ghisalberti, E. L. , Flematti, G. R. , & Smith, S. M. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis . Plant Physiology, 165, 1221–1232. 10.1104/pp.114.240036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda, C. , Guzmán, M. A. , Li, Q. , Villaécija‐Aguilar, J. A. , Martinez, S. E. , Kamran, M. , Khosla, A. , Liu, W. , Gendron, J. M. , Gutjahr, C. , Waters, M. T. , & Nelson, D. C. (2022). KARRIKIN UP‐REGULATED F‐BOX 1 (KUF1) imposes negative feedback regulation of karrikin and KAI2 ligand metabolism in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, 119, e2112820119. 10.1073/pnas.2112820119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisaphaithong, T. , Yanase, M. , Mano, T. , Tanabe, S. , Minami, E. , Tanaka, A. , Hata, S. , & Kobae, Y. (2021). Localized expression of the Dwarf14‐like2a gene in rice roots on infection of arbuscular mycorrhizal fungus and hydrolysis of rac‐GR24 by the encoded protein. Plant Signaling & Behavior, 16, 2009998. 10.1080/15592324.2021.2009998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. , Grosse‐Kunstleve, R. W. , Afonine, P. V. , Moriarty, N. W. , Zwart, P. H. , Hung, L. W. , Read, R. J. , & Adams, P. D. (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallographica. Section D, Biological Crystallography, 64, 61–69. 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W. , Chen, C. , Lei, X. , Zhao, J. , & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46, W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, S. , Holbrook‐Smith, D. , Stogios, P. J. , Onopriyenko, O. , Lumba, S. , Tsuchiya, Y. , Savchenko, A. , & McCourt, P. (2015). Structure‐function analysis identifies highly sensitive strigolactone receptors in Striga . Science, 350, 203–207. 10.1126/science.aac9476 [DOI] [PubMed] [Google Scholar]
- Trott, O. , & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31, 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. , Akiyama, K. , Arite, T. , Takeda‐Kamiya, N. , Magome, H. , Kamiya, Y. , Shirasu, K. , Yoneyama, K. , Kyozuka, J. , & Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Végh, A. , Incze, N. , Fabian, A. , Huo, H. , Bradford, K. J. , Balazs, E. , & Soos, V. (2017). Comprehensive analysis of DWARF14‐LIKE2 (DLK2) reveals its functional divergence from strigolactone‐related paralogs. Frontiers in Plant Science, 8, 1641. 10.3389/fpls.2017.01641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Yao, R. , Du, X. , Guo, L. , Chen, L. , Xie, D. , & Smith, S. M. (2021). Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga . Plant Physiology, 185, 1411–1428. 10.1093/plphys/kiaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M. T. , Nelson, D. C. , Scaffidi, A. , Flematti, G. R. , Sun, Y. K. , Dixon, K. W. , & Smith, S. M. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis . Development, 139, 1285–1295. 10.1242/dev.074567 [DOI] [PubMed] [Google Scholar]
- Waters, M. T. , Scaffidi, A. , Flematti, G. , & Smith, S. M. (2015). Substrate‐induced degradation of the alpha/beta‐fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Molecular Plant, 8, 814–817. 10.1016/j.molp.2014.12.020 [DOI] [PubMed] [Google Scholar]
- Waters, M. T. , Scaffidi, A. , Moulin, S. L. , Sun, Y. K. , Flematti, G. R. , & Smith, S. M. (2015). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell, 27, 1925–1944. 10.1105/tpc.15.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M. T. , Scaffidi, A. , Sun, Y. K. , Flematti, G. R. , & Smith, S. M. (2014). The karrikin response system of Arabidopsis . The Plant Journal, 79, 623–631. 10.1111/tpj.12430 [DOI] [PubMed] [Google Scholar]
- Xu, Y. , Miyakawa, T. , Nakamura, H. , Nakamura, A. , Imamura, Y. , Asami, T. , & Tanokura, M. (2016). Structural basis of unique ligand specificity of KAI2‐like protein from parasitic weed Striga hermonthica. Scientific Reports, 6, 31386. 10.1038/srep31386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Miyakawa, T. , Nosaki, S. , Nakamura, A. , Lyu, Y. , Nakamura, H. , Ohto, U. , Ishida, H. , Shimizu, T. , Asami, T. , & Tanokura, M. (2018). Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga . Nature Communications, 9, 3947. 10.1038/s41467-018-06452-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. , Ming, Z. , Yan, L. , Li, S. , Wang, F. , Ma, S. , Yu, C. , Yang, M. , Chen, L. , Chen, L. , Li, Y. , Yan, C. , Miao, D. , Sun, Z. , Yan, J. , Sun, Y. , Wang, L. , Chu, J. , Fan, S. , … Xie, D. (2016). DWARF14 is a non‐canonical hormone receptor for strigolactone. Nature, 536, 469–473. 10.1038/nature19073 [DOI] [PubMed] [Google Scholar]
- Zhao, L. H. , Zhou, X. E. , Wu, Z. S. , Yi, W. , Xu, Y. , Li, S. , Xu, T. H. , Liu, Y. , Chen, R. Z. , Kovach, A. , Kang, Y. , Hou, L. , He, Y. , Xie, C. , Song, W. , Zhong, D. , Xu, Y. , Wang, Y. , Li, J. , … Xu, H. E. (2013). Crystal structures of two phytohormone signal‐transducing alpha/beta hydrolases: Karrikin‐signaling KAI2 and strigolactone‐signaling DWARF14. Cell Research, 23, 436–439. 10.1038/cr.2013.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, T. J. , Bürger, M. , Tashiro, E. , Kondoh, Y. , Martinez, N. E. , Gormer, K. , Rosin‐Steiner, S. , Shimizu, T. , Ozaki, S. , Mikoshiba, K. , Watanabe, N. , Hall, D. , Vetter, I. R. , Osada, H. , Hedberg, C. , & Waldmann, H. (2013). Boron‐based inhibitors of acyl protein thioesterases 1 and 2. Chembiochem, 14, 115–122. 10.1002/cbic.201200571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Comparison of the AtDLK2 structure predicted by AlphaFold and the experimentally obtained AtDLK2 structure. (a‐b) The predicted AtDLK2 structure has a ligand binding pocket that is disconnected from the protein surface and has a smaller volume, compared to the experimental X‐ray structure of AtDLK2. (c‐d) The positions of the gatekeeper residue Phe167 match well between the predicted and the experimental structures, however, the side chain of Phe200 closes the pocket in the AlphaFold‐predicted structure. The 2mFo − DFc electron density map of the Phe167 and Phe200 side chains in the experimental structure is contoured at 1 σ and shown in blue.
Figure S2: Inhibition of AtDLK2 by pyrrolo‐quinoline‐dione compounds (a) 2, (b) 3, and (c) 4. Enzymatic activity was measured in triplicates as absorbance of para‐nitrophenol at 410 nm as result of enzymatic activity against paranitrophenyl acetate. All values have been corrected for spontaneous para‐nitrophenyl acetate hydrolysis. Error bars represent standard deviation.
Data Availability Statement
The structural coordinates and diffraction data of AtDLK2 have been deposited in the Protein Data Bank under accession code 7TVW.






