Abstract
The bacteriophage λ excisionase (Xis) is a sequence-specific DNA binding protein required for excisive recombination. Xis binds cooperatively to two DNA sites arranged as direct repeats on the phage DNA. Efficient excision is achieved through a cooperative interaction between Xis and the host-encoded factor for inversion stimulation as well as a cooperative interaction between Xis and integrase. The secondary structure of the Xis protein was predicted to contain a typical amphipathic helix that spans residues 18 to 28. Several mutants, defective in promoting excision in vivo, were isolated with mutations at positions encoding polar amino acids in the putative helix (T. E. Numrych, R. I. Gumport, and J. F. Gardner, EMBO J. 11:3797–3806, 1992). We substituted alanines for the polar amino acids in this region. Mutant proteins with substitutions for polar amino acids in the amino-terminal region of the putative helix exhibited decreased excision in vivo and were defective in DNA binding. In addition, an alanine substitution at glutamic acid 40 also resulted in altered DNA binding. This indicates that the hydrophilic face of the α-helix and the region containing glutamic acid 40 may form the DNA binding surfaces of the Xis protein.
Site-specific recombination by bacteriophage λ is a complex process that requires the formation of nucleoprotein complexes composed of specific DNA sites in the phage and bacterial chromosomes and host- and phage-encoded proteins. Integrative recombination between specific attachment sites, attP on the phage DNA and attB on the bacterial chromosome, generates recombinant attR and attL sites flanking the prophage DNA (14). Excision of the prophage is accomplished by site-specific recombination between the attR and attL sites to regenerate the intact host and phage genomes. Both reactions are catalyzed by the phage-encoded protein integrase (Int), assisted by accessory proteins. The host-encoded integration host factor (IHF) is a protein required for both reactions. Excision requires an additional phage-encoded protein called excisionase (Xis). Excision is stimulated by the factor for inversion stimulation (FIS) supplied by the host. The directionality of recombination is determined by the amount of Xis present in the cell. During prophage induction, enough active Xis is present to promote excision (7, 26). The amount of Xis protein present during establishment of lysogeny is limited due to instability of the protein (26). The integration reaction is inhibited by Xis in vitro (1, 17).
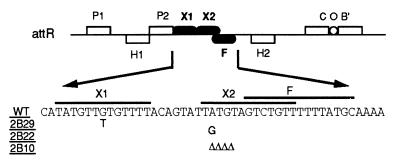
Xis is a sequence-specific DNA binding protein of 72 amino acids (1). Xis recognizes two direct, imperfect 13-bp repeats, designated X1 and X2, on the attR site (see Fig. 1) (28). The FIS binding site, designated F, partly overlaps the X2 site (24, 25). Xis binds DNA cooperatively at the X1 and X2 sites (4) and also binds to X1 cooperatively with FIS at the F site (19, 24). Both Xis and FIS bend DNA upon binding to their specific sites (23). In addition, occupation of X1 by Xis facilitates binding of Int to the P2 site that lies adjacent to the X1 site, presumably through protein-protein interactions (4, 19, 27). The P2 site is a relatively weak Int binding site and is required for excision but not for integration (2, 25).
FIG. 1.
The attR site of bacteriophage λ and nucleotide sequences of wild-type and variant Xis binding sites. The two arm-type binding sites, P1 and P2, and the two core-type sites, C and B′, on the attR region are recognized by Int. IHF binds to the sites designated as H1 and H2. Xis binds to the X1 and X2 sites. FIS binds to the F site. The isolation of the mutant Xis binding sites 2B10, 2B29, and 2B22 has been described by Numrych et al. (18). Δ, deletion of the indicated base.
Xis protein is multifunctional despite its small size. It binds to DNA and interacts with FIS and Int. However, little structural information on the Xis protein is available. Numrych et al. (19) carried out an extensive mutational analysis of Xis and isolated amino acid substitution mutants of Xis with decreased DNA binding affinity in the bacteriophage P22 challenge-phage assays. Their mutations are located in the amino-terminal half of the protein. In contrast, other mutations resulting in defective interaction with Int are at the carboxyl end of the protein (19, 27). No mutants that bound DNA but failed to interact with FIS were isolated. Of the 15 amino acid substitution mutations that decrease DNA binding, 8 change polar residues. Four of these substitutions are clustered between glutamic acid 19 and arginine 26. In addition, another substitution mutant, E40K, showed decreased DNA binding in a challenge-phage assay. Because these polar residues may interact specifically with DNA or FIS, we constructed alanine substitution mutants of each and analyzed their DNA binding properties and their ability to promote excision in vivo.
MATERIALS AND METHODS
Bacterial and phage strains.
Escherichia coli strain DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used for cloning and purification of variant Xis proteins. BL21(DE3)[F− ompT hsdSB(rB−mB−) gal(λ cI857 ind1 Sam7 nin5 lacUV5-T7gene1) dcm] was used for purification of wild-type His-tagged Xis protein. The E. coli strain LE292 {HfrH argE(Am) rpoB galT::(λΔ[int-FII])} and its fis::kan derivative were used for the red colony test. LE292 (fis::kan) was constructed by generalized transduction using phage P1vir grown on E. coli strain MO (fis::kan) (19). A crude extract containing the wild-type Xis protein was prepared from RJ1529 (fis::kan) harboring pPS2-3ΔRS (19). The challenge phage P22xis2B and its derivatives containing variant X1-X2-F sites were used as templates for preparation of DNA in gel-shift assays (see Fig. 1) (18). As a nonspecific DNA for gel-shift assays, a challenge phage, P22 P′123(II), was used (15).
Media, chemicals, and enzymes.
The media and buffers were described previously (19). Antibiotics (Sigma) were added to the media as follows: ampicillin to 50 μg/ml, spectinomycin to 100 μg/ml, and kanamycin to 50 μg/ml. For the red-colony test, timetin (SmithKline Beecham Pharmaceuticals) was used instead of ampicillin at a concentration of 50 μg/ml to prevent the growth of ampicillin-sensitive satellite colonies. Isopropyl-β-d-thiogalactopyranoside (IPTG) was obtained from Sigma and used at the indicated concentrations. T4 DNA ligase, T4 polynucleotide kinase, and restriction endonucleases were obtained from Bethesda Research Laboratories or New England Biolabs. Taq DNA polymerase was obtained from Promega.
Plasmid constructions.
PCR was used to isolate and amplify the xis gene from phage λ DNA. The upstream primer Xis-1 contained an NheI restriction site preceding the initiation codon of the xis gene, and the downstream primer Xis-2 carried an EcoRI site following the xis translation termination codon (Table 1). PCR was carried out using Taq DNA polymerase in a solution containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, and 200 μM concentrations of each deoxynucleoside triphosphate. The fragment was digested with NheI and EcoRI and ligated into the plasmid pET28a (Novagen), which was predigested with the same set of enzymes. After transformation into E. coli strain DH5α, a clone harboring the insert was confirmed by sequencing (22). The resulting clone, pYL-XisHP, was transformed into E. coli BL21(DE3) for expression and purification of the Xis protein.
TABLE 1.
Oligonucleotides used for PCR amplification
| Primer | Sequencea | Description |
|---|---|---|
| Xis-1 | d(CTGAGCTAGCATGTACTTGACACTTCAGGAGTGG) | Upstream primer for xis |
| Xis-2 | d(CTGAGAATTCTCATGACTTCGCCTTCTTCCCATT) | Downstream primer for xis |
| PCKR-p | d(ATCCATCGATGCTTAGGAGG) | Upstream primer for mutagenesis |
| E19A | d(AGAAGCCTTGCAACAGTTCG) | Mutagenic primer for E19A |
| R22A | d(TGAAACAGTTGCTCGATGGGTT) | Mutagenic primer for R22A |
| R23A | d(AACAGTTCGTGCATGGGTTCGGGA) | Mutagenic primer for R23A |
| R26A | d(TCGATGGGTCGCGGAATGCAGGAT) | Mutagenic primer for R26A |
| R27A | d(TGGGTTCGGGCATGCAGGATATTC) | Mutagenic primer for R27A |
| E40A | d(GATGGAAGAGCTTATCTGTTCCAC) | Mutagenic primer for E40A |
| Omnt | d(CGGCATTTTGCTATTCC) | Used in synthesizing gel-shift DNA |
| α-Omnt | d(GATCATCTAGCCATGC) | Used in synthesizing gel-shift DNA |
Underlined sequences are complementary to the xis gene. Bold-faced sequences represent the alanine codons.
For excision assays in vivo, the XbaI-HindIII fragment containing the His-tagged xis gene of the plasmid pYL-XisHP was subcloned into the XbaI-HindIII backbone of the plasmid pCKR101 (15). The resulting plasmid, pYL-XisHR, carried the His-tagged xis gene under control of the Ptac promoter.
Site-directed mutagenesis.
The plasmids containing alanine substitution mutants, pXisE19A, pXisR22A, pXisR23A, pXisR26A, pXisE27A, and pXisE40A, were constructed via site-specific mutagenesis using the modified megaplasmid PCR method (12). Ten picomoles of the mutagenic oligonucleotide and Xis-2 primer (Table 1) were added to the template pYL-XisHR DNA. This reaction amplified short fragments containing desired base-pair changes, which in turn would be used as a megaprimer in subsequence amplifications. After 10 cycles, 12.5 pmol of upstream primer, PCKR-p, was added, and DNA was amplified for 10 more cycles. In the next 10 cycles, 12.5 pmol of Xis-2 was supplied to finish the amplification. The amplified DNA was digested with EcoRI and cloned into the EcoRI backbone of the plasmid pYL-XisHR. Each mutation was confirmed by DNA sequencing.
Expression and purification of His-tagged Xis and its derivatives.
E. coli strains DH5α, harboring derivatives of the pYL-XisHR plasmid, and BL21(DE3), with the pYL-XisHP plasmid, were used for expression of the Xis mutants and the wild-type Xis protein, respectively. Cells were grown to mid-log phase in Luria-Bertani medium containing the appropriate antibiotics. Production of His-tagged proteins was induced by adding IPTG to a final concentration of 1 mM, followed by growth for 3 h at 37°C. After centrifugation, harvested cells were disrupted by treatment in a French pressure cell in 50 mM sodium phosphate buffer, pH 7.4, with 300 mM NaCl. Prior to disruption, 250 μl of protease inhibitor cocktail for bacterial cell extracts (Sigma product no. P8465) was added per gram of cell mass. Cell lysates were centrifuged at 12,000 × g for 30 min, and the supernatants were mixed with nickel-nitrilotriacetic acid column material (Qiagen) saturated in 50 mM sodium phosphate buffer, pH 7.4, with 300 mM NaCl and 10% glycerol. The column was washed extensively, and the His-tagged proteins were eluted by adding 250 mM imidazole in the same buffer. The total protein concentrations were measured using the dye-binding assay (3). The amount of Xis in each protein sample was determined by measuring peaks after scanning the Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel.
Preparation of crude extracts.
An E. coli strain, RJ1529, harboring the plasmid pPS2-3ΔRS (18) was grown at 37°C to mid-exponential phase, and expression of Xis protein was induced by IPTG at a concentration of 1 mM for 1 h. After centrifugation, cell pellets were collected and resuspended in a solution containing 20 mM Tris-HCl (pH 7.4), 100 mM EDTA, 20 mM NaCl, and 10% glycerol. The cell suspension was sonicated using Branson Sonifier Cell Disruptor 200 and centrifuged at 20,000 × g for 1 h. The cleared sonic extract was used as a source of crude Xis protein.
Gel-shift assays.
The DNA fragments used in gel-shift assays were amplified by PCR using phage P22xis2B or its derivatives as templates (Fig. 1). The phage P22 P′123(II), which contained the attL Int arm-type binding sites of λ instead of the Xis binding sites, was used as a source of labeled, nonspecific DNA. A pair of synthetic oligodeoxyribonucleotides, Omnt and α-Omnt (Table 1), were used as primers for amplification. They were annealed to the sequences encompassing the Xis binding sites or Int arm-type binding sites. One of the primers was labeled with [γ-32P]ATP and polynucleotide kinase prior to amplification (21). The PCRs were carried out using Taq DNA polymerase (Promega). The DNA fragments were purified on a Micro Bio-Spin chromatography column 30 (Bio-Rad) and were quantified with a PhosphorImager (Molecular Dynamics).
Aliquots of labeled DNA fragments were incubated with various concentrations of Xis proteins in binding buffer (27.5 mM Tris-HCl [pH 8.0], 33 mM KCl, 25 mM NaCl, 1 mM EDTA, 250 μg of bovine serum albumin [BSA]/ml, 5% glycerol) at room temperature for 30 min. Sonicated calf thymus DNA and FIS protein were added to the reaction as indicated. The final reaction volumes were adjusted to 10 μl. After the entire reaction mixture was loaded and the components were separated by electrophoresis in 5% polyacrylamide gels at room temperature or at 4°C, the gels were dried and exposed to X-ray film for autoradiography.
Excision assay.
The red-colony test (11) was used to assay the ability of Xis derivatives to promote excisive recombination in vivo. The galT gene of E. coli strain LE292 contains a λ prophage that lacks the int and xis genes. When the Int and Xis proteins are supplied from plasmids, the inserted prophage is excised from the chromosome and renders the cells Gal positive. Plasmids containing the wild-type or mutant xis genes were transformed into strain LE292 containing pIntB1, and the cells were spread on MacConkey galactose-timetin-spectinomycin plates containing 1 mM IPTG. The plates were incubated at 30°C, and the time required for the colonies to turn red was used as a measurement of in vivo excisive recombination activity.
Secondary structure predictions.
The secondary structure of the Xis protein was predicted using various algorithms available at the Network Protein Sequence @nalysis web server (http://www.pbil.ibcp.fr/NPSA). The methods used to analyze the structure were DPM (5), DSC (13), GOR IV (9), PHD (20), Predator (8), SIMPA96 (16), and SOPM (10).
RESULTS AND DISCUSSION
His-tagged Xis retains the binding specificity and functions of Xis.
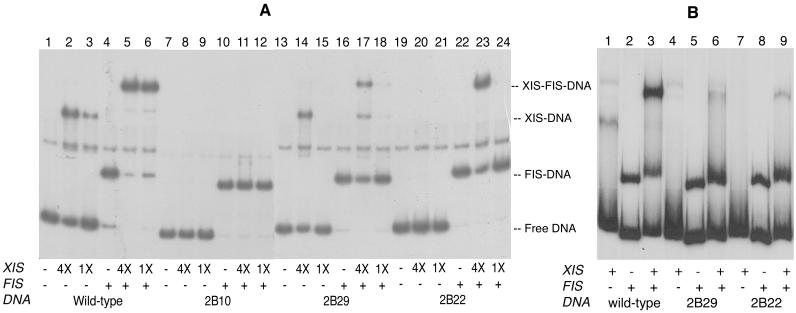
In order to simplify the purification of Xis and its mutant derivatives, we constructed a modified gene that encoded a His tag at the amino terminus of the protein. The xis gene from phage λ was amplified and cloned into the NheI-EcoRI backbone of plasmid pET28a as described in Materials and Methods. The resulting clone, pYL-XisHP, was transformed into E. coli BL21(DE3) for expression and subsequent purification of the His-tagged Xis. After isolating the wild-type His-tagged protein, we analyzed its sequence-specific DNA binding properties in vitro by gel-shift assays. The DNA fragments used in the binding reactions contained either the wild-type X1-X2-F sites or the variant sites shown in Fig. 1. The mutant site 2B10 carries a 4-bp deletion in the X2 site. The other two variant sites, 2B29 and 2B22, have a single-nucleotide substitution in X1 and X2, respectively. Variants were isolated as mutants defective for Xis binding using the challenge-phage system (18).
The purified wild-type His-tagged Xis bound DNA containing the wild-type X1-X2-F sites (Fig. 2A, lanes 2 and 3). Addition of FIS to the reaction facilitated binding of His-tagged Xis to its binding site, indicating that His-tagged Xis interacts cooperatively with FIS in DNA binding (Fig. 2A, lanes 5 and 6). Similar results showing weak cooperativity between FIS and Xis have been reported previously (19, 25). The Xis protein did not bind the deletion variant site 2B10, but FIS protein shifted DNA fragments containing the variant 2B10 (Fig. 2A, lanes 7 to 12). The mutant site 2B29 was bound by His-tagged Xis but with lower affinity than the wild-type site (Fig. 2A, lanes 13 to 18). Mutant 2B22 was bound only when FIS was present (Fig. 2A, lanes 19 to 24).
FIG. 2.
DNA binding specificity of His-tagged Xis (A) and the wild-type Xis (B). The 32P-labeled DNA fragments were about 180 bp long and were amplified from P22 challenge phages containing either wild-type or variant Xis binding sites. A 1X amount of Xis (A) corresponds to 0.1 μM protein. The wild-type Xis protein (B) was in crude extracts prepared from an E. coli strain, RJ1529, containing the plasmid pPS2-3ΔRS (18). Each reaction contained 2.5 nM labeled DNA, and where indicated, 12.5 ng (A) or 7.6 ng (B) of FIS was present. As a nonspecific competitor, 0.25 μg (A) or 1 μg (B) of sonicated calf thymus DNA was added. Electrophoresis was performed at room temperature. A contaminating fragment (probably single-stranded DNA) occurring as an artifact of the PCR migrated between the Xis-DNA complex and the FIS-DNA complex.
The binding patterns of the fusion protein were the same as those of the wild-type Xis protein without the His tag (Fig. 2B), showing that the positively charged His tag was not affecting binding. When crude extracts containing the wild-type Xis protein were added to DNA with Xis binding sites, Xis-DNA complexes were detected only with the wild-type binding site, not with the mutant binding sites 2B29 and 2B22 (Fig. 2B, lanes 1, 4, and 7). If FIS protein was supplied to the reaction, Xis-FIS-DNA complexes were detected in the reactions with all three Xis binding sequences. However, the amounts of the ternary complexes formed with the variant sites, 2B29 and 2B22, were significantly less than with wild-type DNA (Fig. 2B, lanes 3, 6, and 9). Thus, the His-tagged Xis protein binds specifically to its binding sites and interacts cooperatively with the FIS protein.
To measure the recombination activity of the protein in vivo, DNA encoding the His-tagged xis gene was subcloned downstream of the Ptac promoter as described in Materials and Methods. The resultant plasmid, pYL-XisHR, was transformed into LE292 containing the Int-producing plasmid pIntB1 (19). Excision was assayed by the red-colony test (11). The Xis protein produced from the plasmid pYL-XisHR functioned as efficiently as the wild-type Xis protein without the His tag in the excision reaction. Colonies turned red within 24 h on MacConkey-galactose plates containing 1 mM IPTG. It took another 12 h if the host carried a defective fis gene. Taken together with the gel-shift data, this led us to conclude that the His tag does not significantly affect the functions of Xis protein, including DNA binding and cooperative interactions with FIS. All the following data were obtained using His-tagged versions of Xis.
Amino acid residues from Leu 18 to Glu 27 may form an amphipathic α-helix.
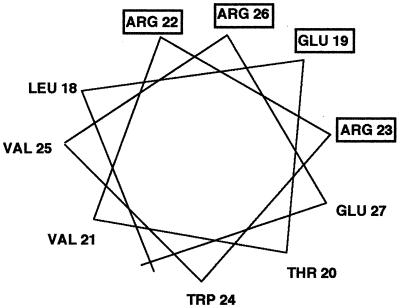
The carboxyl-terminal region of Xis is required for cooperative binding of Int, presumably through protein-protein interactions (19, 27). A nonsense mutant was isolated that encodes an Xis protein containing the amino-terminal 53 amino acids. It bound to DNA containing the X1-X2-F sites and interacted cooperatively with the FIS protein (19). The precise regions of Xis involved in DNA binding or FIS interaction were not localized. Secondary structure prediction algorithms indicated that three regions of the Xis protein could form α-helices (5, 8, 9, 10, 13, 16, 20). Amino acids from residues 5 to 10 were predicted to form the first helix, and the region spanning leucine 18 to glutamic acid 27 was predicted to form the second helix. The third postulated helix was in the region proposed by Numrych et al. (19) to be involved in cooperative interactions with Int. However, Xis lacked recognizable DNA binding motifs, for example, a helix-turn-helix motif (6). We observed that although Xis does not form a canonical helix-turn-helix motif, the second helical region could form a typical amphipathic helix (Fig. 3). Furthermore, Numrych et al. (19) found that several substitutions for the hydrophilic residues in this region resulted in a loss of Xis function in vivo. Those substitutions included the changing of glutamic acid 19 to a lysine, arginine 22 to a histidine, arginine 23 to a glutamine, and arginine 26 to a tryptophan. Those results are consistent with the hypothesis that this region forms an α-helix and the surface-exposed hydrophilic residues may be in direct contact with DNA or FIS.
FIG. 3.
Helical wheel projection of the putative α-helix spanning amino acid residues 18 to 27 in Xis. The residues named in the boxes are the amino acids for which substitutions resulted in defective excision in vivo (19).
Substitutions for polar residues in the putative amphipathic helix change DNA binding.
To gain more information on the function of each of the polar side chains on the putative amphipathic helix, we constructed alanine substitution mutants of the hydrophilic residues. Glutamic acid 19, arginines 23 and 26, and glutamic acid 27 were individually replaced with an alanine residue. The proteins were designated E19A, R23A, R26A, and E27A, respectively. We also constructed a mutant with an alanine substitution at position 22, but we could not detect the protein on a sodium dodecyl sulfate-polyacrylamide gel. This mutant was not analyzed further. Each of the Xis variants was tested for DNA binding and ability to promote excision in vivo. Gel-shift assays were performed using partially purified proteins as described in Materials and Methods.
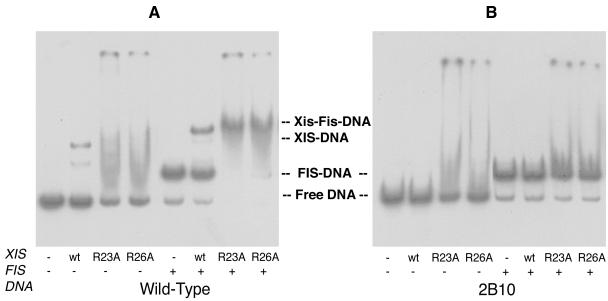
The mutant E27A protein bound DNA containing the X1-X2-F sites with an affinity similar to that of the wild-type protein. It also discriminated between the wild-type and variant Xis binding sites as did the wild-type protein (data not shown). Variants R23A and R26A, however, failed to form specific complexes with DNA in the absence of the FIS protein, even when 50-fold more protein was added to the reactions than in reactions using the wild-type Xis. Mutants R23A and R26A formed a complex with DNA containing wild-type Xis binding sites only in the presence of FIS (Fig. 4A). However, they failed to bind the variant site 2B10 under the same conditions (Fig. 4B). These results indicate that although the latter two mutants have decreased DNA binding affinity, they retain the ability to interact with the FIS protein. They bind to the specific Xis binding site only when FIS is present.
FIG. 4.
DNA binding of XisR23A and XisR26A. The final concentration of wild-type Xis was 0.1 μM, and that of the mutant Xis proteins was 5 μM. Each reaction contained 2.5 nM labeled DNA and 50 ng of sonicated calf thymus DNA. Where indicated, 6.3 ng of FIS was present. Electrophoresis was performed at 4°C.
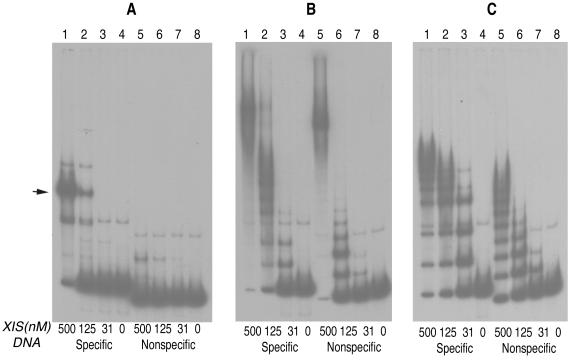
Another variant, E19A, bound DNA differently. The E19A protein did not form a discrete complex with DNA containing the X1-X2-F sites as the wild-type protein did. Instead, it formed complexes that migrated slowly only at high protein concentrations (Fig. 5B, lane 1). Multiple, discrete complexes were formed when the protein was present at lower concentrations (Fig. 5B, lanes 2 and 3). We suggest that the rapidly migrating bands are complexes with the mutant protein bound nonspecifically to DNA. To test if the E19A protein could bind to a sequence lacking the Xis or FIS binding sites, we used DNA containing the P′1-P′2-P′3 Int arm-type binding sites of λ. As shown in Fig. 5B, lanes 4 to 6, the E19A protein also formed multiple discrete complexes with nonspecific DNA. We interpret the multiple bands to be nonspecific complexes with various amounts of the Xis E19A protein bound to a single DNA fragment. We note that the binding affinity of E19A for nonspecific DNA was greater than that of the wild-type Xis (Fig. 5). At a 31 nM concentration, wild-type Xis formed neither specific nor nonspecific complexes (Fig. 5A, lanes 3 and 7). Specific complexes of the wild-type Xis with DNA fragments containing the Xis binding site were detected when 125 nM protein or more was added (Fig. 5A, lanes 1 and 2). Under the same conditions (125 and 500 nM Xis), small amounts of nonspecific complexes were also detected (Fig. 5A, lanes 1, 2, 5, and 6). In contrast, the E19A protein bound nonspecifically to DNA at a concentration of 31 nM (Fig. 5B, lanes 3 and 7).
FIG. 5.
DNA binding patterns of wild-type Xis (A), XisE19A (B), and XisE40A (C). The specific DNA fragments contained the Xis and FIS binding sites, X1-X2-F. The band indicated by an arrow in panel A corresponds to a specific Xis-DNA complex. Nonspecific DNA contained the Int arm-type sites P′1, P′2, and P′3 in place of the Xis and FIS binding sites. The flanking sequences of nonspecific DNA were the same as those of the specific DNA fragment. The final concentration of labeled DNA in each reaction was 1 nM. Nonspecific competitor DNAs were absent from the reactions. Electrophoresis was performed at room temperature. A contaminating fragment migrating slower than free DNA is believed to be single-stranded because it was not retarded by the nonspecific DNA binding mutants.
The ability of each mutant to promote the excision reaction of λ in vivo was assessed by the red-colony test. The results are shown in Table 2. The E27A protein promoted excision as efficiently as the wild-type protein when expression of proteins was induced by IPTG at a concentration of 1 mM. Two variants, R23A and R26A, could excise λ DNA only in the presence of FIS but not as well as the wild type did. These results are consistent with the gel-shift data. It was surprising to find that the E19A protein also promoted excision, albeit with lower efficiency than the wild-type Xis. One explanation for this result is that although the E19A protein binds to nonspecific DNA significantly better than the wild-type Xis, it might continue to form a bent DNA complex when bound to the specific site, thereby stimulating excision. The fact that the E19A protein promotes excision better in the presence of the FIS protein also indicates that it interacts cooperatively with the FIS protein. Thus, with help of FIS, the E19A protein may be effectively recruited to the Xis binding site to bend DNA and to promote the excision reaction.
TABLE 2.
Promotion of excision by Xis mutants in vivoa
| Protein | Level of excision
|
|
|---|---|---|
| With FIS | Without FIS | |
| Wild-type Xis | +++ | ++ |
| E19A | ++ | + |
| R23A | + | − |
| R26A | + | − |
| E27Ab | +++ | ++ |
| E40Ab | +++ | ++ |
| Vector only | − | − |
Excision was measured as described in Materials and Methods and scored as the length of time required for the colonies to change color on MacConkey-galactose plates containing 1 mM IPTG after the plasmid harboring the xis gene was introduced. The level of excision was scored as +++, ++, and + for colonies that turned red after 24, 36, and 48 h of incubation, respectively. Colonies that remained white after 3 days of growth were scored as −.
Turned red more slowly than the wild-type Xis.
Mutant with a substitution of an alanine for glutamic acid 40 binds to nonspecific DNA.
Numrych et al. (19) also isolated a mutant in which glutamic acid 40 was replaced by a lysine. To study the function of this residue in DNA recognition in more detail, we made a His-tagged construct of it. Gel-shift assays showed that the E40A protein, like the E19A protein, bound to both specific and nonspecific DNA fragments (Fig. 5C, lanes 5 to 8). However, the binding pattern for the fragment with specific sequences was different from that for the fragment with nonspecific DNA sequences (Fig. 5C). Although we do not understand the cause of the difference, it might indicate that the E40A protein distinguishes the specific Xis binding sites from random DNA to some extent. The fact that the E40A mutant promoted excision and showed cooperativity with FIS (Table 2) supports this hypothesis. As discussed above for the E19A protein, the E40A protein may also bend DNA when bound to a specific Xis binding site and provides a functional substrate for excision. However, the sequence specificity of the wild-type Xis was significantly relaxed by the substitution for glutamic acid 40, indicating that this residue may also participate, either directly or indirectly, in sequence-specific DNA recognition.
In summary, we constructed mutant Xis proteins with alanine substitutions of polar residues on the putative amphipathic α-helix and glutamic acid 40. Three of the four alanine substitutions, E19A, R23A, and R26A, altered the DNA binding patterns of Xis. One substitution, E27A, which resulted in a change at the carboxyl end of the helix, did not change the DNA binding specificity of Xis. This behavior is consistent with the hypothesis that Xis forms an amphipathic α-helix from leucine 18 to glutamic acid 27, although the latter amino acid residue may not interact with DNA. This study suggests that the amino-terminal, hydrophilic face of the amphiphatic helix may be in close contact with DNA. In particular, glutamic acid 19 appears to play a role, direct or indirect, in conferring DNA binding specificity, and arginines at positions 23 and 26 are required to bind to DNA with high affinity. The finding that a substitution for glutamic acid 40 also increased binding affinity to nonspecific DNA suggests that the region containing glutamic acid 40 may form an additional DNA binding surface on the Xis protein.
We note that the putative helical region from amino acid 18 to 28 and the region carrying glutamic acid 40 are separated by an unusual amino acid sequence containing three consecutive prolines. Thus, the proline residues may play a role in positioning the flanking amino acid residues in a conformation that allows them to interact simultaneously with DNA. We look forward to comparing our analysis to the emerging structural studies. The combination of the two approaches may reveal the exact nature of the functional interactions between the amino acids and binding-site DNA that lead to excision.
ACKNOWLEDGMENTS
We thank R. Johnson for providing the purified FIS protein and strain RJ1529 and Yu-Hong Li for constructing plasmids pYL-XisHP and pYL-XisHR. We also thank R. Johnson and H. Huang for constructive comments on the manuscript.
This work was supported by NIH grant 28717 and KOSEF grant 971-0502-009-2 from the Korea Science and Engineering Foundation.
REFERENCES
- 1.Abremski K, Gottesman S. Purification of the bacteriophage xis gene product required for λ excisive recombination. J Biol Chem. 1982;257:9658–9662. [PubMed] [Google Scholar]
- 2.Bauer C E, Hesse S D, Gumport R I, Gardner J F. Mutational analysis of integrase arm-type binding sites of bacteriophage lambda. J Mol Biol. 1986;121:179–192. doi: 10.1016/0022-2836(86)90273-1. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–253. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Bushman W, Yin S, Thio L, Landy A. Determinants of directionality in lambda site-specific recombination. Cell. 1984;39:699–707. doi: 10.1016/0092-8674(84)90477-x. [DOI] [PubMed] [Google Scholar]
- 5.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 6.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echols H, Guarneros G. Control of integration and excision. In: Hendrix R, Roberts J, Stahl F, Weisberg R, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1983. pp. 75–93. [Google Scholar]
- 8.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 9.Garnier J, Gibrat J F, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 10.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 11.Han Y W, Gumport R I, Gardner J F. Mapping the functional domains of bacteriophage lambda integrase protein. J Mol Biol. 1994;235:908–925. doi: 10.1006/jmbi.1994.1048. [DOI] [PubMed] [Google Scholar]
- 12.Ke S-H, Madison E L. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King R D, Sternberg M J. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landy A. Dynamic, structural, and regulatory aspects of site-specific recombination. Annu Rev Biochem. 1989;58:913–950. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 15.Lee E C, Gumport R I, Gardner J F. Genetic analysis of bacteriophage integrase interactions with arm-type attachment site sequences. J Bacteriol. 1990;172:1529–1538. doi: 10.1128/jb.172.3.1529-1538.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin J M, Robson B, Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 17.Nash H. Integrative recombination of bacteriophage lambda DNA in vitro. Proc Natl Acad Sci USA. 1975;72:1072–1076. doi: 10.1073/pnas.72.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numrych T E, Gumport R I, Gardner J F. A genetic analysis of Xis and FIS interactions with their binding sites in bacteriophage lambda. J Bacteriol. 1991;173:5954–5959. doi: 10.1128/jb.173.19.5954-5963.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Numrych T E, Gumport R I, Gardner J F. Characterization of the bacteriophage lambda excisionase (Xis) protein: the C-terminus is required for Xis-integrase cooperativity but not for DNA binding. EMBO J. 1992;11:3797–3806. doi: 10.1002/j.1460-2075.1992.tb05465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J F, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J F, Moitoso de Vargas L, Skinner S E, Landy A. Protein-protein interactions in a higher-order structure direct lambda site-specific recombination. J Mol Biol. 1987;195:481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J F, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the site-specific recombination pathway. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 26.Weisberg R A, Gottesman M E. The stability of Int and Xis functions. In: Hershey A D, editor. The bacteriophage lambda. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1971. pp. 489–500. [Google Scholar]
- 27.Wu Z, Gumport R I, Gardner J F. Defining the structural and functional roles of the carboxyl region of the bacteriophage lambda excisionase (Xis) protein. J Mol Biol. 1998;281:651–661. doi: 10.1006/jmbi.1998.1963. [DOI] [PubMed] [Google Scholar]
- 28.Yin S, Bushman W, Landy A. Interaction of the lambda site-specific recombination protein Xis with attachment site DNA. Proc Natl Acad Sci USA. 1985;82:1040–1044. doi: 10.1073/pnas.82.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]