Abstract
Escherichia coli possesses two distinct nitrite reductase enzymes encoded by the nrfA and nirB operons. The expression of each operon is induced during anaerobic cell growth conditions and is further modulated by the presence of either nitrite or nitrate in the cells' environment. To examine how each operon is expressed at low, intermediate, and high levels of either nitrate or nitrite, anaerobic chemostat culture techniques were employed using nrfA-lacZ and nirB-lacZ reporter fusions. Steady-state gene expression studies revealed a differential pattern of nitrite reductase gene expression where optimal nrfA-lacZ expression occurred only at low to intermediate levels of nitrate and where nirB-lacZ expression was induced only by high nitrate conditions. Under these conditions, the presence of high levels of nitrate suppressed nrfA gene expression. While either NarL or NarP was able to induce nrfA-lacZ expression in response to low levels of nitrate, only NarL could repress at high nitrate levels. The different expression profile for the alternative nitrite reductase operon encoded by nirBDC under high-nitrate conditions was due to transcriptional activation by either NarL or NarP. Neither response regulator could repress nirB expression. Nitrite was also an inducer of nirB and nrfA gene expression, but nitrate was always the more potent inducer by >100-fold. Lastly, since nrfA operon expression is only induced under low-nitrate concentrations, the NrfA enzyme is predicted to have a physiological role only where nitrate (or nitrite) is limiting in the cell environment. In contrast, the nirB nitrite reductase is optimally synthesized only when nitrate or nitrite is in excess of the cell's capacity to consume it. Revised regulatory schemes are presented for NarL and NarP in control of the two operons.
Escherichia coli possesses two biochemically distinct nitrite reductase enzymes encoded by the nrfABCDEFG and nirBDC operons (4). The NirB nitrite reductase is a soluble siroheme-containing enzyme that uses NADH as an electron donor to reduce nitrite in the cytoplasm. The NrfA nitrite reductase is a membrane-associated respiratory enzyme that couples to the membrane-associated formate-oxidizing enzymes via quinones in order to generate membrane potential. The abundance of each enzyme is elevated during anaerobic cell growth conditions when either nitrate and/or nitrite is present (14). Nitrite, the substrate for the two enzymes, must either be encountered environmentally or generated by the cell from nitrate reduction by one of the three E. coli nitrate reductases.
Expression of the nrfABCDEFG operon (previously described as aeg-93 [3]) and the nirBDC operon is elevated during anaerobic cell growth by the Fnr regulatory protein (1, 10, 14). The addition of nitrite, but not nitrate, is reported to further elevate nrfA expression via either the NarL or NarP response regulators. In contrast, NarL is reported to repress nrfA expression in response to nitrate, whereas NarP cannot (14–16, 23, 24). Expression of the nirB operon differs from nrfA in that NarL and NarP are reported to activate nirB expression in response to nitrate. In contrast, only NarL is reported to activate nirB in response to nitrite (23, 24). The locations of sites for NarL and NarP binding have been proposed for each operon (described below).
It is unknown how the nrfA and nirB operons are expressed in response to either low or intermediate levels of either nitrate or nitrite since the prior gene regulation studies were performed in batch cultures using high levels of each anion. Thus, we examined the steady-state expression of nrfA-lacZ and nirB-lacZ reporter fusions using anaerobic chemostat culture methods under limiting nitrite or nitrate conditions. The findings reveal a differential pattern of nitrite reductase gene expression whereby the nrfA operon is preferentially expressed only at low nitrate concentrations. Maximal nirB expression occurs only at high levels of nitrate. Nitrite, the substrate for each enzyme, was shown to be a less potent regulatory signal for either operon relative to nitrate in contrast to prior reports. The chemostat studies also revealed new roles for the NarL and NarP proteins in the activation and/or repression of the two operons in contrast to prior conclusions derived from batch culture studies. Thus, revised models for the control of the nrfA and nirB nitrite reductase operons are proposed.
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
E. coli MC4100 [F− Δ(argF-lac)U169 araD139 rpsL150 deoC1 relA1 flbB5301 rbsR ptsF25] was used for all chemostat and batch culture experiments unless indicated otherwise (19). The lacZ reporter fusions used to monitor expression of the two-nitrite reductase operons were λHW1 (nrfA-lacZ) and λHW3 (nirB-lacZ). As the fusions were integrated at the lambda att site on the chromosome, each strain is wild type for the nrfABCDEFGH and nirBDC operons. The nrfA-lacZ fusion was constructed by the generation of a DNA fragment containing the nrfABCDEFG control region from position −297 to position +120 relative to the start site of nrfA transcription (17). The resulting DNA fragment was then inserted into plasmid pRS415 (20) to give the nrfA-lacZ operon fusion plasmid designated pHW1. The DNA insert in pHW1 was DNA sequenced to confirm the intended construction (18). The nrfA-lacZ fusion was then transferred onto λRS45 to generate λHW1. A high-titer lysate was then used to introduce the phage into MC4100 in single copy as previously described (20). The nirB-lacZ fusion was constructed by the generation of a DNA fragment containing the nirBDC control region from position −342 to position +222 relative to the start site of transcription using standard PCR protocols (17). The subsequent cloning steps were as described for nrfA-lacZ above. The nrfA-lacZ and nirB-lacZ reporter fusions were introduced into the isogenic wild-type, narL, narP, and narL narP double mutant strains (Table 1) by P1 transduction methods as previously described (13). The narL narP strain HL101 was constructed by P1 transduction of narP::kan Tn10 tet from strain HL101 into RCC70, followed by selection for tetracycline resistance. Strain HL101 was constructed by inserting Tn10 nearby the narP::kan allele of RCC71 by standard methods (19).
TABLE 1.
Strains, plasmids, and phages
| Strain, phage, or plasmid | Parent | Genotype or phenotype | Source or reference |
|---|---|---|---|
| Strains | |||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR | 19 | |
| RCC70 | MC4100 | narL::kan | 2 |
| RCC71 | MC4100 | narP::kan | 2 |
| HL110 | RCC70 | narP::tet narL::kan | This study |
| HL101 | MC4100 | narP::kan Tn10 tet | This study |
| Plasmids | |||
| pHW1 | nrfA-lacZ | This study | |
| pHW3 | nirB-lacZ | This study | |
| pRS415 | lacZ lacY+ lacA+ | 20 | |
| Phages | |||
| M13mp19-100 | M13mp19 | nrfA′ | Laboratory stock |
| λRS45 | lacZ | 20 | |
| λHW1 | λRS45 | nrfA-lacZ lacY+ lacA+ | This study |
| λHW3 | λRS45 | nirB-lacZ lacY+ lacA+ | This study |
Cell growth.
For routine cell growth and plasmid construction, cells were grown in Luria-Bertani liquid or solid medium. For batch cell culture, cells were grown in a glucose (40 mM) minimal medium (5). Where indicated, sodium nitrate (20 mM) or sodium nitrite (2.5 mM) was added to the growth medium after sterilization. Anaerobic cell growth was performed at 37°C in 10-ml anaerobic culture tubes fitted with butyl rubber stoppers (11). Cells grown overnight under identical conditions in the same medium were used for inoculation.
Continuous culture experiments were performed in a Bioflo 3000 Bioreactor (New Brunswick Scientific) fitted with a 2-liter glass vessel and operated at a 1-liter liquid working volume as previously described (25). A modified Vogel-Bonner medium supplemented with glucose (2.25 mM) was used to limit cell growth (i.e., carbon-limiting conditions) (21). During the experiments, the chemostat was maintained at a medium flow rate of 10 ml/min, which corresponds to a cell doubling time of 70 min. Anaerobic culture conditions were maintained by continuously sparging the vessel with oxygen-free nitrogen at a flow rate of 200 ml/min (21). To vary the concentration of nitrate or nitrite in the medium, NaNO3 or NaNO2 was added at the amount indicated after medium sterilization.
When the chemostat was shifted to a new nitrate (or nitrite) concentration, new steady-state levels of gene expression were verified as described earlier (25). The values obtained for each culture condition were independently determined at least twice and there was <10% variation in the β-galactosidase activity. Strain stability and purity was monitored as described elsewhere (21). β-Galactosidase assays were performed as described previously (5), where one unit of β-galactosidase is defined as the hydrolysis of 1 nmol of o-nitrophenyl-β-d-galactopyranoside (ONPG) per min per mg of protein.
Determination of nitrate and nitrite levels.
The concentration of nitrate and nitrite in the culture medium was determined as previously described (25), where the sensitivities were 0.03 and 0.04 μM, respectively.
Materials.
ONPG was purchased from Sigma Chemical Co., St. Louis, Mo.; X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was obtained from International Biotechnologies, Inc., New Haven, Conn.; and the Casamino Acids were from Difco Co., Detroit, Mich. Nitrogen gas was supplied by Arco, Inc. All other chemicals used in this study were of reagent grade.
RESULTS
Effect of nitrate concentration on nrfA-lacZ expression.
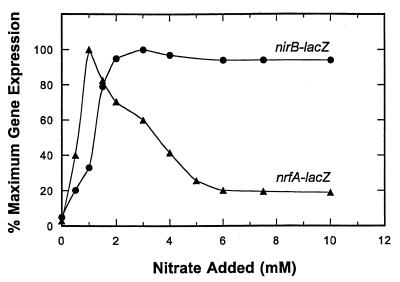
To determine how the nrfABCDEFG operon is expressed in response to different levels of nitrate, a nrfA-lacZ reporter fusion was examined using steady-state chemostat cell culture methods (Fig. 1). In the absence of any added nitrate, a basal level of nrfA-lacZ gene expression (ca. 100 U) was observed. When the steady-state nitrate addition level was increased to 0.5 mM, expression was elevated by 8-fold, while the maximal expression of 25-fold induction was seen at 1 mM nitrate addition (ca. 2,500 U). As the level of nitrate was further elevated, nrfA-lacZ expression was gradually decreased until nitrate additions reached 6 mM (Fig. 1). Under these conditions, gene expression was still fivefold above the basal level seen when no nitrate anion was added. Between the 6 and 40 mM nitrate additions, nrfA-lacZ expression remained unchanged (Fig. 1 and data not shown).
FIG. 1.
Effect of nitrate on nrfA-lacZ and nirB-lacZ expression during anaerobic cell growth. The amount of nitrate present in the medium added to the chemostat vessel was adjusted prior to sterilization (see Materials and Methods). Upon the shift to a new condition, steady state was generally achieved within five residence times. Expression of the nrfA-lacZ fusion (▴) and the nirB-lacZ fusion (●) is relative to the maximum level achieved for each fusion. Maximal nrfA-lacZ expression was 2,500 U versus 18,000 U for nirB-lacZ expression.
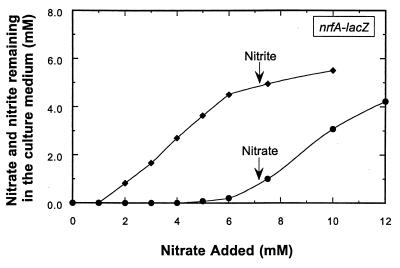
Since the E. coli cells were continually consuming nitrate by reducing it to nitrite via the various nitrate reductase enzymes, the nitrate additions shown in Fig. 1 do not indicate the actual level of nitrate remaining in the culture vessel (25). The chemostat was therefore sampled at each steady-state condition shown in Fig. 1, and the residual level of nitrate or nitrite in the vessel was determined. These values were essentially identical to those previously reported (25; Fig. 2 reproduced here). Nitrite accumulation, first observed when nitrate was added at a level of 1 mM, increased to 5 mM as the nitrate additions were raised from 1 to 6 mM. Maximum nrfA-lacZ expression was achieved only when submicromolar amounts of nitrate were present in the chemostat vessel (ca. 0.2 μM). Therefore, nrfA gene expression is exquisitely sensitive to nitrate as a regulatory signal in contrast to prior reports that nitrite was the primary regulatory signal (1, 10, 14).
FIG. 2.
Levels of nitrate and nitrite accumulated in the chemostat vessel at different concentrations of nitrate addition. Upon the shift to each new condition, steady state was generally achieved within five residence times. The vessel was then sampled, and the concentration of nitrate and nitrite remaining in the growth medium was determined by high-pressure liquid chromatography (HPLC) (Materials and Methods). Symbols: ●, nitrate; ⧫, nitrite.
Effect of nitrate concentration on nirB-lacZ gene expression.
The pattern of nirB-lacZ expression in response to steady-state nitrate additions was different than that for nrfA-lacZ expression (Fig. 1). While nirB-lacZ expression was also lowest (850 U) in the absence of added nitrate, it was then increased by 21-fold as the level of nitrate addition was incrementally raised to 2 mM. The level of nirB-lacZ expression then remained high as the nitrate additions were raised up to 40 mM (Fig. 1 and data not shown). In contrast, maximal nrfA-lacZ expression occurred at 1 mM added nitrate but was reduced to near basal activity at the higher levels of the oxyanion signal, where nirB gene expression was optimal. Therefore, the nirB and nrfA nitrite reductase operons are differentially expressed in response to nitrate.
Effect of narL and narP mutations on nitrate-dependent nirB-lacZ expression.
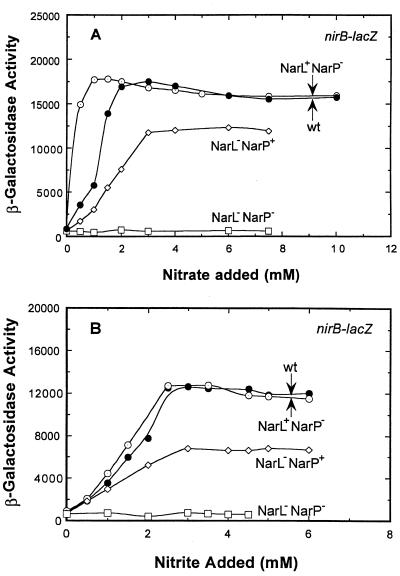
From prior batch culture experiments, both NarL and NarP were reported to activate nirB-lacZ in response to nitrate (23, 24). The chemostat experiments (Fig. 3A) also demonstrate that NarL or NarP can function independently of each other to induce nirB expression in response to nitrate. However, NarL appears to be the superior activator of nirB-lacZ expression (Fig. 3A; compare the NarL+ NarP− strain and the NarL− NarP+ strain). At the highest levels of nitrate tested, NarL-dependent induction was approximately 34-fold, while NarP induction was about 26-fold. Significantly, NarP also appears to somehow antagonize the ability of NarL to activate nirB-lacZ expression at low nitrate levels since, in a NarP− NarL+ strain, nirB-lacZ expression was higher relative to the wild-type strain during low nitrate additions. Similar results were observed for the nrfA operon (see below). Finally, the induction of nirB-lacZ expression was abolished in a strain deleted for both narL and narP.
FIG. 3.
Effect of nitrate (A) and nitrite (B) concentration on nirB-lacZ expression in narL and narP strains. The chemostat was sampled after each steady-state condition was achieved, and the β-galactosidase activity was determined. The fermentor conditions were identical to that used in Fig. 1. Expression levels in the wild-type strain (●), the narL (○), narP (◊), and the narL narP strain (□) are indicated.
Effect of nitrite additions on nirB-lacZ expression.
A nearly identical pattern of nirB-lacZ gene expression was seen when nitrite was used in place of nitrate (Fig. 3B). However, nitrate was a significantly more effective anion signal compared to nitrite (compare the maximal induction of nirB expression in each panel of Fig. 3). Maximal nirB gene expression occurred when the nitrate level in the vessel was still <50 μM, whereas an equivalent induction by nitrite required anion additions of >2 mM (Fig. 2). In addition, the maximal response to nitrite was only 70% of that achieved when nitrate was the inducer (Fig. 3). Finally, the rate that nirB-lacZ expression was increased in response to increasing levels of anion was lower for nitrite than for nitrate.
Either NarL or NarP was able to induce nirB-lacZ expression in response to nitrite (compare the NarL− NarP+ and the NarL+ NarP− strains to the wild-type strain [Fig. 3B]). In contrast, only NarL was reported to respond to the nitrite signal (22). NarL is clearly the more effective activator of nirB-lacZ expression across the entire range of nitrite anion additions (compare the NarL+ NarP− and the NarL− NarP+ strains). NarL also caused a higher final level of gene expression than NarP. Interestingly, when nitrite was present, NarP did not antagonize NarL activation of nirB-lacZ expression to the extent that it did when nitrate was present (Fig. 3). Finally, the induction of gene expression was abolished in a narL narP double mutant. In summary, NarP was the less-effective response regulator for inducing nirB-lacZ expression relative to NarL when either nitrate or nitrite was present as the inducer signal.
Effect of narL and narP mutations on nitrate-dependent nrfA-lacZ expression.
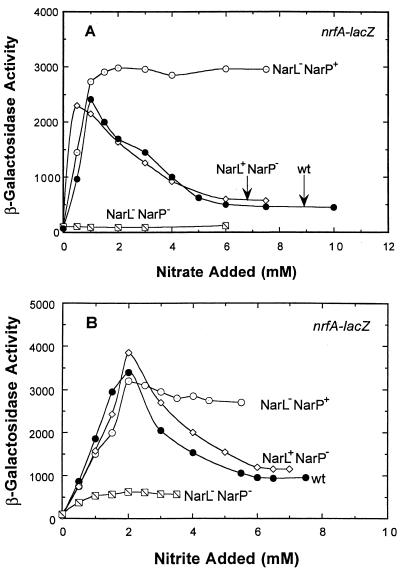
During batch cell culture conditions, nitrite was reported to induce nrfA expression via either the NarL or NarP response regulator proteins, while only nitrate was reported to cause repression of nrfA expression and only by NarL (14, 16, 24). However, when we examined the effect of narL and narP mutations on nrfA-lacZ expression in continuous culture (Fig. 4A), four points were immediately clear. First, either NarL or NarP can activate nrfA-lacZ expression in response to added nitrate. Second, NarL can repress nrfA-lacZ expression when nitrate is present, whereas NarP cannot (compare the NarL+ NarP− strain to the NarL− NarP+ strain and to the wild-type strain). Third, NarL is the more effective response regulator in activating nrfA-lacZ expression at low nitrate levels (as revealed by comparing the NarL+ NarP− and the NarL− NarP+ strains). Lastly, NarP appears to weakly antagonize the ability of NarL to activate nrfA-lacZ expression at low nitrate levels (compare the NarL+ NarP− strain to the wild-type strain). These findings therefore require a reexamination of the regulatory models based on batch culture studies wherein NarL or NarP can activate nrfA expression in response to nitrite only (24). Rather, NarL and nitrate clearly provide the major inputs in controlling of nrfA gene expression.
FIG. 4.
Effect of nitrate (A) and nitrite (B) concentration on nrfA-lacZ expression in narL and narP strains. The chemostat was sampled after each steady-state condition was achieved, and the β-galactosidase activity was determined. The fermentor conditions were identical to that used in Fig. 1. Expression levels in the wild-type strain (●), the narL (○), narP (◊), and the narL narP strain ( ) are indicated.
) are indicated.
Effect of nitrite additions on nrfA-lacZ expression.
We next examined how nitrite additions modulate the steady-state expression of the nrfA-lacZ reporter fusion. Nitrite induced nrfA-lacZ expression in a pattern similar to that shown for nitrate (Fig. 4B). However, a twofold-higher level of nitrite addition was required to elicit the same level of gene expression than nitrate did. The actual level of nitrate present in the growth vessel at maximal nrfA-lacZ expression was over 1,000-fold lower than for nitrite (ca. 0.2 μM versus 1 mM). Thus, nitrate is also the more potent regulatory signal for inducing nrfA gene expression. This finding is in contrast to previous reports in which nitrite was concluded to be the primary signal (14). At saturating levels of nitrite (ca. 6 mM), nrfA-lacZ expression was reduced to an intermediate level that was about 25% of the maximal level seen with nitrite. Finally, either NarL or NarP was able to activate nrfA-lacZ expression independently of the other, while only NarL could repress expression.
Comparison of nrfA-lacZ and nirB-lacZ gene expression in batch culture and in continuous culture.
To directly compare the steady-state chemostat and batch culture methods, the above nrfA-lacZ and nirB-lacZ reporter strains were analyzed in batch culture (Table 2). The batch culture results are in good agreement with previous studies (22). Therefore, any differences between the chemostat and batch culture data cannot be due to medium composition or strain differences between the two methods. It is therefore evident that considerable regulatory information was not revealed by the batch culture studies. Similar conclusions resulted from chemostat studies with the narG and napF operons that encode the two nitrate-regulated nitrate reductase enzymes in E. coli (25).
TABLE 2.
Anaerobic expression of nrfA-lacZ and nirB-lacZ reporter fusions in batch culture expression in wild-type and narL and narP mutant strains
| Relevant phenotype | Expressiona of:
|
|||||
|---|---|---|---|---|---|---|
|
nrfA-lacZ
|
nirB-lacZ
|
|||||
| None | Plus nitrite | Plus nitrate | None | Plus nitrite | Plus nitrate | |
| NarL+ NarP+ | 40 | 320 | 25 | 420 | 2,600 | 3,500 |
| NarL− NarP+ | 35 | 365 | 215 | 210 | 2,060 | 3,100 |
| NarL+ NarP− | 60 | 330 | 25 | 550 | 4,170 | 3,960 |
| NarL− NarP− | 25 | 70 | 25 | 150 | 270 | 205 |
Cultures were grown anaerobically in glucose minimal medium as described in Materials and Methods. Where indicated, sodium nitrite was added at an initial concentration of 2.5 mM and sodium nitrate was added at 20 mM. Expression is presented in β-galactosidase units, which are expressed as nanomoles of ONPG hydrolyzed per minute per milligram of protein. None, no additive.
For the narL or the narP strains growing in the chemostat, it is unknown if the amount of nitrate or nitrite remaining in the chemostat differs significantly from the wild-type strain. This is unlikely under high-anion-addition conditions, but it is less clear when very low to intermediate levels of anion additions are made. For the batch culture experiments, the levels of nitrate remaining and the levels of nitrite accumulated in the vessel at the time of cell sampling for gene expression measurements are unknown, and it is unlikely that steady-state gene expression was achieved.
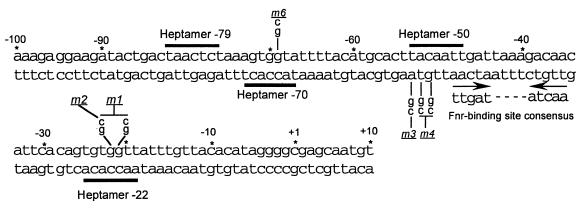
Roles of the −22, −50, and −70 NarL heptamer recognition sites in the activation and repression of nrfA-lacZ expression.
Prior DNA footprinting studies demonstrate that NarL and NarP bind to and protect several NarL heptamer recognition sites located in the nrfA promoter region (8). The nrfA operon has a promoter element containing an activation site consisting of the −79 and −70 heptamers that can be recognized by either NarL or NarP. Additional binding sites for NarL occur at positions −50 and −22. To evaluate how the sites contribute to the activation versus the repression of nrfA gene expression at low, intermediate, and high levels of nitrate, single- and double-base-pair mutations were introduced into the NarL heptamer sites designated −22, −50, and −70, (Fig. 5). The intended mutations were confirmed by DNA sequence analysis, and each altered DNA fragment was then used to construct a nrfA-lacZ reporter fusion for subsequent in vivo chemostat analysis.
FIG. 5.
Location of NarL heptamer recognition sites located at positions −22, −50, −70, and −79 in the nrfA promoter region. The sequence of the nrfA promoter region with the associated NarL heptamer sites (8) is shown relative to the start of nrfA transcription. The location of the Fnr recognition site is indicated by the inverted arrows below the DNA sequence. The mutations introduced into the −22, −50, and −70 heptamer sites are indicated below or above the sequence. The mutated nrfA promoter designated m5 was made by combining the m1 and the m4 mutations.
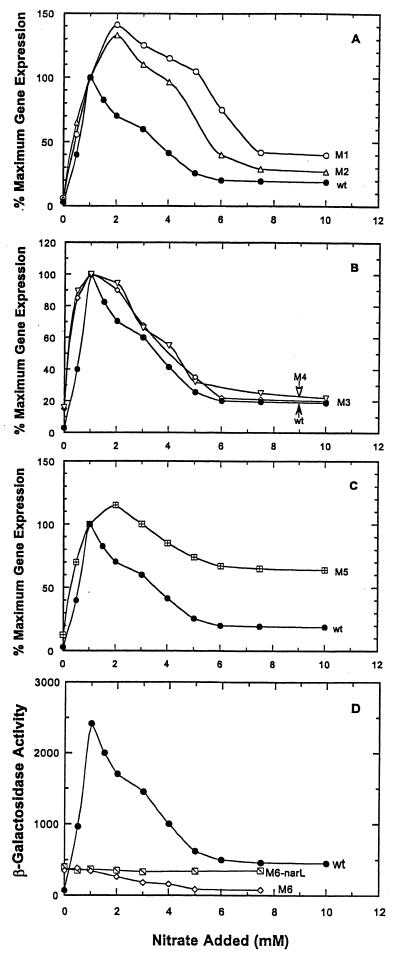
Introduction of a single base change in the −70 NarL heptamer, designated the nrfA m6 promoter mutation, completely abolished the nitrate-dependent induction of nrfA-lacZ expression (Fig. 6D). Interestingly, in the NarL− NarP+ strain, nrfA-lacZ expression remained at a basal level (400 U), whereas in the NarL+ NarP− strain, expression was gradually reduced below the initial basal level as the level of nitrate addition was increased from 0 to 7.5 mM. This indicates that (i) the −70 NarL heptamer site is essential for NarL to function as an activator of nrfA expression and that (ii) other sites are used for NarL to repress expression independently of the −70 site, in contrast to prior interpretations (see Discussion).
FIG. 6.
Effect of −22, −50, and −70 NarL heptamer site mutations on nitrate-dependent nrfA-lacZ expression. The expression of each nrfA-lacZ reporter fusion was evaluated at different levels of nitrate addition as described in Fig. 1. (A) Expression of the nrfA-lacZ m1 and m2 fusions. (B) Expression of the nrfA-lacZ m3 and m4 fusions. (C) Expression of the m5 nrfA-lacZ fusion. (D) Expression of the m6 nrfA-lacZ fusion in wild-type and narL strains.
To further explore the regulatory process, single and double mutations were introduced into the −50 NarL heptamer site to give the altered nrfA promoter fusions designated m3 and m4 (Fig. 5). While NarL was still able to activate and to repress nrfA gene expression, the pattern differed from the wild-type nrfA-lacZ reporter fusion (Fig. 6B). The maximal level of gene expression was equivalent, but the induction occurred at a lower nitrate level in the m3 and m4 reporter strains. Repression of nrfA-lacZ expression was also seen, but it was delayed until higher levels of nitrate additions were made. Thus, the −50 NarL heptamer site appears to fine-tune the activation and repression of nrfA gene expression by narrowing the window of maximal gene expression.
When a single- or double-base change was introduced into the −22 NarL heptamer site to give the m1 or the m2 nrfA-lacZ fusions, a distinct pattern of nrfA-lacZ expression was seen relative to either the −50 or the −70 mutant (Fig. 6A). The onset of activation seen for the m1 and m2 nrfA-lacZ reporter fusions was like the wild-type nrfA-lacZ reporter fusion. However, the maximum level of gene expression was 40% higher, and it occurred only at higher levels of nitrate addition. Third, the subsequent repression of nrfA-lacZ expression by NarL was also delayed until higher nitrate additions. Fourth, the degree of repression was less severe at the highest level of nitrate tested. Therefore, the −22 heptamer site is a critical site for mediating NarL repression of nrfA gene expression. When mutations were introduced into both the −22 and the −50 NarL heptamer sites (designated as the nrfA-lacZ m5 reporter fusion), repression of nrfA-lacZ expression was further impaired relative to when mutations were introduced only in the −22 site (Fig. 6C versus A). Gene expression was nearly derepressed relative to the wild-type strain. These results suggest that both the −22 and the −50 sites are needed for optimal repression of nrfA gene expression. This repression also occurs in the absence of an intact −70 activator site (Fig. 6D).
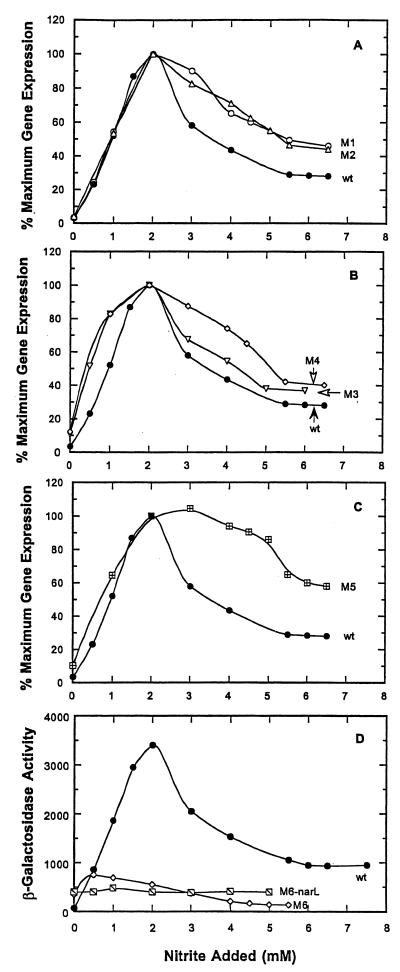
When the m1, m2, m3, m4, m5, and m6 nrfA-lacZ reporter fusions were tested in response to nitrite additions (Fig. 7), similar patterns of nrfA gene expression were seen relative to the nitrate addition experiments (compare these results to those in Fig. 6). It is therefore evident that NarL and NarP regulate nrfA-lacZ expression in response to nitrite addition in the same qualitative way that the nitrate anion signal does in contrast to prior proposals. However, a significantly higher level of nitrite is required to elicit a somewhat inferior response relative to nitrate. These findings also imply that the NarX and/or NarQ sensor transmitters detect each anion in the same general way but where nitrate is the more potent signal.
FIG. 7.
Effect of −22, −50, and −70 NarL heptamer site mutations on nitrite-dependent nrfA-lacZ expression. Effect of −22, −50, and −70 NarL heptamer site mutations on nitrite dependent nrfA-lacZ expression. The expression of each nrfA-lacZ reporter fusion was evaluated at different levels of nitrate addition as described in Fig. 1. (A) Expression of the nrfA-lacZ m1 and m2 fusions. (B) Expression of the nrfA-lacZ m3 and m4 fusions. (C) Expression of the m5 nrfA-lacZ fusion. (D) Expression of the m6 nrfA-lacZ fusion in wild-type and narL strains.
DISCUSSION
The nrfA and nirB operons are expressed in a complementary style.
The chemostat gene expression studies clearly establish a complementary pattern of nitrite reductase gene expression in E. coli (Fig. 1, 3, and 4). When the nitrate level is very low, the cell induces the “first response” nitrite reductase encoded by the nrfA operon to consume nitrite in the environment. It is interesting to speculate that the NrfA nitrite reductase may therefore have a higher (e.g., stronger) affinity for nitrite than does the nirB encoded nitrite reductase. Tests of this prediction must await additional biochemical characterization of the NrfA enzyme.
When the level of nitrate in the environment becomes elevated, the second nitrite reductase encoded by the nirB operon is preferentially synthesized due to transcriptional controls imposed by the Nar regulatory circuit. The cytoplasmic reductase is then the predominant nitrite reductase in the cell since the NrfA enzyme is nearly absent under high-nitrate conditions (Fig. 1)! Apparently, the two-nitrite reductase enzymes and their transcriptional regulatory elements are evolved to function in a complementary way to provide for nitrite reduction under these different conditions (discussed below).
The 25-fold induction of nrf operon expression seen at low concentrations of nitrate was not revealed by previous batch culture studies since those experiments were performed only at saturating levels of the anion (1, 14, 22). Therefore, the steady-state chemostat studies indicate that the nrfA operon must serve a far more important role in cell metabolism than was previously indicated by the modest three- to fourfold change seen in batch culture experiments (Table 2; discussed below). Second, the roles of the two anion inducer molecules on nrfA gene expression (i.e., nitrate and nitrite), as well as the roles for the two response regulatory proteins, NarL and NarP, are far more intricate than previously envisioned. Likewise, nirB operon expression was examined under identical chemostat conditions at low and intermediate levels of nitrate or nitrite (Fig. 3). With low nitrate additions of 1 mM where nrfA-lacZ expression was maximal, nirB expression was only 30% of its final maximal level. With nitrate additions ranging from 1 to 2 mM, both nitrite reductase operons are simultaneously expressed at significant levels.
The conclusions derived from batch culture and chemostat experiments for NarL- and NarP-dependent control of nrfA and nirB gene regulation in response to nitrate and nitrite are summarized in Table 3. From the chemostat studies, new roles for NarL and nitrate in the control of each operon are evident. Also, since NarL and NarP clearly function in more versatile ways in response to either nitrate or nitrite as a regulatory signal, the prior models based on the batch culture studies must be reconsidered (6, 24). New regulatory schemes for the NarL and NarP control of nrfA and nirB operon expression based on the chemostat data shown in Fig. 1 to 7 are depicted in Fig. 8 and 9. One of the more striking points in each model is that the NarP protein is nonessential (i.e., not a major player) and that it can function as an antagonist of NarL in its ability to activate nirB or nrfA operon expression. Rather, NarP appears to fine-tune gene expression under low-nitrate conditions by delaying the regulatory response to nitrate. This antagonizing effect is relatively weak when nitrite is present in place of nitrate. The simplest explanation for these findings is that the cognate sensor transmitter proteins, NarX and/or NarQ, are unable to respond to nitrite in the same way they do for nitrate. The latter anion is a more potent stimulator of NarX kinase activity by 2 to 3 orders of magnitude in concentration relative to nitrite (12). Nitrite also fails to elicit the same steady-state level of NarX-phosphate that nitrate does.
TABLE 3.
Functioning of NarL and NarP in the control of nrfA and nirB operon expression in response to nitrate and nitrite
| Reporter fusion and method | Actiona by the response regulator
|
Source or reference | |||
|---|---|---|---|---|---|
| NarL
|
NarP
|
||||
| Nitrite | Nitrate | Nitrite | Nitrate | ||
| nrfA | |||||
| Batch culture | + | − | + | + | 24 |
| Chemostat culture | +, − | +, − | + | + | This study |
| nirB | |||||
| Batch culture | + | + | None | + | 24 |
| Chemostat culture | + | + | + | + | This study |
+, activation of transcription by either the NarL or the NarP response regulatory protein; −, transcription repression.
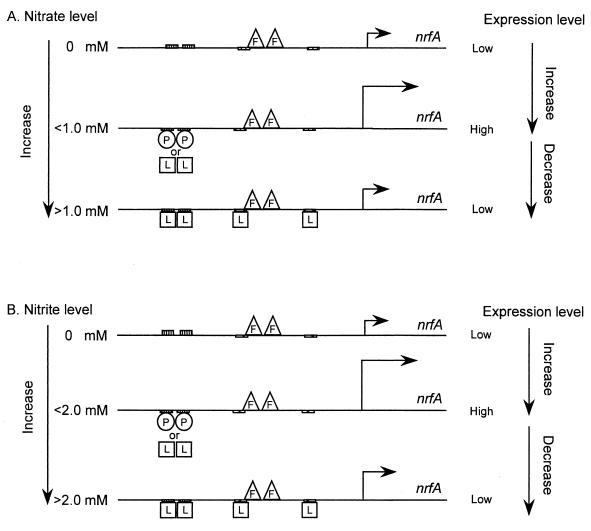
FIG. 8.
Model for NarL- and NarP-dependent control of nrfA operon expression. (A) Nitrate control. The Fnr protein (F) induces nrfA operon expression under anaerobic conditions. In the presence of low levels of nitrate, either NarL (L) or NarP (P) bind at the −70 and −79 heptamer sites (hatched boxes) to activate nrfA gene expression. NarL is the more proficient activator under low-nitrate conditions, where NarP antagonizes NarL binding (Fig. 4A). At elevated-nitrate conditions, additional NarL molecules bind to the flanking NarL heptamer sites at −50 and −22 to repress nrfA operon expression (Fig. 5). NarP is not essential for either the activation or repression process. (B) Nitrite control. A similar regulatory response is observed in response to nitrite except that higher anion levels are required relative to the nitrate (Fig. 4B).
FIG. 9.
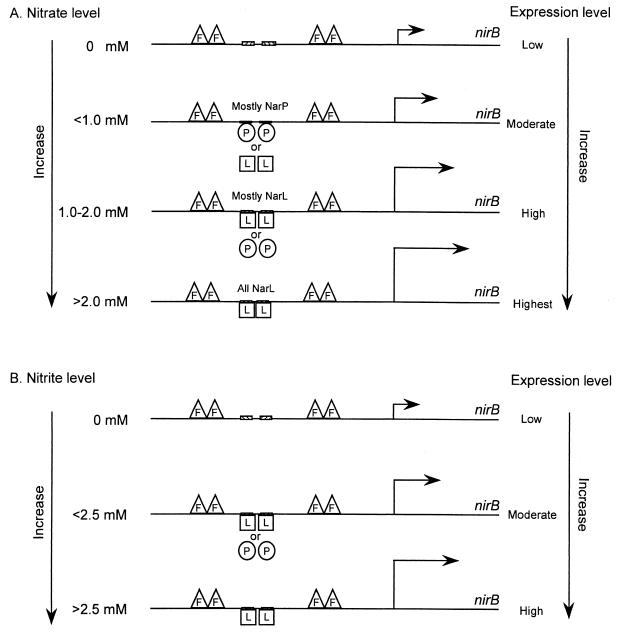
Model for NarL- and NarP-dependent control of nirB operon expression. (A) Nitrate control. During anaerobiosis, the Fnr protein (F) binds at its consensus recognition site, centered at −42, to activate nirB operon expression. When low-nitrate conditions are encountered, NarL protein (L) binds at the two heptamer sites at −74 and −65 to activate nirB expression (8). NarP interaction at the heptamer sites somehow antagonizes NarL function (Fig. 3A). (B) Nitrite control. A similar regulatory response is observed in response to nitrite except that higher anion levels are required relative to nitrate (Fig. 3B).
Revised models for NarL and NarP control of nrfA and nirB gene expression.
As noted above, the nrfA operon has a promoter element containing an activation site consisting of the −79 and −70 heptamers that can be recognized by either NarL or NarP (7). Additional binding sites for NarL are at positions −50 and −22 (8). NarL-phosphate binding at the −50 site presumably interferes with Fnr binding at its site adjacent to the NarL heptamer. Additional NarL binding at the −22 site is envisioned to interfere with RNA polymerase interactions at the nrfA promoter.
A revised regulatory model for the nrfA operon is shown in Fig. 8 based on the chemostat studies with strains defective in narL or narP (Fig. 4). Overall, nrfA gene expression can be modulated by 25-fold by adjusting the balance between the activation and repression events. It is evident that NarL can serve as an activator and as a repressor of nrfA expression in vivo in response to either nitrate or nitrite (Table 3). In the absence of either inducer anion, gene expression is minimal. When sufficient amounts of either anion are detected by the NarX and/or NarQ sensors, both NarL and NarP are phosphorylated where either response regulator can then activate nrfA gene expression. However, the chemostat experiments demonstrate that NarP-phosphate cannot repress nrfA gene expression under high nitrate or nitrite conditions. This is consistent with the in vitro experiments that both NarP-phosphate and NarL-phosphate bind to the −70 and −79 heptamer sites, while only NarL-phosphate bind at the −50 and −22 sites (8). In fact, NarL has the highest affinity for the activation sites at −70 and −79 as previously shown by Darwin et al. (8), whereas NarP exhibits a weaker affinity for these sites. When high-nitrate or -nitrite conditions are encountered, additional NarL-phosphate molecules are made that then bind at the nearby NarL heptamers (Fig. 5) to inhibit nrfA gene expression. As described above, much higher levels of nitrite are required to give the same result that submicromolar amounts of nitrate does (Fig. 4B). Batch culture experiments clearly do not permit the quality of the nitrate or nitrite signals to be deduced (25). Interestingly, the nrfA operon expression pattern is similar to that of the napF operon (25). However, the latter operon has a strikingly different regulatory element where the locations of the NarL and NarP heptamer sequences and the Fnr sites are reversed in relative position (7). Thus, positions of the binding sites alone cannot be used to predict the differential expression patterns for these two operons.
A revised model for NarL-dependent activation of nirB gene expression is shown in Fig. 9. Under low-nitrate conditions, NarP-phosphate binding at its two heptamer recognition sites (8) weakly induces nirB expression (Fig. 3A). Under these conditions NarP also antagonizes the superior activator response by NarL-phosphate. However, when elevated nitrate conditions are encountered, NarL-phosphate overcomes the antagonistic effect of NarP to give optimal induction of nirB expression. NarP-phosphate can clearly also elicit activation of nirB operon expression in response to nitrite (Figure 3B) in contrast to prior conclusions based on batch culture methods (23, 24).
Different environmental conditions for the two-nitrite reductase enzymes?
Since nrf operon expression is optimal only at low-nitrate conditions, while nirB expression is optimal during high-nitrate conditions (Fig. 1), the two nitrite reductase enzymes must serve distinct roles within the cell. The high-nitrite conditions needed for nirB induction are in keeping with the proposed role of the NirB enzyme in detoxification (4). The chemostat experiments also support a second plausible role for the NirB enzyme whereby it recycles NADH by oxidizing it under conditions when excess reducing equivalents are present inside the cell. Such conditions would occur when sufficient energy is generated by nitrate-dependent respiration via the NarG nitrate reductase complex. Use of the NirB enzyme would thus allow the cell to effectively decouple carbon dissimilation from the nitrite respiratory pathways by using a futile cycle for NADH-NAD recycling. It is apparent that the respective regulatory elements for the two nitrite reductase operons are evolved to ensure that each enzyme is synthesized under different environmental conditions.
The differential and complementary patterns for nrfA and nirB expression in response to nitrate or nitrite (Fig. 1, 3, and 4) are similar to the expression patterns for the two nitrate reductase operons encoded by the narGHJI and napAB genes (25). The nirB operon is induced primarily in response to intermediate to high levels of nitrate like the narGHJI nitrate reductase operon (25). The pattern of nrfA operon expression is similar to that seen for the napAB operon where both operons are optimally expressed only during low-nitrate growth conditions (25; this study). This supports a physiological role of the NrfA respiratory enzyme in providing additional membrane potential needed for cellular ATP synthesis in concert with the NapAB respiratory nitrate reductase. Since the NapAB enzyme is periplasmic and apparently of lower proton pumping efficiency due to its inability to form vectoral coupling, NapAB would therefore have a lower efficiency in energy generation relative to the NarG enzyme (4). The NrfA enzyme could thus effectively assist in the NapAB enzyme in energy harvesting under low-nitrate growth conditions. In high-nitrate conditions when the expression of both the nrfA and napA operons is switched off (25; this study) and where the expression of the narG and nirB operons are switched on, the NarG respiratory nitrate reductase appears to be proficient in generating an appropriate proton gradient for the cell. Under these conditions, most of the newly formed nitrite is excreted from the cell and accumulates in the medium (Fig. 2).
Is nitrite a significant physiological inducer of nrfA or nirB gene expression?
From the chemostat studies, it is evident that nitrate is the more potent inducer of nrfA and nirB gene expression than nitrite by at least 2 orders of magnitude in concentration. Submicromolar levels of nitrate (i.e., <0.03 μM nitrate) are sufficient to induce both nrfA and nirB gene expression, as also seen for narG and napF gene expression (25).
Additionally, nitrite was not a coinducer nor an antagonist of the cells' ability to recognize nitrate as a signal based on chemostat studies with the napF and narG operons (25). Therefore, nitrite does not antagonize the cell ability to detect nitrate via NarX and NarQ. The cell exhibits a considerable ability to discriminate between the two structurally related oxyanion molecules (12). Nitrate is also the superior regulatory signal for control of the narG and napF operons in vivo (25).
Differentially expressed genes in E. coli.
These studies provide the third example of differentially expressed pairs of respiratory pathway genes in E. coli. These include two nitrite reductase operons (nrfA and nirB; this study), two nitrate reductase operons (napFDAGHBC and narGHJI) (25), and two cytochrome oxidase operons (cydAB and cyoABCDE) (21). The first two sets of operons are regulated primarily by nitrate and to a lesser extent by nitrite, while the latter set of operons are controlled in response to oxygen availability. The differential and complementary expression of these pairs of operons in response to their environmental signals allows the cell to more effectively compete for respiratory substrates at low concentrations versus when they are in excess. The cell would thus conserve energy by not synthesizing unneeded respiratory enzymes. Finally, the cell does not use an abrupt “switch” to turn on or turn off respiratory gene expression. Rather, the regulatory response can be adjusted over a range of signal concentrations (Fig. 1 and 2).
Since many other enteric and soil bacteria also possess dual sets of nitrate reductase, nitrite reductase, and cytochrome oxidase enzymes such as those present in E. coli (9, 26), it is conceivable that the operons in these organisms are also regulated in similar ways. From the above studies, it is evident that the chemostat approach provides a powerful tool for better understanding the physiological basis of gene expression in response to environmental signals. This is especially relevant when the inducer or repressor molecule is present in low amounts.
ACKNOWLEDGMENT
This study was supported in part by the Public Health Service grant AI21678 from the National Institutes of Health.
REFERENCES
- 1.Bell A, Cole J, Busby S. Molecular genetic analysis of an FNR-dependent anaerobically inducible Escherichia coli promoter. Mol Microbiol. 1990;4:1753–1763. doi: 10.1111/j.1365-2958.1990.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 2.Chiang R C, Cavicchioli R, Gunsalus R P. ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NarQ and NarX sensors affect nitrate- and nitrite-dependent regulation by NarL and NarP. Mol Microbiol. 1997;24:1049–1060. doi: 10.1046/j.1365-2958.1997.4131779.x. [DOI] [PubMed] [Google Scholar]
- 3.Choe M, Reznikoff W S. Identification of the regulatory sequence of anaerobically expressed locus aeg-46.5. J Bacteriol. 1993;175:1165–1172. doi: 10.1128/jb.175.4.1165-1172.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole J. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol Lett. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- 5.Cotter P A, Gunsalus R P. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J Bacteriol. 1989;171:3817–3823. doi: 10.1128/jb.171.7.3817-3823.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin A J, Stewart V. The Nar modulon systems: nitrate and nitrite regulation of anaerobic gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Company; 1996. pp. 333–359. [Google Scholar]
- 7.Darwin A J, Stewart V. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J Mol Biol. 1995;251:15–29. doi: 10.1006/jmbi.1995.0412. [DOI] [PubMed] [Google Scholar]
- 8.Darwin A J, Tyson K L, Busby S J, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Horsmann J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman P S, Cole J, Busby S. Mutational analysis of the nucleotide sequence at the FNR-dependent nirB promoter in Escherichia coli. Nucleic Acids Res. 1989;17:135–145. doi: 10.1093/nar/17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones H M, Gunsalus R P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol. 1987;169:3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee A I, Delgado A, Gunsalus R P. Signal-dependent phosphorylation of the membrane bound NarX two-component sensor transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J Bacteriol. 1999;181:5309–5316. doi: 10.1128/jb.181.17.5309-5316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 14.Page L, Griffiths L, Cole J A. Different physiological roles for two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch Microbiol. 1990;154:349–354. doi: 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- 15.Peakman T, Crouzet J, Mayaux J F, Busby S, Mohan S, Harborne N, Wootton J, Nicolson R, Cole J. Nucleotide sequence, organisation and structural analysis of the products of genes in the nirB-cysG region of the Escherichia coli K-12 chromosome. Eur J Biochem. 1990;191:315–323. doi: 10.1111/j.1432-1033.1990.tb19125.x. [DOI] [PubMed] [Google Scholar]
- 16.Rabin R S, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of b-globin genomic sequences for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen B J, Coulson A R. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 20.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 21.Tseng C P, Hansen A K, Cotter P, Gunsalus R P. Effect of cell growth rate on expression of the anaerobic respiratory pathway operons frdABCD, dmsABC, and narGHJI of Escherichia coli. J Bacteriol. 1994;176:6599–6605. doi: 10.1128/jb.176.21.6599-6605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyson K, Busby S, Cole J. Catabolite regulation of two Escherichia coli operons encoding nitrite reductases: role of the Cra protein. Arch Microbiol. 1997;168:240–244. doi: 10.1007/s002030050494. [DOI] [PubMed] [Google Scholar]
- 23.Tyson K L, Bell A I, Cole J A, Busby S J W. Definition of nitrate and nitrite response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol Microbiol. 1993;7:151–157. doi: 10.1111/j.1365-2958.1993.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 24.Tyson K L, Bell A I, Cole J A, Busby S J W. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol Microbiol. 1994;5:353–360. doi: 10.1111/j.1365-2958.1994.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Tseng C-P, Gunsalus R P. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J Bacteriol. 1999;181:5303–5308. doi: 10.1128/jb.181.17.5303-5308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]