Abstract
COVID-19 is a contagious disease that attacks many organs but the lungs are the main organs affected. The inflammasome activation results in the exacerbation of inflammatory response in infectious disease. The aim of this study is to investigate the formation and activity of the NLRP3 inflammasome complex and the histopathological changes caused by the coronavirus in the lung of deceased persons with COVID-19. In total, 10 corpses; 5 corpses with no history of any infectious diseases and COVID-19 and 5 corpses with the cause of death of COVID-19 were included in this study. Lung tissue samples were harvested during autopsy under safe conditions. Fresh tissues in each group were used to measure the genes expression and proteins level of NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α and a routine hematoxylin and eosin staining was performed for histological assessment. Data are represented as the means ± SD. Statistical significance difference was accepted at a p-value less than 5%. The NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α genes expression and proteins level were elevated in the lung of the COVID-19 group in comparison with the control group. Histological findings presented the increasing number of polymorphonuclear leukocytes, macrophages and also pulmonary fibrosis in the lungs of corpses with the cause of death of COVID-19. High expression of NLRP3 inflammasome components and its relation with the pathophysiology of the coronavirus-infected lung suggested that targeting the NLRP3 inflammasome could be helpful in achieving a more effective treatment in patients with COVID-19.
Keywords: COVID-19, NLRP3, Inflammasome, Lung, Histopathology, Coronavirus
Introduction
It was late in December 2019 that the first cases of COVID-19 appeared in Wu et al. (2020). Up to April 2021, there have been 127 million cases of COVID-19 and more than 2.79 million deaths. The disease is very contagious and it also attacks many organs in the body (Anderson et al. 2020; Larsen et al. 2020). Lungs are the main organs that get involved and show changes in CT scan both in symptomatic and asymptomatic patients (Pan et al. 2020). Alveolar epithelial cells are the main injury site in the lungs in COVID-19 disease (Li and Ma 2020). Observation of pathological findings revealed the presence of exudative inflammation occurring in the early phase of COVID-19 pneumonia (Gallelli et al. 2020). A lung CT scan is the gold standard diagnostic tool for lung complications (Buonsenso et al. 2020). Unfortunately, we still don’t have an effective drug to cure patients with COVID-19 (Brown et al. 2021). It has been shown that the virus can enter host cells through the ACE2 receptor and cause cell involvement (Ni et al. 2020). The presence of the virus in the body leads to sepsis and the creation of a cytokine storm, eventually leading to the failure of organs such as the lungs as the main hosts of the virus (Mangalmurti and Hunter 2020). This cytokine storm has been thought to be the main reason for mortality in COVID-19 patients (Cron 2021). Inflammasomes are protein complexes that initiate and advance the inflammatory cascade. The NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3) inflammasome is the most well-known type of inflammasome and consists of the NLRP3 scaffold, the ASC (the adaptor molecule apoptosis-associated speck-like protein containing a CARD) adaptor, and caspase-1 (Gholaminejhad et al. 2022). Inflammasomes recognize damage signals and a diverse range of viruses, bacteria, stress that result in activation of caspase-1, which subsequently changes pro-interleukin 1β and pro-interleukin 18 into their active form and cause pyroptosis (a form of cell death) (Schroder and Tschopp 2010; Strowig et al. 2012; Zoete et al. 2014; Guo et al. 2015; Mohammed et al. 2020; Ijaz et al. 2020). These inflammasome actions result in the exacerbation of inflammatory response in infectious disease (Rodrigues et al. 2021). There are still many questions about coronavirus that we do not have answers to. Getting to know more and more about the pathogenic process of the virus is needed to end the nightmare that it caused. Accordingly, this study was designed to investigate the formation and activity of the NLRP3 inflammasome complex and the histopathological changes caused by the coronavirus in the lung of deceased persons with COVID-19.
Materials and methods
Study design
In total, 10 corpses; 5 corpses with no history of any infectious diseases and COVID-19 (Control group, mean age 41 years, range between 34 and 50) and 5 corpses with the cause of death of COVID-19 (COVID-19 group, mean age 56 years, range between 49 and 61) that coronavirus infection was confirmed by positive Real-time PCR and/or CT scan at the Hospitals, were included in this study. These corpses were brought to the Legal Medicine Organization of Tehran Province (Kahrizak, Tehran, Iran). The tissue sampling time was less than 10 h after death. Lung tissue samples were harvested during autopsy under safe conditions. The samples were harvested from the peripheral area of the lungs. The fresh specimens were put in the liquid nitrogen storage tank and then were moved to a -80 freezer and stored until the next molecular tests. The specimens for the histopathological test were put in the formaldehyde solution 4% for histological staining. The personal information of all the corpses was preserved and the samples were determined with a special code. This study was approved (ethical code: IR.LMO.REC.1399.055) by the Research Ethics Committee of the legal Medicine Organization, Islamic Republic of Iran (Biomedical Research Ethics Committee).
Real-time polymerase chain reaction (real-time PCR)
Fresh lung tissues from corpses in each group were used to measure the gene expression of NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α. Total RNA was extracted from lung tissue (n = 5 per group, 3-replica) with Trizol reagent according to the manufacturer’s instructions (RiboExTM LS, GeneAll, Korea). BioFact™ RT (South Korea) Synthesis Kit was used for reverse transcription of total extracted RNA to obtain cDNA. Real-time PCR was performed using the reaction mixture contained 5 µl SYBR Green Master Mix (BioFact, South Korea), 0.8 µl primers (Forward + Reverse), 0.8 µl cDNA and 3.4 µl RNase Free dH2O (Invitrogen) in a Real-time PCR instrument (QIAGEN Rotor-Gene, Germany). Samples were incubated at 95 °C for 10 min for initial denaturation and enzyme activation. Then the following three steps were done: denaturation, at 95 °C for 15 s; annealing, at 59–61 °C for 25 s; extension and fluorescence acquiring, at 72 °C for 30 s. The 2−ΔΔCt method was applied for the relative quantification of data and normalization of GAPDH as housekeeping. The obtained data were represented as fold change gene expression compared to the control group. Primer sequences are listed in Table 1.
Table 1.
Primer sequences and product size (base pair = bp)
| sequence | product size (bp) | ||
|---|---|---|---|
| NLRP3 | Forward | GGAGTGGATGGGTTTACTGGAG | 165 |
| Reverse | CGTGTGTAGCGTTTGTTGAGG | ||
| PYCARD (ASC) | Forward | CGTTGAGTGGCTGCTGGATG | 95 |
| Reverse | GCATCTTGCTTGGGTTGGTG | ||
| Caspase-1 | Forward | CCAGCTATGCCCACATCCTC | 201 |
| Reverse | TGTGATGTCAACCTCAGCTCC | ||
| IL-1β | Forward | CAGAAGTACCTGAGCTCGCC | 153 |
| Reverse | AGATTCGTAGCTGGATGCCG | ||
| IL-6 | Forward | CTTCGGTCCAGTTGCCTTCT | 169 |
| Reverse | GATGCCGTCGAGGATGTACC | ||
| TNF-α | Forward | CTCTTCTGCCTGCTGCACTTTG | 135 |
| Reverse | ATGGGCTACAGGCTTGTCACTC | ||
| GAPDH | Forward | GTGGTCTCCTCTGACTTCAAC | 97 |
| Reverse | GGAAATGAGGCTTGACAAAGTGG |
Enzyme-linked immunosorbent assay (ELISA)
To determine the protein level of NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α, the ELISA technique was performed. Briefly, the fresh lung tissue (n = 5 per group, 3-replica) was homogenized and was used for the analysis of the protein levels using human ELISA kits (NLRP3, MBS917009, MyBioSource, USA; ASC, CSB-EL019114HU, CUSABIO, USA; Caspase-1, MBS264676, MyBioSource, USA; IL-1β, 850.006.096, Diaclone, France; IL-6, 950.030.096, Diaclone, France; TNF-α, 950.090.096, Diaclone, France). According to each company’s protocol, the experiments were done.
Histological study
Lung specimens were fixed in formaldehyde solution 4%, buffered, pH 6.9 (Merck, Germany). Lung tissue samples were dehydrated and were embedded in paraffin (Merck, Germany) and sectioned into 5 μm thickness slices with 25 μm intervals. A usual hematoxylin and eosin (H&E) staining was performed for histological assessment. Five sections from each corpse were used for evaluations. Two histopathologists, blinded to the groups, evaluated the sections. The sections were visible under a light microscope (LABOMED, Labo America, Inc. USA). A digital camera (LABOMED, USA) was used to take histophotographs. Total histopathological scoring was done according to the following parameters: hemorrhage, thickened wall, inflammatory cells infiltration, and fibrotic changes with a score of None = 0, Mild = 1, Moderate = 2, and Severe = 3 (Gibson-Corley et al. 2013).
Statistical analysis
Statistical analyses were carried out by GraphPad Prism (version 8.0.0 for Windows, GraphPad Software, San Diego, California USA). We used the t-test for two-group comparisons. Data were represented as the means ± SD. statistical significance difference was accepted in a p-value less than 5%.
Results
Gene expression of NLRP3 inflammasome components and inflammatory cytokines increased in the lung of the COVID-19 group
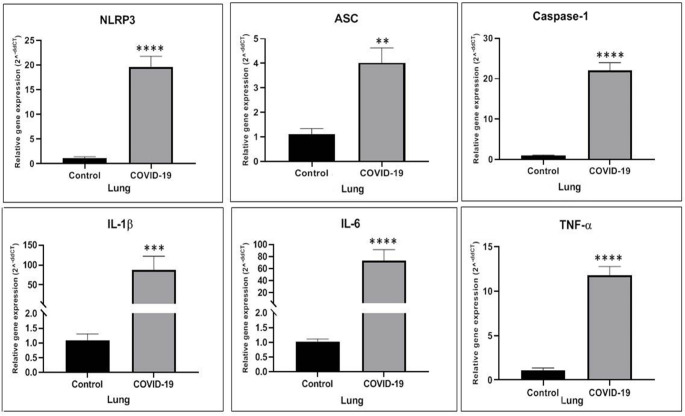
In the present study, we evaluated the level of NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α gene expression by Real-time PCR test. The level of NLRP3 gene expression in the lung significantly increased as compared to the Control group (p˂0.0001). There was a significant difference between the COVID-19 group and the Control group in the level of ASC gene expression (p = 0.002). Compared with the Control group, the level of Caspase-1 was statistically increased in the COVID-19 group (p˂0.0001). Similar results were obtained in the Real-time PCR analysis, where the expression of IL-1β and IL-6 genes significantly raised in the COVID-19 group in comparison to the Control group (p = 0.0012 and p˂0.0001 respectively). Following the Coronavirus disease, the level of TNF-α gene expression in the lung meaningfully raised as compared to the Control group (p˂0.0001) (Fig. 1).
Fig. 1.
The NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α genes expression elevated in the lung of COVID-19 group in comparison to the Control group. * p < 0.05 compared to the Control group, ** p < 0.01 compared to the Control group, *** p < 0.001 compared to the Control group and **** p < 0.0001 compared to the Control group (N = 5 per group, 3 replicates)
The protein level of NLRP3 inflammasome components and inflammatory cytokines increased in the lung of the COVID-19 group
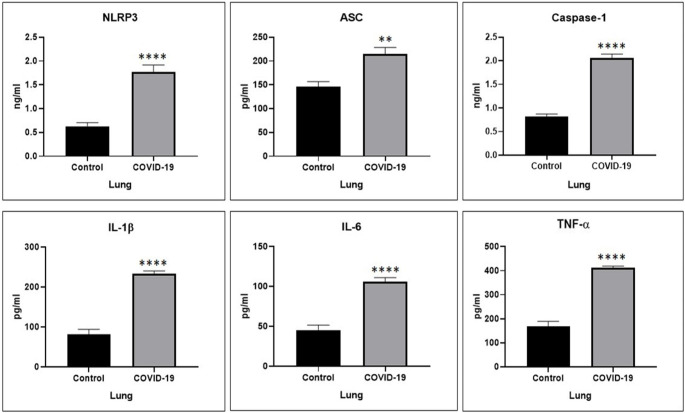
To determine and compare the role of the inflammatory cascade in the lung of COVID-19 and control groups, the level of NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α proteins level was examined, via ELISA test. There was a significant difference between the COVID-19 group and the Control group in the level of NLRP3 protein level (p < 0.0001). The findings of the present study indicated a significant difference in the level of ASC protein level between the two groups (p = 0.0014). COVID-19 caused a meaningful increase in the level of Caspase-1 protein level as compared to the Control group (p < 0.0001). Similar results were obtained in the ELISA analysis, where the level of IL-1β and IL-6 proteins significantly raised in the COVID-19 group in comparison to the Control group (p < 0.0001 and p < 0.0001 respectively). Following the Coronavirus disease, the level of TNF-α protein in the lung significantly increased as compared to the Control group (p < 0.0001) (Fig. 2).
Fig. 2.
The NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α proteins level elevated in the lung of COVID-19 group in comparison to the Control group. * p < 0.05 compared to the Control group, ** p < 0.01 compared to the Control group, *** p < 0.001 compared to the Control group and **** p < 0.0001 compared to the Control group (N = 5 per group, 3 replicates)
Histopathological changes were observed in the lung of the COVID-19 group
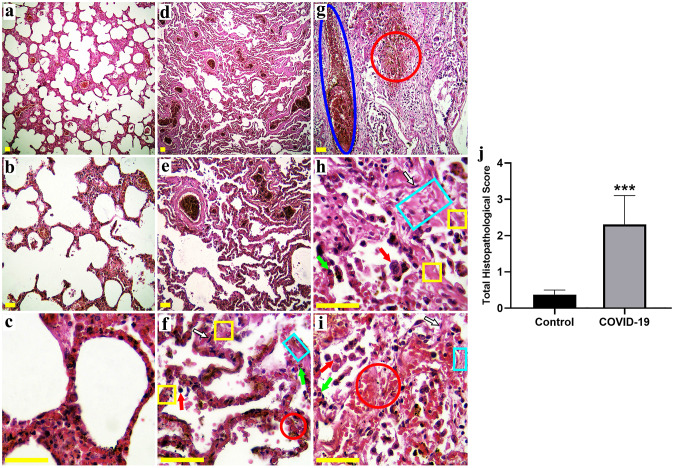
Histological findings of the present study showed an increase in the number of macrophages in the COVID-19 group (Fig. 3f,h,i), and also a higher density of PMNs infiltration was observed in this group (Fig. 3f,h,i) as compared to the Control group (Fig. 3a,b,c). The results revealed that COVID-19 is characterized by the loss of alveolar structures, and high-grade hemorrhage can be seen between the alveoli (Fig. 3f,g,i). The alteration in the structure of alveoli can cause the loss of type II alveolar epithelial cells, which leads to the disturbance of surfactant production. Injury and inflammation of the arterioles, venules or alveolar septal are the main reasons for hemorrhage in the alveoli. Thickened inter-alveolar septa with a high number of fibroblasts showed fibrotic lung changes (Fig. 3f,h,i) that may lead to blood clots and infections of the lung. Congestion and dilated vessels with lots of blood cells (Fig. 3 g) were observed in the COVID-19 group as compared to the Control group. The total histopathological score was meaningfully higher in the COVID-19 group in comparison with the Control group (p < 0.001) (Fig. 3j) (Fig. 3).
Fig. 3.
Photomicrographs showing the pulmonary structure in the Control (a-c) and COVID-19 (d-i) groups. H&E-stained sections showed an increase in the number of macrophages (red arrow) and PMNs infiltration (green arrow) in the COVID-19 group. The results revealed that COVID-19 is characterized by the loss of alveolar structures, and high-grade hemorrhage (red circle) can be seen between the alveoli. Thickened inter-alveolar septa (yellow square) with a high amount of fibroblasts (black-white arrow) showed fibrotic lung changes (light blue rectangular). Congestion and dilated vessels (dark blue ellipsoid) with lots of blood cells were observed in the COVID-19 group. The total histopathological score was significantly higher in the COVID-19 group compared to the Control group (j). (Hematoxylin and eosin stains, a, d ×40, b, e, g ×100, c, f, h, i ×400; Scale bar: All 100 μm), *** p < 0.001 compared to the Control group (N = 5 per group, 5 sections from each corpse)
Discussion
In the present study, we described and compared the NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α genes expression and their proteins level and histological markers in the lungs of COVID-19 and normal corpses. One of the manifestations of several diseases and infections like COVID-19 is the hyperactivation of the inflammasome complex and also it has been proved that different tissues such as lungs, heart, kidneys and liver to be affected in COVID-19 patients (Mokhtari et al. 2020; Russo et al. 2020; Hassanzadeh et al. 2021). Uncontrolled inflammation can lead to an extreme release of cytokines that is called a cytokine storm. This cytokine storm can lead to alveolar damage and finally reduced gas exchange in different ways like increasing the dead cells and proteins and accumulation of fluid in the lungs which is called edema (Tay et al. 2020). Our results showed that the expression of The NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α genes significantly increased in the COVID-19 group as compared to the Control group in the lung. Similar to the findings of this study, the findings of clinical studies by Baines et al. (2011, 2010). Baines et al. (2011). In line with the results of the present study, Toldo et al. in 2021 only showed NLRP3 inflammasome components were increased in formalin 10%-fixed lung tissue by IHC in individuals who passed away from fatal COVID-19, but we showed these in both genes expression and proteins level (by Real-time PCR and ELISA, respectively). In addition, these results only indicated the formation of the NLRP3 inflammasome, and it is not enough to show the inflammatory cascade and its effect on lung histophysiology/histopathology. However, we demonstrated the activity of NLRP3 inflammasome that increased the production and secretion of key proinflammatory/inflammatory cytokines (IL-1b, TNF-a, and IL-6) and their effects on histophysiology/histopathology of lung tissue of deceased individuals through COVID-19 (Toldo et al. 2021). The findings of the present study showed that the level of The NLRP3, ASC, Caspase-1, IL-1β, IL-6 and TNF-α proteins in the COVID-19 group meaningfully increased in comparison with the Control group in the lung. Determining the participation of inflammasome-derived products was one of the main goals of the Huang et al. (2020), Lucas et al. (2020), and Wen et al. (2020) studies in 2020. The results of their studies demonstrated the presence of inflammasome-derived products and also cell death and these findings are in agreement with the results of the present study and exhibit the participation of inflammasome in the COVID-19 disease. Rodrigues et al. (2020). The results of Simpson et al. (2014). Pulmonary fibrosis is a condition that appeared when the lung tissue becomes scarred which is observed in the COVID-19 disease and this scar tissue can damage the normal lung that causes breathing becomes increasingly difficult (George et al. 2020; Spagnolo et al. 2020). White blood cells also called leukocytes are the cells of the immune system that are involved in protecting the body against infectious organisms and PMNs such as neutrophils, eosinophils, basophils, and mast cells are a subtype of them. PMNs are placed in the microvasculature of the lung to respond to inflammatory stimuli (Russo et al. 2020). Histological findings of our study presented the increasing number of PMNs, macrophages and also pulmonary fibrosis in the lungs of corpses with the cause of death of COVID-19. Histopathological findings of Tian et al. (2020), Barton et al. (2020), Xu et al. (2020), and Yao et al. (2020) studies in 2020 demonstrated the diffuse alveolar damage, intra-alveolar hemorrhage, intra-alveolar neutrophil infiltration and increased stromal cells in pulmonary tissues which are in line with the results of the present study. Kristine et al. in 2020 in a histological study investigated the postmortem lung findings from a 37-year-old man who died of COVID-19 (Konopka et al. 2020). The results of their study exhibited the presence of paucicellular mucus plugs, goblet cell metaplasia and thickening of subepithelial basement membranes in cartilaginous and non-cartilaginous airways. Furin is a transmembrane protein family which is expressed in several organs, like the lungs and preventing the expression of it could be a possible way to prevent COVID-19 infection (AbdelMassih et al. 2020). The NLRP3 inflammasome can be activated by microbial pathogens, like opportunistic bacteria, atypical bacteria and viruses in the lung (Abdul-Sater et al. 2010; Wang et al. 2014). It seems that, in this study, the mechanism that can be mentioned for lung complication is the attachment of the virus to ACE2 and disturbance of host cells which leads to infect them and raise the inflammatory responses and cell death (Mokhtari et al. 2020).
Conclusion
Infiltration of PMNs and tissue fibrosis and the inflammatory cascade created due to the high activity of the NLRP3 inflammasome complex are directly related to the pathophysiology of the coronavirus-infected lung and its side effects. It is suggested that further studies and targeting of the NLRP3 inflammasome in patients with COVID-19 could be helpful to the achievement of more effective treatment for this disease.
Acknowledgements
We would like to thank the Legal Medicine Research Center, Legal Medicine Organization, Tehran, Iran for financial supports (registration and grant number: M/99/1083).
Author’s contribution
M Gh: Methodology, Data curation, Investigation, Software, Data analysis, Writing-original draft preparation, M F: Methodology, Study design, B E: Data curation, Writing-original draft preparation, S A M: Validation, Writing-review and editing, S D M: Investigation, Data interpretation, Validation, A L: Writing-original draft preparation, S J M: Methodology, Data curation, M O H: Data analysis, Gh H: Study design, Conceptualization, Supervision, Data interpretation, Validation, Writing-review and editing, Funding acquisition.
Declarations
Conflict of interest
The authors declare that there are no conflicts of interests.
Footnotes
This manuscript is part of topical contents: Viral Infection and Pathogenesis
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- AbdelMassih AF, Ye J, Kamel A, Mishriky F, Ismail H-A, Ragab HA et al (2020) A multicenter consensus: A role of furin in the endothelial tropism in obese patients with COVID-19 infection.Obesity Medicine. :100281 [DOI] [PMC free article] [PubMed]
- Abdul-Sater AA, Saïd-Sadier N, Padilla EV, Ojcius DM. Chlamydial infection of monocytes stimulates IL-1β secretion through activation of the NLRP3 inflammasome. Microbes Infect. 2010;12(8–9):652–661. doi: 10.1016/j.micinf.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Heesterbeek H, Klinkenberg D, Hollingsworth TD. How will country-based mitigation measures influence the course of the COVID-19 epidemic? The lancet. 2020;395(10228):931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127(1):153–160. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. Covid-19 autopsies, oklahoma, usa. Am J Clin Pathol. 2020;153(6):725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Peltan I, Kumar N, Leither L, Webb BJ, Starr N, et al. Hydroxychloroquine versus Azithromycin for Hospitalized Patients with COVID-19. Results of a Randomized, Active Comparator Trial. Annals of the American Thoracic Society. 2021;18(4):590. doi: 10.1513/AnnalsATS.202008-940OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonsenso D, Raffaelli F, Tamburrini E, Biasucci D, Salvi S, Smargiassi A, et al. Clinical role of lung ultrasound for diagnosis and monitoring of COVID-19 pneumonia in pregnant women. Ultrasound in Obstetrics & Gynecology. 2020;56(1):106–109. doi: 10.1002/uog.22055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron RQ (2021) COVID-19 cytokine storm: targeting the appropriate cytokine.The Lancet Rheumatology. [DOI] [PMC free article] [PubMed]
- de Zoete MR, Palm NW, Zhu S, Flavell RA, Inflammasomes Cold Spring Harb Perspect Biol. 2014;6(12):a016287. doi: 10.1101/cshperspect.a016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallelli L, Zhang L, Wang T, Fu F. Severe Acute Lung Injury Related to COVID-19 Infection: A Review and the Possible Role for Escin. J Clin Pharmacol. 2020;60(7):815–825. doi: 10.1002/jcph.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George PM, Wells AU, Jenkins RG (2020) Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy.The Lancet Respiratory Medicine. [DOI] [PMC free article] [PubMed]
- Gholaminejhad M, Jameie SB, Abdi M, Abolhassani F, Mohammed I, Hassanzadeh G. All-Trans Retinoic Acid–Preconditioned Mesenchymal Stem Cells Improve Motor Function and Alleviate Tissue Damage After Spinal Cord Injury by Inhibition of HMGB1/NF-κB/NLRP3 Pathway Through Autophagy Activation. J Mol Neurosci. 2022;72(5):947–962. doi: 10.1007/s12031-022-01977-0. [DOI] [PubMed] [Google Scholar]
- Gibson-Corley KN, Olivier AK, Meyerholz DK. Principles for valid histopathologic scoring in research. Vet Pathol. 2013;50(6):1007–1015. doi: 10.1177/0300985813485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanzadeh N, Ebrahimi B, Moshkdanian G, Hosseinjani E (2021) Cardiac diseases following COVID-19 in children and adults: A narrative review on mechanisms and medical implications.Iraq Medical Journal. ; 5(3)
- Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125(5):1028–1036. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijaz S, Mohammed I, Gholaminejhad M, Mokhtari T, Akbari M, Hassanzadeh G. Modulating pro-inflammatory cytokines, tissue damage magnitude, and motor deficit in spinal cord injury with subventricular zone-derived extracellular vesicles. J Mol Neurosci. 2020;70(3):458–466. doi: 10.1007/s12031-019-01437-2. [DOI] [PubMed] [Google Scholar]
- Konopka KE, Wilson A, Myers JL. Postmortem lung findings in a patient with asthma and coronavirus disease 2019. Chest. 2020;158(3):e99–e101. doi: 10.1016/j.chest.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JR, Martin MR, Martin JD, Kuhn P, Hicks JB. Modeling the Onset of Symptoms of COVID-19. Front public health. 2020;8:473. doi: 10.3389/fpubh.2020.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ma X. Acute respiratory failure in COVID-19: is it “typical”. ARDS? Crit Care. 2020;24:1–5. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, Wong P, Klein J, Castro TB, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N, Hunter CA (2020) Cytokine storms: understanding COVID-19. Immunity. [DOI] [PMC free article] [PubMed]
- Mohammed I, Ijaz S, Mokhtari T, Gholaminejhad M, Mahdavipour M, Jameie B, et al. Subventricular zone-derived extracellular vesicles promote functional recovery in rat model of spinal cord injury by inhibition of NLRP3 inflammasome complex formation. Metab Brain Dis. 2020;35(5):809–818. doi: 10.1007/s11011-020-00563-w. [DOI] [PubMed] [Google Scholar]
- Mokhtari T, Hassani F, Ghaffari N, Ebrahimi B, Yarahmadi A, Hassanzadeh G (2020) COVID-19 and multiorgan failure: A narrative review on potential mechanisms.Journal of molecular histology. :1–16 [DOI] [PMC free article] [PubMed]
- Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):1–10. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Ye T, Sun P, Gui S, Liang B, Li L, et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TS, de Sá KS, Ishimoto AY, Becerra A, Oliveira S, Almeida L, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2020;218(3):e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TS, de Sá KS, Ishimoto AY, Becerra A, Oliveira S, Almeida L et al (2021) Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients.Journal of Experimental Medicine. ; 218(3) [DOI] [PMC free article] [PubMed]
- Russo V, Bottino R, Carbone A, Rago A, Papa AA, Golino P, et al. Covid-19 and heart: from clinical features to pharmacological implications. J Clin Med. 2020;9(6):1944. doi: 10.3390/jcm9061944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Phipps S, Baines KJ, Oreo KM, Gunawardhana L, Gibson PG. Elevated expression of the NLRP3 inflammasome in neutrophilic asthma. Eur Respir J. 2014;43(4):1067–1076. doi: 10.1183/09031936.00105013. [DOI] [PubMed] [Google Scholar]
- Spagnolo P, Balestro E, Aliberti S, Cocconcelli E, Biondini D, Della Casa G, et al. Pulmonary fibrosis secondary to COVID-19: a call to arms? The Lancet Respiratory Medicine. 2020;8(8):750–752. doi: 10.1016/S2213-2600(20)30222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Xiong Y, Liu H, Niu L, Guo J, Liao M, et al. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020;33(6):1007–1014. doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toldo S, Bussani R, Nuzzi V, Bonaventura A, Mauro AG, Cannatà A, et al. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm Res. 2021;70(1):7–10. doi: 10.1007/s00011-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z, et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol. 2014;15(12):1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell discovery. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y-C, Chen C-S, Chan Y-J. The outbreak of COVID-19: an overview. J Chin Med association. 2020;83(3):217. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet respiratory medicine. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Li T, He Z, Ping Y, Liu H, Yu S, et al. A pathological report of three COVID-19 cases by minimally invasive autopsies. Zhonghua bing li xue za zhi = Chinese. J Pathol. 2020;49:E009–E. doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]