Abstract
Using a genetic screen we have identified two chromosomal genes, cusRS (ylcA ybcZ), from Escherichia coli K-12 that encode a two-component, signal transduction system that is responsive to copper ions. This regulatory system is required for copper-induced expression of pcoE, a plasmid-borne gene from the E. coli copper resistance operon pco. The closest homologs of CusR and CusS are plasmid-borne two-component systems that are also involved in metal responsive gene regulation: PcoR and PcoS from the pco operon of E. coli; CopR and CopS from the cop operon, which provides copper resistance to Pseudomonas syringae; and SilR and SilS from the sil locus, which provides silver ion resistance to Salmonella enterica serovar Typhimurium. The genes cusRS are also required for the copper-dependent expression of at least one chromosomal gene, designated cusC (ylcB), which is allelic to the recently identified virulence gene ibeB in E. coli K1. The cus locus may comprise a copper ion efflux system, because the expression of cusC is induced by high concentrations of copper ions. Furthermore, the translation products of cusC and additional downstream genes are homologous to known metal ion antiporters.
Copper ions present a dual challenge to both eukaryotic and prokaryotic cells in that they are useful but can also be lethal. Copper is required in the active sites of many enzymes, including terminal oxidases, monooxygenases, and dioxygenases, and is required for the transport of electrons in several photosynthetic and respiratory pathways. However, copper ions can catalyze harmful redox reactions resulting in oxidation of lipid membranes, damage to nucleic acids, and generation of free radicals from hydrogen peroxide (17, 19). Therefore, a cell must meet its physiological requirement for copper ions while preventing their deleterious effects. Cellular systems involved in the acquisition, sequestration, intracellular distribution, and efflux of copper must respond to changes in the extracellular bioavailability of this element over a wide and dynamic concentration range. An example of how organisms cope with this dichotomy is the chromosomally encoded bacterial copper homeostasis cop system of Enterococcus hirae, which encodes two independently regulated copper-transporting ATPases: one that apparently imports copper into the cell and another that effluxes copper from the cell (41).
Some microbes are able to colonize environments containing concentrations of copper ions that would overwhelm chromosomally encoded copper metabolic systems. Typically, these organisms contain extrachromosomal loci that confer resistance to copper. The best characterized of these loci have been isolated from gram-negative bacteria colonizing agricultural areas contaminated by the repeated application of copper salts as a feed additive, bactericidal agent, or antifungal agent. Copper-resistant strains of Escherichia coli have been isolated from the discharge of an Australian pig farm where the diet of piglets is supplemented with CuSO4 to increase their growth (35). In these strains copper resistance is conferred by the plasmid-borne pco operon (9, 35). Copper-resistant strains of the pathovar Pseudomonas syringae have been isolated from tomato fields in California where solutions containing CuSO4 were applied as an antifungal agent. In these strains copper resistance is provided by the plasmid-borne cop operon (6). Southern blot hybridization studies and sequence analysis have shown that the pco and cop operons are closely related (8, 10). These systems appear to be geographically widespread because similar systems have been found in copper-resistant strains of Xanthomonas campestris pv. vesicatoria from Florida, Oklahoma, and California (38) and enteric bacteria from the United Kingdom (40).
The pco and cop operons carry four related structural genes, pcoABCD and copABCD (10), which are expressed from the upstream, copper-inducible promoters PpcoA and PcopA, respectively (23, 32). These structural genes are not related to known families of cation transport genes, such as those described for E. hirae (41). The structural genes encode periplasmic and membrane proteins; however, despite their similarity, the pco operon enhances copper efflux (8) while the cop operon may lead to copper sequestration (11). These differences might be the result of the different genetic background of each organism. In neither case is the mechanism understood, although it has been proposed that PcoA is a multicopper oxidase (10, 21). The pco operon also encodes an additional gene, pcoE (8), for which a P. syringae homolog has not been found. This gene is expressed from a separate copper-inducible promoter, PpcoE (32). PcoE, a periplasmic protein, is not strictly required for copper resistance in standard growth assays, but it reduces the time required for E. coli strains to recover from copper ion stress (G. P. Munson, F. W. Outten, and T. V. O'Halloran unpublished results).
Both the pco and cop loci also carry two-component signal transduction systems, encoded by pcoRS or copRS, respectively, which are required for the copper-inducible expression of copper resistance (8, 25). Signal transduction systems of this type are common in many microbial systems and comprise a superfamily of conserved proteins (for a review, see reference (18)). PcoS and CopS are homologous to sensor histidine kinases and are predicted to have two cytoplasmic-membrane-spanning domains with peptide loops extending into the periplasm. As copper levels in the medium increase, these kinases are envisioned to phosphorylate their cognate response regulators, PcoR or CopR, converting them to transcription activators (25, 32). While mutations that disrupt pcoR or pcoS abolish copper resistance (8), copper-dependent expression from PpcoA and PpcoE is not completely lost (32). Furthermore, some strains of P. syringae have been shown to carry chromosomal homologs of copRS by DNA hybridization and in vivo transcription of a copRS-regulated promoter (22). This demonstrates that one or more copper-responsive regulators are encoded in the chromosome of each organism. Using a genetic screen, we have identified two genes on the E. coli chromosome, cusRS (ylcA ybcZ), that encode a copper-responsive two-component system. These genes are required for the copper-inducible expression of pcoE and a chromosomal gene, cusC (ybcZ). The cus locus may maintain intracellular copper levels within a safe range, because CusRS activate expression of cusC as the concentration of copper in the medium exceeds a threshold value and the cus locus encodes proteins homologous to known metal ion antiporters.
MATERIALS AND METHODS
Nitrous acid mutagenesis.
E. coli strain DH5α/pCOIV199-D7 was grown overnight at 37°C in 5-ml cultures of Luria-Bertani (LB) medium (Bacto tryptone, 10 g liter−1; yeast extract, 5 g liter−1; NaCl, 5 g liter−1) with ampicillin (100 μg ml−1) and then exposed to the mutagen nitrous acid as described previously (24). Serial dilutions of nitrous acid-treated cells were plated onto LB agar plates with 1 or 2 mM CuSO4 and incubated overnight at 37°C.
Construction of an E. coli genomic library.
Chromosomal DNA was isolated from wild-type E. coli strain DH5α by the CTAB (hexadecyltrimethylammonium bromide) method as described previously (2). The chromosomal DNA was partially digested with Sau3AI, and DNA fragments of 2 to 4 kb were ligated into the BamHI site of the vector pSX34NoHindIII to construct a genomic library.
Nucleotide sequencing.
Both strands of the cus locus were sequenced by the dideoxy method with a CircumVent Thermal Cycle Dideoxy DNA Sequencing Kit (New England Biolabs). The manufacturer's recommended dideoxy termination solutions were altered by reducing the level of unlabeled dATP by 50% in all solutions to increase the incorporation of 35S-labeled dATP. The complete cus sequence was determined by a combination of primer walking with custom oligonucleotides and primers complementary to cloning vectors.
Southern blots.
Chromosomal DNAs were isolated from E. coli strains by the CTAB method as described previously (2), digested with restriction endonucleases, and separated by electrophoresis on TBE (90 mM Tris-borate, 2 mM EDTA, pH 8.0) agarose gels. DNA was depurinated in 0.25 M HCl, and then the acid was neutralized with 0.4 M NaOH. DNA was transferred to positively charged nylon membranes by capillary action with 0.4 M NaOH as the transfer buffer.
DNA probes were generated by PCR using primer pairs CLA (5′ CTGGTGATTT ATGCCGCCAAC TTTA) and CL20 (5′ GCCCGGGCAA TTCTAGAGTA GCGGG), CLC (5′ GAGGTGCCGG ATGGTCAGTA AGCC) and CL01 (5′ TCATCATCGT CGGGCCGGAA AGGAG), and CLS (5′ GGTAACGTCG GATGCGCGGG G) and CL00 (5′ CGTCCAGCCC GCTGATGAAC ATG), with nucleotide solutions supplemented with [α-32P]dGTP and [α-32P]dATP. Labeled probes were purified on nondenaturing acrylamide gels. Denatured probes were hybridized to Southern blots in hybridization buffer (5× SSC [33], 5× Denhardt Solution [33], 1% sodium dodecyl sulfate, and 100 μg of sheared salmon sperm DNA ml−1) at 65°C overnight, washed twice in 2× SSC–0.1% sodium dodecyl sulfate at 65°C, and then rinsed with 2× SSC at 65°C.
Strains, plasmids, and phages.
Strains, plasmids, and lacZ reporter constructs are described in Table 1 and Fig. 2.
TABLE 1.
Strains, phages, and plasmids
| Strain, phage, or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | Wild-type, F−endA1 hsdR17 supE44 thi-1 recA1 deoR gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15 λ− | New England Biolabs |
| DLA | Cus−, mutant of parent strain DH5α | This study |
| DLB | Cus−, mutant of parent strain DH5α | This study |
| DLG | Cus−, ΔcusRS, mutant of parent strain DH5α | This study |
| DLH | Cus−, mutant of parent strain DH5α | This study |
| DLI | Cus−, mutant of parent strain DH5α | This study |
| DLJ | Cus−, ΔcusRS, mutant of parent strain DH5α | This study |
| DLK | Cus−, ΔcusRS, mutant of parent strain DH5α | This study |
| DLN | Cus−, mutant of parent strain DH5α | This study |
| Phages | ||
| λPpcoE-lacZ | Reporter phage carrying PpcoE −70 to +31a cloned upstream of promoterless lacZ of λRS45 | This study |
| λPpcoA-lacZ | Reporter phage carrying PpcoA −576 to +405a cloned upstream of promoterless lacZ of λRS45 | This study |
| λPcusC-lacZ | Reporter phage carrying PcusC −114 to +12a cloned upstream of promoterless lacZ of λRS45 | This study |
| λRS45 | Reporter phage carrying promoterless lacZ | 33a |
| Plasmids | ||
| pCOVI133 | pcoRS cloned into pSX34lacZα | This study |
| pCOIV199-D7 | pcoE-lacZ gene fusion cloned into pRS414 expressed from PpcoE −348 to +186a | This study |
| pPA87 | pcoABCDRSE cloned into pBR322 | 32 |
| pCL27-1 | cus, 2-kb Sau3Al fragment of E. coli chromosome cloned into BamHI site of pSX34NoHindIII | This study |
| pCL115-1 | cus, 6-kb NsiI-BamHI fragment of E. coli chromosome cloned into PstI-BamHI sites of pSX34LacZα | This study |
| pCOIV239-B1 | pcoABCDRSE, 7.5-kb HindIII-BglII fragment from pPA87 cloned into HindIII-BamHI sites of pSX34LacZα | This study |
| pRS414 | Promoterless lacZ cloning vector | 33a |
| pSX34LacZα | Low-copy-number cloning vector | New England Biolabs |
| pSX34NoHindIII | Low-copy-number cloning vector, derivative of pSX34LacZα without lacZα complementation | This study |
Numbering is relative to the transcription start site.
FIG. 2.
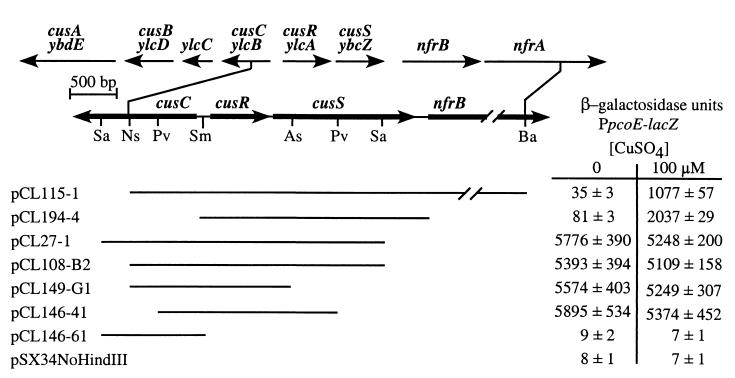
The Cus− phenotype is complemented by cusRS in trans. Open reading frames within and surrounding the cus locus are represented by arrows. Both the proposed cus gene names and the gene designations that were assigned by their positions on the E. coli chromosome are shown. Thin lines represent various DNA fragments of the cus locus carried by the listed plasmids. Restriction endonuclease sites used in the construction of plasmids are shown, except for pCL194-1, which was constructed by cloning of a PCR product. Each plasmid was transformed into E. coli strain DLG/Φ(PpcoE-lacZ), which carries a deletion of cusRS. β-Galactosidase expression was assayed before or 1 h after addition of 100.0 μM CuSO4. Each enzymatic assay was performed in triplicate, and the mean and standard deviation are shown. Abbreviations: Sa, Sau3A1; N, NsiI; P, PvuII; Sm, SmaI; A, AseI; B, BamHI.
Primer extensions.
E. coli strains were grown aerobically to log phase in LB medium at 37°C. Total RNA was isolated with an RNeasy total RNA isolation kit (Qiagen) according to the manufacturer's protocols. E. coli strains induced with copper were exposed to 500 μM CuSO4 for 1 h prior to isolation of RNA. Primer PE3 (5′GGACGCTGAT AATCCGGTGC C), labeled with 32P by T4 polynucleotide kinase, was used with 10 μg of total RNA for primer extension analysis. Primer and RNA were heated at 65°C for 5 min, chilled on ice, and then added to a reaction mixture of Moloney murine leukemia virus reverse transcriptase (New England Biolabs) with nucleotides and incubated at 42°C for 1 h. Sequencing was carried out using labeled primer PE3 as directed in the CircumVent Thermal Cycle Dideoxy DNA Sequencing Kit (New England Biolabs). Primer extension and sequencing reactions were run together on denaturing sequencing gels.
β-Galactosidase assays.
E. coli strains were grown to log phase in A minimal medium [7.6 mM (NH4)2SO4, 33 mM KH2PO4, 60 mM K2HPO4, 1.7 mM Na3C6H5O7 (sodium citrate), 1 mM MgSO4, 0.2% glucose, 5 × 10−5% thiamine] at 37°C with aeration and assayed for β-galactosidase activity as described previously (24) 1 h after metal ion addition. Where appropriate, antibiotics were used at the following concentrations; kanamycin, 20 μg ml−1; chloramphenicol, 20 μg ml−1; and ampicillin 100 μg ml−1.
Nucleotide sequence accession numbers.
The cus nucleotide sequence has been deposited in GenBank under accession number AF245661.
RESULTS
pcoRS are not required for copper-inducible expression of PpcoE.
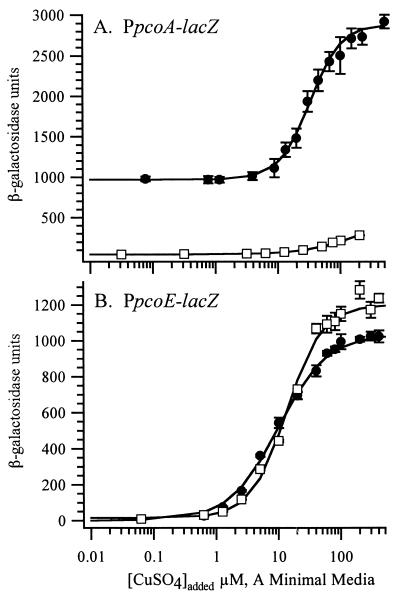
To examine the roles of copper-responsive regulators carried by the plasmid-borne pco operon and the E. coli chromosome, two copper-inducible promoters, PpcoA and PpcoE, were cloned from pco and placed upstream of promoterless lacZ. Reporter constructs were integrated into the chromosome as λ prophages in order to rigorously control for copy number. Basal, not copper-induced, expression of β-galactosidase was less than 50 β-galactosidase units from either promoter in the absence of pco (Fig. 1). When pco was provided in trans, basal expression from PpcoA increased to 900 β-galactosidase units but basal-level expression from PpcoE remained low. Expression from PpcoA increased with increasing concentrations of copper ions in the growth medium, to a maximum of 2,800 β-galactosidase units, and required pco in trans (Fig. 1A). In the absence of pco, copper-inducible expression from PpcoA was less pronounced, increasing from less than 50 β-galactosidase units to no more than 366 β-galactosidase units even at 400 μM CuSO4. Higher concentrations of copper were not assayed because precipitation was observed above 400 μM CuSO4 in A medium at 37°C. Deletion of pcoRS, which encode a two-component system, from the pco operon abolished the effect of pco upon expression from PpcoA (data not shown). This is consistent with a previous study (8) that showed that pcoRS are required for maximum copper-inducible expression from PpcoA.
FIG. 1.
Copper-induced expression from promoter-lacZ fusions in the presence or absence of the copper resistance operon pco. The expression of β-galactosidase from PpcoA-lacZ (A) or PpcoE-lacZ (B) reporter prophage was assayed 1 h after addition of CuSO4 to A minimal growth medium. Circles, E. coli strain DH5α transformed with pCOIV239-B1 carrying the pco operon; squares, E. coli strain DH5α transformed with vector pSX34LacZα. Each data point is the mean of at least three enzymatic assays, with error bars showing the standard deviation of the mean.
In contrast to that from PpcoA, expression from PpcoE was highly induced by the addition of copper ions to the medium, and this induction did not require the pco operon. In both the presence and absence of pco, expression of β-galactosidase from PpcoE increased from less than 16 to over 1,000 β-galactosidase units (Fig. 1B). Copper-inducible expression from PpcoE is highest in the absence of pco and slightly lower in its presence. This demonstrates that a chromosomal factor, or factors, regulates expression of PpcoE either alone or in addition to pco-encoded regulation. We have given this chromosomal regulator(s) the designation Cus for Cu sensing because it detects and mediates a cellular response to increasing concentrations of copper ions. These results contradict those of a previous study that reported that PpcoE was only partially regulated by chromosomal factors (8). Although the reasons for this difference are unclear, it may be the result of the different genetic backgrounds of the strains used or an effect of using plasmid (32) compared to single-copy reporters (this study). In either case it is clear that both PpcoA and PpcoE are activated by Cus in a copper-dependent fashion, although PpcoA is fully activated only when the plasmid-based regulatory system encoded by pcoRS is provided in trans (Fig 1).
Selection of Cus− strains.
Although it is clear from the above results that PpcoA and PpcoE are activated by Cus in the absence of pcoRS, it is not clear whether Cus is required for the activity of PcoRS or whether these systems operate independently. It is also unclear whether PpcoA and PpcoE are regulated by the same or separate chromosomal factors. To address these issues, a selection strategy was devised to isolate Cus− strains so that the chromosomal factor(s) that provides copper-inducible expression to PpcoA and PpcoE could be identified. It was assumed that PpcoE was positively regulated and that disruption of Cus would prevent the copper-inducible expression of a lethal gene product cloned downstream of PpcoE, allowing the survival of Cus− strains on LB agar supplemented with copper ions. Plasmid pCOIV199-D7 carries a gene fusion between pcoE (codons 1 to 20) and lacZ (codons 9 to 1024) whose expression is Cus and copper dependent. Previous studies have shown that fusion of a signal leader sequence like that of PcoE to the amino terminus of β-galactosidase produces a fusion protein that is lethal to E. coli when moderately or highly expressed (3). As expected, parent strain DH5α grew when plated on LB agar supplemented with 1 or 2 mM CuSO4, but strain DH5α/pCOIV199-D7 did not. There was no growth difference between strains DH5α and DH5α/pCOIV199-D7 when plated on LB agar without added CuSO4 because, as shown above, expression from PpcoE is low in the absence of added copper ions.
After exposure to the mutagen nitrous acid, strain DH5α/pCOIV199-D7 was plated onto LB agar supplemented with 1 to 2 mM CuSO4. Colonies that formed after overnight incubation at 37°C were transferred to LB agar with ampicillin to select for the resistance marker of plasmid pCOIV199-D7. This second screen eliminated those strains that survived by loss of the plasmid. Selection for ampicillin resistance was not possible in the presence of copper ions because they catalyze the rapid degradation of ampicillin (5). Ampicillin-resistant strains were then cured of pCOIV199-D7 and infected with a PpcoE-lacZ reporter phage. β-Galactosidase assays were performed on each lysogen with and without inducing levels of CuSO4. Strains that retained copper-inducible β-galactosidase expression were discarded. Presumably these strains had survived the initial selection through mutations that disrupted the lethal gene fusion or PpcoE. With this selection and screening strategy, eight Cus− strains (DLA, DLB, DLG, DLH, DLI, DLJ, DLK, and DLN) were isolated.
Cloning of the cus locus.
The cus locus was isolated from an E. coli plasmid library by screening the library for plasmids that complemented the Cus− phenotype. One plasmid, pCL27-1, that produced a Lac+ phenotype when transformed into Cus− Lac− strain DLG/Φ(PpcoE-lacZ) was obtained. However, expression of β-galactosidase was constitutive, not copper inducible (Fig. 2). The restriction map of the cus locus was determined by using the DNA fragment carried by pCL27-1 as a probe of Southern blots. This facilitated the cloning of a larger, 6-kb NsiI-BamHI DNA fragment that carries the cus locus and restores copper-inducible expression from PpcoE when transformed into DLG/Φ(PpcoE-lacZ) (Fig. 2) and each of the other Cus− strains (data not shown).
A total of 3,538 bp of the cus locus was sequenced (GenBank accession number AF245661) and found to be 100% identical to bases 592305 to 595842 of the E. coli K-12 genome (GenBank accession number AE000162). Sequence analysis revealed that the 6-kb fragment cloned into pCL115-1 carries three complete and two partial open reading frames (Fig. 2). Two of the complete open reading frames encode proteins that are homologous to proteins belonging to the superfamily of two-component signal transduction systems (for a review, see reference (18)). CusR is homologous to phosphate receiver response regulators, and CusS is homologous to sensor histidine kinases. In particular, the closest homologs to CusRS are two-component regulatory systems that are involved in metal-responsive gene regulation. CusR has 83% identity to SilR and 61% identity to both PcoR and CopR. CusS has 56% identity to SilS, 42% identity to CopS, and 38% identity to PcoS. SilRS are carried by a silver-resistant strain of Salmonella enterica serovar Typhimrium that was isolated from a hospital burn ward (15). CopRS are required for the expression of copper resistance genes within the plasmid-borne cop operon of the pathovar P. syringae (25). Similarly, PcoRS are required for the expression of copper resistance genes of the plasmid-borne pco operon in some strains of E. coli (32).
Plasmid subclones were constructed and tested for their ability to restore copper-inducible expression to PpcoE to determine which of the genes carried by pCL115-1 are required to complement the Cus− phenotype. Plasmids carrying cusRS restore copper-inducible expression of β-galactosidase when transformed into Cus− strain DLG/Φ(PpcoE-lacZ) (Fig. 2). In particular, plasmid pCL194-4 carries a DNA fragment with only an additional 143 bp upstream of cusR and 250 bp downstream of cusS, indicating that no other genes are required to complement the Cus− phenotype. Plasmids carrying truncations of cusS (pCL27-1, pCL108-B2, pCL149-G, and pCL146-41) produced constitutive β-galactosidase expression when transformed into strain DLG/Φ(PpcoE-lacZ) (Fig. 2). Constitutive expression required cusR, because it was not observed when strain DLG/Φ(PpcoE-lacZ) was transformed with pCL146-61, a plasmid that carries a truncation of cusR (Fig. 2). In the absence of its cognate histidine kinase, CusR may be gratuitously activated by another histidine kinase, as has been reported for other two-component systems (1, 39). These results show that cusRS, which are deleted in strain DLG (see below), are necessary and sufficient to restore copper-inducible expression from PpcoE when provided in trans.
cusRS are deleted in some Cus− strains.
To determine if the Cus− phenotype is produced by mutations within cusRS, Southern blots of chromosomal DNAs isolated from selected strains were hybridized with probes complementary to cusRS. Nitrous acid, the mutagen used to generate Cus− strains, has been shown to produce large deletions (34) which are amenable to detection by Southern blotting. Radiolabeled probes complementary to the 5′ region of cusR, the 3′ region of cusR and 5′ region of cusS, and the 3′ region of cusS were sequentially hybridized to the same Southern blot (data not shown). In addition, a probe complementary to tonB was used as a control to verify that approximately equivalent amounts of DNA from each strain had been transferred to the blot. The three cus probes did not hybridize to DNAs from Cus− strains DLG, DLJ, and DLK, but each did hybridize to the DNAs from the parent strain DH5α and other Cus− strains. This shows that cusRS are deleted in strains DLG, DLJ, and DLK. In addition, restriction fragments carrying cusR are 2 to 3 kb larger in strains DLH and DLI than in the parent strain DH5α, indicating that an undefined mutation has occurred within or upstream of cusR in these strains. Mutations were not apparent in cusRS from the other three Cus− strains; however, these strains may carry other types of nitrous acid-generated mutations not detectable by this type of analysis, such as base conversions (13). Nevertheless, cusRS are deleted or appear to be altered in five of eight Cus− strains, and the Cus− phenotype of all eight strains is complemented when cusRS are provided in trans (data not shown). This suggests that at least in some strains the Cus− phenotype is produced by mutations within cusRS.
Identification of a chromosomal promoter regulated by cusRS.
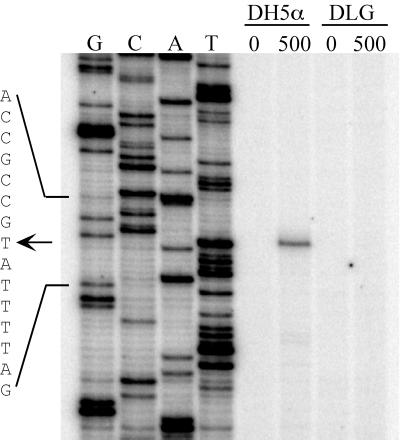
Sequence analysis revealed a divergently encoded open reading frame that begins 157 bp upstream of cusRS (Fig. 2), which, as discussed below, we designated cusC. Because prokaryotic regulators sometimes regulate the expression of nearby genes, we sought to determine if expression of cusC was inducible by copper ions. RNAs were isolated from strains DH5α and DLG grown in medium with and without added copper ions and used in primer extension assays (Fig. 3). A single transcription start site 26 nucleotides upstream of cusC was observed only with RNA from strain DH5α grown in copper-containing medium. A transcript was not observed with RNA isolated from DH5α grown without copper ions, nor was it observed with RNA from strain DLG. This shows that expression of cusC is induced by copper ions and, as shown below, is dependent upon cusRS.
FIG. 3.
Transcription start site mapping of copper-dependent cusC mRNA. Primer extension products of total RNA isolated from wild-type E. coli strain DH5α or ΔcusRS strain DLG grown with or without 500.0 μM CuSO4 added to the growth medium are shown. Lanes G, C, A, and T, products of dideoxy sequencing reactions using the same primer as for primer extension reactions. The sequence shown is that of the noncoding strand of cusC. The arrow indicates the position of the 5′ end of cusC mRNA.
pcoRS and cusRS are independent regulatory systems.
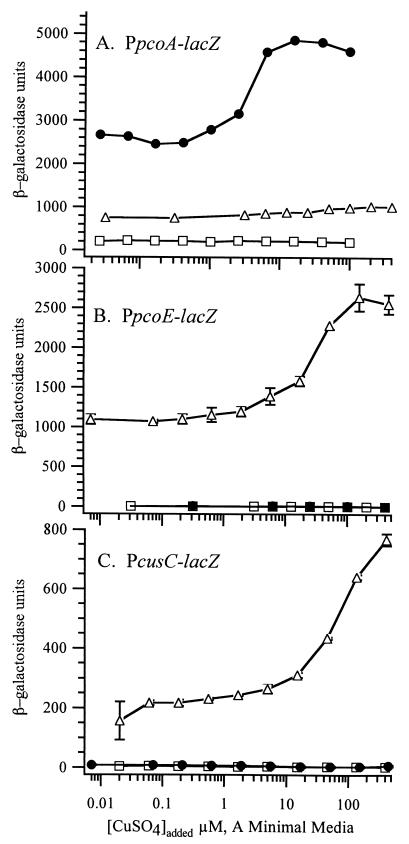
To determine whether CusRS regulate the same promoters as PcoRS or separate promoters, the genes encoding each two-component system were provided in trans in ΔcusRS strain DLG lysogenized with PpcoE-lacZ, PpcoA-lacZ, or PcusC-lacZ reporter prophage. In a ΔcusRS strain pcoRS were able to provide copper-inducible expression of β-galactosidase from PpcoA but not from PpcoE or PcusC (Fig. 4). In contrast, cusRS provided copper-inducible expression to PpcoE and PcusC and increased the basal-level expression of PpcoA (Fig. 4). These results show that PcoRS regulate PpcoA but not PpcoE and PcusC. The promoters PpcoE and PcusC are regulated by CusRS, which also provide a low level of activation to PpcoA. Both regulatory systems also function in the absence of the other, indicating that the two systems are able to operate independently.
FIG. 4.
Complementation of ΔcusRS by cusRS or pcoRS. The expression of β-galactosidase from PpcoA-lacZ (A), PpcoE-lacZ (B), or PcusC-lacZ (C) reporter prophage was assayed after addition of CuSO4 to the growth medium of E. coli strain DLG (ΔcusRS) transformed with plasmids carrying pcoRS (●), cusRS (▵), or a vector control (□). Each point is the mean of at least three enzymatic assays, with error bars showing the standard deviation of the mean.
DISCUSSION
cusRS and pcoRS encode independent, copper-responsive regulatory systems.
In this study we have identified two genes, cusRS, on the chromosome of E. coli K-12 that encode a regulatory system which activates the expression of genes in response to increasing levels of copper ions. Based on homology to characterized systems (31), cusRS appear to encode a two-component signal transduction system with CusS as the membrane-bound histidine kinase and CusR as the cytoplasmic response regulator. These genes are required for the copper-inducible expression of cusC, a chromosomal gene, and pcoE, a gene from the plasmid-borne copper resistance operon pco. Deletion of cusRS abolished copper-inducible expression of the cusC promoter and the plasmid-derived promoter PpcoE. Copper-inducible expression was restored at both of these promoters by providing cusRS in trans but not by providing pcoRS, a two-component system from the pco operon that, by homology, is closely related to cusRS. The cusRS genes also provided a low level of copper-inducible expression to the plasmid-derived promoter PpcoA, but full expression from PpcoA was observed only when pcoRS were provided in trans. Thus, although cusRS and pcoRS encode homologous copper-responsive regulatory systems, they cannot substitute for one another. These results demonstrate that expression of pcoE is dependent upon a chromosomal regulatory system, while other copper resistance genes which are expressed from PpcoA are regulated by pcoRS. Although these two regulatory systems both respond to copper ions, they may have different sensitivities or induction profiles. If so, this may allow the cell to finely tune its response to copper ions.
Copper- and silver-induced promoters are preceded by a copper box.
A conserved palindrome is present upstream of several copper-responsive promoters, including the newly identified PcusC (Fig. 5). This conserved sequence has previously been identified as a copper box, a DNA sequence required for regulation of both PpcoA and PpcoE (32). Removal of the copper box abolishes copper-inducible expression of PpcoA and PpcoE (32). In vitro DNase I footprinting has shown that CopR from P. syringae, a CusR homolog, binds to the copper box upstream of PcopA (26). Thus, the results of transcription studies (32), the homology of CusR to CopR, and our finding that PcusC and PpcoE are CusR-dependent promoters suggest that the copper boxes upstream of these promoters are the binding sites for CusR. Also of note, the copper box is upstream of promoters that are regulated by SilRS, close CusRS homologs from the S. enterica serovar. Typhimurium silver resistance operon, sil (Fig. 5). Given similarities between Cu(I) and Ag(I) coordination chemistry, it is perhaps not surprising to find parallels between the regulation of copper and silver loci.
FIG. 5.
Copper- and silver-inducible promoters are preceded by the palindromic copper box. The upstream regions of E. coli promoters PpcoE, PcusC, and PpcoA; P. syringae promoters PcopA and PcopH; and S. enterica serovar Typhimirium promoters PsilE and PsilS have been aligned to highlight the identity between the palindromic copper boxes, indicated by thick arrows, that are found upstream of these copper- or silver-inducible promoters. The two-component regulatory system that is required for the metal ion-dependent activation of each promoter is shown to the right of the nucleotide sequences. Predicted −35 and −10 hexamers are shown by single and double underlines, respectively. Transcription start sites that have been determined are shown by boldface underlines and by a thin arrow for the transcription start site of PpcoE.
The cus locus may encode a copper ion antiporter.
Analysis of the recently released complete nucleotide sequence of the E. coli K-12 chromosome (7) suggests that the cus locus encodes a detoxification system for copper ions. Immediately downstream of cusC are two large open reading frames, cusBA (ylcD ybdE) (Fig. 2). These genes may encode additional components of a copper ion antiporter. The naming of these genes with the cus mnemonic is suggested by their putative function and homology to other metal resistance systems (discussed below). CusB is predicted to be an integral membrane protein with one transmembrane domain. CusA is predicted to be a 115-kDa integral membrane protein with 12 transmembrane domains and may form a two-channel pump for the export of metal ions and concurrent import of protons, as has been shown for its homolog CzcA (14). The organization and translation products of these three genes are similar to those of other operons that are known or thought to encode metal ion efflux systems. This includes the S. enterica serovar Typhimurium sil locus, which provides resistance to silver ions (15), and two loci from Ralstonia sp. strain CH34: the czc locus, which provides resistance to cobalt, zinc, and cadmium ions (28), and the cnr locus, which confers resistance to cobalt and nickel ions (29). Expression of the genes within these systems is induced by the metal ions for which that they provide resistance (15, 27, 36). The homology of the cus locus to these other systems and our finding that cusC is induced by copper ions suggest that the cus locus encodes a copper efflux system. However, this can be definitively concluded only through biochemical characterization of this locus.
Like its homologs in the sil, czc, and cnr systems, CusC may be associated with the outer membrane. It may even be an outer membrane lipoprotein, because its amino terminus has both a signal export sequence and a cysteine at position 18. This cysteine may be the site for the thiol-ether linkage to diacylglyceride and the linkage to a monoacyl group. If CusC is a lipoprotein, the final steps of its posttranslational processing would require an apoliprotein N-acyltransferase (37). In fact, it has previously been shown that E. coli strains carrying mutations within a gene, cutE, encoding an apolipoprotein N-acyltransferase (16) are sensitive to and accumulate copper ions (30). It is possible that the copper-sensitive phenotype of cutE strains results from the inability of these strains to process CusC. This would be consistent with the prediction that CusC is an outer membrane lipoprotein and a component of a copper efflux system. In contradiction to this scenario, the Cus− strains that we have isolated do not have a copper-sensitive phenotype. However, in this study cus mutations were selected for by plating E. coli strains on medium supplemented with CuSO4. Therefore, it seems possible that this selection strategy also selected for suppressors of a copper-sensitive phenotype. To resolve these uncertainties, future studies will utilize isogenic mutations within the cus locus to characterize the functions of the proteins that it encodes. The subcellular locations of these proteins will be determined by biochemical or immunological methods.
Copper homeostasis and virulence.
A gene allelic to cusC has recently been identified as a virulence gene required for the invasion and pathogenicity of E. coli K1 in a bacterial meninigitis model (20). This gene, ibeB, encodes a protein that is 97% identical to CusC. Disruption of ibeB reduced the ability of E. coli to invade brain microvascular endothelial cells in vitro and the central nervous systems of infant rats in an in vivo model (20). Given that expression of cusC is induced by copper ions and that cusC is within a locus that is homologous to other metal ion efflux systems, it is plausible that copper efflux is critical for virulence in some pathogenic strains of E. coli. Studies have also shown that mutations within a copper-transporting P-type ATPase reduce the virulence of Listeria monocytogenes (12) and that expression of the copper-containing periplasmic enzyme Cu,Zn-superoxide dismutase enhances intracellular survival of E. coli (4). Little is otherwise known about the involvement of copper homeostasis systems in pathogenicity. Like iron, copper may be a resource that the host and invading bacterium compete for. For instance, copper is known to stimulate vascularization (21). Alternatively, copper efflux might afford the microorganism with some defense against host responses such as resistance to reactive oxygen species generated by macrophages. A better understanding of copper transport and metabolism will provide insight into the relationship between copper and pathogenicity, and this may in turn provide new therapeutic targets against bacterial pathogens.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant GM54111 (to T.V.O) and Training Grant GM0806l (to G.P.M. and F.W.O.).
We thank B. Lee and Jim Camakaris for plasmids pRJ1004 and pPA87 carrying the pco operon and for many helpful discussions, R. W. Simons for providing promoterless lacZ reporter vectors, D. Ralston Horvath and J. Bryson for helpful discussions, and New England Biolabs for providing plasmid pSX34LacZα.
ADDENDUM IN PROOF
An unrelated chromosomal copper resistance locus in Escherichia coli has been recently described that encodes CopA, a P-type ATPase cation efflux pump. C. Rensing, B. Fan, R. Sharma, B. Mitra, and B. P. Rosen, Proc. Natl. Acad. Sci. USA 97:652–665, 2000). Expression of this copper efflux pump is not regulated by the CusRS system, but by a MerR-like metalloregulatory protein. (F. W. Outten, C. E. Outten, J. Hale, and T. V. O'Halloran, J. Biol. Chem., in press). Expression of CopA in the Cus− background may contribute to the absence of a copper-sensitive phenotype in Cus− strains.
REFERENCES
- 1.Amemura M, Makino K, Shinagawa H, Nakata A. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J Bacteriol. 1990;172:6300–6307. doi: 10.1128/jb.172.11.6300-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Bassford P J, Jr, Silhavy T J, Beckwith J R. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J Bacteriol. 1979;139:19–31. doi: 10.1128/jb.139.1.19-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battistoni A, Pacello F, Folcarelli S, Ajello M, Donnarumma G, Greco R, Ammendolia M G, Le Touati D, Rotilio G, Valenti P. Increased expression of periplasmic Cu,Zn superoxide dismutase enhances survival of Escherichia coli invasive strains within nonphagocytic cells. Infect Immun. 2000;68:30–37. doi: 10.1128/iai.68.1.30-37.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beard S J, Ciccognani D T, Hughes M N, Poole R K. Metal ion-catalysed hydrolysis of ampicillin in microbiological growth media. FEMS Microbiol Lett. 1992;75:207–211. doi: 10.1016/0378-1097(92)90405-d. [DOI] [PubMed] [Google Scholar]
- 6.Bender C L, Cooksey D A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987;169:470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Brown N L, Barrett S R, Camakaris J, Lee B T, Rouch D A. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol Microbiol. 1995;17:1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown N L, Rouch D A, Lee B T. Copper resistance determinants in bacteria. Plasmid. 1992;27:41–51. doi: 10.1016/0147-619x(92)90005-u. [DOI] [PubMed] [Google Scholar]
- 10.Bryson J W, O'Halloran T V, Rouch D A, Brown N L, Camakaris J, Lee B T O. Chemical and genetic studies of copper resistance in E. coli. In: Karlin K D, Tyeklar Z, editors. Bioinorganic chemistry of copper. New York, N.Y: Chapman & Hall; 1993. pp. 101–109. [Google Scholar]
- 11.Cha J S, Cooksey D A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis M S, Thomas C J. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb Pathol. 1997;22:67–78. doi: 10.1006/mpat.1996.0092. [DOI] [PubMed] [Google Scholar]
- 13.Frankel A D, Duncan B K, Hartman P E. Nitrous acid damage to duplex deoxyribonucleic acid: distinction between deamination of cytosine residues and a novel mutational lesion. J Bacteriol. 1980;142:335–338. doi: 10.1128/jb.142.1.335-338.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg M, Pribyl T, Juhnke S, Nies D H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem. 1999;274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 15.Gupta A, Matsui K, Lo J F, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S D, Gan K, Schmid M B, Wu H C. Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J Biol Chem. 1993;268:16551–16556. [PubMed] [Google Scholar]
- 17.Halliwell B, Gutteridge J M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 19.Hoshino N, Kimura T, Yamaji A, Ando T. Damage to the cytoplasmic membrane of Escherichia coli by catechin-copper (II) complexes. Free Radic Biol Med. 1999;27:1245–1250. doi: 10.1016/s0891-5849(99)00157-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang S H, Chen Y H, Fu Q, Stins M, Wang Y, Wass C, Kim K S. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67:2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan J, O'Halloran T V. Iron metabolism in eukaryotes: Mars and Venus at it again. Science. 1996;271:1510–1512. doi: 10.1126/science.271.5255.1510. [DOI] [PubMed] [Google Scholar]
- 22.Lim C K, Cooksey D A. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;175:4492–4498. doi: 10.1128/jb.175.14.4492-4498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellano M A, Cooksey D A. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988;170:4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Mills S D, Jasalavich C A, Cooksey D A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills S D, Lim C K, Cooksey D A. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet. 1994;244:341–351. doi: 10.1007/BF00286685. [DOI] [PubMed] [Google Scholar]
- 27.Nies D H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nies D H, Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:896–900. doi: 10.1128/jb.171.2.896-900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers S D, Bhave M R, Mercer J F, Camakaris J, Lee B T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991;173:6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronson C W, Nixon B T, Ausubel F M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987;49:579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- 32.Rouch D A, Brown N L. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology. 1997;143:1191–1202. doi: 10.1099/00221287-143-4-1191. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33a.Simons R W, Houman F, Klecknen N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 34.Tessman I. The induction of large deletions by nitrous acid. J Mol Biol. 1962;5:442. doi: 10.1016/s0022-2836(62)80033-3. [DOI] [PubMed] [Google Scholar]
- 35.Tetaz T J, Luke R K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983;154:1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tibazarwa C, Wuertz S, Mergeay M, Wyns L, van der Lelie D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J Bacteriol. 2000;182:1399–1309. doi: 10.1128/jb.182.5.1399-1409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokunaga M, Tokunaga H, Wu H C. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA. 1982;79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voloudakis A E, Bender C L, Cooksey D A. Similarity between copper resistance genes from Xanthomonas campestris and Pseudomonas syringae. Appl Environ Microbiol. 1993;59:1627–1634. doi: 10.1128/aem.59.5.1627-1634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams J R, Morgan A G, Rouch D A, Brown N L, Lee B T. Copper-resistant enteric bacteria from United Kingdom and Australian piggeries. Appl Environ Microbiol. 1993;59:2531–2537. doi: 10.1128/aem.59.8.2531-2537.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wunderli-Ye H, Solioz M. Copper homeostasis in Enterococcus hirae. Adv Exp Med Biol. 1999;448:255–264. doi: 10.1007/978-1-4615-4859-1_23. [DOI] [PubMed] [Google Scholar]