Abstract
The ssyF29 mutation, originally selected as an extragenic suppressor of a protein export defect, has been mapped within the rpsA gene encoding ribosomal protein S1. Here, we examine the nature of this mutation and its effect on translation. Sequencing of the rpsA gene from the ssyF mutant has revealed that, due to an IS10R insertion, its product lacks the last 92 residues of the wild-type S1 protein corresponding to one of the four homologous repeats of the RNA-binding domain. To investigate how this truncation affects translation, we have created two series of Escherichia coli strains (rpsA+ and ssyF) bearing various translation initiation regions (TIRs) fused to the chromosomal lacZ gene. Using a β-galactosidase assay, we show that none of these TIRs differ in activity between ssyF and rpsA+ cells, except for the rpsA TIR: the latter is stimulated threefold in ssyF cells, provided it retains at least ca. 90 nucleotides upstream of the start codon. Similarly, the activity of this TIR can be severely repressed in trans by excess S1, again provided it retains the same minimal upstream sequence. Thus, the ssyF stimulation requires the presence of the rpsA translational autogenous operator. As an interpretation, we propose that the ssyF mutation relieves the residual repression caused by normal supply of S1 (i.e., that it impairs autogenous control). Thus, the C-terminal repeat of the S1 RNA-binding domain appears to be required for autoregulation, but not for overall mRNA recognition.
Protein translocation in Escherichia coli is catalyzed by a preprotein translocase comprising SecA and a SecY-SecE-SecG complex (7, 23). Mutations in the sec genes cause defects in protein export and hence accumulation of precursors of periplasmic and outer membrane proteins within the cell. Studies of extragenic suppressors have revealed a close functional connection between protein export and other cell processes, in particular protein synthesis (16, 30, 31, 37, 38). Thus, two of the extragenic suppressors (called ssy) of the secY24(Ts) mutation impairing preprotein translocation were mapped within genes normally involved in initiation of translation: i.e., infB encoding initiation factor 2 (ssyG) and rpsA encoding ribosomal protein S1 (ssyF) (30, 31). However, the mechanism whereby protein export might be modulated by essential components of the translational apparatus remains obscure. Here, we have characterized the structural and functional changes caused by the ssyF29 mutation in ribosomal protein S1.
Protein S1 is an essential component of the protein synthesis machinery of E. coli and other gram-negative bacteria (25, 34, 35). It plays two well documented roles in translation. First, it is indispensable for efficient recognition and binding of the majority of bacterial and phage mRNAs by the 30S ribosomal subunit during the initiation process (25, 35). In some cases, the S1-mRNA interactions at this stage were shown to involve preferentially single-stranded AU- or U-rich regions which are frequently found within 5′-untranslated mRNA leaders (2, 3, 39, 45). Second, protein S1, like several other ribosomal proteins, down-regulates its own translation (33, 44). However, the mechanism of this autogenous repression remains a puzzle. Other ribosomal proteins that act as translational repressors bind to the ribosome via specific rRNA motifs, and it is believed that they repress translation by also binding to specific motifs on their mRNAs; moreover, frequently, their rRNA and mRNA targets are obviously structurally related (44). In contrast, S1 is attached to ribosomes by means of protein-protein interactions (4), and it uses its RNA-binding ability for binding to various mRNAs without strict sequence specificity (35). Yet, S1 must somehow recognize its own mRNA among all others to act as an autogenous repressor.
Besides these activities, S1 was shown to play a variety of roles during phage infections: it is one of the four integral subunits of the replicases of RNA bacteriophages (reviewed in reference 40), it stimulates the highly specific T4 endoribonuclease RegB (26), and it forms a complex with phage λ β-protein which is involved in recombination (20, 41). This list is not necessarily exhaustive, and this multifunctional protein may play still unknown roles not only in phage-infected cells, but also in uninfected cells. Thus, S1 has been reported to bind specifically to BoxA, the transcriptional RNA antiterminator of the E. coli rRNA operons (19); moreover, according to a recent hypothesis, it might mediate the function of poly(A) tails in mRNAs (14). Therefore, the mechanism whereby the ssyF29 mutation suppresses the secY24(Ts) defect may reflect a change in either translation initiation efficiency or some other function of S1.
The nature of the suppressor ssyF29 mutation has not been characterized. Since this mutation was not revertible and resulted in synthesis of a protein with a reduced apparent molecular weight (about 52,000) compared with that of the wild-type protein (61,000), Shiba et al. supposed that the ssyF mutation was a deletion (31). In this work, we have structurally characterized this mutation and studied how it affects the main activities of protein S1 in translation. We have found an IS10R element insertion in the 3′ region of the mutant rpsA gene interrupting translation and causing synthesis of a truncated S1 lacking 92 C-terminal amino acid residues; hence, the ssyF mutation can be designated as rpsA::IS10R. The central and C-terminal parts of S1 are known to form its RNA-binding domain, which consists of four highly homologous repeats of the so-called S1 motif (5, 35, 36). We show here that despite the loss of the last S1 motif (R4), the protein remains active in vivo for promoting initiation of protein synthesis on natural translation initiation regions (TIRs), whether or not they bear putative S1-binding sites upstream of their Shine-Dalgarno (SD) sequence. In contrast, the truncated S1 appears unable to function as a translational autorepressor within the mutant cell.
MATERIALS AND METHODS
Conventions and abbreviations.
Throughout this work, gene sequences are numbered from the corresponding translation start points, with the first base of the initiation codon being noted as +1. The term “translation initiation region” (TIR) was initially used to designate all mRNA sequence or structure features contributing to the efficiency of translation initiation, whereas the ribosome binding site (RBS) is the RNA region extending from ca. −20 to +15, which is protected from nucleases by the 30S subunit within the initiation complex (17). Here, we use the term TIR to designate not only an mRNA region, but also the corresponding DNA sequence. Exogenous DNA fragments used in this work for driving lacZ translation generally extend beyond the limits of the RBS, and the 5′ extensions contribute much to translation efficiency; hence, these fragments are operationally called here “TIRs.” The SD sequence is a continuous nucleotide stretch complementary to the 3′ end of 16S rRNA (…ACCUCCUUA3′) and located upstream from the start codon. The 5′ untranslated mRNA region (5′ UTR) is also referred to as the mRNA leader.
Bacterial strains and plasmids.
The strains and plasmids used in this work are listed in Table 1. A general technique for replacing a small region of the E. coli chromosome encompassing the lacZ RBS with in-phase DNA fragments harboring TIRs from other genes has been described earlier (8, 11, 43) (Fig. 1). Briefly, strain ENS0 (formerly called HfrG6Δlac12) carries a short chromosomal deletion encompassing the lac promoter, lac operator, and lacZ RBS (nucleotides [nt] −52 to +44), and pEMBLΔ46 is a pEMBL8+ derivative in which a smaller region (−15 to +23) has been replaced by multiple cloning sites. TIRs are inserted in phase with the lac sequence of pEMBLΔ46 and then transferred onto the chromosome of ENS0 by homologous recombination, selecting for a Lac+ phenotype (Fig. 1).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| CAG18478 | MG1655 zbj-1230::Tn10 | 32 |
| IQ646 | ssyF29 ara D139 Δ(argF-lac)U169 rpsL150 relA1 flbB301 deoC1 ptsF25 rbsR | 31 |
| ENS0 | HfrG6lacZΔ12 (HfrG6 lacking lac promoter-RBS) | 8 |
| ENS0-xTIR | HfrG6lacZ::TIR (Lac+ ENS0 derivatives carrying TIR of gene x fused to lacZ | This work |
| Plasmids | ||
| pEMBLΔ46 | Ampr, pEMBL8+ derivative lacking lacZ RBS | 8 |
| pACYC184 | Tetr Cmr; shortly named here pCtr (control) | 6 |
| pJS200 | Cmr, pACYC184 derivative bearing rpsA gene and 252 nt of 5′ flanking sequence | 29 |
| pSP261 | Cmr, pACYC184 derivative bearing cmk-rpsA-hip genes; named here as pS1 | 22 |
FIG. 1.
Construction of E. coli strains in which TIRs originating from various genes are used to drive translation of the chromosomal lacZ gene. The DNA fragment carrying the TIR of interest (solid box) is first cloned in phase with the α-peptide gene (′lacZ′) of pEMBLΔ46, a pEMBL8+ derivative carrying a small deletion encompassing the lacZ RBS. The TIR is then transferred onto the chromosome of ENS0 by homologous recombination between lac sequences (lacI and lacZ) present on both the plasmid and chromosome. The chromosome of ENS0 (Lac−) carries a slightly larger lac deletion than the plasmid, encompassing the lac promoter (Plac) and operator (op).
The ssyF29 mutation was P1 transduced (18) into the ENS0 derivatives described above in two steps. First, by using CAG18478 (32) as the donor strain and selecting for Tetr, Tn10 was introduced near the rpsA gene of IQ646, which bears the ssyF mutation (31) (Table 1). The mutation was then P1 transduced from the resulting strain into the above-described ENS0 derivatives by selecting again for Tetr. Both steps actually yielded a mixture of rpsA+ and ssyF transductants, but the latter were easily identified by their slow growth on agar medium. Moreover, the growth of ssyF cells was restored by introducing plasmid pSP261 (kindly provided by S. Pedersen), a derivative of pACYC184 (6) carrying the rpsA gene under the control of its own promoter system (22).
Preparation of fragments bearing TIRs from individual genes. (i) rplL.
A PCR-generated fragment encompassing the TIR (nt −84 to +87) of the rplL gene coding for the ribosomal protein L7/12 was originally cloned into the pSP73 vector (Promega Biotec) for in vitro studies (2). Here the same fragment was in phase cloned into the HincII site of pEMBLΔ46 to create pEL784. To generate 5′ truncations of this TIR, pEL784 was treated with BamHI, then Bal31 exonuclease, and finally HindIII. The resulting fragments differing in their 5′ ends were then recloned in pEMBLΔ46/HincII, HindIII and transferred onto the chromosome of ENS0 (Fig. 1). The shortest rplL leader obtained by this method comprises 24 nt (see Fig. 3).
FIG. 3.
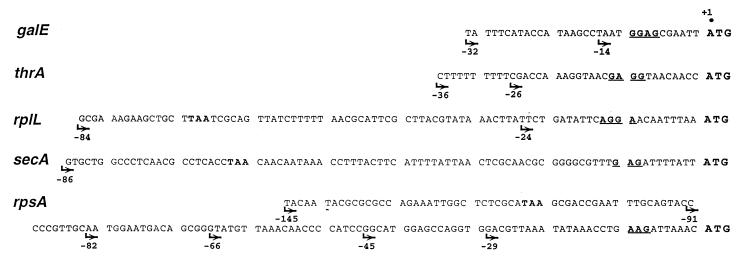
Sequence of DNA fragments used as TIRs in this study. The name of the gene from which each fragment originated is indicated on the left (boldface, italic). Only sequences located upstream of the initiation codon (ATG) are shown in each case. SD sequences are underlined. Arrows indicate the exact 5′ boundaries of the different fragments that have actually been used as TIRs. Note that for rplL, secA, and rpsA, the longest fragments used retain the stop codon from the preceding gene (TAA in boldface).
(ii) rpsA.
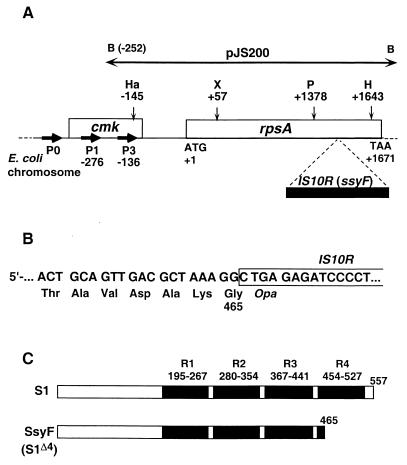
Plasmid pJS200 (29) (gift of J. Schnier) was used as a source of the rpsA sequence. The partial restriction map (28) and transcriptional organization of the rpsA operon (22) are illustrated in Fig. 2A. An HaeII-XmaI fragment encompassing the sequence from −145 to +57 was made blunt-ended with mung bean nuclease and in phase cloned into the HincII site of pEMBLΔ46, generating plasmid pES1145. Derivatives of this plasmid carrying 5′-truncated rpsA leaders were obtained as described for the rplL TIR and named pES191, pES182, pES166, pES145, and pES129: the last two numbers designate the upstream boundary of the TIR in each case (i.e., −91, −82, etc.) (Fig. 3). These TIRs were then transferred onto the chromosome of ENS0 as described. In the resulting strains, the transcription of the rpsA′-′lacZ fusions is driven by the lac promoter (Fig. 1). To create a fusion retaining a genuine rpsA promoter, we inserted an rpsA fragment extending from −252 to +57 (BamHI-XmaI fragment of pJS200) (Fig. 2A) into pEMBLΔ46 and then onto the E. coli chromosome. The resulting fusion carries the strong rpsA P3 promoter (“−35” from −169 to −163, “−10” from −145 to −140) (22) downstream of the lac promoter-operator sequence.
FIG. 2.
(A) General representation of the E. coli chromosome region encompassing genes cmk and rpsA, which encode cytidine monophosphate kinase and ribosomal protein S1, respectively (open boxes). The initiation (ATG) and termination (TAA) codons of rpsA are also indicated. The solid box indicates an IS10R insertion which has been found within the ssyF allele of rpsA (see text). B, Ha, X, P, and H designate restriction sites for the enzymes BamHI, HaeII, XmaI, PstI, and HindIII, respectively. The main promoters responsible for rpsA transcription are noted as P0, P1, and P3 (numbers below P1 and P3 refer to the positions of the corresponding transcription start points) (22). The horizontal, double-arrowed bar shows the rpsA region carried by plasmid pJS200 (29). (B) Sequence of the rpsA-IS10R junction in the ssyF allele showing premature interruption of translation within the inserted sequence. (C) General structure of the 557-residue-long S1 protein (S1) and of the truncated polypeptide encoded by the ssyF allele (SsyF, renamed here S1Δ4). Solid boxes indicate the four S1 motifs (R1 to R4), and numbers indicate the positions of the corresponding amino acids according to Subramanian (35).
(iii) thrA.
A fragment carrying the thrA TIR (−36 to +38) had been obtained previously (8); it contains a stretch of 9 T residues at the 5′ end (Fig. 3) corresponding to oligo(U) sequences in the 5′ UTR of the mRNA. Oligo(U) sequences within mRNA leaders have been proposed to serve as S1 binding sites (3, 45), hence the interest in testing the role of this stretch upon the activity of the thrA TIR. To this end, the pEMBLΔ46 derivative carrying this TIR was treated with TaqI, which cleaves immediately downstream of the oligo(T) stretch, and then made blunt-ended and finally digested with PstI. The truncated TIR was then recloned between the BamHI site (blunt-ended) and the PstI site of pEMBLΔ46.
(iv) galE.
Two galE TIR variants used here (Fig. 3) correspond to nt −32 to +26 and −14 to +26, respectively. Both fragments were obtained as described previously (8).
(v) secA.
A fragment comprising the TIR of the secA gene (nt −97 to +58) was generated from the E. coli chromosomal DNA by PCR with a couple of primers bearing BamHI and HindIII sites convenient for in-phase cloning into pEMBLΔ46.
(vi) Growth of cells and β-galactosidase assays.
Cell growth and β-galactosidase assays were essentially performed as described previously (43). Briefly, cells were harvested in the exponential phase (A600, 0.3 to 0.6) after at least four generations of balanced growth in glycerol-MOPS (morpholinepropanesulfonic acid)-rich medium (21, 43) supplemented with chloramphenicol (34 μg/ml) and, unless otherwise indicated, IPTG (0.2 mM) for lac operon induction. All β-galactosidase activities, measured in sonicated cell extracts, are expressed in nanomoles of o-nitrophenyl-β-d-galactopyranoside (ONPG) hydrolyzed per minute per milligram of total soluble cell proteins.
RNA analysis.
Total RNA from the same cultures used for β-galactosidase assays was isolated and analyzed on Northern blots essentially as described previously (43). As a probe for the rpsA mRNA, we used an equimolar mixture of SmaI-PstI and PstI-PstI fragments encompassing the region +60 to +1244 of the rpsA gene. This region is not present in the rpsA-lacZ mRNA, but it is common to both the wild-type and ssyF alleles of the rpsA gene. The fragments were uniformly 32P labeled with a BRL Multiprime kit. The 23S rRNA was probed with 5′-32P-AAGGTTAAGCCTCACGGTTC, an oligonucleotide complementary to its 3′ region. The membranes were hybridized successively with the rpsA and 23S RNA probes, analyzed with the Fuji BAS 1000 imager to quantify the results, and autoradiographed.
Structural analysis of the rpsA gene from the ssyF29 mutant.
DNA fragments containing the mutant or wild-type rpsA genes were obtained by PCR from chromosomal DNA of the corresponding strains. Forward (−66 to −45, 5′ GTATGTTAAACACCCCATCCG) and reverse (1738 to 1717, 5′ ACGAAACCTGCAATCTGTCAAG) primers from the 5′ and 3′ UTRs of the rpsA gene were designed according to sequences given in references 22 and 28, respectively. The PCR products were compared by restriction analysis and then sequenced to localize the ssyF mutation (see Results).
RESULTS
Nature of the ssyF29 mutation.
The ssyF29 mutation was initially assumed to be a deletion (31). To localize this mutation within the rpsA gene, we have compared the length of PCR fragments amplified from the rpsA chromosomal region of wild-type and ssyF cells by using primers corresponding to the 5′ and 3′ UTRs of the gene. This analysis revealed an insertion of about 1.3 kb within the ssyF allele (not shown). Restriction analysis showed that the insertion was located within a PstI-HindIII fragment (1378 to 1643 in the wild-type gene) (Fig. 2A). This fragment was subcloned in pUC19 and sequenced on both strands by primer walking. The data revealed that the ssyF mutation actually consists of a disruption of the rpsA gene by the transposable element IS10R (12). Whereas one of the recombinant pUC19 plasmids showed several divergences from the published IS10R sequence (1), others did not, indicating that these changes have arisen during PCR amplification.
In the ssyF mutant, the IS10R sequence within the rpsA gene starts just after the rpsA position 1391, and it is flanked by a 9-bp direct duplication of the target site CGCTAAAGG (Fig. 2B). Such a duplication is typical for Tn10 and IS10R insertions (15), although in our case, the 9-bp repeat does not include the hot spot symmetrical consensus sequence 5′-GCTNAGC. The insertion causes premature termination of translation at the very beginning of the inserted sequence (Fig. 2B). As a result, the ssyF mutant produces a truncated form of S1 comprising 465 amino acids instead of 557 for the wild-type protein. According to current knowledge of the S1 structure (5, 35, 36), the central and C-terminal regions of the protein form its RNA-binding domain, which consists of four highly homologous repeats of 72 to 74 amino acids, the S1 motifs R1 to R4 (Fig. 2C). The SsyF protein therefore lacks most of repeat R4 and is renamed here “S1Δ4” to emphasize this fact. The questions of why S1 has evolved this modular organization and what is the functional role of each repeat remain unsettled. However, the ssyF mutant is viable (31), even though it grows much more slowly than the wild-type parent. (At 37°C in fully supplemented MOPS-glycerol medium, its doubling time was estimated as ca. 110 min versus 40 min for rpsA+ cells.) Therefore, the cell can tolerate the loss of repeat R4, and we have exploited this feature to evaluate its role in translation initiation.
Effect of the ssyF mutation upon translation initiation from individual TIRs.
To compare the efficiencies of the wild-type and truncated S1 proteins in translation initiation in vivo, we have constructed series of either rpsA+ or ssyF strains in which the translation of the chromosomal lacZ gene is driven by TIRs originating from a variety of other genes. Strains within each series are isogenic except for the TIR replacement (Fig. 1); in particular, the lac promoter-operator sequences, as well as most of the lac transcribed sequence, are identical in each case, so that strain-to-strain differences in β-galactosidase synthesis essentially reflect the variable efficiencies of the TIRs used. This approach has proven useful for comparing the strengths of various natural or artificial TIRs in vivo (8, 11, 43). Here, we exploited it to assess the effect of the ssyF mutation upon the efficiency of individual TIRs. To provide a control for the S1 overexpression experiments to be described below, cells used for β-galactosidase assays always harbored pACYC184. The presence of this plasmid (referred to here as pCtr, for pControl) (Table 1) does not affect β-galactosidase yield from the different TIRs (not shown).
The choice of the TIRs used in this study (Fig. 3) deserves some comments. Many bacterial mRNAs harbor U- or A/U-rich single-stranded regions upstream of their SD sequence, and in several cases, these regions have been shown to bind S1 during initiation complex formation in vitro. In vivo, these regions also often stimulate translation initiation—hence, the belief that this stimulating effect reflects an improved 30S binding via S1-mRNA interaction (2, 3, 39, 45). It was therefore of interest to investigate how this effect is affected by the removal of repeat R4. To this end, we have selected TIRs from the galE, thrA, and rplL genes, all of which harbor such A/U-rich putative S1 binding sites. Meanwhile, to verify that these regions do stimulate translation, strains carrying truncated forms of the same TIRs lacking these elements were also constructed (Fig. 3). Two additional TIRs, corresponding to the secA and rpsA genes, were also included in this study. As concerns secA, it has been noted that the defect caused by the secY24 mutation can be corrected by elevated concentration of the SecA protein (9). It was therefore plausible that the ssyF mutation suppresses the secY24(Ts) defect indirectly by stimulating translation from the secA TIR—hence the inclusion of this TIR in our study. With regard to the rpsA TIR, it has been used here to evaluate a possible implication of the repeat R4 in autoregulation (see below).
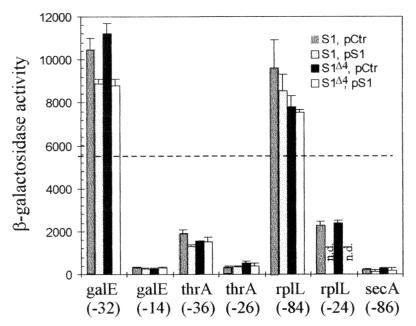
In rpsA+ cells, all three TIRs carrying U- or A/U-rich upstream sequences were efficient in driving translation of the lacZ gene, with the corresponding β-galactosidase levels ranging from ca. 30% (thrA) to 180% (galE) of that obtained with the genuine lacZ TIR (5,600 U) (Fig. 4). This observation is consistent with former results showing that foreign TIRs usually remain functional within the lacZ gene (8). The deletion of the upstream U- or A/U-rich regions caused a 4-fold (rplL) to 30-fold (galE) drop in β-galactosidase synthesis, confirming that these regions indeed stimulate translation (Fig. 4). Remarkably, when the ssyF mutation was introduced into the strains described above, these β-galactosidase levels were hardly affected, implying that the fraction of β-galactosidase in total protein synthesis is insensitive to the ssyF mutation. This conclusion holds true whatever the TIR used, and, in particular, whether U- or AU-rich upstream sequences are present or not (Fig. 4). Thus, the S1Δ4 protein is either as efficient as the wild-type S1 protein in supporting translation initiation, or, if it is not, its efficiency is reduced evenly whatever the TIRs. We conclude that the R4 repeat plays no specific role in the recognition of individual TIRs.
FIG. 4.
Histograms showing the activities of the different TIRs listed in Fig. 3 (except rpsA TIR) as measured by the β-galactosidase activity resulting from their fusion to lacZ (Fig. 1). Each group of four vertical bars (from left to right; S1, pCtr; S1, pS1; S1Δ4, pCtr; S1Δ4, pS1) illustrates the activity of a given TIR when the chromosomal rpsA gene is either wild type or ssyF (i.e., encodes either the full-length [S1] or the truncated [S1Δ4] protein and the cell contains either plasmid pACYC184 [pCtr] or the same plasmid carrying the wild-type rpsA gene [pS1]). Below each group of four bars is indicated the gene from which the TIR originates and the 5′ boundary of the particular fragment used as TIR (Fig. 3). Given β-galactosidase activity (in nanomoles of ONPG hydrolyzed per minute per milligram of total protein) is the average of two to five experiments. n.d., not determined. The horizontal dotted line corresponds to the β-galactosidase expression observed with the genuine lacZ TIR in rpsA+ cells lacking any plasmid (5,600 U) (43).
As concerns the secA TIR, it was quite inefficient in driving lacZ translation in rpsA+ cells (the β-galactosidase level was only 3% of that observed with the genuine lacZ TIR) (Fig. 4), either because it lacks the GG motif which constitutes the core of most SD elements (Fig. 3) or because it can form inhibitory secondary structures. Significantly, this level again remained very nearly the same in ssyF cells. Therefore, the ssyF mutation is unlikely to suppress the secY24 defect by favoring secA translation.
Effect of the ssyF mutation upon translation initiation from the rpsA TIR.
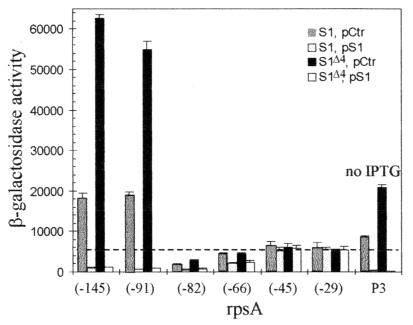
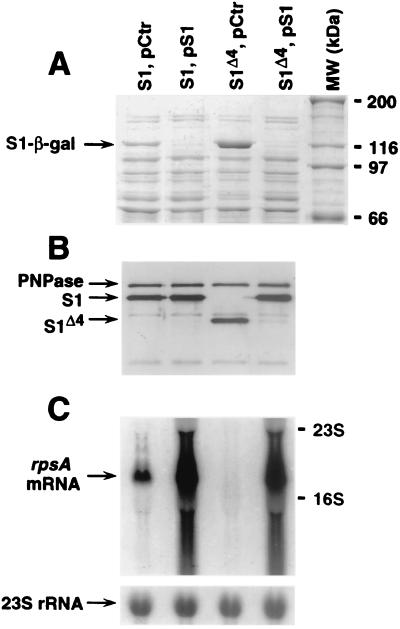
In our initial construct, the rpsA TIR extended from nt −145 to +57 with respect to the start codon (Fig. 2A and 3). This sequence includes the −10 region of the strong rpsA promoter P3, but not its −35 region. Consistently, as for all other fusions described above, β-galactosidase synthesis was strictly dependent upon the presence of IPTG in the growth medium, showing that transcription of the rpsA-lacZ fusion originates exclusively from the lac promoter. The β-galactosidase assay immediately revealed the unique character of the rpsA TIR (Fig. 4 and 5). First, in rpsA+ cells, it was more efficient in driving lacZ translation than any other TIR tested here, or, indeed, than any other TIR previously assayed in this system (8, 11, 43): thus, the β-galactosidase level in this case was over 300% of the level observed with the genuine lacZ TIR (Fig. 5), a result all the more remarkable since the rpsA SD element deviates from the consensus even more than the weak secA SD element (Fig. 3). Second, although already very high, this level was further increased circa threefold by the ssyF mutation. This large overexpression was clearly visible when cell extracts were analyzed on sodium dodecyl sulfate gels (Fig. 6A). The comparison of the β-galactosidase activity observed in this case (ca. 60,000 U) (Fig. 5) with the specific activity of pure β-galactosidase (400,000 U) (10) shows that β-galactosidase represents about 15% of total cell proteins, an amazing value for the product of a single-copy gene.
FIG. 5.
Same as Fig. 4, except that the rpsA TIR has been used. Seven rpsA fragments differing in their 5′ end have been fused to lacZ, and the number below each group of four vertical bars corresponds to the 5′ boundary of the fragment used (Fig. 3). The 5′ boundary of fragment P3, which extends to nt −252, is not shown on Fig. 3. Since it carries the intact rpsAp3 (P3) promoter, it can drive β-galactosidase synthesis in the absence of IPTG. All symbols are defined as in Fig. 4.
FIG. 6.
(A) Total protein extracts of E. coli (10 μg of proteins per lane) separated on a 7.5% Laemmli gel and stained with Coomassie blue. In the experiment shown, the expression of the fusion β-galactosidase (arrowed) is driven by the rpsA TIR (−91). The exact extracts used are indicated above each lane (all symbols are defined as in Fig. 4). MW, standard proteins, with the corresponding molecular mass (kDa) given on the right. (B) Western blot analysis of samples from the same cultures as those described above. Samples (1 μg) were separated on a 10% Laemmli gel. The blot was revealed with polyclonal rabbit antibodies raised against purified S1 (3) mixed with the antibodies against PNPase to detect eventual lane-to-lane differences in total protein loading. Secondary antirabbit horseradish peroxidase-labelled antibodies (Promega) and ECL (enhanced chemiluminescence) reagent (Amersham) were used for detection. The positions of PNPase, S1, and S1Δ4 are marked with arrows. (C) Northern analysis of the rpsA mRNA in the same cultures as in panels A and B. Total RNA was separated on the 1% agarose–formaldehyde gel, blotted, and hybridized essentially as in reference 43. The 32P random-labelled rpsA-specific probe is described in Materials and Methods. The arrows show the positions of the main rpsA mRNA species and 16S and 23S rRNAs, as indicated. The strip below the main panel shows reprobing of the same membrane with a 23S rRNA-specific oligonucleotide probe.
To define the region of the rpsA TIR responsible for its high translational activity and for its stimulation by the ssyF mutation, we created a series of rpsA-lacZ fusions in which the rpsA TIR is progressively shortened from the 5′ side. Deletion of the sequence from −145 to −91 did not bring any significant change, indicating that this upstream region is irrelevant to high translation efficiency or stimulation by ssyF. In contrast, a slightly larger deletion (to −82) dramatically impaired both properties (Fig. 5). Further shortening partly restored the translational activity in rpsA+ cells, but not the ssyF stimulation; in particular, the shortest rpsA TIR used here (nt −29 to +57) (Fig. 3) remained fairly efficient in rpsA+ cells, but was no longer stimulated by ssyF (Fig. 5).
In summary, stimulation of the rpsA TIR by the ssyF mutation requires sequences extending far upstream (i.e., to ca. −90) of the start codon; shorter versions of this TIR are insensitive to the mutation, as are TIRs unrelated to rpsA.
The stimulation of the rpsA TIR by the ssyF mutation is closely related to autogenous control.
It is known that excess S1 can repress its own translation, i.e., that the rpsA gene, like other ribosomal protein operons, is autoregulated (33, 44). To test whether autoregulation can be reproduced in our experimental system, rpsA+ strains carrying the rpsA TIR-lacZ fusions were transformed with the multicopy plasmid pSP261 (22). This pACYC184 derivative (named here “pS1”) (Table 1) carries the rpsA gene under the control of its own promoters; its presence in E. coli cells is known to repress translation of individual copies of the rpsA gene so that the overall level of synthesis of S1 is only slightly higher than in its absence (22, 24) (Fig. 6B). As a control, we also introduced the same plasmid into strains harboring TIRs unrelated to rpsA. In all cases, the presence of the plasmid decreased the growth rate by 10 to 20% at 37°C (at 30°C, this decrease became more pronounced). Moreover, it also caused a small decrease in β-galactosidase expression from all TIRs unrelated to rpsA or from 5′-truncated versions of the rpsA TIR (Fig. 4 and 5). This modest effect, which reflects a small reduction in the synthesis of β-galactosidase compared to other proteins, is obviously not TIR specific and was not investigated further. A completely different result was obtained with the rpsA TIR with the longest 5′ extensions (i.e., to −145 and −91). In this case, β-galactosidase activity dropped more than 20-fold in the presence of the plasmid (Fig. 5). Thus, autogenous repression can be reproduced in our experimental system, provided the rpsA TIR retains regions extending well upstream of the start codon (i.e., to around −90).
Concerning the current ssyF cells, the presence of the plasmid pS1 corrected all phenotypic traits associated with the mutation; thus, the growth rate became indistinguishable from those of rpsA+ cells carrying the same plasmid. The same holds true for β-galactosidase synthesis, whatever the TIR used. In particular, the extremely high activity of long versions of the rpsA TIR which is characteristic of ssyF cells was reduced by nearly 2 orders of magnitude in the presence of the plasmid, down to the level observed in rpsA+ cells (Fig. 5 and 6A).
In summary, the rpsA TIR possesses two specific properties—stimulation by the ssyF mutation and repression by extra rpsA copies—which appear closely related: the former effect can be completely reverted by the latter effect, and both require that the TIR extend at least to ca. −90 upstream of the rpsA start codon.
Effect of the ssyF mutation and S1 overexpression upon expression of the rpsA gene.
To exclude the remote possibility that the lac promoter or operator which drives the transcription of our rpsA TIR-lacZ fusions plays a role in the repression of β-galactosidase synthesis by excess S1, or in its stimulation by the ssyF mutation, we created a similar fusion under the control of a genuine rpsA promoter. To this end, the rpsA fragment fused in phase of lacZ was extended to −252 so as to include a functional P3 rpsA promoter (Fig. 2A). As expected, this particular fusion was unique in allowing β-galactosidase synthesis in the absence of IPTG. Synthesis was, however, twofold less in this case than with the fusion carrying the rpsA TIR extending to −145 in the presence of IPTG. This difference may reflect the lower strength of the rpsA P3 compared to that of the lac promoter. Kajitani and Ishihama (13) similarly reported that, under their assay conditions, the P3 promoter was only 20 to 50% as active as the lacUV5 promoter. Significantly, however, the β-galactosidase synthesis driven by the rpsA P3-TIR combination was stimulated 2.5-fold by the ssyF mutation and depressed more than 20-fold when the pS1 plasmid was present (Fig. 5). These results parallel those observed when transcription is driven by the lac promoter.
Next, we tested the effect of the ssyF mutation and of extra copies of the rpsA gene upon the expression of the rpsA gene itself, by using Western blotting and polyclonal rabbit anti-S1 antibodies. Extracts of rpsA+ and ssyF cells revealed signals corresponding to products of the sizes expected for proteins S1 and S1Δ4, respectively (Fig. 6B, lanes 1 and 3). These signals remained the same after protein synthesis had been inhibited for 4 h, showing that both S1 and S1Δ4 are stable in vivo (not illustrated).
Interestingly, when identical amounts of extracts were compared, the S1Δ4 signal was not more intense than the signal from the wild-type S1. While this observation should be interpreted with some caution, because S1Δ4 presumably lacks some of the epitopes normally recognized by the polyclonal antibodies, it nevertheless suggests that the ssyF mutation does not cause overproduction of S1Δ4 as it does for the S1–β-galactosidase fusion protein, although both are translated from the same rpsA TIR (compare Fig. 6A and B). This difference is not surprising: whereas the β-galactosidase is synthesized from identical mRNAs in both rpsA+ and ssyF cells, this is definitely not the case for the S1 and S1Δ4 proteins, since the corresponding mRNAs differ because of IS10R inserted in the ssyF allele. The insertion might have decreased the rpsA::IS10 mRNA level, reducing S1Δ4 synthesis and neutralizing the effect of TIR stimulation. Such an effect of IS10 is not unprecedented: it has been shown that an IS10-like element, inserted immediately downstream of the rpsO coding region, reduces the rpsO mRNA level and, correspondingly, the ribosomal protein S15 synthesis to about 10% of those of the wild type (42).
To verify whether the IS10 insert does affect the amount of the rpsA mRNA, we compared the steady-state levels of the rpsA mRNA in rpsA+ and ssyF cells by using Northern blot analysis (Fig. 6C). Total RNA from the same cultures which were used for Western blotting (Fig. 6B) was hybridized with labeled rpsA-specific probes, which can hybridize with the rpsA mRNA from both rpsA+ and ssyF cells, but not with the mRNA from rpsA-lacZ fusions (see Materials and Methods). The signals were normalized to those obtained with the 23S rRNA-specific probe as an internal control. The relative positions of the rpsA-specific signals in pCtr lanes (Fig. 6C) show that transcription of the rpsA::IS10 gene stops early in the IS10 sequence. Quantitatively, the intensity of the signal was reduced more than fivefold in ssyF cells compared to that in wild-type cells (Fig. 6C). Since nevertheless, the synthesis of S1Δ4 is comparable to that of the wild-type S1 (pCtr lanes on Fig. 6B), the rpsA mRNA must be translated much more efficiently in mutant cells, indicating significant TIR stimulation.
As concerns plasmid pS1, its introduction caused a large increase in the rpsA mRNA level, but only a small increase in S1 expression (Fig. 6B and C), indicating that translation of individual rpsA mRNA copies is severely inhibited because of autorepression, as for the rpsA TIR-lacZ fusions (Fig. 6A). In ssyF cells, introduction of pS1 resulted in rpsA mRNA and S1 patterns that were almost indistinguishable from those observed for rpsA+ cells carrying the same plasmid; in particular, S1Δ4 expression was now barely detectable (Fig. 6B).
Altogether, these results show directly that the ssyF mutation markedly stimulates rpsA translation, although the overexpression of S1Δ4 does not take place because of the small amount of the rpsA mRNA in mutant cells. We conclude that the ssyF mutation impairs rpsA autoregulation, thus allowing the cell to produce a sufficient amount of S1 from a limited supply of rpsA mRNA.
DISCUSSION
While ribosomal protein S1, the product of the rpsA gene, clearly plays important and multiple functions in E. coli and other gram-negative bacteria (see the introduction), genetic studies of these functions have been impaired by the fact that S1 is essential for viability. Only two nonlethal chromosomal rpsA mutations have been isolated so far, both yielding truncated products lacking the C-terminal region (31, 35; this paper). Here, we have used one of them (ssyF29) to study in vivo the relationship between the structure of the S1 protein and its role in translation initiation. Thus, although the role of truncated S1 as a suppressor of a protein export defect (31) is intriguing, our primary goal here is not to explain this suppression but rather to characterize the properties of the mutant protein.
We have found that ssyF cells carry an IS10R element inserted within rpsA, resulting in an S1 polypeptide lacking the fourth homologous repeat (R4) of the RNA-binding domain (Fig. 2B and C). Thus, the ssyF29 mutation should be designated as rpsA::IS10. Mutation ssyB63, another suppressor from the ssy collection, also corresponds to an IS10 insertion, and the corresponding mutant protein (NusB) should be similarly truncated in its C-terminal region (38). Other ssy suppressors have not been structurally characterized. The SsyF protein (S1Δ4) is particularly intriguing because little is known about the respective roles of the individual S1 motifs. Earlier in vitro studies showed that an S1 polypeptide lacking R3 and R4 could still bind to either poly(U), poly(A), or MS2 RNA. However, with regard to translation initiation, it was only functional toward the two homopolymers and not toward the natural mRNA (36). In contrast, a longer protein (m1-S1) retaining most of R3 could support the in vitro translation of all three RNAs with only a slightly reduced efficiency (by 20 to 30%) compared to that of the full-length protein (35, 36). On this basis, it has been proposed that at least R3 is strictly necessary for the in-phase positioning of natural mRNAs on the ribosomal decoding center, i.e., for the precise fitting of the AUG codon to the P site (36).
Consistent with these in vitro results, R4 is clearly not strictly required for protein synthesis in vivo, since ssyF cells are viable. To gain further insight into the role of R4 in translation initiation from individual genes, we have exploited a genetic system in which the translation of the chromosomal lacZ gene is driven by TIRs from other genes (8) (Fig. 1). The removal of R4 has no effect upon the β-galactosidase synthesis from the diverse TIRs used here (with the marked exception of the rpsA TIR [see below]). In particular, the U- or A/U-rich upstream sequences which stimulate translation in vivo (17, 45; this paper) and in several cases have been shown to constitute strong S1-binding sites in vitro (2, 3, 39; I. Boni, unpublished results) have the same enhancing effect, whether R4 is present or not. These observations argue against a specific role for R4 in the recognition of individual TIRs. Recently, Sacerdot et al. reported that, similarly, the ssyF mutation affected neither the activity of the thrS TIR nor that of a derivative lacking an upstream enhancing sequence (27). Although these authors concluded that S1 does not participate in the recognition of the upstream element, a more logical conclusion is that the R4 motif is not involved in this recognition.
Aside from its role in assisting translation initiation from most or all TIRs, S1 also represses its own translation when present in excess (24, 33). Autorepression is easily reproduced in our experimental system: the β-galactosidase yield from the rpsA TIR is repressed strongly (20-fold) and specifically in the presence of plasmid pS1 bearing the rpsA gene (Fig. 5 and 6A). It is noteworthy, however, that the TIR must retain ca. 90 nt of rpsA sequence upstream of the start codon for repression to take place (the 5′ boundary of the minimally required sequence actually lies between −82 and −91). Obviously, the translational operator extends that far upstream.
The rpsA TIR does not solely differ from all other TIRs tested here in being repressed by excess S1: it is also unique in being markedly stimulated by the ssyF mutation (Fig. 5 and 6A). Just like repression by excess S1, stimulation by ssyF requires that the rpsA TIR extends far upstream of the start codon, with the minimal required extensions being identical in both cases. Moreover, the ssyF stimulation can be completely reverted by excess S1. It seems plausible then that the stimulation by ssyF and the repression by excess S1 correspond to the same phenomenon. Although autorepression is most apparent upon introduction of extra rpsA copies in the cell, it must still take place even when the rpsA gene is present as a single copy. We believe that the ssyF mutation alleviates this residual autorepression, thereby increasing the apparent activity of the TIR. According to this interpretation, the activity of the rpsA TIR is intrinsically extremely high (Fig. 5), but it is reduced circa threefold in cells harboring a single-copy wild-type rpsA gene because of autorepression. Thus, S1Δ4 appears to be unable to repress its own synthesis in the ssyF mutant.
A reasonable interpretation is that repeat R4 is required for formation of the repressed state of the rpsA TIR. Our attempts to confirm this point directly by testing the effect of S1Δ4 overproduction upon the activity of the rpsA TIR in vivo were frustrated by the fact that the ssyF allele seems to be too toxic for propagation on a multicopy plasmid, at least in the genetic context used. However, such an interpretation is supported by observations from S. Pedersen and colleagues (24). These authors found that, whereas the expression of a plasmid-borne rpsA-lacZ fusion was repressed by the presence of pS1 in the same cell, this repression was relieved when pS1 carried a frameshift mutation interrupting normal rpsA translation at codon 395, in the middle of R3. Derepression in this case was as large as that with a mutation interrupting translation at the 15th codon, i.e., at the very beginning of the coding sequence (24). It is clear, therefore, that a protein retaining the first 395 amino acids of S1 (followed by 50 unrelated amino acids resulting from the frameshift) is inactive as a repressor. This observation, which pinpoints the C-terminal region as being required for autoregulation, is compatible with our proposal that S1Δ4 is deficient in this function. In contrast, it seems inconsistent with an earlier result from the same group according to which a polypeptide retaining only the N-terminal region of S1, corresponding to the ribosome binding domain (35), can act as a repressor (33). Fragments carrying the N-terminal region of S1 are known to exchange readily with the ribosome-bound S1 (35); therefore, we consider the possibility that the observed repression (33) was actually mediated by full-length S1 molecules that had been displaced from the ribosome in the presence of a high concentration of the N-terminal domain.
As shown here, the IS10 insertion not only causes the production of the truncated protein S1 lacking R4 repeat, it also markedly reduces the steady-state level of the rpsA mRNA, without decreasing the S1Δ4 level to the same extent. Taking this fact into account, we cannot at present completely exclude an alternative explanation for the loss of the rpsA autogenous control in the ssyF mutant. The scarcity of the rpsA mRNA may reduce the rate of accumulation of S1Δ4 in the cell, consistent with the known slow growth of the ssyF mutant. The steady-state concentration of free S1Δ4 in the ssyF mutant might then be insufficient to form a tight repressor complex and to compete with 30S ribosomes for the rpsA TIR. In vitro experiments are in progress to evaluate an intrinsic capacity and concentration requirements of S1 lacking R4 to repress translation of its own mRNA.
ACKNOWLEDGMENTS
We thank S. Pedersen and J. Schnier for plasmids, K. Ito for strain IQ656 (ssyF29), and A. G. Carpousis for the anti-PNPase antibodies.
This work was supported by the RFBR grant 97-04-48834 to I.V.B. and by an MENRT grant (programme “Microbiologie”) to M.D. I.V.B. has been supported by a “Chercheur Associé” fellowship from CNRS, France.
REFERENCES
- 1.Artamonova V S, Boni I V. The ssyF29 mutation in E. coli ribosomal protein S1 gene which suppresses the defect in protein export machinery results from an insertion of IS10R-element. Bioorg Khim. 1996;22:941–943. [PubMed] [Google Scholar]
- 2.Artamonova V S, Tsareva N V, Boni I V. Regulation of the ribosomal protein L7/12 synthesis: the role of the rplJL intercistronic region as a translational enhancer. Bioorg Khim. 1998;24:530–538. [PubMed] [Google Scholar]
- 3.Boni I V, Issaeva D M, Musychenko M L, Tzareva N V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boni I V, Zlatkin I V, Budowsky E I. Ribosomal protein S1 associates with Escherichia coli ribosomal 30S subunit by means of protein-protein interactions. Eur J Biochem. 1982;121:371–376. doi: 10.1111/j.1432-1033.1982.tb05796.x. [DOI] [PubMed] [Google Scholar]
- 5.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 6.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douville K, Price A, Eicher J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 9.Fandl J P, Cabeli R, Oliver D, Tai P C. SecA suppresses the temperature-sensitive SecY24 defect in protein translocation in Escherichia coli membrane vesicles. Proc Natl Acad Sci USA. 1988;85:8953–8957. doi: 10.1073/pnas.85.23.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler A V, Zabin I. Purification, structure, and properties of hybrid β-galactosidase proteins. J Biol Chem. 1983;258:14354–14358. [PubMed] [Google Scholar]
- 11.Guillerez J, Gazeau M, Dreyfus M. In the Escherichia coli lacZ gene the spacing between the translating ribosomes is insensitive to the efficiency of translational initiation. Nucleic Acids Res. 1991;19:6743–6750. doi: 10.1093/nar/19.24.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halling S M, Simons R W, Way J C, Wash R B, Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci USA. 1982;79:2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kajitani M, Ishihama A. Determination of the promoter strength in the mixed transcription system. II. Promoters of ribosomal RNA, ribosomal protein S1 and recA protein operons from Escherichia coli. Nucleic Acids Res. 1983;11:3873–3888. doi: 10.1093/nar/11.12.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalapos M P, Paulus H, Sarkar N. Identification of ribosomal protein S1 as a poly(A) binding protein in Escherichia coli. Biochimie. 1997;79:493–502. doi: 10.1016/s0300-9084(97)82741-1. [DOI] [PubMed] [Google Scholar]
- 15.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 227–268. [Google Scholar]
- 16.Lee C A, Beckwith J. Suppression of growth and protein secretion defects in Escherichia coli secA mutants by decreasing protein synthesis. J Bacteriol. 1986;166:878–883. doi: 10.1128/jb.166.3.878-883.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy J E G, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 18.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 268–274. [Google Scholar]
- 19.Mogridge J, Greenblatt J. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J Bacteriol. 1998;180:2248–2252. doi: 10.1128/jb.180.8.2248-2252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muniyappa K, Mythili E. Phage λ β protein, a component of general recombination, is associated with host ribosomal S1 protein. Biochem Mol Biol Int. 1993;31:1–11. [PubMed] [Google Scholar]
- 21.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen S, Skouv J, Kajitani M, Ishihama A. Transcriptional organisation of the rpsA operon of Escherichia coli. Mol Gen Genet. 1984;196:135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- 23.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen M D, Sorensen M A, Pedersen S. Isolation and characterization of mutants with impaired regulation of rpsA, the gene encoding ribosomal protein S1 of Escherichia coli. Mol Gen Genet. 1993;240:23–28. doi: 10.1007/BF00276879. [DOI] [PubMed] [Google Scholar]
- 25.Roberts M W, Rabinovitz J C. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J Biol Chem. 1989;264:2228–2235. [PubMed] [Google Scholar]
- 26.Ruckman J, Ringquist S, Brody E, Gold L. The bacteriophage T4 regB ribonuclease. Stimulation of the purified enzyme by ribosomal protein S1. J Biol Chem. 1994;269:26655–26662. [PubMed] [Google Scholar]
- 27.Sacerdot C, Caillet J, Graffe M, Eyermann F, Ehresmann B, Ehresmann C, Springer M, Romby P. The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol Microbiol. 1998;29:1077–1090. doi: 10.1046/j.1365-2958.1998.00995.x. [DOI] [PubMed] [Google Scholar]
- 28.Schnier J, Isono K. The DNA sequence of the gene rpsA of Escherichia coli coding for ribosomal protein S1. Nucleic Acids Res. 1982;10:1857–1865. doi: 10.1093/nar/10.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnier J, Stoffler G, Nishi K. Deletion and insertion mutants in the structural gene for ribosomal protein S1 from Escherichia coli. J Biol Chem. 1986;261:11866–11871. [PubMed] [Google Scholar]
- 30.Shiba K, Ito K, Nakamura Y, Dondon J, Grunberg-Manago M. Altered initiation factor 2 in the cold-sensitive ssyG mutant affects protein export in Escherichia coli. EMBO J. 1986;5:3001–3006. doi: 10.1002/j.1460-2075.1986.tb04598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiba K, Ito K, Yura T. Suppressors of the secY24 mutation: identification and characterization of additional ssy genes in Escherichia coli. J Bacteriol. 1986;166:849–856. doi: 10.1128/jb.166.3.849-856.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1–24. [DOI] [PMC free article] [PubMed]
- 33.Skouv J, Schnier J, Rasmussen M D, Subramanian A R, Pedersen S. Ribosomal protein S1 of Escherichia coli is the effector for the regulation of its own synthesis. J Biol Chem. 1990;265:17044–17049. [PubMed] [Google Scholar]
- 34.Sorensen M A, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J Mol Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A R. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 36.Suryanarayana T, Subramanian A R. Function of the repeating homologous sequences in nucleic acid binding domain of ribosomal protein S1. Biochemistry. 1984;23:1047–1051. doi: 10.1021/bi00301a002. [DOI] [PubMed] [Google Scholar]
- 37.Taura T, Akiyama Y, Ito K. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol Gen Genet. 1994;243:261–269. doi: 10.1007/BF00301061. [DOI] [PubMed] [Google Scholar]
- 38.Taura T, Ueguchi C, Shiba K, Ito K. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol Gen Genet. 1992;234:429–432. doi: 10.1007/BF00538702. [DOI] [PubMed] [Google Scholar]
- 39.Tsareva N, Makhno V, Boni I. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- 40.Van Duin J. The single-stranded RNA bacteriophages. In: Fraenkel-Conrad H, Wagner R, editors. The viruses. New York, N.Y: Plenum; 1988. pp. 116–166. [Google Scholar]
- 41.Venkatesh T V, Radding C M. Ribosomal protein S1 and NusA protein complexed to recombination protein β of phage λ. J Bacteriol. 1993;175:1844–1846. doi: 10.1128/jb.175.6.1844-1846.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano R, Yura T. Suppression of the Escherichia coli rpoH opal mutation by ribosomes lacking S15 protein. J Bacteriol. 1989;171:1712–1717. doi: 10.1128/jb.171.3.1712-1717.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarchuk O, Jacques N, Guillerez J, Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1992;226:581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- 44.Zengel J M, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Deutsher M P. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc Natl Acad Sci USA. 1992;79:2608–2612. doi: 10.1073/pnas.89.7.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]