Abstract
Objective
To assess the performance of rapid antigen tests with unsupervised nasal and combined oropharyngeal and nasal self-sampling during the omicron period.
Design
Prospective cross sectional diagnostic test accuracy study.
Setting
Three public health service covid-19 test sites in the Netherlands, 21 December 2021 to 10 February 2022.
Participants
6497 people with covid-19 symptoms aged ≥16 years presenting for testing.
Interventions
Participants had a swab sample taken for reverse transcription polymerase chain reaction (RT-PCR, reference test) and received one rapid antigen test to perform unsupervised using either nasal self-sampling (during the emergence of omicron, and when omicron accounted for >90% of infections, phase 1) or with combined oropharyngeal and nasal self-sampling in a subsequent (phase 2; when omicron accounted for >99% of infections). The evaluated tests were Flowflex (Acon Laboratories; phase 1 only), MPBio (MP Biomedicals), and Clinitest (Siemens-Healthineers).
Main outcome measures
The main outcomes were sensitivity, specificity, and positive and negative predictive values of each self-test, with RT-PCR testing as the reference standard.
Results
During phase 1, 45.0% (n=279) of participants in the Flowflex group, 29.1% (n=239) in the MPBio group, and 35.4% ((n=257) in the Clinitest group were confirmatory testers (previously tested positive by a self-test at own initiative). Overall sensitivities with nasal self-sampling were 79.0% (95% confidence interval 74.7% to 82.8%) for Flowflex, 69.9% (65.1% to 74.4%) for MPBio, and 70.2% (65.6% to 74.5%) for Clinitest. Sensitivities were substantially higher in confirmatory testers (93.6%, 83.6%, and 85.7%, respectively) than in those who tested for other reasons (52.4%, 51.5%, and 49.5%, respectively). Sensitivities decreased from 87.0% to 80.9% (P=0.16 by χ2 test), 80.0% to 73.0% (P=0.60), and 83.1% to 70.3% (P=0.03), respectively, when transitioning from omicron accounting for 29% of infections to >95% of infections. During phase 2, 53.0% (n=288) of participants in the MPBio group and 44.4% (n=290) in the Clinitest group were confirmatory testers. Overall sensitivities with combined oropharyngeal and nasal self-sampling were 83.0% (78.8% to 86.7%) for MPBio and 77.3% (72.9% to 81.2%) for Clinitest. When combined oropharyngeal and nasal self-sampling was compared with nasal self-sampling, sensitivities were found to be slightly higher in confirmatory testers (87.4% and 86.1%, respectively) and substantially higher in those testing for other reasons (69.3% and 59.9%, respectively).
Conclusions
Sensitivities of three rapid antigen tests with nasal self-sampling decreased during the emergence of omicron but was only statistically significant for Clinitest. Sensitivities appeared to be substantially influenced by the proportion of confirmatory testers. Sensitivities of MPBio and Clinitest improved after the addition of oropharyngeal to nasal self-sampling. A positive self-test result justifies prompt self-isolation without the need for confirmatory testing. Individuals with a negative self-test result should adhere to general preventive measures because a false negative result cannot be ruled out. Manufacturers of MPBio and Clinitest may consider extending their instructions for use to include combined oropharyngeal and nasal self-sampling, and other manufacturers of rapid antigen tests should consider evaluating this as well.
Introduction
Rapid antigen tests show promising performance for the detection of SARS-CoV-2.1 2 3 4 5 The tests require minimal equipment, provide a result within 15-30 minutes, and can be performed in a range of settings without laboratory facilities. Although rapid antigen tests were initially introduced for use by trained professionals, they are currently widely available over the counter. Self-testing, without supervision of a trained professional, lowers the threshold for testing and allows individuals to obtain a result quickly, at their own convenience. This in turn could support early detection and self-isolation of infectious people and reduce community transmission.6
We previously showed that the sensitivity of the Roche/SD Biosensor (Roche Diagnostics) rapid antigen test with unsupervised nasal self-sampling was 78.5% in individuals with symptoms.7 Since the end of November 2021, however, the omicron variant of SARS-CoV-2 rapidly replaced the delta variant. Performance of rapid antigen tests for omicron could be different because of alterations in viral proteins and infection dynamics. Initial studies comparing omicron with delta found similar sensitivities for molecular tests,8 mixed analytical performance of lateral flow devices,9 10 and similar real world sensitivities for rapid antigen tests with sampling and testing by trained professionals.11 12 Additionally, anecdotal concerns were raised about the performance of such tests when applying only nasal sampling because omicron viral particles seem to be more prevalent in the throat than nose. One study showed improved sensitivity of a rapid antigen test with combined throat and nasal sampling by trained professionals.12 Currently, real world data on comparative accuracy of rapid antigen tests with unsupervised nasal self-sampling or combined oropharyngeal and nasal self-sampling are lacking.
We studied the diagnostic accuracy of three widely available commercial rapid antigen tests (see box 1) with unsupervised self-sampling during and after the emergence of omicron, using reverse transcription polymerase chain reaction (RT-PCR) as the reference standard; evaluated whether accuracies of tests with nasal self-sampling changed over time; and quantified whether diagnostic performance was improved with the addition of oropharyngeal to nasal self-sampling.
Box 1. The three studied rapid antigen tests.
The rapid antigen diagnostic tests studied were Flowflex (Acon Laboratories), MPBio (MP Biomedicals), and Clinitest (Siemens-Healthineers).
These tests have been freely distributed by the Dutch Ministry of Health, Welfare, and Sport across various target audiences. Primary schools, secondary schools, universities, institutions caring for vulnerable people, and organisations that aid civilians who cannot afford to buy tests were among those receiving tests from the ministry for distribution to their constituents.
From April 2021 the ministry distributed almost 120 million rapid antigen tests for self-use, of which 10.6 million were Flowflex, 28.7 million MPBio, and 12.4 million Clinitest. In addition, the ministry still has more than 11.7 million Flowflex, 14 million MPBio, and 7.1 million Clinitest tests in stock.
Methods
The study is reported according to the Standards for Reporting Diagnostic Accuracy Studies (STARD) 2015 guidelines.13
Study design and population
This prospective diagnostic test accuracy study was embedded within the Dutch public infrastructure for covid-19 testing. Testing is always by RT-PCR, free of charge, but only available for government approved test indications. During the study period, 21 December 2021 to 10 February 2022, these indications were having any symptom of potential SARS-CoV-2 infection; being identified as having close contact with an index case of SARS-CoV-2 infection; testing positive on any commercially available rapid antigen test after self-sampling at own initiative (confirmatory testers); or having returned from a country listed by the government as high risk.14
Participants were recruited consecutively at three public health service covid-19 test sites: Rotterdam-Rijnmond (Rotterdam), Central and Northeast Brabant (Tilburg), and West-Brabant (Roosendaal). Individuals were eligible if aged 16 years or older and willing and able to sign a digital informed consent form in Dutch. Current analyses only include those who reported any SARS-CoV-2 infection related symptom at the time of sampling, regardless of the reason for visiting the test site.
According to national SARS-CoV-2 pathogen surveillance, during the study period the percentage of infections attributable to omicron increased from 29% (week 51 in 2021) to 99% (week 5 in 2022; >95% BA.1 variant).15 16 17 From 12 January 2022 onwards, omicron accounted for >90% of infections. Most analyses, apart from the time trend analyses, included data from the latter omicron period. We further subdivided that period into a nasal self-sampling only phase (phase 1; omicron present in >90% of surveillance samples) and a combined oropharyngeal and nasal self-sampling phase (phase 2; omicron >99%). Table 1 shows the inclusion dates for each test location and phase.
Table 1.
Baseline characteristics of participants in the period when omicron dominated, stratified by rapid antigen test. Values are numbers (percentages) unless stated otherwise
| Characteristics | Flowflex: nasal (n=620); Rotterdam | MPBio; Tilburg | Clinitest; Roosendaal | ||||
|---|---|---|---|---|---|---|---|
| Nasal (n=820) | OP-N (n=543) | Nasal (n=726) | OP-N (n=653) | ||||
| Inclusion dates omicron period (>90% omicron related infections) | 12 Jan-3 Feb | 12-18 Jan | 26 Jan-10 Feb | 12-23 Jan | 25 Jan-8 Feb | ||
| Mean (SD) age (years); range | 37 (14); 16-77 | 38 (14); 16-85 | 36 (13); 16-72 | 39 (14); 16-81 | 40 (13); 16-87 | ||
| Sex, female | 357 (57.6) | 524 (63.9) | 375 (69.1) | 449 (61.8) | 437 (66.9) | ||
| Self-reported reason for testing*: | |||||||
| Positive self-test result | 279 (45.0) | 239 (29.1) | 288 (53.0) | 257 (35.4) | 290 (44.4) | ||
| Symptoms | 405 (65.3) | 510 (62.2) | 354 (65.2) | 459 (63.2) | 390 (59.7) | ||
| Close contact | 72 (11.6) | 170 (20.7) | 58 (10.7) | 144 (19.8) | 99 (15.2) | ||
| Other | 49 (7.9) | 54 (6.6) | 15 (2.8) | 29 (4.0) | 24 (3.7) | ||
| Vaccination status: | |||||||
| Not vaccinated | 75 (12.1) | 68 (8.3) | 56 (10.3) | 90 (12.4) | 66 (10.1) | ||
| Vaccinated with ≥1 dose | 545 (87.9) | 752 (91.7) | 487 (89.7) | 636 (87.6) | 586 (89.7) | ||
| No of doses received†: | |||||||
| 1 | 38 (7.0) | 86 (11.4) | 40 (8.2) | 52 (8.2) | 40 (6.8) | ||
| 2 | 233 (42.8) | 329 (43.8) | 184 (37.8) | 334 (52.5) | 214 (36.5) | ||
| 3 | 274 (50.3) | 336 (44.7) | 263 (54.0) | 248 (39.0) | 332 (56.7) | ||
| Unknown | 0 (0) | 1 (0.1) | 0 (0) | 2 (0.3) | 0 (0) | ||
| Initial vaccination series†: | |||||||
| BNT162b2 (Pfizer-BioNTech) | 438 (80.4) | 502 (66.8) | 341 (70.0) | 410 (64.5) | 352 (60.1) | ||
| mRNA-1273 (Moderna) | 34 (6.2) | 96 (12.8) | 43 (8.8) | 116 (18.2) | 104 (17.7) | ||
| ChAdOx1-S (Oxford-AstraZeneca) | 32 (5.9) | 88 (11.7) | 51 (10.5) | 62 (9.7) | 82 (14.0) | ||
| Ad26.COV2.S (Janssen/Johnson & Johnson) | 40 (7.3) | 62 (8.2) | 50 (10.3) | 45 (7.1) | 44 (7.5) | ||
| Unknown/other | 1 (0.2) | 4 (0.5) | 2 (0.4) | 3 (0.5) | 4 (0.7) | ||
| Booster vaccine†: | |||||||
| Pfizer | 200 (36.7) | 219 (29.1) | 214 (43.9) | 168 (26.4) | 256 (43.7) | ||
| Moderna | 99 (18.2) | 137 (18.2) | 76 (15.6) | 93 (14.6) | 93 (15.9) | ||
| None | 242 (44.4) | 388 (51.6) | 187 (38.4) | 366 (57.5) | 225 (38.4) | ||
| Unknown | 4 (0.7) | 8 (1.1) | 10 (2.1) | 9 (1.4) | 12 (2.0) | ||
| ≥1 previous SARS-CoV-2 infection | 148 (23.9) | 185 (22.6) | 135 (24.9) | 127 (17.5) | 121 (18.5) | ||
| Timing of most recent SARS-CoV-2 infection: | |||||||
| <2 months | 14 (9.5) | 16 (8.6) | 12 (8.9) | 13 (10.2) | 11 (9.1) | ||
| 2-6 months | 24 (16.2) | 22 (11.9) | 25 (18.5) | 7 (5.5) | 34 (28.1) | ||
| 6-12 months | 62 (41.9) | 84 (45.4) | 50 (37.0) | 40 (31.5) | 33 (27.3) | ||
| >12 months | 48 (32.4) | 63 (34.1) | 48 (35.6) | 66 (52.0) | 43 (35.5) | ||
| Unknown | 0 (0) | 0 (0) | 0 (0) | 1 (0.8) | 0 (0) | ||
| Symptom onset: | |||||||
| Day of sampling | 48 (7.7) | 71 (8.7) | 59 (10.9) | 42 (5.8) | 40 (6.1) | ||
| Day before sampling | 239 (38.5) | 281 (34.3) | 179 (33.0) | 201 (27.7) | 178 (27.3) | ||
| 2 days before sampling | 192 (31.0) | 257 (31.3) | 157 (28.9) | 273 (37.6) | 229 (35.1) | ||
| ≥3 days before sampling | 140 (22.6) | 208 (25.4) | 148 (27.3) | 210 (28.9) | 205 (31.4) | ||
| Unknown | 1 (0.2) | 3 (0.4) | 0 (0) | 0 (0) | 1 (0.2) | ||
| Symptoms (self-reported)‡: | |||||||
| Common cold | 543 (87.6) | 717 (87.4) | 499 (91.9) | 613 (84.4) | 583 (89.3) | ||
| Shortness of breath | 93 (15.0) | 109 (13.3) | 87 (16.0) | 99 (13.6) | 102 (15.6) | ||
| Fever | 143 (23.1) | 141 (17.2) | 120 (22.1) | 162 (22.3) | 157 (24.0) | ||
| Coughing | 310 (50.0) | 378 (46.1) | 283 (52.1) | 386 (53.2) | 350 (53.6) | ||
| Loss of taste or smell | 26 (4.2) | 27 (3.3) | 20 (3.7) | 25 (3.4) | 29 (4.4) | ||
| Muscle aches | 137 (22.1) | 154 (18.8) | 122 (22.5) | 192 (26.4) | 152 (23.3) | ||
| Other | 88 (14.2) | 123 (15.0) | 85 (15.7) | 116 (16.0) | 108 (16.5) | ||
| Experience with self-tests: | 589 (95.2) | 791 (96.7) | 535 (98.5) | 698 (96.3) | 627 (96.2) | ||
| Timing of last self-test§: | |||||||
| <7 days | 501 (85.1) | 661 (83.6) | 461 (86.2) | 588 (84.2) | 545 (86.9) | ||
| 1-4 weeks | 64 (10.9) | 92 (11.6) | 51 (9.5) | 75 (10.7) | 47 (7.5) | ||
| >1 months | 23 (3.9) | 37 (4.7) | 22 (4.1) | 35 (5.1) | 34 (5.4) | ||
| Unknown | 1 (0.2) | 1 (0.1) | 9 (0.2) | 0 (0) | 1 (0.2) | ||
| No of ever performed self-tests: | |||||||
| 1-3 | 110 (18.7) | 171 (21.6) | 90 (16.8) | 193 (27.7) | 126 (20.2) | ||
| 4-6 | 125 (21.2) | 198 (25.1) | 128 (23.9) | 208 (29.8) | 182 (29.1) | ||
| 7-10 | 158 (26.8) | 181 (22.9) | 118 (22.1) | 153 (21.9) | 130 (20.8) | ||
| >10 | 196 (33.3) | 240 (30.4) | 199 (37.2) | 144 (20.6) | 187 (29.9) | ||
Flowflex=Flowflex COVID-19 Antigen Home Test (Acon Laboratories); MPBio=Rapid SARS-CoV-2 Antigen Test Card (MP Biomedicals); Clinitest=CLINITEST Rapid COVID-19 Antigen Test (Siemens-Healthineers); OP-N=combined oropharyngeal and nasal; SD=standard deviation.
Participants could report more than one reason.
Proportion of those vaccinated.
Participants could report more than one symptom. Participants were asked separately whether they had symptoms on the day of study participation. All participants reported symptoms but only 60-65% reported symptoms as a reason for testing.
Proportion of those with experience of a self-test.
Inclusion procedure, specimen collection, and testing
Test site staff asked people visiting one of the participating sites whether they would be willing to participate in the study. If interested, they received information about the study, a test site specific rapid antigen test, and an email with a link to study documentation. Trained test site staff then took a swab sample for routine RT-PCR testing. The RT-PCR sampling method differed slightly across test sites; the Rotterdam and Tilburg sites used oropharyngeal and nasopharyngeal sampling and the Roosendaal site combined oropharyngeal and nasal sampling (see supplementary material 3). At all three sites, samples were tested in an off-site laboratory by RT-PCR on a Cobas 6800 or 8800 platform (Roche Diagnostics International).
During the initial study weeks (in 2021 and the first week in 2022) and during phase 1 (weeks 2 to 3 (MPBio and Clinitest) and weeks 2 to 5 (Flowflex) in 2022), participants received instructions to perform the rapid antigen test at home using only nasal self-sampling according to the manufacturers’ instructions. Participants received one of three tests (see box): Acon Laboratories’ Flowflex COVID-19 Antigen Home Test (Flowflex) in Rotterdam, MP Biomedicals’ Rapid SARS-CoV-2 Antigen Test Card (MPBio) in Tilburg, and Siemens-Healthineers CLINITEST Rapid COVID-19 Antigen Test (Clinitest) in Roosendaal. During phase 2 (weeks 4 to 6 in 2022), participants in Tilburg (MPBio) and Roosendaal (Clinitest) received instructions to perform oropharyngeal and nasal self-sampling with the same swab according to the investigator’s instructions for oropharyngeal sampling plus the manufacturer’s instructions for nasal self-sampling. We did not evaluate the Flowflex test for combined oropharyngeal and nasal self-sampling because the swab provided in the test kits was deemed not suitable for oropharyngeal self-sampling. All tests are CE marked for nasal sampling. MPBio and Clinitest were not CE marked for oropharyngeal and nasal sampling, but after safety checks by the quality team of the West-Brabant Public Health Service, and consultation with in-house in-vitro diagnostic regulation experts and the Medical Research Ethics Committee Utrecht, both tests were considered safe for use with oropharyngeal and nasal sampling.
Participants interpreted their test results visually according to manufacturer’s instructions, and always before they received their RT-PCR result from the public health service. Conversely, the rapid antigen test result was not available to the laboratories that conducted the RT-PCR tests for the public health service. Participants received their RT-PCR result according to the public health services’ routine practice to direct any further management, such as isolation, if applicable.
Participants were asked to complete the study procedures at home as soon as possible, and within three hours of their test site visit. They were asked to first provide informed consent electronically through the participation link in the email, then to perform the self-test, and finally to complete a short online questionnaire (see supplementary material 1). A call centre contacted participants who did not complete this questionnaire within three hours of their test site visit with the request to perform the self-test and complete the questionnaire as soon as possible.
Participants with a negative RT-PCR test result received an email after 10 days to complete a follow-up questionnaire (see supplementary material 2) to capture any infections that were missed by the baseline RT-PCR test.
Outcome measures
Primary outcomes were diagnostic accuracy (sensitivity, specificity, and positive and negative predictive values with corresponding 95% confidence intervals) of each rapid antigen test either with nasal self-sampling or with combined oropharyngeal and nasal self-sampling, and RT-PCR testing as reference. Secondary outcomes were diagnostic accuracies stratified by reason for testing (confirmatory testing after a positive self-test result at one’s own initiative, type of symptoms, close contact with an index case, or other reason), covid-19 vaccination status (no vaccination or vaccinated once, twice, or three times), previous SARS-CoV-2 infection, sex, and age (16-40 years, >40 years).
Statistical analysis
We assessed whether performance of the three rapid antigen tests with nasal self-sampling changed over time during the emergence of omicron, using χ2 tests to assess the sensitivities and specificities of the tests in different inclusion weeks and comparing the sensitivities in the first inclusion week with the sensitivities in the last inclusion week. We chose weekly intervals because the extent of omicron’s contribution to infection in the Netherlands was assessed weekly in the national pathogen surveillance.16
All primary and secondary diagnostic accuracies were also determined after applying a viral load cut-off (≥5.2 log10 SARS-CoV-2 E gene copies/mL). The viral load of each sample was estimated from the cycle threshold value of that sample using formulas based on the results of a previous study (see supplementary material 3). This was the viral load cut-off above which 95% of people with a positive RT-PCR test result had a positive virus culture result based on previous work.2 Furthermore, considering the large influence of confirmatory testers in our study populations, all analyses were repeated stratified by confirmatory testing (yes or no).
Finally, self-reported user experiences with each rapid antigen test and self-reported numbers of infections that may have been missed by baseline RT-PCR testing were assessed.
We performed complete cases analysis because the number of individuals without RT-PCR or rapid antigen test results was low (see fig 1, fig 2, and fig 3). All analyses were performed in R version 4.1.2 (2021-11-01) Bird Hippie.18
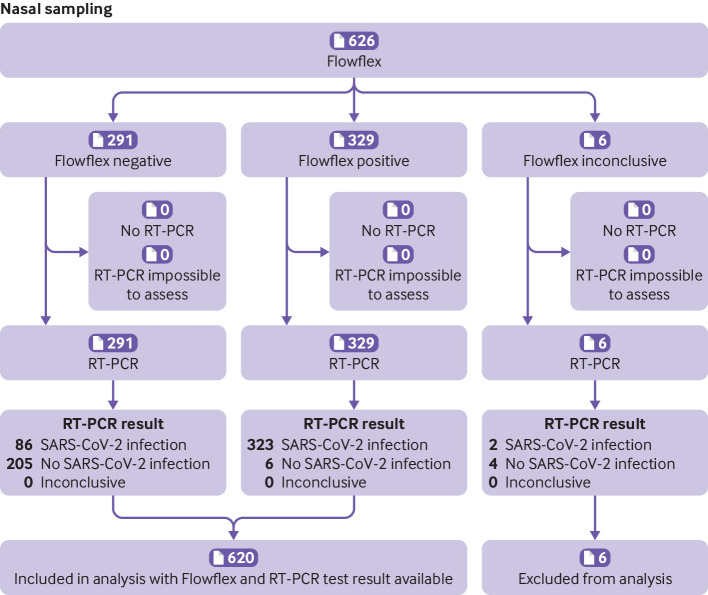
Fig 1.
Flow of participants who used the Flowflex (Acon Laboratories) rapid antigen test with nasal self-sampling during the omicron period, Rotterdam, the Netherlands. Inconclusive represents a combination of tests that showed no control line, test tubes were dropped, and test result was difficult to interpret (eg, faint line). RT-PCR=reverse transcription polymerase chain reaction
Fig 2.
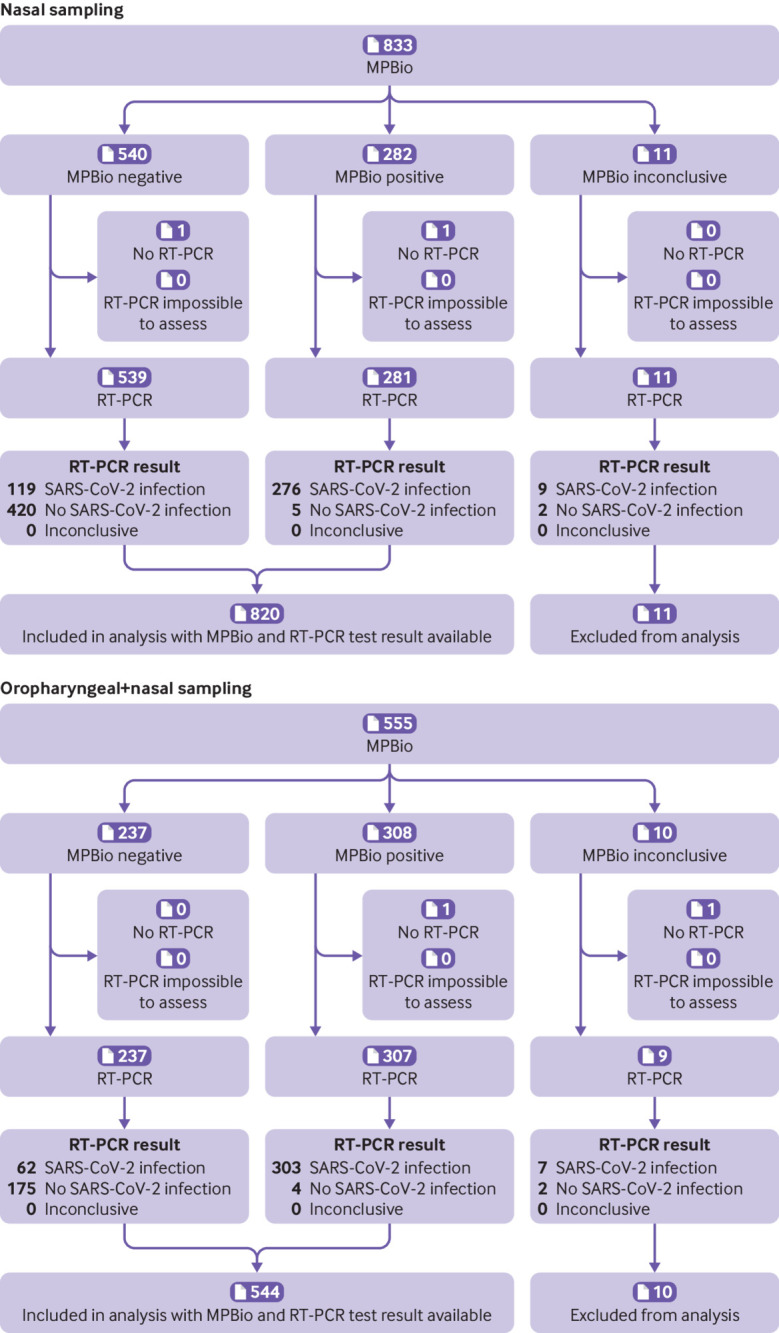
Flow of participants who used the MPBio (MP Biomedicals) rapid antigen test with nasal or combined oropharyngeal and nasal self-sampling, Tilburg, the Netherlands during omicron period. Inconclusive represents a combination of tests that showed no control line, test tubes were dropped, and test result was difficult to interpret (eg, faint line). RT-PCR=reverse transcription polymerase chain reaction
Fig 3.
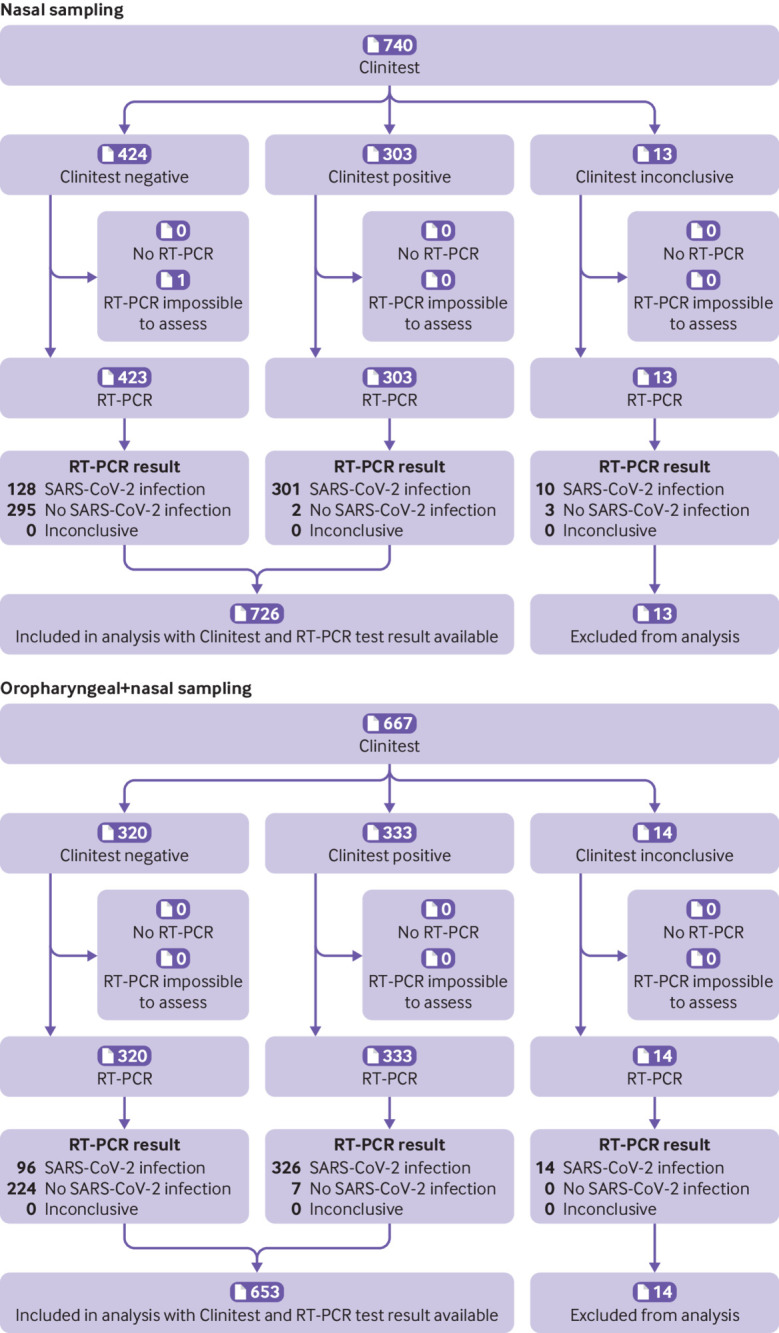
Flow of participants who used the Clinitest (Siemens-Healthineers) rapid antigen test with nasal or combined oropharyngeal and nasal self-sampling, Roosendaal, the Netherlands during omicron period. Inconclusive represents a combination of tests that showed no control line, test tubes were dropped, and test result was difficult to interpret (eg, faint line). RT-PCR=reverse transcription polymerase chain reaction
Sample size calculation
In our previous study of self-testing using the Roche/SD-Biosensor (Roche Diagnostics) SARS-CoV-2 nasal rapid antigen test, a sensitivity of 79% was observed in participants with symptoms.7 For the present study, we conservatively assumed sensitivities of 70% for all three rapid antigen tests irrespective of sampling method, with an error margin of 5%, type I error of 5%, and power of 80%. For each rapid antigen test and for each self-sampling method we therefore aimed for 335 positive RT-PCR test results. Because omicron was emerging at the start of the study in mid-December 2021, we extended the study to ensure accrual of at least 335 positive RT-PCR test results for each rapid antigen test and sampling strategy when omicron accounted for >90% of infections.16
Patient and public involvement
In late 2021 in the Netherlands, the public debate about covid-19 included discussions about the sensitivity of self-testing using commercially available tests. This question became even more urgent with the rapid surge of the omicron variant, and with experts advertising combined oropharyngeal and nasal sampling in national news outlets. Because of the urgency of the study, and the short time from study conception to conduct, we did not include the lay public in study design and implementation.
Results
A total of 3076 individuals participated in the delta-omicron transition phase before phase 1 (see supplementary figure S1) and a further 2199 in phase 1 and 1222 individuals in phase 2 (fig 1, fig 2, and fig 3). Most participants (84.5%, n=5490) performed the rapid antigen test within three hours of visiting the test site. We found no differences in test results overall nor in the RT-PCR test positive group between participants who completed the questionnaire within three hours and those who completed it later on. Supplementary table S1 presents the characteristics of the participants for the delta-omicron transition phase and table 1 for phases 1 and 2.
Overall test accuracies with nasal self-sampling in omicron period
Overall sensitivities were 79.0% (95% confidence interval 74.7% to 82.8%) for Flowflex, 69.9% (65.1% to 74.4%) for MPBio, and 70.2% (65.6% to 74.5%) for Clinitest (table 2, fig 4, fig 5, and fig 6). After applying the viral load cut-off, sensitivities were observed to increase to 85.6% (81.5% to 89.1%), 78.5% (73.8% to 82.8%), and 77.0% (72.4% to 81.2%), respectively (see supplementary figure S2). Specificities were >92%, positive predictive values >94%, and negative predictive values >59% for all three rapid antigen tests in all analyses (table 2), with slightly higher specificities and positive predictive values for MPBio and Clinitest and higher negative predictive values for Flowflex. Supplementary tables S2-S4 show all 2×2 tables.
Table 2.
Diagnostic accuracy variables for the three rapid antigen tests in participants with covid-19 symptoms in the omicron period
| Sampling method | No | RT-PCR test positivity* (%) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Positive predictive value (%) (95% CI) | Negative predictive value (%) (95% CI) | |
|---|---|---|---|---|---|---|---|
| Flowflex | |||||||
| Primary analysis | Nasal | 620 | 66.0 | 79.0 (74.7 to 82.8) | 97.2 (93.9 to 98.9) | 98.2 (96.1 to 99.3) | 70.4 (64.8 to 75.6) |
| Secondary (stratified) analyses: | |||||||
| Viral load cut-off† | Nasal | 620 | 58.2 | 85.6 (81.5 to 89.1) | 92.3 (88.3 to 95.2) | 93.9 (90.8 to 96.2) | 82.1 (77.2 to 86.4) |
| Vaccinated (at least once): | |||||||
| Yes | Nasal | 545 | 64.2 | 78.6 (73.9 to 82.8) | 96.9 (93.4 to 98.9) | 97.9 (95.4 to 99.2) | 71.6 (65.7 to 77.0) |
| No | Nasal | 75 | 78.7 | 81.4 (69.1 to 90.3) | 100 (79.4 to 100) | 100 (92.6 to 100) | 59.3 (38.8 to 77.6) |
| Previous SARS-CoV-2 infection: | |||||||
| Yes | Nasal | 148 | 63.5 | 67.0 (56.6 to 76.4) | 98.1 (90.1 to 100) | 98.4 (91.6 to 100) | 63.1 (51.9 to 73.4) |
| No | Nasal | 472 | 66.7 | 82.5 (77.9 to 86.6) | 96.8 (92.7 to 99.0) | 98.1 (95.7 to 99.4) | 73.4 (66.9 to 79.3) |
| Sex: | |||||||
| Female | Nasal | 357 | 63.9 | 75.0 (68.9 to 80.5) | 98.4 (94.5 to 99.8) | 98.8 (95.9 to 99.9) | 69.0 (61.8 to 75.6) |
| Male | Nasal | 260 | 68.5 | 83.7 (77.4 to 88.8) | 95.1 (88.0 to 98.7) | 97.4 (93.4 to 99.3) | 72.9 (63.4 to 81.0) |
| Age (years): | |||||||
| 16-40 | Nasal | 385 | 66.0 | 79.1 (73.6 to 84.0) | 96.2 (94.4 to 99.2) | 97.6 (94.4 to 99.2) | 70.4 (63.1 to 77.0) |
| >40 | Nasal | 235 | 66.0 | 78.7 (71.4 to 84.9) | 98.8 (93.2 to 100) | 99.2 (95.6 to 100) | 70.5 (61.2 to 78.8) |
| Reason for testing was positive self-test: | |||||||
| Yes | Nasal | 279 | 94.6 | 93.6 (89.9 to 96.2) | 80.0 (51.9 to 95.7) | 98.8 (96.5 to 99.8) | 41.4 (23.5 to 61.1) |
| No | Nasal | 341 | 42.5 | 52.4 (44.0 to 60.8) | 98.5 (95.6 to 99.7) | 96.2 (89.3 to 99.2) | 73.7 (67.9 to 78.9) |
| MPBio | |||||||
| Primary analysis | Nasal | 820 | 48.2 | 69.9 (65.1 to 74.4) | 98.8 (97.3 to 99.6) | 98.2 (95.9 to 99.4) | 77.9 (74.2 to 81.4) |
| OP-N | 543 | 67.2 | 83.0 (78.8 to 86.7) | 97.8 (94.3 to 99.4) | 98.7 (94.3 to 99.4) | 73.7 (67.6 to 79.2) | |
| Secondary (stratified) analyses: | |||||||
| Viral load cut-off† | Nasal | 819 | 41.5 | 78.5 (73.8 to 82.8) | 97.1 (95.1 to 98.4) | 95.0 (91.8 to 97.2) | 86.4 (83.2 to 89.2) |
| OP-N | 543 | 58.0 | 89.8 (86.0 to 92.9) | 89.5 (84.7 to 93.1) | 92.2 (88.6 to 94.9) | 86.4 (81.4 to 90.5) | |
| Vaccinated (at least one): | |||||||
| Yes | Nasal | 752 | 47.7 | 68.2 (63.2 to 73.0) | 99.2 (97.8 to 99.8) | 98.8 (96.5 to 99.7) | 77.4 (73.5 to 81.0) |
| OP-N | 487 | 66.9 | 82.5 (77.9 to 86.5) | 97.5 (93.8 to 99.3) | 98.5 (96.3 to 99.6) | 73.4 (66.9 to 79.2) | |
| No | Nasal | 68 | 52.9 | 86.1 (70.5 to 95.3) | 93.8 (79.2 to 99.2) | 93.9 (79.8 to 99.3) | 85.7 (69.7 to 95.2) |
| OP-N | 56 | 69.6 | 87.2 (72.6 to 95.7) | 100 (80.5 to 100) | 100 (89.7 to 100) | 77.3 (54.6 to 92.2) | |
| Previous SARS-CoV-2 infection: | |||||||
| Yes | Nasal | 185 | 35.7 | 60.6 (47.8 to 72.4) | 100 (96.9 to 100) | 100 (91.2 to 100) | 82.1 (74.8 to 87.9) |
| OP-N | 135 | 65.2 | 85.2 (76.1 to 91.9) | 97.9 (88.7 to 99.9) | 98.7 (92.9 to 100) | 78.0 (65.3 to 87.7) | |
| No | Nasal | 634 | 51.9 | 71.7 (66.5 to 76.5) | 98.4 (96.2 to 99.5) | 97.9 (95.2 to 99.3) | 76.3 (71.8 to 80.5) |
| OP-N | 408 | 67.9 | 82.3 (77.3 to 86.6) | 97.7 (93.5 to 99.5) | 98.7 (96.3 to 99.7) | 72.3 (65.1 to 78.8) | |
| Sex: | |||||||
| Female | Nasal | 524 | 45.0 | 70.3 (64.1 to 76.1) | 99.0 (97.0 to 99.8) | 98.2 (94.9 to 99.6) | 80.3 (75.8 to 84.3) |
| OP-N | 375 | 65.3 | 82.4 (77.1 to 87.0) | 97.7 (93.4 to 99.5) | 98.5 (95.8 to 99.7) | 74.7 (67.5 to 81.0) | |
| Male | Nasal | 295 | 53.9 | 69.2 (61.4 to 76.3) | 98.5 (94.8 to 99.8) | 98.2 (93.7 to 99.8) | 73.2 (66.2 to 79.5) |
| OP-N | 166 | 71.1 | 83.9 (76.0 to 80.0) | 97.9 (88.9 to 99.9) | 99.0 (94.6 to 100) | 71.2 (58.7 to 81.7) | |
| Age (years): | |||||||
| 16-40 | Nasal | 475 | 49.5 | 75.3 (69.3 to 80.7) | 99.2 (97.0 to 99.9) | 98.9 (96.0 to 99.9) | 80.4 (75.4 to 84.8) |
| OP-N | 335 | 68.4 | 82.5 (77.0 to 87.2) | 97.2 (92.0 to 99.4) | 98.4 (95.5 to 99.7) | 72.0 (63.9 to 79.2) | |
| >40 | Nasal | 345 | 46.4 | 61.9 (53.9 to 69.4) | 98.4 (95.3 to 99.7) | 97.1 (91.6 to 99.4) | 74.9 (69.0 to 80.2) |
| OP-N | 208 | 65.4 | 83.8 (76.5 to 89.6) | 98.6 (92.5 to 100) | 99.1 (95.3 to 100) | 76.3 (66.4 to 84.5) | |
| Reasons for testing was positive self-test: | |||||||
| Yes | Nasal | 239 | 94.6 | 83.6 (78.1 to 88.2) | 84.6 (54.6 to 98.1) | 99.0 (96.3 to 99.9) | 22.9 (12.0 to 37.3) |
| OP-N | 288 | 96.2 | 87.4 (82.9 to 91.0) | 90.9 (58.7 to 99.8) | 99.6 (97.7 to 100) | 22.2 (11.2 to 37.1) | |
| No | Nasal | 581 | 29.1 | 51.5 (43.7 to 59.2) | 99.3 (97.9 to 99.8) | 96.7 (90.6 to 99.3) | 83.3 (79.7 to 86.5) |
| OP-N | 255 | 34.5 | 69.3 (58.6 to 78.7) | 98.2 (94.8 to 99.6) | 95.3 (86.9 to 99.0) | 85.9 (80.1 to 90.5) | |
| Clinitest | |||||||
| Primary analysis | Nasal | 726 | 59.1 | 70.2 (65.6 to 74.5) | 99.3 (97.6 to 99.9) | 99.3 (97.6 to 99.9) | 69.7 (65.1 to 74.1) |
| OP-N | 653 | 64.6 | 77.3 (72.9 to 81.2) | 97.0 (93.9 to 98.8) | 97.9 (95.7 to 99.2) | 70.0 (64.7 to 75.0) | |
| Secondary (stratified) analyses: | |||||||
| Viral load cut-off† | Nasal | 711 | 52.0 | 77.0 (72.4 to 81.2) | 97.4 (95.0 to 98.8) | 96.9 (94.3 to 98.6) | 79.6 (75.4 to 83.4) |
| OP-N | 653 | 57.3 | 83.7 (79.5 to 87.3) | 92.8 (89.1 to 95.6) | 94.0 (90.9 to 96.3) | 80.9 (76.2 to 85.1) | |
| Vaccinated (at least one): | |||||||
| Yes | Nasal | 635 | 58.6 | 68.5 (63.6 to 73.2) | 100 (98.6 to 100) | 100 (98.6 to 100) | 69.2 (64.3 to 73.8) |
| OP-N | 586 | 64.7 | 77.0 (72.5 to 81.2) | 98.1 (95.1 to 99.5) | 98.6 (96.6 to 99.6) | 70.0 (64.4 to 75.2) | |
| No | Nasal | 90 | 63.3 | 80.7 (68.1 to 90.0) | 93.9 (79.8 to 99.3) | 95.8 (85.7 to 99.5) | 73.8 (58.0 to 86.1) |
| OP-N | 66 | 65.2 | 79.1 (64.0 to 90.0) | 91.3 (72.0 to 98.9) | 94.4 (81.3 to 99.3) | 70.0 (50.6 to 85.3) | |
| Previous SARS-CoV-2 infection: | |||||||
| Yes | Nasal | 127 | 49.6 | 60.3 (47.2 to 72.4) | 96.9 (89.2 to 99.6) | 95.0 (83.1 to 99.4) | 71.3 (60.6 to 80.5) |
| OP-N | 121 | 49.6 | 66.7 (53.3 to 78.3) | 95.1 (86.3 to 99.0) | 93.0 (80.9 to 98.5) | 74.4 (63.2 to 83.6) | |
| No | Nasal | 599 | 61.1 | 71.9 (66.9 to 76.4) | 100 (98.4 to 100) | 100 (98.6 to 100) | 69.3 (64.1 to 74.2) |
| OP-N | 532 | 68.0 | 79.0 (74.4 to 83.1) | 97.6 (94.1 to 99.4) | 98.6 (96.5 to 99.6) | 68.6 (62.3 to 74.4) | |
| Sex: | |||||||
| Female | Nasal | 449 | 57.7 | 65.3 (59.1 to 71.0) | 98.9 (96.2 to 99.9) | 98.8 (95.8 to 99.9) | 67.6 (61.8 to 73.1) |
| OP-N | 437 | 60.2 | 74.9 (69.2 to 80.0) | 96.6 (92.6 to 98.7) | 97.0 (93.7 to 98.9) | 71.8 (65.6 to 77.5) | |
| Male | Nasal | 276 | 61.2 | 77.5 (70.5 to 83.6) | 100 (96.6 to 100) | 100 (97.2 to 100) | 73.8 (65.8 to 80.7) |
| OP-N | 213 | 73.7 | 80.9 (73.9 to 86.7) | 98.2 (90.4 to 100) | 99.2 (95.7 to 100) | 64.7 (53.6 to 74.8) | |
| Age (years): | |||||||
| 16-40 | Nasal | 385 | 62.1 | 71.1 (64.9 to 76.8) | 99.3 (96.2 to 100) | 99.4 (96.8 to 100) | 67.8 (61.0 to 74.0) |
| OP-N | 339 | 64.9 | 79.1 (73.1 to 84.3) | 96.6 (91.6 to 99.1) | 97.8 (94.3 to 99.4) | 71.4 (63.8 to 78.3) | |
| >40 | Nasal | 341 | 55.7 | 68.9 (61.8 to 75.4) | 99.3 (96.4 to 100) | 99.2 (95.9 to 100) | 71.8 (65.1 to 77.8) |
| OP-N | 313 | 64.5 | 75.2 (68.7 to 81.0) | 97.3 (92.3 to 99.4) | 98.1 (94.4 to 99.6) | 68.4 (60.5 to 75.5) | |
| Tested because of positive self-test result: | |||||||
| Yes | Nasal | 257 | 95.3 | 85.7 (80.7 to 89.8) | 91.7 (61.5 to 99.8) | 99.5 (97.4 to 100) | 23.9 (12.6 to 38.8) |
| OP-N | 290 | 96.6 | 86.1 (81.5 to 89.9) | 80.0 (44.4 to 97.5) | 99.2 (97.1 to 99.9) | 17.0 (7.6 to 30.8) | |
| No | Nasal | 469 | 39.2 | 49.5 (42.0 to 56.9) | 99.6 (98.1 to 100) | 98.9 (94.1 to 100) | 75.3 (70.7 to 79.6) |
| OP-N | 363 | 39.1 | 59.9 (51.3 to 68.0) | 97.7 (94.8 to 99.3) | 94.4 (87.5 to 98.2) | 79.1 (73.8 to 83.8) | |
Flowflex=Flowflex COVID-19 Antigen Home Test (Acon Laboratories); MPBio=Rapid SARS-CoV-2 Antigen Test Card (MP Biomedicals); Clinitest=CLINITEST Rapid COVID-19 Antigen Test (Siemens-Healthineers); OP-N=combined oropharyngeal and nasal; RT-PCR=reverse transcription polymerase chain reaction.
SARS-CoV-2 infections based on RT-PCR test result.
Defined as viral load above which 95% of individuals with a positive RT-PCR test result had a positive viral culture,6 which was 5.2 log10 SARS-CoV-2 E gene copies/mL.
Fig 4.
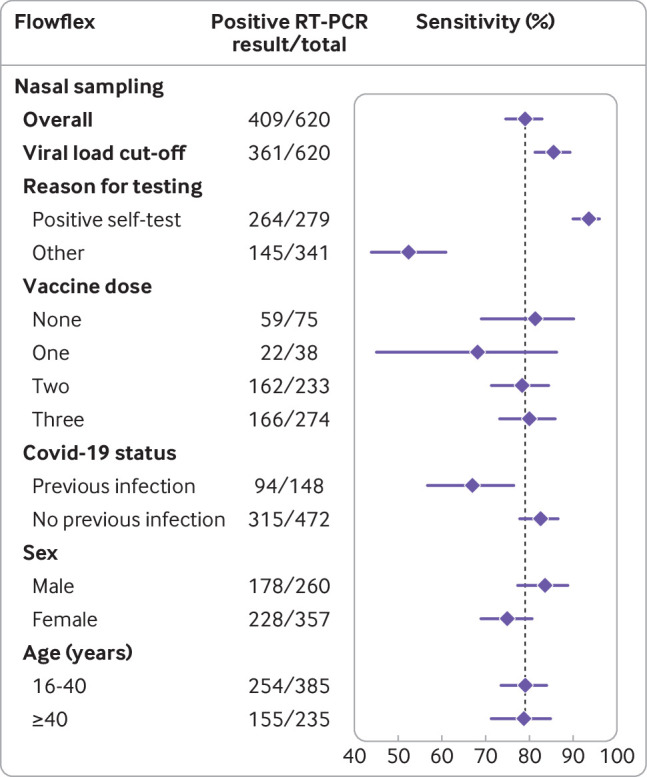
Sensitivities with 95% confidence intervals for the Flexflow (Acon Laboratories) rapid antigen test with nasal self- sampling using reverse transcription polymerase chain (RT-PCR) reaction as reference test, stratified according to covid-19 vaccination status, previous infection status, sex, and age. Vertical line indicates sensitivity of the rapid antigen test in the overall study population
Fig 5.
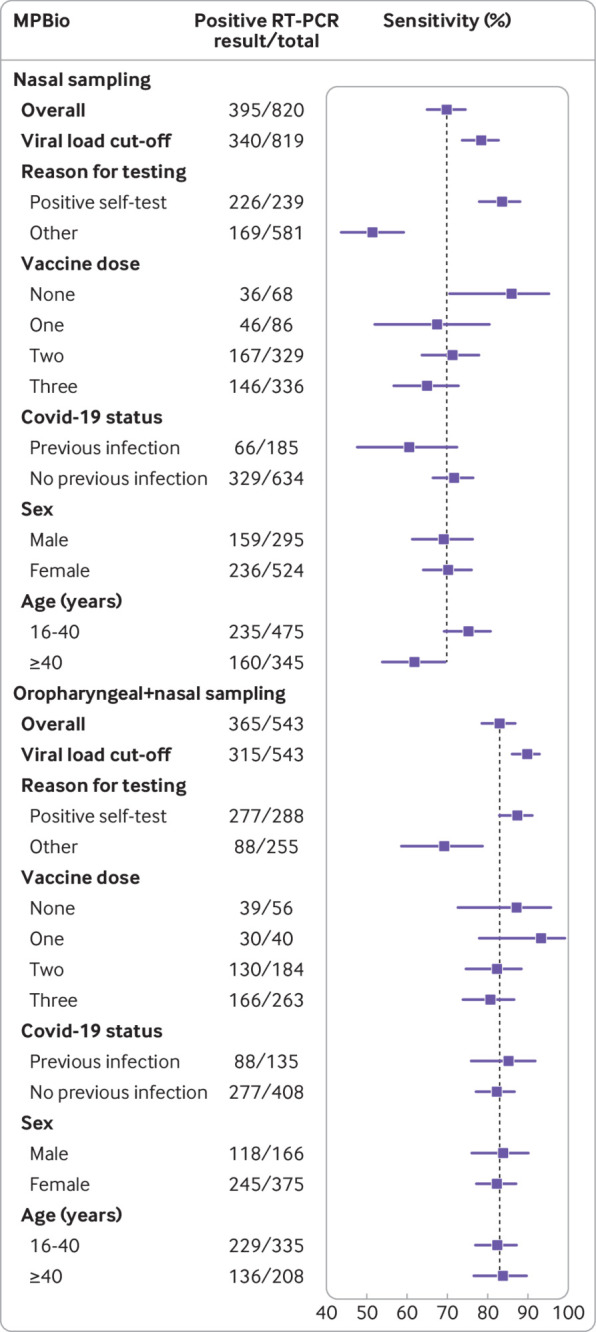
Sensitivities with 95% confidence intervals for the MPBio (MP Biomedicals) rapid antigen test using reverse transcription polymerase chain reaction (RT-PCR) as reference test, stratified according to covid-19 vaccination status, previous infection status, sex, and age, with nasal or combined oropharyngeal and nasal self-sampling. Vertical line indicates sensitivity of the rapid antigen test in the overall study population
Fig 6.
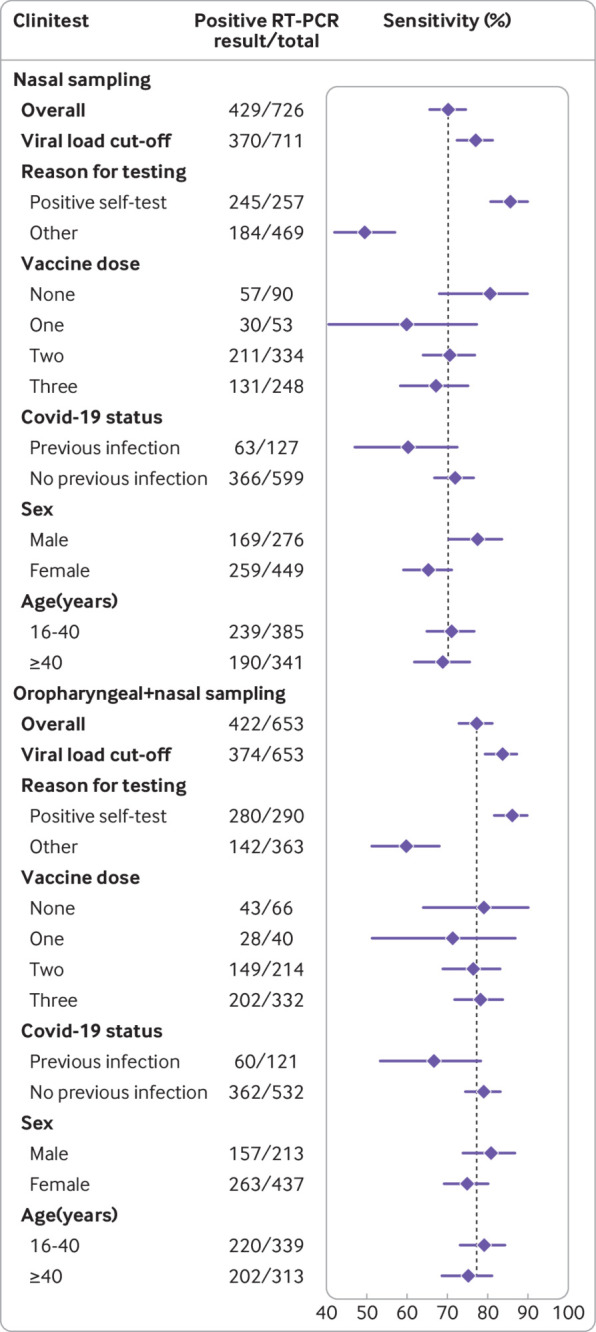
Sensitivities with 95% confidence intervals for the Clinitest (Siemens-Healthineers) rapid antigen test using reverse transcription polymerase chain reaction (RT-PCR) as reference test, stratified according to covid-19 vaccination status, previous infection status, sex, and age, with nasal or combined oropharyngeal and nasal self-sampling. Vertical line indicates sensitivity of the rapid antigen test in the overall study population
Overall test accuracies with oropharyngeal+nasal self-sampling in omicron period
Overall sensitivities were 83.0% (78.8% to 86.7%) for MPBio and 77.3% (72.9% to 81.2%) for Clinitest (table 2, fig 5 and fig 6). After applying the viral load cut-off, sensitivities were observed to increase to 89.8% (86.0% to 92.9%) and 83.7% (79.5% to 87.3%), respectively (see supplementary figure S2). Specificities, positive predictive values, and negative predictive values for both tests were >93%, >96%, and >75%, respectively, in all analyses (table 2). Supplementary tables S5 and S6 show all 2×2 tables.
Stratified test accuracies by self-sampling method
Although there were some differences across the three tests, we found lower sensitivities in participants with previous SARS-CoV-2 infection, women, and those older than 40 years (table 2, fig 4, fig 5, and fig 6). Higher sensitivities were observed after applying the viral load cut-off, but all stratification trends remained similar, except differences in sensitivity for previous SARS-CoV-2 infection status no longer appeared statistically significant for Flowflex (see supplementary figure S2). The largest differences in RT-PCR positivity and performances of the rapid antigen tests were between confirmatory testers and individuals who visited the test site for other reasons (table 2, fig 4, fig 5, and fig 6). In confirmatory testers, sensitivities were 93.6% (89.9% to 96.2%) for Flowflex, 83.6% (78.1% to 88.2%) for MPBio, and 85.7% (80.7% to 89.8%) for Clinitest with nasal self-sampling only, and 87.4% (82.9% to 91.0%) for MPBio and 86.1% (81.5% to 89.9%) for Clinitest with combined oropharyngeal and nasal self-sampling (table 2, fig 4, fig 5, and fig 6). In participants who tested for other reasons, sensitivities were 52.4% (44.0% to 60.8%) for Flowflex, 51.5% (43.7% to 59.2%) for MPBio, and 49.5% (42.0% to 56.9%) for Clinitest with nasal self-sampling only, and 69.3% (58.6% to 78.7%) for MPBio and 59.9% (51.3% to 68.0%) for Clinitest with combined oropharyngeal and nasal self-sampling. Supplementary table S7 presents diagnostic accuracies stratified by all reasons for testing.
Repeating all primary and secondary analyses separately in participants who did or did not undergo confirmatory tests indicated no distinctly different trends in sensitivities across subgroups (see supplementary figure S3 for nasal self-sampling and supplementary figure S4 for combined oropharyngeal and self-sampling). Differences across subgroups were less pronounced in the confirmatory testers, with much higher sensitivities among confirmatory testers in all strata.
Test accuracy changes with nasal self-sampling over time during omicron’s emergence
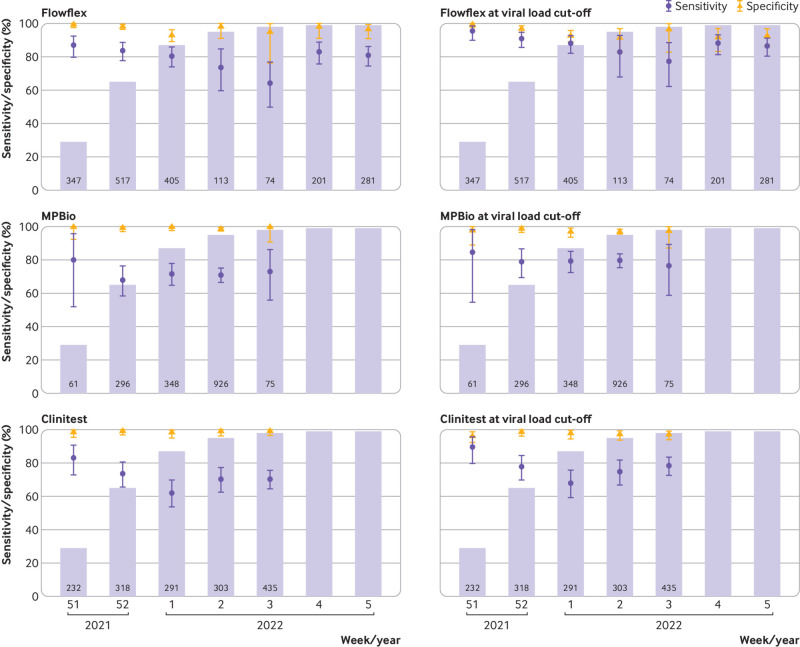
Sensitivities of all three rapid antigen tests were highest during the first week (fig 7) when omicron accounted for 28.6% of infections: 87.0% (79.7% to 92.4%) for Flowflex, 80.0% (51.9% to 95.7%) for MPBio, and 83.1% (72.9% to 90.7%) for Clinitest. With the emergence of omicron, sensitivities were found to decrease to 80.9% for Flowflex (χ2=2.0; P=0.16), 73.0% for MPBio (χ2=0.28; P=0.60), and 70.3% for Clinitest (χ2=5.0; P=0.03). Specificities varied between 93.2% and 99.6% over time. When the viral load cut-off was applied, sensitivities were observed to be higher, but all trends over time remained similar (fig 7).
Fig 7.
Sensitivities and specificities with 95% confidence intervals of Flowflex (Acon Laboratories), MPBio (MP Biomedicals), and Clinitest (Siemens-Healthineeers) with nasal self-sampling using reverse transcription polymerase chain reaction as reference test by week of inclusion, before and after application of a viral load cut-off. Bar charts indicate the percentage of SARS-CoV-2 infections attributable to omicron according to the national pathogen surveillance, while the numbers indicate the number of participants included in each week
Stratification by reason for testing revealed that the changes in sensitivity over time were similar but more pronounced in non-confirmatory testers than in confirmatory testers, although confidence intervals were wide (see supplementary figure S5).
User experiences and 10-day follow-up
Supplementary tables S8 and S9 present information on user experiences and positive RT-PCR test results during the 10-day follow-up period, respectively.
Discussion
This large diagnostic accuracy evaluation of three commercially available SARS-CoV-2 rapid antigen tests (Flowflex, MPBio, and Clinitest) with unsupervised nasal self-sampling by individuals with symptoms showed a decline in overall sensitivities with the emergence of omicron. However, the observed decline was only statistically significant for Clinitest. Sensitivities were observed to increase when the tests (assessed for MPBio and Clinitest only) used combined oropharyngeal and nasal self-sampling instead of nasal self-sampling only. Sensitivities were substantially higher in confirmatory testers (those tested to confirm a positive self-test result) than in those who visited test sites for other reasons. Only the MPBio test with combined oropharyngeal and nasal self-sampling met the World Health Organization’s standards for rapid antigen tests (≥80% sensitivity and ≥97% specificity among individuals with symptoms).19
Comparison with other studies
Our pre-omicron studies, and when less than 5% of participants were confirmatory testers, found sensitivities of 72% to 83% for three different rapid antigen tests when performed by trained professionals, and 78.5% for the Roche/SD Biosensor rapid antigen test with unsupervised nasal self-sampling.7 20 21 The sensitivities we found in the first week of the current study, when delta was still highly dominant, were similar (Flowflex 87%, MPBio 80%, and Clinitest 83%), although the percentage of confirmatory testers was much higher (21% to 24%) than in the previous studies. However, sensitivities declined to 80%, 70%, and 70%, respectively, in the omicron period. Recent studies from the United States and Italy that evaluated rapid antigen tests when omicron was dominant, found comparable sensitivities of 74% (128/173 RT-PCR positive results) and 82% (126/154 RT-PCR positive results), although sampling was performed by professionals and sample sizes were smaller.11 12
We postulate several reasons for the somewhat lower sensitivities with omicron. Firstly, mutations in omicron’s nucleocapsid protein, the target of rapid antigen tests, could influence binding efficiency of antibodies used in the tests. However, analytical sensitivity based on isolated omicron and delta viruses generally appeared similar.9 Secondly, the proportion of confirmatory testers, who have a higher a priori chance of testing positive on the rapid antigen test, could have fluctuated over time and by test site, although our assessment of sensitivity over time did not confirm this hypothesis. Thirdly, a larger proportion of individuals over time have experienced a SARS-CoV-2 infection, which may have affected test performance. Fourthly, during the study period the participating test sites and laboratories experienced increasing requests for tests, exceeding the maximum capacity of the Dutch testing infrastructure. As a result, during those weeks the exposure-testing intervals of participants may have been increased, resulting in somewhat lower viral loads at the time of inclusion in the study.
Confirmatory testers
The largest differences in RT-PCR positivity percentages and performances of the rapid antigen tests were between confirmatory testers and individuals who attended the test sites for other reasons. As expected, RT-PCR positivity percentages were close to 100% in the confirmatory testers and substantially lower (30% to 43%) in the group that tested for other reasons. This agreed with test positivity percentages observed in national surveillance during the study. Logically, a higher proportion of confirmatory testers (93% to 95%) than other testers (76% to 77%) had viral loads above the used viral load cut-off. Adding oropharyngeal to nasal self-sampling was associated with a larger benefit in the group attending for other reasons (10% to 18% increase in sensitivity) than in the group of confirmatory testers (<1% to 4%) because the sensitivities were already high in the latter group.
In the Netherlands, all available SARS-CoV-2 self-tests are lateral flow antigen tests. Previous studies, including our own studies, have shown that antigen tests require a higher viral load to show positivity than molecular tests such as RT-PCR.2 5 7 This was confirmed in the current study: the mean viral load in confirmatory testers was higher than in the non-confirmatory testers. In a post hoc analysis, we assessed the impact of self-testing frequency. Confirmatory testers did have more self-testing experience than non-confirmatory testers (>10 self-tests reported by 37.2% v 30.0% of participants in the Flowflex group, 42.0% v 25.7% in the MPBio group, and 22.5% v 19.6% in the Clinitest group, respectively). If testing experience were to impact sensitivity, a higher sensitivity would be expected in those who had performed more than 10 self-tests compared with those who only performed one to three self-tests. However, in non-confirmatory testers, we found the opposite (51.3% v 73.1% for Flowflex, 47.9% v 54.0% for MPBio, and 41.4% v 55.9% for Clinitest, respectively). These data suggest that inexperienced individuals are as capable as experienced individuals at performing these tests unsupervised at home.
Subgroup analyses
For Flowflex with nasal self-sampling only, sensitivities were significantly lower in participants with a previous SARS-CoV-2 infection (67%) compared with those without (83%). Non-statistically significant differences of >10% were found for MPBio with nasal self-sampling and for Clinitest with nasal self-sampling and combined oropharyngeal and nasal self-sampling. These findings should be interpreted with caution because of the larger uncertainty around these subgroup specific accuracy estimates. In two previous studies, however, we observed similar trends for the BD Veritor (Becton, Dickinson) and Roche/SD Biosensor rapid antigen tests with either professional or self-sampling.5 7 The lower sensitivities of rapid antigen tests in individuals with a previous infection might be explained by generally lower viral loads, with some individuals potentially carrying viral RNA in the absence of a productive infection (ie, no viral antigen production). Another explanation might be that individuals with a previous infection have circulating anti-nucleocapsid protein antibodies,22 which might bind to the nucleocapsid protein produced during the new infection and thereby hamper the binding of monoclonal antibodies against the nucleocapsid protein in the test device. These effects might be test device specific given the variability in the performance across the three rapid antigen tests.5 We also found trends towards slightly higher sensitivities in participants who had not been vaccinated against covid-19 for MPBio and Clinitest with nasal self-sampling, but all confidence intervals overlapped with those who had been vaccinated against covid-10 once, twice, and three times, and no differential impact was observed when combined oropharyngeal and nasal self-sampling was applied. Supplementary material 4 discusses the results for subgroup effects based on sex and age.
Differences in diagnostic performances across subgroups may be explained by differences in the underlying characteristics of these subgroups. For example, diagnostic performance was shown to be affected by confirmatory testing and a previous SARS-CoV-2 infection. Since individuals in these two subgroups are not equally distributed across age and sex groups, the diagnostic performance in age and sex subgroups may be affected as well. We further hypothesise that diagnostic test performance in the epidemic setting mostly depends on SARS-CoV-2 viral load in the body area that is being sampled and on the quality of the sample. Our study provided direct evidence for the former, as sensitivity appeared to greatly improve when using a viral load cut-off. Some of the subgroups that we evaluated may have had lower viral loads on average. For example, the immune responses mounted by vaccinated individuals or individuals with previous SARS-CoV-2 infection may inhibit the virus from replicating. Our empirical data did show lower sensitivities in these groups.
Strengths and limitations of this study
In our diagnostic accuracy study conducted during the emergence of omicron, we compared the performances of rapid antigen tests with nasal self-sampling versus combined oropharyngeal and nasal self-sampling. Additional strengths include the large numbers of participants recruited at multiple test sites, the low percentage of missing values, reference test sampling and rapid antigen test self-testing within a few hours, unsupervised self-testing mimicking the real world context of self-testing, blinding of participants to the reference test result, blinding of laboratory staff to the rapid antigen test result, and the use of a viral load cut-off.
Our study also has some limitations. Firstly, the sample size calculation was based on the primary analysis, and diagnostic accuracy variables are by definition less precise for stratified and weekly analyses. Secondly, we did not determine the virus lineage in individual samples but relied on the national pathogen surveillance data to estimate the weekly prevalence of the omicron variant.16 This surveillance system includes about 2000 random samples from positive samples across the country on a weekly basis. Since regional variations in the Netherlands are small (data not shown), we are confident that omicron accounted for more than 90% of infections in all test sites from 12 January 2022 onwards. Thirdly, the viral load calculations were based on standard curves in a previous study.2 These standard curves were not repeated with each RT-PCR run in this study. The viral loads should therefore be considered as best estimates. Fourthly, the viral load cut-off that we used was the cut-off above which 95% of people with a positive RT-PCR test result had a positive virus culture in our similar previous study.2 Those experiments were done when the alpha variant was dominant, and participants were mostly unvaccinated. However, we believe that this estimate is still more meaningful than using arbitrary cycle threshold value cut-offs of 25 or 30, as is often done.23 24 Fifthly, we did not collect detailed information on the exact timing of RT-PCR sampling and rapid antigen testing. Therefore, the time interval was approximated by assessing the difference between the time a participant was registered at the test site (generally minutes after the RT-PCR sampling) and the time the online questionnaire was opened by the participant. Sixthly, nasal and combined oropharyngeal and nasal self-sampling were conducted in different time periods, but the omicron variant was present in >90% of samples in the national surveillance in both periods. Potentially, the proportion of SARS-CoV-2 infections attributed to the omicron variant may have been higher during the combined oropharyngeal and nasal self-sampling period. As we observed a decline in diagnostic accuracy with increasing dominance of the omicron variant in the nasal self-sampling period, the higher proportion of infections attributed to the omicron variant in the combined oropharyngeal and nasal self-sampling period may have led to an underestimation of the true difference in diagnostic accuracy between both sampling methods. Therefore, we are confident that combined oropharyngeal and nasal self-sampling is superior to nasal self-sampling only in the omicron era. Finally, slight differences in sampling methods (combined oropharyngeal and nasal versus more invasive oropharyngeal and nasopharyngeal) for the reference (RT-PCR) test, might have influenced the results of the study. However, we think that RT-PCR test performance is high regardless of the sampling method used. Indeed, the test site that evaluated the Clinitest used the less invasive combined oropharyngeal and nasal sampling method, but the performance of Clinitest was in fact worse (rather than better) than the performances of the other two tests that used the combined oropharyngeal and nasal sampling method. We therefore do not expect that the sampling method of the reference test substantially impacted our results or their generalisability.
Policy implications
In mid-January 2022, the Dutch government advised all individuals with covid-19 symptoms to do a self-test but advised vulnerable people and those in close contact with vulnerable people to have RT-PCR tests done at the public health service. When a self-test result was negative, individuals were allowed to go to work or school. Our data show that this was associated with a reduction in risk but did not minimise transmission risks because of the likelihood of false negative rapid antigen test results. As per national policy, we recommend that people who test negative by self-test should adhere to general preventive measures, such as applying hand hygiene, ensuring indoor ventilation, and wearing mouth-nose masks in crowed places. In case of a positive self-test result, self-isolation is required, but confirmatory testing seems unnecessary in most situations if the infection rate is high.
Conclusions
We found that the performance of rapid antigen tests with nasal self-sampling declined during the period omicron emerged. We also showed that the performance of rapid antigen tests can be improved by adding oropharyngeal to nasal self-sampling. Therefore, after proper evaluation, manufacturers of rapid antigen tests should consider extending their instructions for use to include combined oropharyngeal and nasal self-sampling. Positive predictive values were high throughout our study, and people with covid-19 symptoms can therefore rely on a positive rapid antigen test result irrespective of SARS-CoV-2 variant dominance or method of self-sampling. Negative predictive values were much lower. Individuals with a negative self-test result should adhere to general preventive measures because a false negative result cannot be ruled out.
What is already known on this topic
Self-testing for SARS-CoV-2 may potentially lower the threshold for testing and would allow individuals to obtain a result quickly and at their own convenience, which in turn could support the early detection of infectious people and reduce community transmission
Real world evidence on the performance of unsupervised nasal and combined oropharyngeal and nasal self-sampling in the omicron variant period is needed to accurately inform end users and policy makers
What this study adds
The sensitivities of three commercially available rapid antigen tests performed with nasal self-sampling decreased during the emergence of omicron, from 87% to 81% for Flowflex, 83% to 76% for MPBio, and 80% to 67% for Clinitest, with only Clinitest reaching statistical significance
Addition oropharyngeal to nasal self-sampling was associated with an improvement in the sensitivity of MPBio from 70% to 83% and Clinitest from 70% to 77% (not done for Flowflex), most notably in individuals who visited the test site for reasons other than to confirm a positive self-test result
Based on these findings, the manufacturers of MPBio and Clinitest may consider extending their instructions for use to include combined oropharyngeal and nasal self-sampling; other manufacturers should consider evaluating this as well
Acknowledgments
We thank the participants and study staff at the participating public health service test sites, participating laboratories, University Medical Center Utrecht, and RIVM for their contributions to the study. A special thanks to Esther Stiefelhagen, Renske Beekes, Sophie Neeleman, Eveline Westergaard, Roel Ensing, Wendy Mouthaan, and Timo Boelsums for their logistic support during the study conduct—written permission was obtained from each to list their names. None of them received any (financial) compensation for their contributions.
Web extra.
Extra material supplied by authors
Supplementary information: Supplementary material 1-4, figures S1-S5, and tables S1-S9
Contributors: KGMM initiated the study. ES, RPV, LH, IKV, WvdB, SDP, EL, MH, RM, CW, IV, CRSN-I, SvdH, JAJWK, JHHMvdW, and KGMM designed the study. ES, RPV, CRSN-I, and KGMM coordinated the study. WvdB, SDP, VFZ, LS, and MK were responsible for laboratory analyses and data processing. ES, RPV, and KGMM verified the underlying data. ES performed the statistical analysis and verified the underlying data in close collaboration with RPV and KGMM. ES, RPV, JHHMvdW, and KGMM drafted the first version of the manuscript. All authors critically read the manuscript and provided feedback. All authors approved the submission of the current version of the manuscript. ES and RPV contributed equally as first authors. JHHMvdW and KGMM contributed equally as senior authors. KGMM is the guarantor. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Funding: This study was funded by the Dutch Ministry of Health, Welfare, and Sport. The funder had no role in the design; collection, analysis, and interpretation of data; writing; and decision to submit the paper for publication.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: support from the Dutch Ministry of Health, Welfare, and Sport for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
The manuscript’s guarantor (KGMM) affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as originally planned (and, if relevant, registered) have been explained.
Dissemination to participants and related patient and public communities: The Dutch Outbreak Management Team that provides guidance to the Ministry of Health, Welfare, and Sport on covid-19 policy has advised, based on the results of this study, that rapid antigen tests can be used in the home setting for detection of a SARS-CoV-2 infection in individuals with symptoms, and that confirmation by a reverse transcription polymerase chain reaction test at a test site is no longer necessary. As such, the results of our study have directly been disseminated and are currently incorporated in the nationwide testing policy. At the time, this change in testing policy was covered by multiple national news outlets.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Ethical approval
Not required because the study was judged by the Medical Research Ethics Committee Utrecht to be outside the scope of the Dutch Medical Research Involving Human Subjects Act (protocol No 21-818 /C). All participants signed an informed consent form before any study procedure.
Data availability statement
Individual participant data collected during the study will be available, after deidentification of all participants. Data will be available to researchers who provide a methodologically sound proposal to achieve the aims in the approved proposal. Proposals should be directed to the corresponding author to gain access to the data. Data requestors will need to sign a data sharing agreement. The study protocol is available upon request by contacting Karel Moons at k.g.m.moons@umcutrecht.nl
References
- 1.RIVM Centrum Infectieziektebestrijding. Status validatie SARS-CoV-2 antigeen sneltesten, 10 Mar 2021 [Dutch]. 2021.
- 2. Schuit E, Veldhuijzen IK, Venekamp RP, et al. Diagnostic accuracy of rapid antigen tests in asymptomatic and presymptomatic close contacts of individuals with confirmed SARS-CoV-2 infection: cross sectional study. BMJ 2021;374:n1676. 10.1136/bmj.n1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brümmer LE, Katzenschlager S, Gaeddert M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: A living systematic review and meta-analysis. PLoS Med 2021;18:e1003735. 10.1371/journal.pmed.1003735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheiblauer H, Filomena A, Nitsche A, et al. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Euro Surveill 2021;26:26. 10.2807/1560-7917.ES.2021.26.44.2100441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Venekamp RP, Veldhuijzen IK, Moons KGM, et al. Detection of SARS-CoV-2 infection in the general population by three prevailing rapid antigen tests: cross-sectional diagnostic accuracy study. BMC Med 2022;20:97. 10.1186/s12916-022-02300-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control (ECDC). Considerations on the use of self-tests for COVID-19 in the EU/EEA. https://www.ecdc.europa.eu/sites/default/files/documents/Considerations-use-of-self-tests-for-COVID-19-in-the-EU-EEA-17-March2021-erratum.pdf; 2021.
- 7. Schuit E, Venekamp RP, Veldhuijzen IK, et al. Accuracy and usability of saliva and nasal rapid antigen self-testing for detection of SARS-CoV-2 infection in the general population: a head-to-head comparison. medRxiv. 2021. 10.1101/2021.12.08.21267452. [DOI]
- 8.Molenkamp R, Igloi Z. Evaluation of Antigen rapid test and PCR test to Omicron variant. 2021.
- 9. Deerain J, Druce J, Tran T, et al. Assessment of the Analytical Sensitivity of 10 Lateral Flow Devices against the SARS-CoV-2 Omicron Variant. J Clin Microbiol 2022;60:e0247921. 10.1128/jcm.02479-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osterman A, Badell I, Basara E, et al. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med Microbiol Immunol 2022;211:105-17. 10.1007/s00430-022-00730-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Michelena P, Torres I, Ramos-García A, et al. Real-life performance of a COVID-19 rapid antigen detection test targeting the SARS-CoV-2 nucleoprotein for diagnosis of COVID-19 due to the Omicron variant. medRxiv. 2022. 10.1101/2022.02.02.22270295. [DOI] [PMC free article] [PubMed]
- 12. Schrom J, Marquez C, Pilarowski G, et al. Direct Comparison of SARS-CoV-2 Nasal RT-PCR and Rapid Antigen Test (BinaxNOW™) at a Community Testing Site During an Omicron Surge. medRxiv. 2022. 10.1101/2022.01.08.22268954. [DOI]
- 13. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD Group . STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. 10.1136/bmj.h5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control (ECDC). Combined indicator: 14-day notification rate, testing rate and test positivity, updated 16 September 2021, weeks 35-36 2021 [updated 16 September 2021]. https://www.ecdc.europa.eu/en/publications-data/combined-indicator-week-36-2021.
- 15.Rijksoverheid. Variants of the corona virus 2021 [updated 2 December 2021]. https://coronadashboard.rijksoverheid.nl/landelijk/varianten.
- 16.Centrum Infectieziektebestrijding RIVM. Variants of the corona virus SARS-CoV-2 [Dutch] 2021 [updated 30 November 2021]. https://www.rivm.nl/coronavirus-covid-19/virus/varianten.
- 17.Centrum Infectieziektebestrijding RIVM. Epidemiologische situatie van SARS-CoV-2 in Nederland [Dutch] 2021 [updated 28 September 2021]. https://www.rivm.nl/sites/default/files/2021-09/COVID-19_WebSite_rapport_wekelijks_20210928_1146_final.pdf.
- 18. Team RCR. A language and environment for statistical computing. R Foundation for Statistical Computing, 2021. [Google Scholar]
- 19.WHO. Use of SARS-CoV-2 antigen-detection rapid diagnostic tests for COVID-19 self-testing WHO; 2022 9 Mar 2022.
- 20. Klein JAF, Krüger LJ, Tobian F, et al. Study Team . Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Med Microbiol Immunol 2021;210:181-6. 10.1007/s00430-021-00710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindner AK, Nikolai O, Kausch F, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur Respir J 2021;57:57. 10.1183/13993003.03961-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allen N, Brady M, Carrion Martin AI, et al. Serological markers of SARS-CoV-2 infection; anti-nucleocapsid antibody positivity may not be the ideal marker of natural infection in vaccinated individuals. J Infect 2021;83:e9-10. 10.1016/j.jinf.2021.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Igloi Z, Velzing J, van Beek J, et al. Clinical Evaluation of Roche SD Biosensor Rapid Antigen Test for SARS-CoV-2 in Municipal Health Service Testing Site, the Netherlands. Emerg Infect Dis 2021;27:1323-9. 10.3201/eid2705.204688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Kampen JJA, van de Vijver DAMC, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021;12:267. 10.1038/s41467-020-20568-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: Supplementary material 1-4, figures S1-S5, and tables S1-S9
Data Availability Statement
Individual participant data collected during the study will be available, after deidentification of all participants. Data will be available to researchers who provide a methodologically sound proposal to achieve the aims in the approved proposal. Proposals should be directed to the corresponding author to gain access to the data. Data requestors will need to sign a data sharing agreement. The study protocol is available upon request by contacting Karel Moons at k.g.m.moons@umcutrecht.nl