Abstract
The molecular mechanisms of regulation of the genes involved in the biosynthesis of cysteine are poorly characterized in Bacillus subtilis and other gram-positive bacteria. In this study we describe the expression pattern of the B. subtilis cysH operon in response to sulfur starvation. A 6.1-kb polycistronic transcript which includes the cysH, cysP, ylnB, ylnC, ylnD, ylnE, and ylnF genes was identified. Its synthesis was induced by sulfur limitation and strongly repressed by cysteine. The cysH operon contains a 5′ leader portion homologous to that of the S box family of genes involved in sulfur metabolism, which are regulated by a transcription termination control system. Here we show that induction of B. subtilis cysH operon expression is dependent on the promoter and independent of the leader region terminator, indicating that the operon is regulated at the level of transcription initiation rather than controlled at the level of premature termination of transcription. Deletion of a 46-bp region adjacent to the −35 region of the cysH promoter led to high-level expression of the operon, even in the presence of cysteine. We also found that O-acetyl-l-serine (OAS), a direct precursor of cysteine, renders cysH transcription independent of sulfur starvation and insensitive to cysteine repression. We propose that transcription of the cysH operon is negatively regulated by a transcriptional repressor whose activity is controlled by the intracellular levels of OAS. Cysteine is predicted to repress transcription by inhibiting the synthesis of OAS, which would act as an inducer of cysH expression. These novel results provide the first direct evidence that cysteine biosynthesis is controlled at a transcriptional level by both negative and positive effectors in a gram-positive organism.
Many bacteria can use sulfate as their principal source of sulfur. Inorganic sulfate is reduced to sulfide by a sequence of enzymatic steps involving ATP sulfurylase, adenosine 5′-phosphosulfate kinase, 3′-phosphoadenosine 5′-phosphosulfate (PAPS) sulfotransferase, and sulfite reductase (for a review, see reference 12). An O-acetyl-l-serine (OAS) (thiol)-lyase incorporates the sulfide, forming the amino acid cysteine. The assimilatory reduction of sulfate and formation of cysteine have been extensively studied in Escherichia coli and Salmonella enterica serovar Typhimurium (12). At least 22 genes required for the transport and reduction of sulfate and its incorporation into cysteine have been identified in these bacteria. Full expression of these genes requires a positively regulatory protein encoded by cysB, sulfur limitation, and a signal of sulfur limitation provided either by O-acetyl-l-serine or N-acetyl-l-serine (NAS), both of which function as internal inducers. CysB protein binds just upstream of the −35 region of positively regulated promoters, where in the presence of inducers it facilitates formation of a transcription initiation complex. Sulfide and thiosulfate provide additional regulation, acting as anti-inducers by inhibiting the binding of CysB protein to cys promoters (12).
In contrast to the knowledge of cysteine biosynthesis in E. coli and S. enterica serovar Typhimurium, there is little information regarding the molecular mechanisms of regulation of the genes involved in the biosynthesis of cysteine in Bacillus subtilis and other gram-positive bacteria. We have previously reported the isolation of the cysH gene of B. subtilis, which encodes PAPS sulfotransferase and whose expression is repressed by cysteine and sulfide and induced by sulfur limitation (14). Analysis of the completed B. subtilis genome revealed that cysH lies in a 6,074-bp fragment, together with six open reading frames (ORFs) (Fig. 1). Three of them belong to the cysteine biosynthetic pathway: cysP encodes a sulfate permease (15), while ylnD and ylnF are involved in the synthesis of siroheme, a cofactor of sulfite reductase (9). The other two ORFs of the operon encode proteins with homology with enzymes of the cysteine biosynthetic pathway: the product of ylnB is similar to sulfate adenylyltransferase, and that of ylnC is similar to adenosine-5-phosphosulfate kinase. The deduced amino acid sequence of the product of ylnE displays homology with the CbiX protein of Bacillus megaterium, a putative precorrin-2-cobalt-chelatase involved in cobalamin biosynthesis (25), and with NirR of Staphylococcus carnosus (20).
FIG. 1.
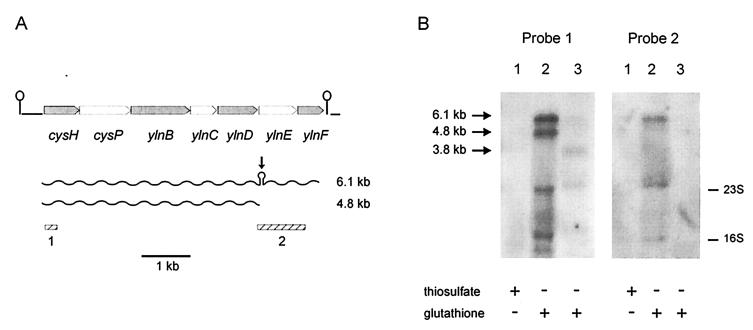
(A) Genetic organization of the B. subtilis cysH operon. The arrows indicate the direction of gene transcription. The secondary structures upstream of cysH and downstream of ylnF represent the putative ρ-independent transcriptional terminators of the pyr operon and the cysH operon, respectively. Hatched bars, DNA fragments used as probes in the Northern blot experiments. The major RNA species observed in the Northern blot experiment shown in panel B are indicated. Vertical arrow, putative site of RNA processing. (B) Northern blot analysis of the cysH operon. B. subtilis JH642 was grown in sulfate-free minimal media supplemented with 0.5 mM sodium thiosulfate (lane 1) or 1 mM glutathione (lane 2) as sulfur sources. Strain MC2620, a JH642 derivative containing a Tn917 insertion in cysH, was grown in sulfur-free minimal media supplemented with 1 mM glutathione (lane 3). Arrows, apparent sizes of the transcripts detected. The 2.4- and 1.3-kb signals correspond to 23S and 16S RNA, not efficiently denatured, which captured cysH mRNA.
Grundy and Henkin (7) reported the presence of 11 copies of a highly conserved motif in the genome of B. subtilis. In all cases, this motif was located in the leader region of putative transcriptional units, upstream of coding sequences that included genes involved in methionine or cysteine biosynthesis. This motif includes an element resembling an intrinsic transcriptional terminator, suggesting that regulation might be controlled at the level of premature termination of transcription. On the basis of mutational analysis of the leader region of the methionine-regulated gene yitJ, Grundy and Henkin (7) proposed a model in which the 5′ portion of the leader forms an antiantiterminator structure, which sequesters sequences required for the formation of an antiterminator, which, in turn, impairs formation of the terminator. The antiantiterminator would be stabilized by the binding of some unknown factor when methionine is available. This set of genes, including the cysH operon, was proposed to form a regulon controlled by a global termination system, which was designated the S box system, as most of the genes are involved in sulfur metabolism (7).
In the work presented here we analyzed the transcription of the cysH operon under a variety of nutritional conditions. We propose that cysteine biosynthesis in B. subtilis is negatively regulated by a still-unknown transcriptional repressor. We also suggest that cysteine downregulates the pathway by inhibiting the synthesis of OAS, a direct cysteine precursor and possibly an inducer of gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. E. coli and B. subtilis strains were routinely grown in Luria-Bertani broth (28). Antibiotics were added to media at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; erythromycin, 1 μg ml−1; spectinomycin, 100 μg ml−1. Spizizen salts (30), supplemented with 0.4% glucose and the required l-amino acids for particular strains, were used as the minimal medium for B. subtilis. In nutritional studies where different sulfur sources were tested, MgSO4 and (NH4)2SO4 were replaced by equimolecular amounts of MgCl2 and NH4Cl, respectively (sulfur-free minimal media). Glutathione (1 mM), Na2S2O3 (0.5 mM), methionine (1 mM), and cysteine supplied as cystine (1 mM) were used as sulfur sources for B. subtilis. Medium was adjusted to pH 6.8 before being autoclaved in experiments testing in vivo effects of OAS to minimize the conversion of OAS to NAS.
TABLE 1.
Bacterial strains
| Strain | Relevant characteristicsa | Source or reference |
|---|---|---|
| B. subtilis | ||
| JH642 | trpC2 pheA1 | Laboratory stock |
| MC530 | JH642 amyE::pMC530 | This study |
| MC374 | JH642 amyE::pMC374 | This study |
| MC328 | JH642 amyE::pMC328 | This study |
| MC276 | JH642 amyE::pMC276 | This study |
| MC181 | JH642 amyE::pMC181 | This study |
| MC311 | JH642 amyE::pMC311 | This study |
| BR151 | lys-3 metB10 trpC2 | BGSCb |
| MC-MET530 | BR151 amyE::pMC530 | This study |
| MC-MET249 | BR151 amyE::pMC249 | This study |
| MC-MET276X | BR151 amyE::pMC276X | This study |
| 1A3 | cysE trpC2 purA26 | BGSC |
| 1A3-530 | 1A3 amyE::pMC530 | This study |
| MC2620 | JH642 cys::Tn917 Emrlac+ | 14 |
| MCD80 | MC2620 p917::Sp Ems Sprlac | This study |
| MCD530 | MCD80 amyE::pMC530 | This study |
| E. coli | ||
| DH5α | supE44 thi-1 ΔlacU169 (φ80lacZΔM15) endA1 recA1 hsdR17 gyrA96 relA1 | Laboratory stock |
Emr and Spr, resistance to erythromycin and spectinomycin, respectively; Ems, susceptibility to erythromycin.
Strain obtained from the Bacillus Genetic Stock Center.
RNA analysis.
Total B. subtilis RNA from cells grown in minimal media with different sulfur sources was isolated as previously described (26). For Northern blot analyses, 10 μg of total RNA was separated under denaturing conditions in a 1.2% agarose gel containing 1.1% formaldehyde and transferred to Hybond C membranes (Amersham). RNA blots were hybridized with 32P-labeled DNA fragments using random hexanucleotides as primers for Klenow DNA polymerase. Probe 1 consisted of a 290-bp HincII-BglII fragment from pBS175 (14) which included the 5′ end of cysH. Probe 2 was a 1,010-bp fragment obtained by PCR amplification from the chromosome of strain JH642 using the oligonucleotides EcoCE and HindCE (Table 2). It included ylnE and the 5′ end of ylnF. Hybridization of the membranes was carried out at 65°C in a solution containing 5× SSPE (0.9 M NaCl, 0.05 M sodium phosphate [pH 7.7], 5 mM EDTA), 0.5% sodium dodecyl sulfate (SDS), 5× Denhardt's solution, and 20 mg of salmon sperm DNA liter−1. Final washing of the membranes was conducted at 65°C in 0.1× SSPE–0.1% SDS. Radioactive signals were detected by exposing the nitrocellulose membranes to X-OMAT film. The size of the cys transcript was determined by comparison with RNA molecular weight standards (Promega). Equivalent loadings of RNA on blots were verified by probing the membranes with probe 3, a 397-bp fragment complementary to the B. subtilis accB gene (16). Probe 3 was obtained by PCR amplification from the chromosome of strain JH642 using the oligonucleotides ACCB1 and ACCB2 (Table 2). Hybridization and washing conditions were the same as described above.
TABLE 2.
Oligonucleotide primers
| Primer | Sequencea |
|---|---|
| CYS116 | ACTCAggtACCGATTATGATACGC |
| CYS269 | GCTaCAAAagCTTGCTGTACTTGAAAAC |
| CYS314 | TTTAATTAAgCtTATAAAAAAAGTCGGG |
| CYS363 | GCTAAAAcaagCTtCAAATCTAAAAACTTATC |
| CYS228 | TGcgcTAAAgctTAAAAAATAAATTCAAAT |
| CYS326 (REV)b | CTcTCgTAAtaggAtCCGACTTTTTTTATAG |
| CYS662 (REV) | CGTTGGAtCcTCCCAATTATC |
| CYS382 (REV) | AGTCCCggAtCCGCTCTTGATAAGTTTTTAG |
| EcoCE | AACATTAGAATTCCGAGTTTAATTGTCATTGG |
| HindCE (REV) | GCAGCGTGCTTGAAGCTTGTGCAGC |
| ACCB1 | AATTTCAAGCTTAGGAGTGCGATTGCCCTGTTAAATATC |
| ACCB2 (REV) | AAGCTTCATGGATCCTACAATGCAGACAACTGTGTTTTC |
Sequences are shown 5′ to 3′. Lowercase letters show variation from the wild-type sequence. Restriction sites are underlined.
REV, primers oriented in reverse complement to the gene sequence.
Genetic techniques.
Plasmid preparations, restriction enzyme digestions, and agarose gel electrophoresis were carried out according to methods described by Sambrook et al. (28). E. coli competent cells were transformed with supercoiled plasmid DNA by using the calcium chloride procedure (28). Transformation of B. subtilis was carried out by the method of Dubnau and Davidoff-Abelson (5). The amy phenotype was assayed, with colonies grown during 48 h in Luria-Bertani starch plates, by flooding the plates with 1% I2-KI solution (29). amy+ colonies produced a clear halo, while amy colonies gave no halo.
Construction of lacZ fusions.
To construct transcriptional fusions of different regions of the 5′ ends of cysH and lacZ, the integrational plasmid pJM116 was used (4). DNA fragments were generated by PCR using the oligonucleotides shown in Table 2. Plasmid pMC2 (14), which contains the wild-type cysH promoter and the complete mRNA leader region, was used as the template DNA. Fusion constructs were verified by DNA sequencing. The cysH-lacZ wild-type fusion contained in pMC530 was constructed using primers CYS116 and CYS662 (Table 2), which generate KpnI and BamHI sites located 530 bp upstream and 23 bp downstream of the translational start of cysH, respectively. For the construction of plasmids pMC374, pMC328, and pMC276, primer CYS662 was used in combination with primers CYS269, CYS314, and CYS363, which generate HindIII sites at 374, 328, and 276 bp upstream from the translational start of cysH, respectively, with the same end point as that of the wild-type fusion. Plasmid pMC181 is a derivative of pMC530 in which the cysH sequences upstream of the EcoRI site located at position −181 of the translational start point were deleted. The xyl promoter (10) from plasmid pRDC9 was cloned into the EcoRI-HindIII sites of pMC276, generating plasmid pMC276X, which contains the S box sequences downstream of the xyl promoter. The fusion contained in plasmid pMC311 was generated using primers CYS116 and CYS326, which generated HindIII and BamHI sites 530 and 311 bp upstream of the cysH translational start, respectively. The fusion contained in plasmid pMC249 was generated using primers CYS228 and CYS382, which generated HindIII and BamHI sites 414 and 249 bp upstream of the cysH translational start, respectively, resulting in deletion of helices 1 to 5 of the S box.
Construction of strain MCD80.
Strain MCD80 was constructed by replacing the Tn917LTV1 insertion of MC2620 (14) with a 2-kb mini-Tn917 element, by recombination with plasmid p917::Sp (31). After transformation of MC2620 (Sps Emr Cmr lac+) with p917::Sp, a Spr Ems Cms colony that no longer expressed lacZ was selected. This strain was named MCD80.
β-Galactosidase assays.
B. subtilis strains harboring cysH-lacZ fusions were grown in sulfur-free minimal media containing either glutathione (as a poor sulfur source) or cysteine (as a rich sulfur source). Cells were collected by centrifugation and resuspended in sulfur-free minimal media supplemented with the appropriate compounds, as described in Results. Samples were taken at 1-h intervals after resuspension and assayed for β-galactosidase activity as previously described (14). The specific activity was expressed in Miller units (MU) (17).
Computer analysis.
RNA secondary structure predictions were made with the MFold program (version 3.0) (34), using the Macfarlane Burnet Centre for Medical Research resources. Nucleotide and protein sequences were analyzed using the BLAST and PSI-BLAST programs (1, 2). DNA sequence pattern searches were done with the SEARCH PATTERN tool of the SubtiList World Wide Web server (19). A neural network-based program was used to find possible transcription promoters (27).
RESULTS
cysH belongs to a 6-kb operon.
The clustering of the cysH, cysP, ylnB, ylnC, ylnD, ylnE, and ylnF genes (Fig. 1A) and their functional relation suggest that these seven genes form a single transcriptional unit. To verify the operon structure and to test whether its transcription is regulated by sulfur limitation, Northern blot analysis was performed. Total cellular RNA was isolated from cultures of strain JH642 (Table 1) grown on glutathione or thiosulfate as sulfur source. The RNA was hybridized with two 32P-labeled probes. Probe 1 included a 290-bp fragment covering the 5′ end of cysH, while probe 2 covered a 1-kb fragment including ylnE and the 5′ end of ylnF (Fig. 1A). The larger RNA species detected with probe 1 also hybridized with probe 2 (Fig. 1B). This mRNA species could correspond to a 6.1-kb transcript starting upstream of cysH and ending downstream of ylnF. These signals were not evident when the cells were grown with thiosulfate as a sulfur source (Fig. 1B, probes 1 and 2, lane 1), confirming previous results of transcriptional regulation of the cysH gene based on β-galactosidase assays (14). In addition, these experiments show that expression of the genes contained in the 6.1-kb operon is clearly coordinately regulated with that of cysH.
Probe 1 also revealed another RNA species of 4.8 kb in conditions of sulfur starvation (Fig. 1B). The differential pattern obtained with both probes could be due to the processing of the 6.1-kb transcript. In fact, analysis of the DNA sequence of the operon using the MFold program (34) predicted the formation of a complex secondary structure that could be a cleavage site for endoribonucleases between ylnD and ylnE (Fig. 1A). Cleavage at this structure could yield RNA species of 4,794 and 1,280 bp. The inability of probe 2 to detect the 1.2-kb RNA species could be due to instability of the transcript (Fig. 1B, probe 2, lane 2).
Induction of cysH expression in response to cysteine starvation.
To monitor the response of the cysH operon to nutritional conditions, a cysH-lacZ transcriptional fusion bearing the complete leader region, postulated to be involved in regulation of sulfur metabolism, was constructed (plasmid pMC530; Fig. 2). Construction of strain MC530 (Table 1), containing an insertion of this fusion into the amyE locus, permitted measurement of β-galactosidase expression without perturbation of the cysH gene function. As shown in Table 3, expression of this fusion was induced by starvation of cysteine. Induction was on the order of 4.9-fold after 4 h of starvation. Starvation for phenylalanine under similar conditions gave no induction, indicating at least partial specificity (data not shown). In cells induced by growth on minimal medium containing a poor sulfur source, such as glutathione, the level of β-galactosidase activity decreased after the addition of cystine to the growth media (data not shown). As shown in Table 3, the strains containing the ectopic lacZ fusions showed somewhat high levels of β-galactosidase activity in the presence of cystine. We have previously reported that a cysH-lacZ fusion introduced into the cysH locus causes sevenfold-llower levels of β-galactosidase activity and is strongly repressed in the presence of cystine (14). Therefore, the chromosomal location of the cysH promoter seems to have some effect on cysH expression.
FIG. 2.
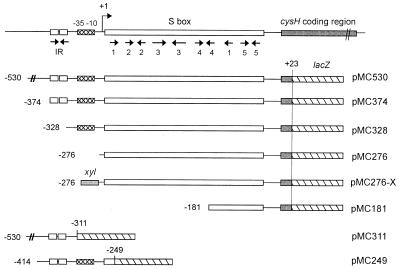
Construction of cysH-lacZ fusions. The 5′ end of the wild-type cysH is shown at the top. The cysH-lacZ fusion in each plasmid is shown from the 5′ end of the cysH promoter upstream region to lacZ. Nucleotide numbers are given starting from the cysH translation initiation site as +1. Crosshatched bars, cysH promoter; numbered arrows, helical regions homologous to S box sequences; hatched bars, lacZ-coding region; IR, putative pyr operon transcriptional terminator (24); xyl, xylose promoter.
TABLE 3.
Induction of cysH-lacZ fusions by cysteine starvation
| Strain | β-Galactosidase
activitya (MU)
|
Induction ratiob | |
|---|---|---|---|
| − Cystine | + Cystine | ||
| MC530 | 2,062 | 420 | 4.9 |
| MCD530 | 1,806 | 510 | 3.5 |
| MC374 | 950 | 210 | 4.5 |
| MC311 | ndc | nd | |
| MC276 | nd | nd | |
| MC181 | nd | nd | |
Cells were grown in sulfur-free minimal media containing cystine; the culture was then washed, split, and grown in the presence (+) or absence (−) of cystine. Samples were taken 4 h after cultures were split.
Induction ratio was defined as the ratio of β-galactosidase activity of cells growing in the absence of cystine to that of cells growing in the presence of cystine.
nd, not detectable.
It has been proposed that the B. subtilis cysH operon forms part of a regulon, together with genes involved in methionine biosynthesis, whose expression is controlled by a global termination system (7). As cysteine is a precursor of methionine (22), it became unclear whether the regulation of the cysH-lacZ fusion in strain MC530, which is a methionine prototroph, was a result of cysteine or methionine starvation. To answer this question, the cysH-lacZ fusion contained in pMC530 was introduced into strain BR151 (Table 1), which is unable to synthesize methionine owing to a mutation in the metB gene, encoding homoserine O-acetyltransferase. This strain was named MC-MET530 (Table 1). Expression of the fusion contained in strain MC-MET530 was induced sixfold in the absence of both methionine and cystine (Table 4). The addition of cystine or cystine plus methionine to the growth media repressed the expression of the cysH-lacZ fusion (Table 4). However, methionine, in the absence of cystine, could only partially repress cysH transcription, reducing β-galactosidase values by 30% (Table 4). These results show that transcription of the cysH operon is tightly repressed by cystine but not by methionine, thus suggesting that regulation of expression of the cysH operon is different from that of other members of the S box, which are strongly repressed by methionine (7).
TABLE 4.
Effect of leader deletions on cysH expression
| Strain | β-Galactosidase
activitya (MU)
|
|||
|---|---|---|---|---|
| − Sulfur | + Cystine | + Metb | Met + cystine | |
| MC-MET530 | 1,034 | 174 | 658 | 119 |
| MC-MET249 | 715 | 290 | 490 | 282 |
| MC-MET276Xc | 192 | 229 | 194 | 241 |
| MC328 | 1,135 | 1,290 | —d | — |
Cells were grown in sulfur-free minimal media containing cystine and methionine; the culture was then washed, split, and grown in the presence (+) or absence (−) of sulfur sources. Samples were taken 4 h after cultures were split.
Met, methionine.
In experiments performed with strain MC-MET276X, 0.2% xylose was added to the growth media.
—, not determined.
YlnE is not required for cysH transcription.
The products of all the ORFs included in the cysH operon, except ylnE, display obvious homology to known proteins involved in cysteine biosynthesis. The deduced amino acid sequence of YlnE shows low similarity to that of NirR of S. carnosus, a protein necessary for transcription of the nir operon, which is involved in nitrite reduction (20). Interestingly, S. carnosus nirR is located between two genes, sirB and sirA, which are similar to ylnD and ylnF of B. subtilis, respectively (20). However, unlike NirR, YlnE does not display homology to the central domain of the transcriptional activator NifA of Bradyrhizobium japonicum (residues 349 to 419), which is involved in an interaction with ς54 (18). To test whether YlnE could be a transcriptional activator, as is its homolog NirR, we constructed strain MCD80, as described in Materials and Methods. This strain, a derivative of MC2620 (Table 1), possesses a Tn917 insertion disrupting cysH that exerts a polar effect on the expression of ylnE, as seen in Northern blot experiments (Fig. 1B, probes 1 and 2, lane 3). MCD80 was transformed with pMC530 to yield MCD530. We have found that the expression levels of the cysH-lacZ fusion contained in ylnE strain MCD530 were essentially the same as those found in ylnE+ strain MC530 growing in minimal media supplemented either with a poor sulfur source or with cystine (Table 3). These results indicate that YlnE is not required for transcriptional regulation of the cysH promoter.
Analysis of the 5′ region of the cysH promoter required for sulfur regulation.
To determine how the cysH operon is regulated at the level of transcription, we attempted deletion analysis of the upstream region of cysH. Several cysH-lacZ transcriptional fusions were constructed using promoterless lacZ vector pJM116 as shown in Fig. 2. One series contained fragments of the cysH upstream region of different lengths, from bp −530, −374, −276, and −181, all of them ending at bp +23 of the cysH start codon (Fig. 2); the fragments were contained in plasmids pMC530, pMC374, pMC276, and pMC181, respectively. The fusion contained in pMC311 covers the cysH upstream region from bp −530 to −311 of the cysH translation initiation site (Fig. 2). These fusions were recombined into the amyE locus of strain JH642. Transformed cells were grown in sulfur-free minimal media containing cystine, and the induction of β-galactosidase activity after cystine starvation was measured. As seen in Table 3, while strains containing plasmids pMC530 and pMC374 showed similar induction levels after sulfur deprivation, strains bearing fusions contained in plasmid pMC181, pMC311, or pMC276 did not show β-galactosidase activity under similar conditions. These results indicate that the sulfur-regulated promoter lies between bp −311 and −276 upstream of the cysH ATG start codon (nucleotides [nt] 341 and 376 of the sequence shown in Fig. 3). The position deduced for the functional promoter coincides with the putative ςA type promoter previously suggested by Grundy and Henkin (7). The transcription initiation site would be located 278 bp upstream of the cysH start codon, thus confirming the previous assumption that the cysH operon contains an RNA leader region with strong homology with the other S box leaders (7).
FIG. 3.
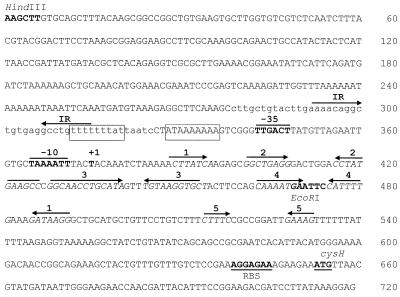
Nucleotide sequence of the cysH upstream region. The −10 and −35 regions, the transcriptional initiation site (+1), and endonuclease restriction sites are in boldface. Numbered arrows, positions of inverted repeats corresponding to the helical regions homologous to S box sequences; lowercase letters, deleted region in plasmid pMC328; boxes, putative cysH operator. The putative cysH ribosome binding site (RBS) and the cysH start point are underlined. IR, putative pyr operon transcriptional terminator (24).
The leader region is not required for cysteine repression of cysH transcription.
It was proposed that the leader region of the S box family folds into two alternative structures (7). One of these structures contains an intrinsic transcriptional terminator (helix 5, Fig. 4), preceded by a complex structure. The pairing of the 3′ segment of helix 1 with sequences on the 5′ side of the terminator results in the formation of an antiterminator structure. Deletion of the terminator involved in premature transcription termination resulted in constitutive, high-level expression of the B. subtilis methionine-controlled yitJ gene (7). To test whether deletion of the putative terminator located in the 5′ end of the cysH gene results in constitutive expression of the operon, we constructed plasmid pMC249, which lacks DNA sequences downstream of helix 1 of the cysH leader region (Fig. 2). Expression of this fusion in the met strain MC-MET249 (Table 1) was repressed by cystine (Table 4). The addition of methionine to the growth medium, in the absence of cystine, resulted in a slight reduction of β-galactosidase activity in a pattern very similar to that observed in strain MC-MET530, which possesses a cysH-lacZ fusion containing the complete S box (Table 4). These results indicate that (i) the response to cysteine starvation was independent of the putative transcription terminator and (ii) the slight repression of cysH transcription mediated by methionine is not related to the S box sequences. The effect of methionine on cysH expression could be attributed to cysteine production derived from the sulfur moiety provided by methionine degradation (32).
FIG. 4.
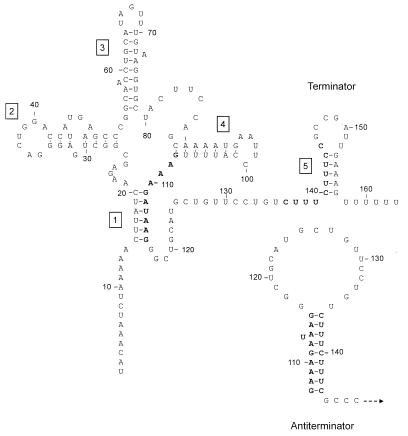
Structural model of cysH leader RNA. Numbers indicate positions relative to the probable transcription initiation site. Helical regions are labeled to correspond with the inverted repeats shown in the sequence of the cysH upstream region (Fig. 2). The cysH leader could form two alternative structures: terminator and antiterminator forms. The terminator form is shown. Helix 1 is the antiantiterminator, while helix 5 is the intrinsic terminator. The antiterminator is derived from pairing sequences on the 3′ side of helix 1 with sequences on the 5′ side of helix 5 (boldface). Dashed arrow, continued transcription.
We also determined that the expression of a lacZ fusion in strain MC-MET276X (Table 1), which contains the cysH putative S box (helices 1 to 5, Fig. 4) downstream of the xyl promoter, instead of the cysH promoter, was not regulated by the sulfur source (Table 4). This result confirms that the cysH promoter is required for sulfur regulation and that neither cysteine nor methionine controls the cysH operon at the level of premature termination of transcription.
To gain insight into the mechanism of regulation of the cysH operon, we introduced a series of deletions upstream of the cysH promoter (data not shown). We found that strain MC328 (Table 1), which contains a cysH-lacZ fusion with a deletion of the 5′ region adjacent to the −35 sequence of the promoter but which comprises the complete S box sequences, shows constitutive high-level expression during growth in the presence or in the absence of cystine (Table 4). This result strongly suggests that the region between nt −96 and −50 of the cysH transcription initiation site may act as a site of interaction for a still-unknown negative regulatory factor. This deleted region (lowercase letters) comprises the middle of a 25-nt symmetric sequence (5′-TTTTTTTATtaatcctATAAAAAAA-3′) (Fig. 3). DNA sequences with twofold rotational symmetry have been reported to act as operator sites that are recognized by symmetric proteins (23). Thus, this DNA sequence positioned between nt −63 and −39 of the transcription initiation site is a good candidate to be the cysH operator site.
OAS is an inducer of expression of the cysH operon.
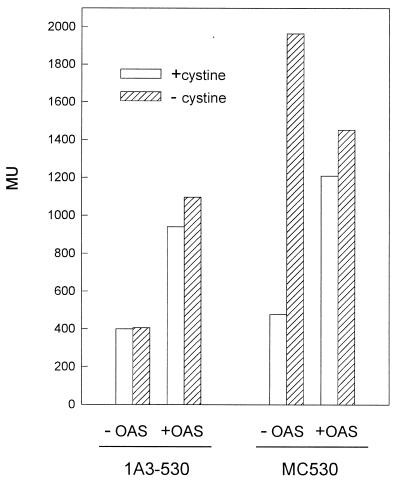
Transcription regulation of the E. coli cysteine regulon occurs at the level of initiation, and the presence of both NAS and the regulator CysB are necessary for transcription activation (11). To test whether OAS is required for expression of the B. subtilis operon, we introduced the cysH-lacZ fusion contained in plasmid pMC530 into strain 1A3, which is an auxotroph for OAS. The resultant strain was named 1A3-530 (Table 1). As seen in Fig. 5, the β-galactosidase levels of strain 1A3-530 are low for both sulfur-starved and cystine-supplemented cells, indicating that in the absence of OAS synthesis expression of cysH is not induced by sulfur deprivation. Addition of OAS increases 2.7-fold the expression of the cysH-lacZ fusion by sulfur-starved cultures of strain 1A3-530 (Fig. 5). This OAS-mediated cysH transcriptional activation is not repressed by cystine (Fig. 5). Thus, the OAS effect on cysH transcription is independent of sulfur starvation and insensitive to cystine repression. It therefore appears that OAS, or its derivative NAS, is necessary for transcription from the cys promoter. These results also indicate that cystine is unable to repress transcription of the cysteine biosynthetic genes in cells which are continuously exposed to the transcriptional activator OAS. This assumption was confirmed using strain MC530 (Table 1), which is able to synthesize OAS. In this case exogenous OAS did not increase the β-galactosidase activity of sulfur-starved cells but reduced, to some extent, the repression of cysH expression by the addition of cystine, causing a 2.5-fold induction of transcription (Fig. 5). Therefore, cysteine may exert a negative effect on the expression of the cysH operon via interference with OAS synthesis rather than directly acting as a repressor (or corepressor) of the transcription of the genes involved in the cysteine biosynthetic pathway.
FIG. 5.
Effect of OAS on cysH expression. Cells of strains MC530 (cysE+) and 1A3-530 (cysE) were grown in sulfur-free minimal media in the presence of cystine. Cells were collected by centrifugation and resuspended in sulfur-free minimal media in the presence or absence of OAS (1 mM). Samples were taken 4 h after resuspension.
DISCUSSION
In this study we have shown that in B. subtilis the genes cysH, cysP, ylnB, ylnC, ylnD, ylnE, and ylnF form part of an operon which is transcriptionally regulated by the sulfur source. This operon, which we named cysH because this gene was the first one functionally characterized in the cluster (14), is induced by sulfur starvation and tightly repressed by cysteine. Deletion analysis of the upstream region of cysH confirmed the suggestion that the sulfur-regulated promoter is located upstream of a 278-nt leader region with strong homology to the S box family (7). It has been reported that this leader region is crucial for regulation of the yitJ gene, which is presumably involved in the synthesis of methionine (7). Analysis of the yitJ gene revealed that its expression is induced by methionine starvation and that induction is independent of the promoter and dependent on the leader region terminator (7). Thus, it was proposed that cysH operon expression could be regulated by a global transcription termination control analogous to that suggested for methionine biosynthesis. In this work we have demonstrated that expression of the cysH operon is not induced by methionine starvation and does not require the S box leader region. This could be due to the less-negative value of predicted free energy of formation (ΔG°) of the cysH terminator (ΔG° = −1.5 kcal mol−1) compared with those of other members of the S box (yitJ terminator ΔG° = −7.6 kcal mol−1; ykrT terminator ΔG° = −18 kcal mol−1). As the cysH antiterminator possesses a considerably negative predicted ΔG° (−8.9 kcal mol−1), termination would be impaired, independently of the available sulfur sources. Therefore, although the cysH operon has been classified as a member of the S box family, its expression is controlled by a mechanism different from that proposed for the S box regulon. In addition, no nucleotide sequence similarity to the leader regions of the S box genes was found in the upstream regions of the B. subtilis yvgR and yvgQ genes, which are similar to the subunits of the E. coli sulfite reductase (13), or ssuB genes, involved in sulfonate utilization (J. R. van der Ploeg and T. Leisinger, Abstr. 10th Int. Conf. Bacilli, abstr. P25, 1999). This suggests that the S box transcriptional antitermination system is not implicated in the control of the first steps of assimilation of organic and inorganic sulfur sources to yield cysteine.
We also report here that the region upstream of the cysH promoter from bp −96 to −50 of the transcription initiation site is required for sulfur repression of the operon. Deletion of this 46-bp region causes the loss of half of a nearly symmetric sequence (Fig. 3), resulting in constitutive expression of a cysH-lacZ fusion (MC328; Table 4) even in the presence of cysteine. This result demonstrates that this region acts as a negative cis element. Therefore, we propose that a cis-acting sequence located upstream of the −35 region of the cysH promoter is a target (an operator) for a transcriptional repressor.
Among the genes cotranscribed with cysH, ylnE shows no obvious homology with genes involved in sulfur utilization. Although the deduced amino acid sequence of YlnE shows homology with the transcriptional activator NirR of S. carnosus, here we demonstrated that its presence is not essential for cysH transcription. We also found that YlnE shows homology with the CbiX protein of B. megaterium (GenBank accession no. CAA04308; 33% identity, 51% homology). In B. megaterium this protein is involved, together with cysG, in cobalt insertion into uroporphyrinogen III to form Co-precorrin-2, a precursor of cobalamin (25). The genes ylnD and ylnF of B. subtilis, which are located upstream and downstream of ylnE, respectively, code for two proteins homologous to the carboxy and amino termini of the product of E. coli cysG, respectively (33). The physical proximity of these three genes in B. subtilis and the fact that no other ORF in its genome displays homology to CbiX suggest that YlnE could participate in cobalamin biosynthesis. Functional characterization of YlnE as well as of the physiological role of the RNA endonucleolytic cleavage that seems to take place between ylnD and ylnE (Fig. 1A) remains to be determined.
Another important conclusion from the present work is that OAS increases the expression of the B. subtilis cysH operon. Moreover, the transcriptional activation of a cysH-lacZ fusion produced by the addition of OAS to a B. subtilis OAS auxotroph was not blocked by the simultaneous addition of cysteine (Fig. 5). In S. enterica serovar Typhimurium cysteine and sulfide almost completely abolish induction of the cysteine regulon by exogenous OAS (21), indicating that both OAS and sulfur limitation are necessary for derepression. Thus, it was suggested that cysteine, sulfide, or some other sulfur metabolite interferes with the effect of the inducer in S. enterica serovar Typhimurium. If OAS is an inducer of cysH expression in B. subtilis, the negative effect of cysteine on cysH transcription could be due to interference with the synthesis of OAS. It is worth noting that expression of the B. subtilis cysE gene coding for serine transacetylase, the enzyme that catalyzes the acetylation of l-serine by acetyl-coenzyme A to give OAS, is repressed by a cysteinyl-tRNA-directed transcription termination mechanism (6).
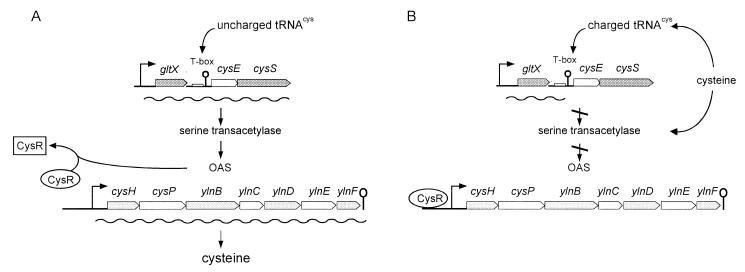
In conclusion, we propose that in the regulation of cysteine biosynthesis the inhibition of transcription of serine transacetylase by cysteine is coupled with a system of negative gene regulation, in which OAS derepresses the expression of the cysH promoter (Fig. 6). In conditions of cysteine starvation the uncharged cysteinyl-tRNA interacts with the leader region of the nascent transcript of cysE (T box; Fig. 6) to promote the formation of the antitermination structure, preventing terminator formation (8). Expression of cysE should result in an increase in the intracellular levels of OAS, which in turn would act as a cysH inducer, possibly by promoting the dissociation of a transcriptional repressor from its operator (CysR; Fig. 6), allowing high-level expression of the operon. When cysteine becomes available the aminoacylation levels of cysteinyl-tRNA should increase; this would promote transcription termination of cysE (Fig. 6). In addition, cysteine could act as an inhibitor of serine transacetylase activity, as has been reported for S. enterica serovar Typhimurium (3), lowering OAS synthesis. The decreased intracellular levels of OAS would allow binding of CysR upstream of the cysH operator, shutting off the transcription of the cysH operon. Further analysis of this control mechanism, as well as the identification of the putative repressor, will be of great interest.
FIG. 6.
A model for regulation of expression of the cysH operon. (A) Transcriptional induction of cysH. In the absence of available sulfur sources, such as cysteine, sulfide, and thiosulfate, uncharged tRNACys would interact with the leader region nascent transcript of cysE, which codes for serine transacetylase, to promote formation of the antitermination structure, preventing terminator formation. cysE would be transcribed, and OAS would be synthesized. OAS would interact with a putative repressor (CysR), inactivating it. RNA polymerase initiates transcription of cys structural genes. (B) Transcriptional repression of cysH. In the presence of available reduced sulfur sources premature termination of the cysE leader is favored, impairing OAS synthesis. The binding of the repressor to the cysH operator prevents transcription of the cys genes.
ACKNOWLEDGMENTS
We gratefully acknowledge R. Raya for the gift of Macaloid clay for RNA extraction, and F. Arigoni for the gift of pRDC9. We thank T. Henkin for helpful advice.
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia Nacional de Promoción Científica y Tecnológica (FONCYT), and Fundación Antorchas. M. C. Mansilla is a fellow from Univ. Nac. de Rosario, D. Albanesi is a fellow from Fundación Antorchas, and D. de Mendoza is a Career Investigator from CONICET.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden L M, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker M A, Kredich N M, Tomkins G M. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J Biol Chem. 1969;244:2418–2427. [PubMed] [Google Scholar]
- 4.Dartois V, Djavakhishvili T, Hoch J A. Identification of a membrane protein involved in activation of the KinB pathway to sporulation in Bacillus subtilis. J Bacteriol. 1996;178:1178–1186. doi: 10.1128/jb.178.4.1178-1186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. 1. Formation and properties of the donor-recipient complex. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 6.Gagnon Y, Breton R, Putzer H, Pelchat M, Grunberg-Manago M, Lapointe J. Clustering and co-transcription of the Bacillus subtilisgenes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J Biol Chem. 1994;269:7473–7482. [PubMed] [Google Scholar]
- 7.Grundy F J, Henkin T M. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 8.Henkin T M. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 9.Johansson P, Hederstedt L. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiology. 1999;145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- 10.Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilisintegration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- 11.Kredich N M. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol Microbiol. 1992;6:2747–2573. doi: 10.1111/j.1365-2958.1992.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 12.Kredich N M. Biosynthesis of cysteine. In: Neidhart F C, Curtiss III R, Ingrahan J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 514–527. [Google Scholar]
- 13.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 14.Mansilla M C, de Mendoza D. l-Cysteine biosynthesis in Bacillus subtilis: identification and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J Bacteriol. 1997;179:976–981. doi: 10.1128/jb.179.3.976-981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansilla M C, de Mendoza D. The Bacillus subtilis cysPgene encodes a sulfate permease related to the inorganic phosphate transporter (Pit) family. Microbiology. 2000;146:815–821. doi: 10.1099/00221287-146-4-815. [DOI] [PubMed] [Google Scholar]
- 16.Marini P, Li S, Gardiol D, Cronan J E, Jr, de Mendoza D. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilisacetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J Bacteriol. 1995;177:7003–7006. doi: 10.1128/jb.177.23.7003-7006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Morett E, Segovia L. The ς54bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilisgenome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 20.Neubauer H, Pantel I, Gotz F. Molecular characterization of the nitrite-reducing system of Staphylococcus carnosus. J Bacteriol. 1999;181:1481–1488. doi: 10.1128/jb.181.5.1481-1488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrowski J, Kredich N. In vitro interactions of CysB protein with the cysJIH promoter of Salmonella typhimurium: inhibitory effects of sulfide. J Bacteriol. 1990;172:779–785. doi: 10.1128/jb.172.2.779-785.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulus H. Biosynthesis of the aspartate family of amino acids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 237–267. [Google Scholar]
- 23.Ptashne M. A genetic switch. 2nd ed. Malden, Ma: Cell Press and Blackwell Scientific Publications; 1992. pp. 33–48. [Google Scholar]
- 24.Quinn C L, Stephenson B T, Switzer R L. Functional organization and nucleotide sequence of the Bacillus subtilispyrimidine biosynthetic operon. J Biol Chem. 1991;266:9113–9127. [PubMed] [Google Scholar]
- 25.Raux E, Lanois A, Rambach A, Warren M J, Thermes C. Cobalamin (vitamin B12) biosynthesis: functional characterization of the Bacillus megaterium cbI genes required to convert uroporphyrinogen III into cobyrinic acid a,c-diamide. Biochem J. 1998;335:167–173. doi: 10.1042/bj3350167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raya R, Bardowski J, Andersen P S, Ehrlich S D, Chopin A. Multiple transcriptional control of the Lactococcus lactis trpoperon. J Bacteriol. 1998;180:3174–3180. doi: 10.1128/jb.180.12.3174-3180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese M G, Harris N L, Eeckman F H. Large scale sequencing specific neural networks for promoter and splice site recognition. In: Hunter L, Klein T E, editors. Biocomputing. Proceedings of the 1996 Pacific Symposium. Singapore, Singapore: World Scientific Publishing Co; 1996. pp. 737–738. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Sekiguchi J, Takada N, Okada H. Genes affecting the productivity of α-amylase in Bacillus subtilisMarburg. J Bacteriol. 1975;121:688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilisby deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinmetz M, Richter R. Plasmid designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 32.van der Ploeg J R, Cummings N J, Leisinger T, Connerton I F. Bacillus subtilisgenes for the utilization of sulfur from aliphatic sulfonates. Microbiology. 1998;144:2555–2561. doi: 10.1099/00221287-144-9-2555. [DOI] [PubMed] [Google Scholar]
- 33.Warren M J, Bolt E L, Roessner C A, Scott A I, Spencer J B, Woodcok S C. Gene dissection demonstrates that the Escherichia coli cysGgene encodes a multifunctional protein. Biochem J. 1994;302:837–844. doi: 10.1042/bj3020837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]