Summary
Different types of immune cells are involved in atherogenesis and may act atheroprotective or atheroprogressive. Here, we describe an in vitro approach to analyze CD11c+ cells and CD11c+-derived ApoE in atherosclerosis. The major steps include harvesting mouse bone marrow, plating cells in culture dishes, treating them with differentiation factors, and collecting cells after removal of undesirable populations. This protocol can be adapted for CD11c+ cells in different contexts, thus, serving as models for different diseases and to analyze cell-specific molecules.
For complete details on the use and execution of this protocol, please refer to Sauter et al. (2021).
Subject areas: Cell biology, Cell culture, Cell isolation, Flow cytometry/Mass cytometry, Health sciences, Immunology, Molecular biology
Graphical abstract
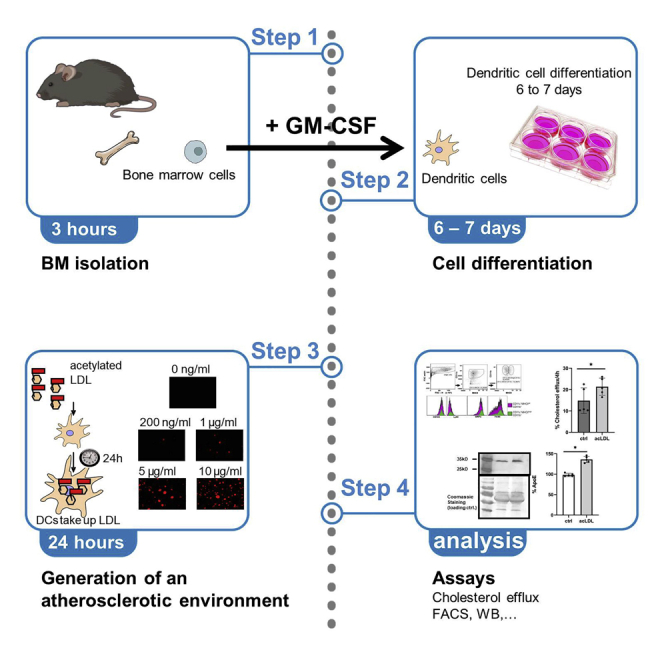
Highlights
-
•
Detailed protocol to isolate bone-marrow-derived dendritic cells (BMDC)
-
•
Exposing BMDC to an atherosclerotic environment in vitro
-
•
Suggestions to analyze BMDC in the context of atherosclerosis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Different types of immune cells are involved in atherogenesis and may act atheroprotective or atheroprogressive. Here, we describe an in vitro approach to analyze CD11c+ cells and CD11c+-derived ApoE in atherosclerosis. The major steps include harvesting mouse bone marrow, plating cells in culture dishes, treating them with differentiation factors, and collecting cells after removal of undesirable populations. This protocol can be adapted for CD11c+ cells in different contexts, thus, serving as models for different diseases and to analyze cell-specific molecules.
Before you begin
The protocol below describes the specific steps for the isolation and differentiation of CD11c+ cells from murine bone marrow and their exposure to an atherosclerotic environment in vitro. Experience in cell culture techniques is required.
Cells isolated from bone marrow can also be differentiated to other cell types, depending on the culture conditions and additives.
Institutional permissions
Experiments involving mice must conform to institutional and governmental guidance. The procedures in this protocol were approved by the Regierungspraesidium Tuebingen (approvals M03/10 and M16/15) and performed in accordance with the Guide for the Care and Use of Laboratory Animals and all other relevant ethical guidelines.
Preparation of cell culture medium
Timing: 30 min
-
1.
Add supplements required for cell culture of BMDC to 430 mL fresh RPMI-medium:
DC medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI – 1640 complete | N/A | 430 mL |
| Fetal Bovine Serum | 10% (v/v) | 50 mL |
| HEPES buffer | 1 mM | 5 mL |
| Penicillin Streptomycin 10,000 U/mL | 1% (v/v) | 5 mL |
| β-Mercaptoethanol | 50 μM | 1.8 μL |
-
2.
pre-warm medium in water bath at 37°C degrees.
DC medium can be stored at 4°C degrees for a maximum of 4 weeks. Check color of medium before usage. If the color changes from yellowish red to purple-pink (which indicates pH changes), discard the medium and make fresh one.
GM-CSF 20 ng/mL needs to be added freshly directly before cells are seeded into the 6 well plate.
Preparation of a 6 well plate for bone marrow isolation
Timing: 10 min
-
3.
Add 70% Ethanol to the first well of the plate (to disinfect the bones).
-
4.
Add approximately 1 mL medium to the second and to the third well (for washing the bones) and 3 mL medium to the fourth well of the plate (for flushing the bone).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Organisms/strains | ||
| C57BL/6J; 6–8 week old female and male mice can be used | The Jackson Laboratory | Stock no. 000664; RRID: IMSR_JAX:000664 |
| Antibodies | ||
| CD103 PerCP/Cy5.5 clone 2E7 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 121416; RRID: AB_2128621 |
| CD11b PE clone M1/70 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 101208; RRID: AB_312791 |
| CD11c APC clone N418 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 117310; RRID: AB_313779 |
| CD172a (SIRPα) PE/Dazzle™ 594 clone P84 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 144016 RRID: AB_2565280 |
| CD45 PB clone 30-F11 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 103125; RRID: AB_493536 |
| F4/80 PE/Cyanine7 clone QA17A29 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 157307 RRID: AB_2832550 |
| I-A/I-E MHCII FITC clone M5/114.15.2 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 107605; RRID: AB_313320 |
| IRDye® 800CW Goat anti-Rabbit IgG (Dilution: 1: 15.000) | LI-COR | Cat. no. 926-32211; RRID: AB_621843 |
| Ly-6C Alexa Fluor® 700 clone 1A8 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 127621; RRID: AB_10640452 |
| Rabbit monoclonal to Apolipoprotein E (Dilution: 1: 5.000) | Abcam | Cat. no. ab183596; RRID: AB_2832971 |
| TCR-b chain Brilliant Violet®510 clone H57-597 (Dilution: 3 μL in 100 μL FACS buffer) | BioLegend | Cat. no. 109233; RRID: AB_2562349 |
| Chemicals, peptides, and recombinant proteins | ||
| 4 × Laemmli sample buffer | Bio-Rad | Cat. no. 1610747 |
| Acetic acid | Merck (Sigma-Aldrich) | Cat. no. A6283 |
| acetylated LDL | Thermo Scientific | Cat. no. L35354 |
| Acrylamide Rotiphorese® Gel 30 | Carl Roth | Cat. no. 3029.1 |
| APS | Carl Roth | Cat. no. 9592.3 |
| Bovine Serum albumin fraction V | Merck (Sigma-Aldrich) | Cat. no. 10735094001 |
| Coomassie Brilliant Blue R-250 protein stain powder | Bio-Rad | Cat. no. 1610400 |
| Dulbecco’s PBS (DPBS) | Gibco | Cat. no. 14190-144 |
| EDTA, 0.5 M, pH 8.0 | Invitrogen | Cat. no. AM9260G |
| Ethidium bromide solution | Sigma-Aldrich | Cat. no. E1510-10ML |
| Fetal bovine serum | Thermo Scientific | Cat. no. 26140-079 |
| Formaldehyde solution 4%, buffered, pH 6.9 | Merck | Cat. no. 1004960700 |
| Glycine | Merck (Sigma-Aldrich) | Cat. no. G8898-500G |
| HEPES buffer | Gibco | Cat. no. 11360-070 |
| NaN3 | Merck (Sigma-Aldrich) | Cat. no. 2002 |
| Invitrogen Agarose UltraPure™ | Thermo Scientific | Cat. no. 16500500 |
| OneComp compensation beads | BD | Cat. no. 552845 |
| PBS without Ca and Mg | Gibco | Cat. no. 14190-169 |
| Penicillin and streptomycin | Sigma-Aldrich | Cat. no. P4333 |
| RBC Lysis Buffer (eBioscience™) | Thermo Scientific | Cat. no. 00-4333-57 |
| Recombinant mouse GMCSF protein | PeproTech | Cat. no. 415-ML-010/CF |
| RPMI 1640 medium | Thermo Scientific | Cat. no. 11875-093 |
| SDS blotting grade | Carl Roth | Cat. no. 0183.1 |
| Skimmed milk powder | AppliChem | Cat. no. A0830 |
| Streptavidin AP conjugate | Merck (Roche) | Cat. no. 11089161001 |
| TEMED | Carl Roth | Cat. no. 2367.4 |
| TRIS | Carl Roth | Cat. no. 4855.2 |
| Tween-20 | Merck (Sigma-Aldrich) | Cat. no. P9416 |
| X-Gal | Merck (Sigma-Aldrich) | Cat. no. 3117073001 |
| Zombie NIR™ Fixable Viability Kit, APC-Fire 750 | BioLegend | Cat. no. 423105 |
| β-Mercaptoethanol | Sigma-Aldrich | Cat. no. M6250 |
| Critical commercial assays | ||
| Cholesterol Efflux Kit (cell based) | Abcam | Cat. no. ab196985 |
| Mouse Apolipoprotein E ELISA Kit | Abcam | Cat. no. ab215086 |
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat. no. 23225 |
| Software and Algorithms | ||
| Cellquest Pro | BD Biosciences | https://www.bd.com/de-de/products/biosciences |
| GraphPad Prism 9.2.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | Wayne Rasband, NIH | https://imagej.nih.gov/ij/ |
| Kaluza Analysis 2.1 | Beckman Coulter | https://www.beckman.de/flow-cytometry/software/kaluza |
| Other | ||
| Cell strainer (70-μm pore size) | Corning | Cat. no. 431751 |
| Dissecting scissors | Fisher Scientific | Cat. no. 08-940 |
| Forceps | Fisher Scientific | Cat. no. 22-327379 |
| Greiner centrifuge tubes, 50 mL, 30 × 115 mm, conical (V) bottom, w/ graduations, I.D. field | Merck | Cat. no. T2318-500EA |
| Needle (27 gauge) | BD Biosciences | Cat. no. 305109 |
| Petri dish | Fisher Scientific | Cat. no. 07-202-011 |
| PVDF membranes (Immun-Blot PVDF Membrane) | Bio-Rad | Cat. no. 1620177 |
| Serological glass pipettes 5 mL, 10 mL, 25 mL, 50 mL | ROTILABO®, Carl Roth | N/A |
| Sterile syringe (10 mL) | BD Biosciences | Cat. no. 309604 |
| Tissue culture plates (6 well) | Corning | Cat. no. 3516 |
Materials and equipment
DC medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI – 1640 complete | N/A | 430 mL |
| Fetal Bovine Serum | 10% (v/v) | 50 mL |
| HEPES buffer | 1 mM | 5 mL |
| Penicillin Streptomycin 10,000 U/mL | 1% (v/v) | 5 mL |
| β-Mercaptoethanol | 50 μM | 1.8 μL |
10× TBS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| TRIS | 200 mM | 24 g |
| NaCl | 1500 mM | 88 g |
| H2O | N/A | 900 mL |
adjust the pH to 7.6 and final volume to 1 L.
TBST buffer (TBS + 0.05% Tween-20)
To make 1 l of TBST wash buffer, add 100 mL of 10× TBS and 500 μL Tween® 20 detergent to 900 mL of water.
Reducing Laemmli buffer
Add 100 μL of β-mercaptoethanol per 900 μL (final concentration of 355 mM).
For best results, do not store sample buffer with β-mercaptoethanol.
10% Polyacrylamide gels for SDS-PAGE
The required 10% separation gel was prepared as described in the following table. In this protocol, 4.5% stacking gels were used.
10% separation gel
| Reagent | Final concentration | Amount |
|---|---|---|
| 30% (w/v) Acrylamide | 10% | 3.3 mL |
| 1,5 M Tris-HCl pH 8.8 | 375 mM | 2.5 mL |
| H2O | N/A | 4.1 mL |
| 10% SDS | 0.1% | 100 μL |
| TEMED | 0.09% | 9 μL |
| 10% APS | 0.03% | 27 μL |
4.5% stacking gel
| Reagent | Final concentration | Amount |
|---|---|---|
| 30% (w/v) Acrylamide | 4.5% | 1.5 mL |
| 0,5 M Tris-HCl pH 6.6 | 375 mM | 2.5 mL |
| H2O | N/A | 5.9 mL |
| 10% SDS | 0.1% | 100 μL |
| TEMED | 0.2% | 20 μL |
| 10% APS | 0.06% | 60 μL |
After pouring the separating gel solution into the gel cassettes, overcoat with isopropanol to ensure a straight, smooth surface. After polymerization of the separating gels, remove the isopropanol, pipette the freshly prepared collection gel solution onto it and insert a comb. After complete polymerization of the collection gel, place the gel cassettes in the electrophoresis chamber that is filled with electrophoresis buffer. After removing the comb, pipette the samples into the gel pockets, which should be rinsed with buffer beforehand.
Step-by-step method details
Bone marrow cell isolation
Timing: 3 h
The following steps provide the procedure of collection and differentiation of bone marrow-derived cells into BMDCs.
Note: Ensure that all the reagents and samples are kept on ice during the entire procedure.
Perform all steps in a laminar flow hood with sterile equipment to maintain sterility.
For one cell culture 6 well microplate, you will need bone marrow from one mouse.
Note: We have found that mice aged 6–8 weeks provide ideal conditions for cultivation of BM derived CD11c+ cells. Young mice provide a good amount of good quality BM cells, while older mice provide more BM cells but the quality is not that good.
CRITICAL: Sterile technique is required for the isolation of BM cells; all tools should be rinsed with ethanol, and procedures should be performed in a laminar flow hood.
-
1.
Euthanize the mice with inhaled anesthetics (here, isoflurane was used: Inhalation of 5.0 relative vol. % in an anesthesia chamber at an oxygen flow of 1 L/min) or CO2, followed by cervical dislocation.
-
2.
Put the mouse abdomen down-faced on a towel sprayed with 70% ethanol. Sterilize the hind legs and back by spraying with 70% (vol/vol) ethanol. Pin the paws to a suitable surface (Styrofoam for example).
-
3.
Using scissors, cut the midline of the back and continue cutting to expose the femurs (see Figure 1A).
-
4.Unpin the right leg and peel the skin off toward the midline.
-
a.Cut the adductor muscles toward the midline and sever the leg between the hip joint and the spine (see Figure 1B).
-
b.Once the leg is detached from the body, separate the femur from the tibia by cutting the knee joint.
-
c.Cut away the paw and remove excess tissue (i.e., muscle and fat) from the bone, which works best by using paper towels and gently rubbing off the tissue.
-
d.Cut the paw off distal to the ankle, and peel off the remaining skin.
-
a.
CRITICAL: Avoid to cut the bones, carefully cut behind the epiphyses!
-
5.
Repeat step 4 for the left leg.
-
6.
Spray both legs with 70% (vol/vol) ethanol.
-
7.Collect femurs and tibiae
-
a.Working on a 70% (vol/vol) ethanol–soaked towel, hold the distal end of the tibia with a forceps so that the knee joint is pointing away from you and the leg is vertical.
-
b.Using the tip of slightly opened scissors, strip the calf tissue by pushing it down toward the knee.
-
c.Grip the tibia and femur with two sets of forceps and gently push against the direction of the knee joint until the tibia breaks right at the knee.
-
d.Remove any hanging tissue and place the tibia on the towel.
-
e.Recover the femur in the same way, snapping the knee off.
-
a.
-
8.
To remove any remaining flesh from the bones, hold the fleshy end of each piece in a dry paper towel and pull the bone with forceps, leaving the flesh in the towel. Then squeeze the remaining fleshy part in the towel and rub it with your fingers (holding it in the towel) to clean the bone off.
-
9.
Collect the bones in a small Petri dish filled with medium on ice.
CRITICAL: If the bone is broken or the soft tissue is inadequately removed, the number of bone marrow cells harvested may be decreased.
Note: DC medium can be stored at 4°C degrees for a maximum of 4 weeks. Check color of medium before usage. If the color changes from yellowish red to purple-pink (which indicates pH changes), discard the medium and make fresh one.
-
10.Disinfect and wash the bones
-
a.Soak the bones in 70% (vol/vol) ethanol in the first well of the prepared 6-well plate for 1 min.
-
b.Place them in a second well filled with 2 mL BMDC culture medium (see Materials and Equipment).
-
c.Move them on to a third well filled with 2 mL BMDC medium.
-
a.
CRITICAL: Broken bones should not be soaked in ethanol.
-
11.
Collect 10 mL fresh BMDC culture medium in a 10-mL syringe and attach a 27-gauge needle.
-
12.
Remove the tibia from the Petri dish and cut it off at the ankle joint at an angle.
-
13.Collect the bone marrow
-
a.Hold the tibia over a fresh 50-mL tube, with the narrow end of the tibia pointing down. Using the needle, gently squirt medium onto the tibia.
-
b.Insert the needle (gently at first) at the top end of the marrow and squirt medium.
-
c.Pull back the needle and insert it again. Squirt with brief, high pressure pushes.
-
d.Push the needle in further until the epiphysis has been crossed and medium comes out at the bottom end of the bone.
-
e.Continue flushing a couple more times; when the bone is white, discard it (Figure 1C).
-
a.
-
14.
Repeat steps 12 and 13 for the femur, cutting off at the hip joint just as the tibia was cut at the ankle joint and using the same 50-mL tube as was used for collection from the tibia.
-
15.
Add an additional 20 mL of BMDC culture medium to the 50-mL tube with a glass pipet and gently suspend the marrow in the BMDC medium with the pipet.
-
16.
Pass the cell suspension through a 70-μm cell strainer into a new 50-mL tube to remove any remaining bone or muscle fragments.
CRITICAL: If performing this step, ensure that the bone marrow is thoroughly suspended before passing through the cell strainer, otherwise cells will be lost.
-
17.
Centrifuge the cell suspension at 250 × g at 22°–23°C for 5 min to pellet the cells. Discard the supernatant carefully aspirating it.
-
18.Lyse the red blood cells (RBC)
-
a.Add 5 mL / Falcon tube of ice-cold lysis buffer for RBC lysis and gently tap the tube with fingers to mix the lysis solution for 5 min.
-
b.Add 10 mL of RPMI medium to dilute the buffer after RBC lysis.
-
a.
-
19.
Centrifuge the cell suspension at 250 × g at 22°–23°C for 5 min to pellet the cells. Discard the supernatant.
-
20.
Resuspend the cells in 18 mL of culture medium and mix well. Optionally: count the cells; usually 2–6 × 107 bone marrow cells can be collected from two tibias and two femurs.
-
21.
Day 0: Seed bone marrow cells of one mouse in one 6 well plate, using 3 mL per well with 10–25 ng/mL of GM-CSF in a 37°C and humidified 5% CO2 incubator.
Note: A cell yield of approximately 1 × 107 cells can be expected. In this protocol, the cells of one mouse were distributed evenly over the 6 wells and were not counted in advance. Cells can alternatively also be counted and seeded at a density of 1,5 × 106 cells per well.
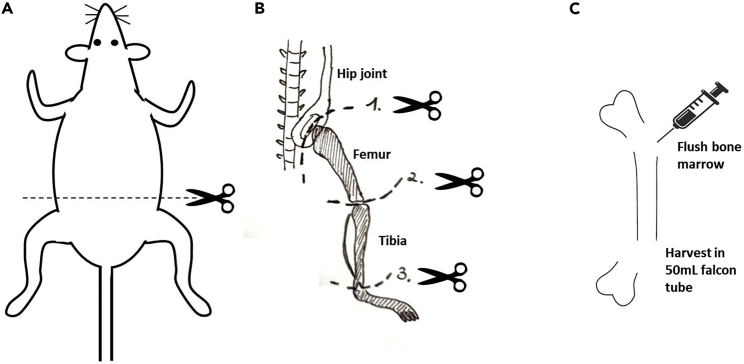
Figure 1.
Isolation of murine femurs and tibiae
(A) Place the mouse face down to the dissecting platform. Cut the skin at the back and expose the legs.
(B) Cut the femur at the hip joint (1.), then take out the whole leg. Separate the femur by cutting the knee joint (2.). Separate the tibia from the paw (3). (C) Cut off the epiphyses on both sides of the bone. Carefully insert a syringe filled with DC medium and flush out bone marrow into a 50mL falcon tube.
Dendritic cell culture and creating an atherosclerotic environment
Timing: 6 days
These steps describe how addition of acetylated LDL creates an atherosclerotic environment for the BMDCs.
-
22.
Day 2, 4, 6: Gently wash cells with pre-warmed PBS and supply them with 3 mL of new, pre-warmed DC-medium (GM-CSF freshly added 20 ng/mL).
-
23.
To create an atherosclerotic environment, add 10 μg/mL acetylated low density lipoprotein (acLDL) or control on day 6.
CRITICAL: Alternatively, also oxidized LDL (oxLDL) can be used. The formation of foam cells has been traditionally studied using acLDL (Landers and Lewis, 1993; Jones et al., 2000). Macrophages that take up acLDL in vitro show phenotypic similarities to foam cells found in atherosclerotic plaques. However, native LDL, oxLDL, and acLDL are each trafficked to different endosomes and accumulate in distinct lysosomal compartments, indicating different endocytic pathways (Wang et al., 2007).
-
24.
Day 7: Collect the dendritic cells as described in the following steps 25–28.
CRITICAL: Be careful to avoid collecting the highly adherent cells (these are considered to be macrophages) by not pipetting too forcefully) The dendritic cells appear as loosely adherent ”heaps” on the highly adherent macrophage layer.
Important: Non-BMDCs can be eliminated by early washing steps, discarding highly adherent cells, and enriching or sorting for CD11c+ cells (Helft et al., 2015).
Note: If the washings and medium changes are not performed carefully, the survival rate and number of the BMDCs will be reduced.
Collecting the dendritic cells
Timing: 30 min
This section describes, how the differentiated BMDCs can be harvested for using them in different assays and analyses.
-
25.
Pre-warm the DC-medium at 37°C in the water bath (GM-CSF does not have to be added).
-
26.
Remove the medium by aspirating.
-
27.Collect the cells
-
a.Pour 2 mL fresh medium over the cells in one well of the 6 well plate with a glass pipette.
-
b.Pipet up and pour over again, repeat this 2 times and finally transfer the medium containing the DCs into a 50 mL Falcon tube.
-
c.Repeat this with all wells of the 6 well plate – pool the cells of one plate in one 50 mL Falcon tube.
-
a.
-
28.
Centrifuge the cell suspension at 250 × g at 22°–23°C for 5 min to pellet the cells. Discard the supernatant.
CRITICAL: The optimal culture period to generate BMDC with GMCSF was reported to be 7–8 days (Inaba et al., 1992). We have followed this recommendation and used these cells for experiments after a maximum of 7 days in culture.
Expected outcomes
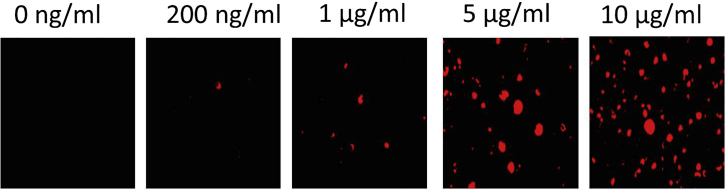
CD11c+ cells accumulate acLDL and secrete ApoE (see Figures 3 and 4). We have shown that the cells are capable of taking up acLDL by incubating them with increasing concentrations of acLDL (Figure 3) A quantification of this uptake could be done via detecting LDL with a specific anti-LDL antibody (for example Anti-LDL antibody GW20089F from Merck) in cell lysates.
Figure 3.
Uptake of acLDL by CD11c+ bone marrow derived cells
BM derived CD11c+ cells were loaded with fluorescently labeled acLDL. With increasing acLDL concentrations the fluorescent signal within the cells is increasing.
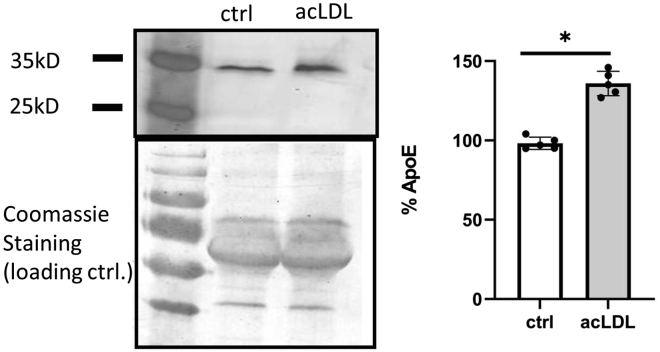
Figure 4.
Secretion of ApoE upon uptake of acLDL by CD11c+ bone marrow derived cells
BM derived CD11c+ cells were loaded with 10 μg/mL acLDL. ApoE was detected in cell culture supernatant via western blot and quantified using ImageJ®. Data are the mean ± SD, ∗P < 0.05.
CD11c+ cells are able to perform cholesterol efflux (Figure 5).
Figure 5.
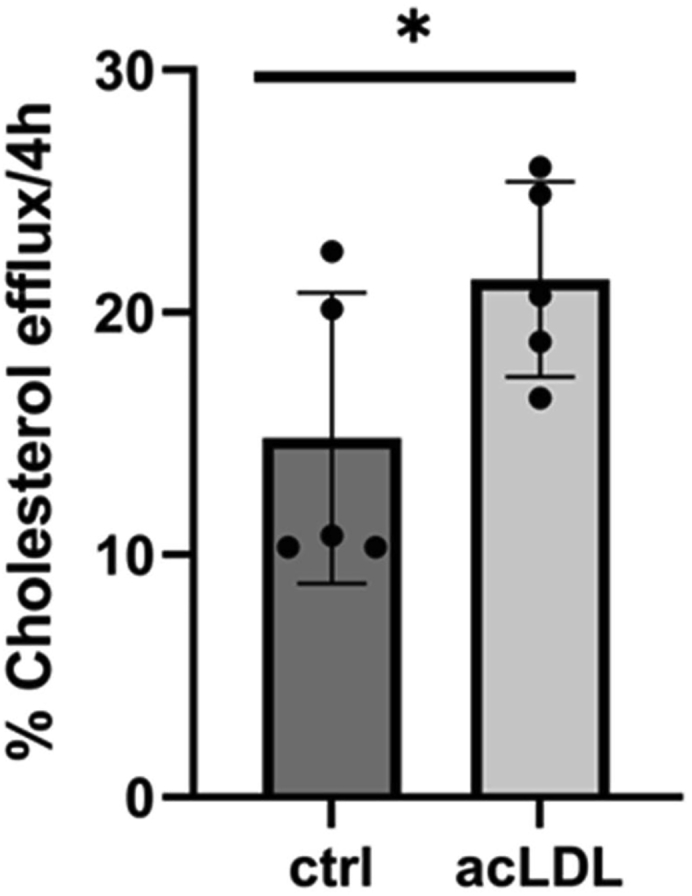
Cholesterol efflux analysis of BM derived CD11c+ cells revealed that cholesterol efflux was significantly enhanced if these cells were exposed to an atherosclerotic environment (loading with acLDL)
Data are the mean ± SD, ∗P < 0.05.
The individual protocols are outlined in “quantification and statistical analysis.”
Quantification and statistical analysis
Flow cytometry
Timing: ∼ 2 h
-
1.
Collect cells as described in the protocol (steps 27–30).
-
2.
Resuspend in FACS buffer at a density of 1 × 106 cells in 50 μL.
-
3.
Incubate with antibodies for 30 min on ice in the dark by adding 3 μL of each antibody. Fill up to 100 μL with FACS buffer.
-
4.
Wash with FACS buffer by centrifugation.
-
5.
Fix the cells in 1% PFA for 15 min on ice and wash again afterwards. Unstained cells and cells treated with unspecific IgG antibodies serve as controls. Fixed cells can be stored at 4°C and analyzed on the next day.
-
6.
Analyze single cell suspensions on a Flow Cytometer. We used FACSCalibur (BD Biosciences) with Cellquest Pro Software and a Cytoflex S 4-laser cytometer (Beckman Coulter, Brea, CA, USA) with Kaluza Analysis 2.1 software (Beckman Coulter). Specific monoclonal antibody binding is expressed as the geometric mean fluorescence intensity (MFI).
Detection of ApoE in cell culture supernatant via western blot
Timing: 2 days and 3 h
-
7.
Incubate cells on culture day 6 with acLDL for 24 h by adding 10 μg/mL acLDL to the wells containing the cells.
-
8.
Wash cells on day 7 and provide them with serum-free medium for 12 h.
-
9.
Harvest cells like described in the protocol (Steps 25–28). Collect supernatant.
-
10.
Determine protein concentration using commercially available kits (for example Pierce™ BCA Protein Assay Kit, Thermo Scientific, cat. no. 23225) as described by the manufacturer.
-
11.
Add 25 μL of 4 × Laemmli SDS sample buffer (Bio Rad) to 100 μL sample and boil for 10 min at 95°C.
Note: Samples can be stored at 4°C for 12 h after this point.
-
12.
Separate by SDS-polyacrylamide gel electrophoresis on 10% polyacrylamide gels (Invitrogen).
Note: Gels can be stored at 4°C for 2–3 days wrapped in wet paper before usage.
-
13.
Collect gels, cut off stacking gel and wash gel by putting it shortly in a tray filled with blotting buffer.
-
14.
Transfer separated protein bands to PVDF-membranes (pre-activated by soaking in methanol for 1 min) using a semi-dry blotter with 7 V for 1 h.
-
15.
After blotting, block with 5% skim milk in Tris-buffered saline 0.05% Tween-20 for one hour at 22°–23°C under gentle shaking.
Note: Blocking buffer can be stored at 4°C for a week.
-
16.
Incubate with primary antibody (Rabbit anti mouse ApoE from Abcam, ab183596) in 5% skim milk in Tris-buffered saline 0.05% Tween-20 at 4°C for 12 h under gentle shaking.
Note: Primary antibody in blocking buffer can be re-used at least 3 times when stored at −20°C in a Falcon tube.
-
17.
Wash the membrane 3 times for 10 min at RT in Tris-buffered saline 0.05% Tween-20 under gentle shaking.
Note: The buffer can be stored at 22°C–24°C for 2–3 weeks.
-
18.
Incubate membrane with secondary antibody (anti rabbit from LI-COR, Bad Homburg, Germany, diluted 1:15000 in 3% Skim milk in Tris-buffered saline 0.05% Tween-20) for 1 h at 22°–23°C in the dark under gentle shaking.
Note: Secondary antibody should not be re-used and always made freshly before usage.
-
19.
Wash the membranes 3 times with Tris-buffered saline 0.05% Tween-20 and let dry.
-
20.
Scan with the Odyssey Infrared Imaging System (LI-COR, Bad Homburg, Germany).
Note: Detection does not have to be performed using LICOR. Chemiluminescence-based detection with HRP-coupled secondary antibodies works well too.
-
21.
After immunodetection, wash membrane twice with Tris-buffered saline 0.05% Tween-20 and stain with 0.1% Coomassie R-250 (Bio-Rad) in methanol/water, 1:1, for 1 min, destain for 20 min in acetic acid/ethanol/water, 1:5:4, wash with water and let air-dry.
Detection of cholesterol efflux
-
22.
Incubate BM derived CD11c+ cells with acLDL on day 6 with 10 μg/mL for 24 h.
-
23.
Analyze cells for their ability to perform cholesterol efflux using a cell-based cholesterol efflux kit by Abcam (ab196985) following the manufacturer`s instructions. Use murine WT serum as cholesterol acceptor.
-
24.
Perform read-out on an ELISA-reader according to the manufacturer’s protocol.
Limitations
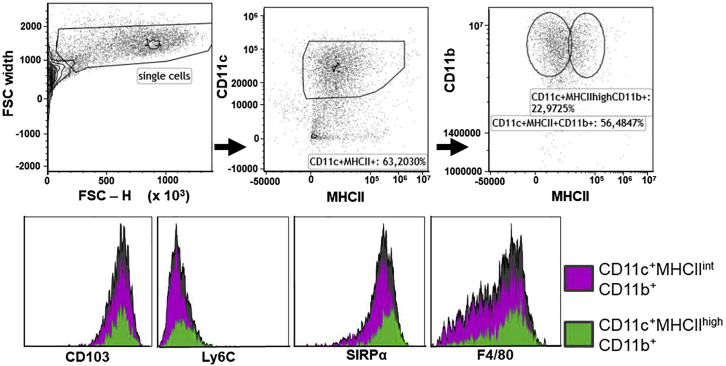
In these protocols, we have described several analytic methods using murine BM-derived CD11c+ cells. Because we have analyzed similar assays on various cells, we consider these protocols to be beneficial. However, it has to be kept in mind that BM cell isolation yields a diverse population of cells, which are not only one distinct cell type but various cells expressing different surface antigens. We characterized these cells using FACS and found 63% of the harvested cells to be CD11c+ MHC-II+ (Figure 2).
Figure 2.
Characterization of BM derived cells via FACS after 6 days of cultivation
Loosely adherent BM derived CD11c+ cells were harvested and incubated with fluorescently labeled antibodies against various surface antigens. 63% of harvested cells were CD11c+ MHC-II+ cells.
Troubleshooting
Problem 1
Bones are broken during the preparation (step 4) and as a result, high cell yields cannot be achieved.
Potential solution
Initially, explant the whole leg and cut the bones only after you already removed surrounding tissue. Make sure you cut the bones carefully within the joints.
Problem 2
Cell numbers achieved from bone marrow are very low in step 20.
Potential solution
Flush the bone marrow from both sides until the bone appears white.
Make sure that the bone marrow is really transferred into the tube and is not sticking at the end of the bone. Do not seed too low cell numbers (in our hands, one 6-well-plate per mouse yielded optimal cell counts).
Problem 3
FACS analysis shows, that the bone marrow cells are not properly differentiated into BMDCs (see section “quantification and statistical analysis”).
Potential solution
Reduce the number of bone marrow cells seeded onto the culture plate. Extend the culturing period of the BMDCs. Increase the concentration of GM-CSF. Use a different lot of fetal bovine serum.
Problem 4
Number of harvested cells is very low.
Potential solution
When harvesting the cells, make sure you rinse the cells properly, not too gentle, three times with the medium before collecting them into the Falcon tube.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Harald F. Langer (harald.langer@uksh.de).
Materials availability
This protocol did not generate unique materials or reagents.
Acknowledgments
We thank Jacob von Esebeck, Anke Constantz, and Sarah Gekeler for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project number 374031971 – TRR 240 and the Volkswagen Foundation (Lichtenberg program) to H.F.L.
Author contributions
M.S. and R.J.S. performed experiments, analyzed and compiled data, and wrote parts of the manuscript. H.F.L. conceived the project, analyzed data, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Manuela Sauter, Email: manuela.sauter@uksh.de.
Harald F. Langer, Email: harald.langer@uksh.de.
Data and code availability
This protocol did not generate unique materials or reagents.
References
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., Reis E Sousa C. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Inaba K., Inaba M., Romani N., Aya H., Deguchi M., Ikehara S., Muramatsu S., Steinman R.M. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.L., Reagan J.W., Willingham M.C. The pathogenesis of foam cell formation. Arterioscler. Thromb. Vasc. Biol. 2000;20:773–781. doi: 10.1161/01.atv.20.3.773. [DOI] [PubMed] [Google Scholar]
- Landers S.C., Lewis J.C. acLDL binding and endocytosis by macrophages and macrophage foam cells in situ. Exp. Mol. Pathol. 1993;59:38–50. doi: 10.1006/exmp.1993.1025. [DOI] [PubMed] [Google Scholar]
- Sauter M., Sauter R.J., Nording H., Lin C., Olbrich M., Autenrieth S., Gleissner C., Thunemann M., Otero N., Lutgens E. Apolipoprotein E derived from CD11c+ cells ameliorates atherosclerosis. iScience. 2021;25(1) doi: 10.1016/j.isci.2021.103677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.D., Kiss R.S., Franklin V., Mcbride H.M., Whitman S.C., Marcel Y.L. Different cellular traffic of LDL-cholesterol and acetylated LDL-cholesterol leads to distinct reverse cholesterol transport pathways. J. Lipid Res. 2007;48:633–645. doi: 10.1194/jlr.M600470-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol did not generate unique materials or reagents.