Abstract
An in silico scan of the partially completed genome sequence of Bordetella pertussis and analyses of transcriptional fusions generated with a new integrational vector were used to identify new potential virulence genes. The genes encoding a putative siderophore receptor, adhesins, and an autotransporter protein appeared to be regulated in a manner similar to Bordetella virulence genes by the global virulence regulator BvgAS. In contrast, the gene encoding a putative intimin-like protein appeared to be repressed under conditions of virulence.
Bordetella pertussis is a strictly human pathogen responsible for whooping cough, an acute respiratory disease particularly severe in young children (18). It produces a number of toxins and adhesins involved in its pathogenicity. The coordinated expression of the virulence genes is controlled by the global sensor and regulator BvgAS (1). The genes under the positive control of BvgAS are called vag (for virulence-activated gene) genes. In response to environmental conditions, such as low temperature or the presence of MgSO4 or nicotinic acid, B. pertussis undergoes a phenotypic modulation. The vag genes are downregulated, while another set of genes called vrg (for virulence-repressed gene) genes is upregulated (19).
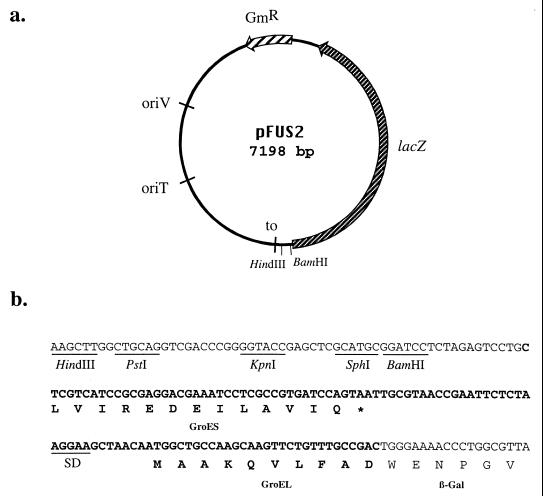
We have scanned the B. pertussis sequences currently available on the Sanger Center website (www.sanger.ac.uk/Projects/B_pertussis) to identify new potential virulence genes (Table 1). We have developed a new suicide vector, pFUS2, for rapid gene inactivation by homologous recombination and generation of transcriptional fusions between the interrupted genes and promoterless lacZ (Fig. 1). pFUS2 was derived from pQE30 (Qiagen, Courtaboeuf, France) by (i) the replacement of the 960-bp EcoRI-blunted BglI fragment containing the tac promoter and the bla gene with a gentamicin resistance cassette, (ii) the insertion into the BamHI and blunted EcoRI sites of the promoterless lacZ from pUTminiTn5lacZ2 (5) contained on a 3-kb BamHI-blunted HindIII fragment, (iii) the insertion of a PCR-amplified 760-bp fragment carrying the RP4 origin of transfer from pJQ200mp18 (27) into the unique XbaI site, and (iv) the insertion into the ClaI and blunted BamHI sites of a PCR-amplified 910-bp fragment containing the very 3′ end of B. pertussis groES, the intergenic groES-groEL region, and the 5′ end of groEL fused to the slightly truncated 5′ end of lacZ.
TABLE 1.
Characteristics of putative B. pertussis proteins
| Protein | Amino acid
sequencea
|
Size (aab) | Closest homolog | GenBank accession no. | % Identityc | |
|---|---|---|---|---|---|---|
| N terminal | C terminal | |||||
| FhaL | MNKHCFKLVH | GASATSRLPP | 4,196 | FHA (B. pertussis) | P12255 | 10 |
| FhaS | AATAILTEIT | PGGSPFSLHR | 2,431 | FHA (B. pertussis) | P12255 | 22 |
| AdhS | MKSRILALAA | AQQLAAALRP | 311 | Adhesin B precursor (Streptococcus parasanguinis) | P31305 | 27 |
| BilA | MNKNIYRVVW | LLLAPVNFPY | 1,308 | Intimin (E. coli) | AAD02815 | 13 |
| BexB | VIGSLIFRET | RAMRRRLHTR | 246 | Transport protein KPSM (E. coli) | P24584 | 30 |
| DrnB | MKYAVVLAAL | DPAAIQRQCG | 246 | Extracellular DNAse (Aeromonas hydrophila) | O31222 | 45 |
| MetC1 | MTDESRHIDT | TQALARAASA | 397 | β-Cystathionase (B. avium) | Q07703 | 79 |
| MetC2 | MKFDTLLTHG | DLDQALRRED | 383 | β-Cystathionase (E. coli) | P06721 | 40 |
| BllY | MAENFQPWEN | MRRGVVKQVD | 370 | 4-Hydroxyphenylpyruvate dioxygenase (Pseudomonas sp.) | P80064 | 54 |
| NprB | MXXPTXKAAR | QAWTDVGVLT | 343 | Extracellular metalloprotease (Erwinia carotovora) | Q99132 | 51 |
| ZnpB | VTAPVPLFTL | VRIRHLMAGA | 421 | Zinc metalloprotease (Campylobacter jejuni) | CAB72997 | 25 |
| Phg | MKPTSILARL | SLNIGYAYRF | 418 | Phg protein (B. pertussis) | CAB38010 | 100 |
| AidB | MVGRSCHRAG | TATAGFRMMF | 903 | Adhesin AidA-I precursor (E. coli) | BAA15189 | 13 |
| SphB1 | MPPPAVPPQL | QLSASLTYRY | 931 | SSP-h1 protease (Serratia marcescens) | BAA33455 | 24 |
| SphB2 | MPSCWAWTAA | TASVVLRWRF | 989 | SSP-h1 protease (S. marcescens) | BAA33455 | 19 |
| SphB3 | MAQASARGMG | TGSLYLKVRF | 1,076 | SSP-h1 protease (S. marcescens) | BAA33455 | 14 |
| BfrD | MKFYSSHPMP | SAMLTFKLSY | 743 | Hydroxamate ferrisiderophore receptor (Pseudomonas aeruginosa) | AAC06215 | 37 |
| BfrE | MEKPLKSLDS | AAVLSFNIKY | 756 | Hydroxamate ferrisiderophore receptor (P. aeruginosa) | AAC06215 | 40 |
Predicted sequences of the putative proteins.
aa, amino acids.
Percentage of identity between the B. pertussis protein and its closest homolog.
FIG. 1.
Map of pFUS2. (a) Main features of the vector. GmR, gentamicin resistance cassette; oriT, RP4 origin of transfer, oriV, ColE1 origin of replication; to, a transcriptional terminator to prevent transcription initiation from vector sequences. (b) Sequence surrounding the multiple cloning site of pFUS2. The portion of the nucleotide sequence corresponding to the 3′ end of groES, the intergenic groES-groEL region, and the 5′ end of groEL is shown in bold. The 3′ end of groES and the 5′ end of groEL have been translated (in bold), as has the beginning of lacZ. SD represents the putative ribosome binding site sequence of groEL. β-Gal, β-galactosidase.
Gene inactivation with pFUS2 was first validated by targeting the known Bvg-activated genes fhaB and ptx, coding for the filamentous hemagglutinin (FHA) and pertussis toxin, respectively (21), and one Bvg-repressed gene, vrg24 (19). Pairs of oligonucleotides were designed to amplify 350- to 550-bp internal fragments of the target genes by PCR. The amplicons were cloned into pFUS2 so that translation of these genes terminated at the groES stop codon, giving rise to transcriptional fusions with lacZ. Gene inactivations were performed in both B. pertussis Tohama I derivatives BPSM (bvg+) (23) and BPLOW (Δbvg). BPLOW carries a chromosomal deletion of bvgAS extending from the first EcoRI site upstream of bvgA to the first EcoRI site within bvgS. This strain was constructed by double homologous recombination using pSS1129 as described previously (28). Both ptx and fhaB fusions were expressed at high levels in BPSM and were modulated by MgSO4 and nicotinic acid (Table 2). The expression of ptx′-′lacZ was very low in BPLOW, and that of fhaB′-′lacZ was undetectable. The expression of the vrg24′-′lacZ fusion in BPSM increased 2.5-fold in the presence of the modulators, and it was slightly higher in the Δbvg background than in the bvg+ background. Altogether, these results confirm the usefulness of pFUS2 to identify vag genes as well as vrg genes. Therefore, we used this vector to target genes encoding new potential virulence factors (Table 1).
TABLE 2.
β-Galactosidase activities of B. pertussis pFus2 integrantsa
| Target gene | Enzyme activity in
integrant
|
|||
|---|---|---|---|---|
| BPSM with:
|
BPLOW with NA | |||
| NA | Mg | Nic | ||
| fhaB | 5,094 ± 22 | 473 ± 68 | 2,133 ± 324 | 0 |
| ptx | 9,220 ± 2,673 | 800 ± 311 | 1,096 ± 218 | 46 ± 22 |
| vrg24 | 207 ± 15 | 490 ± 50 | 496 ± 60 | 295 ± 10 |
| fhaL | 161 ± 16 | 17.5 ± 3.2 | 6 ± 3 | <1 |
| fhaS | 260 ± 62 | 54 ± 10.7 | 20.7 ± 4.5 | 15.6 ± 9 |
| adhS | 9 ± 2 | 10 ± 4.6 | 6 ± 0.7 | <2 |
| bilA | 287 ± 17 | 1,920 ± 291 | 3,680 ± 145 | 1,080 ± 62 |
| bexB | 0 | 31.7 ± 2.4 | <5 | 0 |
| drnB | 39 ± 3 | 31 ± 1 | 30 ± 5 | 32 ± 12 |
| metC1 | 177 ± 24 | 240 ± 65 | 226 ± 14 | 211 ± 54 |
| metC2 | 45 ± 17 | 93 ± 22 | 88 ± 8 | 67 ± 16 |
| bllY | 91 ± 18 | 129 ± 16 | 159 ± 21 | 28 ± 5.8 |
| nprB | 62 ± 8 | 116 ± 22 | 104 ± 4 | 70 ± 15 |
| znpB | 80 ± 12 | 132 ± 40 | 120 ± 30 | 61 ± 20 |
| pgh | 48 ± 13 | 32 ± 8 | 43 ± 16 | 39 ± 20 |
| aidB | 148 ± 56 | 150 ± 22 | 183 ± 27 | 138 ± 56 |
| sphB1 | 1,063 ± 265 | 23 ± 3 | 93 ± 46 | 0 |
| sphB2 | 0 | 0 | 0 | <2 |
| sphB3 | <5 | 5 ± 2 | <2 | <4 |
| bfrD | 789 ± 308 | 10 ± 1 | 144 ± 36 | 60.5 ± 17 |
| bfrE | 13 ± 4 | 10.1 ± 1 | 9.3 ± 3.2 | 16.3 ± 1.4 |
Three independent clones of each recombinant strain were grown for 3 days at 37°C on Bordet Gengou (Gibco) agar plates containing 10% sheep blood without (NA), or with the modulating agents MgSO4 (50 mM) (Mg) or nicotinic acid (20 mM) (Nic). The cells were scraped from the plates and suspended in phosphate-buffered saline. The cell density was estimated by optical density measurements at 600 nm (OD600). Then the cells were broken by passage through a French pressure cell. β-Galactosidase activities were determined by measuring the initial rates of o-nitrophenyl-β-d-galactopyranoside hydrolysis at 420 nm and calculated according to the following formula: (ΔA420/minutes × 1,000)/OD600 × volume [in milliliters] of cell lysate in the reaction mixture). The results are given as means ± standard deviations.
Adhesins.
Two genes code for putative proteins homologous to FHA. One of them, called fhaL (for FHA-like, large), corresponds to the largest open reading frame (ORF) of the entire genome. The gene encoding the other FHA-like protein, named FhaS (for FHA-like, small), harbors two frameshifts, and its 5′ region is not included in the current contig. Both genes appeared to be well expressed, albeit at much lower levels than fhaB (Table 2). Interestingly, both are regulated by Bvg and thus qualify as vag genes. The reasons for this apparent redundancy are not clear. FHA is of great importance to B. pertussis pathogenicity. Therefore, the bacterium may maintain backup gene copies. Alternatively, the poorly expressed copies might act as reservoirs for homologous recombination with the master gene to generate antigenic diversity, similar to the pilin genes in Neisseria (16). It is also conceivable that the three proteins play related but distinct functions, similar to the antigen 85 complex of Mycobacterium tuberculosis (2). Alternatively, the bacterium progressively sheds obsolete copies.
The gene encoding a putative signal peptide-bearing protein homologous to a salivary streptococcal adhesin (13) was identified and called adhS. This gene is part of an operon also including genes encoding a permease and an ATPase, and the corresponding protein shares limited similarity with periplasmic components of ATP-binding cassette transporters. It is therefore unclear whether it should be classified as an adhesin. This gene was expressed at a very low level and apparently was not modulated.
A gene was identified coding for a protein whose closest homolog is the enteropathogenic Escherichia coli intimin, and it was therefore called bilA (for Bordetella intimin-like). However, this putative protein is somewhat longer than intimin, and it lacks the two disulfide-bonded cysteines shown to be essential for the binding activity of intimin (12). The expression of bilA was very high in the Δbvg background and was dramatically upregulated by nicotinic acid and MgSO4 in BPSM. These features indicate that bilA is a vrg gene. The roles of the vrg genes in B. pertussis infection remain mysterious (1, 22). Recent evidence suggests that these genes may be expressed upon entry into eukaryotic cells or at high bacterial cell densities (25). The product of bilA has been recently identified in Bordetella bronchiseptica and has been shown to be involved in colonization in a rabbit model (K. E. Stockbauer, B. Fuchslocher, J. F. Miller, and P. A. Cotter, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. B-184, 2000). It will be interesting to determine whether the protein is involved in cytoskeleton rearrangements in the host cell, like intimin is (6).
Capsule.
The B. pertussis genome contains a complete operon for the biosynthesis of a polysaccharide capsule. Earlier literature reports have suggested that the bacterium is capsulated (20). A gene in the 5′ region of this operon, named bexB (for Bordetella capsule export gene B) based on the usual nomenclature for other species, was targeted by pFUS2. No expression of bexB was detectable in either BPSM or BPLOW, but surprisingly a low but significant level of activity was observed in the presence of MgSO4. It is possible that the appropriate environmental conditions have not been met for optimal gene expression.
Enzymes.
An ORF was found which encodes a putative signal peptide-bearing protein homologous to secreted DNases of several pathogens (3). We called this gene drnB (for DNase of Bordetella). Its expression was rather low and was not influenced by modulation.
The B. pertussis genome analysis revealed two ORFs encoding homologs of β-cystathionase. These genes were named metC1 and metC2 based on the nomenclature for other species. The product of metC1 is highly similar to Bordetella avium osteotoxin, which metabolizes cystine into an osteoblast-toxic molecule (14). metC1 was expressed at a fairly high level but was not Bvg regulated. In contrast, metC2 was upregulated twofold in the presence of MgSO4 or nicotinic acid, and its expression was slightly higher in the Δbvg background than in the bvg+ background, reminiscent of vrg24. Therefore, metC2 might be a vrg gene. The small amplitude of modulation may reflect the indirect effect of modulating agents on vrg expression in B. pertussis. Whereas the transcription of vag genes is directly activated by binding of the BvgA regulator to their promoter regions, that of the vrg genes is indirectly regulated by a Bvg-dependent repressor (24).
The sequence analysis also uncovered an ORF encoding a protein homologous to the Legionella pneumophila legiolysin, which in fact is a dioxygenase (17, 30). After inactivation of the B. pertussis bllY (for Bordetella legiolysin) gene, no strong modulation was observed. However, the β-galactosidase activity of that fusion in the Δbvg background was lower than in the bvg+ background.
The B. pertussis chromosome contains several genes coding for proteases other than housekeeping proteases. Two of them, called nrpB (for neutral protease of Bordetella) and znpB (for Zn protease of Bordetella) in agreement with the names given to homologs in other species, were selected for inactivation by pFUS2. Both were expressed but were not regulated by Bvg.
Autotransporters.
The genes for several autotransporters, in addition to BrkA, Tcf, pertactin, and Vag-8 (4, 8, 9, 11), were also identified in the genome of B. pertussis. Five of them (phg, aidB, sphB1 [for serine protease homolog of Bordetella], sphB2, and sphB3) were targeted with pFUS2. The expression levels of sphB2 and sphB3 were very low or undetectable, while phg, aidB, and sphB1 were better expressed. In addition, sphB1 was strongly activated by Bvg, making it a new vag gene.
Iron metabolism.
A number of genes encoding potential siderophore and heme receptors were uncovered. In particular, an outer membrane protein of unknown function (26) was found to be homologous to TonB-dependent siderophore receptors of various bacterial species. Its gene is followed by a second gene, coding for a 56.7% identical protein. The two genes, called bfrD and bfrE (for Bordetella ferrisiderophore receptor), are separated by approximately 200 bp. Expression of bfrE was very low and did not appear to be Bvg regulated. In contrast, bfrD was expressed at a high level at a Bvg-dependent fashion, making it the first vag gene involved in iron acquisition in Bordetella. Interestingly, siderophore production is repressed by Bvg in certain B. bronchiseptica strains but not in B. pertussis (15). This divergent regulation may relate to considerable differences between the two species in the ability to survive outside of their hosts (1).
In conclusion, pFUS2 has allowed us to uncover several new Bvg-regulated genes. Undoubtedly, additional members of the Bvg regulon remain to be found. In fact, BvgAS appears to control various aspects of Bordetella physiology in addition to virulence, including the production of a cytochrome (7), a porin (10), and cell wall hydrolase(s) (29). Further characterization of the Bvg regulon will greatly benefit from the development of transcriptomic and proteomic tools.
Acknowledgments
We thank K. Timmis for the kind gift of pUTminiTn5lacZ2 and Alain Baulard and Jean Dubuisson for critical reading of the manuscript. The technical help of E. Fort, E. Fontaine, and A. Levard is acknowledged.
F. J.-D. is Chargé de Recherche CNRS. This work was supported by INSERM, Institut Pasteur de Lille, Ministère de l'Education Nationale de la Recherche et de la Technologie, and Région Nord-Pas de Calais.
REFERENCES
- 1.Akerley B J, Miller J F. Understanding signal transduction during bacterial infection. Trends Microbiol. 1996;4:141–146. doi: 10.1016/0966-842x(96)10024-x. [DOI] [PubMed] [Google Scholar]
- 2.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosisin cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 3.Chang M C, Chang S Y, Chen S L, Chuang S M. Cloning and expression in Escherichia coli of the gene encoding an extracellular deoxyribonuclease (DNase) from Aeromonas hydrophila. Gene. 1992;122:175–180. doi: 10.1016/0378-1119(92)90046-r. [DOI] [PubMed] [Google Scholar]
- 4.Charles I G, Dougan G, Pickard D, Chatfield S, Smith M, Novotny P, Morrissey P, Fairweather N F. Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc Natl Acad Sci USA. 1989;86:3554–3558. doi: 10.1073/pnas.86.10.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Lorenzo V, Timmis K. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ezzel J W, Dobrogosz W J, Kloos W E, Manclark C R. Phase-shift markers in the genus Bordetella: loss of cytochrome d-629 in phase IV variants. Microbios. 1981;31:171–181. [PubMed] [Google Scholar]
- 8.Fernandez R C, Weiss A A. Cloning and sequencing of a Bordetella pertussisserum resistance locus. Infect Immun. 1994;62:4727–4738. doi: 10.1128/iai.62.11.4727-4738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn T M, Amsbaugh D F. Vag8, a Bordetella pertussis bvg-regulated protein. Infect Immun. 1998;66:3985–3989. doi: 10.1128/iai.66.8.3985-3989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn T M, Li Z, Kocsis E. Identification of a Bordetella pertussis bvg-regulated porin-like protein. J Bacteriol. 1995;177:805–809. doi: 10.1128/jb.177.3.805-809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn T M, Stevens L A. Tracheal colonization factor: a Bordetella pertussissecreted virulence determinant. Mol Microbiol. 1995;16:625–634. doi: 10.1111/j.1365-2958.1995.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 12.Frankel G, Candy D C A, Fabiani E, Adu-Bobie J, Gil S, Novakova M, Phillips A D, Dougan G. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect Immun. 1995;63:4323–4328. doi: 10.1128/iai.63.11.4323-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganeshkumar N, Hannam P M, Kolenbrander P E, McBride B C. Nucleotide sequence of a gene coding for a saliva-binding protein (SsaB) from Streptococcus sanguis12 and possible role of the protein in coaggregation with actinomyces. Infect Immun. 1991;59:1093–1099. doi: 10.1128/iai.59.3.1093-1099.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry-Weeks C R, Spokes J, Thompson J. β-Cystathionase from Bordetella avium. J Biol Chem. 1995;270:7695–7702. doi: 10.1074/jbc.270.13.7695. [DOI] [PubMed] [Google Scholar]
- 15.Giardina P C, Foster L-A, Musser J M, Akerley B J, Miller J F, Dyer D W. bvg repression of alcaligin synthesis in Bordetella bronchisepticais associated with phylogenetic lineage. J Bacteriol. 1995;177:6058–6063. doi: 10.1128/jb.177.21.6058-6063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas R, Meyer T F. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 17.Hacker J, Wintermeyer E, Ludwig B, Fischer G. Analysis of virulence factors of Legionella pneumophila. Zentbl Bakteriol. 1993;278:348–358. doi: 10.1016/s0934-8840(11)80851-0. [DOI] [PubMed] [Google Scholar]
- 18.Hewlett E L. Pertussis: current concepts of pathogenesis and prevention. Pediatr Infect Dis J. 1997;16:S78–S84. doi: 10.1097/00006454-199704001-00002. [DOI] [PubMed] [Google Scholar]
- 19.Knapp S, Mekalanos J J. Two trans-acting regulatory genes (vir and mod) control antigenic modulation in Bordetella pertussis. J Bacteriol. 1988;170:5059–5066. doi: 10.1128/jb.170.11.5059-5066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson G M. Modified technique for staining capsules of Haemophilus pertussis. J Lab Clin Med. 1940;25:435–438. [Google Scholar]
- 21.Locht C. Molecular aspects of Bordetella pertussispathogenesis. Int Microbiol. 1999;2:137–144. [PubMed] [Google Scholar]
- 22.Martinez de Tejada G, Cotter P A, Heininger U, Camilli A, Akerley B J, Mekalanos J J, Miller J F. Neither the Bvg− phase nor the vrg6 locus of Bordetella pertussisis required for respiratory infection in mice. Infect Immun. 1998;66:2762–2768. doi: 10.1128/iai.66.6.2762-2768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menozzi F D, Mutombo R, Renauld G, Gantiez C, Hannah J H, Leininger E, Brennan M J, Locht C. Heparin-inhibitable lectin activity of the filamentous hemagglutinin adhesin of Bordetella pertussis. Infect Immun. 1994;62:769–778. doi: 10.1128/iai.62.3.769-778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkel T J, Barros C, Stibitz S. Characterization of the bvgR locus of Bordetella pertussis. J Bacteriol. 1998;180:1682–1690. doi: 10.1128/jb.180.7.1682-1690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merkel, T. J., and J. M. Keith. The regulation of gene expression in Bordetella pertussis by quorum sensing. Int. J. Med. Microbiol., in press.
- 26.Passerini di Rossi B N, Friedman L E, Gonzales Flecha F L, Castello P R, Franco M A, Rossi J P F C. Identification of Bordetella pertussisvirulence-associated outer membrane proteins. FEMS Microbiol Lett. 1999;172:9–13. doi: 10.1111/j.1574-6968.1999.tb13442.x. [DOI] [PubMed] [Google Scholar]
- 27.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 28.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 29.Tuomanen E, Schwartz J, Sande S. The vir locus affects the response of Bordetella pertussisto antibiotics: phenotypic tolerance and control of autolysis. J Infect Dis. 1990;162:560–563. doi: 10.1093/infdis/162.2.560. [DOI] [PubMed] [Google Scholar]
- 30.Wintermeyer E, Rdest U, Ludwig B, Debes A, Hacker J. Characterization of legiolysin (lly), responsible for haemolytic activity, colour production and fluorescence of Legionella pneumophila. Mol Microbiol. 1991;5:1135–1143. doi: 10.1111/j.1365-2958.1991.tb01886.x. [DOI] [PubMed] [Google Scholar]