Abstract
A large gene cluster for the Clostridium cellulovorans cellulosome has been cloned and sequenced upstream and downstream of the cbpA and exgS genes (C.-C. Liu and R. H. Doi, Gene 211:39–47, 1998). Gene walking revealed that the engL gene cluster (Y. Tamaru and R. H. Doi, J. Bacteriol. 182:244–247, 2000) was located downstream of the cbpA-exgS genes. Further DNA sequencing revealed that this cluster contains the genes for the scaffolding protein CbpA, the exoglucanase ExgS, several endoglucanases of family 9, the mannanase ManA, and the hydrophobic protein HbpA containing a surface layer homology domain and a hydrophobic (or cohesin) domain. The sequence of the clustered genes is cbpA-exgS-engH-engK-hbpA-engL-manA-engM-engN and is about 22 kb in length. The engN gene did not have a complete catalytic domain, indicating that engN is a truncated gene. This large gene cluster is flanked at the 5′ end by a putative noncellulosomal operon consisting of nifV-orf1-sigX-regA and at the 3′ end by noncellulosomal genes with homology to transposase (trp) and malate permease (mle). Since gene clusters for the cellulosome are also found in C. cellulolyticum and C. josui, they seem to be typical of mesophilic clostridia, indicating that the large gene clusters may arise from a common ancestor with some evolutionary modifications.
Clostridium cellulovorans (ATCC 35296) (19), an anaerobic, mesophilic, and spore-forming bacterium, produces extracellular polysaccharolytic multicomponent complexes called the cellulosome (1, 8), which has the ability to degrade cellulose, xylan, mannan, and pectin (19, 21). The C. cellulovorans cellulosome (3) consists of three major subunits, CbpA, P100, and P70, and several minor subunits (10, 16). We have previously cloned and sequenced several cellulosomal subunits, i.e., the scaffolding protein CbpA (18), the endoglucanases EngB (4, 17) and EngE (20), and the exoglucanase ExgS (9). More recently, we have completely sequenced the engL gene cluster, which consists of five different open reading frames (ORFs) containing a cellulosomal ManA-encoding sequence (21).
In a recent 16S rRNA gene analysis of polysaccharolytic clostridia, C. cellulovorans was classified in group I of the phylogenetic tree (13) while most cellulolytic clostridia, such as C. cellulolyticum, C. josui, C. papyrosolvens, and C. thermocellum, belonged to the same cluster (group III) (7). Although C. cellulovorans was located far from the other cellulolytic clostridia in the phylogenetic tree, the gene clusters of the C. cellulovorans cellulosome (22) seem similar to those of C. cellulolyticum (2) and C. josui (6, 7). Since a large gene cluster in C. cellulolyticum (cipC-celF-celC-celG-celE-ORFX-celH-celJ- celK) has recently been reported (2), such a gene cluster seems to be specific for mesophilic clostridia and did not occur in the thermophilic bacterium C. thermocellum. Furthermore, recent data obtained with C. cellulovorans, C. cellulolyticum, C. josui, and C. acetobutylicum revealed that all of these gene clusters begin with the scaffoldin gene, followed by a gene encoding a family 48 cellulase (2). It is of interest to determine the chromosomal organization of the genes of the cellulosome complex, since it may provide information concerning the number of genes, the transcriptional regulation, the coordinate expression, and the evolutionary relationship of the genes in the complex.
In this paper, we describe the large gene cluster around the cbpA and exgS genes of C. cellulovorans. We also analyzed the amino acid sequences of the corresponding proteins and compared them with those of other proteins. Furthermore, this large gene cluster also codes for a small 25-kDa protein, hydrophobic protein A (HbpA), that showed homology with hydrophobic domains (HBDs or type I cohesins) in CbpA (18). The role of HbpA is still not understood, but it may function in a manner similar to that reported for OlpA of C. thermocellum (1) and ORFXp of C. cellulolyticum (11). The occurrence of this small HbpA may be widespread among mesophilic clostridia that produce cellulosomes.
Cloning and DNA sequencing of the gene cluster.
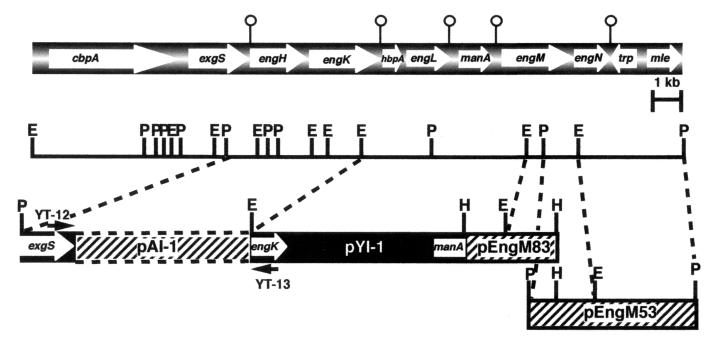
The major gene cluster of the cellulosome consists of nine genes, as shown in Fig. 1. We have cloned and sequenced the cbpA-exgS gene cluster (9) and the engL gene cluster (pYI-1) harboring five different ORFs, i.e., engK-hbpA-engL-manA-engM (21). Since it was expected that the engL gene cluster might be located downstream of the cbpA-exgS gene cluster, we cloned the region between exgS and engK by gene walking. As shown in Fig. 1, the internal fragment between exgS and engK was amplified by PCR with two synthesized oligonucleotides, YT-12 (5′-CTGATATGAACGGTGATGGAAAAG-3′), corresponding to exgS, and YT-13 (5′-CCACCAGTTAATGTAGTTGGCA-3′), corresponding to engK. As a result, a 4.6-kb PCR fragment (pAI-1) was obtained and cloned into the pCR2.1 vector with a TA cloning kit (Invitrogen) and then sequenced (Fig. 1). The DNA sequence of the pAI-1 fragment contained the engH and engK genes. No potential transcription terminator was observed between engH and engK, while a large potential terminator (14) was seen after engK. This observation indicated that the engH and engK genes might be encoded by an operon. Likewise, since no repeat elements were observed between cbpA and exgS and between hbpA and engL, they appear to be encoded as operons; large transcriptional terminators were found between exgS and engL. There is a potential transcriptional terminator downstream of manA, indicating that manA is a monocistronic gene. In fact, ManA production is repressed by cellobiose (21) while the three major cellulosome subunits are expressed in the presence of cellobiose (10). Thus, it will be of extreme interest to study the regulation of expression of these putative operons. One might expect coordinated expression of the operons for the enzymatic subunits with the cbpA-exgS operon.
FIG. 1.
Restriction enzyme map of a cellulosomal gene cluster. The genes coding for CbpA, ExgS, EngH, EngK, HbpA, EngL, ManA, EngM, and EngN are shown at the top. The pin-like marks indicate palindromes. E, H, and P indicate EcoRI, HindIII, and PstI restriction sites, respectively.
To obtain the complete engM gene, Southern hybridization analysis with a partial engM fragment of pYI-1 as a probe was carried out. Either HindIII or PstI digestion of C. cellulovorans chromosomal DNA gave a 3.3- or 4.6-kb fragment which was associated with the probe (data not shown). As a result of screening by colony hybridization with the same probe, we cloned two kinds of plasmids that were named pEngM83 (3.3-kb HindIII fragment) and pEngM53 (4.6-kb PstI fragment), respectively (Fig. 1). The DNA sequence of these fragments contained four ORFs. The first ORF coded for EngM; the second ORF, named engN, encoded only the N-terminal amino acid sequence of family 9 cellulases. The last two ORFs coded for proteins that were homologous to transposase (trn) and malate permease (mle), respectively (Fig. 1), and these two genes flanked the cellulosome gene cluster at the 3′ end. On the other hand, the gene cluster was flanked at the 5′ end by the noncellulosomal gene cluster nifV-orf1-sigX-regA (S. Karita and R. H. Doi, unpublished data; 18). There are three cellulosomal genes that are unlinked to the major gene cluster and unlinked to each other, i.e., engB (17), engE (20), and engY-pelA (22).
The engN gene is an anomaly, since the coding sequence, which has been checked several times in all three reading frames, indicated that EngN does not have a complete catalytic domain. Repeated sequencing experiments indicate strongly that engN is a truncated gene. Furthermore, no duplicated sequence (DS) is present in the coding sequence. The cloned engN gene also does not express any endoglucanase activity in Escherichia coli, while the other enzymatic genes are expressed in E. coli as active enzymes. Since engN is flanked by engM and the transposase gene (Y. Tamaru and R. H. Doi, unpublished data), there does not appear to have been some accidental deletion during cloning.
Amino acid sequences encoded by the gene cluster.
The cellulosomal subunits of C. cellulovorans are summarized in Table 1. We have previously characterized several cellulosomal subunits, i.e., CbpA (18), EngE (20), ExgS (9), EngB (4, 5), and ManA (21). Four family 9 cellulases, i.e., EngH, EngK, EngL, and EngM, have been found in the gene cluster. EngK and EngM belong to subfamily E1 in family 9, while EngH and EngL belong to subfamily E2 in family 9. Also, except for EngL, family 9 cellulases in the gene cluster contain a cellulose-binding domain (CBD). EngH contains a family IIIc CBD, while EngK and EngM have a family IV CBD.
TABLE 1.
Cellulosomal subunits of C. cellulovorans
| Gene product | Modular structurea | No. of residuesb | Mol wtbc | Reference or source; GenBank accession no. |
|---|---|---|---|---|
| EngE | (SLH)3-GH5-X-DS | 1,030 | 111,796 | 20; AF105331 |
| EngK | CBDIV-Ig-GH9-DS | 892 | 97,024 | This study; AF132735 |
| EngM | CBDIV-Ig-GH9-DS | 876 | 96,373 | This study; AF132735 |
| ExgS | GH48-DS | 727 | 80,485 | 9; U34793 |
| EngH | GH9-CBDIII-DS | 715 | 79,321 | This study; U34793 |
| EngL | GH9-DS | 522 | 57,629 | 21; AF132735 |
| EngB | GH5-DS | 441 | 48,823 | 5; M37456 |
| ManA | DS-GH5 | 425 | 47,156 | 21; AF132735 |
| Cbp | CBD-SLH-(HBD)2-SLH-(HBD)6-(SLH)2-HBD | 1,848 | 189,149 | 18; M73817 |
| HbpA | SLH-HBD | 240 | 24,930 | 21; AF132735 |
Catalytic modules are shown in boldface. Module abbreviations: CBDIV, family IV cellulose-binding domain; GH9, family 9 glycosyl hydrolase; Ig, immunoglobulin-like domain; X, unknown domain.
Includes signal sequence.
Molecular weights were determined from the peptide sequences.
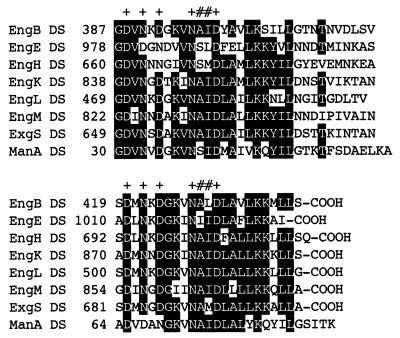
The presence of DSs (or dockerins), each sequence consisting of about 22 amino acids, is one of the tell-tale signs of a cellulase enzyme belonging to the cellulosome. The cellulosomal gene products are all characterized by the presence of a DS, usually at the C terminus of the protein, although the DS of ManA is located at its N terminus (Fig. 2). Although a DAL or DAI motif is conserved in the DSs from C. cellulolyticum and C. josui and an NST motif is conserved in those from C. thermocellum (7), this motif of C. cellulovorans is replaced by NAI. Since the cohesin-dockerin interaction in Clostridium species is a species-specific phenomenon (12), the C. cellulovorans NAI motif may be essential as a recognition code for binding specificity. Furthermore, the linkage of the DS to the catalytic domain may have a special structure since, almost invariably, when these enzyme subunits are expressed in E. coli, a protease in E. coli cleaves off the DS and leaves a still-active catalytic domain. This suggests strongly that a protease-accessible structure is present between the catalytic domain and DS domains of C. cellulovorans cellulosomal enzymes.
FIG. 2.
Alignment of the DSs of cellulosomal subunits of C. cellulovorans. Amino acids which are conserved in at least five of the eight sequences are highlighted. Identical amino acid residues are highlighted. Pluses indicate amino acid residues involved in calcium binding. Residues suspected of serving as selectivity determinants are indicated by pound signs.
DNA sequence of hbpA and domain structure of HbpA.
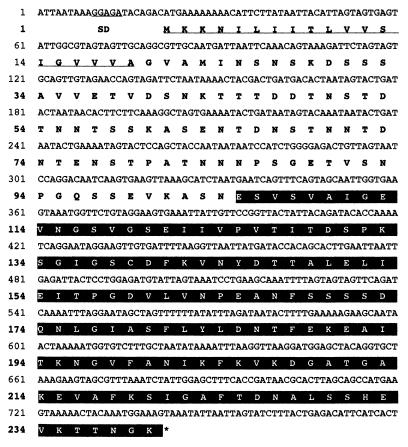
Figure 3 shows the complete nucleotide sequence of the hbpA structural gene along with its flanking regions. The hbpA gene consists of 720 nucleotides encoding a protein of 240 amino acids with a predicted molecular weight of 24,930. The putative initiation codon (ATG) is preceded by a spacing of 7 bp and by a typical ribosome-binding sequence, AGGAG, which is homologous to the consensus Shine-Dalgarno sequence. Downstream of the TAA translation termination codon, a transcription terminator was not observed, suggesting that hbpA and engL are in an operon.
FIG. 3.
Nucleotide and deduced amino acid sequences of hbpA and HbpA, respectively. The Shine-Dalgarno (SD) and signal peptide sequences are underlined. The stop codon is indicated by an asterisk. The amino acids of the HBD are highlighted.
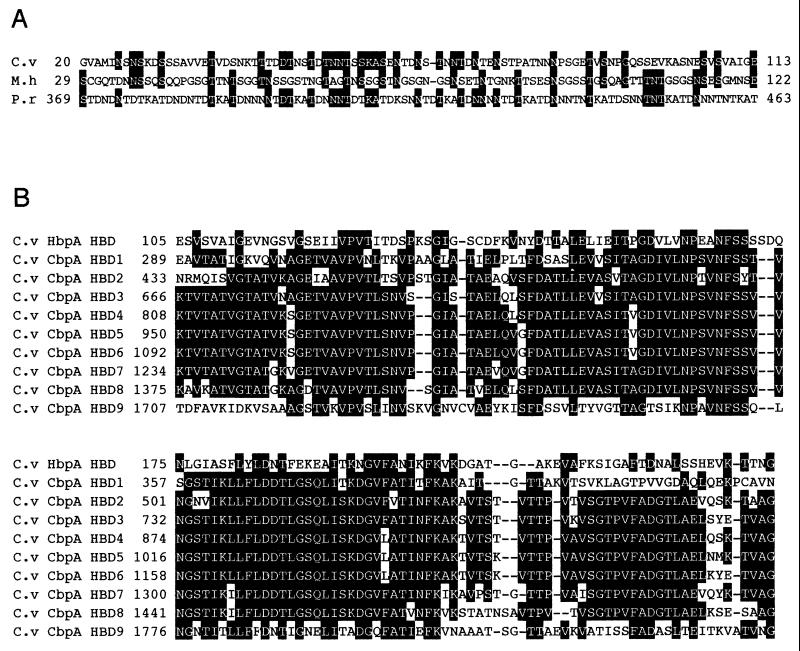
The N-terminal amino acid sequence of HbpA exhibits a typical signal peptide and consensus sequence (Val-X-Ala) (23), where the predicted cleavage site is located between positions 19 (Ala) and 20 (Gly) (Fig. 3). The N-terminal region of HbpA (residues 20 to 104) contains a surface layer homology (SLH) domain which shows homology with S-layer proteins from Mycoplasma hyorhinis (18.5% identity and 84.5% similarity among 103 amino acids; accession no. P29228) and Plasmodium reichenowi (26.5% identity; 91.6% similarity among 83 amino acids; accession no. Z30339) (Fig. 4A). The SLH sequences vary among different surface layer proteins but can be recognized as SLH domains by a few conserved identical amino acids (15).
FIG. 4.
(A) Alignment of the N-terminal region of HbpA from C. cellulovorans (C.v) with the corresponding proteins from M. hyorhinis (M.h) and P. reichenowi (P.r). (B) Alignment of the C-terminal region of HbpA with HBDs of CbpA from C. cellulovorans (C.v). Identical amino acids are highlighted. Gaps left to improve the alignment are indicated by dashes. The numbers refer to amino acid residues at the start of the respective lines; all sequences are numbered from Met-1 of the peptide.
Also, the N terminus of HbpA has several potential O-glycosylation sites. Since it does not contain a DS, HbpA most likely does not bind to CbpA and is not part of the cellulosome. The C-terminal region (residues 105 to 240) shows 32 to 37% identity with HBDs of CbpA (18) (Fig. 4B), while this region has about the same identity with type I cohesins of other Clostridium species (data not shown). Furthermore, the whole HbpA sequence reveals 29.6% identity and 86.2% similarity to C. cellulolyticum ORFXp (11) (Fig. 5). The presence of the N-terminal SLH domain suggests that HbpA is a cell surface-bound protein with some function in cellulosome assembly, as postulated previously for a similar protein, ORFXp, from C. cellulolyticum (11). It was postulated that the cohesin in ORFXp acts as a temporary binding station for cellulosomal enzymes that are destined for CipA during the assembly of the cellulosome (11). A significant difference between C. cellulolyticum ORFXp and C. cellulovorans HbpA is the absence of an SLH domain in ORFXp. The presence of the glycosylation sites suggests that HbpA can be glycosylated, while ORFXp is highly glycosylated (11). Thus, the occurrence of this small, hydrophobic protein may be widespread among mesophilic clostridia that produce cellulosomes.
FIG. 5.
Alignment of C. cellulovorans (C.v) HbpA with C. cellulolyticum (C.c) ORFXp. The gap left to improve the alignment is indicated by a dash. Identical and similar amino acid residues are indicated by asterisks and dots, respectively. The numbers refer to amino acid residues at the start of the respective lines; all sequences are numbered from Met-1 of the peptide.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to GenBank under accession no. U34793 and AF132735.
Acknowledgments
This research was supported in part by grant DE-FG03-92ER20069 from the U.S. Department of Energy.
REFERENCES
- 1.Bayer E A, Shimon L J W, Shoham Y, Lamed R. Cellulosomes—structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 2.Belaich J P, Belaich A, Fierobe H P, Gal L, Gaudin C, Pages S, Reverbel-Leroy C, Tardif C. The cellulolytic system of Clostridium cellulolyticum. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers; 1999. pp. 479–487. [Google Scholar]
- 3.Doi R H, Park J-S, Liu C-C, Malburg L M, Tamaru Y, Ichi-Ishi A, Ibrahim A. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles. 1998;2:53–60. doi: 10.1007/s007920050042. [DOI] [PubMed] [Google Scholar]
- 4.Foong F, Hamamoto T, Shoseyov O, Doi R H. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J Gen Microbiol. 1991;137:1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- 5.Foong F C-F, Doi R H. Characterization and comparison of Clostridium cellulovorans endoglucanases-xylanases EngB and EngD hyperexpressed in Escherichia coli. J Bacteriol. 1992;174:1403–1409. doi: 10.1128/jb.174.4.1403-1409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujino T, Karita S, Ohmiya K. Nucleotide sequence of the celB gene encoding endo-1,4-β-glucanase-2, ORF1 and ORF2 forming a putative cellulase gene cluster of Clostridium josui. J Ferment Bioeng. 1993;76:243–250. [Google Scholar]
- 7.Kakiuchi M, Isui A, Suzuki K, Fujino T, Fujino E, Kimura T, Karita S, Sakka K, Ohmiya K. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180:4303–4308. doi: 10.1128/jb.180.16.4303-4308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamed R, Bayer E A. The cellulosome concept: exocellular and extracellular enzyme factor centers for efficient binding and cellulolysis. In: Aubert J-P, Béguin P, Millet J, editors. Biochemistry and genetics of cellulose degradation. San Diego, Calif: Academic Press, Inc.; 1988. pp. 101–116. [Google Scholar]
- 9.Liu C-C, Doi R H. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene. 1998;211:39–47. doi: 10.1016/s0378-1119(98)00081-x. [DOI] [PubMed] [Google Scholar]
- 10.Matano Y, Park J-S, Goldstein M A, Doi R H. Cellulose promotes extracellular assembly of Clostridium cellulovorans cellulosomes. J Bacteriol. 1994;176:6952–6956. doi: 10.1128/jb.176.22.6952-6956.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagès S, Bélaïch A, Fierobe H-P, Tardif C, Gaudin C, Bélaïch J-P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181:1801–1810. doi: 10.1128/jb.181.6.1801-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagès S, Bélaïch A, Bélaïch J-P, Morag E, Lamed R, Shoham Y, Bayer E A. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Protein. 1997;29:517–527. [PubMed] [Google Scholar]
- 13.Rainey F A, Stackebrandt E. 16 S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 15.Sára M, Sleytr U B. S-layer proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoseyov O, Doi R H. Essential 170 kDa subunit for degradation of crystalline cellulose of Clostridium cellulovorans cellulase. Proc Natl Acad Sci USA. 1990;87:2192–2195. doi: 10.1073/pnas.87.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoseyov O, Hamamono T, Foong F, Doi R H. Cloning of Clostridium cellulovorans endo-1,4-β-glucanase genes. Biochem Biophys Res Commun. 1990;169:667–672. doi: 10.1016/0006-291x(90)90382-w. [DOI] [PubMed] [Google Scholar]
- 18.Shoseyov O, Takagi M, Goldstein M, Doi R H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A (CbpA) Proc Natl Acad Sci USA. 1992;89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sleat R, Mah R A, Robinson R. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984;48:88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamaru Y, Doi R H. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J Bacteriol. 1999;181:3270–3276. doi: 10.1128/jb.181.10.3270-3276.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamaru Y, Doi R H. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal ManA. J Bacteriol. 2000;182:244–247. doi: 10.1128/jb.182.1.244-247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamaru Y, Liu C-C, Malburg L, Doi R H. The Clostridium cellulovorans cellulosome and non-cellulosomal cellulases. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers; 1999. pp. 488–494. [Google Scholar]
- 23.von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]