Abstract
The predictive modeling literature for biomedical applications is dominated by biostatistical methods for survival analysis, and more recently some out of the box machine learning approaches. In this article, we show a presentation of a machine learning method appropriate for time-to-event modeling in the area of prostate cancer long-term disease progression. Using XGBoost adapted to long-term disease progression, we developed a predictive model for 118 788 patients with localized prostate cancer at diagnosis from the Department of Veterans Affairs (VA). Our model accounted for patient censoring. Harrell’s c-index for our model using only features available at the time of diagnosis was 0.757 95% confidence interval [0.756, 0.757]. Our results show that machine learning methods like XGBoost can be adapted to use accelerated failure time (AFT) with censoring to model long-term risk of disease progression. The long median survival justifies and requires censoring. Overall, we show that an existing machine learning approach can be used for AFT outcome modeling in prostate cancer, and more generally for other chronic diseases with long observation times.
Keywords: machine learning, xgboost, survival analysis, predictive modeling
INTRODUCTION
Historically, predictive models have been dominated by classical statistical approaches. In recent years, access to an increasing volume and diversity of biomedical data, as well as advances in computational capabilities have generated significant interest in application of machine learning for biomedical research.1 Machine learning techniques offer several advantages over statistical approaches: they can account for large numbers of independent variables and multiple outcomes, making fewer assumptions regarding the shape of the predictor and hazard functions. Machine learning approaches also have drawbacks when they do not include censoring information. For chronic diseases with long observation timeframes, loss to follow-up is of serious concern. Statistical approaches account for these situations by capturing the observation window and censoring time. In this article, we present a domain-aware predictive modeling approach based on XGBoost2 for prostate cancer disease progression using machine learning that considers time-to-event with censoring for a direct comparison with traditional biostatistical approaches.
RELATED WORK
Prostate cancer is the leading malignancy in male veterans3 and the second in United States males.4 Annually, over 13 000 new cases are diagnosed in the veteran population.3 Fortunately, 80% of new prostate cancer diagnoses are non-metastatic. In this population, treatment options to improve long-term survival and maximize quality of life are extremely complex,5 and there is significant need for data driven predictive models to inform the course of therapy. Over the years, several models of prostate cancer survival have been developed to address the need for survival prediction tools, nomograms, and calculators available to both patients and physicians. The Memorial Sloan Kettering nomograms6 are widely used in the prostate cancer community and were built using data from 10 000 prostate cancer cases. The PREDICT prostate7 tool used multivariate Cox models applied to 10 089 non-metastatic prostate cancer cases in England and externally validated on 2546 cases in Singapore. These tools were mostly developed using traditional biostatistical approaches that have dominated predictive modeling since the 1980s. Recently, the availability of the MIMIC dataset8 has resulted in more machine and deep learning approaches being used out of the box for biomedical predictive modeling.9 However, adapting machine and deep learning techniques to time to event with censoring has not received as much attention from the research community, and methods for doing so are still being investigated.10 Outcome modeling with random survival trees (RSFs)11 is a machine learning non-parametric alternative to the traditional Cox proportional hazards model. Due to the nature of the random forest it is based on, RSF is more sensitive to unbalanced datasets, such as those used for this analysis, compared to XGBoost. DeepSurv12 is an implementation of a Cox proportional hazards model using a deep neural network.13 discusses adapting generative adversarial networks for time to event modeling with censoring. In ref.,14 the investigators used XGBoost without time to event with censoring for mortality prediction 10 years after diagnosis in a 76 693 patient cohort recruited as part of a multicenter project in the United States. The studies discussed in this section also focus on a singular outcome (mortality).
METHODS
This study has been approved by the VA Central Institutional Review Board (IRB).
Cohort selection
Our cohort consisted of patients with localized prostate cancer at diagnosis from the Department of Veterans Affairs (VA) Corporate Data Warehouse (CDW) cancer registry. We defined localized prostate cancer using the tumor, node, metastases (TNM) staging system: having clinical N0 and M0. We included only patients with a Gleason score clinical between 6 and 10, and restricted our study to individuals with a baseline prostate-specific antigen (PSA) between 1 and 100. We also excluded patients without a biopsy confirmed diagnosis and those who had another primary cancer either before or after the prostate cancer diagnosis.
Predictors
The 13 predictors measured at or before primary prostate cancer diagnosis, fell in 2 categories: (1) A set of independent variables abstracted from the CDW cancer registry characterizing patient demographics (age at diagnosis, race, ethnicity) and disease staging (Gleason score clinical, AJCC stage group, SEER summary stage and a computed stage value from the registry (TNM) variables); (2) PSA values, aggregated into minimum, maximum, average, density, standard deviation values across the 5-year period prior to diagnosis. In addition, we included the last PSA before diagnosis adjudicated over the last year, the penultimate PSA over the last 5 years, and their rate of change.
Outcome
The outcome was a composite in days from primary diagnosis, consisting of cancer-related death from the National Death Index as well as registry documentation, a PSA >50, and inpatient and outpatient ICD codes indicative of metastatic disease. In our dataset, more than half of the population was censored, therefore we could not calculate a median survival/disease progression, and we reported survival in 5-year intervals.
Censoring
We censored all patients 1 year after their last PSA value or December 31, 2017, whichever came first.
Computational methods
We used a well-known machine learning algorithm, XGBoost15 adapted for accelerated failure time (AFT).2 The modification uses XGBoost to fit a good tree ensemble τ(x). It expresses the AFT model of the form:
where Y is the output label, x is the input feature vector. Since the noise is a random variable, the goal of xgboost is maximizing ln(Y) by fitting a good tree ensemble τ(x). Unlike traditional survival analysis methodologies that compute a hazard ratio, our machine learning algorithm calculated time to event survival. The model training and 5-fold cross-validation used 80% of the dataset. The remaining 20% was used solely for independent testing of the trained model to compute the performance metrics. We performed grid search to determine the max_depth, learning_rate, aft_loss_distribution_scale, min_child_weight, n_estimators hyperparameters needed for the model. Our step by step methodology followed the approach detailed in ref.16
Explainability
For improving the explainability of the machine learning approach, we derived feature importance measures using a tree solution for SHAP (SHapley Additive explanation),17 a method that is both consistent and accurate.18 The SHAP value of each feature for each prediction is the marginal contribution of that feature to the output. In addition to averaging these contributions for all patients in our dataset, we also generated instance level predictions for 2 scenarios: a patient with more aggressive disease at diagnosis and one with lower grade disease.
Metrics
To determine the discrimination power of our algorithms, we used 2 metrics: the modified Harrell’s c-statistic, and the accuracy. We modified the Harrell’s c-statistic calculation19 to use the time to event survival predictions generated by our model instead of the risk to generate the list of concordant pairs and ties.
RESULTS
Cohort selection
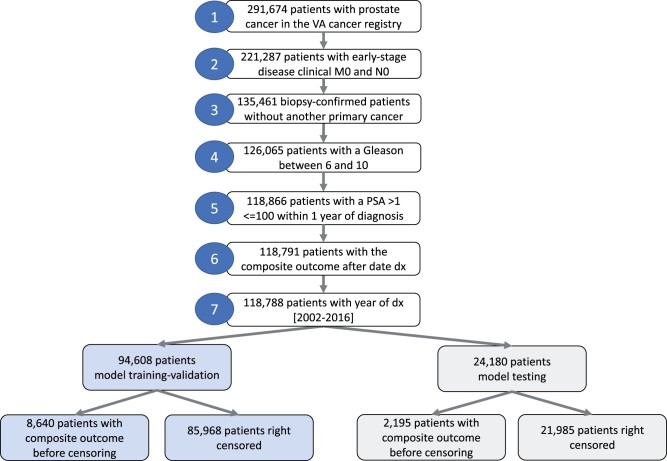
Figure 1 below shows the cohort selection procedure.
Figure 1.
Cohort selection.
Population
Table 1 shows the population demographics in detail. Our cohort is dominated by 55–74 years olds, consistent with previous studies and current screening guidelines. The dates of diagnosis span a period of 14 years from 2002 to 2016. Our population consists of a higher percentage of African Americans compared to the VA overall.20 The Gleason scores, stage, and PSA at diagnosis are consistent with localized disease. More than 50% of the population is outcome-free at 5 years with 14.19% observed outcome-free 10 years after diagnosis.
Table 1.
Population demographics
| Train-validation set (n = 94 608) | Test set (n = 24 180) | ||
|---|---|---|---|
| Demographics | |||
| Age at diagnosis | |||
| Patient count (% of total) | |||
| <55 | 5745 (6.07%) | 1540 (6.36%) | |
| 55–64 | 36 323 (38.39%) | 9231 (38.17%) | |
| 65–74 | 39 674 (41.93%) | 10 134 (41.91%) | |
| ≥75 | 12 866 (13.59%) | 3275 (13.54%) | |
| Race | |||
| Patient count (% of total) | |||
| African-American | 27 239 (28.79%) | 7038 (29.1%) | |
| East Asian | 185 (0.19%) | 47 (0.19%) | |
| South Asian | 28 (0.02%) | 5 (0.02%) | |
| Polinesian/Hawaian | 153 (0.16%) | 36 (0.14%) | |
| White | 63 903 (67.54%) | 16 264 (67.26%) | |
| Other/unknown | 3100 (3.27%) | 790 (3.26%) | |
| Ethnicity | |||
| Patient count (% of total) | |||
| Hispanic or Latino | 5268 (5.56%) | 1318 (5.45%) | |
| Not Hispanic or Latino | 89 340 (94.43%) | 22 862 (94.54%) | |
| Staging variables | |||
| T-stage | |||
| Patient count (% of total) | |||
| 1A | 726 (0.76%) | 188 (0.19%) | |
| 1B | 419 (0.44%) | 119 (0.49%) | |
| 1C | 64 691 (68.37%) | 16 597 (68.63%) | |
| 1 | 335 (0.35%) | 79 (0.32%) | |
| 2A | 10 087 (10.66%) | 2549 (10.54%) | |
| 2B | 4230 (4.47%) | 1115 (4.61%) | |
| 2C | 8588 (9.07%) | 2167 (8.96%) | |
| 2 | 3749 (3.96%) | 936 (3.87%) | |
| 3 | 1598 (1.68%) | 385 (1.59%) | |
| 4 | 181 (0.19%) | 44 (0.18%) | |
| missing | 4 (0%) | 1 (0%) | |
| Gleason score | |||
| Patient count (% of total) | |||
| 6 | 38 937 (41.15%) | 9919 (41.02%) | |
| 7 | 39 828 (42.09%) | 10 332 (42.72%) | |
| 8 | 9223 (9.74%) | 2280 (9.42%) | |
| 9 | 6078 (6.42%) | 1509 (6.24%) | |
| 10 | 542 (0.57%) | 140 (0.57%) | |
| PSA at diagnosis | 9.777 (10.844) | 9.706 (10.733) | |
| mean (std) | |||
| Outcome | |||
| Composite outcome | |||
| Patient count (% of total) | |||
| <5 years | 5545 (5.86%) | 1419 (5.86%) | |
| 5–10 years | 2501 (2.64%) | 632 (2.61%) | |
| >10 years | 594 (0.62%) | 144 (0.59%) | |
| Censoring | |||
| Patient count (% of total) | |||
| <5 years | 39 766 (42.03%) | 10 197 (42.17%) | |
| 5–10 years | 32 768 (34.63%) | 8331 (34.45%) | |
| >10 years | 13 434 (14.19%) | 3457 (14.29%) |
Performance metrics
The model shows robust performance from the validation to the test dataset. The test c-index is 0.757, with a narrow confidence interval. Table 2 shows the accuracy and c-index metrics on the validation and test datasets.
Table 2.
Performance metrics
| Validation accuracy [95% CI] | Validation c-index [95% CI] | Test accuracy [95% CI] | Test c-index [95% CI] |
|---|---|---|---|
| 0.889 [0.888, 0.889] | 0.764 [0.759, 0.768] | 0.891 [0.890, 0.891] | 0.757 [0.756, 0.757] |
Explainability
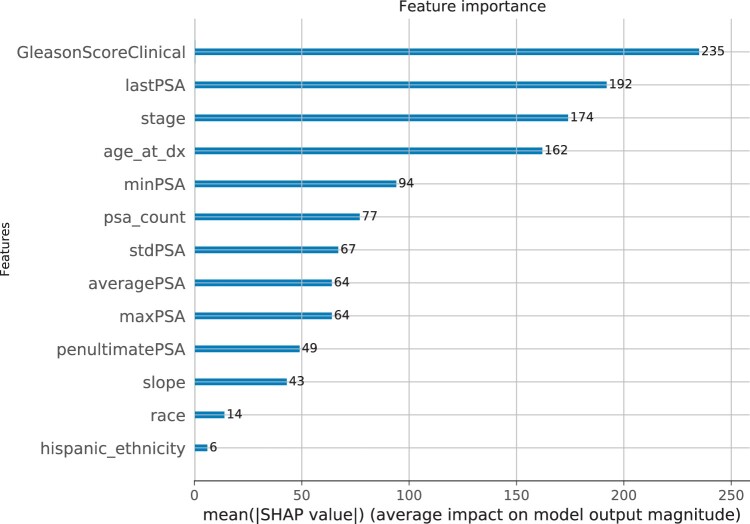
Figure 2 shows the average impact of the independent variables on model output magnitude. The Gleason score is the most important overall feature, and the Hispanic/Latino ethnicity has the lowest impact.
Figure 2.
SHAP predictor importance (average impact on model output magnitude).
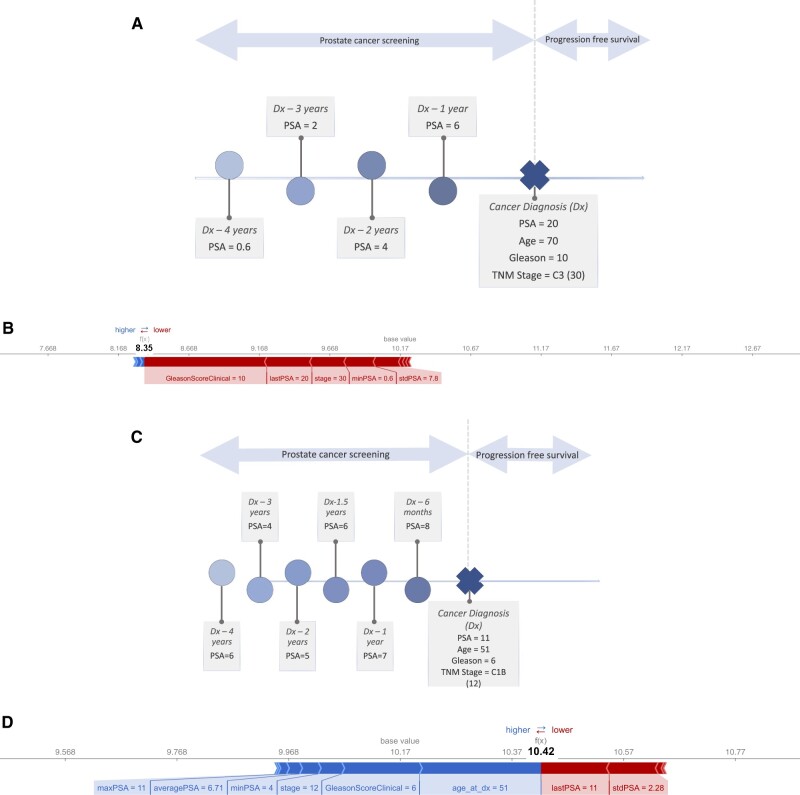
At an individual patient level, the feature importance varies, as shown in Figure 3. For a hypothetical patient 1 (Figure 3a and 3b), with more aggressive disease at diagnosis, the higher Gleason score, PSA and stage reduce the survival time (f(x)) from the average (base value).
Figure 3.
(A) Instance level predictor importance for hypothetical patient 1 with more aggressive disease at diagnosis—Hypothetical patient 1 timeline. (B) Instance level predictor importance for hypothetical patient 1 with more aggressive disease at diagnosis—Outcome prediction. (C) Instance level predictor importance for hypothetical patient 2 with less aggressive disease at diagnosis—Hypothetical patient 2 timeline. (D) Instance level predictor importance for hypothetical patient 2 with less aggressive disease at diagnosis—Outcome prediction.
For hypothetical patient 2 (Figure 3c and 3d), with less aggressive disease at diagnosis, the age, Gleason score, stage, minimum, maximum, and average PSA values before diagnosis drive an increase in the survival, while the last PSA and the standard deviation of the PSA values contribute to a reduction.
DISCUSSION
Overall, we show an approach that uses an existing machine learning method adapted for AFT with censoring to model long-term risk of disease progression. In our study, 14% of the population is censored more than 10 years after diagnosis indicating that loss to follow-up is an important consideration. Especially when they align more closely with traditional biostatistical methods, the value proposition of machine learning over other approaches are multifold. First, unlike Cox, we are not making any assumptions regarding the contributions of predictors in our survival function over time. Secondly, machine learning algorithms like XGBoost are well-suited computationally for problems with large numbers of predictors. Our model was intentionally simpler with only 13 predictors, but our methods are scalable to problems with hundreds of independent variables. Machine learning approaches also provide better incorporation of interaction terms: while they can be included in regression, this process is cumbersome as the range of possibilities is broad, even for pairwise interactions. Additionally, having the capability to analyze the predictor importance, will allow us to factor in these hundreds of predictors across the entire electronic health record. Since SHAP values depend on the choice of predictors, their degree of collinearity and ultimately the type of machine learning algorithm, correctly contextualizing the predictor importance given the clinical problem, will allow us to find the most relevant ones.
The predictor importance plots in Figures 2 and 3 highlight another benefit of machine learning models. Whereas in traditional regression models, the coefficients for each feature are static, our modeling paradigm allows for recommendations tailored to the individual patient. Traditional biostatistical approaches try to account for more personalized predictions using subgroup analysis, capability also accounted for by our machine learning approach that generates the decision tree splits as part of the algorithm training.
Our c-statistic is lower than other studies of long-term outcomes for prostate cancer patients: 0.847 and 0.8.14 We hypothesize this is due to a number of factors including (1) our prediction happens at the time of diagnosis not accounting for any treatment modalities that could significantly impact disease progression, as reported in other studies.14,21 The long follow-up time creates a long window of opportunity for patients with similar profiles at diagnosis to follow different treatment arms, and therefore have different outcomes; (2) our intent was to build a simple model using a large cohort with long follow-up times, that would enable clinicians to make informative decisions with the limited data available to them at the time of diagnosis. Consequently, we used a small set of predictors compared to other studies,14 not accounting for the medical history, physical activity or socio-economic status; (3) unlike other studies that focus on a single outcome, mortality (cancer related or all-cause), we used a composite that consists of cancer mortality and metastatic disease. In our scenario, 2 patients with similar profiles at diagnosis, could have significant times to outcome if one is missing an ICD code indicating metastasis; (4) our dataset includes patients diagnosed over a period of 14 years, time in which coding standards changed for our independent and dependent variables; (5) training also suffered from class imbalance problems because the cohort who had the outcome before censoring was about 10% of the total number of patients.
CONCLUSION
Machine and deep learning methodologies that can capture the full complexities of datasets are both needed and underrepresented in the literature and current biomedical practice. Our methods scale to complex problems with thousands of predictors and are applicable to a variety of chronic conditions with long observation periods, for which loss to follow-up is an important consideration. In the current data-rich biomedical environment, domain-specific computational methods such as machine and deep learning that scale to large longitudinal datasets hold the promise for future discoveries.
FUNDING
This research was supported by award MVP017 from the Million Veteran Program, Office of Research and Development, Veterans Health Administration.
This research used resources of the Knowledge Discovery Infrastructure at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725 and the Department of Veterans Affairs Office of Information Technology Inter-Agency Agreement with the Department of Energy under IAA No. VA118-16-M-1062.
AUTHOR CONTRIBUTIONS
ID proposed the study design, methods, wrote the first draft and oversaw subsequent revisions of the paper. ID and IG generated the data used for this study. All authors contributed to iterative study design and manuscript editing.
ACKNOWLEDGMENTS
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. This publication does not represent the views of the Department of Veteran Affairs, the Department of Energy or the United States Government. This project used data from the Center of Excellence for Mortality Data Repository, Joint Department of Veterans Affairs (VA) and Department of Defense (DoD) Suicide Data Repository–National Death Index (NDI). http://vaww.virec.research.va.gov/Mortality/Overview.htm; Extract <November 20, 2020>.
The authors also wish to acknowledge the support of the larger partnership. Most importantly, the authors would like to thank and acknowledge the veterans who chose to get their care at the VA.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
Contributor Information
Ioana Danciu, Advanced Computing for Health Sciences Group, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA; Department of Biomedical Informatics, Vanderbilt University, Nashville, Tennessee, USA.
Greeshma Agasthya, Advanced Computing for Health Sciences Group, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA.
Janet P Tate, Department of Veterans Affairs Connecticut Healthcare System, West Haven, Connecticut, USA; Yale School of Medicine, New Haven, Connecticut, USA.
Mayanka Chandra-Shekar, Advanced Computing for Health Sciences Group, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA.
Ian Goethert, Advanced Computing for Health Sciences Group, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA.
Olga S Ovchinnikova, Advanced Computing for Health Sciences Group, Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA.
Benjamin H McMahon, Theoretical Biology Group, Los Alamos National Laboratory, Los Alamos, New Mexico, USA.
Amy C Justice, Department of Veterans Affairs Connecticut Healthcare System, West Haven, Connecticut, USA; Yale School of Medicine, New Haven, Connecticut, USA; Yale School of Public Health, New Haven, Connecticut, USA.
Data Availability
Final datasets underlying this study cannot be shared outside the VA, except as required under the Freedom of Information Act (FOIA), per VA policy. However, upon request through the formal mechanisms in place and pending approval from the VHA Office of Research Oversight (ORO), a de-identified, anonymized dataset underlying this study can be created and shared.
REFERENCES
- 1. Goecks J, Jalili V, Heiser LM, Gray JW.. How machine learning will transform biomedicine. Cell 2020; 181 (1): 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnwal A, Cho H, Hocking TD. Survival Regression with Accelerated Failure Time Model in XGBoost. ArXiv200604920 Cs Stat, 2020. http://arxiv.org/abs/2006.04920 Accessed March 05, 2021.
- 3. Zullig LL, Sims KJ, McNeil R, et al. Cancer incidence among patients of the United States veterans affairs (VA) healthcare system: 2010 update. Mil Med 2017; 182 (7): e1883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CDCBreastCancer. Prostate Cancer Statistics. Centers for Disease Control and Prevention, 2021. https://www.cdc.gov/cancer/prostate/statistics/index.htm Accessed August 14, 2021.
- 5. Wilt TJ, Jones KM, Barry MJ, et al. Follow-up of prostatectomy versus observation for early prostate cancer. N Engl J Med 2017; 377 (2): 132–42. [DOI] [PubMed] [Google Scholar]
- 6.Prostate Cancer Nomograms | Memorial Sloan Kettering Cancer Center. https://www.mskcc.org/nomograms/prostate Accessed August 16, 2021.
- 7. Thurtle DR, Greenberg DC, Lee LS, Huang HH, Pharoah PD, Gnanapragasam VJ.. Individual prognosis at diagnosis in nonmetastatic prostate cancer: Development and external validation of the PREDICT Prostate multivariable model. PLoS Med 2019; 16 (3): e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson AE, Stone DJ, Celi LA, Pollard TJ.. The MIMIC Code Repository: enabling reproducibility in critical care research. J Am Med Inform Assoc 2018; 25 (1): 32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vock DM, Wolfson J, Bandyopadhyay S, et al. Adapting machine learning techniques to censored time-to-event health record data: a general-purpose approach using inverse probability of censoring weighting. J Biomed Inform 2016; 61: 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS.. Random survival forests. Ann Appl Stat 2008; 2 (3): 841–60. [Google Scholar]
- 12. Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y.. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol 2018; 18 (1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chapfuwa P, Tao C, Li C, et al. Adversarial time-to-event modeling. Proc Mach Learn Res 2018; 80: 735–44. [PMC free article] [PubMed] [Google Scholar]
- 14. Bibault J-E, Hancock S, Buyyounouski MK, et al. Development and validation of an interpretable artificial intelligence model to predict 10-year prostate cancer mortality. Cancers 2021; 13 (12): 3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. In: Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 13–17, 2016: 785–794; San Francisco, CA, USA. doi: 10.1145/2939672.2939785. [DOI]
- 16. Survival Analysis with Accelerated Failure Time—xgboost 1.5.2 Documentation. https://xgboost.readthedocs.io/en/stable/tutorials/aft_survival_analysis.html Accessed April 10, 2022.
- 17. Lundberg SM, Erion GG, Lee S-I. Consistent Individualized Feature Attribution for Tree Ensembles. 2018. https://arxiv.org/abs/1802.03888v3 Accessed August 19, 2021.
- 18. Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems, Vol. 30. 2017. https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html Accessed August 19, 2021.
- 19. Harrell FE. Evaluating the Yield of Medical Tests | JAMA | JAMA Network. https://jamanetwork.com/journals/jama/article-abstract/372568 Accessed March 06, 2021. [PubMed]
- 20. Veteran Population—National Center for Veterans Analysis and Statistics. https://www.va.gov/vetdata/veteran_population.asp Accessed September 02, 2021.
- 21. Hamdy FC, Donovan JL, Lane JA, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016; 375 (15): 1415–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Final datasets underlying this study cannot be shared outside the VA, except as required under the Freedom of Information Act (FOIA), per VA policy. However, upon request through the formal mechanisms in place and pending approval from the VHA Office of Research Oversight (ORO), a de-identified, anonymized dataset underlying this study can be created and shared.