Abstract
Nonalcoholic fatty liver disease (NAFLD) is an epidemic affecting 30% of the US population. It is characterized by insulin resistance, and by defective lipid metabolism and mitochondrial dysfunction in the liver. SLC25A34 is a major repressive target of miR-122, a miR that has a central role in NAFLD and liver cancer. However, little is known about the function of SLC25A34. To investigate SLC25A34 in vitro, mitochondrial respiration and bioenergetics were examined using hepatocytes depleted of Slc25a34 or overexpressing Slc25a34. To test the function of SLC25A34 in vivo, a hepatocyte-specific knockout mouse was generated, and loss of SLC25A34 was assessed in mice maintained on a chow diet and a fast-food diet (FFD), a model for NAFLD. Hepatocytes depleted of Slc25a34 displayed increased mitochondrial biogenesis, lipid synthesis, and ADP/ATP ratio; Slc25a34 overexpression had the opposite effect. In the knockout model on chow diet, SLC25A34 loss modestly affected liver function (altered glucose metabolism was the most pronounced defect). RNA-sequencing revealed changes in metabolic processes, especially fatty acid metabolism. After 2 months on FFD, knockouts had a more severe phenotype, with increased lipid content and impaired glucose tolerance, which was attenuated after longer FFD feeding (6 months). This work thus presents a novel model for studying SLC25A34 in vivo in which SLC25A34 plays a role in mitochondrial respiration and bioenergetics during NAFLD.
SLC25A34 is a poorly characterized family member of the SLC25 family that consists of 53 inner mitochondrial membrane proteins shown and/or predicted to function as solute carriers involved in the transport of biomolecules (eg, amino acids, nucleotides, carboxylates, keto acids, other substrates) across the inner mitochondrial membrane.1,2 Fourteen SLC25 family members are involved in metabolic diseases, and 15 are uncharacterized. To date, a single study in yeast suggests that SLC25A34 is homologous to an oxaloacetate carrier, Oacp1, involved in leucine biosynthesis.3,4 In mammals, leucine is considered an essential amino acid that is not synthesized naturally and must be obtained via diet, suggesting that SLC25A34 plays an alternate role.5 Oxaloacetate, a biomolecule produced in mitochondria during the Krebs cycle, is converted to reducing equivalents that can feed into a myriad of biochemical pathways which regulate energy metabolism. Thus, SLC25A34 may play a role in energy metabolism in mammals.
The liver is the central organ of energy metabolism, which regulates the metabolism of lipids and glucose to maintain energy homeostasis.6,7 Energy homeostasis is tightly regulated by the energy stores and key molecular players in insulin-responsive organs, all of which influence glucose, lipid, and mitochondrial homeostasis and thus help maintain energy homeostasis. Dysregulation of energy homeostasis is a hallmark of several metabolic diseases and is rampant in nonalcoholic fatty liver disease (NAFLD). NAFLD is a metabolic disorder that is currently a worldwide epidemic. Nearly 30% of the US population has NAFLD, which is exacerbated by the presence of risk factors such as obesity, type 2 diabetes, hypertension, and hypertriglyceridemia.8 In fact, the hepatic manifestation of metabolic syndrome is NAFLD, which presents in the form of insulin resistance and defects in lipid metabolism and mitochondrial dysfunction.9 NAFLD encompasses a broad spectrum of liver disorders beginning with hepatic steatosis and can progress to nonalcoholic steatohepatitis (NASH), hepatic fibrosis, cirrhosis, and hepatocellular carcinoma.8 NAFLD is predicted to be the leading cause of liver transplantation in the future.10 Alarmingly, there are no regulatory-approved pharmaceutic alternatives currently available to treat NAFLD except lifestyle management, and thus it poses a significant clinical burden.
SLC25A34 has been identified as a novel and strongly regulated target of miRNA-122 (miR-122) in which mice lacking miR-122 develop hepatic steatosis, hepatitis, and hepatocellular carcinoma.11, 12, 13 Intriguingly, altered Slc25a34 expression has been reported in mouse models that have altered energy metabolism (Table 1).14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 Moreover, there are conflicting observations of SLC25A34 expression in humans; one report shows up-regulation in NASH and simple steatosis,26 whereas a different report shows unchanged and down-regulated SLC25A34 in steatosis NASH and normal NASH,27 respectively. Metabolic dysfunction is a common theme uniting these studies, particularly altered glucose metabolism and fatty acid metabolism. Therefore, based on these observations, SLC25A34 is speculated to play a role in energy homeostasis.
Table 1.
Slc25a34 Dysregulation in Mouse Models Involving Metabolic Disorders
| Slc25a34 | Context | Metabolic relevance | GEO profile (ref.) |
|---|---|---|---|
| Down | MCD + HFD versus normal chow | NAFLD | GDS4883 (14) |
| Down | HFD versus normal chow | NAFLD | GDS4830 (15) |
| Down | Gpr120 knockout versus WT | FAM | GDS4830 (15) |
| Down | n3-PUFA–depleted diet versus normal chow | FAM | GDS4796 (16) |
| Down | Scd1 knockout on low fat, high carbohydrate diet versus chow | FAM | GDS1517 (17) |
| Down | Sucrose diet versus Palatinose diet | GM | GDS5435 (18) |
| Down | ob/ob mice versus WT lean | MD | GDS5258 (19) |
| Down | Ppar knockout versus WT | MD | GDS3748 (20) |
| Down | Trim24 knockout HCC versus normal liver | MD | GDS3087 (21) |
| Up | Metformin supplemented HFD + MCD versus MCD + HFD | NAFLD | GDS4883 (14) |
| Up | Mcad1 knockout versus WT, when fasted | FAM | GDS4546 (22) |
| Up | Npc1 knockout mice (Niemann-Pick Type 1 model), 55 vs 20 days old | FAM | GDS4427 (23) |
| Up | Gpa1 knockout (Gaucher disease model) versus control | FAM, GM | GDS4162 (24) |
| Up | Fasted versus fed | Bioenergetics | GDS3135 (25) |
FAM, fatty acid metabolism; GEO, Gene Expression Omnibus; GM, glucose metabolism; HCC, hepatocellular carcinoma; HFD, high-fat diet; MCD, methionine-choline–deficient diet; MD, metabolic disorder; n3-PUFA, n-3 polyunsaturated fatty acid; NAFLD, nonalcoholic fatty liver disease; WT, wild type.
In the current work, the metabolic role of SLC25A34 was investigated in both primary hepatocytes and a novel animal model. Initially, Slc25a34 was depleted and overexpressed in wild-type (WT) primary hepatocytes. Hepatocytes depleted of Slc25a34 displayed increased mitochondrial biogenesis, lipid synthesis, and ADP/ATP ratio, whereas Slc25a34 overexpression had the opposite effect. Second, a hepatocyte-specific Slc25a34 knockout mouse was generated to determine how SLC25A34 affects liver metabolism. Loss of SLC25A34 for up to 7 months modestly affected liver function, with altered glucose metabolism as the most pronounced defect. RNA-sequencing (RNA-seq) revealed widespread changes in metabolic processes, especially fatty acid metabolism. Finally, hepatocyte-specific Slc25a34 knockout mice were fed a fast-food diet (FFD), a model of NAFLD. After 2 months on FFD, knockout mice had a more severe phenotype, with increased lipid content in the liver and impaired glucose tolerance. Unexpectedly, the transcriptomic analysis revealed attenuation of lipogenesis programs that were observed after longer FFD feeding (6 months). Thus, SLC25A34 influenced energy metabolism at a basal level, and its effect is most pronounced during NAFLD-induced liver injury. The data suggest a model in which SLC25A34 deletion initially predisposes the liver for metabolic distress but, surprisingly, accelerates metabolic adaptation and protects the liver after extended injury.
Materials and Methods
Mouse Strains and Husbandry
The Institutional Care and Use Committee of the University of Pittsburgh approved all the experiments using mice. Optimice cages (Animal Care Systems, Centennial, CO) with Sani-Chip Coarse bedding (P.J. Murphy, Montville, NJ) were used for housing the mice. The mice were provided ad libitum access to water and standard mouse chow (Purina ISO Pro Rodent 3000, LabDiet, St. Louis, MO) in a 12-hour light and 12-hour dark cycle. Huts and running wheels were provided for enrichment. All animals were euthanized between 9:00 am and noon daily. Male WT C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) and Mir122 germline knockout (Mir122-KO) mice were used.11,12 Slc25a34 knockout and hepatocyte-specific knockout mice were generated in-house.
Generation of Slc25a34 Floxed Mice
The Slc25a34 gene contains 5 exons, but the intron/exon junctions are all in the same reading frame. It was therefore not possible to target an exon whose deletion, after Cre-mediated recombination, would create a frameshift and a null allele. Instead, the targeting strategy was to flank exon 1 and the promoter (conserved region as defined on phylogenetic sequence conservation) with LoxP sites. The conditional knockout allele was generated directly in mouse zygotes by using CRISPR/Cas9 technology following the Easi-CRISPR strategy in which long single-stranded DNA serves as a template and is injected with pre-assembled Cas9 ribonucleoprotein complexes.28 In essence, LoxP sites were inserted at the site of the SpyCas9 targeting sequence, and EcoRI restriction sites were introduced adjacent to the LoxP site to facilitate genotyping. Briefly, fertilized C57BL/6J embryos produced by natural mating were microinjected in pronuclei with a mixture of 0.3 μM EnGen Cas9 protein (M0646T; New England Biolabs, Ipswich, MA), Slc25a34-guide3, Slc25a34-guide6 (21.23 ng/μL each), and a long single-stranded oligonucleotide “Slc25a34-flox-Megamer” (10 ng/μL) (Integrated DNA Technologies, Coralville, IA). The injected zygotes were cultured overnight, and embryos that developed to the two-cell stage were transferred to oviducts of pseudopregnant CD1 female recipients. Mice carrying the correctly targeted allele were identified by using PCR, and the sequence of the targeted allele was verified according to Sanger sequencing. Gene targeting produced heterozygous mice with a floxed Slc25a34 allele as well as heterozygotes with a knockout allele, presumably generated by spontaneous recombination.29,30 Founder mice were backcrossed to C57BL/6J for at least five generations and bred to homozygosity.
The single guide RNA templates were generated by PCR and were used for the synthesis of the single guide RNAs using the HiScribe T7 Quick High Yield RNA Synthesis Kit (E2050S; New England Biolabs).29 The single guide RNAs were purified by using the MEGAclear Kit (Thermo Fisher Scientific, Pittsburgh, PA) following the manufacturer’s instructions. The target sequence single guide RNAs are 5′-GAGAGAATAGGGCTATAATCTGG-3′ (Slc25a34-guide3) and 5′-AAACTGACACGCCCAACCCAGGG-3′ (Slc25a34-guide6). The sequence of the long single-stranded oligonucleotide Slc25a34-flox-Megamer, including EcoRI (bold) and LoxP sites (underlined), is as follows: 5′-AGGCAGAGGCGGGCGGATCTCTGTGCGTCTATGGCCACCTGATCTACATAGCTAGTTTCAGGCCAGCCAGGGTTATATAGAGAGACCTCGCTTCAAAGAAAAAAGATATTAAGTGAATAAACCTTTGAGAGAGATGAGAGAGAATAGGGCTATAGAATTCATAACTTCGTATAATGTATGCTATACGAAGTTATATCTGGCAGAGGTTAAAACGGCACAGTTGCTGGTTGGCCACGCTTACAGCCAAGACACCACAAGCCTAGGGCTTTTAGCTGGAGCCCTACAGGGACAGGAGATGGAACAGGAAGTCTAGAGGACAAGGAGGGGTGCCCTCCCCTACTCTAGCTTGGCTCTTTCTTCCTGCCCACTGTCCTTCCAGTCACAAGTGACCTTTCCTCCAGATCCCATGGCCCAGGCCCGCCCTGGCCAGCAGCAGCTGGGATTGGTCTGAGCCAGAGGAGGTGTGTCCAGATTCCCATAGCCCTGCTGATGAACAGCAGGTGGAGTTCCAAGACAACCAGGGGCCACCAGCCAGTGGCAGGATCTCTGCCCTCTAGACATGGTGCTGGCCCCCTTTGATCCCCAGCCCTGTGACCATGCAGAGGTACAGACTGCCTAGAGACTACCATCTATAGCCTGTGCCATGAAGCCAACTCAGGCACAGATGGCTCCTGCCATGGATTCGAGGGAGATGGTGTCCCCGGCTGTGGACCTGGTGCTGGGTGCCTCAGCCTGCTGCCTGGCCTGTGTATTCACCAACCCCCTGGAAGTGGTAAAGACCCGTCTACAACTGCAGGGGGAACTGCAGGCCCCAGGCACCTACCCACGGCCCTACCGGGGCTTTGTGTCTTCTGTTGCAGCCGTGGCCCGGGCAGATGGGCTATGGGGCCTGCAGAAGGGGCTGGCTGCTGGCCTTCTCTACCAGGGCCTCATGAATGGTGTCCGTTTCTACTGCTATAGCCTGGCATGCCAGGCTGGCCTCACCCAACAACCAGGCGGCACTGTGGTCGCAGGCGCTGCGGCTGGGGCATTGGGGGCCTTCGTGGGGAGTCCTGCTTACCTGGTAAGTGTCTTCTCCTTCCTTTATAGCTCCGTGGCCAACGTGACCAGTGGACTTTGTGGCCCTGGGAATTCATAACTTCGTATAATGTATGCTATACGAAGTTATGTTGGGCGTGTCAGTTTGGGAGGGGACTGTTCGCTGGGAATATCTGGCTCAGACCTGAAATCTGCCCTCACTCTGACCCTTGCTTCTTTGGGAGCCATGTCCACTGGTCTTTGCTGGGAGAGGGAGCAGGTTATGGAGTAGGTAGATTCTAGTCCTCGCTTGCCTGTGGTGG-3′.
Genotyping Slc25a34 Floxed and Knockout Mice
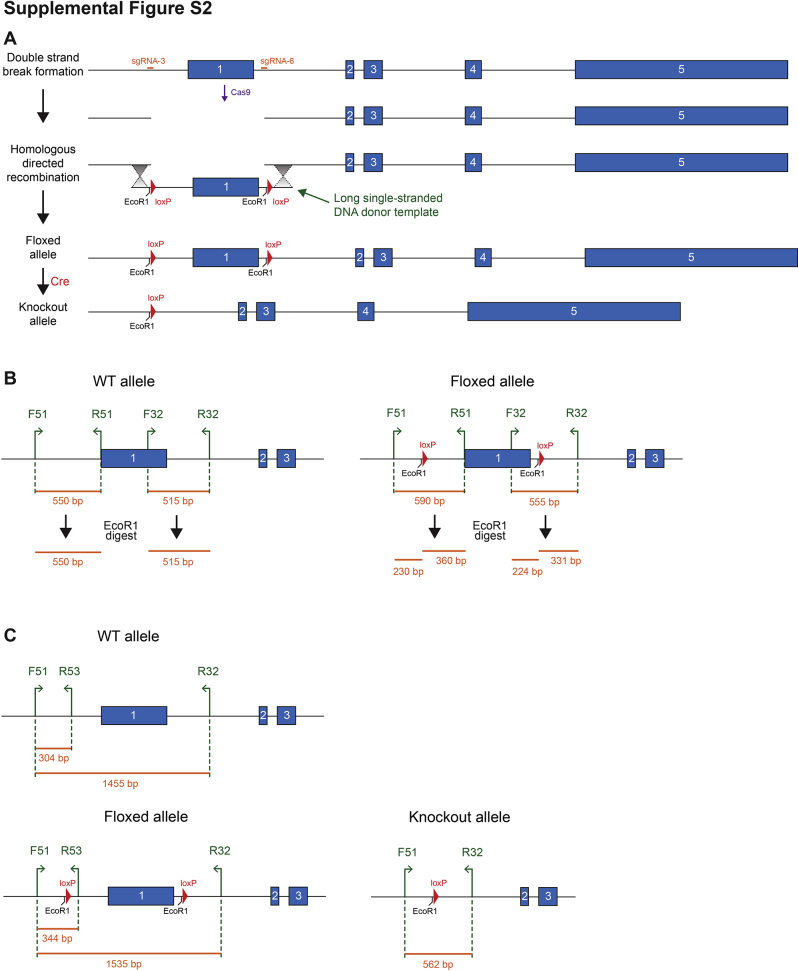
To confirm the presence of a floxed Slc25a34 allele, DNA was amplified with oligos F51 and R51 (to target to amplify the 5′ loxP site) and digested with EcoR1. Primer sequences are provided (Table 2). The WT allele generates a 550 bp fragment that lacks an EcoR1 site, whereas the floxed allele generates a 590 bp fragment upon amplification and 230 bp and 260 bp fragments after digestion. Similarly, to confirm the presence of the 3′ loxP site, DNA was amplified with oligos F32 and R32. The WT allele generates a 515 bp fragment that lacks an EcoR1 site, whereas the floxed allele generates a 555 bp fragment upon amplification and 224 bp and 331 bp fragments after digestion. Identification of the knockout allele was confirmed by a three-primer PCR using oligos F51, R53, and R32, which produces unique fragments for the WT allele (304 and 1455 bp), the floxed allele (344 and 1535 bp), and the knockout allele (562 bp).
Table 2.
Primers and TaqMan Probes (Both from Thermo Fisher Scientific) for Gene Expression, mtDNA Copy Number, and Genotyping Assays
| Application | Gene/locus | Name | Sequence or TaqMan identifier |
|---|---|---|---|
| Gene expression | 18S Rrna | 18S-fwd | 5′-TGACGGAAGGGCACCACCAG-3′ |
| 18S Rrna | 18S-rev | 5′-TCGCTCCACCAACTAAGAACGGC | |
| Acly | Acly-fwd | 5′-GGAGTCAAATCCTGGCTAAAACCTC-3′ | |
| Acly | Acly-rev | 5′-TTTCGTCCACACCCACAAGCAG-3′ | |
| B2m | B2M-fwd | 5′-TTCAGTATGTTCGGCTTCCC-3′ | |
| B2m | B2m-rev | 5′-CCTGGTCTTTCTGGTGCTTG-3′ | |
| Cpt1a | Cpt1a-fwd | 5′-CTCAGTGGGAGCGACTCTTCA-3′ | |
| Cpt1a | Cpt1a-rev | 5′-GGCCTCTGTGGTACACGACAA-3′ | |
| Gapdh | Gapdh-fwd | 5′-AAGGTCGGTGTGAACGGATTTGG-3′ | |
| Gapdh | Gapdh-rev | 5′-CGTTGAATTTGCCGTGAGTGGAG-3′ | |
| Ppargc1a | Ppargc1a-fwd | 5′-GTCATTCGGGAGCTGGATGG-3′ | |
| Ppargc1a | Ppargc1a-rev | 5′-CAACCAGAGCAGCACACTCT-3′ | |
| Ppargc1b | Ppargc1b-fwd | 5′-CACAAAGCGACTCTCTCTTGGCA-3′ | |
| Ppargc1b | Ppargc1b-rev | 5′-GGCTTGTATGGAGGTGTGTGGT-3′ | |
| Pprc | PRC-fwd | 5′-TCGAGTCTGTTGGAGCAGTTT-3′ | |
| Pprc | PRC-rev | 5′-GGGAATGTCAATGCCTGAGTTT-3′ | |
| Srebf1 | Srebf1-fwd | 5′-TGGTTGTTGATGAGCTGGAG-3′ | |
| Srebf1 | Srebf1-rev | 5′-AAAGACAAGGGGCTACTCTGGGAG-3′ | |
| Tfam | Tfam-fwd | 5′-GCCCGAGCGATGGCG-3′ | |
| Tfam | Tfam-rev | 5′-CAGACAAGACTGATAGACGAGGG-3′ | |
| 18S Rrna | TaqMan Probe | ID# Mm03928990_m1 | |
| Slc25a34 | TaqMan Probe | ID# Mm01350550_m1 | |
| mtDNA copy number | Mt-Co1 | TaqMan Probe | ID# Mm04225243_g1 |
| Tfrc | TaqMan Probe | ID# 4458366 | |
| Genotyping | Slc25a34 | Slc25a34-F32 | 5′-CTGCTGGCCTTCTCTACCAG-3′ |
| Slc25a34 | Slc25a34-F51 | 5′-CCCTCCCTTGCATTCACTCC-3′ | |
| Slc25a34 | Slc25a34-R32 | 5′-TTTGGGACTGTGTTGCCACT-3′ | |
| Slc25a34 | Slc25a34-R51 | 5′-GTCTTGGAACTCCACCTGCT-3′ | |
| Slc25a34 | Slc25a34-R53 | 5′-AAAGCCCTAGGCTTGTGGTG-3′ |
mtDNA, mitochondrial DNA.
Liver-specific Slc25a34 Knockout Mice
To specifically delete Slc25a34 in hepatocytes, 2-month–old Slc25a34 floxed mice were injected intraperitoneally with 1.25 × 1011 viral particles of AAV8–throxine-binding globulin promoter (TBG)–Cre virus, which targets >95% of hepatocytes31 and generates a hepatocyte-specific Slc25a34 knockout (Slc-HKO). Slc25a34 floxed mice injected with AAV8-TBG-GFP were used as WT control. AAV8-TBG-Cre and AAV8-TBG-GFP were a gift from James M. Wilson (viral prep 107787-AAV8 and 105535-AAV8, respectively; Addgene, Watertown, MA).
Fast-Food Diet
Mice were maintained on an FFD for 2 months (short-term FFD) or 6 months (long-term FFD) before functional testing and euthanasia. The mice were given an adjusted-calorie diet in which 42% of the calories are from fat (TD.88137; Envigo, Indianapolis, IN) along with high-fructose/high-glucose drinking water (d-glucose, 18.9 g/L; d-fructose, 23.1 g/L), as described elsewhere.32 Mice were weighed at least weekly.
Hepatocyte Isolation
Primary hepatocytes were isolated by using a two-step collagenase perfusion.33 After induction of general anesthesia, a catheter was inserted into the portal vein or inferior vena cava, and 0.3 mg/mL collagenase II (Worthington, Lakewood, NJ) was circulated through the liver. Digested livers were placed in Dulbecco’s modified Eagle’s medium (DMEM)-F12 with 15 mmol/L HEPES (Corning, Corning, NY) + 5% fetal bovine serum (MilliporeSigma, St. Louis, MO), passed through a 70 μm cell strainer and washed twice by using low-speed centrifugation (55 × g for 2 minutes) to remove nonparenchymal cells. Hepatocyte viability, determined by trypan blue staining, was approximately 80%.
Hepatocyte Transfection
Hepatocytes from 2-month–old WT C57BL/6J mice were seeded at 40,000 viable cells per well in 24-well Primaria Cell Culture plates (Corning) or 96-well plates in growth media: DMEM-F12 with 15 mmol/L HEPES (Corning), 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), Antibiotic-Antimycotic Solution (Corning), and ITS Supplement (containing 5 μg/mL insulin, 5 μg/mL transferrin, and 5 ng/mL sodium selenite; Roche, Indianapolis, IN). After 16 hours, growth media was replaced with fresh media containing 0.5% fetal bovine serum and lacking Antibiotic-Antimycotic Solution, and cells were transfected with 40 nm Slc25a34 Stealth siRNA (MSS242504; Thermo Fisher Scientific) or corresponding control siRNA; alternatively, cells were transfected with 250 ng of a mouse Slc25a34 overexpression plasmid (pCMV6-mSlc25a34- Myc-DDk tag, MR224667; OriGene, Rockville, MD) or empty vector. Six hours after transfection, media was replaced with fresh growth media containing 0.5% fetal bovine serum. Transfected primary hepatocytes were maintained for 72 hours before further analysis.
Mitochondrial Stress Test
Approximately 10,000 cells of primary hepatocytes were seeded in the wells of Seahorse XF 96-well cell culture microplates (Seahorse Bioscience, Billerica, MA) and transfected as described in Hepatocyte Transfection. Approximately 72 hours after transfection, culture media was replaced with serum-free buffered DMEM with 5 mmol/L glucose for 3 hours. Cells were washed with unbuffered DMEM containing 5 mmol/L glucose and 2 mmol/L pyruvate and then placed in the same medium for 1 hour in an incubator without carbon dioxide for degassing. Cellular oxygen consumption rate (OCR) measurements were collected after each cycle of 3 minutes of mixing and 2 minutes of wait time, followed by measurements for 3 minutes with the Seahorse analyzer. The entire assay was completed in 12 cycles in which DMEM, oligomycin (2 μmol/L), carbonyl cyanide 4- (trifluoromethoxy)phenylhydrazone (2.5 μmol/L), and rotenone with antimycin A (10 μmol/L each) were injected after cycles 3, 5, 7, and 9, respectively. Non-mitochondrial OCR was measured after the addition of rotenone and antimycin A. After OCR measurements were completed, the cell mass in each well was estimated by crystal violet assay (A550) in each well. OCR values from each well were normalized by cell mass to yield a final corrected OCR in pMol O2/minute/A550. The non-mitochondrial OCR was subtracted from the total OCR to yield the mitochondrial OCR. The proton leak OCR was obtained by subtracting the non-mitochondrial OCR from the post-oligomycin OCR, whereas the maximal OCR was calculated by subtracting the non-mitochondrial OCR from the post–carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone OCR. OCR studies were repeated with primary hepatocytes from at least three mice.
Glycolysis Stress Test
Approximately 10,000 cells of primary hepatocytes were seeded in the wells of Seahorse XF 96-well cell culture microplates and transfected as described previously. Approximately 72 hours after Slc25a34 siRNA knockdown or overexpression, cells were washed with un-buffered DMEM with l-glutamine in the absence of glucose for 3 hours and placed for 1 hour in an incubator without carbon dioxide for degassing. Wells were read for extracellular acidification rate (ECAR) after every cycle of 3 minutes of mixing and 2 minutes of wait time, followed by measurements for 3 minutes with the Seahorse analyzer. The entire assay was completed in 12 total cycles in which glucose (25 mmol/L), oligomycin (2 μmol/L), 2-deoxy-glucose (100 mmol/L), and rotenone/antimycin A (10 μmol/L each) were injected after cycles 3, 5, 7, and 9, respectively. Non-glycolytic ECAR was measured after cycle 12.
After ECAR measurements were completed, the cell mass in each well was estimated by crystal violet assay. ECAR values from each well were normalized by cell mass to yield a final corrected ECAR in pMol O2/minute/A550. The non-glycolytic ECAR was subtracted from the post-glucose ECAR to provide the glycolysis ECAR. Next, the non-glycolytic ECAR was subtracted from the post-oligomycin ECAR to provide the glycolysis capacity ECAR. Subtracting the glycolysis ECAR from the glycolysis capacity ECAR yielded the glycolysis spare reserve capacity. ECAR studies were repeated with primary hepatocytes from at least three mice.
mtDNA Copy Number
Approximately 72 hours after Slc25a34 siRNA knockdown or overexpression, DNA extraction was performed by using an organic-solvent extraction as described previously.34 Briefly, the sample in ice-cold lysis buffer underwent dounce homogenization, followed by Proteinase K digestion at 55°C for 3 hours. Nonsoluble fractions were removed by centrifugation, and DNA in the supernatant was extracted by using the standard phenol-chloroform-isoamyl alcohol method. DNA was precipitated by using 3 M sodium acetate and isopropanol at −20°C. The DNA pellet was washed twice with 70% ethanol. Finally, the extracted DNA pellet was air-dried, resuspended in nuclease-free water, and quantified. Relative amounts of mitochondrially encoded cytochrome C oxidase I (Mt-Co1) DNA and transferrin receptor (Tfrc) genomic DNA were determined by quantitative PCR using TaqMan probes with a StepOnePlus System (Thermo Fisher Scientific). Probe identifiers are listed in Table 2. Mitochondrial DNA copy number was determined by the ratio of Mt-Co1 to Tfrc.35
Oil Red O Staining and Quantification
Approximately 72 hours after Slc25a34 siRNA knockdown or overexpression, Oil Red O (ORO) staining was performed in primary hepatocytes as described previously.36 Briefly, hepatocytes were washed with phosphate-buffered saline (PBS), fixed with 10% formalin, acclimatized with 100% propylene glycol, and stained with 1% ORO (MilliporeSigma) for 1.5 hours. After staining, cells were washed with 75% propylene glycol and deionized water, before drying the plate overnight. ORO-stained lipid particles in the hepatocytes were extracted with 100% isopropanol and quantified colorimetrically at A515 on an Infinite 200 PRO plate reader (Tecan, Baldwin Park, CA). To quantify ORO in liver tissue, ORO-stained liver sections were imaged for lipid droplets (fluorescence) and cell/tissue structures (brightfield). Lipid count, lipid intensity, and tissue area covered by lipids were determined with NIS-Elements Advanced Research software 5.21 (Nikon Instruments, Melville, NY).
ADP/ATP Assay
Approximately 72 hours after Slc25a34 siRNA knockdown or overexpression, the ADP/ATP ratio was determined with the ApoSENSOR ADP/ATP Ratio Bioluminescence Assay Kit (BioVision, Milpitas, CA) as described previously.37 The ADP level is measured by its conversion to ATP, which is subsequently detected by using the same reaction. Briefly, the freshly harvested hepatocytes were lysed by using nucleotide-releasing buffer to measure the ATP levels first. After this luminesce reaction stabilizes, ADP is measured by converting the ADP to ATP by ADP-converting enzyme, now indicating the levels of ADP. Bioluminescence was measured by using the Tecan Infinite 200 PRO plate reader.
RNA Isolation and Quantitative Reverse Transcriptase PCR
Total RNA was isolated from liver tissues or cultured hepatocytes by using TRIzol (Thermo Fisher Scientific) and subjected to the quantitative RT-PCR to quantify the expression of protein-coding genes. Total RNA was first reverse-transcribed into complementary DNA using M-MLV Reverse Transcriptase (RNase H; Thermo Fisher Scientific) and quantitative PCR performed with SYBR Green PCR master mix (Thermo Fisher Scientific), Bullseye EvaGreen PCR Master Mix (Midwest Scientific, St. Louis, MO), or TaqMan probes (Thermo Fisher Scientific) with TaqMan Universal Master Mix II (Thermo Fisher Scientific). Primer sequences and TaqMan probe identifiers are described in Table 2. Reactions were performed by using a StepOnePlus system (Thermo Fisher Scientific). Relative expression was calculated by using the ΔΔCT method.
Mouse Euthanasia and Liver/Blood Isolation
Mice were fasted for 16 hours before euthanasia. At the time of euthanasia, blood was collected by retro-orbital bleeding or cardiac puncture, and serum was immediately stored at −80°C in opaque tubes. Liver was collected, weighed, and flash-frozen for nucleic acid and protein isolation. A portion of each liver was fixed in 10% formalin and embedded in paraffin and/or freshly frozen in optimal cutting temperature compound (Sakura Finetek, Torrance, CA) for histologic analysis.
Liver Histology
Formalin-fixed, paraffin-embedded liver tissue was stained with hematoxylin and eosin, periodic acid-Schiff, and picrosirius red; frozen liver sections in optimal cutting temperature compound were stained with ORO. All staining was performed by the Pittsburgh Liver Center Clinical Biospecimen Repository and Processing Core. Histologic scoring for steatosis, lobular inflammation, hepatocyte ballooning, and fibrosis was performed as described previously.38
Liver Triglycerides
Liver triglycerides were determined as described previously.39 Briefly, frozen liver tissue was homogenized and lipids extracted in chloroform:methanol (2:1), followed by acidification with H2SO4 and phase separation by centrifugation. Triglyceride levels were measured by using the colorimetric Infinity Triglyceride Reagent (Thermo Fisher Scientific).
Mitochondrial Isolation from Liver
Approximately 200 mg of liver was harvested, washed with cold PBS, and dounce homogenized (five strokes) using 2 mL Kimble Kontes Dounce Tissue Grinders (Thermo Fisher Scientific). Mitochondria were isolated from liver tissues by using the Mitochondrial Isolation kit for tissue following the manufacturer’s instructions (89801; Thermo Fisher Scientific). Mitochondrial protein lysates of the isolated mitochondrial fractions were prepared by using SDS-based lysis buffer (50 mmol/L Tris-HCl, pH 8.0, 10 mmol/L EDTA, 1% SDS) containing protease/phosphatase inhibitors (1X RIPA Buffer).
Body Weight and Liver Weight to Body Weight
Livers from experimental animals were excised, washed in PBS, and weighed. The percentage of the weight occupied by the liver was determined by dividing the liver weight by the total body weight of the mouse.
Liver Enzymes
Serum was collected from experimental animals. The level of circulating serum enzymes, including alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase, as well as cholesterol and triglyceride levels, were determined by the University of Pittsburgh Medical Center Clinical Laboratory.
Glucose and Insulin Tolerance Tests
Glucose and insulin tolerance tests were performed as described previously.40,41 Mice were fasted for 8 hours (4:00 am to 12:00 pm). For the glucose tolerance test, glucose was injected intraperitoneally at 2 g/kg body weight (using 200 mg/mL d-glucose in sterile PBS; MilliporeSigma), and blood glucose levels were measured before glucose administration and every 15 to 30 minutes for up to 240 minutes by tail vein blood collection. Blood glucose measurements were performed with Accu-Chek Aviva Plus strips using an Accu-Chek Aviva glucometer (Roche). For the insulin tolerance test, human regular insulin (Humulin R, Eli Lilly and Company, Indianapolis, IN) was injected intraperitoneally at 0.75 U/kg body weight (dissolved in sterile PBS), and blood glucose levels were measured before insulin administration and every 15 to 30 minutes for 120 minutes by tail vein blood collection. Mice were provided food at the end of the test.
Oroboros Respirometry and Citrate Synthase Activity
Mitochondrial respiratory capacity was assessed as previously described.42 Briefly, mitochondrially enriched liver lysates were prepared by homogenizing liver at the time of dissection in STE buffer (250 mmol/L sucrose, 10 mmol/L Tris-HCl, 1 mmol/L EDTA) with a glass vessel and Teflon pestle, after which the homogenate was centrifuged at 800 × g for 10 minutes at 4°C, yielding a mitochondrially enriched supernatant. Respiration was measured at 37°C in the O2K using MiR05 buffer (Oroboros Instruments, Innsbruck, Austria). Respiratory capacity was assessed following sequential injection of the following: pyruvate (5 mmol/L), malate (2 mmol/L), ADP (2 mmol/L), glutamate (10 mmol/L), succinate (10 mmol/L), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (titrations of 0.5 μmol/L until maximal respiration achieved), and antimycin A (2.5 μmol/L). Oxygen consumption rates were normalized to total protein. Citrate synthase activity was measured by using the same mitochondrially enriched lysate as above, where lysate was added to a reaction mixture consisting of 0.25% Triton X-100, 0.31 mmol/L acetyl-CoA, 0.1 mmol/L 5,5′-dithiobis (2-nitrobenzoic acid), 0.1 mol/L triethanolamine, 1 mmol/L EDTA, 1 mol/L Tris-HCl, and addition of 5 mmol/L oxaloacetate to initiate the reaction. Citrate synthase activity was measured as the change in absorbance at 412 nm measured every 10 seconds over a 2-minute period and normalized to protein concentration.
Western Blot Analysis
Whole liver and mitochondrial protein lysates were prepared from 50 to 200 mg of frozen tissue by using 1X RIPA Buffer. Lysates were transferred to a fresh 1.5-mL tube and centrifuged at 18,400 × g for 10 minutes at 4°C to remove clear supernatant to a new 1.5-mL tube while disposing of the pellet. Samples were stored at −80°C until utilization or determination of protein concentration via bicinchoninic acid protein assay (Thermo Fisher Scientific) to ensure equal protein concentrations for subsequent assays. For Western blot analysis, 50 to 200 μg of total protein was loaded onto 8% to 12% SDS-PAGE gels. Proteins were transferred to Immobilon-P polyvinylidene difluoride membrane (IPVH00010; MilliporeSigma) either for 1 hour at 100 V at 4°C or overnight at 35 V and 4°C. After transfer, the membranes were blocked in either 5% nonfat dry milk or 5% bovine serum albumin dissolved in bovine lacto transfer technique optimizer (Blotto) (0.15 mol/L NaCl, 0.02 mol/L Tris [pH 7.5], 0.1% Tween in distilled H2O), followed by incubation with the following antibodies: anti-SLC25A34 (17557-1-AP; Thermo Fisher Scientific), anti–β-actin (ab8227; Abcam, Cambridge, United Kingdom), anti-SREBP1 (Sc13551; Santa Cruz Biotechnology, Dallas, TX), anti-TOMM20 (PA5-52843; Thermo Fisher Scientific), anti–acetyl-CoA carboxylase (3676), anti–phospho-acetyl-CoA carboxylase (Ser79) (11818), anti–ATP-citrate lyase (4332), anti–phospho-ATP-citrate lyase (Ser455) (4331), and anti–fatty acid synthase (3180). Membranes were washed for 1 hour at room temperature before incubation with species-specific secondary antibodies: goat anti-rabbit horseradish peroxidase (7074P2) or goat anti-mouse horseradish peroxidase (PI31444; Thermo Fisher Scientific). Antibodies were from Cell Signaling Technologies (Danvers, MA) unless specified otherwise. Membranes were exposed to SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) for 1 to 2 minutes, and bands reflective of target proteins were viewed by using a ChemiDoc imaging system (Bio-Rad, Hercules, CA). Bands were quantified with ImageJ software version 1.52a (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
RNA-Seq and Analysis
Paired-end RNA-seq (150 bp) was performed on Slc-HKO and WT control mice maintained on short-term FFD or chow diet (n = 3 mice per genotype per diet). Library preparation, sequencing, and alignment were performed by Novogene (Durham, NC). Low-quality reads and adapter sequences were filtered out by using Fastp (version 0.20.1).43 Reads were aligned to the mouse reference genome mm10 using RNA STAR software 2.6.1d.44 Read counts were quantified by FeatureCounts (version 1.5.0-p3),45 using the ENSEMBL annotation of mm10. The read counts were normalized to the total count of aligned reads for each sample library. The raw RNA-seq data were submitted to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo; accession number GSE186314). Significantly expressed genes were analyzed by using Ingenuity Pathway Analysis version 01-10 (QIAGEN Silicon Valley, Redwood, CA). Canonical signaling pathways and upstream regulators were predicted based on changes in downstream gene expression patterns. Enriched biological processes (gene ontology terms) and Kyoto Encyclopedia Gene and Genome pathways were determined compared with the Mus musculus reference gene list using DAVID software version 6.8.46
Microscopy
Micrographs were captured with a Nikon TiU fluorescent microscope equipped with a Photometrics CoolSNAP ES2 monochrome camera (fluorescent images) or Nikon DS-Fi2 color camera (non-fluorescent images) (Photometrics, Tucson, AZ). Images were processed with Nikon NIS-Elements Advanced Research software 5.21 (Nikon Instruments), ImageJ software version 1.52a, and Adobe Photoshop 23.2 (Adobe, San Jose, CA).
Statistical Analysis
Data are expressed as means ± SEM. Statistical significance was calculated by using a two-tailed t-test with GraphPad Prism 9.3 (GraphPad Software, La Jolla, CA), as appropriate. P < 0.05 was considered significant.
Results
Slc25a34 Affects Mitochondrial Respiration and Bioenergetics in Primary Hepatocytes
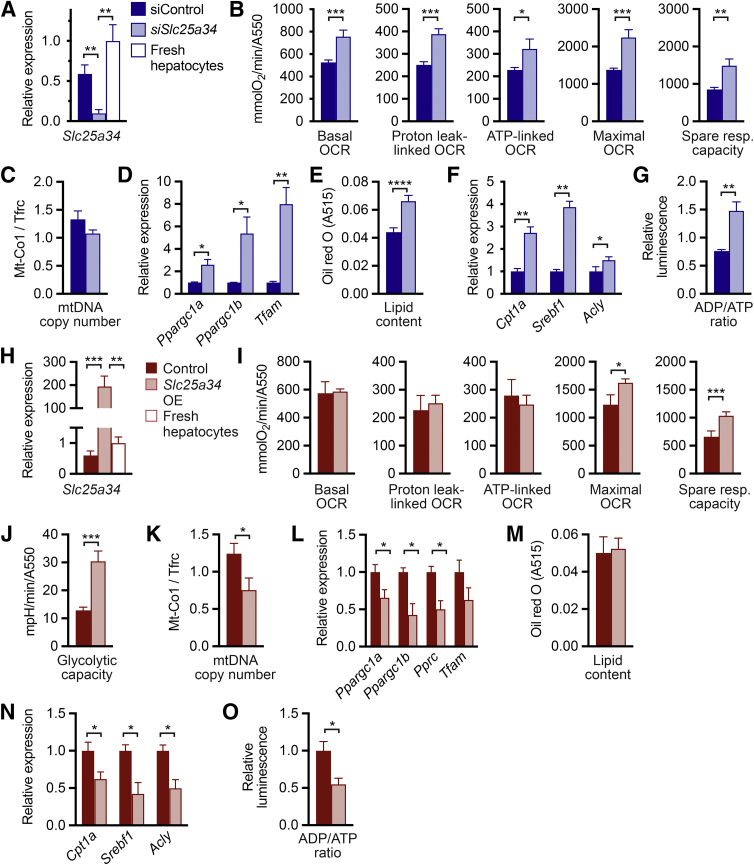
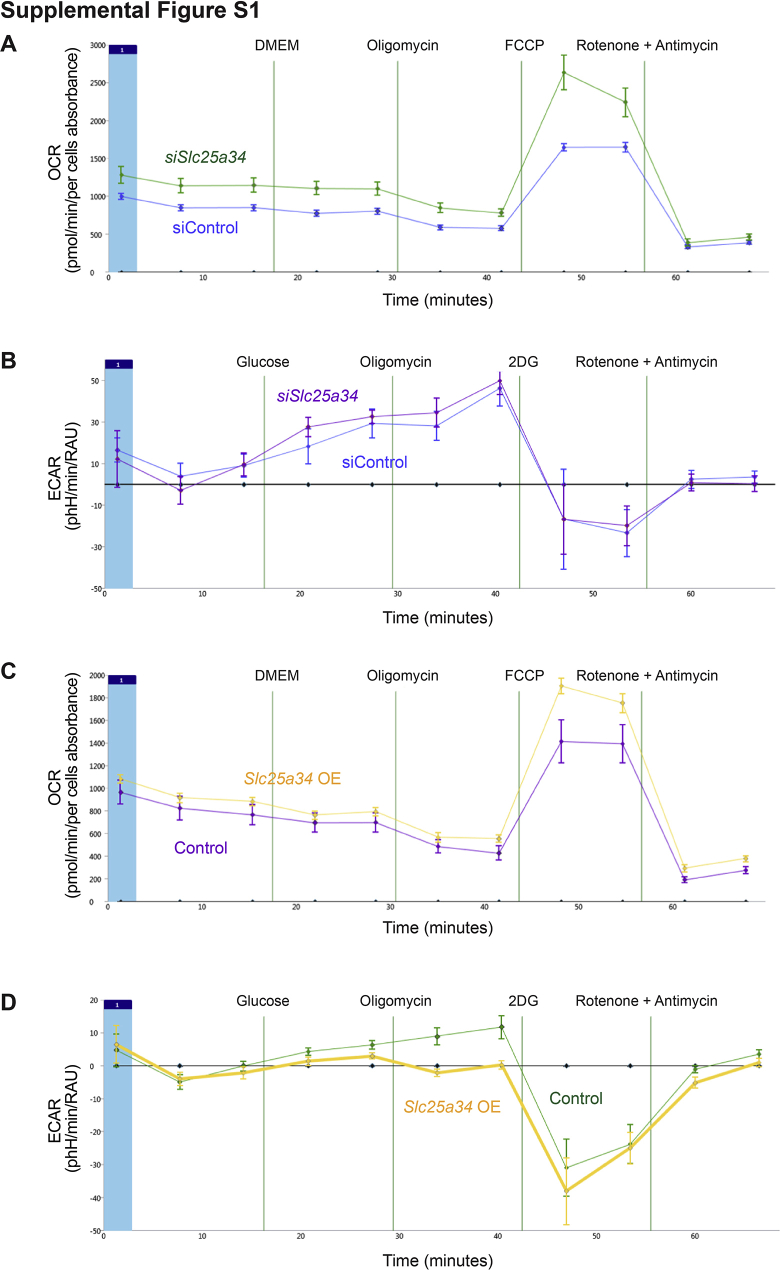
To understand how SLC25A34 affects mitochondrial function and bioenergetics, primary hepatocytes from WT mice were seeded on tissue culture plastic. Transfection with Slc25a34 siRNA resulted in an 80% reduction of Slc25a34 compared with that of control siRNA (Figure 1A). A mitochondrial stress test was performed to determine whether Slc25a34 deletion affects cellular respiration (Figure 1B and Supplemental Figure S1A). Basal mitochondrial OCR was elevated in Slc25a34-depleted hepatocytes and following oligomycin treatment, which inhibits mitochondrial ATP synthase. Slc25a34 depletion increased both ATP synthase–linked and proton leak–linked mitochondrial OCR. Similarly, after carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone treatment, maximal mitochondrial respiration and spare respiratory capacity were increased.
Figure 1.
Knockdown and overexpression of Slc25a34 alter mitochondrial respiration and bioenergetics in primary hepatocytes. A–G: Primary hepatocytes isolated from 2-month–old wild-type C57BL/6 mice were transfected with control siRNA (siControl) or siRNA against Slc25a34 (siSlc25a34) and harvested 72 hours later. A: Graph shows the relative expression of Slc25a34 of cultured hepatocytes after siRNA-mediated knockdown compared with siControl and hepatocytes from fresh liver. B: Mitochondrial stress test showing basal oxygen consumption rate (OCR), proton leak–linked OCR, ATP-linked OCR, maximal OCR, and spare respiratory (resp.) capacity. Representative OCR data are shown in Supplemental Figure S1A. C: Mitochondrial DNA (mtDNA) copy number, defined by ratio of the mitochondrial-encoded gene (Mt-Co1) and nuclear-encoded gene (Tfrc). D: Relative expression of genes involved in mitochondrial biogenesis. E: Lipid content of Oil Red O–stained hepatocytes stained after extraction and quantification at A515. F: Relative gene expression of genes involved in lipid regulation. G: ADP/ATP ratio. H–O: Primary hepatocytes isolated from 2-month–old wild-type C57BL/6 mice were transfected with control plasmid (Control) or plasmid encoding Slc25a34 (Slc25a34 overexpression [OE]) and harvested 72 hours later. H: Graph shows relative Slc25a34 in cultured hepatocytes after Slc25a34 overexpression and hepatocytes from fresh liver. I: Mitochondrial stress test. Supplemental Figure S1C provides primary OCR data. J: Glycolytic capacity determined by extracellular acidification rate. Supplemental Figure S1D provides primary data. K: mtDNA copy number. L: Relative expression of genes involved in mitochondrial biogenesis. M: Lipid content of Oil Red O–stained hepatocytes. N: Relative gene expression of genes involved in lipid regulation. O: ADP/ATP ratio. Graphs represent means ± SEM. n = 4 to 5 (A); data are representative of 3 to 4 experiments with n = 10 replicates per experiment (B); n = 3 (D, F, K, L, and N ); n = 8 to 9 (E); n = 4 (G); n = 4 to 6 (H); data are representative of 3 to 4 experiments with n = 7 to 11 replicates per experiment (I); data are representative of 3 to 4 experiments with n = 7 replicates per experiment (J); n = 5 (M); n = 4 (O). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Relative gene expression is normalized to B2m. ECAR, extracellular acidification rate.
To determine how SLC25A34 affects mitochondrial respiration, key determinants of bioenergetics were investigated. First, mitochondrial DNA copy number (an indicator of mitochondrial mass) was equivalent in control and Slc25a34-depleted hepatocytes (Figure 1C), whereas master regulators of mitochondrial biogenesis (Pgc1α, Pgc1β, and Tfam) were increased by twofold to eightfold (Figure 1D), suggesting that differences in the quality of mitochondria are responsible for increased basal mitochondrial respiration.47 Second, because the mitochondrial DNA copy number did not change, increased maximal mitochondrial respiration and spare respiratory capacity could be driven by glycolysis or lipid levels. Glycolysis was equivalent in control and Slc25a34-depleted hepatocytes (Supplemental Figure S1B). However, Slc25a34 depletion increased lipid content by 50% (Figure 1E), which is supported by 1.5-fold to fourfold overexpression of lipogenesis genes (Srebf1 and Acly) and the rate-limiting step of β-oxidation (Cpt1a) (Figure1F).48,49 Third, an increase in ATP synthase–linked mitochondrial respiration suggests that ATP synthase is more active in hepatocytes depleted of Slc25a34. This could be explained by increased ADP levels, which cause ATP synthase to work harder to attain equilibrium. Consistent with this idea, the ADP/ATP ratio was twofold higher in Slc25a34-depleted hepatocytes (Figure 1G).
To further understand how SLC25A34 affects mitochondrial function, Slc25a34 was overexpressed in WT primary hepatocytes (Figure 1H). Mitochondrial respiration was largely unchanged, although maximal mitochondrial respiration and spare respiratory capacity were increased (Figure 1I and Supplemental Figure S1C). However, glycolytic capacity increased threefold after Slc25a34 overexpression (Figure 1J and Supplemental Figure S1D). Key determinants of mitochondrial metabolism were investigated again to determine mechanisms of bioenergetic regulation. Overexpression of Slc25a34 resulted in decreased mitochondrial DNA copy number (Figure 1K) and 40% to 60% reduction of key mitochondrial biogenesis genes (Ppargc1a, Ppargc1b, Pprc, and Tfam) (Figure 1L). Lipid levels were equivalent (Figure 1M), although fatty acid synthesis (Srebf1 and Acly) and β-oxidation (Cpt1a) genes were reduced by 50% (Figure 1N). Finally, the ADP/ATP ratio was reduced by 50% (Figure 1O), suggesting that Slc25a34 overexpression decreases ADP levels. Together, the in vitro data show that SLC25A34 affects mitochondrial respiration and energy production. Slc25a34 depletion creates an energy-generating state that increases ATP synthase activity, mitochondrial biogenesis, and lipid synthesis. In contrast, Slc25a34 overexpression reduces energy generation (mitochondrial mass, regulators of fatty acid synthesis, and ATP synthase activity) and promotes a switch from a more efficient mode (mitochondrial respiration) to a less efficient mode (glycolysis) of energy production.
Generation of a Hepatocyte-specific Slc25a34 Knockout Mouse
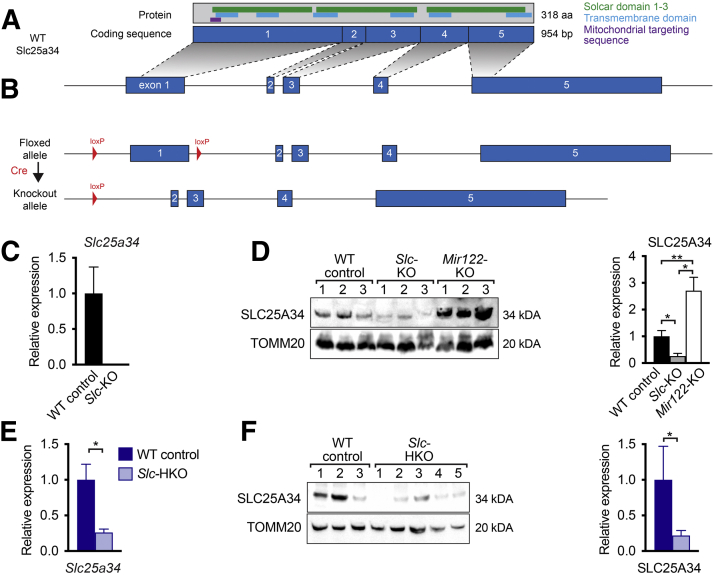
WT SLC25A34 protein contains 318 amino acids and comprises five transmembrane domains, three solute carrier (Solcar) domains, and a putative mitochondrial targeting sequence (Figure 2A). Using CRISPR/Cas9 and homology-directed repair, loxP sites were inserted into the promoter and intron 1 of Slc25a34 genomic DNA to generate a floxed allele (Figure 2B and Supplemental Figure S2A). After Cre recombinase–mediated recombination, the promoter and first half of the protein (amino acids 1 to 152) are deleted, including Solcar 1, the first three transmembrane domains, and the putative mitochondrial targeting sequence. Thus, if a truncated mRNA is transcribed (eg, by an alternative start site), the resulting protein would lack mitochondrial specificity. Gene targeting produced heterozygous mice with a floxed Slc25a34 allele as well as heterozygotes with a knockout allele, presumably generated by template-independent nonhomologous DNA end-joining.29,30 Floxed and knockout alleles were verified by DNA sequencing, and founder mice were backcrossed to C57BL/6J for at least five generations and bred to homozygosity (Supplemental Figure S2, B and C).
Figure 2.
Gene targeting disrupts Slc25a34 expression in the liver. A: Schematic depicting wild-type (WT) Slc25a34 gene structure, coding sequence, and protein domains. B: Simplified schematic of the floxed Slc25a34 allele (loxP sites are highlighted) and knockout allele after Cre-mediated recombination. A detailed scheme is shown in Supplemental Figure S2. C: Slc25a34 mRNA expression in livers from whole body knockouts (Slc-KO), normalized to Gapdh. D: Slc25a34 protein expression in mitochondrial fractions from WT control, Slc-KO, and Mir122-KO mice. Representative western blot is shown (left), and relative expression normalized to TOMM20 and quantified by densitometry (right). E: Slc25a34 mRNA expression in livers from hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice, normalized to 18S. F: Slc25a34 protein expression in mitochondrial fractions from WT control and Slc-HKO mice. Representative western blot is shown (left), and relative expression normalized to TOMM20 and quantified by densitometry (right). Graphs represent means ± SEM. n = 3 (C and D); n = 4 (E); n = 3 to 5 (F). ∗P < 0.05, ∗∗P < 0.01.
Homozygous whole body Slc25a34 knockout mice (Slc-KO) were fertile and produced viable offspring. Slc-KO livers were devoid of Slc25a34 mRNA, as determined by RT-PCR using primers targeted within exon 1 (Figure 2C). Despite the absence of mRNA, a weak protein signal was observed at the same molecular weight as SLC25A34 in Slc-KO, likely due to nonspecificity of the antibody (Figure 2D). Nonetheless, the SLC25A34 antibody recognizes full-length protein, as shown by elevated SLC25A34 in liver lysates from Mir122-KO mice (Figure 2D).11 To specifically delete Slc25a34 in hepatocytes from 2-month–old floxed mice, Cre recombinase was delivered by adeno-associated virus driven by the thyroxine-binding globulin (TBG) promoter, AAV8-TBG-Cre, which targets >95% of hepatocytes,31 generating a hepatocyte-specific Slc25a34 knockout (Slc-HKO). As a control, Slc25a34 floxed mice were injected with AAV8-TBG-GFP, which does not affect Slc25a34 expression (WT control). One month after injection, Slc25a34 mRNA and protein from Slc-HKO livers were reduced 75% compared with WT control (Figure 2, E and F). Collectively, the Slc25a34 targeting strategy yielded both alleles for whole body knockout and floxed alleles for a tissue-specific knockout. Injection of floxed mice with AAV8-TBG-Cre effectively deletes hepatic Slc25a34. To eliminate potentially confounding effects of whole body deletion on the liver, all subsequent experiments used Slc-HKO and WT control mice.
Glucose Homeostasis Is Perturbed in Slc-HKO Mice
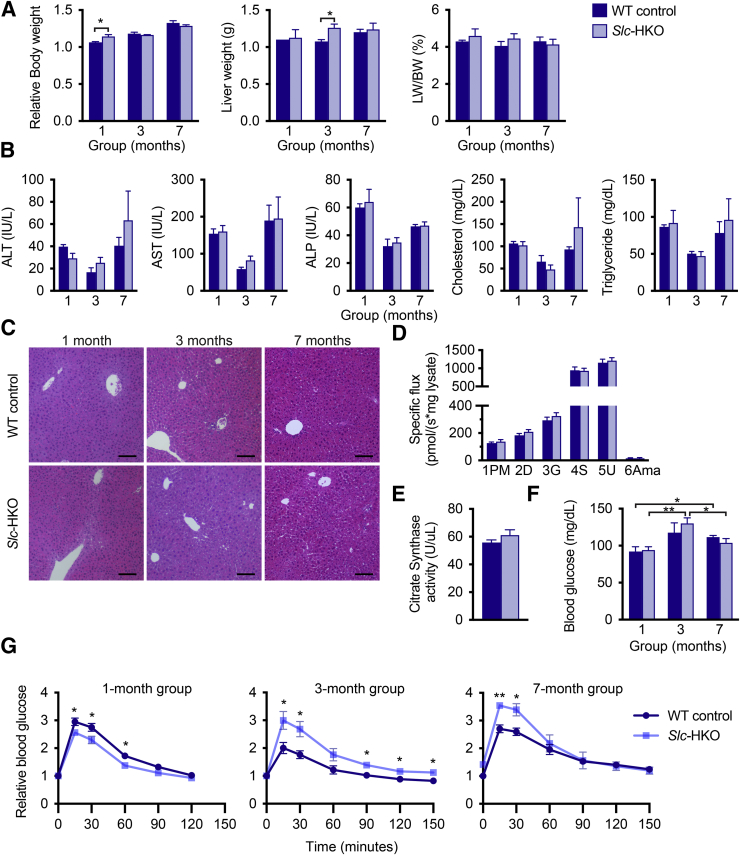
Slc-HKO and WT control mice were examined 1, 3, and 7 months after AAV8 injection to determine the effect of Slc25a34 deletion in vivo (Supplemental Figure S3A). Relative body weight of Slc-HKO mice increased slightly after 1 month; liver weight modestly increased in the 3-month group while the ratio of liver weight to body weight was equivalent in each group (Figure 3A). Alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, and serum cholesterol and triglyceride levels were equivalent (Figure 3B). Slc-HKO and WT control livers appeared similar according to hematoxylin and eosin staining (Figure 3C). Although mitochondrial OCR increased in primary hepatocytes depleted of Slc25a34 (Figure 1B), respiration was equivalent in mitochondrial preparations from Slc-HKO and WT control livers (Figure 3D). Furthermore, citrate synthase activity was comparable between Slc-HKO and WT controls, indicating equivalent mitochondrial mass, which is consistent with in vitro data (Figure 3E).
Figure 3.
SLC25A34 deletion leads to moderate defects in glucose homeostasis. Eight-week–old Slc25a34 floxed mice were injected with AAV8-TBG-GFP or AAV8-TBG-Cre to generate wild-type (WT) control and hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice, respectively, and analyzed after 1, 3, or 7 months. A: Relative body weight, liver weight, and percentage of liver weight to body weight (LW/BW) of Slc-HKO and WT control mice from each group. B: Analysis of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), cholesterol, and triglyceride levels. C: Representative hematoxylin and eosin staining reveals equivalent liver architecture in Slc-HKO and WT control mice. D: Respiration in mitochondrial preparations per milligram of protein from Slc-HKO and WT control mice in the 3-month group, measured by Oroboros. E: Citrate synthase activity in the 3-month group. F: Blood glucose level in mice after fasting for 16 hours. G: Glucose tolerance test: mice were fasted for 8 hours, injected with 2 g/kg glucose, and glucose level determined for up to 2.5 hours. Graphs represent means ± SEM. n = 4 to 5 (A and C); n = 3 to 5 (B); n = 7 (D and E); n = 4 to 7 (F and G). ∗P < 0.05, ∗∗P < 0.01. Scale bars = 100 μm (C). Labels along the x axis refer to the substrate-uncoupler-inhibitor titration protocols performed. Numbers 1–6 refer to the sequence: 1PM, pyruvate and malate; 2D, adenosine diphosphate; 3G, glutamate; 4S, succinate; 5U, Uncoupler (FCCP, mitochondrial oxidative phosphorylation uncoupler); 6Ama, antimycine A.
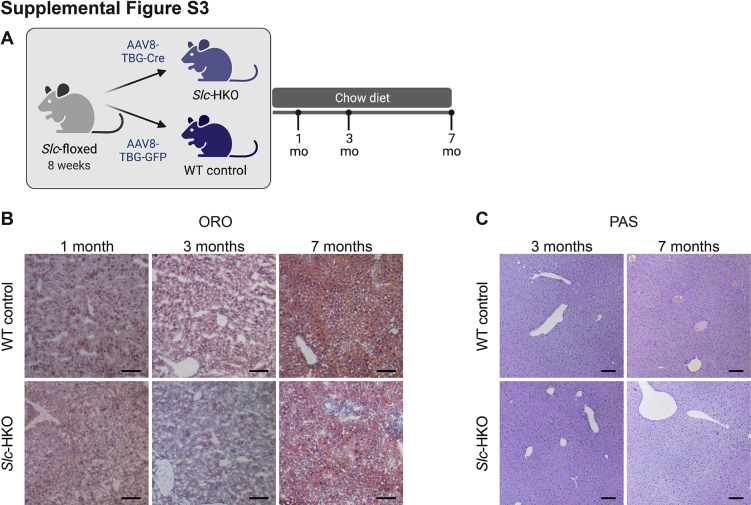
To evaluate glucose metabolism, blood glucose levels were first measured after fasting for 16 hours, which depletes energy store50; blood glucose levels were equivalent between Slc-HKO and WT control mice at each time point (Figure 3F). Hepatic lipids and glycogen were equivalent in Slc-HKO and WT controls (Supplemental Figure S3, B and C). Next, a glucose tolerance test was performed. Slc-HKO and WT control mice were fasted for 8 hours, injected with 2 g/kg glucose, and blood glucose levels were measured over 2.5 hours. Blood glucose increased twofold to threefold in WT control mice in each group immediately after injection and declined to baseline by 2 hours, suggesting normal insulin release and glucose clearance (Figure 3G). Glucose tolerance varied with age in Slc-HKO mice. In the 1-month group, glucose was modestly reduced in Slc-HKO mice compared with WT control mice, indicating rapid release of insulin and glucose clearance. Blood glucose levels were elevated in Slc-HKO mice in the 3-month group and in the 7-month group, suggesting slower insulin release or impaired glucose clearance. Together, the data suggest that hepatocyte-specific loss of Slc25a34 primarily affects glucose homeostasis through 7 months in unstressed conditions.
De novo Lipogenesis Is Dysregulated in Slc-HKO Livers
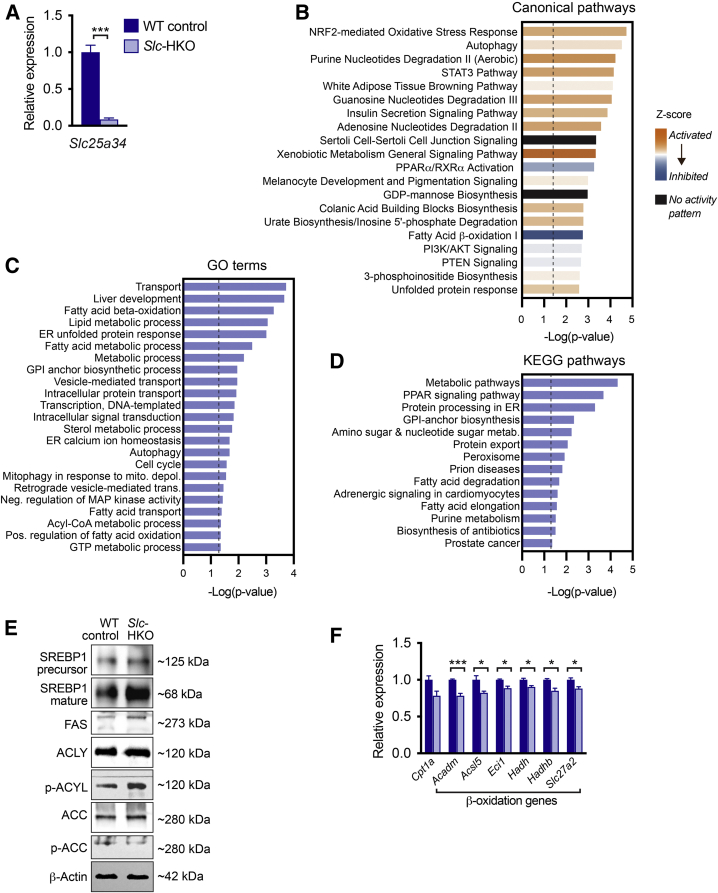
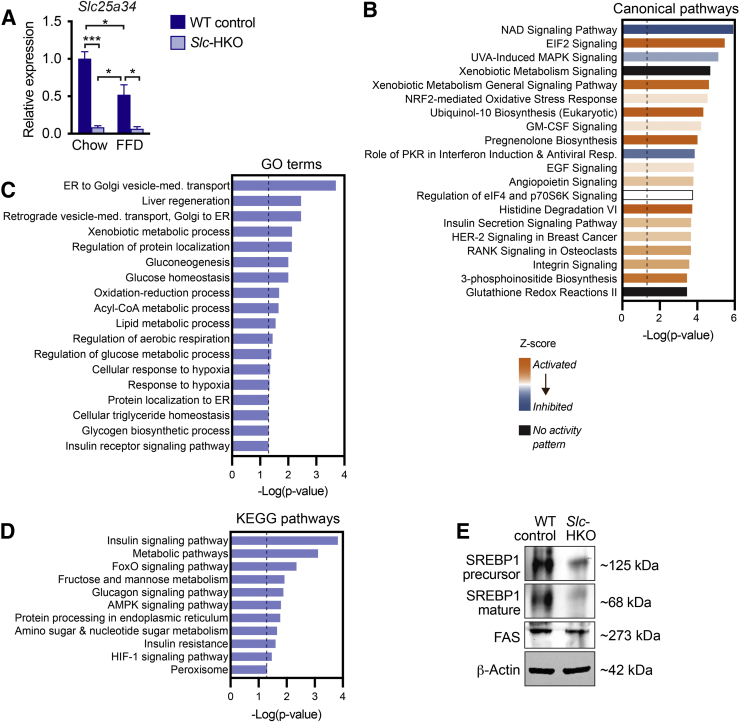
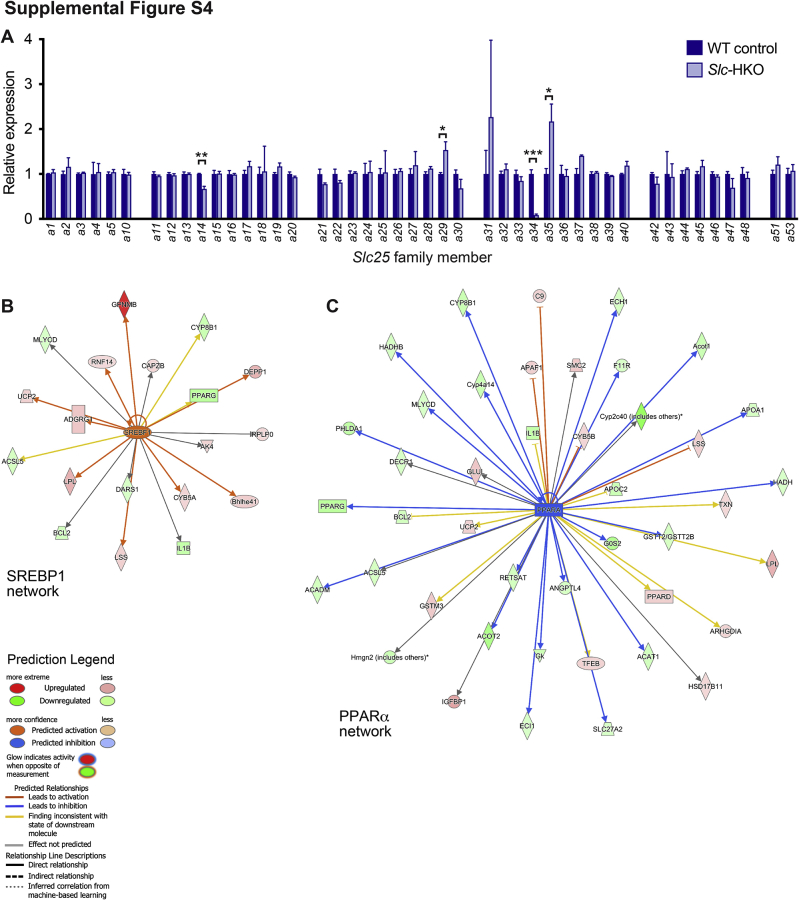
To determine how the loss of SLC25A34 affects the liver transcriptome, RNA-seq was performed on livers from Slc-HKO and WT control mice 3 months after AAV8 infection. Slc25a34 expression was reduced 90% in knockout livers; although expression of most other Slc25 family members remained unchanged, modest changes in Slc25a14, Slc25a29, and Slc25a35 were detected (Figure 4A and Supplemental Figure S4A). Compared with WT control, 1007 genes were differentially expressed in Slc-HKO (Supplemental Table S1). Ingenuity Pathway Analysis of differentially expressed genes revealed dysregulation of several pathways, including metabolic and metabolically relevant pathways (Figure 4B). Nine of the top 20 upstream regulators have strong metabolic relevance (Table 3). Gene ontology and Kyoto Encyclopedia Gene and Genome pathway analysis identified alterations in gene pools relevant to fatty acid metabolism, including β-oxidation, fatty acid transport, fatty acid degradation and elongation, acyl-co-A metabolic process, and peroxisome proliferator-activated receptor (PPAR) signaling (Figure 4C). Thus, the transcriptomic analysis underscores a role for SLC25A34 in a wide range of metabolic processes, particularly fatty acid metabolism.
Figure 4.
Transcriptomic analysis of hepatocyte-specific Slc25a34 knockout (Slc-HKO) livers. A: RNA isolated from Slc-HKO and wild-type (WT) control livers 3 months after AAV8 injection and subjected to RNA-sequencing. Slc25a34 expression was reduced in Slc-HKO. B: Ingenuity Pathway Analysis identified altered metabolic and metabolically relevant pathways in Slc-HKO. Bars represent activity patterns based on Z scores. C and D:Slc-HKO livers are characterized by additional alterations in biological processes [C, Gene ontology (GO) terms] and pathways [D, Kyoto Encyclopedia Gene and Genome (KEGG) pathways]. E: Western blot analysis of liver lysates from Slc-HKO and WT control mice. Each lane represents a pooled protein from multiple mice. F: Genes involved in β-oxidation were reduced in Slc-HKO. For A and F, absolute read count values for each gene were normalized to total read counts obtained per mouse and presented as relative expression of Slc-HKO compared with WT control. Error bars represent SEM. n = 3 (A); n = 3 to 4 (E). ∗P < 0.05, ∗∗∗P < 0.001. Dashed lines in B, C, and D represent −Log(P value) = 1.3, the threshold for significance.
Table 3.
Differential Activation of Upstream Regulators by Slc-HKO Livers Compared with WT Controls (Chow Diet, 3 months)
| Upstream regulator | Predicted activation state | Activation Z score | P value of overlap |
|---|---|---|---|
| PPARA | Inhibited | −2.9 | 2.97 × 10−07 |
| SLC27A2 | Activated | 3.1 | 6.59 × 10−06 |
| XBP1 | Activated | 3.8 | 1.68 × 10−05 |
| CLPP | Activated | 2.5 | 2.75 × 10−04 |
| ERN1 | Activated | 2.2 | 2.24 × 10−03 |
| PCK1 | Activated | 2.0 | 2.58 × 10−03 |
| KLF15 | Inhibited | −2.6 | 4.85 × 10−03 |
| LPL | Inhibited | −2.2 | 6.63 × 10−03 |
| ELL2 | Activated | 2.6 | 8.85 × 10−03 |
| SREBF1 | Activated | 2.0 | 9.04 × 10−03 |
| miR-223 | Activated | 2.7 | 1.46 × 10−02 |
| miR-30c-5p (and other miRNAs with seed GUAAACA) | Inhibited | −2.3 | 1.46 × 10−02 |
| MEX3A | Activated | 2.2 | 1.81 × 10−02 |
| PTPRR | Inhibited | −2.3 | 1.82 × 10−02 |
| ZBTB7B | Inhibited | −2.2 | 2.50 × 10−02 |
| MYCN | Activated | 2.2 | 2.59 × 10−02 |
| TP63 | Activated | 2.8 | 3.93 × 10−02 |
| miR-10 | Inhibited | −2.4 | 4.27 × 10−02 |
| TP73 | Activated | 2.2 | 4.88 × 10−02 |
“Upstream Regulator” is defined by Ingenuity Pathway Analysis as any molecule that can affect the expression of other molecules. Nineteen upstream regulators were identified (Z score less than −2 and >2; P value of overlap <0.05). Bold indicates metabolic relevance.
Slc-HKO, hepatocyte-specific Slc25a34 knockout; WT, wild type.
Key molecular players of de novo lipogenesis were investigated to determine how SLC25A34 affects fatty acid metabolism. Sterol-responsive element binding protein-1 (SREBP1) establishes a lipogenic transcriptional program after nuclear translocation.51,52 At the protein level, precursor SREBP1 was unchanged in Slc-HKO livers, but mature SREBP1 increased (Figure 4D). Fatty acid synthase, a gene target of SREBP1 and the limiting enzyme of fatty acid synthesis pathways, was also increased. Ingenuity Pathway Analysis further revealed activation of numerous SREBP1 gene targets (Supplemental Figure S4B). Consistent with the induction of a lipogenic program, gene signatures indicated reduced β-oxidation. For example, Cpt1a, an enzyme that transports fatty acids into mitochondria for β-oxidation, trended downward in Slc-HKO livers, and genes encoding the rate-limiting steps of β-oxidation were attenuated (Acadm, Acsl5, Eci1, Hadh, Hadhb, and Slc27a2) (Figure 4E). Finally, the expression of genes in the PPARα network, which promotes β-oxidation, was inhibited (Supplemental Figure S4C).
To further understand how loss of SLC25A34 influences de novo lipogenesis, ATP citrate lyase (ACLY) and acetyl Co-A carboxylase (ACC), rate-limiting enzymes of fatty acid synthesis and β-oxidation, were investigated (Figure 4E).51 ACLY was expressed at equivalent levels in Slc-HKO and WT control livers, but phosphorylated ACLY was increased in knockouts. Phosphorylation of ACLY increases its catalytic activity and promotes acetyl Co-A formation. ACC, whose activity is inhibited by phosphorylation, converts acetyl Co-A to malonyl Co-A. ACC was expressed at similar levels, but phosphorylated ACC was reduced in Slc-HKO, suggesting elevated malonyl Co-A that can inhibit carnitine palmitoyltransferase 1 and serve as a precursor for fatty acid synthesis. Thus, SLC25A34 deletion alters rate-limiting enzymes of fatty acid metabolism, ACLY, and ACC, which leads to fatty acid synthesis and β-oxidation inhibition.
SLC25A34 Deletion Exacerbates NAFLD Phenotype after Short-Term FFD
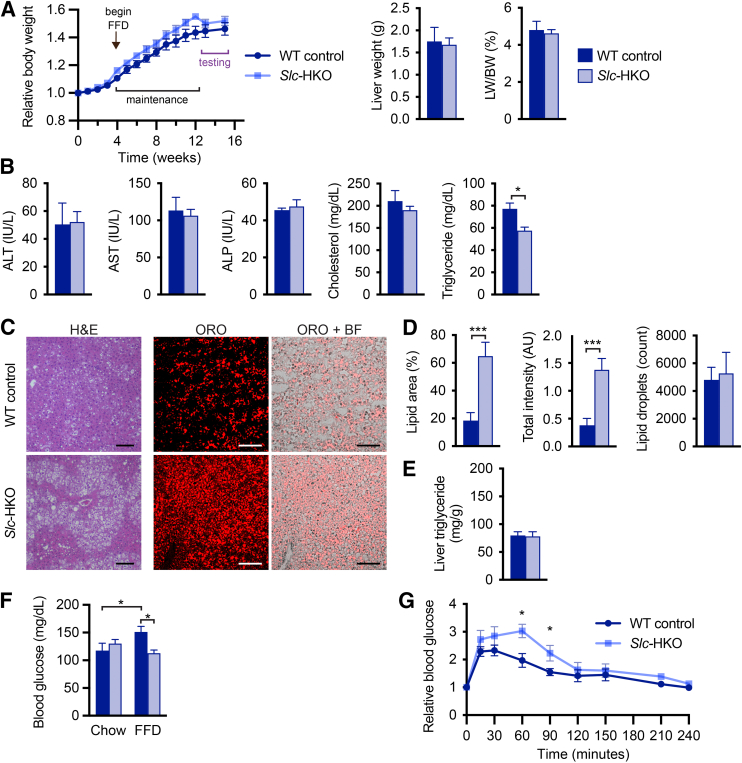
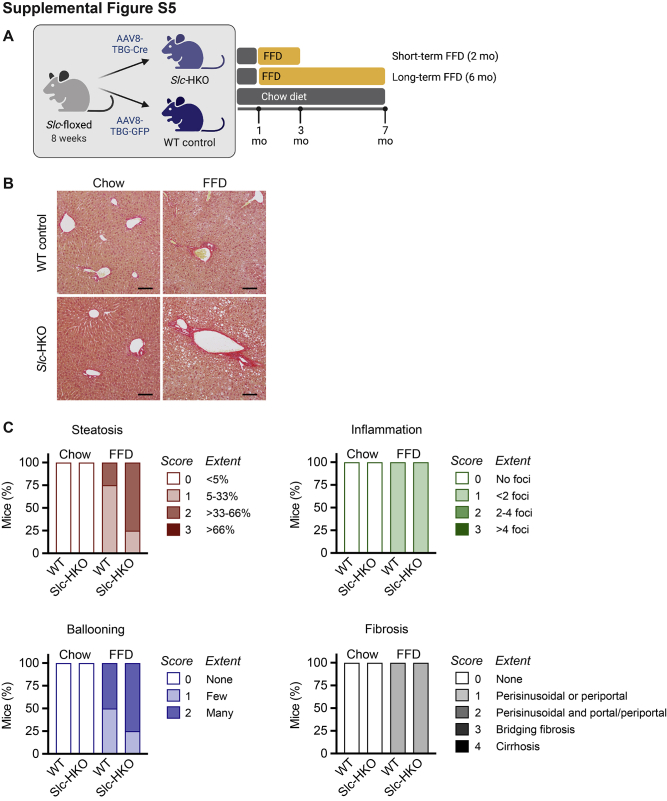
Slc-HKO mice are characterized by dysregulated glucose homeostasis and molecular signatures associated with altered fatty acid metabolism, features that are common in patients with NAFLD.53 To determine whether SLC25A34 deletion exacerbates the NAFLD phenotype, Slc-HKO and WT control mice were maintained on chow for 1 month after AAV8 injection and then provided an FFD for 2 months to induce hepatic steatosis, referred to hereafter as “short-term FFD” (Supplemental Figure S5A).32,54 SLC25A34 deletion did not alter body weight (Figure 5A). Liver weight, the ratio of liver to body weight, and levels of the serum biomarkers alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase were comparable (Figure 5, A and B). Serum cholesterol was also equivalent, but triglycerides were reduced 25% in Slc-HKO mice (Figure 5B). Histologic scoring of liver sections (hematoxylin and eosin, ORO, and picrosirius red) revealed a modest increase in steatosis, lobular inflammation, hepatocyte ballooning, and fibrosis associated with short-term FFD (Figure 5C and Supplemental Figure S5, B and C). The degree of inflammation, ballooning, and fibrosis was similar in WT and Slc-HKO livers on each diet. However, steatosis was elevated in Slc-HKO on an FFD, which is supported by larger lipid vacuoles in the Slc-HKO mice (Figure 5C and Supplemental Figure S5C). The area covered by lipids increased 3.5-fold in Slc-HKO and stain intensity 3.7-fold, but the number of lipid droplets was unchanged (Figure 5D). Liver triglyceride levels were equivalent in each strain (Figure 5E). Thus, mice lacking SLC25A34 had increased hepatic lipid accumulation when maintained on short-term FFD.
Figure 5.
Nonalcoholic fatty liver disease phenotype is aggravated in hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice after being fed a short-term fast-food diet (FFD). A: Eight-week–old Slc25a34 floxed mice injected with AAV8-TBG-GFP or AAV8-TBG-Cre to generate wild-type (WT) control and Slc-HKO mice, respectively, were maintained on chow diet for 1 month and then switched to FFD for 2 months (short-term FFD) before analysis. Relative body weight during FFD feeding; liver weight and percentage of liver weight to body weight (LW/BW) at the time of euthanasia. B: Analysis of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), cholesterol, and triglyceride levels. C: Representative hematoxylin and eosin (H&E) and Oil Red O (ORO) staining reveal lipid accumulation in Slc-HKO and WT control livers. D: Quantification of lipid area, total lipid intensity, and number of lipid droplets. E: Quantification of liver triglycerides. F: Blood glucose level in mice after fasting for 16 hours. Slc-HKO and WT control mice on short-term FFD compared with mice maintained on chow for 3 months. G: Glucose tolerance test: mice were fasted for 8 hours, injected with 2 g/kg glucose, and glucose level was determined for 4 hours. Graphs represent means ± SEM. n = 4 to 10 (A); n = 4 (B, C, D, and E); n = 4 to 7 (F); n = 4 to 5 (G). ∗P < 0.05, ∗∗∗P < 0.001. Scale bars = 100 μm (C). AU, arbitrary units; BF, brightfield.
Glucose metabolism was evaluated after short-term FFD. After fasting for 16 hours, blood glucose was reduced 25% in Slc-HKO mice, compared with WT control mice (Figure 5F). Furthermore, although blood glucose levels in WT control mice increased 28% on short-term FFD (versus chow), glucose levels remained unchanged in Slc-HKO mice on short-term FFD (versus chow). Glucose tolerance testing was performed next (Figure 5G). In WT control mice, blood glucose increased 2.3-fold after glucose injection and returned to baseline after 3.5 hours, indicating impaired glucose tolerance (compare with findings in 3-month chow mice in Figure 3G that returned to baseline by 2.0 hours). In Slc-HKO mice, blood glucose levels increased 2.7-fold after injection and remained 25% to 50% higher than levels in the WT control mice over 1.5 hours before returning to baseline by 3.5 hours; this suggests reduced insulin sensitivity and impaired glucose homeostasis. Overall, the data suggest that SLC25A34 deletion leads to increased lipid accumulation and aberrations in glucose homeostasis after short-term FFD administration.
Altered Lipid Metabolism in Slc-HKO Livers after Short-Term FFD Compared with Chow
RNA-seq was performed to determine how the loss of SLC25A34 affects the liver transcriptome in the NAFLD model. Intriguingly, Slc25a34 expression was reduced nearly 50% in chow diet compared with WT controls, indicating down-regulation of Slc25a34 during NAFLD-induced stress (Figure 6A). Compared with WT control mice after short-term FFD, 1139 genes were differentially expressed in Slc-HKO (Supplemental Table S2). Ingenuity Pathway Analysis of these differentially expressed genes revealed dysregulation of pathways related to metabolism (Figure 6B). Gene ontology analysis showed alterations relevant to glucose homeostasis, gluconeogenesis, glycogen biosynthesis, triglyceride homeostasis, and insulin receptor signaling pathway (Figure 6C). In addition, Kyoto Encyclopedia Gene and Genome pathway–based analysis revealed alterations in gene pools of metabolically dysregulated pathways, such as insulin, AMPK, and HIF1, and metabolism of fructose, mannose, and carbohydrate digestion (Figure 6D). Thus, the transcriptomic analysis underscores a role for SLC25A34 in a wide range of metabolic processes related to NAFLD.
Figure 6.
Transcriptomic analysis livers from hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice fed a short-term fast-food diet (FFD). A: RNA isolated from Slc-HKO and wild-type (WT) control livers after short-term FFD and subjected to RNA-sequencing. Slc25a34 expression after short-term FFD compared with corresponding chow-fed mice (3-month group). B: Ingenuity Pathway Analysis identified altered metabolic and metabolically relevant pathways in Slc-HKO versus WT after short-term FFD. Bars represent activity patterns based on Z scores. C and D:Slc-HKO livers are characterized by alterations in biological processes [C, gene ontology (GO) terms and pathways [D, Kyoto Encyclopedia Gene and Genome (KEGG) pathways]. E: Western blot analysis of liver lysates from Slc-HKO and WT control mice. Each lane represents a pooled protein from multiple mice harvested after short-term FFD. Error bars represent SEM. n = 3 (A); n = 3 to 4 (E). ∗P < 0.05, ∗∗∗P < 0.001. Dashed lines in B, C, and D represent −Log(P value) = 1.3, the threshold for significance. SREBP1, sterol-responsive element binding protein-1.
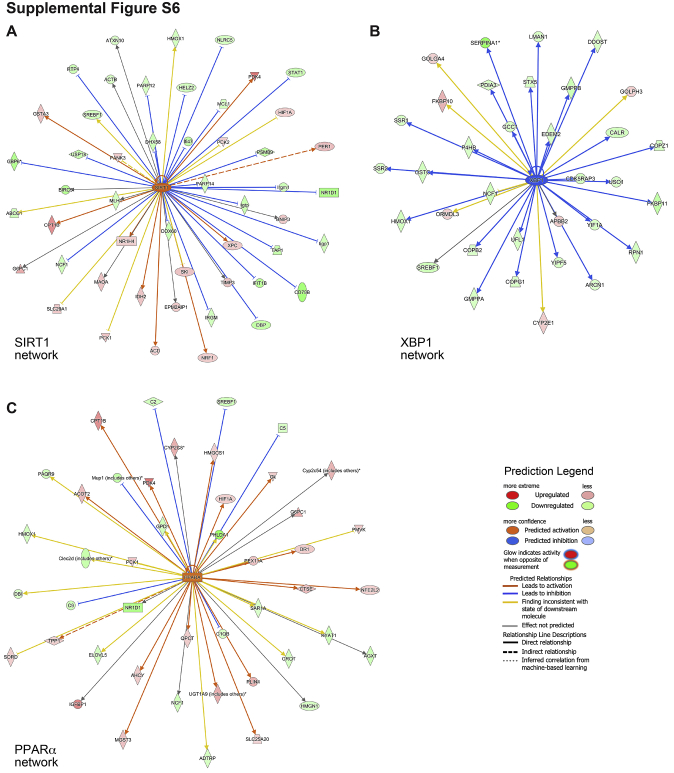
Seven of the top 20 upstream regulators identified in Slc-HKO livers have a significant role in FFD pathogenesis (Table 4). For example, sirtuin 1, which has a protective role in NAFLD, is predicted to be activated (Supplemental Figure S6A).55 XBP1, which promotes hepatic lipogenesis, is predicted to be inhibited (Supplemental Figure S6B). Although not included in the top 20 list, the PPARα pathway, which promotes β-oxidation, is predicted to be activated in Slc-HKO livers (Supplemental Figure S6C). Moreover, consistent with expression patterns leading to decreased lipid accumulation, protein levels of SREBP1 and fatty acid synthase, which promote lipogenesis, were decreased (Figure 6E). Taken together, the molecular profile of Slc-HKO mice on short-term FFD suggests a decrease in the lipid levels, which contrasts with the phenotype of increased lipid accumulation.
Table 4.
Differential Activation of Upstream Regulators by Slc-HKO Livers Compared with WT Controls (Short-term FFD)
| Upstream regulator | Predicted activation state | Activation Z score | P value of overlap |
|---|---|---|---|
| ITPR2 | Inhibited | −2.3 | 1.70 × 10−10 |
| PNPT1 | Activated | 3.5 | 5.33 × 10−10 |
| IRF7 | Inhibited | −4.9 | 1.98 × 10−09 |
| SIRT1 | Activated | 4.0 | 3.14 × 10−09 |
| XBP1 | Inhibited | −3.4 | 8.83 × 1−09 |
| NKX2-3 | Activated | 3.1 | 1.41 × 10−08 |
| PIK3CG | Activated | 2.9 | 1.81 × 10−08 |
| IGRM1 | Activated | 4.3 | 1.04 × 10−07 |
| TRIM24 | Activated | 4.4 | 2.76 × 10−07 |
| G protein alpha | Inhibited | −3.5 | 3.07 × 10−07 |
| miR-21 | Activated | 2.6 | 6.01 × 10−07 |
| STAT1 | Inhibited | −4.3 | 6.89 × 10−07 |
| ZC3H12C | Activated | 2.5 | 1.24 × 10−06 |
| IFNAR | Inhibited | −4.0 | 2.12 × 10−06 |
| IRF3 | Inhibited | −4.5 | 4.42 × 10−06 |
| G protein alpha | Inhibited | −2.5 | 8.21 × 10−06 |
| IFNL1 | Inhibited | −3.8 | 1.01 × 10−05 |
| NR1I3 | Activated | 2.9 | 1.25 × 10−05 |
| PTGER4 | Activated | 3.3 | 1.33 × 10−05 |
| ACKR2 | Activated | 3.2 | 2.29 × 10−05 |
A total of 105 Upstream Regulators were identified (Z score less than −2 and >2; P value of overlap <0.05), and the top 20 are shown. Bold indicates metabolic relevance.
FFD, fast-food diet; Slc-HKO, hepatocyte-specific Slc25a34 knockout; WT, wild type.
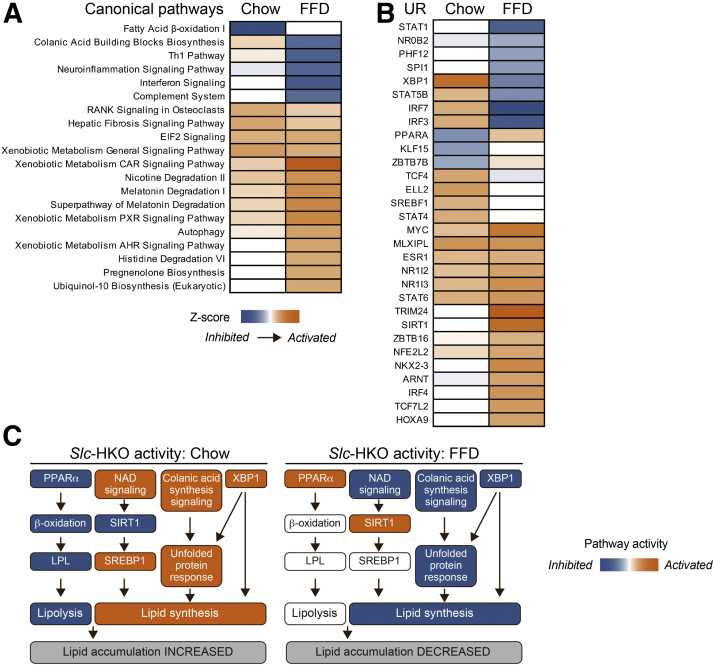
To determine why lipid metabolism is more severely affected when Slc-HKO mice are maintained on FFD than chow, gene signatures were compared on each diet. The comparison focused on canonical signaling pathways (Figure 7A) and upstream regulators (Figure 7B) differentially altered by Slc-HKO livers on chow and FFD. Four notable pathways associated with lipid metabolism consistently differed between Slc-HKO on short-term FFD compared with the chow diet (Figure 7C). First, the PPARα pathway promotes β-oxidation and lipoprotein lipase expression, which leads to lipid catabolism.49 PPARα gene targets, β-oxidation, and lipoprotein lipase are inhibited in Slc-HKO mice on chow, whereas PPARα gene targets are activated, and β-oxidation and lipoprotein lipase are unchanged in FFD. Second, NAD signaling regulates sirtuin 1, a master regulator of energy and mitochondrial metabolism, which can inhibit SREBP1 to abrogate lipogenesis.56 In chow-fed Slc-HKO mice, NAD signaling and SERBP1 were activated and sirtuin 1was inhibited, while a reversal of pathway activations was seen in FFD mice. Third, colanic acid induces unfolded protein response signaling, which alters mitochondrial dynamics and can induce lipid synthesis.57, 58, 59 Colanic acid and unfolded protein response signaling were activated in Slc-HKO mice on chow but reduced or inhibited with an FFD. Fourth, XBP1 is classically known for its role in unfolded protein response but also plays an essential role in the promotion of hepatic lipogenesis.60 XPB1 is predicted to be activated in Slc-HKO mice on chow but inhibited in FFD.
Figure 7.
Diet-dependent effects on the hepatocyte-specific Slc25a34 knockout (Slc-HKO) liver transcriptome. A and B: Ingenuity Pathway Analysis predictions of canonical pathways (A, Z score less than −2 and >2; and P value of overlap <0.05) and upstream regulators (B, Z score less than −2 and >2; and P value of overlap <0.01) that are activated or inhibited after a short-term fast-food diet (FFD) compared with chow (3-month group). Heatmaps show activity for Slc-HKO versus wild-type (WT) control. (C) Integrated network activity that affects lipid accumulation by Slc-HKO livers on short-term FFD or chow. LPL, lipoprotein lipase; PPARα, peroxisome proliferator-activated receptor α; SIRT1, sirtuin 1; SREBP1, sterol-responsive element binding protein-1; UR, upstream regulator.
Intriguingly, the pathway predictions contrast with functional observations. For example, chow-fed Slc-HKO mice did not accumulate lipids, whereas pathway analysis suggests activation of lipogenesis programs. Furthermore, FFD-fed Slc-HKO livers accumulated lipids, but pathway analysis indicates programs for reduced lipid accumulation. The data suggest the possibility that the gene signatures reflect a protective response to the physiological consequences of SLC25A34 deletion. For instance, Slc-HKO livers responded to FFD-mediated lipid accumulation by inhibiting lipogenesis programs.
SLC25A34 Deletion Partially Rescues NAFLD Phenotype after Long-Term FFD
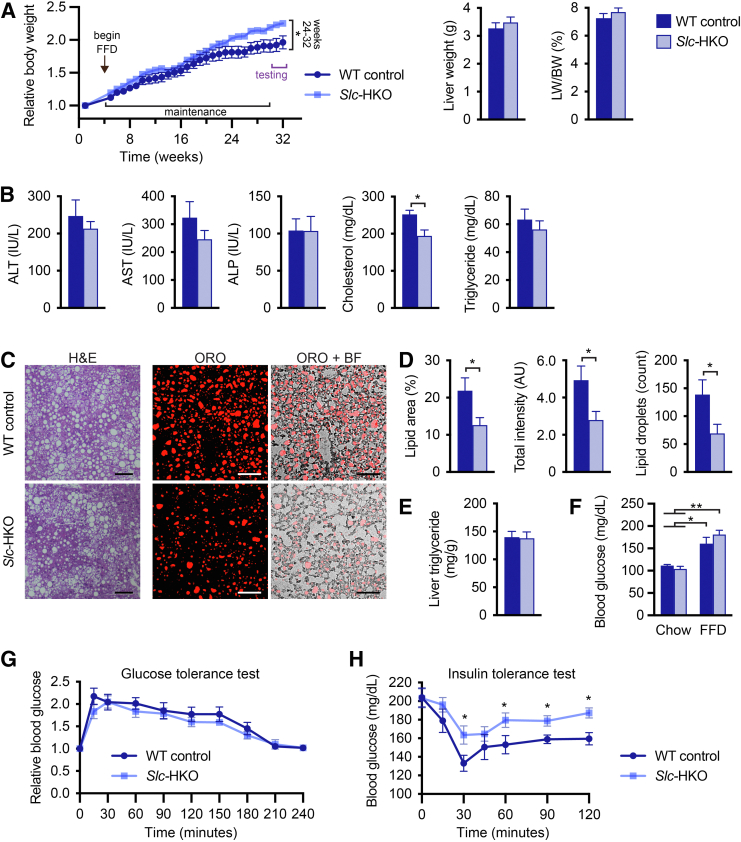
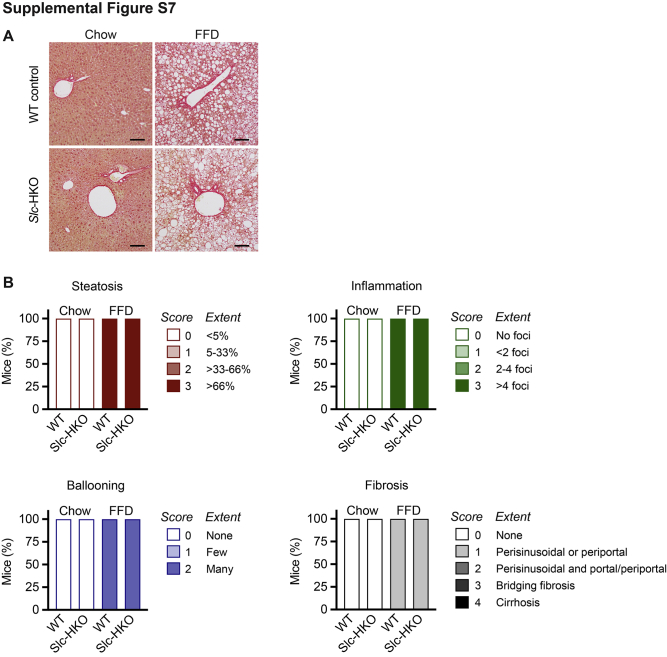
To determine whether gene signatures in Slc-HKO livers facilitate a protective response to short-term FFD-induced stress, mice were maintained on FFD for 6 months (long-term FFD) (Supplemental Figure S5). Compared with body weight of WT control mice, body weight of Slc-HKO mice increased up to 15% by 6 months; the liver weight and liver weight to body weight ratio were equivalent (Figure 8A). Levels of the serum markers alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase were comparable (Figure 8B). Serum triglyceride levels were similar, and serum cholesterol decreased by 23% in Slc-HKO mice (Figure 8B). Histologic scoring of liver sections (hematoxylin and eosin, ORO, and picrosirius red) revealed increased steatosis, lobular inflammation, hepatocyte ballooning, and fibrosis associated with long-term FFD (Figure 8C and Supplemental Figure S7, A and B). The degree of steatosis, inflammation, ballooning, and fibrosis scored similarly in WT and Slc-HKO livers on each diet. However, after FFD, the total lipid area and ORO uptake were reduced by 40% in Slc-HKO, and lipid droplet size between 400 and 1000 μm2 decreased by 60% (Figure 8D). Unexpectedly, liver triglyceride content was unchanged in each strain (Figure 8E). Thus, in contrast to WT control mice, mice lacking SLC25A34 are partially protected from lipid accumulation associated with long-term FFD.
Figure 8.
Improved nonalcoholic fatty liver disease phenotype in hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice after a long-term fast-food diet (FFD). A: Eight-week–old Slc25a34 floxed mice injected with AAV8-TBG-GFP or AAV8-TBG-Cre to generate wild-type (WT) control and Slc-HKO mice, respectively, were maintained on chow diet for 1 month and then switched to FFD for 6 months (long-term FFD) before analysis. Relative body weight during FFD feeding; liver weight and percentage of liver weight to body weight (LW/BW) at the time of euthanasia. B: Analysis of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), cholesterol, and triglyceride levels. C: Representative hematoxylin and eosin (H&E) and Oil Red O (ORO) staining reveal lipid accumulation in Slc-HKO and WT control livers. D: Lipid area, intensity, and number are reduced in Slc-HKO livers. E: Quantification of liver triglycerides. F: Blood glucose level in mice after fasting for 16 hours. Slc-HKO and WT control mice on short-term FFD compared with mice maintained on chow for 7 months. G: Glucose tolerance test: mice were fasted for 8 hours, injected with 2 g/kg glucose, and glucose level was determined for 4 hours. H: Insulin tolerance test: mice were fasted for 4 hours, injected with 0.75 U/kg insulin, and blood glucose was monitored over 2 hours. Graphs represent means ± SEM. n = 6 (A and G); n = 4 to 6 (B, C, D, and F); n = 5 (E); n = 5 to 6 (H). ∗P < 0.05, ∗∗P < 0.01. Scale bars = 100 μm (C). AU, arbitrary units; BF, brightfield.
Finally, glucose homeostasis was evaluated after long-term FFD. After a 16-hour fast, blood glucose levels increased 60% to 80% after long-term FFD compared with chow, but Slc-HKO and WT control glucose levels were comparable on each diet (Figure 8F). Glucose tolerance test results revealed equivalent levels of blood glucose in Slc-HKO and WT control mice throughout the challenge (Figure 8G), which represents improved glucose regulation by Slc-HKO; comparison with short-term FFD is shown in Figure 5G. Finally, an insulin tolerance test was performed in which mice were fasted for 4 hours, injected with insulin, and blood glucose monitored (Figure 8H). In Slc-HKO mice, blood glucose levels remained higher than in WT control mice over the monitoring period, indicating reduced insulin sensitivity. Taken together, long-term FFD Slc-HKO mice displayed improvement in de novo lipogenesis and glucose homeostasis, despite increased body weight and impaired insulin resistance.
Discussion
SLC25A34 is a poorly characterized inner mitochondrial membrane protein belonging to a family of substrate transporters.1,2 SLC25A34 is believed to be a homologue of yeast oxaloacetate carrier (Oac1p).3,4 SLC25A34 has been identified as a target of miR-122 in the mammalian system11; mice lacking miR-122 develop metabolically dysregulated conditions in the liver, thus raising the possibility that a subset of its targets plays a role in liver metabolism.12 Conflicting reports show up-regulation and down-regulation of SLC25A34 in patients with NASH, suggesting that SLC25A34 plays a complex role in liver pathogenesis.26,27 The current work investigates SLC25A34 in liver metabolism in primary hepatocytes and a new hepatocyte-specific Slc25a34 knockout model.
Depletion and overexpression of Slc25a34 in WT primary hepatocytes revealed an inverse relationship between Slc25a34 and the ADP/ATP ratio, a determinant of the energy state that guides downstream metabolic changes, suggesting that alteration of Slc25a34 creates two distinctly different metabolic states. Reduced levels of Slc25a34 generated more energy (lower ADP/ATP ratio), whereas increased levels of Slc25a34 reduced energy generation (higher ADP/ATP ratio). Slc25a34 manipulation altered drivers of energy generation, including mitochondrial mass, the transcriptional program for mitochondrial biogenesis, lipid synthesis, and β-oxidation. At the functional level, Slc25a34 depletion increased mitochondrial respiration, and overexpression increased glycolytic capacity. Thus, manipulation of SLC25A34 in vitro alters multiple facets of energy metabolism, strongly indicating a role for SLC25A34 in energy homeostasis.
A hepatocyte-specific Slc25a34 knockout mouse was generated to investigate SLC25A34 function in vivo. Slc-HKO and WT control mice were maintained on chow diet and FFD to model liver stress in NAFLD. On the chow diet, Slc-HKO mice presented minimal changes to liver physiology, although distinct differences in glucose metabolism and de novo lipogenesis were found. During chronic stress by FFD, Slc-HKO mice displayed changes in lipid content, de novo lipogenesis, fasted blood glucose levels, and glucose handling. The extent of these changes transformed over both time and continued injury. In the glucose tolerance test of mice on chow or short-term FFD, Slc-HKO mice displayed elevated circulating glucose levels after injection, and the return to baseline glucose level was impaired (compared with WT control mice). However, after long-term FFD, circulating glucose levels were comparable in Slc-HKO and WT control mice. The degree of hepatic steatosis followed a similar pattern in mice lacking SLC25A34. Although lipid accumulation measured by ORO uptake in tissue sections was nearly fourfold higher in Slc-HKO livers after short-term FFD, lipid accumulation was reduced by 60% after long-term FFD. Interestingly, liver triglyceride levels were equivalent in WT and Slc-HKO mice after short- and long-term FFD. It is unclear why these data conflict with the variation in neutral lipids detected by ORO, although discrepancies in liver triglycerides and lipid droplet area, for example, have been reported.61 Male mice were used for all of the studies, and additional research is necessary to determine whether SLC25A34 affects the liver in a sex-specific manner. Taken together, in the context of glucose homeostasis and lipid metabolism, SLC25A34 loss exacerbates the degree of liver injury early on (during aging and short-term FFD) and ultimately promotes liver healing or adaptation (during long-term FFD).
Slc-HKO livers were investigated to determine molecular mechanisms by which SLC25A34 could affect lipid metabolism. RNA-seq and western blot analysis revealed significant reprogramming of fatty acid metabolism in chow-fed Slc-HKO mice. Lipogenesis signaling was induced, as seen by increased expression of mature SREBP1, SREBP1 gene targets, and increased expression of phosphorylated ACLY. Moreover, β-oxidation signaling was reduced, as seen by down-regulation of Cpt1a, depression of the PPARα network, and reduced phosphorylated ACC. In addition to directly affecting lipogenesis and β-oxidation, SLC25A34 may affect lipid secretion. After short-term FFD, hepatic lipid levels increased in Slc-HKO mice, compared with WT mice, while serum triglyceride levels decreased. Decreased serum triglyceride levels are frequently associated with decreased very-low-density lipoprotein secretion.62 Thus, future work should determine whether SLC25A34 loss impairs the secretion of lipids, specifically apolipoprotein B-100 and very-low-density lipoprotein. Overall, the data suggest that SLC25A34 deletion promotes a net increase of lipids in chow-fed mice, which would increase de novo lipogenesis. However, chow-fed Slc-HKO and WT control mice had equivalent levels of hepatic lipids, and it is only when hepatocytes were stressed, such as by short-term FFD (or cell culture of primary hepatocytes), that lipid accumulation occurred. Thus, loss of SLC25A34 is tolerated in the absence of stress, which primes Slc-HKO mice for lipid accumulation when subjected to short-term FFD stress.
Increased lipid accumulation in Slc-HKO mice maintained on short-term FFD is surprising. Both fatty acid synthase and mature SREBP1 were reduced in Slc-HKO mice, implying abrogation of the lipogenic program. RNA-seq revealed pathway activity leading to reduced de novo lipogenesis (eg, increased lipolysis stimulated by PPARα-driven β-oxidation and lipoprotein lipase; and reduced lipid synthesis via inhibition of NAD signaling, colonic acid synthesis signaling, and XBP1 signaling). However, when mice were subjected to an extended period of stress by long-term FFD feeding, lipid accumulation was ameliorated in Slc-HKO livers. Taken together, there is a pattern in which gene signatures in chow-fed Slc-HKO livers (eg, increased lipid accumulation) are observed after short-term FFD, and gene signatures in short-term FFD Slc-HKO (eg, reduced lipid accumulation) are observed after long-term FFD. It is possible that SLC25A34 plays a dual role, such that its loss first exacerbates liver injury and subsequently facilitates liver protection over time and/or chronic stress.
The data present a model in which SLC25A34 attenuates mitochondrial mass and energy production. During normal homeostasis and the early stages of fatty liver disease, SLC25A34 activity restricts lipid accumulation (via decreased lipogenesis and increased β-oxidation) and contributes to the maintenance of hepatic glucose sensitivity. This explains why the loss of SLC25A34 led to increased lipid accumulation and impaired glucose handling on chow and short-term FFD. Notably, Slc25a34 expression was reduced by 50% after short-term FFD in WT livers, suggesting that reduced SLC25A34 activity may accelerate disease progression in WT mice. The role of SLC25A34 in advanced NAFLD is unclear because the loss of SLC25A34 after long-term FFD counterintuitively improved glucose sensitivity. How SLC25A34 function changes in a complex NAFLD microenvironment should be investigated, including its role in liver cirrhosis and hepatocellular carcinoma. It will also be important to determine how SLC25A34 expression changes during the initiation and progression of metabolically dysregulated chronic conditions in patients. Considering the role of SLC25A34 in energy homeostasis, it is unclear how differential SLC25A34 expression in patients affects energy homeostasis in unstressed and stressed conditions.26,27 Studies should determine whether increased levels of SLC25A34 accelerate pathogenesis or protects the liver from injury.
Acknowledgments
We thank Tyler Slomiak and Abigayle Wagner for mouse breeding and genotyping, Dr. Chunming Bi and Zhaohui Kou of the Transgenic and Gene Targeting Core (University of Pittsburgh School of Medicine, Department of Immunology) for microinjection of zygotes and production of Slc25a34 floxed mice, and Michael Reynolds for assistance with bioenergetic measurements. The Mir122-KO mice were kindly provided by Dr. Kalpana Ghoshal.
Footnotes
Supported by grants from the NIH (R01 DK103645), Commonwealth of Pennsylvania, and University of Pittsburgh Physicians Foundation (A.W.D); the Hemophilia Center of Western Pennsylvania and NIH (R01HL133003-01A1) (S.S.); NIH (R01 DK122990) (B.B.); and NIH-NIBIB training grant (T32 EB001026) entitled “Cellular Approaches to Tissue Engineering and Regeneration” (S.R.W.). Additional support provided by the Pittsburgh Liver Research Center Clinical Biospecimen Repository and Processing Core (P30 DK120531); the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource, which is supported in part by award P30CA047904 from the NIH; and the Center for Metabolism and Mitochondrial Medicine, which is supported in part by the Pittsburgh Foundation (MR2020 109502).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2022.06.002.
Supplemental Data
Supplemental Figure S1.
Effect of Slc25a34 knockdown and overexpression on mitochondrial respiration and glycolysis in primary hepatocytes. Primary hepatocytes isolated from 2-month–old wild-type C57BL/6 mice were transfected with either control siRNA (siControl) or siRNA against Slc25a34 (siSlc25a34) (A, B) or control plasmid (Control) or plasmid encoding Slc25a34 (Slc25a34 overexpression [OE]) (C, D) and assessed 72 hours later. A, C: Representative mitochondrial stress test after Slc25a34 knockdown or OE, conducted with Seahorse extracellular flux analyzer. B, D: Representative glycolytic rate assay profile after Slc25a34 knockdown or OE. Data are representative of three to four experiments with n = 7 to 11 replicates per experiment. Error bars represent ± SD. 2DG, 2-deoxy-glucose; DMEM, Dulbecco’s modified Eagle’s medium; ECAR, extracellular acidification rate; FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; OCR, oxygen consumption rate; RAU, raw absorbance units.
Supplemental Figure S2.
Detailed overview of Slc25a34 gene targeting and genotyping. A: Wild-type (WT) Slc25a34 genomic DNA was cleaved by Cas9 at the specific single guide RNA-3 and single guide RNA-6 target sequences located upstream and downstream of exon 1, respectively. Boxes represent exons 1 to 5. Homology-directed repair of the cleaved WT allele with a synthetic long single-stranded DNA donor template introduced EcoR1 and loxP sites to generate a floxed allele. Cre-mediated recombination produced a knockout allele. B: To confirm the presence of a floxed Slc25a34 allele, DNA was amplified with oligos F51 and R51 (to target to amplify the 5′ loxP site) and digested with EcoR1. Similarly, to confirm the presence of the 3′ loxP site, DNA was amplified with oligos F32 and R32. Fragment sizes are shown. C: Identification of the knockout allele was confirmed by using a three-primer PCR using oligos F51, R53, and R32, which produces unique fragments for the WT, floxed, and knockout alleles.
Supplemental Figure S3.
Characterization of hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice on chow diet. A: Experimental design: 8-week–old Slc25a34 floxed mice were injected with AAV8-TBG-GFP or AAV8-TBG-Cre to generate wild-type (WT) control and Slc-HKO mice, respectively. Mice were maintained on chow diet for 1, 3, or 7 months before functional testing and euthanasia. B: Oil Red O (ORO) staining of Slc-HKO and WT control livers revealed equivalent lipid content in each group. C: Periodic acid-Schiff (PAS) staining of Slc-HKO and WT control livers after 3 and 7 months revealed comparable intrahepatic glycogen. n = 4 to 7 (B); n = 4 (C). Scale bars: 100 μm (B and C). The graphic in panelA was created in BioRender.com (2021; Toronto, ON, Canada).
Supplemental Figure S4.
Differential expression of Slc25 family members and networks by hepatocyte-specific Slc25a34 knockout (Slc-HKO) livers. A: Expression of Slc25 family members. Absolute read count values for each gene were normalized to total read counts obtained per mouse and presented as relative expression of Slc-HKO compared with wild-type (WT) control. B: Ingenuity Pathway Analysis revealed activation of the sterol-responsive element binding protein-1 (SREBP1) network (Srebf1 is the gene encoding SREBP1 protein) in Slc-HKO (Z score, 2.03; P value of overlap 9.0 × 10−3). (C) Ingenuity Pathway Analysis showed inhibition of the peroxisome proliferator-activated receptor α (PPARα) network in Slc-HKO (Z score, –2.90; P value of overlap 3.0 × 10−7). Graphs represent means ± SEM. n = 3 (A). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Supplemental Figure S5.
Histologic scoring after short-term fast-food diet (FFD). A: Experimental design to model nonalcoholic fatty liver disease with short-term and long-term FFD. Eight-week–old Slc25a34 floxed mice injected with AAV8-TBG-GFP or AAV8-TBG-Cre to generate wild-type (WT) control and hepatocyte-specific Slc25a34 knockout (Slc-HKO) mice, respectively, were maintained on chow diet for 1 month and then switched to FFD for 2 months (short-term FFD) or 6 months (long-term FFD) before functional testing and euthanasia. Mice on chow diet (described earlier in Supplemental Figure S3A) were used for comparison with short- and long-term FFD. B: Representative picrosirius red staining of WT and Slc-HKO livers after short-term chow and FFD. C: Histologic scoring of steatosis, lobular inflammation, hepatocyte ballooning, and fibrosis. Steatosis score refers to the surface area evaluated on low- to medium-power examination. Lobular inflammation refers to the number of foci as observed at 200× magnification. Ballooning refers to ballooned hepatocytes. Fibrosis refers to the type and degree of fibrosis in the liver. After short-term FFD, WT and Slc-HKO livers (fibrosis score of 1) displayed mild portal or periportal fibrosis. n = 4 to 5 (C). Scale bars = 100 μm (B). The graphic in panelA was created in BioRender.com (2021; Toronto, ON, Canada).
Supplemental Figure S6.
Activation or inhibition of networks in hepatocyte-specific Slc25a34 knockout (Slc-HKO) livers after short-term fast-food diet. A, B, C: Ingenuity Pathway Analysis revealed activation of the sirtuin 1 (SIRT1) network (A, Z score, 4.1; P value of overlap 1.1 × 10−13); inhibition of the XBP1 network (B, Z score, –3.4; P value of overlap 8.8 × 10−9); and activation of the peroxisome proliferator-activated receptor α (PPARα) network (C, Z score, 2.3; P value of overlap 5.5 × 10−6) in Slc-HKO after short-term FFD.
Supplemental Figure S7.
Histologic scoring after long-term fast-food diet (FFD). A: Representative picrosirius red staining of wild-type (WT) and hepatocyte-specific Slc25a34 knockout (Slc-HKO) livers after long-term chow and FFD. B: Histologic scoring of steatosis, lobular inflammation, hepatocyte ballooning, and fibrosis. Steatosis score refers to the surface area evaluated on low- to medium-power examination. Lobular inflammation refers to the number of foci as observed at 200× magnification. Ballooning refers to ballooned hepatocytes. Fibrosis refers to the type and degree of fibrosis in the liver. After long-term FFD, WT and Slc-HKO livers (fibrosis score of 1) displayed mild portal or periportal fibrosis. n = 4 to 6 (B). Scale bars = 100 μm (A).
References
- 1.Palmieri F., Monné M. Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta. 2016;1863:2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Ruprecht J.J., Kunji E.R.S. The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem Sci. 2020;45:244–258. doi: 10.1016/j.tibs.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmieri L., Vozza A., Agrimi G., De Marco V., Runswick M.J., Palmieri F., Walker J.E. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J Biol Chem. 1999;274:22184–22190. doi: 10.1074/jbc.274.32.22184. [DOI] [PubMed] [Google Scholar]
- 4.Marobbio C.M., Giannuzzi G., Paradies E., Pierri C.L., Palmieri F. alpha-Isopropylmalate, a leucine biosynthesis intermediate in yeast, is transported by the mitochondrial oxalacetate carrier. J Biol Chem. 2008;283:28445–28453. doi: 10.1074/jbc.M804637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedroso J.A., Zampieri T.T., Donato J., Jr. Reviewing the effects of L-leucine supplementation in the regulation of food intake, energy balance, and glucose homeostasis. Nutrients. 2015;7:3914–3937. doi: 10.3390/nu7053914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trefts E., Gannon M., Wasserman D.H. The liver. Curr Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuppan D., Schattenberg J.M. Non-alcoholic steatohepatitis: pathogenesis and novel therapeutic approaches. J Gastroenterol Hepatol. 2013;28(Suppl 1):68–76. doi: 10.1111/jgh.12212. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed M. Non-alcoholic fatty liver disease in 2015. World J Hepatol. 2015;7:1450–1459. doi: 10.4254/wjh.v7.i11.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Younossi Z.M. Non-alcoholic fatty liver disease—a global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 11.Hsu S.-H., Delgado E.R., Otero P.A., Teng K.-Y., Kutay H., Meehan K.M., Moroney J.B., Monga J.K., Hand N.J., Friedman J.R., Ghoshal K., Duncan A.W. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016;64:599–615. doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu S.-H., Wang B., Kota J., Yu J., Costinean S., Kutay H., Yu L., Bai S., La Perle K., Chivukula R.R., Mao H., Wei M., Clark K.R., Mendell J.R., Caligiuri M.A., Jacob S.T., Mendell J.T., Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu S.-H., Wang B., Kutay H., Bid H., Shreve J., Zhang X., Costinean S., Bratasz A., Houghton P., Ghoshal K. Hepatic loss of miR-122 predisposes mice to hepatobiliary cyst and hepatocellular carcinoma upon diethylnitrosamine exposure. Am J Pathol. 2013;183:1719–1730. doi: 10.1016/j.ajpath.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kita Y., Takamura T., Misu H., Ota T., Kurita S., Takeshita Y., Uno M., Matsuzawa-Nagata N., Kato K.-I., Ando H., Fujimura A., Hayashi K., Kimura T., Ni Y., Otoda T., Miyamoto K.-I., Zen Y., Nakanuma Y., Kaneko S. Metformin prevents and reverses inflammation in a non-diabetic mouse model of nonalcoholic steatohepatitis. PLoS One. 2012;7:e43056. doi: 10.1371/journal.pone.0043056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichimura A., Hirasawa A., Poulain-Godefroy O., Bonnefond A., Hara T., Yengo L., et al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature. 2012;483:350–354. doi: 10.1038/nature10798. [DOI] [PubMed] [Google Scholar]
- 16.Pachikian B.D., Essaghir A., Demoulin J.-B., Neyrinck A.M., Catry E., De Backer F.C., Dejeans N., Dewulf E.M., Sohet F.M., Portois L., Deldicque L., Molendi-Coste O., Leclercq I.A., Francaux M., Carpentier Y.A., Foufelle F., Muccioli G.G., Cani P.D., Delzenne N.M. Hepatic n-3 polyunsaturated fatty acid depletion promotes steatosis and insulin resistance in mice: genomic analysis of cellular targets. PLoS One. 2011;6:e23365. doi: 10.1371/journal.pone.0023365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers M.T., Groen A.K., Oler A.T., Keller M.P., Choi Y., Schueler K.L., Richards O.C., Lan H., Miyazaki M., Kuipers F., Kendziorski C.M., Ntambi J.M., Attie A.D. Cholestasis and hypercholesterolemia in SCD1-deficient mice fed a low-fat, high-carbohydrate diet. J Lipid Res. 2006;47:2668–2680. doi: 10.1194/jlr.M600203-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Keyhani-Nejad F., Irmler M., Isken F., Wirth E.K., Beckers J., Birkenfeld A.L., Pfeiffer A.F.H. Nutritional strategy to prevent fatty liver and insulin resistance independent of obesity by reducing glucose-dependent insulinotropic polypeptide responses in mice. Diabetologia. 2015;58:374–383. doi: 10.1007/s00125-014-3423-5. [DOI] [PubMed] [Google Scholar]
- 19.Anthérieu S., Le Guillou D., Coulouarn C., Begriche K., Trak-Smayra V., Martinais S., Porceddu M., Robin M.-A., Fromenty B. Chronic exposure to low doses of pharmaceuticals disturbs the hepatic expression of circadian genes in lean and obese mice. Toxicol Appl Pharmacol. 2014;276:63–72. doi: 10.1016/j.taap.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Ren H., Aleksunes L.M., Wood C., Vallanat B., George M.H., Klaassen C.D., Corton J.C. Characterization of peroxisome proliferator-activated receptor alpha–independent effects of PPARalpha activators in the rodent liver: di-(2-ethylhexyl) phthalate also activates the constitutive-activated receptor. Toxicol Sci. 2010;113:45–59. doi: 10.1093/toxsci/kfp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khetchoumian K., Teletin M., Tisserand J., Mark M., Herquel B., Ignat M., Zucman-Rossi J., Cammas F., Lerouge T., Thibault C., Metzger D., Chambon P., Losson R. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 22.Herrema H., Derks T.G.J., van Dijk T.H., Bloks V.W., Gerding A., Havinga R., Tietge U.J.F., Müller M., Smit G.P.A., Kuipers F., Reijngoud D.-J. Disturbed hepatic carbohydrate management during high metabolic demand in medium-chain acyl-CoA dehydrogenase (MCAD)-deficient mice. Hepatology. 2008;47:1894–1904. doi: 10.1002/hep.22284. [DOI] [PubMed] [Google Scholar]
- 23.Alam M.S., Getz M., Safeukui I., Yi S., Tamez P., Shin J., Velázquez P., Haldar K. Genomic expression analyses reveal lysosomal, innate immunity proteins, as disease correlates in murine models of a lysosomal storage disorder. PLoS One. 2012;7:e48273. doi: 10.1371/journal.pone.0048273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mistry P.K., Liu J., Yang M., Nottoli T., McGrath J., Jain D., Zhang K., Keutzer J., Chuang W.-L., Mehal W.Z., Zhao H., Lin A., Mane S., Liu X., Peng Y.Z., Li J.H., Agrawal M., Zhu L.-L., Blair H.C., Robinson L.J., Iqbal J., Sun L., Zaidi M. Glucocerebrosidase gene-deficient mouse recapitulates Gaucher disease displaying cellular and molecular dysregulation beyond the macrophage. Proc Natl Acad Sci U S A. 2010;107:19473–19478. doi: 10.1073/pnas.1003308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai Y., Hashida H., Kadota K., Minami M., Shimizu K., Matsumoto I., Kato H., Abe K. Up-regulation of genes related to the ubiquitin-proteasome system in the brown adipose tissue of 24-h-fasted rats. Biosci Biotechnol Biochem. 2008;72:139–148. doi: 10.1271/bbb.70508. [DOI] [PubMed] [Google Scholar]
- 26.Arendt B.M., Comelli E.M., Ma D.W., Lou W., Teterina A., Kim T., Fung S.K., Wong D.K.H., McGilvray I., Fischer S.E., Allard J.P. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology. 2015;61:1565–1578. doi: 10.1002/hep.27695. [DOI] [PubMed] [Google Scholar]
- 27.Lake A.D., Novak P., Fisher C.D., Jackson J.P., Hardwick R.N., Billheimer D.D., Klimecki W.T., Cherrington N.J. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39:1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quadros R.M., Miura H., Harms D.W., Akatsuka H., Sato T., Aida T., Redder R., Richardson G.P., Inagaki Y., Sakai D., Buckley S.M., Seshacharyulu P., Batra S.K., Behlke M.A., Zeiner S.A., Jacobi A.M., Izu Y., Thoreson W.B., Urness L.D., Mansour S.L., Ohtsuka M., Gurumurthy C.B. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18:92. doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier S., Gingras S., Green D.R. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity. 2015;42:18–27. doi: 10.1016/j.immuni.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song A.J., Palmiter R.D. Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 2018;34:333–340. doi: 10.1016/j.tig.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhushan B., Molina L., Koral K., Stoops J.W., Mars W.M., Banerjee S., Orr A., Paranjpe S., Monga S.P., Locker J., Michalopoulos G.K. Yes-associated protein is crucial for constitutive androstane receptor-driven hepatocyte proliferation but not for induction of drug metabolism genes in mice. Hepatology. 2021;73:2005–2022. doi: 10.1002/hep.31521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhushan B., Banerjee S., Paranjpe S., Koral K., Mars W.M., Stoops J.W., Orr A., Bowen W.C., Locker J., Michalopoulos G.K. Pharmacologic inhibition of epidermal growth factor receptor suppresses nonalcoholic fatty liver disease in a murine fast-food diet model. Hepatology. 2019;70:1546–1563. doi: 10.1002/hep.30696. [DOI] [PubMed] [Google Scholar]
- 33.Overturf K., Al-Dhalimy M., Tanguay R., Brantly M., Ou C.N., Finegold M., Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 34.Guo W., Jiang L., Bhasin S., Khan S.M., Swerdlow R.H. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9:261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J., Kim J., Mikami T. Exercise-induced lactate release mediates mitochondrial biogenesis in the hippocampus of mice via monocarboxylate transporters. Front Physiol. 2021;12:736905. doi: 10.3389/fphys.2021.736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquez M.P., Alencastro F., Madrigal A., Jimenez J.L., Blanco G., Gureghian A., Keagy L., Lee C., Liu R., Tan L., Deignan K., Armstrong B., Zhao Y. The role of cellular proliferation in adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells. Stem Cells Dev. 2017;26:1578–1595. doi: 10.1089/scd.2017.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva D.F., Selfridge J.E., Lu J., Lezi E., Roy N., Hutfles L., Burns J.M., Michaelis E.K., Yan S., Cardoso S.M., Swerdlow R.H. Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum Mol Genet. 2013;22:3931–3946. doi: 10.1093/hmg/ddt247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.-C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J., Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 39.Edmunds L.R., Huckestein B.R., Kahn M., Zhang D., Chu Y., Zhang Y., Wendell S.G., Shulman G.I., Jurczak M.J. Hepatic insulin sensitivity is improved in high-fat diet-fed Park2 knockout mice in association with increased hepatic AMPK activation and reduced steatosis. Physiol Rep. 2019;7:e14281. doi: 10.14814/phy2.14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alquier T., Poitout V. Considerations and guidelines for mouse metabolic phenotyping in diabetes research. Diabetologia. 2018;61:526–538. doi: 10.1007/s00125-017-4495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benedé-Ubieto R., Estévez-Vázquez O., Ramadori P., Cubero F.J., Nevzorova Y.A. Guidelines and considerations for metabolic tolerance tests in mice. Diabetes Metab Syndr Obes. 2020;13:439–450. doi: 10.2147/DMSO.S234665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edmunds L.R., Xie B., Mills A.M., Huckestein B.R., Undamatla R., Murali A., Pangburn M.M., Martin J., Sipula I., Kaufman B.A., Scott I., Jurczak M.J. Liver-specific Prkn knockout mice are more susceptible to diet-induced hepatic steatosis and insulin resistance. Mol Metab. 2020;41:101051. doi: 10.1016/j.molmet.2020.101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 46.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P.A., Hou X., Hao S. Mitochondrial biogenesis in neurodegeneration. J Neurosci Res. 2017;95:2025–2029. doi: 10.1002/jnr.24042. [DOI] [PubMed] [Google Scholar]
- 48.Ipsen D.H., Lykkesfeldt J., Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol Life Sci. 2018;75:3313–3327. doi: 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura M.T., Yudell B.E., Loor J.J. Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Jensen T.L., Kiersgaard M.K., Sørensen D.B., Mikkelsen L.F. Fasting of mice: a review. Lab Anim. 2013;47:225–240. doi: 10.1177/0023677213501659. [DOI] [PubMed] [Google Scholar]
- 51.Ameer F., Scandiuzzi L., Hasnain S., Kalbacher H., Zaidi N. De novo lipogenesis in health and disease. Metabolism. 2014;63:895–902. doi: 10.1016/j.metabol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Sanders F.W., Griffin J.L. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol Rev Camb Philos Soc. 2016;91:452–468. doi: 10.1111/brv.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charlton M., Krishnan A., Viker K., Sanderson S., Cazanave S., McConico A., Masuoko H., Gores G. Fast food diet mouse: novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am J Physiol Gastrointest Liver Physiol. 2011;301:G825–G834. doi: 10.1152/ajpgi.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding R.-B., Bao J., Deng C.-X. Emerging roles of SIRT1 in fatty liver diseases. Int J Biol Sci. 2017;13:852–867. doi: 10.7150/ijbs.19370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schug T.T., Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakrabarti A., Membrez M., Morin-Rivron D., Siddharth J., Chou C.J., Henry H., Bruce S., Metairon S., Raymond F., Betrisey B., Loyer C., Parkinson S.J., Masoodi M. Transcriptomics-driven lipidomics (TDL) identifies the microbiome-regulated targets of ileal lipid metabolism. NPJ Syst Biol Appl. 2017;3:33. doi: 10.1038/s41540-017-0033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., Hotamisligil G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han B., Sivaramakrishnan P., Lin C.-C.J., Neve I.A.A., He J., Tay L.W.R., Sowa J.N., Sizovs A., Du G., Wang J., Herman C., Wang M.C. Microbial genetic composition tunes host longevity. Cell. 2017;169:1249–1262.e13. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurczak M.J., Lee A.-H., Jornayvaz F.R., Lee H.-Y., Birkenfeld A.L., Guigni B.A., Kahn M., Samuel V.T., Glimcher L.H., Shulman G.I. Dissociation of inositol-requiring enzyme (IRE1[alpha])-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem. 2012;287:2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al Rijjal D., Liu Y., Lai M., Song Y., Danaei Z., Wu A., Mohan H., Wei L., Schopfer F.J., Dai F.F., Wheeler M.B. Vascepa protects against high-fat diet-induced glucose intolerance, insulin resistance, and impaired [beta]-cell function. iScience. 2021;24:102909. doi: 10.1016/j.isci.2021.102909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minehira K., Young S.G., Villanueva C.J., Yetukuri L., Oresic M., Hellerstein M.K., Farese R.V., Jr., Horton J.D., Preitner F., Thorens B., Tappy L. Blocking VLDL secretion causes hepatic steatosis but does not affect peripheral lipid stores or insulin sensitivity in mice. J Lipid Res. 2008;49:2038–2044. doi: 10.1194/jlr.M800248-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.