Abstract
Background
Immune checkpoint blockade (ICB) has revolutionized cancer immunotherapy. However, most patients with cancer fail to respond clinically. One potential reason is the accumulation of immunosuppressive transforming growth factor β (TGFβ) in the tumor microenvironment (TME). TGFβ drives cancer immune evasion in part by inducing regulatory T cells (Tregs) and limiting CD8+ T cell function. Glycoprotein-A repetitions predominant (GARP) is a cell surface docking receptor for activating latent TGFβ1, TGFβ2 and TGFβ3, with its expression restricted predominantly to effector Tregs, cancer cells, and platelets.
Methods
We investigated the role of GARP in human patients with cancer by analyzing existing large databases. In addition, we generated and humanized an anti-GARP monoclonal antibody and evaluated its antitumor efficacy and underlying mechanisms of action in murine models of cancer.
Results
We demonstrate that GARP overexpression in human cancers correlates with a tolerogenic TME and poor clinical response to ICB, suggesting GARP blockade may improve cancer immunotherapy. We report on a unique anti-human GARP antibody (named PIIO-1) that specifically binds the ligand-interacting domain of all latent TGFβ isoforms. PIIO-1 lacks recognition of GARP-TGFβ complex on platelets. Using human LRRC32 (encoding GARP) knock-in mice, we find that PIIO-1 does not cause thrombocytopenia; is preferentially distributed in the TME; and exhibits therapeutic efficacy against GARP+ and GARP- cancers, alone or in combination with anti-PD-1 antibody. Mechanistically, PIIO-1 treatment reduces canonical TGFβ signaling in tumor-infiltrating immune cells, prevents T cell exhaustion, and enhances CD8+ T cell migration into the TME in a C-X-C motif chemokine receptor 3 (CXCR3)-dependent manner.
Conclusion
GARP contributes to multiple aspects of immune resistance in cancer. Anti-human GARP antibody PIIO-1 is an efficacious and safe strategy to block GARP-mediated LTGFβ activation, enhance CD8+ T cell trafficking and functionality in the tumor, and overcome primary resistance to anti-PD-1 ICB. PIIO-1 therefore warrants clinical development as a novel cancer immunotherapeutic.
Keywords: immunotherapy, CD8-positive T-lymphocytes, programmed cell death 1 receptor, transplantation immunology
Background
Immune checkpoint blockade (ICB) targeting programmed cell death protein 1 (PD-1) or its ligand PD-L1 has emerged as an effective approach in cancer immunotherapy.1 PD-1 is expressed on T, natural killer (NK), B, and myeloid cells,2–5 and its expression on activated T cells restores homeostasis to prevent hyperactivation.6 Persistent PD-1 expression by antigen-stimulated T cells (as in the settings of chronic viral infections or cancers) leads to T cell dysfunction or exhaustion, which can be functionally reversed by PD-L1 blockade in vivo.7 8 However, primary and adaptive resistance to anti-PD-L1 ICB commonly results in treatment failure, yielding inadequate clinical outcomes. Understanding the mechanisms underlying resistance is imperative to develop more effective ICB strategies and broaden their clinical applicability.
Transforming growth factor β (TGFβ) is a master regulator of immune tolerance and plays an important role in maintaining immune homeostasis.9 TGFβ is richly expressed in the tumor microenvironment (TME) where cancers can use its signaling axis to mediate immune evasion, especially in late stage disease.10 Indeed, TGFβ accumulation mediates ICB resistance in multiple cancer types including urothelial carcinoma, metastatic colon cancer, and triple negative breast cancer (TNBC).11–13 However, selective targeting of TGFβ is difficult due to its spatially and temporally dependent functions. Long-term inhibition of TGFβ can have inflammatory, autoimmune, and cardiovascular side effects.14 Furthermore, a PD-L1-directed TGFβ trap showed promise in early phase clinical trials15 but ultimately failed to demonstrate superiority to anti-PD-1 alone for non-small cell lung cancer, likely due to saturation by active TGFβ present in the circulation. Therefore, novel strategies are needed to preferentially target TGFβ within the TME.
Glycoprotein-A repetitions predominant (GARP, encoded by Lrrc32) is a cell surface docking and activating receptor for all three isoforms of latent TGFβ (LTGFβ1, –2 and −3). It is expressed on platelets, Tregs, activated B cells, mesenchymal stromal cells, and cancer cells.16–21 The strategic expression of GARP on the cell surface of various cells in the TME suggests that it may facilitate an increased local concentration of TGFβ, resulting in TGFβ-dependent immune evasion. Furthermore, GARP overexpression is a negative prognostic factor for clinical outcomes.19 Using a monoclonal antibody (mAb) that blocks the release of active TGFβ1 from the GARP-LTGFβ1 complex, other groups were able to improve the therapeutic effect of anti-PD-1 therapy by enhancing antitumor CD8+ T cell cytotoxicity.22 23 This antibody (ABBV-151) is under evaluation in a phase I clinical trial in combination with anti-PD-1 therapy for multiple types of solid tumors (NCT03821935). However, circulating platelets may serve as a sink for ABBV-151 due to their high surface expression of the GARP-LTGFβ1 complex, and thus reduce the accumulation and action of ABBV-151 in the TME.
In this study, we evaluated the role of the GARP-TGFβ axis in modulating the immune landscape and ICB responsiveness in human cancers. We found that LRRC32 and LRRC32-TGFB-related gene signatures were elevated in ICB non-responders, especially in patients with immune-excluded tumors.11 We generated, characterized, and humanized an anti-GARP antibody that specifically binds to human ligand-free GARP but not the GARP-LTGFβ complex (called 'PIIO-1'), and lacks mouse cross-reactivity. Since platelets, unlike Tregs, only express the GARP-LTGFβ complex, PIIO-1 is functionally a Treg-selective anti-GARP antibody that eliminates platelet binding and risk of platelet-related toxicities such as thrombocytopenia. We generated and utilized a human LRRC32 knock-in (hLRRC32KI) mouse model to test preclinical efficacy, safety, and mechanisms of action studies. We found that PIIO-1 has single agent activity against multiple GARP- tumor models and can augment anti-PD-1 ICB. PIIO-1 broadly reduces canonical TGFβ signaling in tumor-infiltrating immune cells, prevents T cell exhaustion, and enhances CD8+ T cell migration into the TME in a C-X-C motif chemokine receptor 3 (CXCR3)-dependent manner.
Methods
The Cancer Genome Atlas database analysis
LRRC32 expression values were obtained from The Cancer Genome Atlas (TCGA) using RNA-seq data available in the cBioPortal database (https://www.cbioportal.org/) and further integrated with the Immune Landscape of Cancer data24 using patient IDs. Comparison of each parameter in the Immune Landscape of Cancer between the top 1/3 (LRRC32 high) versus the bottom 1/3 expression groups (LRRC32 low) was implemented by an independent t-test.
Generation of anti-human GARP (hGARP) antibodies
The generation of anti-hGARP antibody has been described previously.19 BALB/c mice was immunized with recombinant human GARP (R&D Systems) in Freund’s complete adjuvant and followed by boosting with SP2/0-hGARP cells for 2–3 times. Splenic B cells with high anti-GARP antibody titers from the immunized mice were fused to SP2/0 cells in the presence of polyethylene glycol. Hybridoma selection was done in HAT medium and cloning was done by limiting dilution assay.
Latency-associated peptide competition binding assay
1×105 Jurkat-hGARP cells were incubated with 400 ng human recombinant LTGFβ1 (R&D) and murine IgG1 isotype control (mIgG1) or anti-GARP antibodies at indicated concentration for 30 min at 37 ℃. Cells were washed with phosphate-buffered saline (PBS) twice and flow cytometry was performed using an anti-latency-associated peptide (LAP) antibody (eBioscience) to determine cell surface expression.
In vivo models. hLRRC32KI mice received mIgG1or PIIO-1 (200 µg) intravenously (i.v.) every other day for three treatments. Indicated organs were collected on day 5. The single cell suspension was prepared, followed by staining and flow cytometry analysis.
TNBC Model. 4T1-hGARP (1×105 cells) was injected into the fourth mammary fat pad of 6–8 weeks old female BALB/c mice. Antibodies were given intraperitoneally (i.p.) at day 7 post tumor injection and continued once every 3 days for five injections. Critical parameters were measured include tumor growth, body weight, survival, lung metastasis, TGFβ level in the sera. To study antitumor memory response, mice with complete rejection of the tumors were then rechallenged with 4T1-wild type (WT; 5×105 cells), followed by monitoring of tumor growth and overall survival time.
Bladder Cancer Model. MB-49 (1×105 cells) was injected subcutaneously (s.c.) on the right flank of hLRRC32KI male mice. mIgG1 or PIIO-1 were given i.p. every 3 days on indicated days. Indicated tissues were then collected 24 hours after the last treatment. To study the efficacy of combination therapy, PIIO-1 (200 µg) and anti-PD-1 antibody were delivered (100 µg) every 3 days i.p. post MB-49 injection. PIIO-1 started on day 4 for six doses and anti-PD-1 antibody started on day 10 for four doses. Tumors were monitored daily. Mice which rejected tumor completely in indicated groups were then rechallenged with MB-49 (1×105) s.c. Tumor growth and overall survival time were monitored.
MB-49 (1×105 cells) were injected s.c. on the right flank of hLRRC32KI male mice. Anti-CD8a antibody (200 µg, i.p) was delivered on day 4, 6, 8, 11 and 14. PIIO-1 was given at 200 µg, i.p. on day 5, 8, 11 and 14. Tumor growth was monitored. To study the roles of T cell migration in the antitumor activity, FTY720 (2 mg/kg) was given on day 6 every 2 days for six doses. PIIO-1 was delivered (200 µg, i.p) on day 6, 9, 12, and 15. Experiment ended on day 17 with tumors and other organs analyzed. The roles of CXCR3 were also evaluated in the MB-49 model, with blocking anti-CXCR3 antibody and PIIO-1 (200 µg, i.p, each) given on day 5 post-MB-49 injection every 3 days for four treatments. Tumor growth was then monitored, with end-of-experiment analysis performed on day 16.
Tumor sizes were measured by longest width and length in mm and reported as tumor areas (width×length). For 4T1, treatment began when tumor area was around 30 mm2 (≈75 mm3 tumor volume) and for MB-49, treatment began when tumor area was around 12–24 mm2 (≈18–48 mm3).
High dimensional flow cytometry analysis, multiplex immunofluorescence microscopy
Antibody staining and high dimensional spectral flow cytometry analysis (Cytek) were performed as detailed previously.25 26 Multiplex immunofluorescence (IF) was done using Vectra Polaris.25 Detailed methods including spatial analysis provided in online supplemental file.
jitc-2022-005433supp001.pdf (1.2MB, pdf)
RNA-seq alignment, preprocess, and analysis
Sequencing was outsourced to Macrogen and performed on an Illumina Hiseq6000. Reads were aligned to the GRCm38 reference using the Hisat2 (V.2.0.5), and read counts were determined using featureCounts (V.1.5.0-p3) software. Raw read counts were used for differentially expressed gene (DEG) analysis based on the DESeq2 package. The enrichment analyses of gene ontology (GO) terms were performed via the R package clusterProfiler (V.3.18.0). Gene Set Enrichment Analyses (GSEA) (V.4.0.3) was implemented for enrichment analysis and visualization. Deconvolution was performed using TIMER V.2.0. Detailed methods provided in online supplemental file.
Statistical analysis
The Student’s t-test was used to compare continuous variables between two groups such as control vs treatment. Kaplan-Meier curves were used to visualize survival and the log rank test was used to quantify significance. Tumor growth curve analysis was performed using repeated measures analysis of variance. All data are presented as mean±SEM. P values less than 0.05 were considered statistically significant. Tukey or Sidak procedures were used for multiple testing correction.
Results
High LRRC32-TGFB expression in human cancers correlates with unfavorable TME and poorer clinical response to ICB
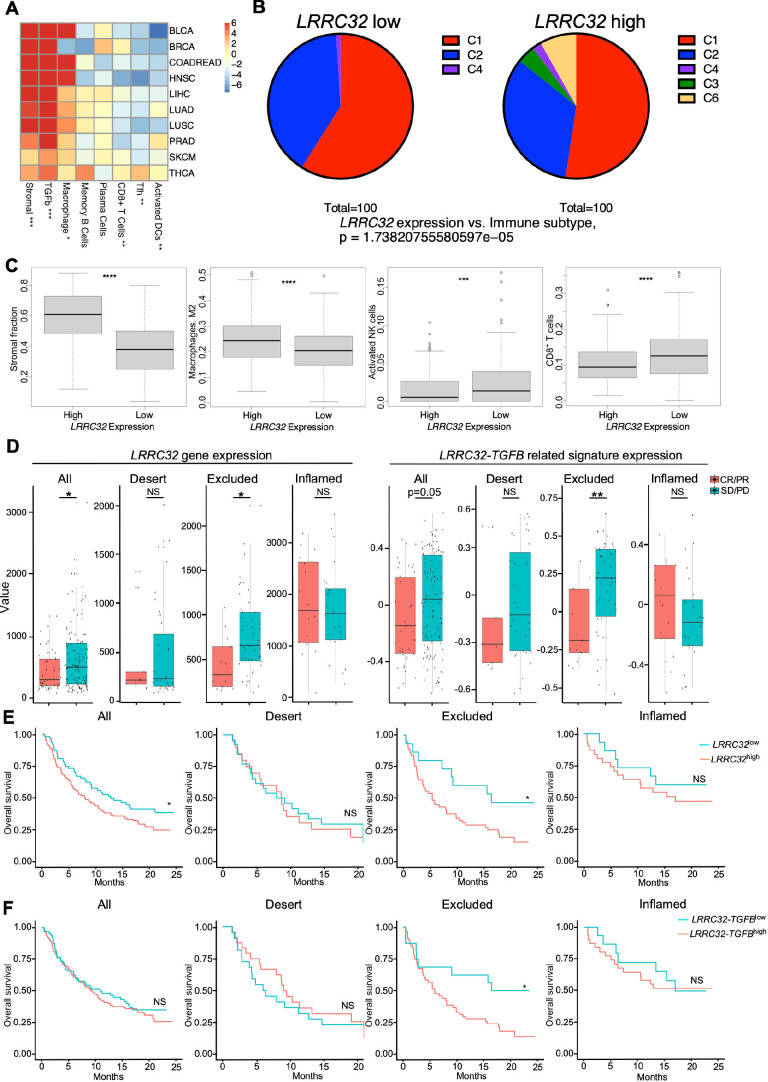
To understand the immunological and clinical implication of GARP expression in cancer, we first mined The Immune Landscape of Cancer database,24 which developed a global immunoprofiling classification by bulk transcriptomic analysis of over 10 000 patients from TCGA. The wound healing classification (C1) reflects an induced expression of genes related to angiogenesis. The interferon γ (IFNγ) dominant classification (C2) contains the highest population of type 1 macrophages (M1) and CD8+ T cells, with high T cell receptor density. Increased T helper (Th) 17 and Th1 related genes, and reduced tumor cell proliferation were included in the inflammatory classification (C3). A low Th1/high type 2 macrophage (M2) response phenotype characterized the lymphocyte depleted classification (C4). The immunologically quiet classification (C5) shows the lowest lymphocyte infiltration and highest M2 response. The TGFβ dominant classification (C6) represents tumors with the highest TGFB gene signature. In 10 common types of solid tumors including bladder and breast cancers, we found that GARP expression positively correlated with tumors rich for stromal, TGFβ, and macrophage signatures and negatively correlated with tumors with T follicular helper (Tfh) signatures, memory B cells, plasma cells, and activated dendritic cells (DCs) (figure 1A). The negative correlation between GARP expression and immune cells (such as Tfh, B cells, plasma cells, and activated DCs) suggested that GARP-rich TME is unfavorable for the generation of tertiary lymphoid structures in the tumors, although this conclusion requires further histological study. Within the lung squamous cell carcinoma cohort, GARPhigh tumors had greater TGFβ dominant immune signatures and lower activated NK cells, CD8+ T cells and IFNγ signatures compared with GARPlow tumors (figure 1B, C). We next evaluated the significance of LRRC32 expression and LRRC32-TGFB related signatures on patient response to immunotherapy in metastatic urothelial cancer.11 We defined the LRRC32-TGFB related signature using genes involved in the TGFβ activation process such as αV integrins. LRRC32 expression and LRRC32-TGFB related signatures are higher in patients who did not respond to anti-PD-L1 ICB (atezolizumab) (figure 1D). Elevated LRRC32 gene signature expression was predominantly observed in immune-excluded tumors, and we found that high LRRC32 expression (figure 1E) and high LRRC32-TGFB related gene signature (figure 1F) significantly correlated with worse overall survival in these patients. Therefore, we conclude that high LRRC32-TGFB expression in human cancers correlates with an unfavorable TME and poorer clinical response to anti-PD-L1 ICB, and that GARP is a biologically relevant target for cancer immunotherapy.
Figure 1.
Impact of LRRC32 gene expression on immune landscape in human cancers and ICB responsiveness. (A–C) TCGA analysis. (A) Correlation of LRRC32 expression level with indicated immune pathways in multiple human cancer types. Values in each cell indicates t-statistics comparing the top 1/3 vs the bottom 1/3 LRRC32 expression groups. Numbers in the heatmap indicated t-statistics result, and >2 or < −2 are statistically significant in a positively or negatively correlated manner, respectively. (B) Correlation of immune subtypes between LRRC32 expression and non-small cell lung cancer. (C1. Wound healing. C2. IFN-γ dominant. C3. Inflammatory. C4. Lymphocyte depleted. C6. TGF-β dominant) (C) Box plots comparing related immune pathway enrichment in human lung cancers between high and low LRRC32 gene expression groups. Statistical significance was determined using t-tests for A and C, and Fisher’s exact test for B. (D–F) Bulk RNA-seq data analysis of pre-treatment tumor samples from 167 patients with metastatic urothelial cancer (mUC) who received atezolizumab in phase two clinical trial (IMvigor210). (D) Box plots comparing the expression of LRRC32 gene (left) and LRRC32-TGFB related signatures (right, defined in online supplemental methods) in all types, immune-desert, excluded and inflamed tumors. Responders (CR/PR) in red and non-responders (SD/PD) in blue. (E, F) Kaplan-Meier survival plots comparing overall survival probability (y-axis) and follow-up time (months, x-axis) in all cancer types including immune-desert, excluded, inflamed tumors. Groups were split by high (red) and low (green) expression levels of LRRC32 gene (E) or LRRC32-TGFB related gene signatures (F). Significance was determined by using t-test for D, and log rank tests for E, F. *p<0.05; **p<0.01, ***p<0.001, ****p<0.0001. CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
Anti-GARP antibody PIIO-1 blocks the formation of GARP-LTGFβ1 complex
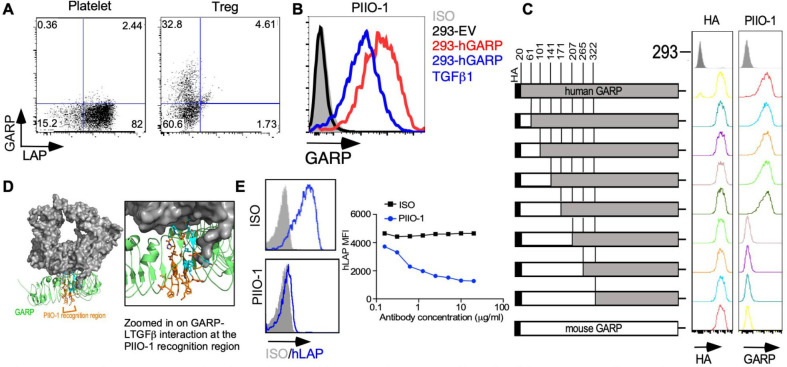
To generate anti-GARP mAbs, mice were immunized with recombinant hGARP, followed by boosting with irradiated whole myeloma hGARP-expressing SP2/0 cells. We characterized the binding of seven antibodies recognizing hGARP using flow cytometry. While all clones recognized hGARP on Tregs, only one failed to recognize hGARP on platelets (PIIO-1; online supplemental figure S1A). GARP is known to exist biochemically in three major forms: ligand-free membrane-bound GARP; membrane-bound GARP-LTGFβ complex; and soluble GARP (released after proteolytic cleavage).27 Tregs express both ligand-free and complexed GARP on their cell surface, whereas platelets only express the complexed form.20 22 Since PIIO-1 only recognized GARP on Tregs but not platelets, we infer that it binds the ligand-free form of GARP (figure 2A). To confirm this prediction, we transfected HEK293FT cells with plasmids expressing hGARP with or without TGFβ1 to create cells expressing either ligand-free GARP (293-hGARP) or the GARP-LTGFβ complex (293-hGARP-TGFβ1) (figure 2B and online supplemental figure S1B). We generated a series of HA-tagged murine GARP (mGARP)/hGARP chimeras through standard PCR-based cloning techniques and determined that PIIO-1 binds an epitope corresponding to amino acids 171–207 on hGARP (figure 2C), which is the known site for LTGFβ binding22 (figure 2D). Using a competition binding assay, we found that LTGFβ1 blocked PIIO-1 binding to GARP (online supplemental figure S1C). Importantly, we found that the expression of cell surface LAP decreases in the presence of PIIO-1 in a dose-dependent manner, indicating that PIIO-1 prevents complex formation between GARP and exogenous LTGFβ1 (half-maximal inhibitory concentration (IC50)=653.4 ng/mL) (figure 2E). In summary, we generated a unique mAb that specifically binds to ligand-free GARP at the LTGFβ1 binding site and blocks the formation of the GARP-LTGFβ1 complex. This antibody specifically targets GARP on Tregs and other cells expressing the ligand-free form of GARP, but does not recognize the TGFβ-GARP complex on platelets.
Figure 2.
In vitro characterization of anti-GARP antibody PIIO-1. (A) GARP expression on human regulatory T cells and platelets was evaluated by flow cytometry after staining with PIIO-1 at 10 µg/mL. (B) 293 FT cell line was transfected with empty vector (EV), human GARP (hGARP)-expressing vector only or co-transfected with hGARP and latent TGFβ1 expression vectors. GARP expression on indicated cell line was detected by flow cytometry after staining with PIIO-1 at 10 µg/mL. (C) Human GARP sequence was replaced by murine GARP according to the schematic diagram to generate the chimeric constructs of human and murine GARP that were tagged with HA (hemagglutinin) epitope. Transfection efficiency was determined using anti-HA antibody. All constructs were transfected into 293 FT cells. (D) Crystal structure of the GARP (green)-LTGFβ (gray) complex (PDB DOI: 10.2210/pdb6GFF/pdb). The region of PIIO-1 recognition is orange and the residues interacting with LTGFβ are cyan. LTGFβ occludes approximately 30% of the potential antibody binding site and may sterically or allosterically restrict access of the antibody to GARP in the LTGFβ-complexed state. Modeling was carried out using Pymol. (E) Jurkat cell line, made to overexpress hGARP, was incubated with LTGFβ1 along with mIgG1 or PIIO-1 at indicated concentration for 30 min at 37℃. Human LAP expression was detected by flow cytometry. All data are representative of 2–6 independent experiments. GARP, Glycoprotein-A repetitions predominant; LAP, latency-associated peptide.
Targeting GARP on tumor cells enhanced PD-1 blockade efficacy in TNBC
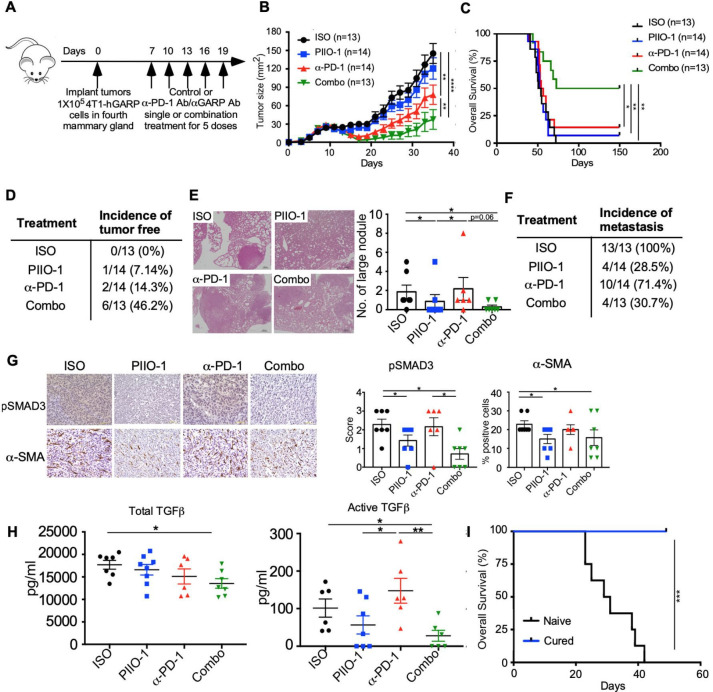
Since PIIO-1 can bind GARP on Tregs, we next addressed whether combining PIIO-1 with anti-PD-1 ICB could augment efficacy by shifting an unfavorable TME toward a phenotype sensitive to immunotherapy. We implanted murine triple negative mammary gland cancer cells stably expressing hGARP (4T1-hGARP)19 orthotopically into BALB/c mice. Mice with established tumors (day 7) were treated with single or combination therapies of PIIO-1 (200 µg/mouse) and anti-PD-1 (150 µg/mouse) every 3 days (experimental schema in figure 3A). Combination therapy with PIIO-1 and anti-PD-1 slowed tumor growth and prolonged overall survival, resulting in complete response in 46% of mice (figure 3B–D). Furthermore, lung metastasis in mice treated with PIIO-1 (either single agent or combination therapy) were significantly reduced (figure 3E, F). To assess the impact of PIIO-1 on TGFβ downstream signaling in the TME, we stained tumors collected at endpoint for phosphorylated SMAD3 (pSMAD3) and α-smooth muscle actin (α-SMA). SMAD3 is phosphorylated following TGFβ activation; α-SMA, a marker of cancer-associated fibroblasts (CAFs), is induced by TGFβ activation. CAFs contribute to primary therapeutic resistance and are an emerging target for cancer immunotherapy.28 We found that both pSMAD3 and α-SMA were reduced in the TME after PIIO-1 treatment (figure 3G), suggesting that local TGFβ signaling was effectively blunted. Both total and active TGFβ were decreased in circulation following combination treatment (figure 3H). Finally, mice that experience complete response following combination treatment (figure 3C) were completely protected against rechallenge from the wild-type 4T1 (4T1-WT) tumor cells, demonstrating that PIIO-1 promotes antitumor memory response (figure 3I). Taken together, these results demonstrate that combining PIIO-1 with anti-PD-1 leads to enhanced antitumor efficacy and antitumor memory, and this is likely mediated by a downregulation of TGFβ activity in the TME.
Figure 3.
PIIO-1 enhanced antitumor efficacy of anti-PD-1 ICB in GARP+ TNBC. (A) Experimental scheme. BALB/c mice were injected with 1×105 4T1-hGARP cells in the mammary fat pad, followed by i.p. injection of 200 µg/mouse of PIIO-1 and/or 150 µg/mouse anti-PD-1 every 3 days. (B) Primary tumor growth curve. (C) Overall survival of mice. (D) Summary of the incidence of tumor-free mice among groups. (E) Lungs were collected and sectioned at experimental end point, then stained with H&E. Representative images from each group of mice are shown. Scale bar, 20 µm. The numbers of visible lung metastatic nodules are quantified. (F) Summary of the incidence of metastasis among groups. (G) Tumors were collected at end point and stained by IHC for pSMAD3, α-SMA. Representative images of tumor tissues from four groups of mice are shown (left). Scale bar, 50 µm. Quantification of the IHC staining is shown (right). (H) Sera from each mouse was collected at end point. Total and active TGFβ level in the sera were assessed by ELISA. (I) Mice with tumor regression following combination therapy were monitored for 300 days, then rechallenged with 5×105 wild-type 4T1 mammary tumor in contralateral mammary fat pad. Naive BALB/c mice without pre-exposure to tumor were used as control. Shown is the overall survival. Tumor curve analysis was performed using repeated measures 2-way analysis of variance. Overall survival is analyzed by log-rank (Mantel-Cox) test. (E, G) were analyzed by paired t-test according to the tumor collection time points. Other data were analyzed by two-tailed Student’s t-test. B, C were corrected for multiple testing using the Tukey procedure. All data are presented as mean±SEM. *p<0.05, **p<0.005, ***p<0.001, ****p<0.0001.
Targeting TGFβ-GARP signaling modulates immune homeostasis and promotes the differentiation of antitumor effector CD8+ T cells in the TME
Next, we generated a hLRRC32KI mouse wherein the extracellular domains of mouse GARP are replaced with the corresponding human GARP domains in the germline (online supplemental figure S2A–C). This model allows us to assess pre-clinical safety and efficacy of PIIO-1. In the platelets of hLRRC32KI mice, hGARP associates with mouse LAP as effectively as mGARP (online supplemental figure S2D). I.p. injections of PIIO-1 were well tolerated without causing significant thrombocytopenia or overt toxicity (online supplemental figure S2E, F). All PIIO-1 treated mice had stable weight for at least 20 days without evidence of cardiac failure such as fluid overload and shortness of breath. To assess the impact of PIIO-1 on the immune compartment in non-tumor bearing hLRRC32KI mice, we injected PIIO-1 or mIgG1 (200 µg/mouse each) i.v. every 2 days for three treatments, followed by tissue harvest, single cell isolation, and immune phenotyping (online supplemental figure S3A). PIIO-1 treatment was associated with increased cellularity of peripheral lymph nodes (pLNs) and elevated frequency of CD8+ T cells (online supplemental figure S3B, C). In addition, we saw reduced Tregs in the pLNs following PIIO-1 treatment (online supplemental figure S3D), consistent with TGFβ’s known role in inducing and maintaining Treg lineage.9 29Corresponding to attenuated Treg function and reduced active TGFβ,30–33 PIIO-1 increased Ki67 expression and tumor necrosis factor α (TNFα) production by CD8+ T cells in pLNs (online supplemental figure S3E, F). No difference in immune cell composition was observed in other organs, such as spleen, thymus, mesenteric LN or peripheral blood (data not shown).
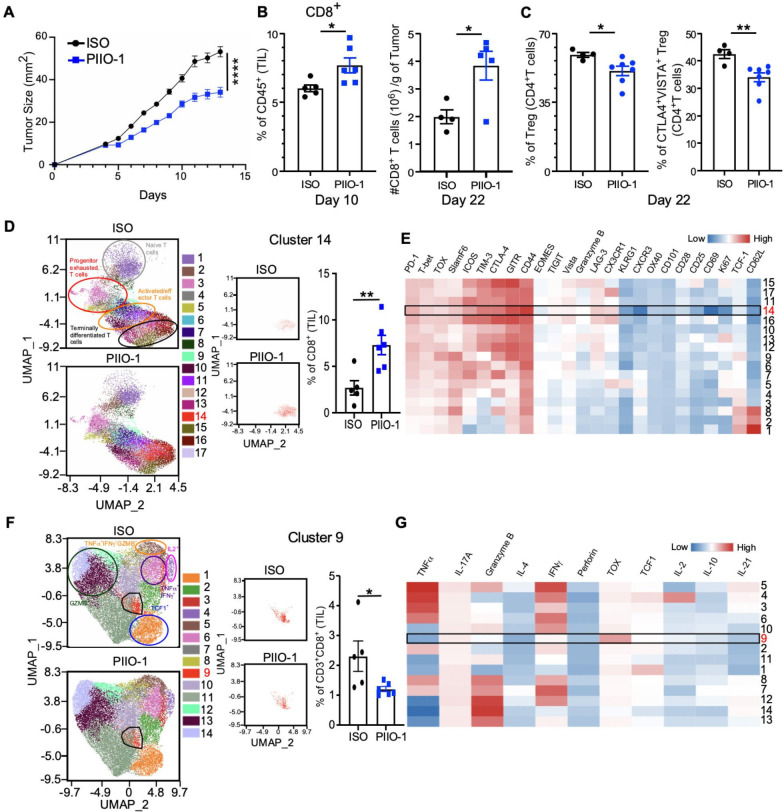
Next, we implanted hLRRC32KI mice s.c. with MB-49 murine urothelial carcinoma, an immunologically ‘lukewarm’ tumor that only partially responds to anti-PD-1 therapy.25 Starting 4 days after tumor implant, PIIO-1 or mIgG1 was administered i.p. every 3 days for four total treatments. PIIO-1-treated mice showed a significant delay in tumor growth (figure 4A). Since murine MB-49 does not express human GARP, this observed antitumor activity must be attributed to an increased antitumor immune response. Thus, in a separate experiment, we treated day 6 MB-49 tumors every 3 days with PIIO-1 or mIgG1 for two (short-term) or six (longer-term) total doses. We harvested tumors 24 hours after final treatment, and isolated tumor-infiltrating lymphocytes (TILs) for analysis by high dimensional spectral flow cytometry. Short term PIIO-1 increased the frequency of CD8+ T cells in the TME (figure 4B, left), and this effect was augmented following longer-term treatment (figure 4B, right). Longer-term exposure to PIIO-1 also decreased both Treg frequency (figure 4C, left) and suppressive function, as indicated by downregulation of CTLA4 and VISTA34 35 (figure 4C, right).
Figure 4.
PIIO-1 monotherapy modulates CD8+ T cells in the TME and confers protection against cancer in hLRRC32KI mice. (A) 1×105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. PIIO-1 or isotype control (ISO) was administered (200 µg/mouse, i.p.) every 3 days for a total of four treatments starting on day 4. Shown is one representative tumor growth curve. (B) 1×105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. PIIO-1 was delivered (200 µg/mouse, i.p.) on days 6 and 9. Tumors were collected on day 10 and tumor-infiltrating leucocytes were stained and analyzed by flow cytometry. Frequency of CD8+ T cells as a proportion of live CD45+ lymphocytes is shown (left). A similar experiment was performed wherein mice were treated with PIIO-1 for a total of 6 treatments starting on day 6. CD8+ TILs were quantified on day 22 (right). (C) Frequency of total Tregs (CD25+Foxp3+) (left) and CTLA4+VISTA+ Tregs (right) in tumor-infiltrating CD4+ T cells in the indicated treatment groups. (D) Differential expression analysis of cluster frequency of CD8+ TILs between ISO and PIIO-1 treated TILs. UMAP dimension reduction of tumor-infiltrating CD8+ T cells from B after staining with 33 markers and high dimensional spectral flow cytometry analysis. Data shown is gated on live CD45+CD3+CD8+ T cells, subsampled on 5000 cells per sample. Unsupervised clustering analysis was done using FlowSOM algorithm with an elbow method approach for cluster number determination. (E) Heatmap of D showing the relative expression levels of indicated markers by each cluster. (A–E) n=4–6 mice per group. (F) Differential expression analysis of cytokine production by CD8+ TILs between ISO and PIIO-1 treated tumors. 1×105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. PIIO-1 or IOS was administered every 3 days for a total of 4 treatments starting on day 5. Tumors were collected on day 17. Intracellular stain for 17 cytokine panel was done, followed by spectral flow cytometry and analysis of CD45+CD3+CD8+ T cells. (G) Cytokine level in panel (F) indicated by heatmap showing expression intensity of cytokines by each CD8+ T cell cluster. Tumor curve analysis was performed using repeated measures two-way analysis of variance. Cluster differences were measured by two-tailed Student’s t test. Data are represented as mean±SEM. *P<0.05, **p<0.01, ****p<0.0001.
To examine the effect of PIIO-1 on CD8+ T cells in the TME at the single cell level, we used a 33-marker T cell exhaustion panel for high dimensional spectral flow.25 26 We performed dimension reduction using the Uniform Manifold Approximation and Projection (UMAP) approach, which allowed the data to be displayed in two dimensions (figure 4D, E). We then performed unsupervised clustering analysis using FlowSOM to partition the data and allow for differential expression analysis between groups. This analysis identified 17 distinct clusters, one of which (cluster 14) was significantly enriched in CD8+ T cells from PIIO-1-treated mice. Cluster 14 displayed elevated expression of activation markers including LAG-3, CD44, GITR, TIM-3, and PD-1 (figure 4D, E), but not TOX, a transcription factor associated with terminal exhaustion.36 Our data support the hypothesis that PIIO-1 induces CD8+ T cell effector differentiation (figure 4D, orange circled population in UMAP) and blocks T cell exhaustion. Indeed, with prolonged PIIO-1 treatment (starting on day 5 for 4 doses), there was a decrease in a terminally exhausted population (cluster 9), as indicated by its TOXhigh status with little or no effector cytokine production (including IL-2, IL-21, TNFα, IFNγ and others; figure 4F, G). Taken together, our data suggest a multifaceted effect of PIIO-1, wherein it simultaneously shifts immunologically lukewarm tumors toward a proinflammatory state with increased CD8+ T cell infiltration, while promoting activation and preventing terminal exhaustion of these TILs. To support these results, we found that enforced expression of hGARP in MB-49 cells (MB-49-hGARP) resulted in higher frequency of tumor-infiltrating CD8+ T cells with an exhausted phenotype compared with empty vector transfected MB-49 (cluster 9; online supplemental figure S4A, B).
Next, we analyzed MB-49 tumors spatially using multiplex IF imaging. Tumors were stained with CD45, CD8, α-SMA and partitioned into tumor interior, intermediate I, intermediate II and exterior regions. CD8+ T cell density increased in the intermediate II region indicating enhanced intratumoral infiltration after PIIO-1 treatment (online supplemental figure S5A). In the interior regions of mIgG1 treated tumors, α-SMA+ cell density negatively correlated with CD8+ T cell density. PIIO-1 treatment decreased the magnitude of this negative correlation (online supplemental figure S5B), suggesting that blocking the GARP-TGFβ axis decreased stromal formation and increased T cell infiltration. By applying spatial two-point correlation analysis, we found that CD8+ T cells co-localize more frequently in both the interior and intermediate II regions of PIIO-1 treated tumors, compared with controls (online supplemental figure S5C, D). In summary, treatment of MB-49 with PIIO-1 alters CD8+ T cell intratumoral infiltration kinetics and mediates functional and spatial changes to their phenotype.
Anti-GARP antibody enhances anti-PD-1 ICB against GARP-negative tumors
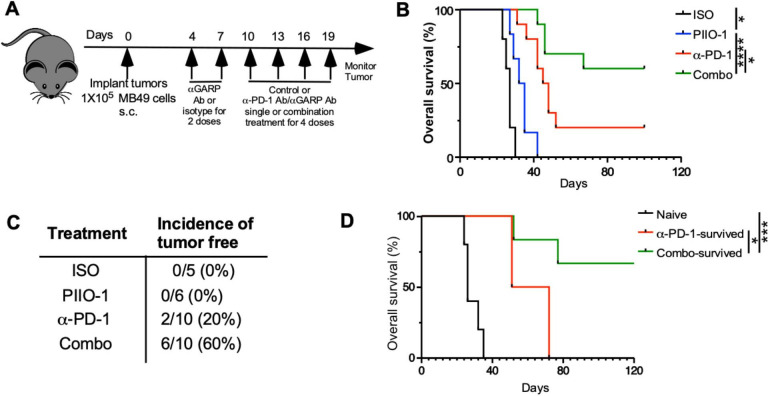
Mechanistically, PD-1 blockade targets progenitor exhausted CD8+ T cells in the TME, which persistently express TCF-1 and SlamF6 with low levels of PD-1 and TIM-3.37 These cells undergo a robust proliferation following anti-PD-1 treatment resulting in differentiation toward an effector phenotype, which induces tumor clearance. Since PIIO-1 monotherapy significantly reduced CD8+ T cell exhaustion in the TME, we evaluated whether it could potentiate the antitumor activity of anti-PD-1 ICB. We treated hLRRC32KI mice bearing subcutaneous day 4 MB-49 tumors sequentially using PIIO-1 (200 µg/mouse; six doses) and anti-PD-1 antibody (100 µg/mouse; four doses started day 10) (figure 5A). While single agent PIIO-1 modestly prolonged overall survival compared with control (figure 5B), combination therapy with anti-PD-1 resulted in complete tumor response in 60% of mice (figure 5C). Finally, when we rechallenged cured mice with MB-49 cells, those mice that previously received combination therapy had better antitumor memory function (figure 5D), indicating that PIIO-1 impacts favorably the generation of antitumor immunological memory.
Figure 5.
PIIO-1 potentiates preclinical antitumor activity of anti-PD-1 antibody against bladder cancer. (A) Experimental scheme. 1x105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. PIIO-1 (200 µg/mouse, i.p.) and anti-PD-1 antibody (100 µg/mouse, i.p.) were administered every 3 days. PIIO-1 started on day 4 for six doses and anti-PD-1 antibody injection began on day 10 for four doses. (B) Represented overall survival of mice treated with ISO (n=5), PIIO-1 (n=6), anti-PD-1 Ab (n=10) or combination of anti-PD-1 Ab and PIIO-1 (n=10). (C) Summary of therapeutic efficacy based on complete response. (D) PIIO-1 and anti-PD-1 generated better antitumor memory responses. Mice rendered tumor-free by indicated treatment were rechallenged with live MB-49 subcutaneously. The overall survival was compared. Tumor-free naïve mice were used as control. Overall survival is analyzed by log-rank (Mantel-Cox) test. (B, D) Were corrected for multiple testing using the Turkey procedure. All data are presented as mean±SEM. *P<0.05, ***p<0.001, ****p<0.0001.
We also tested PIIO-1 and anti-PD-1 combination therapy against murine Lewis Lung Carcinoma (LLC) and CMT-167 lung cancer models, both of which are immunologically cold tumors and are resistant to anti-PD-1 ICB. Day 8 LLC tumors in hLRRC32KI mice were treated every 3 days with single or combination therapy (PIIO-1 200 µg±anti-PD-1 100 µg; four doses total). The combination of PIIO-1 and anti-PD-1 was most effective in slowing LLC growth when compared with anti-PD-1 monotherapy (online supplemental figure S6A). These results were recapitulated in the CMT-167 model wherein adding PIIO-1 overcame the anti-PD-1 resistance seen in CMT-167, which correlated with increased CD8+ T cells in the TME (online supplemental figure S6B, C).
Humanized PIIO-1 blunts canonical TGFβ signaling in tumor-infiltrating immune cells and promotes proinflammatory TME
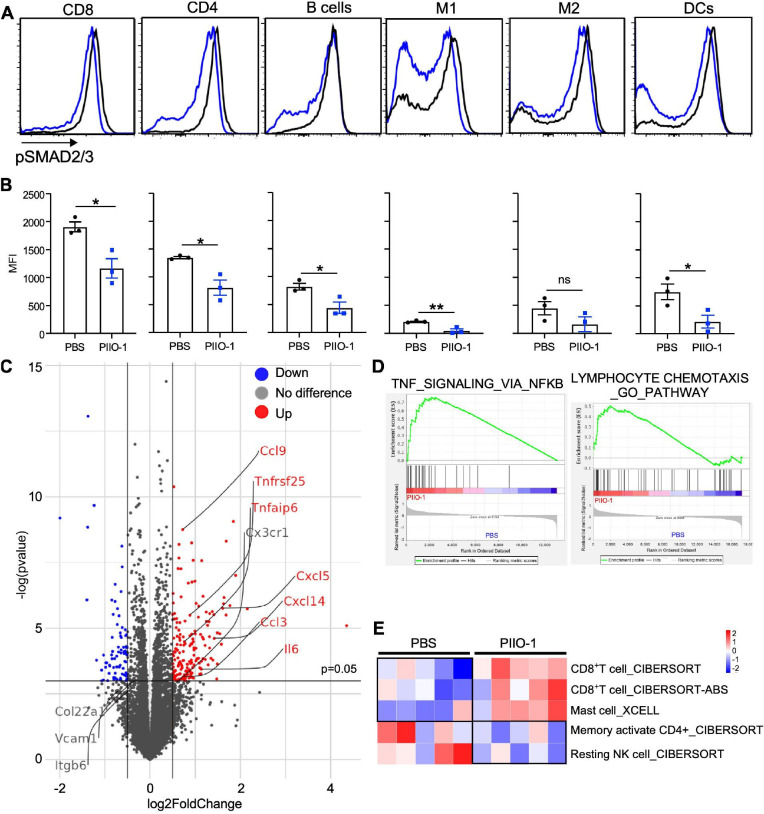
We next humanized PIIO-1 by fusing its complementarity determining regions of the variable domains with the remainder of the chain from human IgG4. The humanized PIIO-1 has identical affinity to the parental antibody for human GARP (Kd, 1–3 nM) and it had similar mono-agent antitumor efficacy in MB-49 tumor model (data not shown). Moreover, PIIO-1 treatment of MB49-bearing tumors resulted in decreased pSMAD2/3 signaling in major tumor-infiltrating immune cell subsets including T, B cell, macrophages, and DCs (figure 6A, B), as well as T and B cells in the dLN (online supplemental figure S7A, B). Interestingly, on a per cell basis, tumor infiltrating CD8+ T cells had the highest TGFβ signaling activity indicated by pSMAD level (figure 6B). To determine the immune cell target of PIIO-1, we injected it into tumor bearing hLRRC32KI mice. Twenty-four hours later, tumors, dLNs, and spleens were harvested, and single cell suspensions were analyzed for cell surface binding of PIIO-1. We found that PIIO-1 only recognizes cells in the tumor and the dLN but not in the spleen (online supplemental figure S7C). Tregs were the major cell population that bound PIIO-1 in the dLN (online supplemental figure S7C). The preferential targeting of PIIO-1 to tumors and the dLNs, but not the spleen underscores the favorable biodistribution of this antibody.
Figure 6.
PIIO-1 attenuates canonical TGFβ signaling pathway in tumor-infiltrating immune cells and rejuvenates antitumor immunity in hLRRC32KI mice. (A) 1×105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. Humanized PIIO-1 (200 µg/mouse, i.p.) were administered on days 18 and 20. Tumors were collected on day 21. TILs were isolated and stained for intracellular pSMAD2/3 with indicated cell linage markers, followed by flow cytometry analysis. (B) Quantification of panel A. (C–E) 1×105 MB-49 cells were injected s.c. on the right flank of hLRRC32KI male mice. Humanized PIIO-1 (200 µg/mouse, i.p.) was delivered on days 6 and 9. Tumors were collected on day 10. Single cell suspension and RNA isolation were prepared, and then subjected to bulk RNA sequencing. (C) Volcano plot of transcript expression. Differential gene expression was shown in red (up) or blue (down). Representative transcripts such as Ccl3, Ccl9, Cxcl14, Cxcl15, Il6 and Tnfrsf25 were indicated. (D) GSEA of differential expression genes between tumors treated with PBS and PIIO-1. (E) Comparison of TILs between PBS and PIIO-1 treated tumors based on deconvolution of bulk RNA sequencing data. Data were performed using two-tailed Student’s t-test, data in (B) represented mean±SEM. *p<0.05, **p<0.01.
Antitumor function of cytotoxic CD8+ T cells requires both lytic function and proinflammatory cytokine production (eg, TNFα and IFNγ). In addition, TGFβ is known to dampen CD8+ T cell function and migration into the TME.38 39 To gain further insight into the mechanism of action of PIIO-1, we performed bulk transcriptome analysis of day 10 MB-49 tumors in hLRRC32KI mice treated with PBS or PIIO-1 on days 6 and 9. mRNA expression analysis revealed that proinflammatory cytokine (eg, Tnf super family, Il6) and chemokine (eg, Ccl3, Ccl9, Cxcl14, Cxcl15) transcripts were increased in the PIIO-1-treated tumors (figure 6C), consistent with the ability of PIIO-1 to induce a proinflammatory TME. GSEA showed a similar phenotype with increased TNF-NFκB signaling and lymphocyte chemotaxis in PIIO-1-treated tumors (figure 6D). The deconvolution analysis of tumor bulk mRNA sequencing data demonstrated enrichment of CD8+ T cells, mast cells and activated NK cells in the TME after PIIO-1 administration (figure 6E). TGFβ can block mast cell activation through inhibiting its expression of high affinity IgE receptor (FcεRI).40 In summary, we conclude that treatment with single agent PIIO-1 remodels an immunosuppressive TME and shifts toward improved immune fitness with a rich pro-inflammatory cytokine milieu and abundance of effector lymphocytes.
Humanized PIIO-1 enhanced antitumor immunity by facilitating CD8+ T cell recruitment into tumors through CXCR3
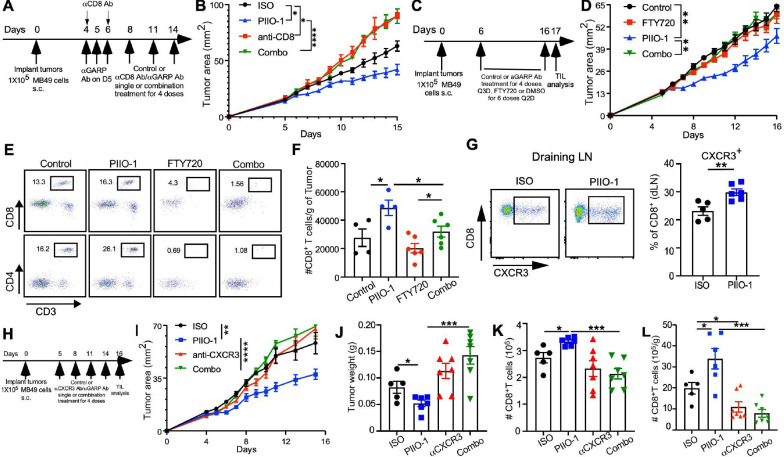
We next addressed the roles of CD8+ T cells in the protective immunity elicited by PIIO-1 and the potential underlying mechanisms. Depleting CD8+ T cells completely ablated the antitumor effects of PIIO-1 against MB-49 tumors (figure 7A, B), underscoring the importance of CD8+ T cells in PIIO-1-mediated tumor control. To determine if antitumor immunity is dependent on continuing migration of activated T cells from the dLNs to the tumor, we blocked T cell egress from dLNs using S1P receptor agonist FTY720 (figure 7C). We found that FTY20 abrogated the antitumor efficacy of PIIO-1 and effectively blocked T cell infiltration (figure 7D–F), indicating that expansion of pre-existing CD8+ T cells in the TME alone was unlikely a contributing factor for the PIIO-1 antitumor activity. Consistent with chemokine-mediated CD8+ T cell migration, we found that the CXCR3+ CD8+ T cell population was enriched in the dLN after PIIO-1 administration (figure 7G), likely due to attenuated TGFβ signaling.38 Blocking CXCR3 during PIIO-1 treatment (figure 7H) completely abolished the antitumor activity of PIIO-1 (figure 7I, J), which correlated with reduced CD8+ T cell (and not Treg) recruitment to the TME (figure 7K, L and online supplemental figure S8). Collectively, by blocking TGFβ activation within the TME, PIIO-1 promotes antitumor CD8+ T cell immunity, in part through increased CXCR3-dependent T cell trafficking into the tumor.
Figure 7.
PIIO-1 promotes antitumor activity that is dependent on CD8+ T cells and CXCR3. (A, B) CD8-dependence of antitumor activity. (A) Experimental scheme. (B) Tumor growth curve of mice treated with indicated conditions (ISO, n=5; PIIO-1, n=5; anti-CD8, n=3; Combo, n=5). (C–F) Antitumor activity of PIIO-1 depends on active egress of lymphocytes from the secondary lymphoid tissues. (C) Experimental scheme. (D) Tumor growth curve of mice treated with indicated conditions (ISO, n=4; PIIO-1, n=4; FTY720, n=6; combo, n=6). (E) The frequencies of CD8+ and CD4+ T cells in the peripheral blood of indicated groups of mice. (F) Absolute number of CD8+ T cells in the tumors. (G) Impact of PIIO-1 on CXCR3+ CD8+ T cells in the dLNs. MB-49 bearing hLRRC32KI mice were treated with 2 courses of PIIO-1or ISO, followed by analysis of CXCR3 expression on CD8+ T cells in the dLN. (H–L) Antitumor effect of PIIO-1 requires CXCR3. (H) Experimental scheme. (I) Tumor growth curve of mice treated with indicated conditions (ISO, n=5; PIIO-1, n=5; FTY720, n=7; combo, n=7). (J) Tumor weight on day 17. (K) Absolute number of CD8+ T cells in the dLN. (L) Absolute number of CD8+ T cells in tumors. Tumor growth analysis was performed using repeated measures two-way analysis of variance. Other data were compared using two-tailed Student’s t-test. (B, D, I) were corrected for multiple testing using the Sidak procedure. Quantitative data are presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Discussion
A key challenge in the field of immuno-oncology is primary and adaptive immune resistance to ICB seen in the majority of patients with cancer, including those with pancreatic cancer, ovarian cancer and most TNBCs.41–43 One underlying mechanism of primary and acquired ICB resistance in advanced malignancies relates to the accumulation of active TGFβ in the TME, which drives immune dysfunction by multiple mechanisms such as inducing Tregs, excluding and inhibiting the function of effector CD8+ T cells,12 44 and limiting effector T cell migration into the TME.45 However, targeting TGFβ has proven difficult to do for the treatment of human diseases due to pleotropic functions that are highly context dependent.46 47 Using a GARP-specific mAb that blocks LTGFβ binding to Tregs, tumor cells and other cell types in the TME without affecting circulating platelets, we have accomplished tumor-selective targeting of the GARP-TGFβ pathway, as well as antitumor activity in multiple preclinical tumor models.
PIIO-1 offers advantages over other technologies that attempt to drug the TGFβ pathway. It only targets GARP-expressing cells, which are primarily found in the TME, unlike agents that block TGFβ systemically such as anti-TGFβ antibodies and small molecule inhibitors against TGFβ signaling receptors.48 It differs from existing anti-GARP antibodies such as ABBV-151 under clinical evaluation in several aspects. First, PIIO-1 binds to ligand-free GARP and blocks the binding of GARP to all LTGFβ isoforms.22 23 Second, platelets express abundant GARP-LTGFβ1 complex due to their high levels of autocrine LTGFβ1.20 Antibodies targeting the GARP-LTGFβ1 complex (such as ABBV-151) pose a potential risk for platelet-related side effects49; the unique epitope targeted by PIIO-1 (free GARP) ablates this risk. Third, the preferential targeting of PIIO-1 to tumors and dLNs underscores the favorable biodistribution of PIIO-1 over ABBV-151, which likely distributes non-selectively in the peripheral blood, bone marrow, and spleen.
PIIO-1 monotherapy successfully modulated the TME by reducing active TGFβ signaling and associated stromal formation, and enhanced accumulation of effector CD8+ T cells within the tumor. Furthermore, combination of PIIO-1 and anti-PD-1 therapy showed robust antitumor activities against GARP- tumors in humanized GARP knock-in mice. Mechanistic studies uncovered several intriguing biological insights related to the roles of GARP in the TME. The increased migration of CD8+ T cells to the TME in response to PIIO-1 is perhaps expected since there was evidence for reduced stromal formation and therefore less immune exclusion.11 Migration was likely also supported by increased chemokine production in the TME and the ability of TGFβ1 to suppress expression of CXCR3 on CD8+ T cells.38 We confirmed that PIIO-1 promotes CXCR3+ CD8+ T cells in the tumor dLNs. Interestingly, we found that CXCR3 is not required for Tregs to migrate into the TME. Therefore, increased CD8+ T cell migration over Tregs into the TME shall translate into reduction of Tregs proportionally, which appeared to be indeed the case. Importantly, PIIO-1 treatment curtailed CD8+ T cell exhaustion. Using a chronic viral infection model, Gabriel et al recently reported that TGFβ1 maintains progenitor exhausted T cells via suppressing mTOR activity, eventually leading to a more terminally exhausted CD8+ T cell state.50 In our study, we used single cell high dimensional flow cytometry to demonstrate that PIIO-1 treatment significantly blocked formation of terminally exhausted CD8+ T cells in the TME, as indicated by high TOX expression and little-to-no expression of effector cytokines. Thus, by blocking active TGFβ production within the TME and dLNs, PIIO-1 augments CD8+ T cell biology in two ways–first, it promotes priming and migration of antigen-specific T cells in the dLNs, and second, it attenuates CD8+ T cell exhaustion in the TME.
We previously reported that platelets are the major source of active TGFβ through GARP-mediated latent TGFβ maturation.20 27 Since PIIO-1 does not block platelet GARP-LTGFβ axis, we conclude that targeting GARP in the non-platelet compartment is sufficient to induce antitumor activity. Alternatively, extravasated tumor-infiltrating platelets, unlike circulating platelets, may also be a target of PIIO-1; this hypothesis is under active investigation using tissue-based spatial technology.
In conclusion, we generated, humanized and characterized a unique anti-GARP antibody that blocks activation of all isoforms of LTGFβ in the TME. Using our human LRRC32 knock-in mice and multiple preclinical tumor models, we demonstrated the potential drugability of the GARP-LTGFβ pathway for cancer immunotherapy. By doing so, we unraveled several new biological aspects of GARP, including how it contributes to immune exclusion, ICB resistance, CD8+ T cell exhaustion, and CD8+ T cell migration into the TME. Thus, PIIO-1 warrants further clinical development as a promising immunotherapeutic agent against advanced cancers with ICB resistance, both as a monotherapy and in combination with ICBs.
Acknowledgments
We thank the following individuals for sharing reagents used in this work: P. Howe (MUSC), G. Xin (OSU), and X. Li (Cedars-Sinai Medical Center). We thank the past and present members of the Li laboratory for stimulating discussions and support during the entire course of the study. The authors would also like to acknowledge C. Jayaprakash (OSU) and R. Grewal (Nationwide Children’s Hospital) for their assistance in the spatial analysis of the data, JD Huck (Yale) for structural illustration of PIIO-1 epitope. Ohio State Innovation Foundation (OSIF) has filed a provisional application on the technology described in the manuscript.
Research reported in this publication was supported by The Ohio State University Comprehensive Cancer Center (OSUCCC) and the National Institutes of Health (NIH) under grant P30CA016058. This research was made possible through resources, expertise, and support provided by the Pelotonia Institute for Immuno-Oncology (PIIO), which is funded by the Pelotonia community and the OSUCCC. We thank the PIIO and the Immune Monitoring and Discovery Platform for their contributions, especially relating to high dimensional flow cytometry analysis and multiplex immunofluorescence microscopy.
Footnotes
Twitter: @t_kwon92, @DrGattiMays
Contributors: ZL and AL conceived the idea, designed the study, and wrote the manuscript. AL, N-JS, XW, BPR, MV, HK, KBC, YW and KD performed the experiments. DTG, CB, KH, DCa, QM, DCh, MG-M, JD and YY participated in the discussion of the data and helped with steering the direction of the study. YC, TO, DCa, GDG and KD assisted in data analysis, statistics, and interpretation. All authors approved the manuscript. ZL supervised the entire study, and is the guarantor of this publication.
Funding: Research in the Li laboratory is supported by multiple NIH grants: R01CA213290, R01CA262069, R01CA255334 and R01AI077283 (Z. Li). AL is supported by the Pelotonia Graduate Fellowship Program from The Ohio State University Comprehensive Cancer Center-James (N/A).
Competing interests: ZL serves as a member of scientific advisory boards for Alphamab, Hengenix, HanchorBio and Ikonisys, and owns the intellectual property right of PIIO-1. KH reports paid consulting or advisory roles for Perthera, Iovance Biotherapeutics, BMS, Geneplus, and Mirati Therapeutics, Beigene, Lyell and Abbvie. He has received research funding from BMS, Mirati Therapeutics, Spectrum pharmaceuticals Adaptimmune, Genentech/Roche, GSK, Amgen, Iovance Biotherapeutics, Abbvie, Beigene and OncoC4, paid to his institution. DC reports consulting or advisory roles to Abbvie, AstraZeneca, BMS, Boehringer-Ingelheim, Daiichi-Sankyo, Eisai, Flame Biosciences, G1 Therapeutics, Genentech, Gritstone, Glaxo-Smith Kline, Iovance, Janssen, Jazz, JNJ, Merck, Merck KGgA, Mirati, Novartis, Novocure, OncoCyte, OncoHost, Pfizer, Regeneron, Roche China, Sanofi, Seattle Genetics. MG-M was site PI for a Phase II clinical trial evaluating bintrafusp alfa, which was funded by EMD Serono. Trial is now closed and there is no additional research support.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available on reasonable request. The sequencing data and processed expression matrix have been deposited at the Gene Expression Omnibus with access number GSE202153.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The study did not use human subjects. Mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of The Ohio State University, following the established guidelines. (ID#: 2019A00000075-R1).
References
- 1.Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 2019;176:677. 10.1016/j.cell.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 2.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000;192:1027–34. 10.1084/jem.192.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan M, Arooj S, Wang H. NK cell-based immune checkpoint inhibition. Front Immunol 2020;11:167. 10.3389/fimmu.2020.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol 2018;18:153–67. 10.1038/nri.2017.108 [DOI] [PubMed] [Google Scholar]
- 5.Strauss L, Mahmoud MAA, Weaver JD, et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol 2020;5:1863. 10.1126/sciimmunol.aay1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 2010;236:219–42. 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber DL, Wherry EJ, Masopust D, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439:682–7. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- 8.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med 2016;8:328rv4. 10.1126/scitranslmed.aad7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreau JM, Velegraki M, Bolyard C, et al. Transforming growth factor-β1 in regulatory T cell biology. Sci Immunol 2022;7:eabi4613. 10.1126/sciimmunol.abi4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell 2008;134:392–404. 10.1016/j.cell.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 13.Hammerl D, Martens JWM, Timmermans M, et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat Commun 2021;12:5668. 10.1038/s41467-021-25962-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhurst RJ. Targeting TGF-β signaling for therapeutic gain. Cold Spring Harb Perspect Biol 2017;9:22301. 10.1101/cshperspect.a022301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind H, Gameiro SR, Jochems C, et al. Dual targeting of TGF-β and PD-L1 via a bifunctional anti-PD-L1/TGF-βRII agent: status of preclinical and clinical advances. J Immunother Cancer 2020;8:e000433. 10.1136/jitc-2019-000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace CH, Wu BX, Salem M, et al. B lymphocytes confer immune tolerance via cell surface GARP-TGF-β complex. JCI Insight 2018;3:99863. 10.1172/jci.insight.99863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrillo-Galvez AB, Cobo M, Cuevas-Ocana S. Mesenchymal stromal cells express GARP/LRRC32 on their surface: effects on their biology and immunomodulatory capacity. Stem Cells 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szepetowski P, Ollendorff V, Grosgeorge J, et al. DNA amplification at 11q13.5-q14 in human breast cancer. Oncogene 1992;7:2513–7. [PubMed] [Google Scholar]
- 19.Metelli A, Wu BX, Fugle CW, et al. Surface expression of TGFβ docking receptor GARP promotes oncogenesis and immune tolerance in breast cancer. Cancer Res 2016;76:7106–17. 10.1158/0008-5472.CAN-16-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachidi S, Metelli A, Riesenberg B, et al. Platelets subvert T cell immunity against cancer via GARP-TGFβ axis. Sci Immunol 2017;2:7911. 10.1126/sciimmunol.aai7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salem M, Wallace C, Velegraki M, et al. Garp dampens cancer immunity by sustaining function and accumulation of regulatory T cells in the colon. Cancer Res 2019;79:1178–90. 10.1158/0008-5472.CAN-18-2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liénart S, Merceron R, Vanderaa C, et al. Structural basis of latent TGF-β1 presentation and activation by GARP on human regulatory T cells. Science 2018;362:952–6. 10.1126/science.aau2909 [DOI] [PubMed] [Google Scholar]
- 23.de Streel G, Bertrand C, Chalon N, et al. Selective inhibition of TGF-β1 produced by GARP-expressing Tregs overcomes resistance to PD-1/PD-L1 blockade in cancer. Nat Commun 2020;11:4545. 10.1038/s41467-020-17811-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2019;51:411–2. 10.1016/j.immuni.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 25.Kwon H, Schafer JM, Song N-J, et al. Androgen conspires with the CD8+ T cell exhaustion program and contributes to sex bias in cancer. Sci Immunol 2022;7:eabq2630. 10.1126/sciimmunol.abq2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song N-J, Allen C, Vilgelm AE, et al. Treatment with soluble CD24 attenuates COVID-19-associated systemic immunopathology. J Hematol Oncol 2022;15:5. 10.1186/s13045-021-01222-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metelli A, Wu BX, Riesenberg B, et al. Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci Transl Med 2020;12:4860. 10.1126/scitranslmed.aay4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kakarla S, Song X-T, Gottschalk S. Cancer-associated fibroblasts as targets for immunotherapy. Immunotherapy 2012;4:1129–38. 10.2217/imt.12.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor FOXP3. Science 2003;299:1057–61. 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- 30.Oh SA, Li MO. TGF-β: guardian of T cell function. J Immunol 2013;191:3973–9. 10.4049/jimmunol.1301843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu E, Chia CPZ, Chen W, et al. T cell receptor-regulated TGF-β type I receptor expression determines T cell quiescence and activation. Immunity 2018;48:745–59. 10.1016/j.immuni.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu J, Yue W, Le-Le Z, et al. Mesenchymal stem cells inhibit T cell activation by releasing TGF-β1 from TGF-β1/GARP complex. Oncotarget 2017;8:99784–800. 10.18632/oncotarget.21549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmadzadeh M, Rosenberg SA. TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol 2005;174:5215–23. 10.4049/jimmunol.174.9.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jie H-B, Schuler PJ, Lee SC, et al. CTLA-4⁺ regulatory T cells increased in cetuximab-treated head and neck cancer patients suppress NK cell cytotoxicity and correlate with poor prognosis. Cancer Res 2015;75:2200–10. 10.1158/0008-5472.CAN-14-2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lines JL, Pantazi E, Mak J, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res 2014;74:1924–32. 10.1158/0008-5472.CAN-13-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott AC, Dündar F, Zumbo P, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 2019;571:270–4. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beltra J-C, Manne S, Abdel-Hakeem MS, et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 2020;52:825–41. 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunderson AJ, Yamazaki T, McCarty K, et al. TGFbeta suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat Commun 2020;11:1749. 10.1038/s41467-020-15404-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutet M, Gauthier L, Leclerc M, et al. TGFbeta signaling intersects with CD103 integrin signaling to promote T-lymphocyte accumulation and antitumor activity in the lung tumor microenvironment. Cancer Res 2016;76:1757–69. 10.1158/0008-5472.CAN-15-1545 [DOI] [PubMed] [Google Scholar]
- 40.Gomez G, Ramirez CD, Rivera J, et al. TGF-beta 1 inhibits mast cell Fc epsilon RI expression. J Immunol 2005;174:5987–93. 10.4049/jimmunol.174.10.5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morad G, Helmink BA, Sharma P, et al. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021;184:5309–37. 10.1016/j.cell.2021.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol 2021;16:223–49. 10.1146/annurev-pathol-042020-042741 [DOI] [PubMed] [Google Scholar]
- 43.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20:25–39. 10.1038/s41577-019-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto M, Kamphorst AO, Im SJ, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med 2018;69:301–18. 10.1146/annurev-med-012017-043208 [DOI] [PubMed] [Google Scholar]
- 45.Chow MT, Ozga AJ, Servis RL, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 2019;50:1498–512. 10.1016/j.immuni.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan Y, Zhang D, Xu C, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med 2018;10:5488. 10.1126/scitranslmed.aan5488 [DOI] [PubMed] [Google Scholar]
- 47.Ravi R, Noonan KA, Pham V, et al. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFbeta enhance the efficacy of cancer immunotherapy. Nat Commun 2018;9:741. 10.1038/s41467-017-02696-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derynck R, Turley SJ, Akhurst RJ. TGFbeta biology in cancer progression and immunotherapy. Nat Rev Clin Oncol 2021;18:9–34. 10.1038/s41571-020-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parichatikanond W, Luangmonkong T, Mangmool S, et al. Therapeutic targets for the treatment of cardiac fibrosis and cancer: focusing on TGF-β signaling. Front Cardiovasc Med 2020;7:34. 10.3389/fcvm.2020.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabriel SS, Tsui C, Chisanga D, et al. Transforming growth factor-β-regulated mTOR activity preserves cellular metabolism to maintain long-term T cell responses in chronic infection. Immunity 2021;54:1698–714. 10.1016/j.immuni.2021.06.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005433supp001.pdf (1.2MB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available on reasonable request. The sequencing data and processed expression matrix have been deposited at the Gene Expression Omnibus with access number GSE202153.