SUMMARY
The homeodomain is found in hundreds of transcription factors that play roles in fate determination via cell-autonomous regulation of gene expression. However, some homeodomain-containing proteins (HPs) are thought to be secreted and penetrate neighboring cells to affect the recipient cell fate. To determine whether this is a general characteristic of HPs, we carried out a large-scale validation for intercellular transfer of HPs. Our screening reveals that intercellular transfer is a general feature of HPs, but it occurs in a cell-context-sensitive manner. We also found the secretion is not solely a function of the homeodomain, but it is supported by external motifs containing hydrophobic residues. Thus, mutations of hydrophobic residues of HPs abrogate secretion and consequently interfere with HP function in recipient cells. Collectively, our study proposes that HP transfer is an intercellular communication method that couples the functions of interacting cells.
Graphical Abstract
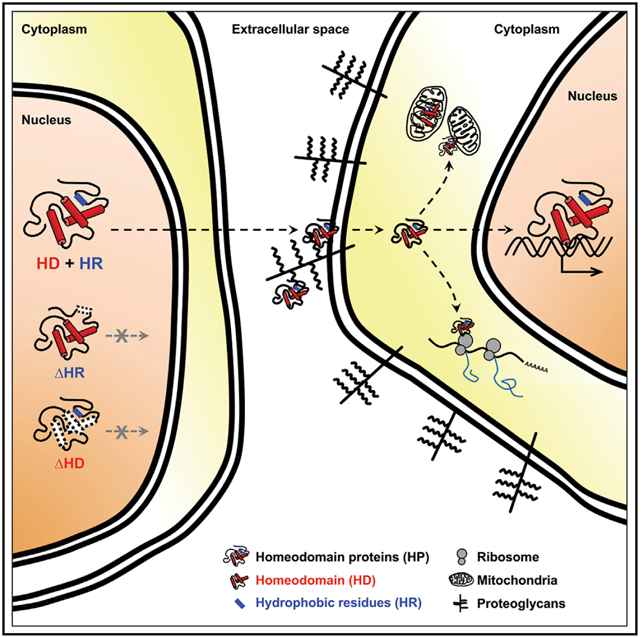
In Brief
Lee et al. evaluate capabilities of homeodomain proteins (HPs) for transfer between cells. They find that intercellular transfer is a general but cell-context-sensitive property of HP. Intercellular HP transfer can be an unconventional way for the cells to communicate with neighboring cells that associate structurally and functionally.
INTRODUCTION
Homeodomain proteins (HPs) are characterized by a highly conserved ~60 amino acids DNA binding domain, which is encoded by a DNA sequence termed the homeobox (Bürglin, 2011; Gehring et al., 1990). There are more than 200 human genes containing homeobox sequences. A comprehensive classification based on phylogenetic analysis of homeobox sequences, chromosomal location, and domain composition subdivides HPs into 11 classes—ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS, ZF, and CERS (Holland et al., 2007).
The HPs play important roles in the development of organisms, especially in the specification of identity of embryonic regions along the body axes (Pearson et al., 2005). Given the possession of the homeodomain, HPs have been believed to function as transcription factors in cell-autonomous manners. Surprisingly, a group of HPs, including Emx1, Emx2, Engrailed-2 (En2), Hoxa5, Hoxb4, Hoxc8, Knotted1, Otx2, Pax6, and Vax1, have shown to be detectable in the cells without mRNA expression (Lucas et al., 1995; Prochiantz and Di Nardo, 2015; Spatazza et al., 2013). This is mediated by intercellular transfer of the HPs from the cells expressing the HP mRNA to those that do not express. The HPs are therefore suggested to play roles as autocrine and paracrine signaling factors because of the secretion and penetration properties.
Although the cell-penetrating capacity of the homeodomain was discovered over a quarter century ago (Joliot et al., 1991), the physiological importance of intercellular HP transfer was verified only recently. En2 is expressed in the optic tectum in chick and superior colliculus in mammals by forming anterior-low and posterior-high concentration gradient. En2 then moves into retinal ganglion cell (RGC) axons, which grow into the optic tectum area to form topographic retinotectal connections, and supports ephrinA/EphA-dependent repulsive response of axon growth cones by regulating local translation of mRNA in the RGC axons (Brunet et al., 2005; Wizenmann et al., 2009; Yoon et al., 2012). In substantia nigra of the midbrain, the exogenous En2 was found to protect dopaminergic neurons not only by increasing the translation of mRNA encoding mitochondrial proteins but also by protecting the cells from DNA damages (Alvarez-Fischer et al., 2011; Rekaik et al., 2015). Vax1 is secreted from the cells in the optic stalk and ventral hypothalamus, and it enters into RGC axons to promote local mRNA translation, which induces the axonal growth toward the midline during the formation of optic chiasm (Kim et al., 2014). It was also shown that Otx2 transfer to parvalbumin (PV)-positive cortical interneurons is related with the critical period of synaptic plasticity in mammalian visual cortex (Sugiyama et al., 2008). The exogenous Otx2 also transferred from the photoreceptors to the bipolar cells in mouse retina mobilized into the mitochondria to facilitate mitochondrial ATP synthesis (Kim et al., 2015). Recently, the exogenous Otx2 in the PV interneurons was further found to involve in epigenetic changes of gene expression, which is necessary for closing the critical period (Apulei et al., 2019).
This unconventional form of HP trafficking is supposedly mediated by the homeodomain (HD), which includes amino acid sequences involved in secretion (Sec) and cell penetration (Pen) (Maizel et al., 1999). Given the cell-penetrating activity, the Pen sequence is utilized to deliver cell-impermeable polar macromolecules, such as small interfering RNA (siRNA) and therapeutic proteins, into the cells (Pooga and Langel, 2015). The Sec and Pen sequences are conserved in HPs; however, it is still unclear whether all HPs move through this unconventional protein-trafficking pathway. In this study, we performed a large-scale testing for the secretion and transfer of HPs using in vitro and in vivo systems and proved that most HPs possess a potential to travel between cells. We also found that not only the HD but also the hydrophobic residues locating outside the HD are necessary for the secretion of the HPs. Alterations of those external residues do not affect the transcription factor activities of HPs, allowing us to investigate the roles of HP transfer separately from cell-autonomous events regulated by HP-dependent gene expression regulation. Furthermore, the information of mobile HPs found in this study can be utilized to understand the physiological relevances of the HP transfer in various multicellular organisms and to cure various diseases associated with the reduced HP level by external supply of the recombinant HPs.
RESULTS
Global Analysis of HP Secretion
To evaluate HP secretability, we transfected human embryonic kidney (HEK) 293T cells with pCAGIG-V5-HP-IRES-EGFP plasmid constructs, which contain human HP cDNA open reading frames (ORFs) with N-terminal V5 tag, an internal ribosome entry site (IRES), and enhanced green fluorescent protein (EGFP) (Figure 1A). We designed these constructs such that the two resulting proteins (i.e., HP and EGFP) are transcribed as a single transcript but independently translated in same cells. We then collected serum-free growth media from the transfected cells and attempted to detect secreted HPs and EGFP by dot blotting (DB). We were, however, unable to detect the majority of HPs as well as co-expressed EGFP in the growth media (Figure 1B).
Figure 1. Global Analysis of HP Secretion.
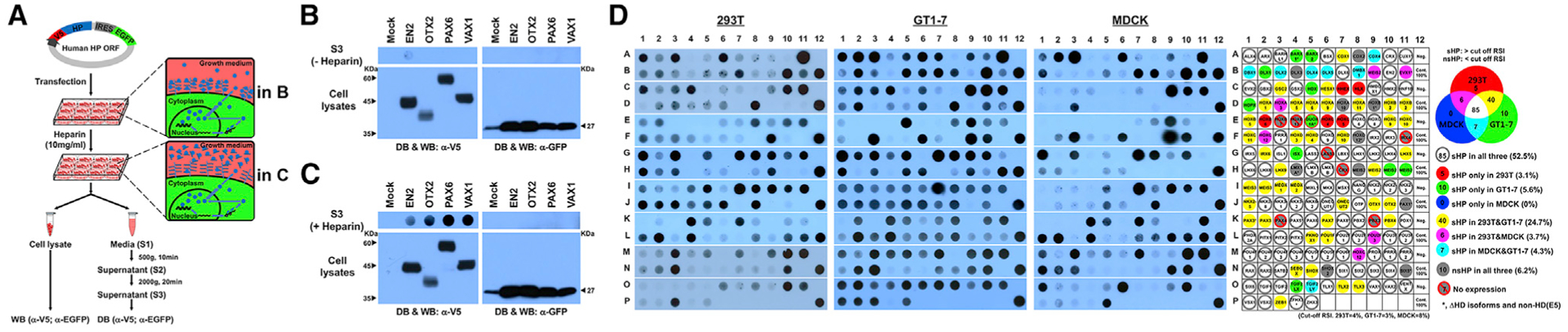
(A) Schematic diagram depicts experimental procedures to detecting secreted HPs.
(B) V5 HPs and EGFP, which are transcribed in same mRNA and translated independently, in the growth media of 293T cells were detected by dot blotting (DB) with anti-V5 and anti-GFP antibodies, respectively. Those proteins expressed in the cells were also detected by western blotting (WB).
(C) Alternatively, the growth media were added with heparin (10 mg/mL) for 3 h, and V5-HPs and EGFP in the growth media were detected.
(D) DB images for V5-HPs in 293T, GT1–7, and MDCK cell growth media treated with heparin (same DB images are provided together with corresponding WB images in Data S1). Secretability of each HP is provided in the virtual DB images in the rightmost column (classification method is provided in Figure S1). Dot colors represent secretion observed in corresponding cell lines.
Vax1 is known to move into retinal axons upon binding extra-cellular sugar chains of heparan sulfate proteoglycans (HSPGs) (Kim et al., 2014). Otx2 also exhibits strong affinity for the chondroitin 6-sulfate (C6S) sugar chains of proteoglycans on the surface of parvalbumin (PV)-positive cortical interneurons (Beurdeley et al., 2012; Miyata et al., 2012). These results suggest our inability to detect HPs in the growth media might be due to their entrapment by cell surface proteoglycan sugar chains. In support of this idea, we could observe a significant increase in HPs in the growth media, but not their co-expressed EGFP, when we added excess free heparin to the growth media to make it compete with cell surface proteoglycans for HP binding (Figure 1C).
Thus, we incubated the HP-transfected 293T, mouse hypothalamic GT1–7 neuronal, and Madin-Darby canine kidney (MDCK) epithelial cells in the growth medium containing free heparin for 3 h prior to the collection of the growth media. We then analyzed the components of growth media trapped in DBs using anti-V5 and anti-GFP antibodies to detect the V5-tagged HP and EGFP proteins, respectively, in the growth media. The absence of EGFP signal in the DBs confirmed the HP signals we observed in duplicated DBs were not just results of non-specific protein release (Figure 1D; Data S1; EGFP results are not shown).
To compare secretion efficiencies of HPs in each cell line, we measured the chemiluminescent intensity of each dot (DHx refers to the amount of HP secreted from the cells; Hx represents the HP clone number) and divided the dot intensity value by its western blotting (WB) band intensity value (WHx refers to total amount of HP expressed in the cells) to normalize the influence of amounts of HPs expressed in the cells on the dot intensities. This DB/WB ratio is designated as a secretion value of HPx (SHx). We then subtracted the secretion value of co-expressed EGFP (SGx), which was obtained by dividing dot intensity (DGx) by WB band intensity (WGx) for EGFP in the same sample, from the SHx value. This produced the specific secretion index for each HP (SIx). Finally, we calculated the relative secretion index (RSIx) for each HP by dividing each SIx by the SIx of PITX1 (SIPITX1), because PITX1 was secreted constantly in all three cell lines and used as a standard for every transfection batch (RSIx = SIx/SIPITX1; Table S1).
We found the RSIs for human HPs are distributed largely in two distinct domains. Although the RSI values of most HPs fall within a normal distribution, roughly 13% (GT1–7), 16% (293T), and 40% (MDCK) of HPs fall much lower (Figure S1A). We classified the HPs in this low RSI group to non-secretory HPs (nsHPs) (left of the dotted lines in Figure S1A) and the rest as secretory HPs (sHPs) (right of the dotted lines in Figure S1A).
Except for 7 HPs that were not expressed and 1 non-HD protein DUOX1, we compared the secretion efficiencies of 162 HPs. These included 151 independent and 11 redundant HP clones. We found 10 of the 162 HPs were undetectable in the media of all three cell lines (i.e., 293T, GT1–7, and MDCK). Five of those 10 nsHPs lack some or all of the HD, and the remaining five (i.e., CDX2, DLX3, HOXA10, MEIS2, and SHOX2) have intact HDs (gray circles in Figure 1D; Data S1). We found evidence of secretion for the remaining 152 HPs in at least one cell line, indicating that secretion is a general feature of HPs. Only 85 HPs (52.5%), however, were secreted from all three cell lines (white circles in Figure 1D; Figures S1B and S1C; Table S1). Of the HPs previously reported to be secreted, EN2, HOXC8, PAX6, and VAX1 were secreted from all three cell lines, and HOXA5 and OTX2 showed higher secretion efficiencies in 293T and GT1–7 than in MDCK cells (Figure 1D; Table S1). The results therefore suggest that HP secretion is a cell-context-sensitive phenomenon.
We also investigated whether secretion is a feature of any specific HD subclass by examining the ratio of sHPs to nsHPs in each subclass. We, however, found that none of the major HD subclasses (having more than 10 HP members) could be classified as exclusively sHPs or nsHPs in any cell context (Figure S2A). We also found the HD subclasses show varying levels of secretion efficiency. In terms of cell-type-independent sHP content, POU subclass (79%) is highest and ANTP-HOXL subclass (19%) is lowest (Figure S2B). Within the ANTP subclass, the NKL subfamily, which includes the sHPs EN2, EMX1, and VAX1, shows higher secretion frequency than the HOXL subfamily, which includes the sHPs HOXA5, HOXB4, and HOXC8 (Figure S2C).
Based on the screening results of V5-HPs, we next evaluated the secretabilities of endogenous HPs. We tested with 6 endogenous HPs, which are expressed in 293T, HeLa, GT1–7, and/or MDCK cells and can be recognized specifically by the antibodies available in the laboratory. In support of our screening results, endogenous sHPs, including DLX2, EN2, PAX6, PROX1, and VSX1, were secreted from the cells expressing them, whereas nsHP SHOX2 was not (Figure S3A). Furthermore, the SI values of endogenous EN2, PAX6, and VSX1, which are expressed in multiple cell lines, are not significantly different from those of V5-tagged HPs overexpressed in the same cells, except for EN2 in GT1–7, PAX6 in MDCK, and VSX1 in GT1–7 cells (Figure S3B).
Evaluation of Intercellular Transfer of HP in Cultured Cells
We next investigated the transfer of HPs in cultured HeLa cells. Given the translation of HP and EGFP proteins from a single transcript, we classified the cells containing HP without EGFP as the recipients of HPs (Figures 2A and 2B). We observed evidence of intercellular HP transfer in HeLa cells transfected with sHP DNA constructs, but not nsHP constructs (Figure 2C; Data S2). We also found the transfer of HPs mediates proteoglycans in cell surface, because it is interfered in the presence of free heparin in the growth medium (Figure 2B, rightmost column).
Figure 2. Intercellular Transfer of HPs in HeLa Cells.
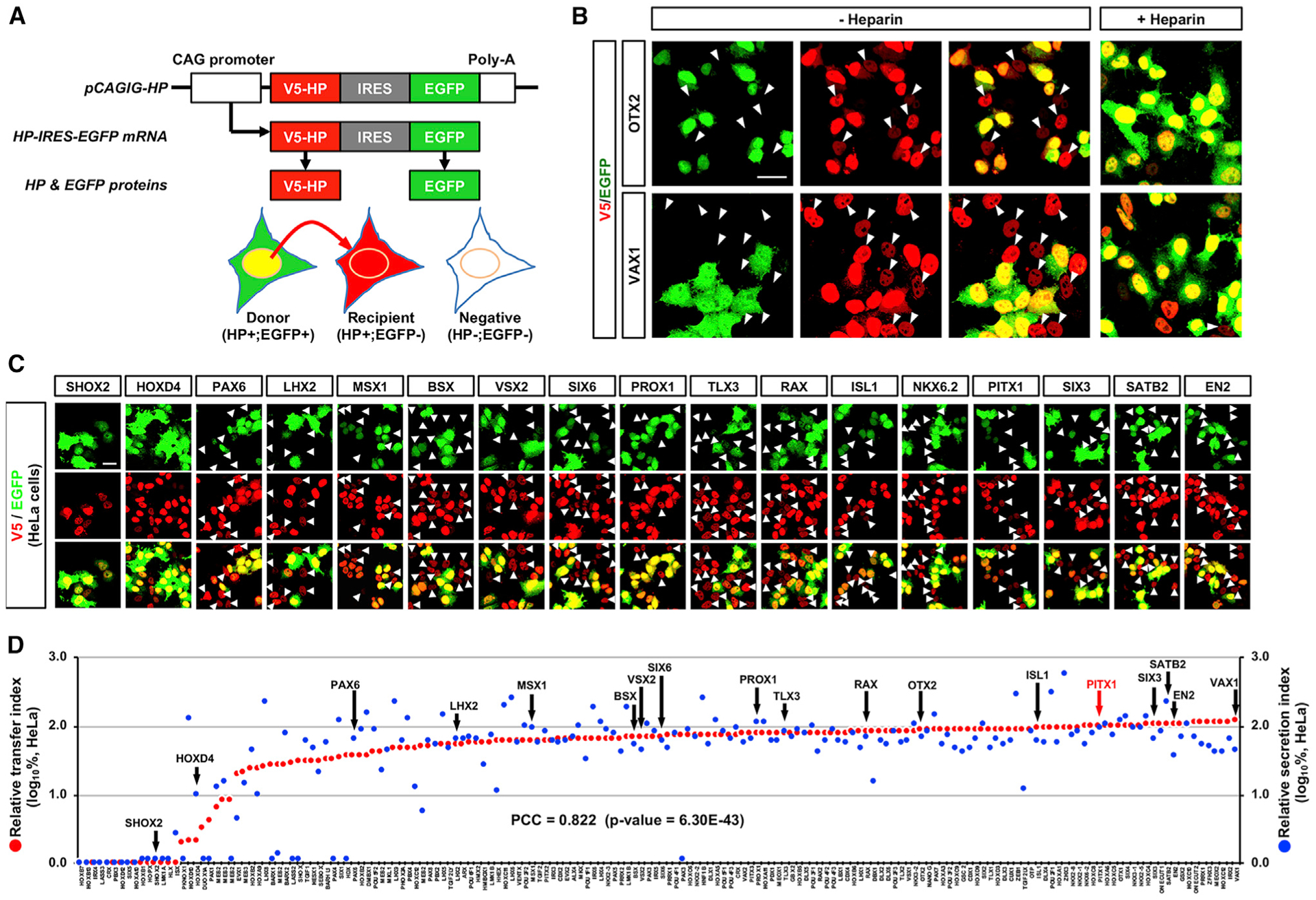
(A) Schematic diagram depicts the method determining intercellular transfer of HP. In principle, V5-HP (red) and EGFP (green) should be expressed in same cells (HP+;EGFP+; named as “donor”), because they are encoded in a same pCAGIG-HP DNA construct, transcribed in a single mRNA, and translated independently. Therefore, cells that express V5-HP alone can be determined as that they received it from the donor cells.
(B) HeLa cells, which were transfected with pCAGIG-V5-OTX2 (top) and pCAGIG-V5-VAX1 (bottom) plasmid DNA, respectively, were co-immunostained with mouse anti-V5 and chick anti-GFP antibodies. Arrowheads indicate the cells expressing V5-OTX2 and V5-VAX1 without EGFP. To test the effects of heparin on the transfer, we also added heparin (10 mg/mL) in the growth media. This interferes with the transfer of OTX2 and VAX1 (rightmost column).
(C) Representing immunostaining images of HeLa cells expressing V5-HPs. Arrowheads indicate cells containing V5-HPs (red) without EGFP (green).
(D) Relative secretion index (RSI) (y axis in right) and relative transfer index (RTI) (y axis in left) of each HP are displayed in blue and red dots, respectively. Values in the y axes are averages (n = 3). Correlation between RTI and RSI was determined by obtaining Pierson correlation coefficient (PCC), which is shown in the graph.
Scale bars, 25 μm.
To quantify this intercellular transfer, we calculated the transfer index (TIx) by counting the number of HP-only cells in an area of each culture slide and dividing it by the number of total HP-expressing cells in the same area. The resulting TIx values were then divided by TIPITX1 to obtain a relative transfer index (RTIx). A ranked list of these RTIs reveals a minor cluster of nsHPs and a major cluster of sHPs (Figures 2D and S1A; Table S2). All previously identified sHPs, including VAX1 ranks top RTI score, were found in the major cluster, whereas nsHPs, including SHOX2, were found in the minor cluster. We also found a strong correlation between the RTI and RSI for HPs (Pierson correlation coefficient [PCC] = 0.822), suggesting HP secretion and penetration are strongly associated events.
Interestingly, the internalization of nsHP SHOX2 was as much efficient as that of sHPs EN2 and OTX2 when they were added directly into the growth medium of HeLa cells (Figure S4). The results suggest that all HPs are competent for the internalization, whereas their secretion is dependent of cellular environment. Collectively, our results suggest that the secretion is a more determining step than the internalization in the intercellular transfer of HP.
Evaluation of Intercellular Transfer of HP in Mouse Brain
We also validated our screening results in vivo by electroporating pCAGIG-V5-HP-IRES-EGFP DNA constructs into embryonic mouse brains at 13.5 dpc (days post-coitum, equivalent to embryonic day 13.5 [E13.5]). These 56 HPs include 52 sHPs and 4 nsHPs, which are classified according to the results in GT1–7 hypothalamic neuronal cells (Figure 1D; Table S1). Using the same approach that we applied to the cultured cells (Figure 2A), we looked in E16.5 brain sections for evidence of HP transfer from HP;EGFP double-positive mouse brain cells to HP-positive but EGFP-negative cells (Figures 3A and 3B; Table S3).
Figure 3. Intercellular Transfer of HPs in Mouse Embryonic Brain.
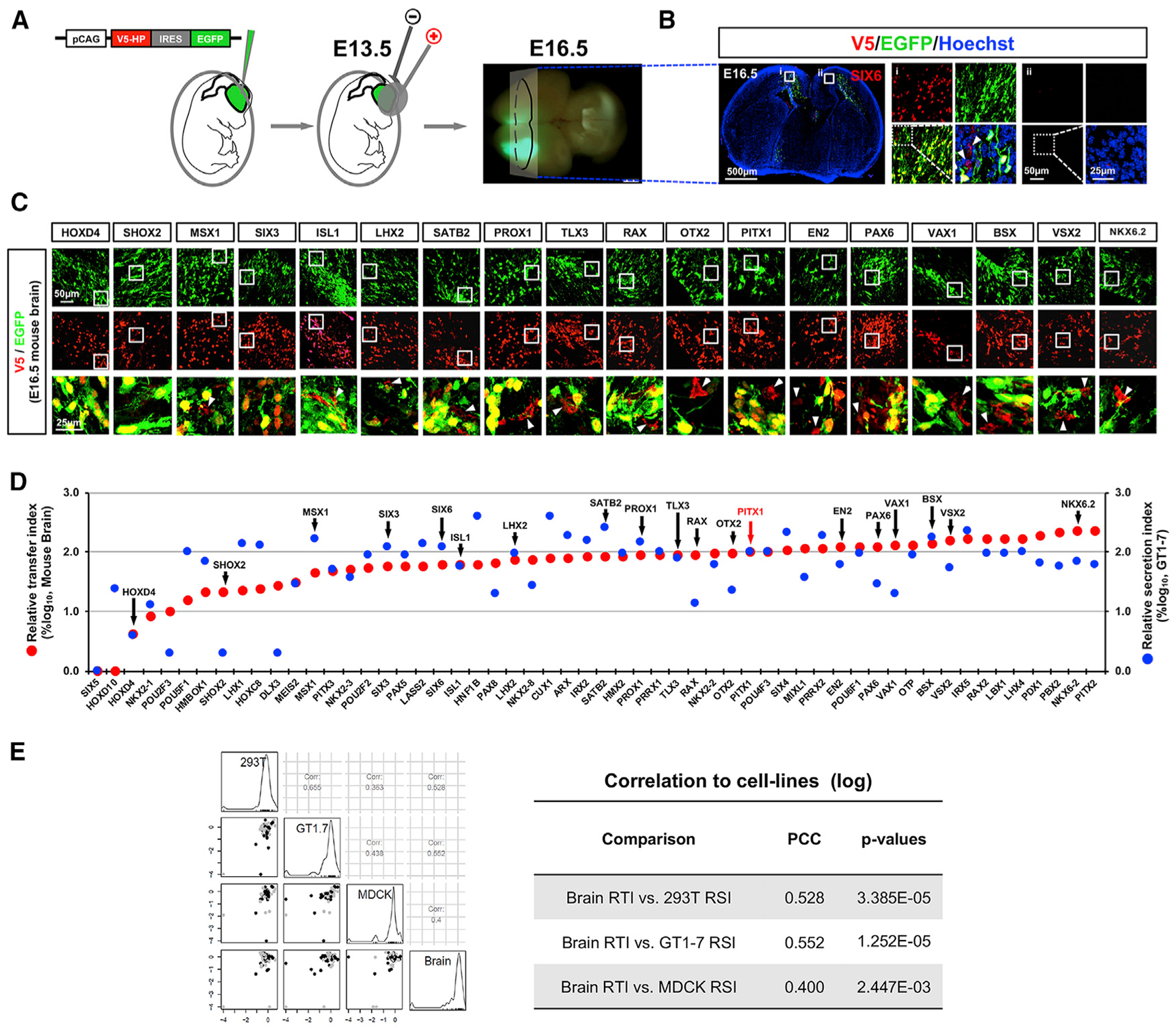
(A) To assess intercellular transfer of HPs in mouse embryonic brain, pCAGIG-V5-HP plasmid DNAs were electroporated into E13.5 mouse brains. The brains were collected from the electroporated mouse embryos at E16.5 after confirming successful expression of the constructs by detecting green fluorescence emitted by EGFP.
(B) Sections of E16.5 mouse brains were stained with antibodies against V5 (red) and GFP (green) for comparison of the distribution of cells expressing V5-HP and EGFP in the brains. The images are the sections of brain electroporated with pCAGIG-V5-SIX6.
(C) Immunostaining images of representing samples. The images in the bottom row are magnified merge images of boxed areas in top two rows.
(D) The RTI of each HP in mouse brain and RSI of each HP in GT1–7 cells are displayed in red and blue circles, respectively (n = 3).
(E) Correlation between RTI of brain and RSI of corresponding cell lines is determined by examining PCC, and the scores are shown in the table.
None or very few cells in mouse brains expressing nsHPs (DLX3, POU2F3, SHOX2, and SIX5) had HP without EGFP, supporting the results we obtained in GT1–7 cells. All sHPs tested in mouse brains led to neighboring cells positive for the HPs without any accompanying EGFP (Figures 3C and 3D; Data S3). The RTI of sHPs in the brain were correlated moderately with the RSI of GT1–7 cells but weakly with the RSI of 293T and MDCK cells, reaffirming that the intercellular transfer is a cell-context-sensitive phenomenon (Figure 3E).
Evaluation of HP Transfer in HP::EGFP BAC Transgenic Mouse Retina
To validate the intercellular transfer of endogenous HPs in vivo, we next examined the distribution of HPs in adult mouse retinas that contain bacterial artificial chromosomes (BAC) encoding EGFP in mouse HP genomic DNA (Heintz, 2004). We tested with 6 HP::EGFP BAC transgenic (TG) mouse strains confirmed to express EGFP in specific retinal cell populations (Siegert et al., 2009). We did this with the goal of observing the transfer of HPs from HP;EGFP double-positive donor cells to EGFP-negative recipient cells (Figure 2A, diagram). We detected sHPs (e.g., Lhx2, Prox1, Satb2, Six3, Six6, and Vsx2) in both EGFP-positive and EGFP-negative cells in the corresponding HP::EGFP BAC-TG mouse retinas (Figure 4).
Figure 4. Intercellular Transfer of Endogenous HPs in Mouse Retina.

Eye sections of P30 HP::EGFP-BAC TG mice, which express EGFP encoded in the genomic DNA sequence of corresponding mouse HP genes, were obtained, and distributions of HPs were compared with those of EGFP by immunostainings. EGFP is believed to represent the expression pattern of corresponding HP genes. The cells expressing HPs without EGFP (pointed by arrowhead), therefore, can be considered as potential recipient cells of the HPs.
Scale bars, 25 μm.
The EGFP signals in postnatal day 30 (P30) Lhx2::EGFP BAG TG mouse retinas were co-detectable in the middle of inner nuclear layer (INL), implicating the expression of EGFP in Lhx2-positive Müller glia (de Melo et al., 2012). In contrast, EGFP signal was not detectable in Lhx2-positive cells in the lower part of INL, suggesting no or very low EGFP expression in Lhx2-positive amacrine cell (AC) subsets (Figure 4, leftmost column; Balasubramanian et al., 2014; Kim et al., 2017). The results implicate Lhx2 in the AC population could be expressed by other regulatory element(s) lacking in the Lhx2 BAC clone or obtained by intercellular transfer.
In P30 Prox1::EGFP BAC TG mouse retinas, we were not only able to detect the co-expression of EGFP and Prox1 in the top half of the INL, but we could also see Prox1 alone, without EGFP co-expression, in the bottom half of the INL (Figure 4, second column from the left). Given the expression of Prox1 in AC subsets in the bottom INL as well as horizontal cells (HZs) and bipolar cells (BPs) in the top INL of mouse retina (Dyer et al., 2003), the results suggest that Prox1 in the AC population could be expressed by other regulatory element(s) lacking in the Prox1 BAC clone or obtained by intercellular transfer.
About a half of Satb2-positive retinal cells, which are glycinergic AC subsets and non-glycine/non-GABA (nGnG) ACs (Kay et al., 2011), co-express EGFP in P30 Satb2::EGFP BAC TG mice (Figure 4, third column from the left). EGFP was not detectable in Neurod6-positive nGnG ACs, whereas GlyT1-positive glycinergic ACs express EGFP partly (data not shown), suggesting that Sabt2 in the nGnG ACs can be originated exogenously or expressed by different regulatory element(s).
EGFP signals were observed in AC subsets and HZs of P30 Six3::EGFP and Six6::EGFP BAC TG mice, consistent to their mRNA expression patterns (Conte et al., 2010; Inoue et al., 2002; Figure 4, second and third columns from the right). Interestingly, about 21% and 38% of Six3- and Six6-positive AC subsets are EGFP-negative, respectively, whereas HZs expressing those HPs are entirely EGFP positive. The results therefore suggest a potential intercellular transfer of Six3 and Six6 to those EGFP-negative AC subsets.
Vsx2 was specifically expressed in BPs in adult mouse retina (Passini et al., 1997), and so was the EGFP. However, the number of Vsx2-positive cells in P30 Vsx2::EGFP BAC TG mouse retina was larger than that of EGFP, implicating the presence of Vsx2(+);EGFP(−) cells in the retina. Indeed, 18% of Vsx2(+) cells were absent EGFP (Figure 4, rightmost column), suggesting a potential transfer of Vsx2 to these BPs from neighboring Vsx2(+);EGFP(+) cells.
Despite their presences in EGFP-negative retinal cells, exogeneity of these HPs remains undetermined. BAC transgenes are often missing regulatory element(s) for encoded genes and cannot reproduce the patterns of endogenous gene expression identically (Heintz, 2004). Therefore, the presence of those exogenous HPs in the mouse retinal cells should be validated further by comparing the distributions of the HPs and their transcripts in those cells.
Hydrophobic Amino Acid Residues outside the HD Support HP Secretion
It has been suggested that the Sec motif, which is a linker region between the second and third helices of the HD, is responsible for the secretion of En2 (Maizel et al., 1999). Given that the Sec motif is shared by all HDs, we were surprised to find no or very low chances of secretion for HPX/HOPX, which has only a PRD class HD without any other functional domain (De Toni et al., 2008; Figure 1D, D1 position in DB; Data S1; Table S1). When we further examined the secretabilities of various isolated HDs, we also found no evidence of secretion in any of the three cell lines (Figure S5A; only the 293T results are shown). In contrast, the isoforms of BARX1, EVX1, PAX3, and ZFHX3 lacking their HDs can be secreted in certain cell contexts (Figure 1D, marked with an asterisk; Data S1; Table S1). Thus, we tested the secretion of sHPs, of which HDs were deleted. The results indicate that OTX2, PAX6, and PROX1 were still capable to be secreted without the HD, whereas VAX1 should have its HD for secretion (Figure S5B). Together, although our results suggest the HD is important, it is not sufficient for secretion. Instead, domains and motifs outside the HD seem to be also necessary for HP secretion.
We thus tried to identify these motifs, which are located outside the HD and are shared among the sHPs, by multiple sequence alignment (Figure 5A; see STAR Methods for details). We found hydrophobic amino acids, including phenylalanine (Phe) (F), leucine (Leu) (L), and methionine (Met) (M), are commonly detectable in the same locations of the sHPs outside the HD (Figure 5B, boxed in red dots). To validate the importance of these residues in secretion, we replaced those amino acids with an acidic amino acid glutamate (Glu) (E), which was observed frequently in the corresponding positions of the nsHPs (Figure 5B, bottom row). For instance, changing Leu residues at 143rd and 191st positions in EN2 to Glu reduced the amount of the mutated EN2 in the growth media (Figures 5C and 5D). These seem to be a general rule, as L (or F) to E mutants of other sHPs, including OTX2(L220E), OTX2(F258E), PAX6(L130E), and VAX1(L305E), also showed reduced ability for secretion (Figures 5C and 5D). Conversely, mutant versions of HOXD4 and SHOX2, in which Ser233 and Pro305 is replaced with Leu, were secreted from 293T cells (Figures 5C and 5D).
Figure 5. Identification of Hydrophobic Amino Acids Supporting the Secretion of HPs.
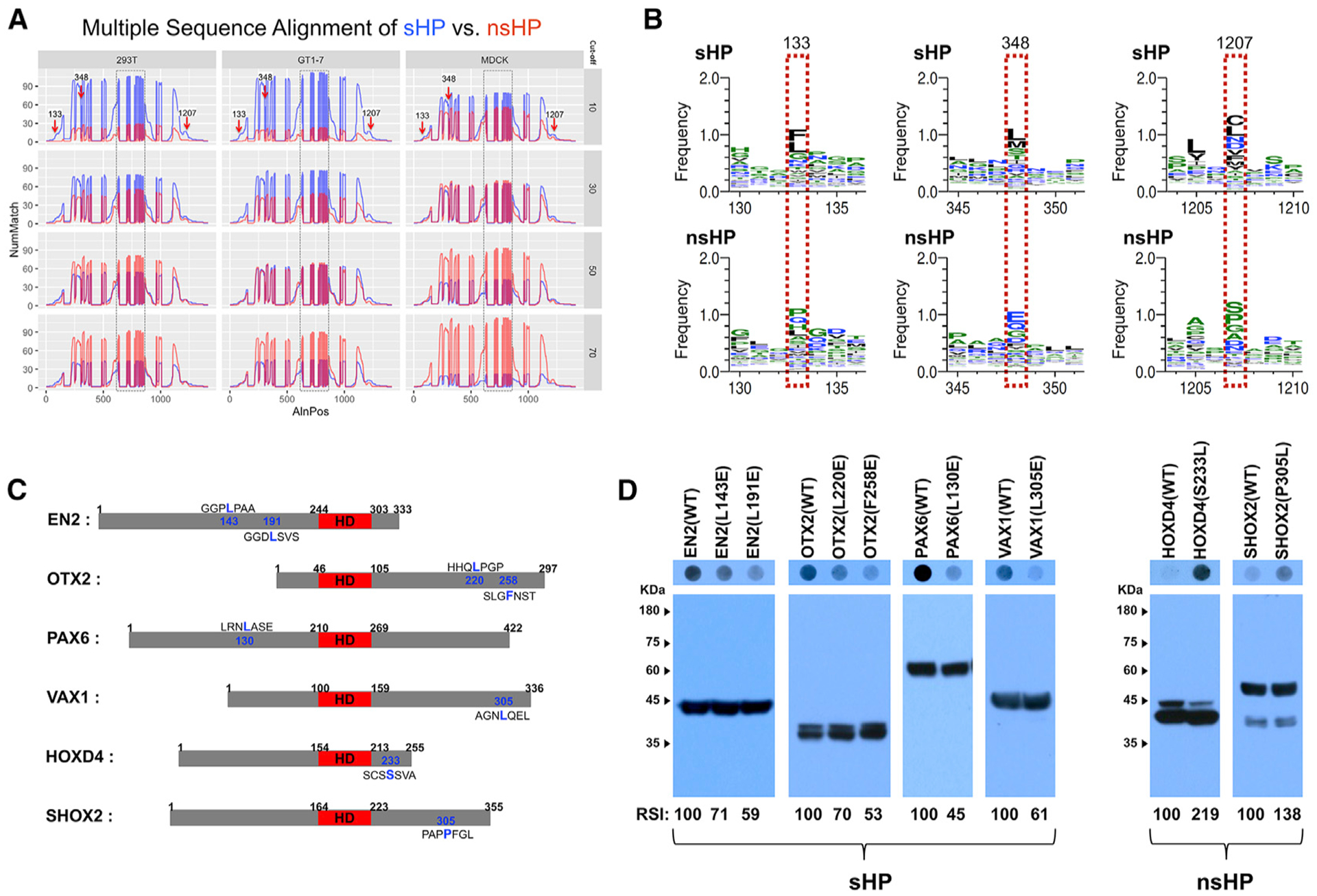
(A) Multiple sequence alignment, which is described in STAR Methods, identified shared motifs in sHPs (arrows). y axis, number of matches (NumMatch); x axis, aligned positions (AlnPos). Boxed areas are HDs, which are aligned from 626 to 849 in the plots and include gaps of unshared amino acid sequences as well as consensus motifs.
(B) Amino acid sequences in the positions pointed by arrows in (A) are different between sHP (top) and nsHP (bottom). Amino acids in each position are displayed in the order of abundance (top to bottom).
(C) Diagrams that show the locations of the amino acids, which are predicted to be shared among sHPs and nsHPs, respectively. The HDs are colored in red.
(D) Secretion abilities of those mutated proteins were determined by DB with anti-V5 antibody (top), and relative levels of HPs in the transfected cells were examined by WB with anti-V5 antibody (bottom). The scores below the WB images are average RSI values comparing with the secretion indexes of WT HPs (n = 3).
The secretion-defective sHPs, including EN2(L191E), OTX2 (F258E), PAX6(L130E), and VAX1(L305E), were transferred less efficiently than wild-type (WT) HPs (Figures 6A and 6B). Conversely, the secretion-promoting mutations of HOXD4 and SHOX2, such as HOXD4(S233L) and SHOX2(P305L), increased their abilities to transfer between HeLa cells (Figures 6A and 6B). Similar to the results in the cultured cells, the hydrophobic residues outside the HD were also important for intercellular HP transfer in mouse brain (Figures 6C and 6D). Collectively, our results suggest that, rather than depending solely on the HD, HP secretion is determined by each HP’s three-dimensional structure, which is determined by residues outside the HD, such as those hydrophobic amino acids. Thus, mutations or post-translational modifications onto those sites could change the secretability of the HPs.
Figure 6. Hydrophobic Amino Acid Residues Outside the HD Are Important for Intercellular Transfer of HPs.
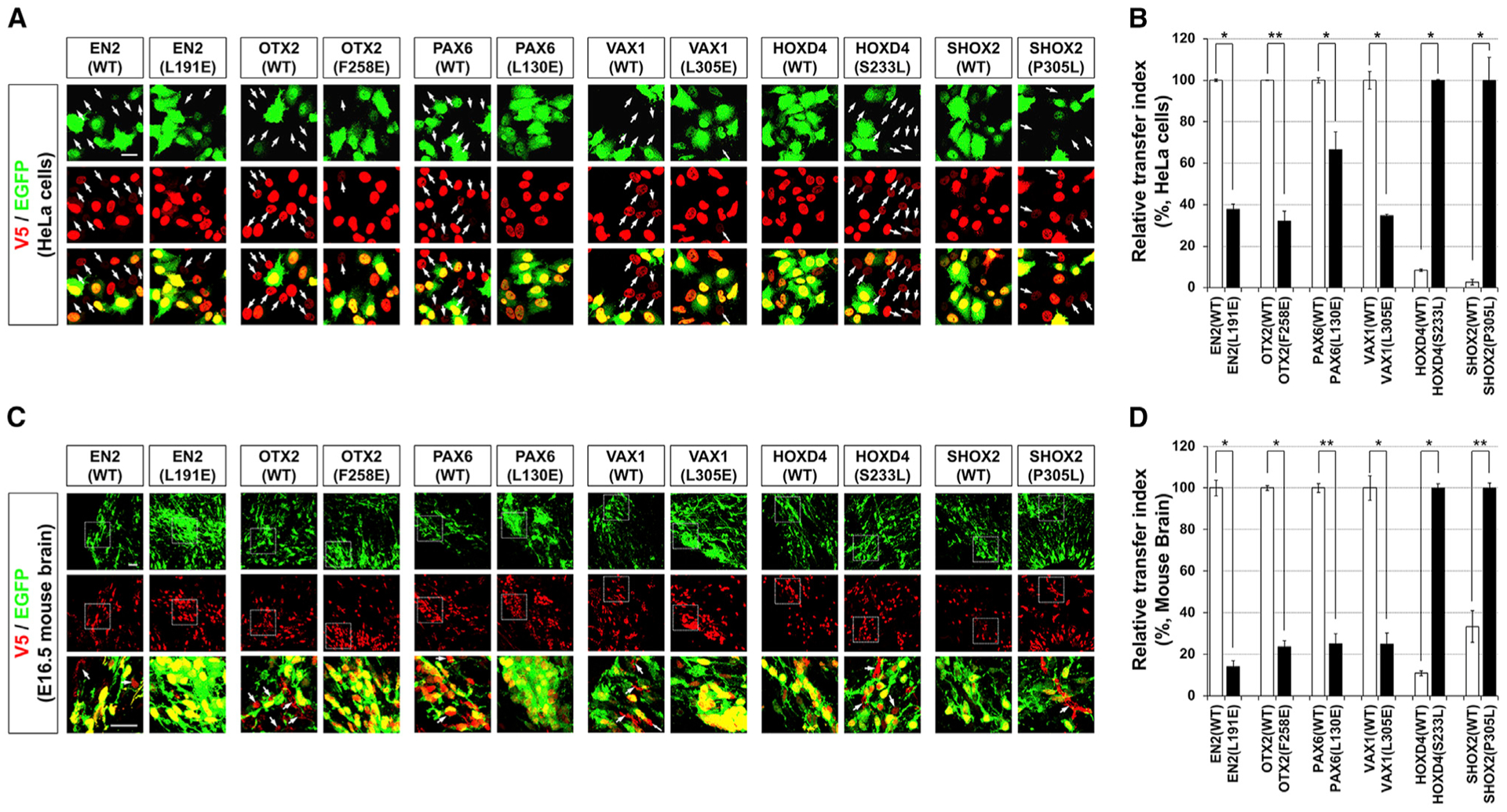
(A and C) Intercellular transfer of WT and mutant HPs between HeLa cells (A) and mouse embryonic brain cells (C) were evaluated as described in Figures 2A and 3A, respectively. Arrows indicate the cells have V5-HPs without EGFP.
(B and D) RTIs of WT and mutant HPs in HeLa cells (B) and mouse embryonic brain cells (D) are shown in graphs. Values in the y axes are averages (n = 3), and error bars denote SDs (*p < 0.01; **p < 0.005; ANOVA).
Scale bars, 25 μm.
Secretion-Defective VAX1(L305E) Is Selectively Defective of Non-Cell-Autonomous Activity
Despite the significant effects on the secretion, the L-to-E mutations did not change intracellular distribution of the sHPs. All L-to-E mutant HPs were detectable in the nucleus (Figure 6A, middle row). Furthermore, OTX2(F258E) and PAX6(L130E) carry transcription factor activities, which induce luciferase reporter expression at downstream of OTX2 target CRX promotor and tandem PAX6 binding sequences, respectively, as strong as WT OTX2 and PAX6 (Figures S6A and S6B). VAX1(L305E) also activates the expression of luciferase, of which transcription is regulated by a VAX1 target transcription factor 7-like 2 (TCF7l2) gene upstream sequence (Vacik et al., 2011), as significantly as WT VAX1 (Figure S6C). These results suggest that the hydrophobic residues enhance the secretion without affecting the transcription activities of the sHPs.
We next compared the ability of VAX1(L305E) with WT VAX1 to inducing retinal axon growth, which is dependent of intercellular transfer of VAX1 (Kim et al., 2014). The V5-VAX1, which was overexpressed in 293T cells, was detectable in the axons projecting from co-cultured mouse retinal explants (Figures 7A, second column, 7B, and 7C). However, V5-VAX1(L305E) proteins expressed in 293T cells were neither detectable in the retinal axons nor promoted the axonal growth (Figures 7A, third column, 7B, and 7C). However, recombinant V5-VAX1(L305E) added to the growth media of retinal explants were detectable in the retinal axons and induced the axonal growth as efficiently as WT VAX1 (Figures 7D, third column, 7E, and 7F). On the contrary, recombinant V5-VAX1(WF/SR), which can be secreted but cannot cross cell membrane (Kim et al., 2014), was not detectable in the retinal axons, and it failed to induce the axonal growth (Figures 7D, rightmost column, 7E, and 7F). Collectively, our results suggest that VAX1(L305E) performs normal transcription factor function in the nucleus, but it is uncapable for the secretion. Consequently, VAX1(L305E) selectively loses a function in the neighboring cells by failing intercellular transfer.
Figure 7. L305E Mutation Impairs Retinal Axon Growth Induced by Secreted VAX1.
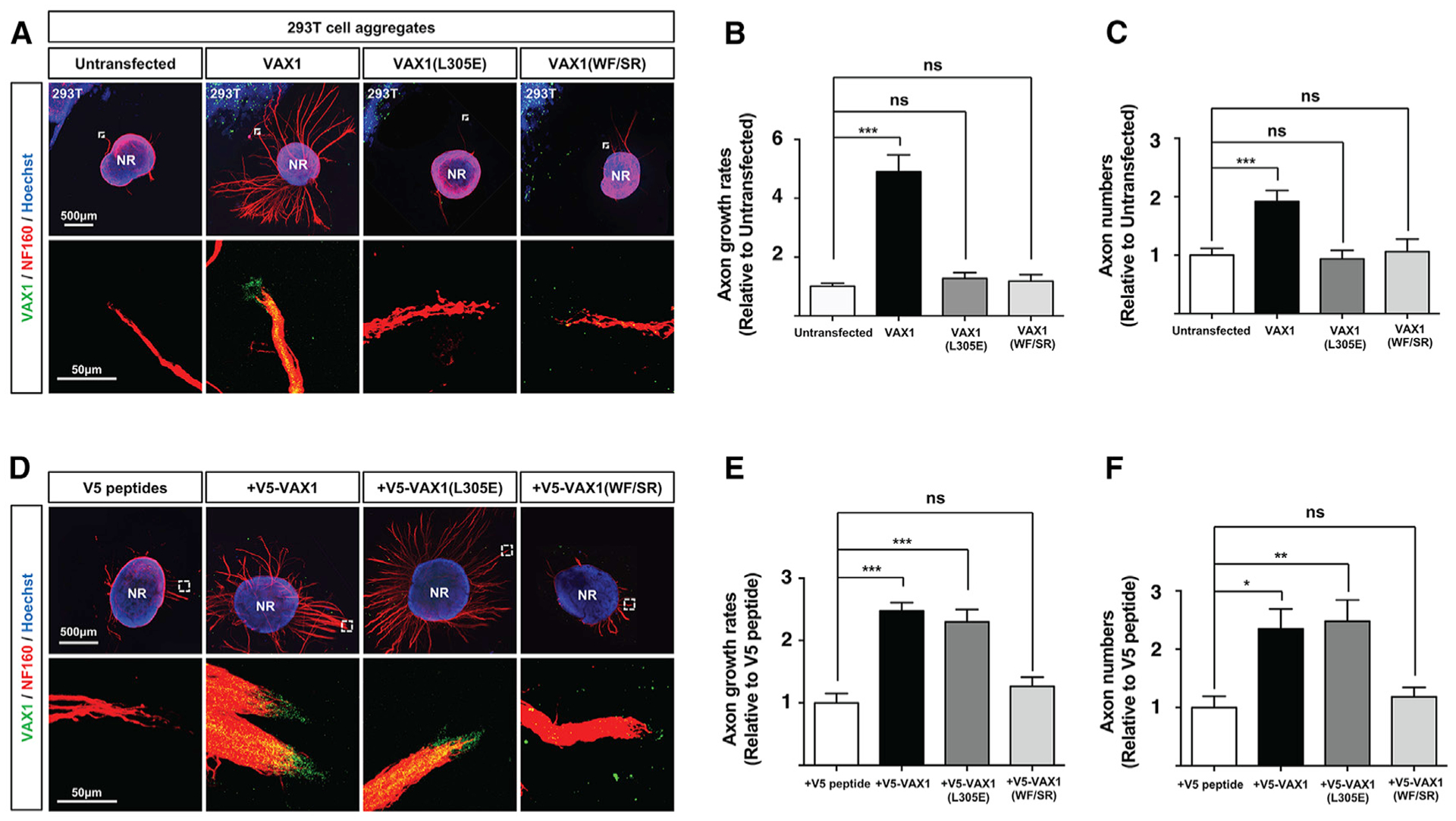
(A) 293T cells overexpressing V5-VAX1, V5-VAX1(L305E), or V5-VAX1(WF/SR) were co-cultured with E13.5 mouse retinal explants (neural retina [NR]) for 48 h (see details in STAR Methods).
(D) Alternatively, the retinal explants were treated with recombinant V5-tagged WT and mutant VAX1 proteins (50 ng/mL) for 48 h. The explants were then stained with a rabbit anti-Vax1 antibody (green) and a mouse anti-NF160 antibody (red). Nuclei of explant cells were counterstained with Hoechst 33342 (blue). The images in bottom rows are magnified versions of the areas marked by dotted boxes in top rows.
(B and E) RGC axon length were plotted as relative values to the length of RGC axons grown from the retinal explants, which were co-cultured with untransfected 293T cell aggregates (B) or treated with V5 peptides (E).
(C and F) Relative numbers of axon bundles projecting from retinal explants were indirectly measured by counting the pixels containing NF160 immunofluorescence in RGC axons between 20 μm and 40 μm from the rim of the explants.
The values are averages (n = 6, three independent experiments), and error bars denote SDs (*p < 0.01; **p < 0.005; ***p < 0.001; ANOVA).
DISCUSSION
Intercellular transfer of HPs is an evolutionarily conserved cell-to-cell communication pathway that has been proven in various multicellular organisms (Lucas et al., 1995; Prochiantz and Di Nardo, 2015; Spatazza et al., 2013). By transferring HPs, donor cells can trigger HP-dependent cellular events in a way that allows the recipient cells to skip the signaling cascades, inducing their own HP gene expression. Interestingly, imported HPs sometimes even have different functions than endogenous HPs, which mainly regulate gene transcription in the nucleus. The effects of exogenous Vax1 and En2 are mediated by local mRNA translation in the axoplasm of RGC (Alvarez-Fischer et al., 2011; Brunet et al., 2005; Kim et al., 2014; Yoon et al., 2012). Alternatively, exogenous Otx2 moves into mitochondria to facilitate mitochondrial activities (Kim et al., 2015). Such target differences between endogenous and exogenous HPs may arise from intracellular locations where the HPs first appear. Endogenous HPs are mainly captured by karyopherins, which are co-translational chaperones that bind basic amino acids comprising the nuclear localization signal (NLS) (Görlich and Kutay, 1999), during translation of their mRNA. Thus, endogenous HPs are delivered into the nucleus after they are synthesized. In contrast, exogenous HPs are not subject to co-translational capture by karyopherins. These exogenous HPs, therefore, more likely interact with other cellular components, such as the translation machinery and with mitochondrial proteins, after they penetrate a recipient cell (Alvarez-Fischer et al., 2011; Kim et al., 2014, 2015; Yoon et al., 2012). The specific binding partners with which they interact in the recipient cells would thus be important in determining which cellular events exogenous HPs involve. For this reason, future studies should catalog the various HP-interacting proteins and mRNA in cytoplasm to improve our understanding of the functions of exogenous HPs, including sHPs identified in this study.
Most of the studies of exogenous HPs have been performed in neurons, which make contacts with HP donor neurons and glia. This suggests that HP transfer happens preferentially between cells that interact directly by a structure like the synapse. Reduction and loss of HP transfer prevent axonal growth (Kim et al., 2014; Wizenmann et al., 2009), functional maturation (Sugiyama et al., 2008), and survival of recipient neurons (Alvarez-Fischer et al., 2011; Kim et al., 2015). Given our finding that intercellular transfer is a feature of most HPs, cells in non-neural tissues may also have exogenous HPs for their growth, differentiation, and survival of associating cells. If these are proven in future studies, intercellular transfer can be acceptable as a general process likely diversifies the functional roles HPs play in the processes of development and tissue maintenance, which require orchestrated growth and function of cells that are coupled in structure and function.
Although we have shown that secretion is a general feature of HPs, the molecular mechanisms of secretion still remain unclear. HP secretion may require HPs to form certain structures that can be recognizable by secretion machinery, because HD-only proteins are not secreted (Figure S5). We found hydrophobic residues are important for the secretion of several HPs (Figure 5). The hydrophobic amino acids tryptophan (W) and phenylalanine (F) in the HD were suggested to interact with the fatty acid chains of membrane lipids during membrane penetration (Christiaens et al., 2002). Therefore, the hydrophobic amino acids outside the HD may also support secretion via interactions with membrane lipids or by intra- and intermolecular hydrophobic protein-protein interactions. However, future studies that identify HP secretion intermediates, such as carrier proteins or lipids, will help to reveal the structural importance of those residues.
Despite the importance of the hydrophobic residues in the secretion of EN2, OTX2, PAX6, and VAX1 (Figure 5), no significant correlation of the mutations on those residues with human diseases has been reported. Furthermore, the changes of L305 to polar Gln (Q) or nonpolar Ala (A) did not affect the secretion of OTX2 and VAX1 (data not shown), implicating the replacement to amino acids by negatively charged amino acids is necessary to impair intercellular transfer of those HPs. However, the chances to have the acidic amino acids, such as Glu (E) and Asp (D), on the hydrophobic residues are very limited (i.e., 4 codon triplets for Glu and Asp among 64 combinations). The chances are still 8/64, even including the replacement to serine and threonine, which are also abundant in the residues of nsHPs and can be charged negatively by phosphorylation. Furthermore, given the intact transcription factor activity of VAX1(L305E), OTX2(F258E), and PAX6(L130E), the development, which is generally dependent of transcription factor activity of the HPs, might be normal in humans carrying the negative amino acids on those hydrophobic residues (even homozygously). Therefore, individuals having those substitutions on L305 of VAX1 might require additional mutations on the proteins that cooperate with the mutant VAX1 to show the phenotypes caused by axon growth defects. This was supported by the reports that only minor population exhibited agenesis of corpus callosum among those having the clefts in the palate and/or lip by VAX1 mutations (Slavotinek et al., 2012). Thus, comprehensive genomic analyses of human population are necessary to provide the answers for physiological importance of intercellular HP transfer regulated by those hydrophobic residues.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jin Woo Kim (jinwookim@kaist.ac.kr).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
HP::EGFP BAC TG mice were obtained from MMRRC. All experiments done with the mice are performed according to approved institutional animal care and use committee (IACUC) protocols (KAIST IACUC 13–130) of Korea Advanced Institute of Science and Technology (KAIST).
In utero electroporation
In utero electroporation was performed as described previously (Lim et al., 2015). Briefly, timed-pregnant C57BL/6J mice (E13.5) were anesthetized with isoflurane (0.4 L/min of oxygen and isoflurane vaporizer gauge 3 during surgery operation). After surgical intervention to expose uterine horn, each plasmid combined with 2 μg/ml of Fast Green (F7252, Sigma, USA) was injected into the lateral ventricle of the embryonic brain using pulled glass. After injection, 50 V electroporation with the ECM830 electroporator (BTX-Harvard apparatus) in five electric pulses of 100 ms at 900-ms intervals was done.
Gateway cloning of human HP ORFs
We obtained 170 human HP cDNAs from human ORFeome v7.0 developed by Dana-Farber Cancer Research Institute (DFCRI) and human ORFeome developed by Johns Hopkins University (Table S1). The HP ORFs were cloned into pCAGIG-V5 vector using the Gateway cloning technology (Invitrogen) for the expression in cultured cell-lines and mouse embryonic brain.
Cell culture, transfection, and dot blot analysis
293T, HeLa, GT1–7, and MDCK cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). 293T cells were then transfected with Polyethylenimine (PEI), and HeLa, GT1–7, and MDCK cells were transfected with Genjet plus DNA in vitro transfection reagent (Signagen) following the manufacturer’s manual. The growth media of the transfected cells were replaced with FreeStyle serum-free media (GIBCO BRL) after 6h of transfection, and were collected as described in Figure 1A. 500 μl of the media were added into each well of the GE Whatman Dot-Blot 96 Well Plate System, which was applied with vacuum to aspirate the media across the PVDF membrane. The membranes, which captured proteins and nuclei acids in the growth media, were then blotted with anti-V5 and anti-GFP antibodies. In parallel, the transfected cells were lysed in RIPA buffer (PBS with 0.1% SDS), and supernatants were collected for western blot analyses.
METHOD DETAILS
Immunostaining
Mouse embryonic brains and eyes were isolated for subsequent fixation in PBS containing 4% paraformaldehyde (PFA) for 1 h. The samples were then moved into 20% sucrose/PBS solution for subsequent incubation 4°C for 16 h in prior to cryopreservation in TissueTek O.C.T. compound for freezing. The frozen samples were then cryosectioned onto the slide glass by the thickness between 12 μm and 20 μm. Alternatively, HeLa cells cultured on the coverslips were fixed in 4% PFA/PBS for 20 min. The tissues on the slides and HeLa cells on the coverslips were then incubated in blocking solution (PBS including 10% normal donkey serum and 0.1% Triton X-100) at room temperature for 1 h. The cells were further incubated in blocking solution including primary antibodies without Triton X-100 at 4°C for 16 h, and subsequently with fluorophore-conjugated secondary antibodies recognizing the primary antibodies (the antibody information is available in Key Resources Table). Fluorescent images of the IHC signals were then obtained by Olympus FV1000 confocal microscope.
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-DLX2 antibody | Milipore | RRID: AB_2093141 |
| Rabbit monoclonal anti-EGF Receptor (EGFR) antibody | Cell Signaling Technology | RRID: AB_2246311 |
| Mouse monoclonal anti-EN2 antibody | Santa Cruz Biotechnology | Cat# SC293311 |
| Rabbit monoclonal anti-GFP antibody | Santa Cruz Biotechnology | RRID: AB_641123 |
| Chick polyclonal anti-GFP antibody | Abcam | RRID: AB_300798 |
| Goat polyclonal anti-LHX2 (N-20) antibody | Santa Cruz Biotechnology | RRID: AB_2135663 |
| Mouse monoclonal anti-NF160 (2H3) antibody | DSHB | RRID: AB_531793 |
| Rabbit polyclonal anti-PAX6 antibody | BioLegend | RRID: AB_2565003 |
| Rabbit polyclonal anti-PROX1 antibody | Milipore | Cat# ABN278 |
| Goat polyclonal anti-PROX1 antibody | R and D Systems | RRID: AB_2170716 |
| Mouse monoclonal anti-SATB2 antibody | Abcam | RRID: AB_882455 |
| Mouse monoclonal anti-SHOX2 antibody | Santa Cruz Biotechnology | RRID: AB_2188563 |
| Rabbit polyclonal anti-SIX3 antibody | Novus | Cat# NBP2–21662 |
| Rabbit polyclonal anti-SIX6 antibody | Novus | RRID: AB_11031172 |
| Mouse monoclonal anti-V5 antibody | GenWay Biotech | RRID: AB_10280227 |
| Rabbit polyclonal anti-VAX1 antibody | (Kim and Lemke, 2006) | N/A |
| Mouse monoclonal anti-VSX1 antibody | Santa Cruz Biotechnology | Cat# SC393699 |
| Mouse monoclonal anti-VSX2 (E-12) antibody | Santa Cruz Biotechnology | RRID: AB_10842442 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gateway LR Clonase II Enzyme mix | Invitrogen | 11791–020 |
| Penicillin-Streptomycin | Invitrogen | 15140–122 |
| Polyethylenimine (PEI) | Polysciences | 23966 |
| GenJet In Vitro DNA Transfection Reagent (Ver. II) | Signagen | SL100489 |
| Heparin | Millipore | 375095–500KUCN |
| Protease inhibitor | Millipore | M535142 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | 34580 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Scientific | 34095 |
| V5 peptide (GKPIPNPLLGLDST) | ANYGEN | N/A |
| BD Monolight D-Luciferin Potassium Salt | BD | 556878 |
| 2,2,2-Tribromoethanol (Avertin) | Sigma | T48402 |
| Hoechst33342 | Invitrogen | H1399 |
| Experimental Models: Cell Lines | ||
| 293T (human embryonic kidney cells, female) | ATCC | RRID:CVCL_0063 |
| GT1–7 (mouse hypothalamic GnRH neurons, female) | ATCC | RRID:CVCL_0281 |
| HeLa (human cervical cancer cells, female) | ATCC | RRID:CVCL_0030 |
| MDCK (canis kidney cells, female) | ATCC | RRID:CVCL_0422 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Lhx2::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_030843-UCD |
| Mouse: Prox1::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_031006-UCD |
| Mouse: Satb2::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_030125-UCD |
| Mouse: Six3::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_034607-UCD |
| Mouse: Six6::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_031167-UCD |
| Mouse: Vsx2::EGFP BAC TG (male, analyzed at post-natal day 30) | MMRRC | RRID:MMRRC_011391-UCD |
| Recombinant DNA | ||
| pCAG-V5-HP-IRES-EGFP recombinant DNA used in this study are listed in Table S1. | This paper | N/A |
| pGL3_basic | Promega | E1751 |
| pSV-β-Galactosidase Control Vector | Promega | E1081 |
| Software and Algorithms | ||
| Fluoview 4.0 | Olympus Corporation | N/A |
| GraphPad Prism v5.0 | GraphPad software | N/A |
| Multi Gauge V3.0 | Fujifilm Life Science | N/A |
| Other | ||
| DMEM/High glucose | Hyclone | SH30243 |
| FreeStyle 293 Expression Medium | GIBCO | 12338–018 |
| Fetal Bovine Serum | GIBCO | 12484010 |
| Fetal Bovine Serum | Capricorn | FBS-22A |
| Protein G Sepharose® 4 Fast Flow | GE Healthcare | 17–0618-01 |
| O.C.T. Compound | Scigen | 4583 |
| Fluorescence Mounting Medium | Dako | S3023 |
| EndoFree Plasmid Maxi Kit | QIAGEN | 12362 |
| Amicon Ultra-0.5 Centrifugal Filter Unit | Millipore | UFC501096 |
Recombinant V5-HP affinity purification
293T cells expressing V5-tagged HP were lysed in a buffer consisting of 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 20% glycerol, 4mM MgCl2, 0.4mM EDTA, and a protease inhibitor cocktail (Millipore). Supernatant fractions of the cells obtained after centrifugation at 13,200 rpm for 10 min were incubated with anti-V5 antibodies at 4°C for 16 h, and then with protein-G Sepharose (GE Healthcare) beads for 2 h. The protein-G Sepharose immune complexes were washed five times with a wash buffer (20 mM Tris-HCl (pH 7.9), 150 mM NaCl, 2mM MgCl2, 0.2mM EDTA, 0.1% NP40, and a protease inhibitor) before V5-tagged HPs were eluted from the protein-G Sepharose beads in the wash buffer containing 0.25 mg/ml V5 peptide. The V5 peptide were then removed by Amicon Ultra centrifugal filter device (Millipore).
Retinal explants and axon growth analysis
Retinal explants were prepared as it was described in our previous report (Kim et al., 2014). Briefly, retinas were isolated from E13.5 mouse embryos and explants were added to a collagen mixture and positioned on plates coated with poly-L-lysine (10 μg/ml) and laminin (10 μg/ml). The explants were then incubated at 37°C for 1h to allow gelling before adding Neurobasal medium containing B27 supplement (Invitrogen). The explants were cultured for 48h before treating with proteins or co-cultured with 293T cell aggregates for 48h.
QUANTIFICATION AND STATISTICAL ANALYSIS
Multiple sequence alignment identifying conserved motifs
To examine sequence features associated with HP secretion, we collected HP protein sequences from UniProt database (The UniProt Consortium, 2017). The multiple sequence alignment was then built by using the MUSCLE (Multiple Sequence Comparison by Log- Expectation) software with the default option, which is designed to give the most accurate gapped alignment (Edgar, 2004). We split the MSA (multiple sequence alignment) results to two sub-MSAs for sHP and nsHP sequences with different RSI cut-offs, respectively, and calculate the sequence profiles, which represent the amino acid frequencies at each MSA column. To identify the distinctive MSA columns discriminating sHP and nsHP sequences, the difference between two sub-MSAs at the MSA column i was calculated by using the JS-divergence defined as
where pi(k) and qi(k) represent the frequencies of the amino acid k at the MSA column i in the sub-MSAs for sHP and nsHP sequences, respectively. λ is 0.5, and ri(k) is defined as (pi(k)+qi(k)) = 2. The MSA columns, at which the number of aligned sHP and nsHP sequences are ≥ 10 in the sub-MSAs and the JS-divergence is > 1, are selected as the distinctive MSA columns. The sequence patterns at 133th, 348th, and 1207th positions are shown as the sequence logos (Crooks et al., 2004), where the heights represent the relative entropy.
Statistical analysis
Statistical tests were performed using Prism Software (GraphPad; v5.0) measurement tools. All data from statistical analysis are presented as the mean ± STE. Comparison between two groups was done by unpaired Student’s t test, and the differences among multiple groups were determined by analysis of variance (ANOVA) with Tukey’s post-test used to determine the significant differences among multiple groups. P values < 0.01 were considered as statistically significant results.
Supplementary Material
Highlights.
Intercellular transfer is a general feature of homeodomain proteins (HPs)
HP secretion is a cell-context-sensitive event
Hydrophobic residues outside of the homeodomain support the secretion of HP
ACKNOWLEDGMENTS
We thank Dr. Alain Prochiantz for valuable comments to the manuscript. We also thank MMRRC and GENSAT project for providing BAC-EGFP transgenic mice. This work was supported by the National Research Foundation of Korea (NRF) grants (NRF-2009-00424, NRF- 2017R1A2B3002862, and NRF-2018R1A5A1024261) funded by Korean Ministry of Science and ICT (MSIT), South Korea.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.06.056.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alvarez-Fischer D, Fuchs J, Castagner F, Stettler O, Massiani-Beaudoin O, Moya KL, Bouillot C, Oertel WH, Lombès A, Faigle W, et al. (2011). Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat. Neurosci 14, 1260–1266. [DOI] [PubMed] [Google Scholar]
- Apulei J, Kim N, Testa D, Ribot J, Morizet D, Bernard C, Jourdren L, Blugeon C, Di Nardo AA, and Prochiantz A (2019). Non-cell Autonomous OTX2 Homeoprotein Regulates Visual Cortex Plasticity Through Gadd45b/g. Cereb. Cortex 29, 2384–2395. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Bui A, Ding Q, and Gan L (2014). Expression of LIM-homeodomain transcription factors in the developing and mature mouse retina. Gene Expr. Patterns 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurdeley M, Spatazza J, Lee HH, Sugiyama S, Bernard C, Di Nardo AA, Hensch TK, and Prochiantz A (2012). Otx2 binding to perineuronal nets persistently regulates plasticity in the mature visual cortex. J. Neurosci 32, 9429–9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet I, Weinl C, Piper M, Trembleau A, Volovitch M, Harris W, Prochiantz A, and Holt C (2005). The transcription factor Engrailed-2 guides retinal axons. Nature 438, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR (2011). Homeodomain subtypes and functional diversity. Subcell. Biochem 52, 95–122. [DOI] [PubMed] [Google Scholar]
- Christiaens B, Symoens S, Verheyden S, Engelborghs Y, Joliot A, Prochiantz A, Vandekerckhove J, Rosseneu M, and Vanloo B (2002). Tryptophan fluorescence study of the interaction of penetratin peptides with model membranes. Eur. J. Biochem 269, 2918–2926. [DOI] [PubMed] [Google Scholar]
- Conte I, Marco-Ferreres R, Beccari L, Cisneros E, Ruiz JM, Tabanera N, and Bovolenta P (2010). Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development 137, 2307–2317. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Miki K, Rattner A, Smallwood P, Zibetti C, Hirokawa K, Monuki ES, Campochiaro PA, and Blackshaw S (2012). Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc. Natl. Acad. Sci. USA 109, 4657–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toni A, Zbinden M, Epstein JA, Ruiz i Altaba A, Prochiantz A, and Caillé I (2008). Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural Dev. 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, and Oliver G (2003). Prox1 function controls progenitor cell population and horizontal cell genesis in the mammalian retina. Nat Genet. 34, 53–58. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Müller M, Affolter M, Percival-Smith A, Billeter M, Qian YQ, Otting G, and Wüthrich K (1990). The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329. [DOI] [PubMed] [Google Scholar]
- Görlich D, and Kutay U (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Heintz N (2004). Gene expression nervous system atlas (GENSAT). Nat. Neurosci 7, 483. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, and Bruford EA (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, and Kageyama R (2002). Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development 129, 831–842. [DOI] [PubMed] [Google Scholar]
- Joliot A, Pernelle C, Deagostini-Bazin H, and Prochiantz A (1991). Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 88, 1864–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW, and Sanes JR (2011). Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat. Neurosci 14, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Min KW, Kang KH, Lee EJ, Kim HT, Moon K, Choi J, Le D, Lee SH, and Kim JW (2014). Regulation of retinal axon growth by secreted Vax1 homeodomain protein. eLife 3, e02671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, and Lemke G (2006). Hedgehog-regulated localization of Vax2 controls eye development. Genes Dev. 20, 2833–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim SJ, Sohn YI, Paik SS, Caplette R, Simonutti M, Moon KH, Lee EJ, Min KW, Kim MJ, et al. (2015). Mitochondrial protection by exogenous Otx2 in mouse retinal neurons. Cell Rep. 13, 990–1002. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lim S, Ha T, Song YH, Sohn YI, Park DJ, Paik SS, Kim-Kaneyama JR, Song MR, Leung A, et al. (2017). The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 α-enhancer activity. eLife 6, e21303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JS, Kim WI, Kang HC, Kim SH, Park AH, Park EK, Cho YW, Kim S, Kim HM, Kim JA, et al. (2015). Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat. Med 21, 395–400. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, and Hake S (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Maizel A, Bensaude O, Prochiantz A, and Joliot A (1999). A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development 126, 3183–3190. [DOI] [PubMed] [Google Scholar]
- Miyata S, Komatsu Y, Yoshimura Y, Taya C, and Kitagawa H (2012). Persistent cortical plasticity by upregulation of chondroitin 6-sulfation. Nat. Neurosci 15, 414–422. [DOI] [PubMed] [Google Scholar]
- Passini MA, Levine EM, Canger AK, Raymond PA, and Schechter N (1997). Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J. Comp. Neurol 388, 495–505. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, and McGinnis W (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet 6, 893–904. [DOI] [PubMed] [Google Scholar]
- Pooga M, and Langel Ü (2015). Classes of cell-penetrating peptides. Methods Mol. Biol 1324, 3–28. [DOI] [PubMed] [Google Scholar]
- Prochiantz A, and Di Nardo AA (2015). Homeoprotein signaling in the developing and adult nervous system. Neuron 85, 911–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekaik H, Blaudin de Thé FX, Fuchs J, Massiani-Beaudoin O, Prochiantz A, and Joshi RL (2015). Engrailed Homeoprotein Protects Mesencephalic Dopaminergic Neurons from Oxidative Stress. Cell Rep. 13, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, and Roska B (2009). Genetic address book for retinal cell types. Nat. Neurosci 12, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Chao R, Vacik T, Yahyavi M, Abouzeid H, Bardakjian T, Schneider A, Shaw G, Sherr EH, Lemke G, et al. (2012). VAX1 mutation associated with microphthalmia, corpus callosum agenesis, and orofacial clefting: the first description of a VAX1 phenotype in humans. Hum. Mutat 33, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatazza J, Di Lullo E, Joliot A, Dupont E, Moya KL, and Prochiantz A (2013). Homeoprotein signaling in development, health, and disease: a shaking of dogmas offers challenges and promises from bench to bed. Pharmacol. Rev 65, 90–104. [DOI] [PubMed] [Google Scholar]
- Sugiyama S, Di Nardo AA, Aizawa S, Matsuo I, Volovitch M, Prochiantz A, and Hensch TK (2008). Experience-dependent transfer of Otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell 134, 508–520. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 (D1), D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacik T, Stubbs JL, and Lemke G (2011). A novel mechanism for the transcriptional regulation of Wnt signaling in development. Genes Dev. 25, 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizenmann A, Brunet I, Lam J, Sonnier L, Beurdeley M, Zarbalis K, Weisenhorn-Vogt D, Weinl C, Dwivedy A, Joliot A, et al. (2009). Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron 64, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, and Holt CE (2012). Local translation of extranuclear lamin B promotes axon maintenance. Cell 148, 752–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


