Abstract
We identified in Listeria monocytogenes a gene encoding a protein homologous to MecA, a regulatory protein acting with ClpC and ComK in the competence pathway of Bacillus subtilis. In L. monocytogenes, MecA is involved, along with ClpC and ClpP, in the downregulation of a 64-kDa secreted protein. In B. subtilis, the MecA protein of L. monocytogenes behaves as a regulatory protein, controlling the transcription of comK and comG. Complete or disrupted ComK homologues were also found in L. monocytogenes. However, we failed to detect competence in various strains of L. monocytogenes, including those with intact ComK. Our results suggest that the functions of MecA in the saprophytes L. monocytogenes and B. subtilis have presumably diverged in response to their respective ecological niches.
The gram-positive bacterium Listeria monocytogenes is a food-borne pathogen widely spread in the environment, where it survives hostile conditions, presumably due to a rapid stress response. Among the stress proteins characterized for L. monocytogenes, the Clp ATPases are members of the HSP-100–Clp family belonging to a highly conserved class of universal molecular chaperones involved in the stress resistance and virulence of this pathogen (4, 14, 16). A homologue of ClpC (formerly designated MecB) in Bacillus subtilis acts both as a general stress protein and as a regulatory factor controlling the expression of competence (8). In the competence pathway, ClpC forms a complex with MecA to negatively regulate ComK (19), a transcriptional activator controlling the late competence genes required for the binding, processing, and internalization of transforming DNA (2). In the absence of ComS, MecA recruits ComK to the ClpC-ClpP proteolytic complex. When ComS is present, MecA is degraded by this complex (18, 19). The identification of ClpC (11, 12) and ClpP (3) in L. monocytogenes suggested that these proteins might have functions similar to those of their B. subtilis homologues. In this work, we searched in L. monocytogenes for MecA and other homologues of the B. subtilis competence pathway.
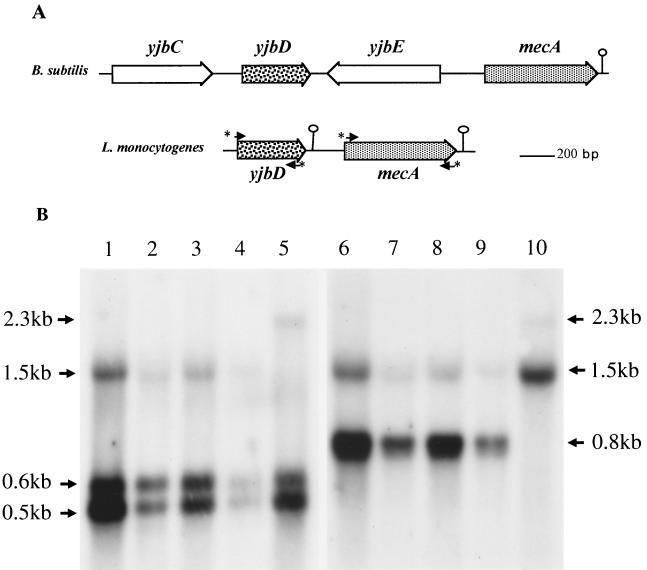
The mecA locus in L. monocytogenes.
A 3-kb fragment was cloned and sequenced from an XbaI-digested genomic library constructed in pUC19 from L. monocytogenes (strain LO28) and screened with a 584-bp intragenic B. subtilis mecA probe by colony hybridization at low stringency (50°C). We found an open reading frame (ORF) (GenBank accession no. AF103794) encoding a putative protein of 217 amino acids and showing 49% and 42% identities with MecA of B. subtilis and Bacillus firmus, respectively, and 33% identity with YpbH of B. subtilis, a MecA homologue of unknown function. Another ORF, located upstream, encodes a putative protein of 144 amino acids and 82% identical to YjbD of B. subtilis, of unknown function (Fig. 1A). Alignment of the MecA sequences of B. subtilis and L. monocytogenes revealed two conserved domains, at the N terminus (up to residue 78) and the C terminus (after residue 125), with 74 and 45% identities, respectively, and separated by a variable spacer region (data not shown). In B. subtilis, the N-terminal domain binds ComK and ComS and the C-terminal domain is involved in binding ClpC (10). MecA orthologues have also been identified in The Institute for Genomic Research genome database for several species of gram-positive bacteria, including Streptococcus pyogenes, Streptococcus pneumoniae, Streptococcus mutans, and Staphylococcus aureus (10). Using specific primers for mecA and yjbD, we showed by PCR that these genes are highly conserved and adjacent in strains of L. monocytogenes (EGD-E, ATCC 19115, ATCC 19111, CNL880203, CHUT861141, CNL895793, CNL895795, INRA119, and INRA85), Listeria ivanovi (ATCC 19119 and SLCC2379), Listeria innocua (ATCC 33090, CHUT861158, and INRA86), Listeria seeligeri (CHUT860478, CHUT861166, and CHUT861167), and Listeria welshimeri (CHUT860477) (data not shown). These strains were previously described (20).
FIG. 1.
(A) MecA regions in B. subtilis and L. monocytogenes LO28. Small arrows with asterisks indicate the positions of the primers used for PCR amplification of the mecA and yjbD regions. (B) Northern blot analysis of total RNA extracted from strain LO28 grown in BHI broth at 37 and 42°C. RNA samples were separated and hybridized with an intragenic yjbD probe (403 bp) (lanes 1 to 5) or mecA probe (584 bp) (lanes 6 to 10). These probes were obtained by PCR from the chromosomal DNA of LO28: yjbD, primers 5′-CCCGATAAGGAGTGTGAATG-3′ and 5′-GCGCTTCACGTAGTTGATACG-3′; mecA, primers 5′-CCCTTCATTGTCAATGAC-3′ and 5′-ACTAACGGCATTGTCAATG-3′. Lanes: 1 and 6, 37°C, exponential phase; 2 and 7, LO28, 37°C, stationary phase; lanes 3 and 8, LO28, 42°C, exponential phase; 4 and 9, LO28, 42°C, stationary phase; 5 and 10, mecA mutant, 37°C, exponential phase. The locations and sizes of mRNAs are indicated by arrows.
Transcriptional analysis of the mecA locus.
Northern blot analysis of total RNA from strain LO28 grown in brain heart infusion (BHI) broth at 37 and 42°C was performed with intragenic probes for mecA and yjbD. These two genes are strongly expressed during the exponential growth phase at 37°C (Fig. 1B, lanes 1 and 6) and more weakly expressed in the stationary phase (lanes 2 and 7). Transcription of these genes was not induced at an elevated temperature in the exponential phase (Fig. 1B, lanes 3 and 8) or the stationary phase (lanes 4 and 9), in contrast to that of clpC (12). We found two mecA transcripts of ∼0.8 and ∼1.5 kb and three yjbD transcripts of ∼0.5, ∼0.6, and ∼1.5 kb. The detection of a 1.5-kb transcript by both probes strongly suggests that mecA and yjbD are cotranscribed as an operon. This notion was confirmed by the results of a transcriptional analysis of a mecA::aphA-3 mutant of LO28 (described below) showing an 0.8-kb increase in the size of the larger transcript (2.3 kb), corresponding to the insertion of the kanamycin resistance cassette (aphA-3) into mecA (Fig. 1B, lanes 5 and 10). The 0.8-kb mecA transcript is also increased in size by the aphA-3 insertion into mecA (Fig. 1B, lane 10), yielding a 1.5-kb transcript.
Construction and phenotypic analysis of a mecA mutant.
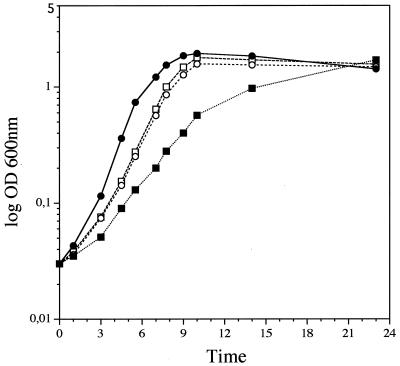
A mecA mutant (mecA::aphA-3′) was constructed from LO28 by deletion of a 225-bp internal fragment (nucleotides 199 to 423) and insertion of a promoterless aphA-3′ gene into the mecA gene using a previously described procedure (3). Using the same strategy, we repeatedly failed to obtain a yjbD mutant of this strain; such a mutation might be lethal for the bacterium. Then, the mecA mutant was complemented using plasmid pAT18 (17) harboring mecA and its promoter region (1,123 bp). Controls included mutant and wild-type LO28 transformed with pAT18. There was no difference between the mecA mutant and the wild-type bacterium with regard to morphology during the exponential and stationary growth phases at 4, 30, 37, and 42°C; motility at 22°C; hemolysis on blood agar plates; and metabolic profiles on API strips (Biomerieux, Marcy l'Etoile, France). However, the exponential growth of the mecA mutant in BHI broth at 37°C was much slower than that of the wild-type bacterium. As measured by optical density, the generation time was almost twice that of the wild-type strain (1 h versus 0.5 h) but ultimately reached a similar value at the end of the exponential growth phase (Fig. 2). No difference in bacterial growth was observed between LO28 and the mutant transformed or not transformed by pAT18 alone (data not shown), indicating that the multicopy plasmid itself does not restrict bacterial growth. Transformation of the mecA mutant with pAT18-mecA partially restored bacterial growth at 37°C. A similar growth curve was found for LO28 transformed with pAT18-mecA, suggesting that large amounts of MecA might be responsible for this apparent incomplete restoration of growth (Fig. 2). The same growth curves were obtained by measuring CFU, showing a good correspondence between viability and optical density (data not shown). So, the absence of MecA in L. monocytogenes did not result in a loss-of-viability phenotype, in contrast to the situation for B. subtilis (5). We also constructed, with the same strategy (3), a double mecA clpC mutant from a previously described clpC mutant of LO28 (11); the double mutant displayed a phenotype similar to that of the clpC mutant at 42°C. (data not shown).
FIG. 2.
Growth of L. monocytogenes LO28 mecA mutant at 37°C and complementation. Symbols: ●, LO28/pAT18; ○, LO28/pAT18-mecA; ■, LO28 mecA mutant/pAT18; □, LO28 mecA mutant/pAT18-mecA. Bacteria were grown in BHI medium, and the optical density (OD) at 600 nm was measured at various intervals.
MecA, ClpC, and ClpP of L. monocytogenes downregulate a 64-kDa secreted protein.
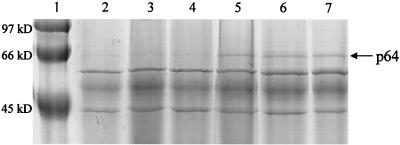
As MecA of B. subtilis is a negative regulatory protein, we compared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis the total and secreted protein profiles of mecA mutant and wild-type bacteria by using a procedure previously described (13). We found overexpression of a 64-kDa protein (p64) in the culture supernatant of the mecA mutant compared to LO28 (Fig. 3, lanes 5 and 2, respectively). An internal fragment of digested p64 was separated on DEAE-C18 and sequenced as VISEPAVTTPVTLSD (J. D'Allayer, Institut Pasteur, Paris, France). According to the Listeria genome database (strain EGD-E), this fragment corresponds to an ORF encoding a putative protein of 569 amino acids (63.4 kDa). This ORF was amplified by PCR and sequenced in LO28 (GenBank accession no. AF282221), and the deduced protein showed 100% identity with p64 from strain EGD-E. The p64 protein contains a signal sequence and two repeated domains with significant identity (∼32%) to pXO1-88, a putative protein of unknown function encoded by virulence plasmid pXO1 of Bacillus anthracis. The expression of p64 was further examined with clpC and clpP mutants (3, 11) to determine whether MecA, ClpC, and ClpP had a common role in the regulation of this protein, as for B. subtilis ComK. The amount of p64 was also increased in these mutants (Fig. 3, lanes 6 and 7) but not in two other mutants (prfA and oppA) from strain LO28, used as controls (Fig. 3, lanes 3 and 4). These results suggest that MecA might be a regulatory protein, acting with ClpC and ClpP to downregulate a 64-kDa secreted protein.
FIG. 3.
A secreted 64-kDa protein accumulates in the culture supernatant of L. monocytogenes in the absence of MecA, ClpC, or ClpP. Bacteria were cultured in BHI medium until mid-log growth phase, and supernatants were collected and filtered. Supernatant protein extracts were prepared by trichloroacetic acid precipitation as previously described (13). Extracts with equal protein concentrations were applied to a sodium dodecyl sulfate–10% polyacrylamide gel and subjected to electrophoresis. Lane 1, molecular size marker; lane 2, LO28; lane 3, LO28 oppA mutant (unpublished data); lane 4, LO28 prfA mutant (12); lane 5, LO28 mecA mutant; lane 6, LO28 clpC mutant; lane 7, LO28 clpP mutant.
L. monocytogenes MecA inhibits comK and comG transcription in B. subtilis.
We studied the function of L. monocytogenes MecA in B. subtilis by using two B. subtilis strains, QB4673 and QB4842, in which transcriptional lacZ fusions with the promoter region from B. subtilis comK or comG were integrated as single copies at the amyE locus (8). Into these two strains, we introduced as a single copy in the thrC locus the mecA gene of L. monocytogenes LO28 under the control of the xylose-inducible promoter. We then disrupted mecA of B. subtilis by allelic replacement and integration of an aphA-3 cassette, yielding strains QB8066 and QB8067 (for the methodology used, see reference 9). When bacteria were grown at 37°C in Luria-Bertani broth without xylose (mecA null mutant background), transcription of comK and comG was strongly induced (reaching in about 3 h ∼600 to 700 U mg of protein−1; in contrast, transcription was strongly downregulated by the addition of xylose (10-fold decrease) (data not shown). These results indicate that L. monocytogenes MecA is functional in the competence regulatory cascade of B. subtilis.
Identification of comK in L. monocytogenes.
The previous results led us to search for comK in L. monocytogenes. From the Listeria genome database (strain EGD-E), we found in EGD-E a comK-like truncated gene cleaved into two parts and separated by a 42-kb region containing several ORFs encoding phage-related products. Some were very similar to ORFs from the recently sequenced bacteriophage A118 of L. monocytogenes, which was also found to be integrated in a comK-like gene (6). The comK-like truncated gene from strain LO28 was sequenced (GenBank accession no. AF191727) and was almost identical to that in EGD-E. Upstream from ′comK, the last gene of the phage was int, encoding a protein of 472 amino acids and 95% identical to the putative integrase of bacteriophage A118. The inactivation of comK might therefore explain the previous failure to demonstrate competence in LO28 (12). So, we screened by Southern blotting with intragenic probes for ′comK and int the 18 strains of Listeria used here. comK was present in all strains tested, whereas int was present in only 5 of 10 L. monocytogenes strains, including EGD-E, and was absent from the other Listeria strains, except for 1 of 3 strains of L. innocua. comK from three integrase-negative strains was sequenced. The ComK proteins of L. monocytogenes strains ATCC 19115 and CNL895793 (GenBank accession no. AF191725 and AF191724, respectively) were almost identical proteins of 202 amino acids (99% identity), sharing 32% identity with ComK of B. subtilis. ComK of L. seeligeri strain CHUT860478 (GenBank accession no. AF191726) was 199 amino acids long and 33% identical to ComK of B. subtilis.
Competence tests in L. monocytogenes.
Two L. monocytogenes strains with complete comK and their isogenic mecA deletion mutants, constructed as described above, were tested for competence in a two-step nutrient shiftdown procedure previously described (7), except for the composition of the competence minimal medium; this medium was adapted to Listeria by the addition (per liter) of l-leucine (100 mg), l-isoleucine (100 mg), l-valine (100 mg), l-methionine (100 mg), l-arginine (100 mg), l-cysteine (100 mg), l-histidine (100 mg), riboflavin (4 mg), biotin (4 mg), thiamine (1 mg), and thioctic acid (0.01 mg). Plasmid pMK4 was added to cultures in exponential or stationary growth phase at a concentration of 1 or 10 μg ml−1, and transformants were selected on BHI agar supplemented with chloramphenicol at 10 μg ml−1. B. subtilis strain 168, used as a control, was efficiently transformed with pMK4, even in the competence medium adapted for Listeria, but we repeatedly failed to transform Listeria, suggesting that this microorganism is not competent under the conditions tested. It remains possible that Listeria requires unusual conditions for competence. However, it is important to recall that high-level natural transformation of B. subtilis could be demonstrated only for a few strains isolated following extensive UV and X-ray mutagenesis (1, 15). The highly transformable strain 168 was then chosen for most studies. Thus, the situation for L. monocytogenes might be reminiscent of that for B. subtilis, with a cryptic DNA uptake apparatus presumably allowing only a very low level of natural transformation in its natural habitat.
From the Listeria genome sequence database, we found homologues for most of the early competence proteins (AbrB, CodY, DegU, SpoOKA, SpoOKB, SpoOKC, SpoOKD, and SpoOKE) and late competence proteins (ComC, ComEA, ComEB, ComEC, ComER, ComFA, ComFC, ComGA, ComGB, ComGC, ComGD, and ComGF) of B. subtilis, which form the surface structure capable of internalizing DNA. However, several homologues for early and late competence proteins involved in the competence of B. subtilis are absent in L. monocytogenes, including ComQ, ComS, ComX, ComFB, ComGE, and ComGG. The alternative is that these proteins may be involved in other, still-undefined functions in L. monocytogenes, presumably related to the transduction of environment signals. Our results suggest that the saprophytic species L. monocytogenes and B. subtilis have developed and adapted similar and complex regulatory cascades in response to specific environmental signals encountered in their respective ecological niches.
Acknowledgments
We thank J. L. Beretti for technical assistance in protein analysis, P. Velge for providing some Listeria strains, A. Charbit for critical reading of the manuscript, P. Trieu-Cuot for providing vector pAT18, and G. Rapoport (part of this work was carried out in his laboratory).
E.B. received a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie. This work was supported by INSERM, The University of Paris V, and two grants from the European Commission (contracts ERBCHRXCT 94-0451 and CT980036).
We also thank the European Listeria Genome Consortium, composed of Philippe Glaser, Alexandra Amend, Fernando Baquero-Mochales, Patrick Berche, Helmut Bloecker, Petra Brandt, Carmen Buchrieser, Trinad Chakraborty, Alain Charbit, Elisabeth Couvé, Antoine de Daruvar, Pierre Dehoux, Eugen Domann, Gustavo Dominguez-Bernal, Lionel Durant, Karl-Dieter Entian, Lionel Frangeul, Hafida Fsihi, Francisco Garcia del Portillo, Patricia Garrido, Werner Goebel, Nuria Gomez-Lopez, Torsten Hain, Joerg Hauf, David Jackson, Jurgen Kreft, Frank Kunst, Jorge Mata-Vicente, Eva Ng, Gabriele Nordsiek, José Claudio Perez-Diaz, Bettina Remmel, Matthias Rose, Christophe Rusniok, Thomas Schlueter, José-Antonio Vazquez-Boland, Hartmut Voss, Jurgen Wehland, and Pascale Cossart.
REFERENCES
- 1.Burkholder P R, Giles N H., Jr Induced biochemical mutations in B. subtilis. Am J Bot. 1947;34:345–348. [PubMed] [Google Scholar]
- 2.Dubnau D. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene. 1997;192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 3.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol Microbiol. 2000;35:1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn J, Bylund J, Haines M, Higgins M, Dubnau D. Inactivation of mecA prevents recovery from the competent state and interferes with cell division and the partitioning of nucleoids in Bacillus subtilis. Mol Microbiol. 1995;18:755–767. doi: 10.1111/j.1365-2958.1995.mmi_18040755.x. [DOI] [PubMed] [Google Scholar]
- 6.Loessner M J, Inman R B, Lauer P, Calendar R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol. 2000;35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 7.Msadek T, Dartois V, Kunst F, Herbaud M L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 8.Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair S, Derre I, Msadek T, Gaillot O, Berche P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol Microbiol. 2000;35:800–811. doi: 10.1046/j.1365-2958.2000.01752.x. [DOI] [PubMed] [Google Scholar]
- 10.Persuh M, Turgay K, Mandic-Mulec I, Dubnau D. The N- and C-terminal domains of MecA recognize different partners in the competence molecular switch. Mol Microbiol. 1999;33:886–894. doi: 10.1046/j.1365-2958.1999.01544.x. [DOI] [PubMed] [Google Scholar]
- 11.Rouquette C, de Chastellier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 12.Rouquette C, Ripio M T, Pellegrini E, Bolla J M, Tascon R I, Vazquez-Boland J A, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 15.Spizizen J. Transformation of biochemically deficient strains of B. subtilis by deoxyribonuclease. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer DNA from Escherichia coli to gram-positive bacteria. Gene. 1991;102:99–104. doi: 10.1016/0378-1119(91)90546-n. [DOI] [PubMed] [Google Scholar]
- 18.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turgay K, Hamoen L W, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- 20.Van Langendonck N, Bottreau E, Bailly S, Tabouret M, Marly J, Pardon P, Velge P. Tissue culture assays using Caco-2 cell line differentiate virulent from non-virulent Listeria monocytogenes strains. J Appl Microbiol. 1998;85:337–346. doi: 10.1046/j.1365-2672.1998.00515.x. [DOI] [PubMed] [Google Scholar]