If asked to identify a favorite paper, I would choose “General nature of the genetic code for proteins” (12). The work is purely genetic and focuses on the rIIA and rIIB genes of bacteriophage T4. It is especially popular with geneticists because it reports “about eighty independent r mutants, all suppressors of FC O, or suppressors of suppressors, or suppressors of suppressors of suppressors.” Armed with only toothpicks and logic, the authors made an impressive number of fundamental discoveries, all described in six beautifully written pages. They characterized frameshift mutations, and they provided compelling evidence that the genetic code was triplet in nature and degenerate and that genes are read from a fixed starting point. In addition, and more to the point of this commentary, they described the first experimental use of a gene fusion.
Actually, Crick et al. (12) did not isolate the gene fusion employed. Benzer and Champe (5) isolated this fusion, which is called deletion r1589. (Unfortunately, such free exchange of ideas and reagents is less common today.) In wild-type T4, the rIIA and rIIB genes are adjacent, and they are transcribed independently from the same DNA strand. Deletion r1589 fuses rIIA to rIIB, resulting in a hybrid gene. This hybrid gene produces a hybrid protein that contains a nonfunctional, amino-terminal fragment of rIIA fused to a large, functional, carboxy-terminal fragment of rIIB.
Today fusions like rIIA-rIIB are often called translational fusions (in contrast to transcriptional fusions; see below) because the production of hybrid protein with functional rIIB activity depends upon the transcription and translation start signals of rIIA.
Crick et al. (12) used r1589 to demonstrate a fixed starting point for gene expression. Frameshift mutations are chain terminating, because they alter the reading frame, and sooner or later, a nonsense codon is encountered in the incorrect frame. In wild-type T4, frameshift mutations in rIIA never affect rIIB expression because the genes are transcribed and translated independently. In r1589, frameshift mutations in rIIA prevent the expression of rIIB activity because they prevent synthesis of the rIIA-rIIB hybrid protein. This proves that in r1589 the synthesis of rIIB activity is driven by the transcription and translation start signals in rIIA. In their original description of r1589, Benzer and Champe (5) used the same logic to show that nonsense mutations are chain terminating and that nonsense suppressors fix the problem. The power and utility of the gene fusion approach is emphasized by the fact that at the time these papers were written, no one could detect the rII proteins. Indeed, the only evidence indicating that these genes coded for proteins was the very existence of frameshift and nonsense mutations. Accordingly, with this experimental system, there was no other way with this experimental system to address the issues of starting point, chain termination, and nonsense suppression.
In a similar time frame, Jacob and Monod were accumulating evidence to support their operon hypothesis. As stated by Hayes (18) in his remarkably comprehensive text on bacterial and phage genetics: “A prediction of the operon hypothesis is that, while deletions involving the operator and the first gene of the sequence should inactivate all the genes of the operon, a proximal extension of the deletion might physically connect these genes to another operon so that they would be activated again, but their activity would now be responsive to a different biochemical control.” This is exactly what was done by Ames, Hartman, and Jacob (1) with the his operon of Salmonella enterica serovar Typhimurium. Deletions that removed the 5′ end of the his operon and part of the first gene, hisG, affected the synthesis of all nine genes in the operon. Pseudorevertants, which expressed all of the genes except hisG, were selected by demanding growth on histidinol. Histidinol is converted to histidine by the last enzyme in the biosynthetic pathway, which is specified by the second gene in the operon, hisD. One class of pseudorevertants contained a second deletion that fuses hisD and the rest of the his operon to an unknown upstream promoter.
Today fusions like these hisD fusions are often called transcriptional fusions. Such fusions do not result in the production of a hybrid protein but rather just place the gene(s) in question under the control of a different promoter.
LAC FUSIONS: THE EARLY YEARS
Shortly after the work of Ames et al. (1), Beckwith (2) isolated the first lac operon fusions. Starting with an Escherichia coli strain carrying a strongly polar mutation early in the first gene in the operon, lacZ, he sought mutations that would restore expression of the second gene in the operon, lacY, by demanding growth on melibiose at high temperatures (the Lac permease is promiscuous, and the melibiose permease of E. coli K-12 is temperature sensitive). One class of mutations that answered this selection was deletions that fused lacY to an unknown upstream promoter.
Jacob, Ullman, and Monod (21) took lac fusions one step further by fusing lacY to the purE operon. This was done by using a variation of the Beckwith approach. To avoid complications caused by deleting essential genes, they started with a merodiploid strain with the following genotype: F′ lacA+Y+Z+IS tsx+ pur+/lacIS tsx. (The lacIS allele specifies superrepressor, which does not respond to inducer.) Selection was made simultaneously for growth on melibiose and resistance to phage T6. Deletions that extend into the lac operon and remove the lacIS and tsx (the gene for the T6 receptor) genes on the episome can answer this selection. Among these one was found that fused lacY to purE. In this strain synthesis of Lac permease is repressed by the addition of excess adenine.
Maxime Schwartz (30) might best sum up the reaction of the bacterial genetics community to the purE-lacY+ fusion. At the time this fusion was isolated, he was a graduate student with Monod, and he learned of this fusion directly from an excited Jacob. Maxime writes: “To tell the truth, I was not impressed. From what I had just been taught at the university the possibility of isolating such fusions was an obvious corollary of what was known of operons. For me, the operon was a fact. For Francois Jacob, it was still a theory.”
Part of the reason for this less than enthusiastic endorsement of genetic fusions stems from the method used to isolate them. What the rIIA-rIIB, hisD, and lacY fusions all have in common is the fact that they were created by deletion events. Accordingly, the genes in question, e.g., rIIA and rIIB or purE and lacY, must be relatively close to one another in the chromosome, and they must be transcribed from the same DNA strand. These requirements severely limited the general utility of the method, and it left many people thinking that fusions were very interesting but esoteric.
In order to make genetic fusions more generally useful, scientists had to learn to manipulate genes more precisely. Cuzin and Jacob (13) took the first step in this direction. In strains carrying a chromosomal lac deletion they showed that F′lac with a temperature-sensitive replication defect (FTSlac) could be integrated at several different places in the chromosome by selecting Lac+ at high temperatures. This was evidenced by the fact that the Hfr strains so formed could transfer various regions of the chromosome at high frequency. In effect, this method allows the transposition of the lac genes to different chromosomal locations. We know today that this transposition is a process mediated by IS elements normally present in F.
Two postdocs in Jacob's lab, Jon Beckwith and Ethan Signer, refined the FTSlac method by figuring out a way to direct transposition of the lac genes to particular chromosomal locations (3). Influenced by the Campbell model (7) for λ integration, Beckwith and Signer reasoned that FTSlac integration into a chromosomal gene would destroy that gene. Assuming that FTSlac integration was mutagenic, they could then direct integration into a particular gene by selecting simultaneously for Lac+ and loss of target gene function at high growth temperatures. They demonstrated their refined method by directing FTSlac to the gene now called tonB using resistance to phage T1 as the selection. Today this seems obvious. However, they had this impressive insight years before the characterization of insertion mutations and the discovery of IS elements in bacteria.
The tonB gene was an especially clever choice because it lies between the attachment site for φ80 and the genes of the trp operon. With lac transposed into tonB, Beckwith and Signer (3) were able to isolate φ80lac specialized transducing phage using the same logic and basically the same methods that Morse, Lederberg, and Lederberg (26) used to isolate λgal. In effect, again with just toothpicks and logic, Beckwith and Signer (3) had cloned the lac operon. As noted by Miller (25), “In my opinion, this work was really the beginning of genetic engineering, since it stimulated a search for many different transducing phages, and for different ways of obtaining them, and put the spotlight on the advantages of cloning genes.”
Integration of φ80lac at att80 transposes the lac genes to a chromosomal location near tonB. Franklin, Dove, and Yanofsky (15) had shown that many mutations conferring resistance to phage T1 were tonB deletions, some of which extended into the trp operon on one side and/or att80 on the other. The fact that lac was now at att80 provided a means to isolate deletions that entered the lac operon. Moreover, some of these deletions generated trp-lac fusions (4). These deletions and trp-lac fusions were used extensively to help elucidate operon structure and detailed regulatory mechanisms for both trp and lac (for example, see the chapters by Beckwith, Miller, and Bassford et al. in reference 24). This work clearly demonstrated the utility of fusions for the genetic analysis of gene regulation.
I first encountered the Beckwith, Signer, and Epstein paper (4) as a young graduate student in the early 1970s. It had enormous impact on me. These guys could do practically anything they wanted with DNA, and this seemed a useful skill to acquire. Moreover, it didn't require fancy equipment or time in the cold room. I wasn't sure that I understood exactly how they did all these transpositions, deletions, and fusions, but I knew I wanted to learn.
MAKING LAC FUSIONS MORE GENERALLY USEFUL
When I started as a postdoc in Jon Beckwith's lab, Malcolm Casadaban was just finishing his Ph.D. thesis. Casadaban (8) had devised a general method for making lac fusions, and I had to learn it from him before he left the lab. What I remember most about my many conversations with Malcolm is the question, “Can you see this in your head, or do I have to draw it out?” This “vision thing” was the first new skill I had to acquire.
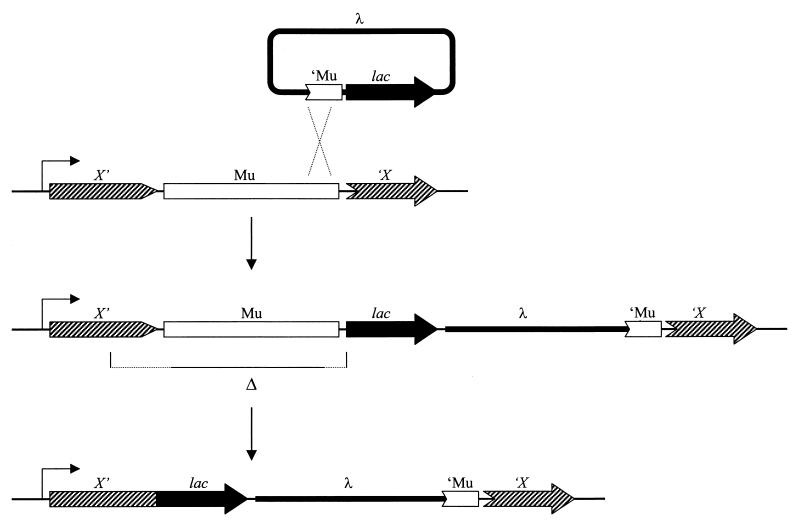
Casadaban's method (8), which is basically a three-step protocol, could be applied to any nonessential gene for which there was a selection or screen for loss of function. The first step utilizes the transposable bacteriophage Mu (named for mutator) (35). Using a lac deletion strain and phage Mu cts (carries a temperature-sensitive repressor) (20), one isolated a lysogen carrying the Mu cts prophage in the target gene of interest, gene X. This was done by selecting or screening for lysogens with an X− phenotype.
The second step uses homologous recombination to transpose the lac genes to the chromosomal region of interest defined by the Mu insertion. For this step, Malcolm constructed a λ specialized transducing phage in which the genes and cis-acting sites required for normal lysogenization had been deleted and which carried instead a piece of the left end of the Mu genome and a promoterless lac operon. When this λ phage infects the gene X::Mu cts lysogen, it will integrate preferentially by homologous recombination between the Mu DNA sequences. If the Mu prophage was in the proper orientation, then the promoterless lac genes will be positioned nearby and downstream from the promoter of gene X. However, in the double λ Mu lysogen, transcription of lac is blocked by the presence of the Mu cts prophage. (Can you see this in your head? Do I have to draw it out? [Fig. 1]).
FIG. 1.
The Casadaban method (8) for constructing lac fusions. Adapted from the Journal of Molecular Biology (8) with permission of the publisher.
In the final step, the double λ Mu lysogen is exposed to high growth temperatures to inactivate the Mu repressor, and Lac+ survivors are selected. Deletions that remove the lethal Mu prophage and fuse the lac genes to the promoter for gene X answer this selection. Nearly all of these are transcriptional fusions.
To obtain translational fusions that synthesize a gene X-LacZ hybrid protein, Casadaban (8) used a trick first developed by Muller-Hill and Kania (27). A strongly polar nonsense mutation near the 5′ end of the lacZ gene was introduced into the λlacMu phage. Double λ Mu lysogens of this lacZ mutant phage yield fewer Lac+ survivors at high temperatures. In this case the deletion that removed the Mu prophage must also enter lacZ, remove the nonsense codon, and fuse lacZ to the target gene in the correct reading frame.
Although outdated, it is instructive to see how Casadaban's three-step method (8) for making lac fusions builds on and extends the early work with fusions. Moreover, it provides a beautiful example of genetic engineering without restriction enzymes. To test your skills, try and follow the many steps involved in the construction of λp1(209), the most useful λlacMu phage. It's not easy.
Despite the fact that successful application of the three-step method required some experience with bacterial and phage genetics, it provided a powerful experimental tool to many scientists. The impact was large. Indeed, everyone who wanted to construct fusions used the lac deletion strain MC4100 that Casadaban built for this work. Today, MC4100 is one of the most commonly used “wild-type” strains.
As a postdoc with Stanley Cohen, Casadaban used a clever combination of genetics and recombinant DNA technology to build a specialized Mu-lac transducing phage that could generate transcriptional fusions in a single step (10). Subsequently, Casadaban and Chou (9) also constructed a Mu-lac phage that could generate translational fusions. Now all one had to do to get fusions was to mix a lysate of this phage with a suspension of E. coli Δlac cells and then select simultaneously for Lac+ and loss of target gene function. These advances made lac fusions generally available, and the increased use of lac fusions in turn stimulated the development of novel applications of fusion technology (see below).
LAC FUSIONS TODAY
Mu-lac was the first example of a fusion-generating transposable genetic element. Today there are many such elements (34). For example, there are specialized λ phages that integrate using the transposition machinery of Mu to generate fusions (6), and there are derivatives of Tn5 that generate lac fusions (23). The advent of PCR and the availability of complete genome sequences further simplify the construction process, and these advances extend the method to all manner of organisms. Fusions can now be made to any target gene very precisely without any prior knowledge of target gene function or mutant phenotypes (17).
USES OF LAC FUSIONS
We used to keep a list of novel uses for lac fusions (33), but this has become unwieldy. In any event this is not the place for a comprehensive listing. To illustrate the extreme utility and impact of this experimental tool, I will divide these uses into three general categories.
Regulation of gene expression.
Because β-galactosidase assays are easy, transcriptional and translational lac fusions have been used to study each and every step in gene expression from the initiation of transcription to stability of the protein product. Moreover, because the “lore” of lac genetics is vast, fusions have been used to identify regulatory genes and regulatory mutations. Fusions are so commonly employed today that it is rare to see a paper dealing with gene regulation in the Journal of Bacteriology that doesn't contain experiments that use them.
Tagging the gene and its product.
Unknown genes in complex regulons can be discovered by searching a population of random lac fusions for those that are regulated in common fashion. Kenyon and Walker (22) first demonstrated this using Casadaban's Mu-lac phage to identify new genes in the SOS regulon of E. coli.
The phenomenal success of lac fusions in bacteria inspired investigators working with more-complex organisms. A beautiful example of this is the development of “enhancer traps” in Drosophila (19, 28). Indeed, O'Kane and Gehring (28) credit the prokaryotic work in their first sentence, an intellectual courtesy that is not always extended by our “eukaryotic” colleagues.
Translational fusions tag an amino-terminal fragment of the target gene product with β-galactosidase, and by exploiting the properties of LacZ, these amino-terminal sequences could be purified. Today there are many different, more sophisticated “epitope tags.” However, the origins of this entire technology can be traced directly to LacZ translational fusions (31).
Cell biology.
Protein secretion was one of the first three-dimensional problems to be successfully addressed in bacteria, and LacZ translational fusions provided the experimental tool that opened the door. The cytoplasmic enzyme β-galactosidase can be directed to different cellular compartments by gene fusion (32). This fact coupled with the novel phenotypes that can occur when lacZ is fused to genes that specify exported proteins provided the means to genetically characterize the signal sequence and to identify the genes for components of the cellular protein secretion machinery (14).
Again success in bacteria prompted similar experiments in eukaryotic cells. This was first done by Hall et al. (16) who showed that β-galactosidase could be targeted by gene fusion to the nucleus in yeast.
Success with β-galactosidase has also prompted the search for new and better reporter genes. The most famous of these is certainly green fluorescent protein (11). Since this reporter works in living cells, it can be used to watch proteins move. A dramatic example of this is the oscillating behavior of the MinC protein in E. coli (29).
CONCLUSION
In a review I wrote with Beckwith long ago on the uses of lac fusions (33), the last sentence was “Given then, the wide range of uses to which lac fusions have been put and the development of new fusion systems with different uses, we imagine that the genetic fusion approach will be even more fruitful in the future.” We were right then, and I think the prediction still holds.
ACKNOWLEDGMENTS
I thank N. Ruiz for help with Fig. 1 and the National Institutes of General Medical Sciences for continued support.
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Ames B N, Hartman P E, Jacob B F. Chromosomal alterations affecting the regulation of histidine biosynthetic enzymes in Salmonella. J Mol Biol. 1963;8:23–42. doi: 10.1016/s0022-2836(63)80016-9. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith J R. A deletion analysis of the lac operator region in E. coli. J Mol Biol. 1964;8:427–430. doi: 10.1016/s0022-2836(64)80206-0. [DOI] [PubMed] [Google Scholar]
- 3.Beckwith J R, Signer E R. Transposition of the Lac region of Escherichia coli. J Mol Biol. 1966;19:254–265. doi: 10.1016/s0022-2836(66)80003-7. [DOI] [PubMed] [Google Scholar]
- 4.Beckwith J R, Signer E R, Epstein W. Transposition of the Lac region of E. coli. Cold Spring Harbor Symp Quant Biol. 1966;31:393–401. doi: 10.1101/sqb.1966.031.01.051. [DOI] [PubMed] [Google Scholar]
- 5.Benzer S, Champe S P. A change from nonsense to sense in the genetic code. Proc Natl Acad Sci USA. 1962;48:1114–1120. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer E, Silhavy T J, Weisemann J M, Weinstock G M. λ placMu: a transposable derivative of bacteriophage lambda for creating lacZ protein fusions in a single step. J Bacteriol. 1984;158:1084–1093. doi: 10.1128/jb.158.3.1084-1093.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell A. Episomes. Adv Genet. 1962;11:101–145. [Google Scholar]
- 8.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban M J, Chou J. In vivo formation of gene fusions encoding hybrid β-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci USA. 1984;81:535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Crick F H C, Barnett L, Brenner S, Watts-Tobin R J. General nature of the genetic code for proteins. Nature (London) 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 13.Cuzin F, Jacob F. Délétions chromosomiques et intégration d'un épisome sexuel F-lac+chez Escherichia coli K-12. C R Acad Sci. 1964;258:1350–1352. [Google Scholar]
- 14.Danese P N, Silhavy T J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Franklin N C, Dove W F, Yanofsky C. The linear insertion of a prophage into the chromosome of E. coli shown by deletion mapping. Biochem Biophys Res Commun. 1965;18:910–923. [Google Scholar]
- 16.Hall M N, Hereford L, Herskowitz I. Targeting of E. coli β-galactosidase to the nucleus in yeast. Cell. 1984;36:1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- 17.Hand, N. J., and T. J. Silhavy. A practical guide to the construction and use of lac fusions in Escherichia coli. Methods in Enzymol., in press. [DOI] [PubMed]
- 18.Hayes W. The genetics of bacteria and their viruses. New York, N.Y: J. Wiley & Sons, Inc.; 1968. p. 724. [Google Scholar]
- 19.Hiromi Y, Kuroiwa A, Gehring W J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 20.Howe M M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973;54:93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- 21.Jacob F, Ullman A, Monod J. Deletions fusion-mant l'operon lactose et un operon purine chez Escherichia coli. J Mol Biol. 1965;42:511–520. [Google Scholar]
- 22.Kenyon C J, Walker G C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroos L, Kaiser D. Construction of Tn lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H, Reznikoff W S. The operon. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1978. p. 234. [Google Scholar]
- 25.Miller J H. Discovering molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 26.Morse M L, Lederberg E M, Lederberg J. Transduction in Escherichia coli K-12. Genetics. 1956;41:142–156. doi: 10.1093/genetics/41.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller-Hill B, Kania J. lac repressor can be fused to β-galactosidase. Nature. 1974;249:561–563. doi: 10.1038/249561a0. [DOI] [PubMed] [Google Scholar]
- 28.O'Kane C J, Gehring W J. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raskin D M, de Boer P A J. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz M. Gene fusions in bacteria. In: Scaife J, Leach D, Galizzi A, editors. Genetics of bacteria. London, United Kingdom: Academic Press; 1985. pp. 65–84. [Google Scholar]
- 31.Shuman H A, Silhavy T J, Beckwith J R. Labeling of proteins with β-galactosidase by gene fusion. Identification of a cytoplasmic membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1980;255:168–174. [PubMed] [Google Scholar]
- 32.Silhavy T J, Casadaban M J, Shuman H A, Beckwith J R. Conversion of β-galactosidase to a membrane-bound state by gene fusion. Proc Natl Acad Sci USA. 1976;73:3423–3427. doi: 10.1073/pnas.73.10.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silhavy T J, Beckwith J. Uses of lac fusions for the study of biological problems. Microbiol Rev. 1985;49:398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slauch J M, Silhavy T J. Genetic fusions as an experimental tool. Methods Enzymol. 1991;204:213–248. doi: 10.1016/0076-6879(91)04011-c. [DOI] [PubMed] [Google Scholar]
- 35.Taylor A L. Bacteriophage-induced mutation in E. coli. Proc Natl Acad Sci USA. 1963;50:1043–1051. doi: 10.1073/pnas.50.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]