Abstract
Purpose:
To develop a deep learning model that generates consistent, high-quality lymph node clinical target volumes (CTV) contours for head and neck cancer (HNC) patients, as an integral part of a fully automated radiation treatment planning workflow.
Methods and Materials:
Computed tomography (CT) scans from 71 HNC patients were retrospectively collected and split into training (n = 51), cross-validation (n = 10), and test (n = 10) data sets. All had target volume delineations covering lymph node levels Ia through V (Ia-V), Ib through V (Ib-V), II through IV (II-IV), and retropharyngeal (RP) nodes, which were previously approved by a radiation oncologist specializing in HNC. Volumes of interest (VOIs) about nodal levels were automatically identified using computer vision techniques. The VOI (cropped CT image) and approved contours were used to train a U-Net autosegmentation model. Each lymph node level was trained independently, with model parameters optimized by assessing performance on the cross-validation data set. Once optimal model parameters were identified, overlap and distance metrics were calculated between ground truth and autosegmentations on the test set. Lastly, this final model was used on 32 additional patient scans (not included in original 71 cases) and autosegmentations visually rated by 3 radiation oncologists as being “clinically acceptable without requiring edits,” “requiring minor edits,” or “requiring major edits.”
Results:
When comparing ground truths to autosegmentations on the test data set, median Dice Similarity Coefficients were 0.90, 0.90, 0.89, and 0.81, and median mean surface distance values were 1.0 mm, 1.0 mm, 1.1 mm, and 1.3 mm for node levels Ia-V, Ib-V, II-IV, and RP nodes, respectively. Qualitative scoring varied among physicians. Overall, 99% of autosegmented target volumes were either scored as being clinically acceptable or requiring minor edits (ie, stylistic recommendations, <2 minutes).
Conclusions:
We developed a fully automated artificial intelligence approach to autodelineate nodal CTVs for patients with intact HNC. Most autosegmentations were found to be clinically acceptable after qualitative review when considering recommended stylistic edits. This promising work automatically delineates nodal CTVs in a robust and consistent manner; this approach can be implemented in ongoing efforts for fully automated radiation treatment planning.
Introduction
The use of intensity modulated radiation therapy (IMRT) techniques provides the ability to conform radiation dose distributions to the targets while sparing nearby normal tissues. Since clinical implementation, IMRT has required practitioners to manually define clinical target volumes (CTVs) and normal tissue organs at risk (OARs); these are both necessary to optimize dose distributions. This manual process is time-consuming and subject to significant inter- and intraobserver variabilities1,2 with reports suggesting that head and neck cancers (HNC) target delineation is both the most time-consuming anatomic site (taking physicians up to 2–3 hours) and subject to the largest variabilities.3–5 Several consensus delineation guidelines have emerged to reduce this variability6–8; yet significant variability persists, especially when delineating the head and neck lymph node levels and low-risk target volumes.
Automatic segmentation of HNC CTVs has been proposed as a solution for expediting the delineation process, promising improved efficiency and consistency in target delineations. Researchers have studied atlas-based autosegmentation, which takes advantage of deformable image registration to map well-defined delineations on 1 atlas to a new patient’s image for autosegmentation.9–14 A challenge with atlas-based autosegmentation is that it relies on the algorithm’s ability to accurately register different patients’ anatomies (a single atlas to a new patient), which can result in inaccuracies due to variable anatomy and low contrast of lymph node regions on computed tomography (CT) scans. To address these inaccuracies, authors proposed that multiatlas-based autosegmentation captures a wider range of anatomic variations resulting in an improvement in the resulting segmentations.11,12,15–18 Multiatlas-based autosegmentation typically uses 8 to 14 patients (atlases) with consensus or peer-reviewed manual contours. A limitation to the number of atlases is that each must be registered to the new patient, resulting in a computationally expensive process. In addition, whereas multiatlas-based autosegmentation provides more reference patient scans than single-atlas autosegmentation algorithms do, it only captures anatomic differences for a small sample of patients (atlases).
Deep learning-based autosegmentation algorithms have achieved impressive success in medical imaging segmentation tasks, with convolutional neural networks (CNNs) being the most popular algorithms used in deep learning.19,20 Researchers have used these networks to autosegment many anatomic sites using computed tomography, magnetic resonance imaging, positron emission tomography, x-rays, and ultrasound images resulting in rapid development and quick translation to the field of radiation oncology. Several studies have focused on normal tissue21–36 and CTV37–40 autosegmentation for HNC radiation therapy. In previous deep learning-based CTV autosegmentation work, researchers developed algorithms to autodelineate targets based on their risk (high-risk37,38 or low-risk37,39,40 CTVs) but lacked the ability to autodelineate individual lymph node levels. Therefore, in the present study, we developed a deep learning model to automatically segment targets based on lymph node level combinations commonly used in HNC radiation therapy. Our model’s architecture uses an ensemble of models and test-time augmentations to improve its generalizability for new patients. To the best of our knowledge, this is the first deep learning-based autosegmentation model for use with individual lymph node level target volumes for radiation treatment planning for HNCs. Unique to this work is that the resulting automatic segmentations can be implemented to administer radiation therapy for HNC at a large majority of subsites. Our hypothesis is that a vast majority of autosegmented lymph node target volumes can be used for radiation therapy treatment planning without user edits.
Methods and Materials
Patient data
Radiation therapy simulation CT scans and clinically approved contours for 71 patients with HNC previously treated at The University of Texas MD Anderson Cancer Center were used in this study under an institutional review board-approved protocol. Each patient had lymph node level Ia-V, Ib-V, and II-IV and retropharyngeal (RP) node level target volume contours (CTV_LN_II_IV, CTV_LN_Ia_V, CTV_LN_Ib_V, and CTV_LN_RP, respectively). These lymph node levels were previously contoured manually or autosegmented using an in-house multiatlas-based algorithm, visually inspected, and approved (as “clinically acceptable without requiring edits”) by a radiation oncologist specializing in HNC. This patient cohort included a variety of HNC disease scenarios, including different primary sites, lymph node statuses (negative vs positive), and lymph node locations (none vs ipsilateral vs bilateral).
Generation of ground-truth contours
In a previous study,17 a multiatlas-based autosegmentation tool was developed by our group using 2 separate atlases that independently autosegmented structures in the left and right lymph node levels. A radiation oncologist (B.M.B.) with more than 10 years of experience treating HNC manually delineated each individual lymph node level target volume on 20 patients’ radiation therapy simulation CT scans. In this previous analysis, manually delineated target volumes consisted of a combination of lymph node levels, except for the retropharyngeal node target volumes, which were delineated according to our clinical practice. These atlases were then used to autosegment the target volumes for 115 HNC patients who subsequently presented to our institution for radiation therapy. The resulting autosegmentations were scored by the same radiation oncologist on a 5-point scale (5: perfect, indistinguishable from physician-drawn contours for dose-volume histogram-based planning; 4: within acceptable interphysician variation for planning purposes; 3: good, needs minor edits to be used for planning purposes; 2: fair, needs significant edits to be used for planning purposes; and 1: poor, large areas need minor or major edits, is unusable for planning purposes). For the present study, 51 of the 115 cases with scores of 4 or 5 for all autosegmented target volumes were collected and combined with the 20 manually contoured cases scored by a radiation oncologist as clinically acceptable without requiring edits to generate a well-curated high-quality ground truth contour data set.
The 71 cases were randomly placed in 3 groups: training (51), cross-validation (10), and final test (10) sets. The training and cross-validation sets were used for training and optimal hyperparameter selection as well as identifying the most favorable postprocessing strategy. Postprocessing strategy was defined by using the trained model and predicting on cross-validation set cases to determine postprocessing steps, including morphology operations such as erosion/dilation with varying filter sizes and dimensionality (ie, 2D vs 3D). The final test set was held until the best model parameters and strategies were identified.
Deep learning-based autosegmentation
Due to the limited number of well-curated training cases available for this study, a model was designed that could predict lymph node target volumes regardless of neck laterality, essentially doubling our training data. Our approach to autosegment HNC lymph node target volumes is described in detail in the following subsections.
Data preparation
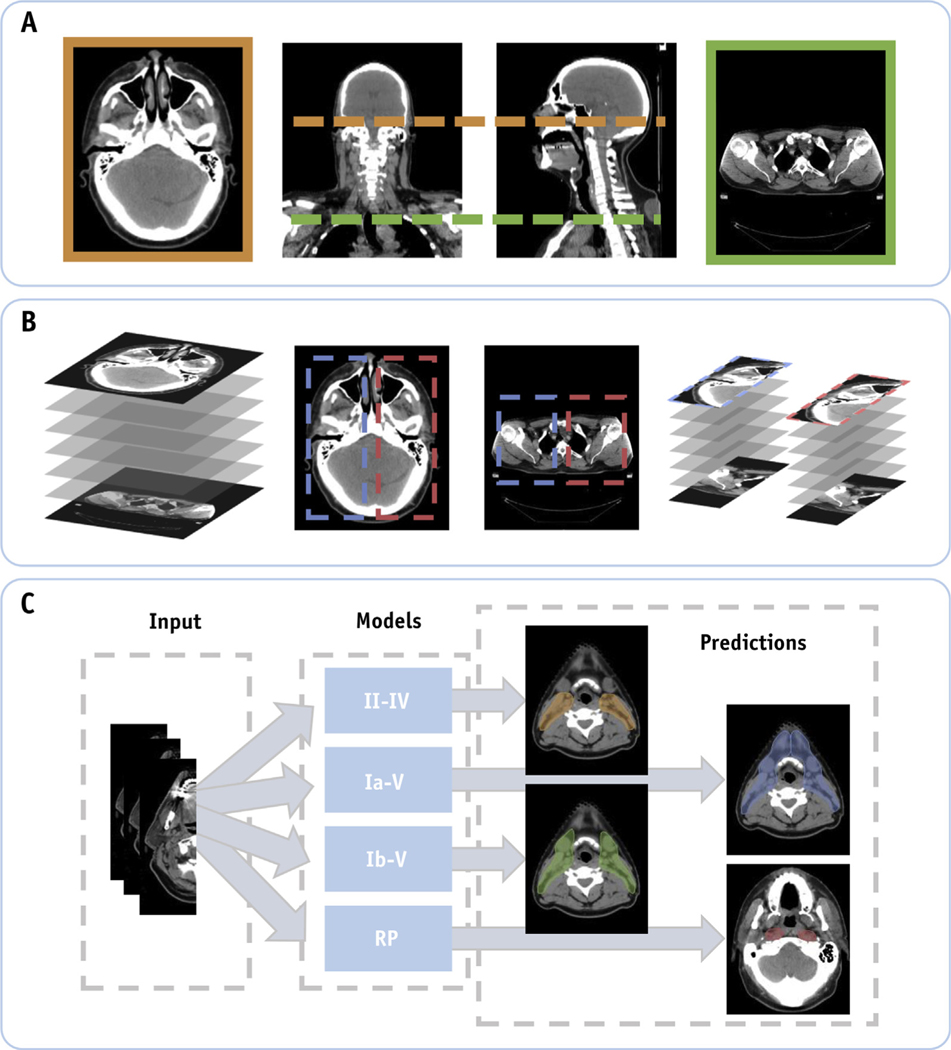
Our model’s input generation is depicted in Figure 1. Similar to our previous work,39 the CT scan field of view was reduced in the craniocaudal direction by identifying (1) the most caudal extent of the fusion of the sphenoid bone and basilar part of the occipital bone as the CT scan’s field of view’s most cranial CT slice and (2) the most cranial extent of the sternum as the field of view’s most caudal CT slice. Normalizing the field of view across patients is performed automatically by training the Xception model in Keras using a TensorFlow backend to classify individual CT slices. This is described in more detail in our previous work.23
Fig. 1.
(A) Computed tomography scans of patients with head and neck cancer are normalized in the craniocaudal extent by automatically cropping out slices below and above predefined anatomic markers. (B) Identification of the left and right neck lymph node regions using computer vision techniques. Here the training data were doubled by performing a horizontal flip of the resulting input data. (C) Our deep learning model is trained using the unilateral input data to automatically segment individual lymph node target volumes.
Once the craniocaudal field of view is identified, the body contour is defined automatically using previously developed in-house software.41 The body contour is then used to reduce the input volume by identifying a bounding box about the most cranial slice to extract the CT image’s input volumes for both the left and right lymph node regions. The resulting CT image volume of interest (VOI) and its corresponding target volume masks are resized using bilinear interpolation to a predefined volume size (64 × 128 × 64) for use as inputs in our deep learning model. Lastly, image intensities were transformed using our clinic’s head and neck CT window/level settings (−350, 350 Hounsfield Units) to have values from 0 to 1 (ie, −350 = 0 and 350 = 1) as was done in our previous work.39
Architecture and training parameters
A hyperparameter search was performed to identify the optimal parameters (eg, resolution steps, kernel size) on a modified 3D U-Net architecture.39 Our U-Net model uses a residual function (short-connections) similar to that described by Milletari et al42 and uses batch normalization43 after each 3D convolutional layer. The same architecture is used to train 5 separate model weights (random initialization of weights) focused on identifying patterns to autosegment each individual target volume (CTV_LN_II_IV, CTV_LN_Ia_V, CTV_LN_Ib_V, and CTV_LN_RP) using the same input volume (preprocessed CT scan as previously described in the Data Preparation section). These models are used in an ensemble approach to further improve the confidence in the resulting segmentation.
The model was trained using the Adam optimizer with a learning rate of 0.001 and early stopping regularization to avoid overfitting of the models. Typical data augmentations (eg, translation, rotation) were used during training. Herein a new overlap loss is introduced that incorporates the Dice similarity coefficient (DSC) loss and false-negative Dice (FND) loss to penalize missed target volumes during training. The generalized “DSC + FND” loss is defined as
in which the first term is the multiclass (C) DSC loss described by Milletari et al,42 and the second term is the multiclass FND component introduced herein. For the DSC loss, the sums run over N voxels of the predicted probabilities and the ground truth binary volume for all classes, whereas for the FND loss, the numerator calculates the sums over N voxels between the complement () of the predicted probabilities and the ground truth binary volume are only calculated for nonbackground classes (here, background is defined by C = 0). The rationale for introducing this loss function is that the DSC loss leads to systematic underestimation of the predicted volumes compared with the cross-validation ground-truth volumes.
Postprocessing and ensemble approach
When generating a prediction on a new patient, an ensemble approach and test-time augmentations are used to further improve the resulting segmentations. Our ensemble approach uses the 5 trained model weights for each region of interest and applies random shifts (total, 13) about the center of the input image to generate 13 probability maps for each trained model. These shifts ranged from ±3, ±10, and ±5 for the z-, y-, and x-directions, respectively. The probability maps for all models and their corresponding test-time augmentations (total, 65) are then shifted back to the original input space and averaged on a per-voxel basis. The resulting averaged probability map for the target volume is then converted into a binary mask by thresholding individual voxels’ probabilities to have a value greater than or equal to 0.5. The resulting binary mask then goes through a postprocessing step to ensure the removal of holes and/or keep the largest autosegmented volume.
Quantitative and qualitative evaluation
During our quantitative evaluation of our model’s autosegmentations, the ground-truth contours are compared with the autosegmented target volumes using overlap and distance metrics, as overlap metrics alone can be less sensitive to larger volumes. These metrics include the DSC, FND, false-positive Dice, volumetric similarity (VS), mean surface distance (MSD), and Hausdorff distance (HD), which are defined in Equations 2 to 7:
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
in which TP, FN, and FP are the numbers of true-positive, false-negative, and false-positive voxels, respectively; Auto and Truth represent the autosegmentation and ground-truth contours, respectively, and da,b is a vector with all the minimum Euclidean distances from each surface point in volume “a” to volume “b.” The FND and FPD are good metrics that help quantify potential near misses and overtreatment, respectively. Because the target volumes used to train the model were generated as a combination of lymph node levels, our quantitative analysis focuses on the target volume as a whole and not explicitly on individual lymph node levels.
To confirm our quantitative analysis results, we conducted a multi-institutional qualitative review of the proposed model’s autosegmentations using a separate set of HNC patient scans (32 new cases) from the 71 cases used to train, cross-validate, and test our models. In this analysis, 3 radiation oncologists (BMB, ASG, and HDS, ordered alphabetically) each with more than 10 years of experience treating HNC visually inspected each target volume on a slice-by-slice basis for each patient. Each individual target volume was scored using a 3-point scale (clinically acceptable without requiring edits, requiring minor edits, and requiring major edits). Acceptable autosegmentations are those that do not require any edits and can be used “as is.” Autosegmentations requiring minor edits are contours that can be manually edited and corrected quicker than 2 minutes and/or are acceptable for clinical use if a CTV-to-planning target volume margin of 5 mm is used suggesting that the edits are stylistic in nature. Autosegmentations requiring major edits are those that are not acceptable for clinical use and are believed to clinically affect the likelihood of cure, adverse events, or locoregional control.
Results
Quantitative evaluation
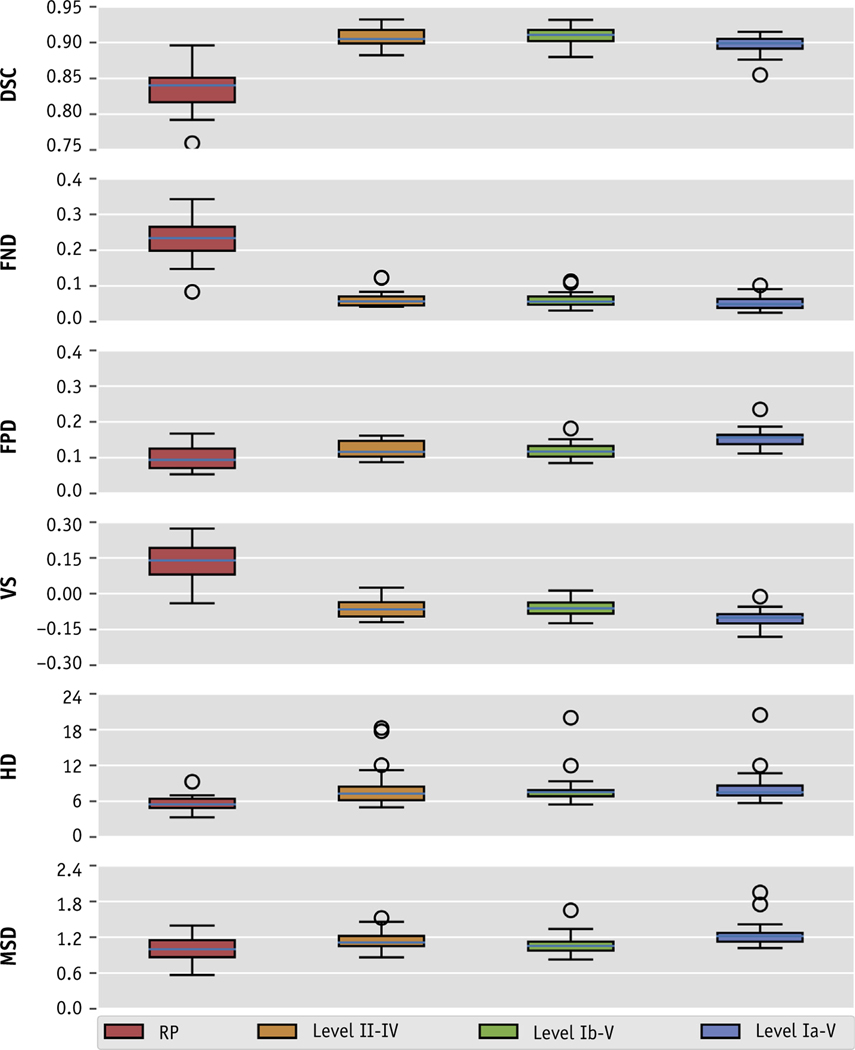
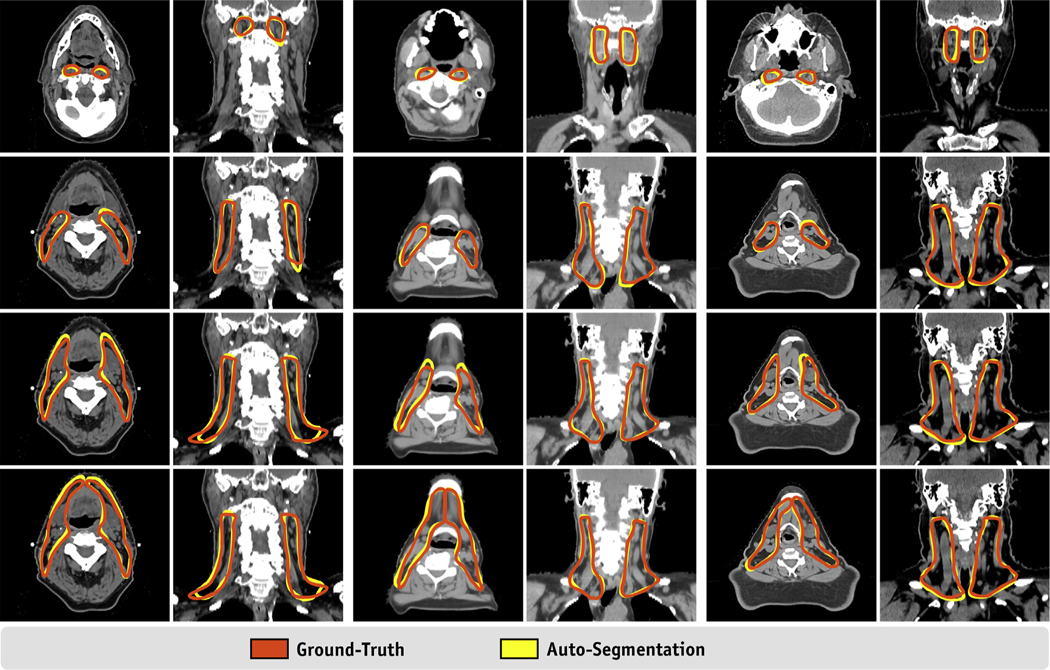
Quantitative results for our ensemble model segmentations are shown in Figure 2 and are summarized in Table 1. The mean (± standard deviation [SD]) DSC values between the final segmentations and the ground truth were 0.843 ± 0.030, 0.907 ± 0.013, 0.909 ± 0.013, and 0.897 ± 0.014 for CTV_LN_RP, CTV_LN_II_IV, CTV_LN_Ib_V, and CTV_LN_Ia_V, respectively. The mean FND/FPD values for these 4 volumes were 0.234/0.099, 0.063/0.123, 0.062/0.120, and 0.053/0.154, respectively. The mean VS values were 0.135, −0.060, −0.057, and −0.101, respectively. The mean (±SD) MSD and HD values were 1.0 ± 0.2 mm and 5.5 ± 1.3 mm, 1.1 ± 0.2 mm and 8.4 ± 3.7 mm, 1.1 ± 0.2 mm and 8.1 ± 3.1 mm, and 1.3 ± 0.2 mm and 8.6 ± 3.1 mm, respectively. When comparing the ensemble model results with each individual model’s segmentations (ensemble - others), we noticed a mean (± SD) improvement of 0.01 ± 0.01 for the DSC. We observed similar slight improvements as demonstrated by mean (± SD) reductions in the MSD and HD across all volumes of −0.1 ± 0.2 mm and −0.7 ± 2.8 mm, respectively. A visual comparison of the ground-truth and autosegmented target volumes is shown in Figure 3.
Fig. 2.
Box plot of the distributions of overlap and distance metrics in a comparison of the ground-truth and autosegmented volumes for each neck lymph node target volume. The boxplots are representative of individual metric’s interquartile range, whereas the whiskers denote values within 1.5 interquartile range, and the outliers (circles) are values that are found outside of this range. Abbreviations: DSC = dice similarity coefficient; FND = false negative dice; FPD = false positive dice; HD = hausdorff distance; MSD = mean Surface distance; VS = volume similarity.
Table 1.
Summary of quantitative evaluation between auto-segmented and ground-truth target volumes
| DSC | FND | FPD | VS | HD (mm) | MSD (mm) | |
|---|---|---|---|---|---|---|
|
| ||||||
| RP | 0.834 ± 0.030 | 0.234 ± 0.064 | 0.099 ± 0.033 | 0.135 ± 0.082 | 5.5 ± 1.3 | 1.0 ± 0.2 |
| Level II-IV | 0.907 ± 0.013 | 0.063 ± 0.023 | 0.123 ± 0.023 | −0.060 ± 0.038 | 8.4 ± 3.7 | 1.1 ± 0.2 |
| Level Ib-V | 0.909 ± 0.013 | 0.062 ± 0.021 | 0.120 ± 0.023 | −0.057 ± 0.036 | 8.1 ± 3.1 | 1.1 ± 0.2 |
| Level Ia-V | 0.897 ± 0.014 | 0.053 ± 0.019 | 0.154 ± 0.027 | −0.101 ± 0.037 | 8.6 ± 3.1 | 1.3 ± 0.2 |
Abbreviations: DSC = dice similarity coefficient; FND = false negative dice; FPD = false positive dice; HD = hausdorff distance; MSD = mean Surface distance; RP = retropharyngeal; VS = volume similarity.
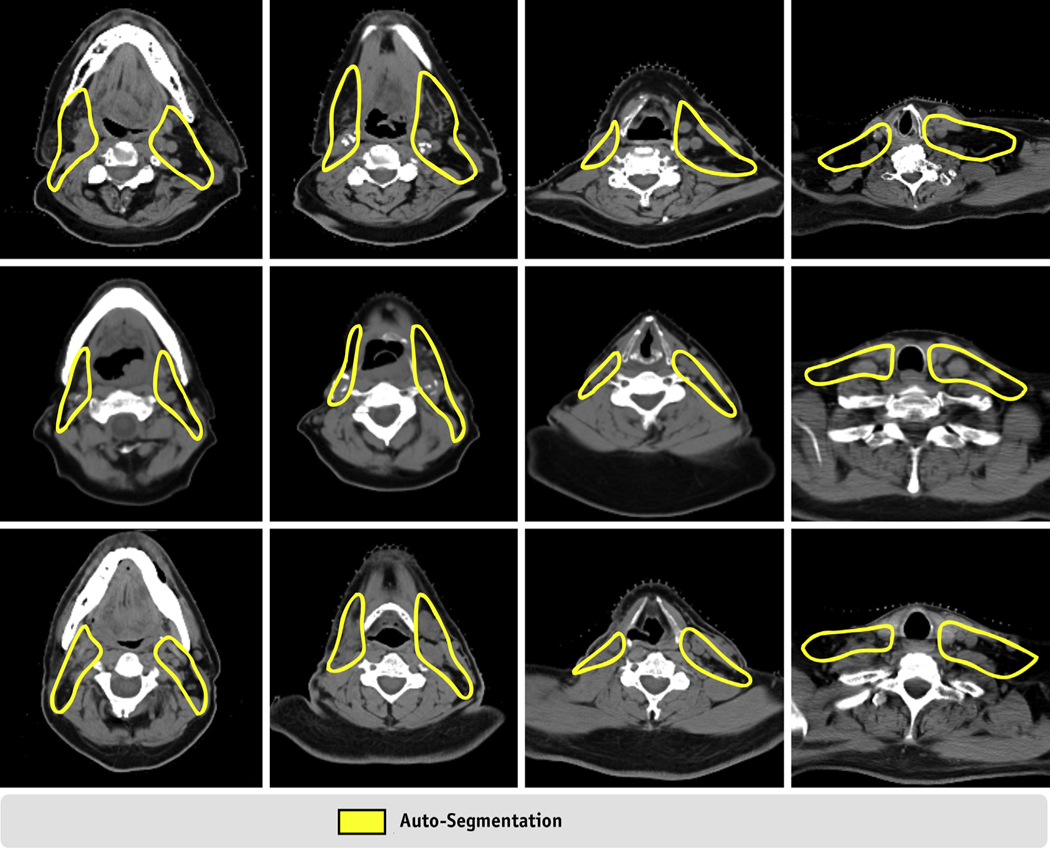
Fig. 3.
Visual comparison of the ground-truth and auto-segmented neck lymph node (LN) target volumes.
The model generated all regions of interest with a mean (± SD) time of 6.0 ± 0.6 minutes using an NVIDIA RTX 2080 graphics processing unit and 32 cores. This time was significantly reduced when a user decided to only include both RP nodes and unique target combinations for both neck sides (mean [± SD] time of 3.2 ± 0.4 minutes when autosegmenting both RP nodes and targets for levels Ia-V and Ib-V for the right and left neck lymph nodes, respectively).
Qualitative evaluation
Physician slice-by-slice review and scoring of the autosegmentations are listed in Table 2. The 3 physicians reviewed 256 target volumes each (8 target volumes for each of the 32 cases). Seven (22%) of these patients previously underwent neck surgery, which included unilateral or bilateral dissection. Of the 768 target volumes reviewed, 438 (57%) were scored as acceptable as is, 323 (42%) were scored as requiring minor edits (ie, stylistic recommendations, <2 minutes), and 7 (1%) required major edits. When considering individual reviewer scores, reviewer 1 scored 96%, 4%, and 0% of autodelineated target volumes as acceptable, minor, and major, respectively, reviewer 2 scored 63%, 38%, and 0% of autodelineated target volumes as acceptable, minor, and major, respectively, and reviewer 3 scored 12%, 85%, and 3% of autodelineated target volumes as acceptable, minor, and major, respectively.
Table 2.
Qualitative scores for 32 cases separated by postoperative status
| Nonpostoperative (n = 25) |
Postoperative (n = 7) |
|||||
|---|---|---|---|---|---|---|
| Scores |
Scores |
|||||
| 1 | 2 | 3 | 1 | 2 | 3 | |
|
| ||||||
| Reviewer 1 | ||||||
| Ia-V right | 25 | 0 | 0 | 4 | 3 | 0 |
| Ia-V left | 25 | 0 | 0 | 7 | 0 | 0 |
| Ib-V right | 25 | 0 | 0 | 4 | 3 | 0 |
| Ib-V left | 25 | 0 | 0 | 7 | 0 | 0 |
| II-IV right | 25 | 0 | 0 | 4 | 3 | 0 |
| II-IV left | 25 | 0 | 0 | 7 | 0 | 0 |
| RP right | 25 | 0 | 0 | 7 | 0 | 0 |
| RP left | 25 | 0 | 0 | 7 | 0 | 0 |
| Reviewer 2 | ||||||
| Ia-V right | 14 | 11 | 0 | 4 | 3 | 0 |
| Ia-V left | 14 | 11 | 0 | 4 | 3 | 0 |
| Ib-V right | 14 | 11 | 0 | 4 | 3 | 0 |
| Ib-V left | 14 | 11 | 0 | 4 | 3 | 0 |
| II-IV right | 14 | 11 | 0 | 4 | 3 | 0 |
| II-IV left | 14 | 11 | 0 | 4 | 3 | 0 |
| RP right | 21 | 4 | 0 | 5 | 2 | 0 |
| RP left | 21 | 4 | 0 | 5 | 2 | 0 |
| Reviewer 3 | ||||||
| Ia-V right | 0 | 25 | 0 | 0 | 5 | 2 |
| Ia-V left | 0 | 24 | 1 | 0 | 7 | 0 |
| Ib-V right | 0 | 25 | 0 | 0 | 5 | 2 |
| Ib-V left | 1 | 23 | 1 | 0 | 7 | 0 |
| II-IV right | 2 | 23 | 0 | 0 | 6 | 1 |
| II-IV left | 4 | 21 | 0 | 1 | 6 | 0 |
| RP right | 9 | 16 | 0 | 1 | 6 | 0 |
| RP left | 11 | 14 | 0 | 2 | 5 | 0 |
Individual cases were reviewed on a slice-by-slice basis by 3 radiation oncologists each having more than 10 years of HNC experience.
Auto-segmentation scores: 1 = clinically acceptable without requiring edits; 2 = requiring minor edits (ie, stylistic recommendations, <2 minutes); 3 = requiring major edits.
Abbreviation: HNC = head and neck cancer.
Discussion
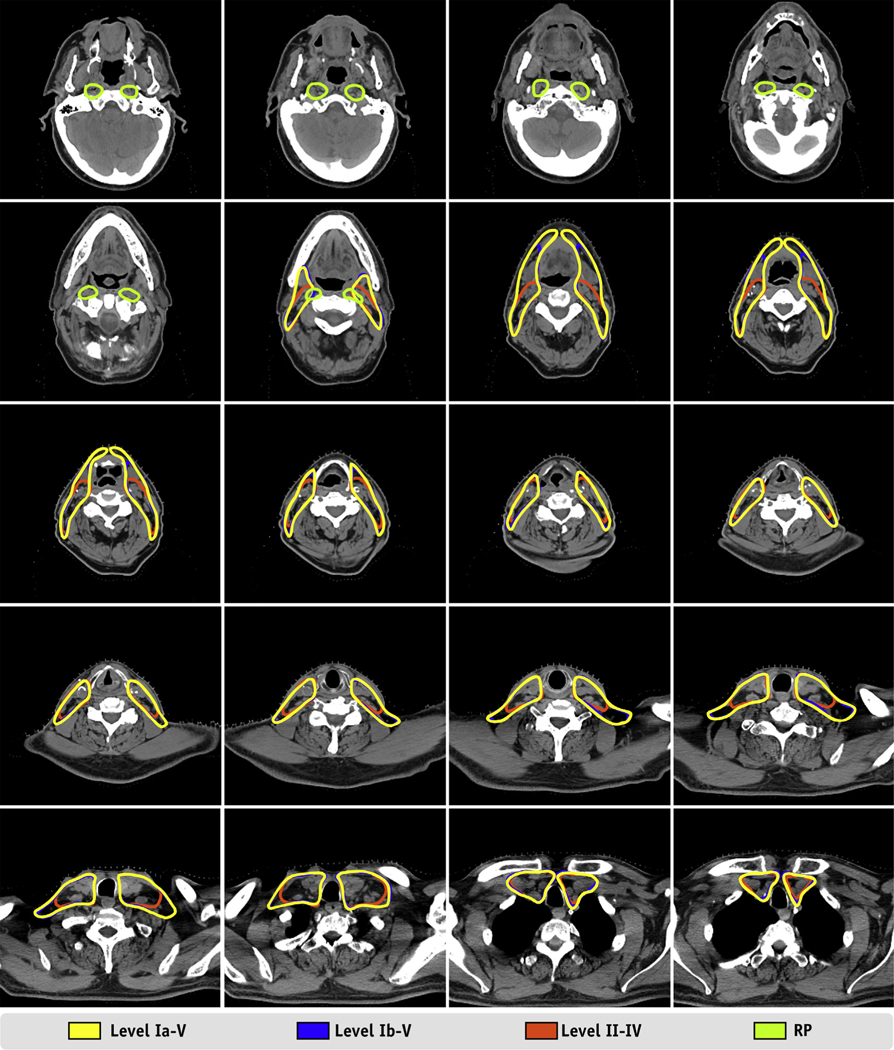
In this study, we developed an ensemble model to automatically delineate nodal CTVs for patients with HNC undergoing radiation treatment planning using a novel deep learning-based approach. The metrics (DSC and HD) and clinical acceptability (based on clinician review of the lymph node target volume autosegmentations) demonstrated excellent performance and are promising for clinical usability, with greater than 99% of the autosegmentations scored as acceptable or requiring only stylistic edits. The resulting model can autosegment multiple CTV level options, allowing radiation oncologists to choose patient-specific target volumes based on lymph node level involvement and clinical history (Fig. 4).
Fig. 4.
Example results from a randomly selected case from our test set. Twenty axial slices from a computed tomography scan of a 57-year-old male patient with base of tongue cancer show the auto-segmented lymph node target volumes. The axial slices are evenly sampled and distributed from the cranial extent of the retropharyngeal lymph nodes to the caudal extent of the level IV lymph node.
Manual delineation of CTVs for HNC radiation therapy remains a challenging and time-consuming task for radiation oncologists. Researchers have proposed several atlas-based methods for head and neck lymph node autosegmentation.9,10,13,16,17,44 Teguh et al44 developed a multiatlas-based approach that resulted in a mean DSC of 0.67 for lymph node levels. Similarly, Yang et al17 reported a median DSC value of 0.778 using a multiatlas-based approach that used the Simultaneous Truth and Performance Level Estimation algorithm. More recently, investigators have developed deep learning models to autosegment lymph node CTVs.37,40,45 Also, Men et al37 developed an end-to-end deep 2-dimensional deconvolutional neural network trained with 184 nasopharyngeal cancer patients who resulted in a mean DSC value of 0.826 for the low-risk CTV. Our group39 reported similar results (mean DSC value, 0.816 for 75 test cases) using a 3-dimensional fully convolutional neural network trained with 210 oropharyngeal cancer patients. More recently, Wong et al40 reported a mean DSC value of 0.72 for neck CTVs using a commercial system (Limbus Contour) with a U-Net—based model. Our approach is different from previous work in that we developed a model that can provide a variety of target volumes for the same patient (ie, the radiation oncologist can choose lymph node level coverage). This makes comparison of our results with those of previously published studies of lymph node target volume autosegmentation difficult to interpret. Furthermore, the greatest benefit of our ensemble approach was a systematic reduction of the number of slices (mode of 1 slice vs 2 slices from the ground truth) in the caudal edges of the target volumes. We expect the caudal extent to exhibit the largest variability in our input data, so it is natural to expect for individual models to be more sensitive to larger deviations in these regions. Using this ensemble approach, our model can define this caudal edge with more confidence than single model autosegmentations, which leads to better generalization compared with ground truth contours.
Although our autosegmentations and ground-truth volumes agreed well in most cases, we noticed that the deep learning model failed to produce acceptable target volumes for the patients who had prior neck surgery. Figure 5 shows CT images from 3 patients who underwent neck dissection. In these cases, physician review suggested minor or major edits at lymph node level II/III where the right neck contours failed to provide appropriate coverage posteriorly in all 3 cases (only CTV_LN_Ib_V autosegmentation is shown in the figure, but we observed similar results for CTV_LN_Ia_V and CTV_LN_II_IV). After noticing consistent undercontouring in these regions during qualitative analysis, we reviewed the cases used to train our models and found that none presented with large resections as observed in these 3 cases. Clearly, when a patient underwent neck dissection, the deep learning-based algorithm lacks the prior knowledge to confidently produce acceptable delineations. Whether similar anatomic scenarios (ie, prior parotidectomy, glossectomy, oral cavity resection) can have similar effects on the model’s ability to produce reliable target volumes remains unknown, and further testing may be warranted to identify additional failure modes in autosegmentations. Inclusion of outlier cases, such as those with prior surgeries, during training of the model could help overcome deficiencies in these scenarios, but this remains to be evaluated.
Fig. 5.
Computed tomography images of 3 patients with auto-segmentations requiring minor edits. All 3 patients (1 per row) had their neck dissection before radiation therapy. In these cases, the auto-segmented volumes were undercontoured between lymph node levels II and III as shown in columns 2 and 3. Whereas target volumes for neck lymph node levels Ib-V are shown in this figure, auto-segmentations for levels II-IV and Ia-V were subject to similar undercontouring in these regions. RP node target volumes were unaffected in this clinical presentation.
Qualitative evaluation showed that 99% of autosegmented target volumes were within acceptable ranges by board-certified head and neck cancer radiation oncologists with either no edits or only stylistic edits. Overall, this demonstrates the use (and safety) of these autosegmented volumes; fundamentally, 99% of them could be used without risk by the treating physicians. However, it also points out, again, that individual physicians have their own contouring styles. Overall, reviewer 1 scored the autosegmentations more favorably than reviewers 2 and 3. Because reviewer 1 generated the manual contours used to train the autosegmentation model, it is not surprising that the autosegmentations were more consistent with reviewer 1’s delineation style. Reviewers 2 and 3 scored the large majority of volumes as “minor edits;” these were described as safe, acceptable in a typical peer review QA of target volumes, and stylistic in nature. When asked for detailed feedback, reviewers 2 and 3 considered the autodelineated target volumes to be slightly generous toward the posterior digastric muscle, scalene muscle, sternocleidomastoid muscle, and/or, occasionally, adjacent parotid. The retropharyngeal node target volumes were scored as having the most “acceptable” scores (72%) among the 3 reviewers; examples of “minor edits” from reviewers 2 and 3 included deleting contours on the most caudal slice or edits to the posterior border (<3 mm in size) of the volumes for some slices. Only 1 reviewer provided recommendations for “major” edits to some of the autosegmented target volumes of 3 cases. Two of these patients had previously received neck dissections (both shown in Fig. 5). For these cases, each individual reviewer provided different scores for the target volumes (ie, these received scores of “acceptable,” “minor,” and “major”). The third case’s contours that received “major edits” scores were scored as “acceptable” by the other reviewers. This qualitative evaluation does highlight the stylistic preferences of different treating physicians, which will be a significant challenge for any automated system.
Automating the delineation of target volumes in HNC radiation therapy has several potential benefits. First, if automatically generated target volume delineations are consistent and accurate, it could lead to the standardization of target volume delineations, which are among the largest sources of uncertainty in radiation treatment planning. Second, consistent, systematic target volume delineations using automated models could lead to increased quality of clinical data. Authors have reported that noncompliant target volume delineations in clinical trials can significantly affect patient outcomes and the quality of clinical trial data. Third, if an autosegmentation model is clinically implemented and validated by positive patient outcomes, it could be scaled to provide low-resource clinics around the world with high-quality delineations. Several ongoing efforts are increasing access to high-quality radiation therapy in low- and middle-income countries through automation. For example, the Radiation Planning Assistant (RPA, https://rpa.mdanderson.org) is being developed at The University of Texas MD Anderson Cancer Center to fully automate the radiation treatment planning process with no or minimal user interventions.45–48 For such a system to be effective in reducing the workload at busy clinics with limited resources, including radiation oncologists, high-quality automatic contouring is essential.23 The present study showed that deep learning can achieve this, and we are integrating these tools to autosegment normal tissue and target volumes into the RAP system. Specifically, the RPA workflow is currently designed so that the radiation oncologist first identifies which lymph node levels should be contoured. After they are automatically contoured, the radiation oncologist contours the gross tumor volumes and reviews and, if necessary, edits the nodal level contours. The RPA then automatically generates a volume-modulated arc therapy plan. This is only reasonable with access to a very robust automated contouring solution such as that described herein. The target volume delineation model in the present work could be integrated into such systems to reliably and consistently generate high-quality lymph node clinical target volumes for HNC radiation therapy in the vast majority of cases.
Our study had a few limitations. First, a single radiation oncologist manually contoured the target volume delineations used to train our model. This individual is subspecialized in HNCs, with more than 10 years of clinical experience; however, practice pattern variations in target volume delineation may not be appreciated. Second, all cases evaluated (both quantitatively and qualitatively) were HNC patients previously given treatment at a single institution. Thus, our patients may not sufficiently represent the large variability in anatomic presentations for HNC observed across different populations. Another limitation is that the model was trained to autosegment target volumes that were a combination of lymph node levels (typically treated in head and neck cancers) and did not allow for individual lymph node level autosegmentation.
Conclusion
We developed a fully automated artificial intelligence approach to autodelineating nodal CTVs for patients with intact HNC. The vast majority of autosegmentations were clinically acceptable after qualitative review when considering recommended stylistic edits. This work is promising in that it automatically delineates high-quality CTVs in a robust and reliable manner. This approach can be implemented in ongoing efforts for fully automated radiation treatment planning for HNC.
Supplementary Material
Acknowledgments—
The authors acknowledge the support of the High Performance Computing facility at the University of Texas MD Anderson Cancer Center for providing computational resources (including consulting services) that have contributed to the research results reported in this paper. The authors thank Peter and Linda Hrebec for their generous philanthropic support of this work. They also thank Don Norwood from the Department of Scientific Publications at the University of Texas MD Anderson Cancer Center for editing this work.
This work was presented in part as an oral presentation at the 2020 Multidisciplinary Head and Neck Cancers Symposium, February 28, 2020, Phoenix, Arizona.
This work was supported by NIH/NCI grants L30 CA242657 (to C.E.C.) and UH2/UH3 CA202665 (to B.M.B. and L.E.C.).
Footnotes
Disclosures: Drs. Cardenas, Beadle, Yang, Rhee, Netherton, Gay, Zhang, and Court are part of the Radiation Planning Assistant Project (https://rpa.mdanderson.org).
Research data are not available at this time.
References
- 1.Moghaddasi L, Bezak E, Marcu LG. Current challenges in clinical target volume definition: Tumour margins and microscopic extensions. Acta Oncol 2012;51:984–995. [DOI] [PubMed] [Google Scholar]
- 2.Yee Chang AT, Tan LT, Duke S, et al. Challenges for quality assurance of target volume delineation in clinical trials. Front Oncol 2017;7:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong TS, Tome WA, Harari PM. Heterogeneity in head and neck IMRT target design and clinical practice. Radiother Oncol 2012;103: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segedin B, Petric P. Uncertainties in target volume delineation in radiotherapy—Are they relevant and what can we do about them? Radiol Oncol 2016;50:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Multi-Institutional Target Delineation in Oncology Group. Human-computer interaction in radiotherapy target volume delineation: A prospective, multi-institutional comparison of user input devices. J Digit Imaging 2011;24:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee AW, Ng WT, Pan JJ, et al. International guideline for the delineation of the clinical target volumes (CTV) for nasopharyngeal carcinoma. Radiother Oncol 2017;126:25–36. [DOI] [PubMed] [Google Scholar]
- 7.Grégoire V, Evans M, Le QT, et al. Delineation of the primary tumour clinical target volumes (CTV-P) in laryngeal, hypopharyngeal, oropharyngeal and oral cavity squamous cell carcinoma: AIRO, CACA, DAHANCA, EORTC, GEORCC, GORTEC, HKNPCSG, HNCIG, IAG-KHT, LPRHHT, NCIC CTG, NCRI, NRG Oncology, PHNS, SBRT, SOMERA, SRO, SSHNO, TROG consensus guidelines. Radiother Oncol 2017;126:3–24. [DOI] [PubMed] [Google Scholar]
- 8.Hansen CR, Johansen J, Samsøe E, et al. Consequences of introducing geometric GTV to CTV margin expansion in DAHANCA contouring guidelines for head and neck radiotherapy. Radiother Oncol 2018;126: 43–47. [DOI] [PubMed] [Google Scholar]
- 9.Stapleford LJ, Lawson JD, Perkins C, et al. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int J Radiat Oncol Biol Phys 2010;77:959–966. [DOI] [PubMed] [Google Scholar]
- 10.Gorthi S, Duay V, Houhou N, et al. Segmentation of head and neck lymph node regions for radiotherapy planning using active contour-based atlas registration. IEEE J Sel Top Signal Process 2009;3:135–147. [Google Scholar]
- 11.Han X, Hoogeman MS, Levendag PC, et al. Atlas-based autosegmentation of head and neck CT images. Med Image Comput Comput Assist Interv 2008;11(Pt 2):434–441. [DOI] [PubMed] [Google Scholar]
- 12.Sjöberg C, Lundmark M, Granberg C, et al. Clinical evaluation of multi-atlas based segmentation of lymph node regions in head and neck and prostate cancer patients. Radiat Oncol 2013;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen A, Deeley MA, Niermann KJ, et al. Combining registration and active shape models for the automatic segmentation of the lymph node regions in head and neck CT images. Med Phys 2010;37:6338–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng CC, Shapiro LG, Kalet IJ. Head and neck lymph node region delineation with image registration. Biomed Eng Online 2010;9:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Commowick O, Grégoire V, Malandain G. Atlas-based delineation of lymph node levels in head and neck computed tomography images. Radiother Oncol 2008;87:281–289. [DOI] [PubMed] [Google Scholar]
- 16.Daisne JF, Blumhofer A. Atlas-based automatic segmentation of head and neck organs at risk and nodal target volumes: A clinical validation. Radiat Oncol 2013;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Beadle BM, Garden AS, et al. Auto-segmentation of low-risk clinical target volume for head and neck radiation therapy. Pract Radiat Oncol 2014;4:e31–e37. [DOI] [PubMed] [Google Scholar]
- 18.Haq R, Berry SL, Deasy JO, et al. Dynamic multiatlas selection-based consensus segmentation of head and neck structures from CT images. Med Phys 2019;46:5612–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosmin M, Ledsam J, Romera-Paredes B, et al. Rapid advances in auto-segmentation of organs at risk and target volumes in head and neck cancer. Radiother Oncol 2019;135:130–140. [DOI] [PubMed] [Google Scholar]
- 20.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in auto-segmentation. Semin Radiat Oncol 2019;29:185–197. [DOI] [PubMed] [Google Scholar]
- 21.Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys 2017; 44:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W, Huang Y, Zeng L, et al. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys 2019;46:576–589. [DOI] [PubMed] [Google Scholar]
- 23.Rhee DJ, Cardenas CE, Elhalawani H, et al. Automatic detection of contouring errors using convolutional neural networks. Med Phys 2019;46:5086–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong N, Gou S, Yang S, et al. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys 2018;45:4558–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolov S, Blackwell S, Mendes R, et al. Deep learning to achieveclinically applicable segmentation of head and neck anatomy for radiotherapy. Available at: http://arxiv.org/abs/1809.04430. Accessed July 27, 2020.
- 26.Hänsch A, Schwier M, Morgas T, et al. Comparison of different deep learning approaches for parotid gland segmentation from CT images. Available at: 10.1117/12.2292962. Accessed July 27, 2020. [DOI]
- 27.Lin L, Dou Q, Jin YM, et al. Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology 2019;291:677–686. [DOI] [PubMed] [Google Scholar]
- 28.Liang S, Tang F, Huang X, et al. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur Radiol 2019;29: 1961–1967. [DOI] [PubMed] [Google Scholar]
- 29.Chan JW, Kearney V, Haaf S, et al. A convolutional neural network algorithm for automatic segmentation of head and neck organs at risk using deep lifelong learning. Med Phys 2019;46:2204–2213. [DOI] [PubMed] [Google Scholar]
- 30.Men K, Geng H, Cheng C, et al. Technical note: More accurate and efficient segmentation of organs-at-risk in radiotherapy with convolutional neural networks cascades. Med Phys 2019;46: 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong N, Gou S, Yang S, et al. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med Phys 2019;46: 2669–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer A, Thor M, Haq R, et al. Deep learning-based auto-segmentation of swallowing and chewing structures in CT. Available at: 10.1101/772178v1. Accessed July 27, 2020. [DOI]
- 33.Tappeiner E, Pröll S, Hönig M, et al. Multi-organ segmentation of the head and neck area: An efficient hierarchical neural networks approach. Int J Comput Assist Radiol Surg 2019;14:745–754. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Zhao L, Wang M, et al. Organ at risk segmentation in head and neck CT images using a two-stage segmentation framework based on 3D U-Net. IEEE Access; 2019;7. 10.1109/access.2019.2944958. Accessed July 27, 2020. [DOI] [Google Scholar]
- 35.Mlynarski P, Delingette H, Alghamdi H, et al. Anatomically consistent segmentation of organs at risk in MRI with convolutional neural networks. Available at: http://arxiv.org/abs/1907.02003. Accessed July 27, 2020. [DOI] [PMC free article] [PubMed]
- 36.Tang H, Chen X, Liu Y, et al. Clinically applicable deep learning framework for organs at risk delineation in CT images. Nat Mach Intell 2019;1:480–491. [Google Scholar]
- 37.Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol 2017;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardenas CE, McCarroll RE, Court LE, et al. Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int J Radiat Oncol Biol Phys 2018;101:468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardenas CE, Anderson BM, Aristophanous M, et al. Auto-delineation of oropharyngeal clinical target volumes using 3D convolutional neural networks. Phys Med Biol 2018;63:215026. [DOI] [PubMed] [Google Scholar]
- 40.Wong J, Fong A, McVicar N, et al. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020;144:152–158. [DOI] [PubMed] [Google Scholar]
- 41.Court LE, Kisling K, McCarroll R, et al. Radiation planning assistant - A streamlined, fully automated radiotherapy treatment planning system. J Vis Exp 2018;e57411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milletari F, Navab N, Ahmadi S-A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. Available at: 10.1109/3DV.2016.79. Accessed July 27, 2020. [DOI]
- 43.Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. Available at: https://arxiv.org/abs/1502.03167. Accessed October 22, 2020.
- 44.Teguh DN, Levendag PC, Voet PWJ, et al. Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int J Radiat Oncol Biol Phys 2011;81:950–957. [DOI] [PubMed] [Google Scholar]
- 45.McCarroll RE, Beadle BM, Balter PA, et al. Retrospective validation and clinical implementation of automated contouring of organs at risk in the head and neck: A step toward automated radiation treatment planning for low- and middle-income countries. J Glob Oncol 2018; 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kisling K, Zhang L, Shaitelman SF, et al. Automated treatment planning of postmastectomy radiotherapy. Med Phys 2019;46:3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kisling K, Zhang L, Simonds H, et al. Fully automatic treatment planning for external-beam radiation therapy of locally advanced cervical cancer: A tool for low-resource clinics. J Glob Oncol 2019;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kisling K, Johnson JL, Simonds H, et al. A risk assessment of automated treatment planning and recommendations for clinical deployment. Med Phys 2019;46:2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.