Abstract
We report two novel genosensors for the quantification of SARS-CoV-2 nucleic acid using glassy carbon electrodes modified with a biocapture nanoplatform made of multi-walled carbon nanotubes (MWCNTs) non-covalently functionalized with avidin (Av) as a support of the biotinylated-DNA probes. One of the genosensors was based on impedimetric transduction offering a non-labelled and non-amplified detection of SARS-CoV-2 nucleic acid through the increment of [Fe(CN)6]3-/4- charge transfer resistance. This biosensor presented an excellent analytical performance, with a linear range of 1.0 × 10−18 M - 1.0 × 10−11 M, a sensitivity of (5.8 ± 0.6) x 102 Ω M−1 (r2 = 0.994), detection and quantification limits of 0.33 aM and 1.0 aM, respectively; and reproducibilities of 5.4% for 1.0 × 10−15 M target using the same MWCNTs-Av-bDNAp nanoplatform, and 6.9% for 1.0 × 10−15 M target using 3 different nanoplatforms. The other genosensor was based on a sandwich hybridization scheme and amperometric transduction using the streptavidin(Strep)-biotinylated horseradish peroxidase (bHRP)/hydrogen peroxide/hydroquinone (HQ) system. This genosensor allowed an extremely sensitive quantification of the SARS-CoV-2 nucleic acid, with a linear range of 1.0 × 10−20 M - 1.0 × 10−17 M, detection limit at zM level, and a reproducibility of 11% for genosensors prepared with the same MWCNTs-Av-bDNAp1 nanoplatform. As a proof-of-concept, and considering the extremely high sensitivity, the genosensor was challenged with highly diluted samples obtained from SARS-CoV-2 RNA PCR amplification.
Keywords: SARS-CoV-2, Impedimetric biosensor, Amperometric biosensor, Carbon nanotubes, Avidin, Nanotechnology
1. Introduction
The coronavirus disease nineteen (COVID-19) is an emerging human infectious disease produced by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wunsch et al., 2022). SARS-CoV-2 belongs to the Coronaviridae family, whose genome is composed by a single-strand and positive RNA, which comprises genes encoding for the main structural proteins as the spike surface glycoprotein (S gene), small envelope (E gene), matrix (M gene), nucleocapsid (N gene), RNA-dependent RNA polymerase (RdRp gene), and several still unidentified non-structural open reading frames, and also includes the 3′UTR (Wu et al., 2020). The outbreak started in China at the end of 2019 and was rapidly spread across the world due to the easy and fast human-to-human transmission, causing a large number of deaths, the collapse of medical systems even in developed countries, and severe economic consequences for billions of persons worldwide (Seyedalinaghi et al., 2022).
The golden standard technique for viral RNA detection is the real-time reverse transcription polymerase chain reaction (rRT-PCR) (Chu et al., 2020). In spite of the great advantages of this technique, mainly connected to the selectivity and sensitivity, it presents several drawbacks due to the cost of the instruments, the requirement of centralized use, the need for trained personnel, and the time taken for obtaining the results. Therefore, developing accurate methodologies to detect the SARS-CoV-2 RNA is highly required to make possible the rapid diagnosis of COVID-19.
Electrochemical biosensors represent an interesting alternative for the detection of RNAs (Kudr et al., 2021; Luong et al., 2021). Since 1960, they have been widely used for the quantification of different biomarkers mainly due to the well-known advantages connected with portability, miniaturization, sensitivity, requirement of small volumes of sample, friendly use, and very competitive analytical performance (Eguílaz et al., 2019; Min et al., 2021; Novodchuk et al., 2020). The advent of nanomaterials largely contributed to the development of innovative and versatile electrochemical sensors (Bakirhan et al., 2020; Mujica et al., 2020; Rivas et al., 2017). In this sense, we have reported the importance of using multi-walled carbon nanotubes (MWCNTs) non-covalently functionalized to build electrochemical (bio)sensors (Eguílaz et al., 2016; Gutierrez et al., 2017, 2019; Mujica et al., 2021; Ortiz et al., 2019; Primo et al., 2014). In particular, the use of avidin (Av) as non-covalent functionalization agent, made possible the development of successful hydrogen peroxide (Gutierrez et al., 2019) and glucose (Gallay et al., 2020) biosensors by drop-coating of the resulting Av-functionalized MWCNTs (MWCNTs-Av hybrid) at glassy carbon electrodes (GCE) and further immobilization of biotinylated horseradish peroxidase (bHRP) or biotinylated glucose oxidase (bGOx), respectively. Recently, we demonstrated the usefulness of MWCNTs-Av hybrid as biocapture platform for the development of a label-free, non-amplified, and highly sensitive impedimetric BRCA1 biosensor through the stable and specific anchorage of the biotinylated DNA probe in solution (Mujica et al., 2022).
Kashefi-Kheyrabadi et al. (2022) reported an interesting electrochemical sensor based on the use of a four-way junction hybridization to simultaneously detect SARS-CoV-2 spike (S) and open reading frame (ORF1ab) genes in 60 min. The proposed sensor was prepared by immobilizing at a gold nanoneedle structured electrode a thiolated universal DNA-hairpin able to hybridize with the target RNA and two adapters, one of them modified with a redox probe, either ferrocene or methylene blue (MB). In the presence of the target, these redox probes approach to the electrode and allow the highly sensitive detection of S and ORF1ab genes. Kumar et al. (2021) proposed a printed circuit board (PCB) electrode and a portable miniPCR for the electrochemical biosensing of SARS-CoV-2 RNA in wastewater at fM level, using the response of the intercalated MB as analytical signal. Peng et al. (2021) described the fM detection of SARS-CoV-2 RNA with an electrochemical biosensor where the target triggers the catalytic hairpin and initiates the DNA polymerization, producing a large amount of long ssDNA that interacts with [Ru(NH3)6]3+. Hwang et al. (2021) reported a label-free, real-time, low-cost, and high throughput biosensor for the detection of SARS-CoV-2 nucleic acid at nM level using interdigitated Pt/Ti electrodes immobilized at a glass substrate modified with 3-aminopropyltriethoxysilane. The increment of the capacitance due to the strengthened spatial charge distribution once the recognition event took place, was used as the analytical signal. Zhao et al. (2021) reported a highly sensitive super sandwich-type electrochemical biosensor based on p-sulfocalix[8]arene (SCX8) functionalized reduced graphene oxide (RGO). They proposed the use of a capture probe immobilized at Au@Fe3O4 nanoparticles, a label probe immobilized at a nanocomposite made of Au@SCX8-RGO-toluidine blue (TB) to obtain the sandwich hybrid, and a secondary probe to produce long concatemers. The analytical signal was obtained from the changes in the currents of TB (redox probe) once the supramolecular multistructure was transferred to screen printed carbon electrodes (SPCE). Hussein et al. (2021) proposed the use of SPCE modified with carbon nanotubes (CNTs)-WO3 to imprint the polymeric matrix with the viral complementary binding sites, reporting a detection limit of 57 pg mL−1. Alafeef et al. (2020) reported a paper-based electrochemical sensor chip using a graphene-based platform modified with gold nanoparticles (AuNPs) capped with four highly specific antisense oligonucleotides (ssDNA) for the fast, low-cost, simple, sensitive, and selective detection of SARS-CoV-2 RNA, without any amplification. The success of this strategy was due to the combination of the advantages associated with the conductivity of graphene, the amplification of the analytical signal by AuNPs, and the selection of ssDNAs able to simultaneously target two regions of SARS-CoV-2 RNA.
In this work, we propose two genosensors for the quantification of SARS-CoV-2 nucleic acid with impedimetric and amperometric transductions, using GCE modified with a biocapture nanoplatform made of MWCNTs non-covalently functionalized with Av as a support of the biotinylated DNA probes. In the following sections we report the optimization of the hybridization and biosensing conditions, and the analytical performance of the resulting genosensors.
2. Material and methods
2.1. Reagents
Multi-walled carbon nanotubes (MWCNTs) (outer diameter: 6–13 nm, length: 2.5–20 μm, purity >98%), bovine serum albumin (BSA), and hydroquinone (HQ) were purchased from Sigma-Aldrich. Ethanol and sodium chloride (NaCl) were obtained from J.T. Baker and Mallinckrodt, respectively. Potassium ferrocyanide (K4Fe(CN)6) and potassium ferricyanide (K3Fe(CN)6) were acquired from Merck. Polysorbate 20 (Tween 20) were purchased from Parafarm. Hydrogen peroxide (30% v/v aqueous solution) was obtained from Cicarelli. Avidin (Av) from egg white (catalog number A-2667), streptavidin (Strep) (catalog number 434302), and biotinylated horseradish peroxidase (bHRP) (catalog number 432040) were supplied by Invitrogen. DNA sequences, purchased from GenScript company, are as follows:
2.1.1. Label-free genosensor
Biotinylated DNA probe (bDNAp): 5′-biotin-C6-TTTTTTTTTT-GGA-AGC-GAC-AAC-AAT-TAG-TTT-TTA-GGA-ATT-TAG-CAA-AAC-CAG-CTA-CTT-TAT-CAT-TGT-AGA-TGT-CA-3′
2.1.2. Sandwich hybridization-based genosensor
Biotinylated DNA probe 1 (bDNAp1): 5’-biotin-C6-GGA-AGC-GAC-AAC-AAT-TAG-TTT-TTA-GGA-ATT-C6-3’
Biotinylated DNA probe 2 (bDNAp2): 5’-ATT-GTA-GAT-GTC-AAA-AGC-CCT-GTA-TAC-GAC-C6-biotin-3’
2.1.3. Common to both genosensors
DNA copy from SARS-CoV-2 RNA (target): 5’-GTC-GTA-TAC-AGG-GCT-TTT-GAC-ATC-TAC-AAT-GAT-AAA-GTA-GCT-GGT-TTT-GCT-AAA-TTC-CTA-AAA-ACT-AAT-TGT-TGT-CGC-TTC-C-3’
Scrambled sequence: 5’-TGA-TGC-GCA-CTT-TAG-GGG-TCA-CGA-GCA-CCG-TCG-CCC-TGC-TAG-TTG-GGG-TAG-CCC-GGA-AGC-CCC-CAG-CCG-GTG-GTG-ATA-GGA-A-3’
Ultrapure water (ρ = 18.2 MΩ cm) from a Millipore-MilliQ system was used for preparing all aqueous solutions.
2.2. Apparatus
A classical three-electrodes system was used for electrochemical measurements: glassy carbon electrode (GCE) as working electrode, a platinum wire as counter electrode, and Ag/AgCl, 3 M NaCl (BAS), as reference electrode. Electrochemical impedance spectroscopy (EIS) was carried out with a PGSTAT30 potentiostat (Metrohm). The impedimetric measurements were performed using 1.0 × 10−3 M [Fe(CN)6]3-/4- as redox marker (prepared in 0.050 M phosphate buffer solution pH 7.40 with 0.500 M NaCl (PB–NaCl)). The EIS parameters were the following: amplitude: 0.010 V, frequency range: from 1.0 × 10−2 to 1.0 × 106 Hz, and working potential: 0.200 V. All EIS spectra were analyzed and fitted using the Z-view program. All amperometric recordings were obtained with a TEQ_04 potentiostat.
Sonication was performed by a TB04TA Testlab ultrasonic cleaner of 40 kHz frequency and 160 W ultrasonic power. The centrifugation process was carried out with a Olto Alresa centrifuge model Digicen 21.
2.3. Preparation of MWCNTs-Av-bDNAp
The procedure to prepare MWCNTs-Av-bDNAp was performed as described previously by Mujica et al. (2022). Briefly, a given amount of MWCNTs was mixed with 1.250 mg mL−1 avidin (prepared in 50/50 v/v ethanol/water) to obtain 0.625 mg MWCNTs per mL of avidin solution. The mixture was sonicated for 15 min using a sonication bath and then, centrifuged at 10,000 rpm for 5.0 min. The supernatant containing MWCNTs-Av hybrid was used for further work as platform to immobilize the bDNAp or bDNAp1.
To obtain MWCNTs-Av-bDNAp or MWCNTs-Av-bDNAp1, 180 μL of 150 ppm bDNAp or bDNAp1 (prepared in PB-NaCl) and 180 μL of MWCNTs-Av hybrid were placed in a stirrer and left to mix for 60 min to finally obtain the corresponding biorecognition platform (either MWCNTs-Av-bDNAp or MWCNTs-Av-bDNAp1) (Fig. S1).
2.4. Construction of the label-free impedimetric genosensor (GCE/MWCNTs-Av-bDNAp/BSA)
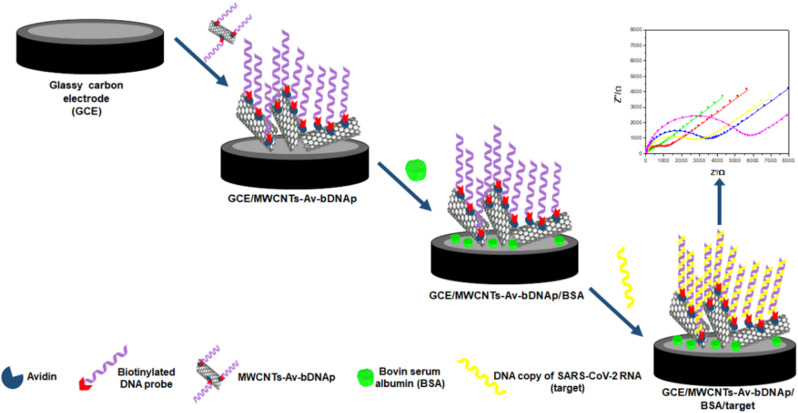
Before modification, GCEs were polished with 1.0, 0.3, and 0.05 μm alumina slurries for 2 min each, rinsed with ultrapure water and dried with nitrogen stream. GCE/MWCNTs-Av-bDNAp was prepared by dropping 10 μL of MWCNTs-Av-bDNAp onto GCE. Once dried, after solvent evaporation at room temperature, the resulting GCE/MWCNTs-Av-bDNAp electrode was washed with PB-NaCl, and dipped into 2.0% w/v BSA solution to avoid non-specific adsorptions (GCE/MWCNTs-Av-bDNAp/BSA). After washing with the same buffer solution, the hybridization took place at GCE/MWCNTs-Av-bDNAp/BSA in the presence of the target solution for 60 min at room temperature. After washing with PB-NaCl, the electrode was immersed in the redox indicator solution to perform the impedimetric measurements. The different steps for the preparation of the genosensor are shown in Fig. 1 .
Fig. 1.
Schematic representation of the steps followed during the construction of the impedimetric genosensor.
2.5. Construction of sandwich-hybridization based amperometric genosensor (GCE/MWCNTs-Av-bDNAp1)
The sensor was prepared in a similar way as GCE/MWCNTs-Av-bDNAp, just using MWCNTs-Av-bDNAp1 instead of bDNAp. After solvent evaporation at room temperature, the resulting sensor GCE/MWCNTs-Av-bDNAp1 was washed with PB-NaCl and dipped into 2.0% w/v BSA solution. After washing with the same buffer solution, the hybridization took place at GCE/MWCNTs-Av-bDNAp1/BSA in the presence of the target solution for 60 min at room temperature. After washing with PB-NaCl, GCE/MWCNTs-Av-bDNAp1/BSA/target was coated with 20 μL of 25 ppm bDNAp2 for 60 min to perform the sandwich hybridization (GCE/MWCNTs-Av-bDNAp1/BSA/target/bDNAp2). Once the hybrid was formed, the surface was coated with 10 μL of 100 μg mL−1 Strep for 30 min to allow the binding of bDNAp2-Strep (GCE/MWCNTs-Av-bDNAp1/BSA/target/bDNAp2/Strep). After rinsing with 0.050 M phosphate buffer pH 7.40, bHRP was placed onto the surface for 15 min (10 μL, 1.0 mg mL−1). The resulting GCE/MWCNTs-Av-bDNAp1/BSA/target/bDNAp2/Strep/bHRP was rinsed with Tween 20, and then immersed in 0.050 M phosphate buffer pH 7.40 containing 0.500 mM HQ as redox mediator. The analytical signal was obtained by amperometry in the presence of 0.100 mM hydrogen peroxide at a potential of −0.100 V.
2.6. Determination of SARS-CoV-2 nucleic acid after amplification by asymmetric PCR
A control DNA fragment of 203 bp derived from the RdRp gene of SARS-CoV-2, was obtained by conventional PCR amplification. PCR assays were also performed with a 20-fold molar excess of either, forward (F3) or reverse (B3) primers to enrich the synthesis of both, sense or antisense single strand DNA fragments, which has the same sequence as target or capture DNA, respectively. Briefly, conventional PCR amplification was performed in a final volume of 25 μL with 1 U of recombinant Taq DNA polymerase (Pegasus, Productos Bio-Lógicos, Quilmes, Argentina), 1.5 mM MgCl2, 200 μM dNTP, 5 μM of forward (5'-TGCTTCAGTCAGCTGATG-3') and reverse (5'-TTAAATTGTCATCTTCGTCCTT-3') primers. For asymmetric PCR, one of the primers was used at 0.25 μM concentration instead of 5 μM, yielding a 20 molar excess of one of them. The reaction was carried out during 30 cycles in a MultiGene™ OptiMax thermal cycler, with the following thermal profile: 94 °C, 1 min, 55 °C, 30 s, 72 °C, 1 min, for the denaturation, hybridization, and elongation steps, respectively. An initial denaturation step was included at the beginning of the reaction (95 °C, 5 min) and a final elongation step at the end of the cycles (72 °C, 5 min).
The PCR products were then analyzed by 1.5% agarose gel electrophoresis during 2 h at 50 mA, in 1X TAE buffer (TAE: Tris-sodium acetate 40 mM pH 8.0, EDTA 1 mM) and visualized under UV radiation upon staining with SYBR Safe (Sigma-Aldrich).
The PCR samples were denatured in a boiling water bath for 5 min followed by immediate cool-down in an ice-bath.
3. Results and discussion
3.1. Impedimetric genosensor for the quantification of SARS-CoV-2 nucleic acid
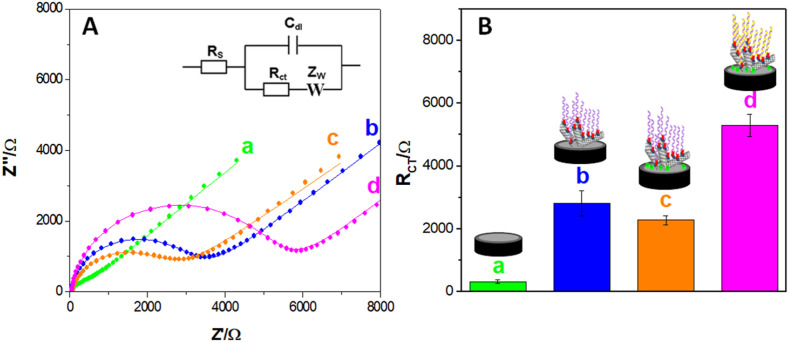
Fig. 2A displays Nyquist plots obtained after each step during the construction of the impedimetric genosensor (GCE (a), GCE/MWCNTs-Av-bDNAp (b), GCE/MWCNTs-Av-bDNAp/BSA (c), and once the hybridization event took place (GCE/MWCNTs-Av-bDNAp/BSA/target (d)). The symbols correspond to the experimental results while the solid lines correspond to the fitting with the model. In this case, the results were fitted with the Randles circuit, where Rs is the solution resistance, Rct the redox marker charge transfer resistance, Cdl the double-layer capacitance, and W the impedance of Warburg. Very well-defined spectra are observed in all cases, showing an excellent agreement with the model. Fig. 2B displays the bars plot for the Rct obtained from the spectra displayed in Fig. 2A. At GCE, the Rct is (3.2 ± 0.5) x 102 Ω and it increases to (2.8 ± 0.4) x 103 Ω after the immobilization of MWCNTs-Av-bDNAp due to: i) the partial blockage of the electrode surface by the protein that supports the MWCNTs and by the non-conductive DNA layer, and ii) the electrostatic repulsion between the negatively charged sugar phosphate-backbone of the nucleic acid and the negatively charged redox indicator. The incorporation of BSA at GCE/MWCNTs-Av-bDNAp (to minimize non-specific interactions), produces a small decrease of Rct mainly due to partial shielding of the negative charges of the phosphate groups that decreases the electrostatic repulsion (Rct = (2.3 ± 0.1) x 103 Ω). The sequence-specific biorecognition of the SARS-CoV-2 nucleic acid produces a significant increment of Rct ((5.3 ± 0.4) x 103 Ω) due to the increased repulsion and surface blockage as a consequence of the duplex formation.
Fig. 2.
(A) Nyquist plots obtained after the different steps during the construction of the impedimetric genosensor: a) GCE, b) GCE/MWCNTs-Av-bDNAp, c) GCE/MWCNTs-Av-DNAp/BSA, d) GCE/MWCNTs-Av-DNAp/BSA/target. Inset: equivalent circuit used to fit the impedance spectra. (B) Bars plot for the charge transfer resistances (Rct) obtained from the Nyquist spectra shown in (A). Hybridization time: 60 min. Target concentration: 1.0 × 10−15 M. Redox marker: 1.0 × 10−3 M [Fe(CN)6]3−/[Fe(CN)6]4−; Frequency range: 10 KHz to 10 mHz; Potential amplitude: 10 mV; Working potential: 0.200 V. Supporting electrolyte: 0.050 M phosphate buffer solution pH 7.40 with 0.500 M NaCl (PB–NaCl).
We investigated the influence of the hybridization time for 1.0 × 10−15 M target (Fig. S2). The Rct of the redox marker increases with the hybridization time up to 60 min due to the enhancement of the blockage and the electrostatic repulsion produced by the increasing amount of the duplex generated at GCE/MWCNTs-Av-bDNAp/BSA surface. After 60 min, the available bDNAp for hybrid formation was saturated, and Rct slightly decreased. Therefore, 60 min was selected as the optimal hybridization time.
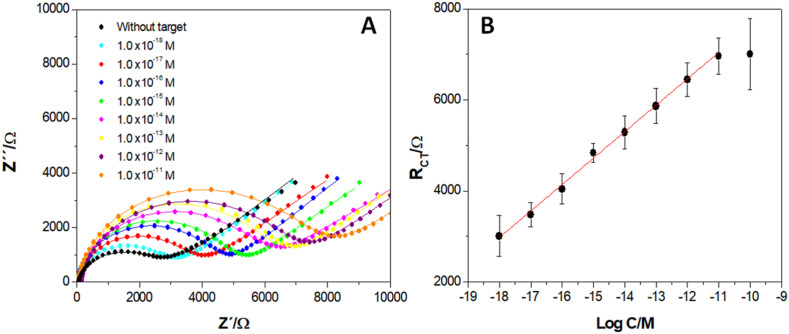
Fig. 3A displays the Nyquist plots obtained for GCE/MWCNTs-Av-bDNAp/BSA in the absence and presence of increasing concentrations of target, from 1.0 × 10−18 M to 1.0 × 10−11 M. Fig. 3B depicts the calibration plot obtained from the Nyquist diagrams displayed in Fig. 3A. A very-well defined impedimetric response is obtained even for aM level of target, with a linear relationship between Rct and the log of target concentration in the whole range evaluated. The sensitivity was (5.8 ± 0.6) x 102 Ω M−1 ((20 ± 2) x 103 Ω M−1 cm−2) (r2 = 0.994), while the detection and quantification limits were 0.33 aM and 1.0 aM, respectively (calculated as 3s/S and 10s/S, respectively, where s is the standard deviation of the blank signal and S the sensitivity).
Fig. 3.
(A) Nyquist plots obtained for GCE/MWCNTs-Av-bDNAp/BSA in the absence and presence of different target concentrations. (B) Calibration plot obtained from the results shown in (A). Other conditions as in Fig. 2.
The genosensor demonstrated to be highly reproducible. The reproducibility for 1.0 × 10−15 M target was 5.4% using 5 different GCEs modified with the same MWCNTs-Av-bDNAp nanoplatform (average Rct = (5.0 ± 0.3) x 103 Ω) and 6.9% using 12 different GCEs modified with three nanoplatforms independently prepared (average Rct = (4.8 ± 0.2) x 103 Ω). We also challenged the genosensor with a scrambled DNA sequence. Fig. S3 depicts the Rct of the redox marker obtained at GCE/MWCNTs-Av-bDNAp/BSA before (a) and after the interaction with 1.0 × 10−15 M target (b) and 1.0 × 10−13 M scrambled sequence (c). Almost no change in the Rct was observed in the presence of the scrambled sequence, even two orders of magnitude more concentrated than the target ((2.3 ± 0.1) x 103 Ω for GCE/MWCNTs-Av-bDNAp-BSA versus (2.0 ± 0.3) x 103 Ω for GCE/MWCNTs-Av-bDNAp-BSA-scrambled), demonstrating the efficient sequence-specific recognition of our genosensor.
3.2. Sandwich-hybridization based amperometric genosensor for the quantification of SARS-CoV-2 nucleic acid
In order to improve even more the detection level and selectivity of the SARS-CoV-2 nucleic acid quantification, we also developed a genosensor based on sandwich type hybridization (Fig. 4 A). In this scheme, the transduction of the biorecognition event was performed by amperometry from the reduction of the redox mediator (HQ) in the presence of hydrogen peroxide. bHRP reduces the hydrogen peroxide and the Fe(III) of the hemoprotein is oxidized to Fe(IV). In turn, HQ reduces the oxidized enzyme, while being oxidized to benzoquinone, which is finally reduced at the electrode surface, generating the analytical signal.
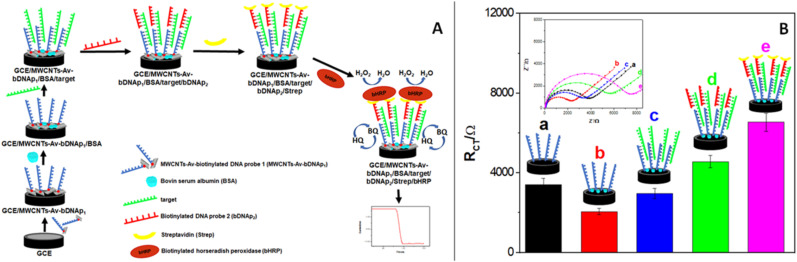
Fig. 4.
(A) Schematic representation of the steps followed during the construction of the sandwich hybridization-based amperometric genosensor. (B) Bars plot for the Rct obtained after the different steps during the construction of the amperometric genosensor: a) GCE/MWCNTs-Av-bDNAp1, b) GCE/MWCNTs-Av-bDNAp1/BSA, c) GCE/MWCNTs-Av-bDNAp1/BSA/target, d) GCE/MWCNTs-Av-bDNAp1/BSA/target/bDNAp2, e) GCE/MWCNTs-Av-bDNAp1/BSA/target/bDNAp2/Strep. Target concentration: 1.0 × 10−15 M. Hybridization time bDNAp1/target and target/bDNAp2: 60 min. Strep interaction time: 30 min. Inset: Nyquist plots obtained after the different steps. Other conditions as in Fig. 2.
The efficiency of the construction of the genosensor was evaluated by EIS from the changes in Rct of the redox mediator. Fig. 4B displays the bars plot for the Rct obtained after the different steps during the sandwich hybridization and Strep incorporation. The inset depicts the corresponding Nyquist diagrams. As expected, according to the results obtained for the impedimetric genosensor, the Rct significantly increases once GCE is modified with MWCNTs-Av-bDNAp1 ((3.2 ± 0.6) x 102 Ω versus (3.4 ± 0.3) x 103 Ω), decreases after the incorporation of BSA ((2.1 ± 0.2) x 103 Ω), and increases after hybridization with 1.0 × 10−15 M DNA target ((3.0 ± 0.3) x 103 Ω). Further incorporation of the bDNAp2 produced an additional increment in the Rct due to the enhanced surface blockage and the electrostatic repulsion with the redox marker ((4.6 ± 0.3) x 103 Ω). The bioaffinity interaction with Strep also produced an increment in the Rct due to the non-conductive nature of the protein ((6.5 ± 0.5) x 103 Ω). A decrease in Rct is obtained after the association of the biotinylated horseradish peroxidase (bHRP) due to the presence of the iron in the heme group (not shown).
We evaluated the influence of hydrogen peroxide and HQ concentrations and the working potential on the analytical performance of the genosensor (not shown). We compared the amperometric response obtained at −0.100 V in the presence of 0.50 mM HQ for additions of 0.10 mM and 1.00 mM hydrogen peroxide. The best compromise between the time required for baseline stabilization and the time necessary to reach the stationary current, was obtained with 0.10 mM hydrogen peroxide. Regarding the HQ concentration, two values were evaluated, 0.25 and 0.50 mM, being 0.50 mM the one that made possible the fast regeneration of the enzyme and stabilization of the baseline; higher concentrations produced higher baseline currents and slower stabilization. Thus, 0.50 mM HQ was the selected concentration value. The working potential was another important parameter, being −0.100 V the one that allowed to obtain a sensitive signal, with reasonable time to stabilize the baseline without oxygen interference. Therefore, the analytical signal was obtained by amperometry at −0.100 V in the presence of 0.50 mM HQ for one addition of 0.10 mM hydrogen peroxide.
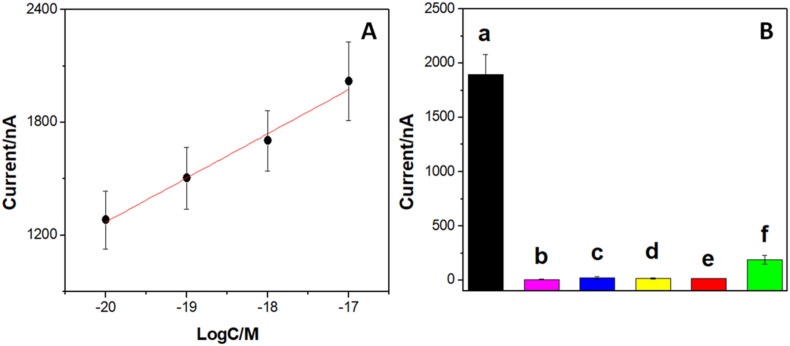
Fig. 5A depicts the calibration plot obtained from the amperometric responses and the inset shows the amperometric responses obtained in the absence and presence of different target concentration. A fast and well-defined response is obtained for all the used concentrations, with a linear relationship between 1.0 × 10−20 M and 1.0 × 10−17 M and a sensitivity of (2.4 ± 0.3) x 102 nA M−1 ((8 ± 1) x 103 nA M−1 cm−2) (r2 = 0.99). In addition, it is important to remark the extremely low detection and quantification limits, with values of 3.3 × 10−21 M, and 10 × 10−21 M, respectively. The reproducibility for 3 genosensors prepared with the same MWCNTs-Av-bDNAp1 platform was 11%.
Fig. 5.
(A) Calibration plot obtained for the amperometric genosensor. Inset: amperometric response obtained in the absence and presence of different target concentrations. bHRP interaction time: 15 min. Working potential: -0.100 V Hydrogen peroxide concentration: 0.10 mM. HQ concentration: 0.50 mM. Other conditions as in Fig. 4 (B). Bars plot for the currents obtained from amperometric experiments using GCE/MWCNTs-Av-bDNAp1/BSA under different conditions: a) in the presence of 1.0 × 10−17 M target; (b) in the absence of the target; (c) in the absence of bDNAp2 for 1.0 × 10−17 M target; (d) in the absence of Strep for 1.0 × 10−17 M target; (e) in the presence of the 1.0 × 10−17 M scrambled sequence instead of the target; and (f) in the presence of non-biotinylated HRP for 1.0 × 10−17 M target. Other conditions as in Fig. 5A.
Different controls were performed to demonstrate that the analytical signal was really due to the sandwich hybridization. Fig. 5B displays the currents obtained under different experimental conditions. In this figure, the current obtained at the optimal biosensor in the presence of 1.0 × 10−17 M target (a) is included for comparison. As expected, in the absence of the target (b), there is no signal since the sandwich hybridization does not occur. If bDNAp2 is not present, the addition of 1.0 × 10−17 M target did not give response since the sandwich hybridization cannot take place (c). When Strep is not immobilized at the biosensor, there is no response since bHRP cannot be incorporated (d). Moreover, there is no response when the target is replaced by a scrambled sequence, demonstrating the high selectivity of the biorecognition process (e). If bHRP is replaced by non-biotinylated HRP, the current is 10% of the value obtained in the presence of bHRP due to minimal non-specific enzyme interaction (f). These results clearly demonstrate that the proposed sandwich hybridization scheme is successful and allows a very sensitive and selective biosensing of SARS-CoV-2 nucleic acid.
Table S1 compares the analytical characteristics of our biosensors with the ones for the most relevant SARS-CoV-2 nucleic acid electrochemical biosensors. It can be seen that our genosensors present highly competitive analytical performances, with comparable or even better detection limits than most of the reported biosensors.
3.3. Proof-of-concept of possible analytical applications
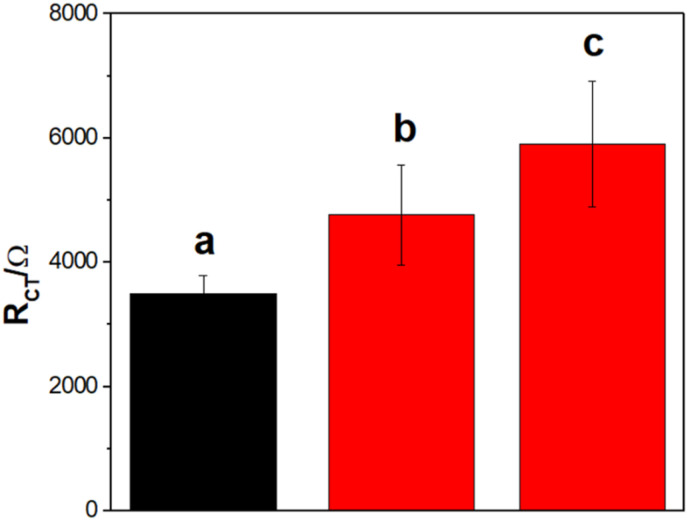
As a proof-of-concept of the possible analytical applications of the proposed biosensor, we used samples obtained from asymmetric PCR of SARS-CoV-2 RNA. Taking into account the excellent sensitivity of the developed genosensors, we evaluated the response of diluted PCR DNA sequence obtained with a 20-fold molar excess of the forward primer (F3), containing the target RdRp DNA sequence. Fig. 6 shows the Rct obtained for GCE/MWCNTs-Av-bDNAp/BSA in the absence (a) and presence of PCR samples diluted 1010 (b) and 104 (c) times. Even after 1010 times dilution, it was possible to detect the presence of SARS-CoV-2 nucleic acid, whereas no specific detection was obtained when a 20-fold molar excess of the reverse primer (B3) was used as control (data not shown). These results demonstrate the enormous potential of our genosensor for further analytical applications either for direct quantification of SARS-CoV-2 nucleic acid or for the association of the biosensor and conventional asymmetric PCR with a considerably lower number of cycles.
Fig. 6.
Proof-of-concept of the potential analytical application of the genosensor. Rct obtained for GCE/MWCNTs-Av-bDNAp/BSA before (a) and after (b, c) hybridization with PCR samples diluted with PB-NaCl 1010 (b) and 104 (c) times. Other conditions as in Fig. 2.
4. Conclusions
The proposed genosensors demonstrate the importance of the functionalization of MWCNTs with Av to obtain, in a simple way, biocapture nanoplatforms able to be used for the development of genosensors via the specific anchoring of bDNA probes. The impedimetric genosensor is an innovative and original alternative that allows the highly sensitive quantification of SARS-CoV-2 nucleic acid at aM levels without any amplification scheme. The sandwich hybridization-based amperometric biosensor made possible an extremely high enhancement of sensitivity reaching detection limits at zM levels. The detection of SARS-CoV-2 nucleic acid in highly diluted PCR samples revealed the high potential of the proposed genosensors for: i) further developments based on their combination with PCR schemes using a considerably lower number of amplification cycles, with the advantages of a drastic decrease of the assay time and cost, and an additional confirmation of positive cases; and ii) the direct quantification of SARS-CoV-2 RNA even without involving any amplification technique.
CRediT authorship contribution statement
Michael López Mujica: Methodology, Formal analysis, Investigation, Visualization. Alejandro Tamborelli: Methodology, Formal analysis, Investigation, Visualization. Andrés Castellaro: Formal analysis, Investigation. Danilo Barcudi: Formal analysis, Investigation. María D. Rubianes: Validation, Project administration. Marcela C. Rodríguez: Validation, Project administration. Héctor A. Saka: Validation, Project administration. José L. Bocco: Conceptualization, Resources, Writing – review & editing, Funding acquisition. Pablo R. Dalmasso: Conceptualization, Validation, Writing – review & editing, Supervision, Funding acquisition. Gustavo A. Rivas: Conceptualization, Validation, Resources, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the financial and institutional support from ANPCyT-FONCyT (IP COVID-19–615, PICT 2016–1261, PICT 2018–03862, PICT 2019–01114, PICT 2015–0543, PICT 2018–4591), CONICET (PIP 2016), Universidad Tecnológica Nacional (PID PAECBCO0008294TC), and Universidad Nacional de Córdoba (SECyT-UNC 2018–2022). MLM and AT acknowledge their doctoral fellowships from CONICET. DB and AC are post-doctoral fellowships from CONICET and ANPCyT-FONCyT, respectively.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biosx.2022.100222.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alafeef M., Dighe K., Moitra P., Pan D. ACS Nano. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakirhan N.K., Topal B.D., Ozcelikay G., Karadurmus L., Ozkan S.A. Crit. Rev. Anal. Chem. 2020;52(3):519–534. doi: 10.1080/10408347.2020.1809339. [DOI] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krichnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguílaz M., Dalmasso P.R., Rubianes M.D., Gutierrez F., Rodríguez M.C., Gallay P.A., López Mujica M.E.J., Ramírez M.L., Tettamanti C.S., Montemerlo A.E., Rivas G.A. Curr. Opin. Electrochem. 2019;14:157–165. [Google Scholar]
- Eguílaz M., Gutierrez A., Rivas G.A. Sensor. Actuator. B Chem. 2016;216:629–637. [Google Scholar]
- Gallay P., Eguílaz M., Rivas G. Biosens. Bioelectron. 2020;148:111764. doi: 10.1016/j.bios.2019.111764. [DOI] [PubMed] [Google Scholar]
- Gutierrez F.A., González-Domínguez J.M., Ansón-Casaos A., Hernández-Ferrer J., Rubianes M.D., Martínez M.T., Rivas G.A. Sensor. Actuator. B Chem. 2017;249:506–514. [Google Scholar]
- Gutierrez F.A., Rubianes M.D., Rivas G.A. Anal. Chim. Acta. 2019;1065:12–20. doi: 10.1016/j.aca.2019.03.022. [DOI] [PubMed] [Google Scholar]
- Hwang C., Park N., Kim E.S., Kim M., Kim S.D., Park S., Kim N.Y., Kim J.H. Biosens. Bioelectron. 2021;185 doi: 10.1016/j.bios.2021.113177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H.A., Kandeil A., Gomaa M., Mohamed El Nashar R., El-Sherbiny I.M., Hassan R.Y.A. ACS Sens. 2021;6(11):4098–4107. doi: 10.1021/acssensors.1c01614. [DOI] [PubMed] [Google Scholar]
- Kashefi-Kheyrabadi L., Nguyen H.V., Go A., Baek C., Jang N., Lee J.M., Cho N.H., Min J., Lee M.H. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. Trends Anal. Chem. 2021;136 doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M.S., Nandeshwar R., Lad S.B., Megha K., Mangat M., Butterworth A., Knapp C.W., Knapp M., Hoskisson P.A., Corrigan D.K., Ward A.C., Kondabagil K., Tallur S. Sensor. Actuator. B Chem. 2021;343 [Google Scholar]
- Luong A.D., Buzid A., Vashist S.K., Luong J.H.T. Curr. Opin. Electrochem. 2021;30 doi: 10.1016/j.coelec.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Sempionatto J.R., Teymourian H., Wang J., Gao W. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112750. [DOI] [PubMed] [Google Scholar]
- Mujica M.L., Gallay P.A., Perrachione F., Montemerlo A.E., Tamborelli L.A., Vaschetti V., Reartes D., Bollo S., Rodríguez M.C., Dalmasso P.R., Rubianes M.D., Rivas G.A. J. Pharm. Biomed. Anal. 2020;189 doi: 10.1016/j.jpba.2020.113478. [DOI] [PubMed] [Google Scholar]
- Mujica M.L., Rubianes M.L., Rivas G.A. Sensor. Actuator. B Chem. 2022;357 [Google Scholar]
- Mujica M.L., Zhang Y., Gutiérrez F., Bédioui F., Rivas G. Microchem. J. 2021;160 [Google Scholar]
- Novodchuk I., Bajcsy M., Yavuz M. Carbon. 2020;172:431–453. [Google Scholar]
- Ortiz E., Gallay P., Galicia L., Eguílaz M., Rivas G.A. Sensor. Actuator. B Chem. 2019;292:254–262. [Google Scholar]
- Peng Y., Pan Y., Sun Z., Li J., Yi Y., Yang J., Li G. Biosens. Bioelectron. 2021;186 doi: 10.1016/j.bios.2021.113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primo E.N., Oviedo B., Sánchez C., n Rubianes M.D., Rivas G.A. Bioelectrochemistry. 2014;99:8–16. doi: 10.1016/j.bioelechem.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Rivas G.A., Rodriguez M.C., Rubianes M.D., Gutierrez F.A., Eguilaz M., Dalmasso P.R., Primo E.N., Tettamanti C., Ramirez M.L., Montemerlo A., Gallay P., Parrado C. Appl. Mater. Today. 2017;9:566–588. [Google Scholar]
- Seyedalinaghi A.A., Karimi A., Mojdeganlou H., Alilou S., Mirghaderi S.P., Noori T., Shamsabadi A., Dadras O., Vahedi F., Mohammadi P., Shojaei A., Mahdiabadi S., Janfaza N., Lonbar A.K., Mehraeen E., Sabatier J.M. Health Sci. Rep. 2022;5 doi: 10.1002/hsr2.516. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunsch K., Kienberger K., Niessner C. Int. J. Environ. Res. Publ. Health. 2022;19(4):2250. doi: 10.3390/ijerph19042250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Sensor. Actuator. B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.