Abstract
Well-structured and publicly accessible databases represent core resources for modern data-rich research as they consolidate filed-specific knowledge and highlight both best practices and challenges faced by each discipline. Further impactful growth of nanomaterials databases requires concerted efforts between database stewards, researchers, funding agencies, and publishers.
Modern nanotechnology research generates rapidly growing collections of experimental data on physical, chemical, and biological properties of nanomaterials. Projects such as nanoHUB,1 caNanoLab,2 or Nanomaterial Registry,3 have been developed predominantly by using manual data collection and curation efforts. Significant investment has been made in nanotechnology and many well-funded nanoscience research laboratories (for instance, hundreds of projects in nanotechnology have been funded by the NIH per NIH Reporter, https://projectreporter.nih.gov/). In contrast, the size of the current nanomaterials databases remains relatively small, with a few thousands of entries at best. This observation reflects a known discrepancy between data generated in the researchers’ laboratories and those available from electronic repositories; indeed, it has been estimated that as little as 12% of research data across biomedical domain have been deposited in electronic databases.4
Due to the diversity of nanomaterials and lack of current practices and tools for data sharing by direct deposition, current nanomaterial databases are not only small but also they often lack standards in the types of nanomaterials properties collected and systematically represented. This gap between data generation and shared data access calls for urgent actions to standardize, structure, and facilitate data collection and deposition into public databases. We argue that the concerted effort between research community, funding agencies, and journal publishers is needed to substantially increase the flow of data into public databases. Furthermore, this effort should be guided by the principles of data science as applied to nanomaterials research. We posit that the growth of publicly accessible databases will stimulate knowledge-based rational design of novel MNPs using nanomaterial modeling and informatics approaches.
Data science and data cycle of nanomaterials.
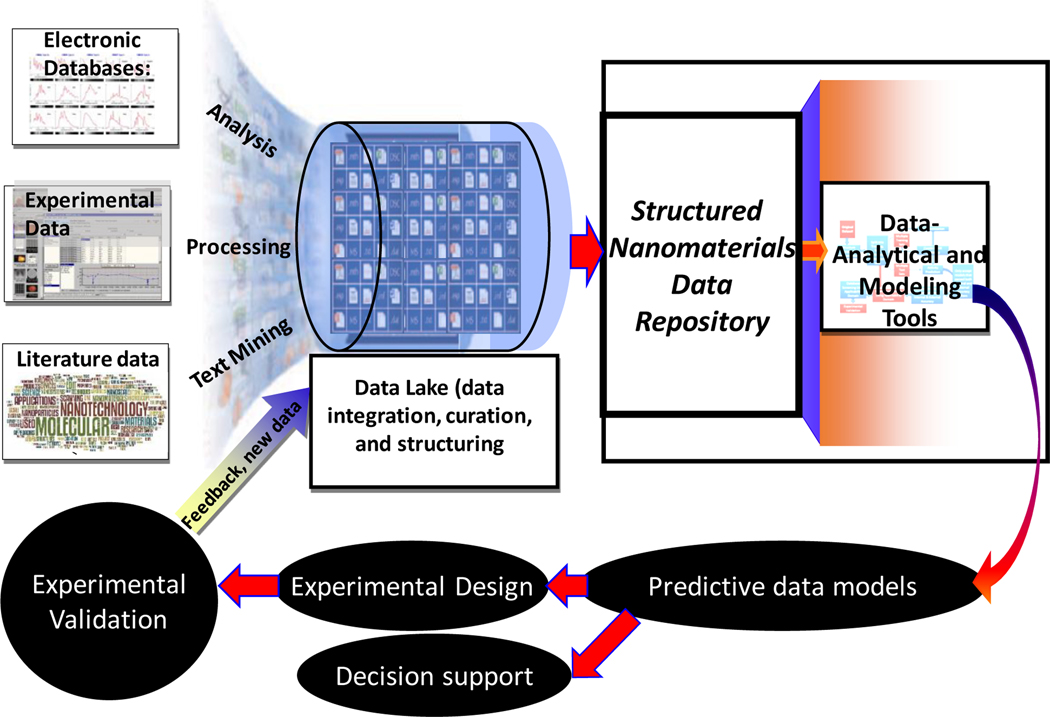
The power of data to catalyze new, rationally designed studies, which is the essence of data science (https://datascience.nih.gov/), requires deep understanding of all aspects of data flow, including initial generation, curation, sharing, organization, and predictive analytics to guide new experiments. In addition to capturing key elements of this data flow, the nanomaterials data cycle (Figure 1) emphasizes the dual role of experimental scientists as both data depositors and beneficiaries of the integrated data collections and models. However, it also underlines the critical role of databases as unifying engines of research progress5 that reflect best practices and approaches to data generation, organization, and modeling, and at the same time bring together experimental and computational scientists. Below, we review the key elements of the modern nanomaterial data cycle and emphasize the need to transform current practices in data collection to enrich nanomaterial databases (by tedious processing of published scientific literature) to community-driven direct deposition of research data into shared databases. We argue that such direct deposition would promote data sharing and dissemination in fulfillment of funding agencies’ requirements and provide data-driven support for experimental research.
Figure 1.
Data cycle in nanomaterials research. The cycle embeds the recently defined FAIR (Findable, Accessible, Interoperable, Reusable) Data Principles14 and includes the following distinct components: (i) diverse sources of data on nanomaterials such as electronic databases, experimental laboratory collections, and research papers; (ii) Data Lake, which consolidates data on nanomaterials from different sources to enable their curation and harmonization; (iii) Nanomaterial data repository, where curated data from Data Lake are organized based on certain ontology such as MIAN6 and accessible for reuse; (iv) Data modeling tools that enable the intelligent analysis of trends in data; (v) data models that can be explored for (vi) identification or design of novel nanomaterials with the desired properties or employed to support regulatory decisions to enable or forbid the manufacturing of industrial nanomaterials; (vii) experimental validation of model-based predictions and new data generation, which completes the nanomaterial data cycle.
Growth of nanomaterials databases, data sharing, and reproducibility crisis
The organized push to structure nanomaterial data has a relatively short history illustrated by the efforts on the part of the National Institutes of Health (NIH) to create curated databases for storing the emerging research results. First came caNanoLab, which was initiated in 2007 (https://cananolab.nci.nih.gov), followed by the formation of the Nanotechnology Working Group in 2008 (https://nciphub.org/groups/nanowg). This initial activities were followed by formulating ‘minimal information about nanomaterials’ (MIAN) ontology6 and the construction of the Nanomaterial Registry (https://www.nanomaterialregistry.org/) in 2010 and the ASTM guideline for nanomaterial data structuring in ISA-TAB-Nano’s template becoming available in 2013.7 In parallel, a Unified Descriptor System8 and ontologies9 have been constructed that would render any collation of data searchable and available for tool development or overlay to extract important metadata regarding questions of importance to the scientific community. Many researchers working in the area of nanomaterial informatics have joined the NCI caBIG Nanotechnology Working Group (https://nciphub.org/groups/nanowg) to develop common formats and ontology (see http://www.nano-ontology.org/) for diverse nanomaterials. The ISA-TAB-Nano effort was critical to enable the data accumulation and exchange between different databases.
While the NIH adopted a data sharing policy in 2003, it only started requiring nanomaterials research data deposition in designated registries in 2014. Specifically, a recent request for applications from the National Cancer Institute10 stipulated, for the first time, that all data generated in projects supported by the funded grants should be deposited either into CaNanolab (https://cananolab.nci.nih.gov/caNanoLab/#/) or Nanomaterial Registry (NR; https://www.nanomaterialregistry.org/).
Several curated databases are currently in use or under construction, each of which has a specific objective in serving the research community (Table 1). All of the available databases offer a strong foundation to support data analytics and the use of data for decision support but currently house small quantities of data. This could be, in part, due to high inefficiency of data sharing and data collection processes.
Table 1.
Key databases for nanomaterials
| Public Databases | Records | Comments |
|---|---|---|
| NBI Knowledgebase nbi.oregonstate.edu | 222 | Curated, in-house data |
| caNanoLab cananolab.nci.nih.gov | 1,226 | Curated, specific to cancer research |
| NanoWerk Nanomaterial Database www.nanowerk.com/nanomaterial-database.php | 2,515 | Curated, only commercially-available materials |
| Nanomaterial Registry www.nanomaterialregistry.org | 2,031 | Curated, validated data, broad resources |
| Nanoparticle Information Library (NIL) nanoparticlelibrary.net | 88 | Crowd-sourced, publicly available |
| Chemical Effects in Biological Systems (CEBS) www.niehs.nih.gov/research/resources/databases/cebs/index.cfm | 9,815 | Incorporates the National Toxicology Program data; only 4 nanomaterials |
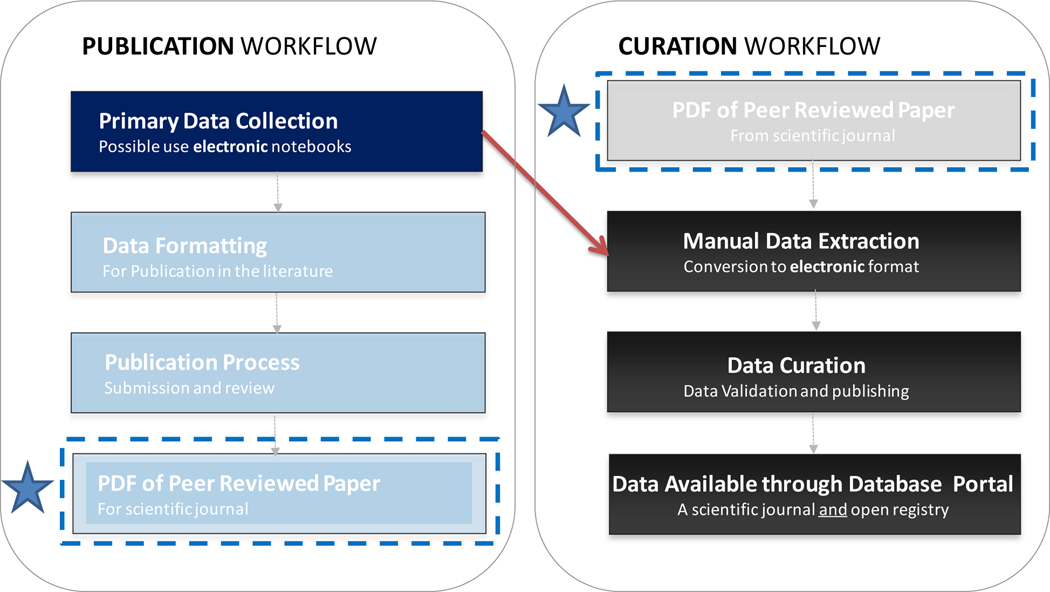
Indeed, Figure 2 illustrates the current flow of data from its collection in the laboratory through publication, extraction, curation and entry into a consolidated database. The inefficiency of this approach is potentially self-evident; but to elaborate, the current process of curation and deposition into the Nanomaterial Registry of data characterizing a single nanomaterial sample in a published paper can take up to 90 minutes. The curator has to locate all pertinent characterization data and meta-data within the study, interpret any biological or environmental assays performed, ensure the study integrity and, finally, manually enter all pieces of information into the database. Although the curator is assisted by an electronic curation tool with smart features, like drop down menus and controlled value relationships, this process is laborious and time consuming.
Figure 2.
Streamlining and expediting the database growth by direct deposition of the experimental data. Greyed boxes show current elements of data capture and processing that would become unnecessary if the direct data deposition is enabled.
The inefficiency of this process of collecting data from published papers and transmitting them into an electronic format suitable for database deposition is especially evident if one recognizes that in most research laboratories the data is initially collected and stored in electronic format. However, under the current practices the (originally, electronic) laboratory data is converted into a journal format such as pdf and submitted, most often, as supplementary material for a paper. Submitting this data to a public database in the same original electronic format would eliminate a significant portion of the manual actions involved in acquiring data from published materials. The potential for transcription errors in transferring the data from a publication into an electronic database by a curator would also be minimized.
The accumulation of highly curated data in public repositories with detailed descriptions of the experimental conditions used to measure nanomaterial properties may also contribute to solving the “reproducibility crisis” described in the authoritative Nature editorial.11 With the recognition that reproducibility of research results is extremely challenging, Nature, in 2013, began requesting full method descriptions, with no content limit, in order to maximize experimental information and the possibility for peer reproduction.11 The Nature Nanotechnology editorial concerning the reproducibility initiative was published in 2014;13 it stated that the journal “will be encouraging authors to deposit data in repositories” and recommended, “that authors choose repositories that provide expert curation to ensure the data are discoverable and can be linked to the paper”. It is hard to overestimate the importance of support by scientific journals for establishing FAIR (Findable, Accessible, Interoperable, Reusable) Data Principles14 to maximize the data utility for accelerating nanotechnology research.
Databases that accept and curate primary data depositions from experimental laboratories represent an invaluable means of flagging incomplete, suspicious, inconsistent, or irreproducible data. Indeed, the variability of synthetic approaches, storage, and handling can change many key physico-chemical and biological characteristics of nanomaterials. Thus, it is critical to capture and report all specific details of the experiments. A post-submission curation process, with the help of the MIAN6,12 ontology and standardized data format developed by ISA-TAB-Nano, would identify duplicative materials and compare their characteristics or highlight inconsistency of experimental nanomaterial characterization. Thus, data sharing through deposition into nanomaterial databases would significantly contribute to solving the reproducibility crisis.
The importance of data analytics for the experimental decision support
Recent developments in the field of nanoinformatics and nanomaterial modeling suggest that, similar to the transformative effects of cheminformatics and bioinformatics on chemistry and biology, respectively, nanoinformatics is poised to transform nanomaterial research from an empirical, trial-and-error driven to data- and knowledge driven. There is a growing list of examples describing the use of nanoinformatics approaches to understand and forecast the relationship between structure and biological activity or toxicity of nanomaterials. Some recent, exciting developments in nanoinformatics were reviewed in the Beilstein J. of Nanotechnology in 2015 (summarized in the editorial by Liu and Cohen15). Several studies have demonstrated that nanoinformatics approaches can successfully elucidate and forecast structure-property relationships for nanomaterials,16–21 and even guide the experimental discovery of novel nanomaterials with the desired properties.22 Importantly, the feasibility of such studies is predicated on the availability of large amounts of high quality, curated data on nanomaterials structure and physico-chemical and biological properties that should be easily accessible in public databases.
Very recently, Oh et al.23 demonstrated the power of intelligent text mining of nanotechnology publications to extract data on the cellular toxicity of cadmium-containing semiconductor quantum dots. Using random forest datamining approach frequently employed in both bio- and cheminformatics, the authors developed models predicting toxicity of engineered nanomaterials from their physico-chemical properties and experimental conditions. This study illustrates the power of modern methods for data capture from the literature; however, it also illuminates the problem of inefficiency or lack of data sharing that could be achieved by direct deposition into public databases.
Streamlining nanomaterials data collection
There are numerous research databases and respective informatics tools in many data-rich fields that have evolved from small data collections to large, wells-structured databases with robust underlying ontologies. Significant examples are provided by the Protein Data Bank24 supporting basic and applied research in structural and functional proteomics, or Pubchem25 supporting research in chemical biology and drug discovery. Both databases provide streamlined data deposition capabilities and facilitate the development of instructional data models to guide focused experimental research. These examples of robust databases established in mature data-rich disciplines show the feasibility of implementing informatics-driven strategies for prospective data collection in nanomaterial science. Importantly, the time to act is now, when the quantity of nanomaterials data is relatively small but the growth of data and associated development of nanoinformatics models is imminent. Building the effective data submission and initial processing capabilities at this point is thus a luxury that many other fields, with decades of cumulative data to manage, have not been afforded.
An excellent model for data sharing has been created and nurtured by the structural biology community where the Protein Data Bank (PDB)24 has been functioning as a primary depository of x-ray crystallography or NMR macromolecular structural data for decades. This unique database was renamed as the Research Collaboratory for Structural Biology (RCSB) to reflect the role that the database plays in catalyzing research on protein structure determination and structure-function studies. The protein community has developed a valuable robust system of mutual dependency between research funding, publication of research results in peer-reviewed journals, and open data sharing via the PDB. Indeed, publishing research results is an ineluctable attribute of basic research; however, the absolute majority of journals would not accept a paper reporting a novel protein structure without a notice from the PDB that the coordinates have been deposited with the database. As the success of grant applications is substantially facilitated by strong publication records of the investigators, and new research activities are impossible without funding, these journal stipulations provide strong incentives to researchers to share protein coordinates obtained in their laboratories with the research community served by the RCSB. Thus, data sharing is an integral part of the structural biology research culture and a standard practice of conducting and publishing research in protein crystallography making the database the centerpiece of structural biology.
Plausible trends are beginning to emerge in nanotechnology but they need to be cultivated and coordinated. Funding agencies such as NIH are beginning to provide special funding to support the development of databases. We also notice much greater openness of journals in recognizing the importance of data as a scientific commodity, which is helpful for developing data deposition formats (this trend is illustrated by the recently launched Scientific Data journal, http://www.nature.com/sdata/). As these important trends mature, we foresee that the culture of data sharing via the deposition into freely accessible nanomaterial databases will be widely adopted by the nanotechnology community but special efforts are needed to stipulate and accelerate this process.
Several important developments will facilitate the acceptance and implementation of data science principles in nanotechnology. Nano-ontologies (e.g., http://www.nano-ontology.org/) and minimal ENMs characterization standards require further development, publicity, and acceptance by the community of researchers. Database developers should facilitate data deposition by providing user-friendly tools for curation and validation as is common for databases such as PDB. Data accuracy and validation are important, as the direct data deposition process should not lower data quality; thus, it is envisioned that data will be deposited concurrently with the acceptance of the respective peer-reviewed manuscript by the journal, which mimics current practices for protein structure deposition to the PDB. Furthermore, deposition streamlining continues to require data curators to evaluate data for completeness, compliance with the deposition standards, and proper annotation.
Funding agencies, such as NIH, should reinforce and specify their data sharing policies by requiring data upload to public databases, such as NIH-funded Nanomaterial Registry or CaNanoLab, as a condition for the award. The longevity and sustainability of these databases should also be addressed. It is acknowledged by the NIH leaders that funding and sustainability of major databases should be given prime attention.5 The growth of nanomaterials databases will position them for competitive funding by sources such as Big Data to Knowledge (BD2K) programs at the NIH (https://datascience.nih.gov/). Additional funding models considered in the aforementioned publication5 by the leading NIH administrators such as “fee for service” or “subscription” merit further consideration.
It is also important that nanotechnology journals promote policies and practices exemplified by those established in structural biology. Specifically, journals need to implement editorial policies that require data deposition into the same curated nanomaterial databases supported by the funding agencies as a prerequisite for paper acceptance for publication.
Researchers in nanomaterials are in a unique position with respect to established fields in which there is a long history of producing large datasets. We have the luxury of evolving an approach to database structure and analytical tools that parallels the generation of data. This can provide real-time feedback into manufacturing, study design, data collection, and interpretation creating an enormous potential for crucial gap filling. Applying the principles of data science and research to nanotechnology will continuously refine the approaches to standardized data collection, representation, organization, comparison and modeling enabling rational, data-driven design of novel nanomaterials with the desired properties. We conclude that the communities of researchers, journal publishers, funding agencies and database managers must commit to the shared vision outlined above if we wish to achieve rapid and impactful outcomes for nanomaterials research and development.
Acknowledgements.
The authors would like to thank NIBIB, NIEHS, and NCI within the National Institutes of Health for funding the development of Nanomaterials Registry under contract HHSN2682010000022C. In addition, AT acknowledges support from NIH grants 5U54CA198999 and U01CA207160.
Footnotes
Competing financial interests: Authors declare no competing financial interests.
References
- 1.Madhavan K. et al. nanoHUB.org: cloud-based services for nanoscale modeling, simulation, and education. Nanotechnol. Rev. 2, 107–117 (2013). [Google Scholar]
- 2.Morris SA, Gaheen S, Lijowski M, Heiskanen M. & Klemm J. Experiences in supporting the structured collection of cancer nanotechnology data using CaNanoLab. Beilstein J. Nanotechnol. 6, 1580–1593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills KC, Murry D, Guzan KA & Ostraat ML Nanomaterial registry: database that captures the minimal information about nanomaterial physico-chemical characteristics. J. Nanoparticle Res. 16, 2219 (2014). [Google Scholar]
- 4.Read KB et al. Sizing the Problem of Improving Discovery and Access to NIH-Funded Data: A Preliminary Study. PLoS One 10, e0132735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne PE, Lorsch JR & Green ED Perspective: Sustaining the big-data ecosystem. Nature 527, S16–7 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Ostraat ML, Mills KC, Guzan KA & Murry D. The Nanomaterial Registry: facilitating the sharing and analysis of data in the diverse nanomaterial community. Int. J. Nanomedicine 8 Suppl 1, 7–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas DG et al. ISA-TAB-Nano: a specification for sharing nanomaterial research data in spreadsheet-based format. BMC Biotechnol. 13, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rumble J, Freiman S. & Teague C. Towards a Uniform Description System for Materials on the Nanoscale. Chem. Int. 37, 3–7 (2015). [Google Scholar]
- 9.Thomas DG, Pappu RV & Baker NA NanoParticle Ontology for cancer nanotechnology research. J. Biomed. Inform. 44, 59–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Institute of Health. Centers of Cancer Nanotechnology Excellence (CCNE) (U54) - See more at: http://grants.nih.gov/grants/guide/rfa-files/RFA-CA-14-013.html#sthash.MGnBJpfD.dpuf.
- 11.Editorial. Journals unite for reproducibility. Nature 515, 7–7 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Ostraat ML, Mills KC & Guzan KA The Nanomaterial Registry: Opportunities and challenges in informatics. in 2012 IEEE International Conference on Bioinformatics and Biomedicine Workshops 884–888 (IEEE, 2012). doi: 10.1109/BIBMW.2012.6470258 [DOI] [Google Scholar]
- 13.Editorial. Joining the reproducibility initiative. Nat. Nanotechnol. 9, 949–949 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu R. & Cohen Y. Nanoinformatics for environmental health and biomedicine. Beilstein J. Nanotechnol. 6, 2449–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourches D, Pu D. & Tropsha A. Exploring quantitative nanostructure-activity relationships (QNAR) modeling as a tool for predicting biological effects of manufactured nanoparticles. Comb. Chem. High Throughput Screen. 14, 217–225 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Fourches D. & Tropsha A. Quantitative Nanostructure-Activity Relationships: from Unstructured Data to Predictive Models for Designing Nanomaterials with Controlled Properties. in Nanotoxicology: Progress toward Nanomedicine (eds. Monteiro-Riviere NA & Lang Tran C) (CRC Press, 2014). [Google Scholar]
- 18.Fourches D. et al. Quantitative nanostructure-activity relationship modeling. ACS Nano 4, 5703–5712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu K, Natarajan B, Morkowchuk L. & Breneman C. From drug discovery QSAR to predictive materials QSPR: the evolution of descriptors, methods, and models. in Informatics for materials science and engineering: data-driven discovery for accelerated experimentation and application (ed. Rajan K) 385–422 (Butterworth-Heinemann, 2013). [Google Scholar]
- 20.Suh C. & Rajan K. Virtual Screening and QSAR Formulations for Crystal Chemistry. QSAR Comb. Sci. 24, 114–119 (2005). [Google Scholar]
- 21.Informatics for Materials Science and Engineering. Data-driven Discovery for Accelerated Experimentation and Application. (Elsevier, 2013). [Google Scholar]
- 22.Fourches D. et al. Computer-aided design of carbon nanotubes with the desired bioactivity and safety profiles. Nanotoxicology 10, 374–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh E. et al. Meta-analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 11, 479–86 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Berman HM et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolton EE, Wang Y, Thiessen PA & Bryant SH PubChem: Integrated Platform of Small Molecules and Biological Activities. in Annual Reports in Computational Chemistry Volume 4 4, 217–241 (American Chemical Society, 2008). [Google Scholar]