Abstract
We used the amphipathic styrene maleic acid (SMA) co-polymer to extract cytochrome c oxidase (CytcO) in its native lipid environment from S. cerevisiae mitochondria. Native nanodiscs containing one CytcO per disc were purified using affinity chromatography. The longest cross-sections of the native nanodiscs were 11 nm × 14 nm. Based on this size we estimated that each CytcO was surrounded by ~100 phospholipids. The native nanodiscs contained the same major phospholipids as those found in the mitochondrial inner membrane. Even though CytcO forms a supercomplex with cytochrome bc1 in the mitochondrial membrane, cyt. bc1 was not found in the native nanodiscs. Yet, the loosely-bound Respiratory SuperComplex factors were found to associate with the isolated CytcO. The native nanodiscs displayed an O2-reduction activity of ~130 electrons CytcO−1 s−1 and the kinetics of the reaction of the fully reduced CytcO with O2 was essentially the same as that observed with CytcO in mitochondrial membranes. The kinetics of CO-ligand binding to the CytcO catalytic site was similar in the native nanodiscs and the mitochondrial membranes. We also found that excess SMA reversibly inhibited the catalytic activity of the mitochondrial CytcO, presumably by interfering with cyt. c binding. These data point to the importance of removing excess SMA after extraction of the membrane protein. Taken together, our data shows the high potential of using SMA-extracted CytcO for functional and structural studies.
Keywords: Bioenergetics, Proton transfer, Membrane protein, Energy conservation
1. Introduction
Functional studies of membrane proteins typically require isolation of the protein of interest at a sufficiently high concentration. The most widely used approach is solubilization in detergents. In most cases it is possible to find a detergent that fulfils the criteria for a sufficiently pure, structurally intact and, when relevant, active protein. However, different detergents may be optimal for different experimental methods and changes in function may be introduced during the removal of the native lipids. In addition, small and/or loosely bound protein subunits (components) may dissociate along with the native lipids. To restore more native-like conditions, amphipathic polymers (known as “amphipols”) may be used to replace the detergent [1]. An even more native-like environment can be re-created by reconstitution of the detergent-purified protein into lipid vesicles [2] or in lipid nanodiscs using a surrounding membrane scaffolding protein [3], which allows functional studies under near-native conditions. However, all these methods involve isolation of the membrane protein of interest using detergent as an intermediate step. Recently, a new technique has emerged that allows extraction and isolation of the membrane protein of interest together with a disc of native lipids [4] using amphipathic styrene maleic acid (SMA) co-polymer (for a recent review, see [5]) without the use of any detergent. This approach has been used to isolate a number of different membrane-bound proteins [6–18], including proton pumps, photosynthetic systems, and ion channels. The procedure has also been used to isolate a secondary transporter from E. coli for studies using negative stain electron microscopy [19]. The isolated protein-lipid native nanodiscs (we use here the nomenclature suggested by Dörr et al. [5]) are surrounded by a ring of the SMA co-polymer with a thickness of ~1 nm [12]. The total diameter of the native nanodiscs has been found in the range of 10–24 nm, indicating that there is a large degree of variability in the size of the isolated systems and the lipid-protein ratio. In one recent study [8], the SMA co-polymer was shown to disrupt the Saccharomyces (S.) cerevisiae inner mitochondrial membrane, resulting in the formation of native nanodiscs. The extract was shown to display O2-reduction activity, catalyzed by the CytcO that was present as one of many components in the mixture of all proteins that were extracted from the native inner mitochondrial membrane.
The mitochondrial CytcO is a multisubunit membrane-bound enzyme located in the cristae of the inner mitochondrial membrane. The S. cerevisiae CytcO is composed of 11 subunits where subunits I-III form the functionally active core of the enzyme. The CytcO catalyzes oxidation of water-soluble cyt. c and reduction of dioxygen to water in the last step of the respiratory chain. Electrons are donated from the more positive (p) side, and protons used in the O2-reduction are taken up from the more negative (n) side, resulting in a charge separation across the membrane. Furthermore, the process is linked to proton pumping across the membrane. The S. cerevisiae CytcO binds four redox-active cofactors. Electrons from cyt. c are first donated to copper A (CuA) and are then transferred consecutively to heme a and the catalytic site, which consists of heme a3 and copper B (CuB). When O2 reacts with the four electron-reduced CytcO, electrons are transferred in a number of distinct steps forming intermediates that have been identified and characterized in spectroscopic studies (for review, see [20–22]). First, O2 binds to the reduced heme a3 resulting in formation of state A. Next, an electron is transferred from heme a to the catalytic site, the O-O bond is broken, and protons are transferred internally within the catalytic site resulting in formation of a ferryl state that, for historical reasons, is called peroxy (P). Formation of this P state triggers proton uptake from the n-side solution to the catalytic site forming the ferryl state, F, which is also linked to fractional electron transfer from CuA to heme a. In the final step of the reaction the electron from the CuA-heme a equilibrium is transferred to the catalytic site where water is formed, leaving the CytcO in the oxidized state (O).
Results from recent studies showed that two additional proteins, called the respiratory supercomplex factors (Rcf) associate with the S. cerevisiae CytcO in the native membranes [23–25]. These factors were suggested to facilitate interactions with the cytochrome (cyt.) bc1 complex in the cyt. bc1-CytcO supercomplex. The Rcfs are presumably loosely bound to the CytcO and can also associate with the cyt. bc1 complex. In a recent study we showed that removal of the regulatory Rcf1 protein results in significant changes in the reaction of the reduced CytcO with O2 [26] and it is therefore important to make sure that this protein co-purifies with the CytcO upon isolation of the enzyme. When using “weak” detergents such as e.g. digitonin, the S. cerevisiae CytcO typically co-purifies with the cyt. bc1 complex in a supercomplex (see above). This supercomplex can be dissociated using detergents such as DDM, but use of this detergent may also results in dissociation of weakly bound components (subunits) such as the Rcf1 polypeptide. Consequently, future studies of the purified CytcO would benefit from development of new methods to isolate the S. cerevisiae CytcO along with all its regulatory factors, but without cyt. bc1.
Here, we used the SMA co-polymer to isolate native nanodiscs containing a single CytcO from S. cerevisiae in each disc. The isolated CytcO contained both Rcf1 and Rcf2, was fully active, and the kinetics of ligand binding and reaction of the reduced CytcO with O2 were similar to those observed with intact membranes. Another advantage of the new approach to isolate CytcO is that the native nanodiscs contained all the major lipid components typically found in the yeast inner mitochondrial membrane. Consequently, the method allows for investigation of CytcO function in an environment that is similar to that in the native membranes. Our data also show that excess SMA inhibits the CytcO, which points to the importance of removing excess of the polymer after purification.
2. Results and discussion
2.1. Isolation of the native nanodiscs
The S. cerevisiae mitochondria containing His-tagged (7-His on Cox13) CytcO were treated with 2% SMA (styrene:maleic acid at a ratio of 3:1) and the CytcO-containing native nanodiscs were isolated via Ni-NTA affinity chromatography as described in Materials and Methods. As shown in Fig. S1 A similar SDS-PAGE patterns were observed for the CytcO-containing native nanodiscs as for the detergent-purified CytcO. The three bands corresponding to molecular weights of 40 kDa, 33 kDa and 22 kDa [27] represent the core subunits I, II and III, respectively. The remaining bands are also very similar for the two samples. The bands above 55 kDa represent contaminations or only partly dissociated CytcO complexes, seen both with the SMA native nanodiscs and with the detergent-purified CytcO. The presence of the loosely-bound Rcf1 and Rcf2 proteins in the CytcO-native nanodiscs was shown using Western blotting of the SDS-PAGE gels followed by immunostaining with specific anti-Rcf1 and Rcf2 antibodies, respectively (Fig. S1B,C). The bright (light) band on the blot area of Fig. S1B indicates the position of Rcf1, which is visualized by chemiluminescence. In the case of Rcf2, some cross-reactivity of the anti-Rcf2 antibodies was observed for bands with lower molecular weight, where a band corresponding to Rcf2 was positioned at 25 kDa (Fig. S1C).
2.2. Size and shape of the native nanodiscs
Negative stain Electron Microscopy (EM) was used to characterize the size and shape of the SMA-CytcO native nanodiscs (Fig. 1). Fig. 1A shows that the majority of purified SMA-CytcO native nanodiscs distribute in a monodisperse fashion on the grid, a requirement for accurate analysis of particles by EM classification and reconstruction. A small portion of particles form large aggregates, which might be caused by sporadic adverse effect of the uranyl formate stain, and these were excluded from 2D image analyses. Reference-free 2D classification of the particles shows multiple views of the SMA-CytcO native nanodiscs (Fig. 1B) that are similar in size and shape to projections of the crystal structure of CytcO from bovine heart (PDB: 3X2Q) without bound lipids (Fig. 1C). As shown in Fig. 1B and C, the CytcO accounts for the majority of the mass in the SMA-CytcO native nanodiscs, with surrounding lipids contributing to a slight increase in particle diameter relative to the forward projections. A histogram of particle diameters (Fig. 1D) shows a bimodal distribution, arising from the disparate dimensions observed for the particles depending on their orientation on the grid. On the basis of the analysis outlined above, we conclude that the native nanodiscs contained one CytcO per disc and that the approximate dimensions of the discs were 11 nm × 14 nm.
Fig. 1.
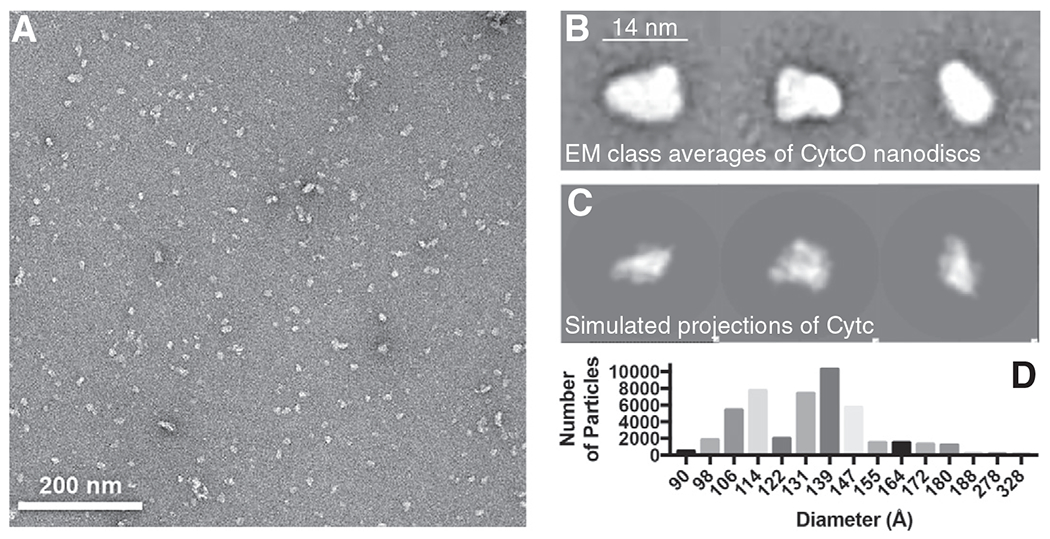
Negative stain EM of CytcO-SMA native nanodiscs. (A) Representative negative stain EM image of CytcO-SMA native nanodiscs showing regularly sized and monodisperse complexes. (B) Representative 2D class averages of CytcO-SMA native nanodisc particles. Each class average contains ~100 particles. (C) Projections of crystal structure of bovine CytcO in three different views, scaled to the same pixel size as in panel (B). (D) Distribution of particle sizes obtained for 46,764 CytcO-SMA native nanodiscs particles.
Fig. S2 shows a size-exclusion chromatography (SEC) analysis of the SMA-CytcO native nanodiscs. From the position of the major cytochrome-containing peak, the size of the particles was estimated to be in the range equivalent to 600–700 kDa, i.e. a relatively broad band. In order to investigate whether this distribution represents a dynamic equilibrium or a distribution of particles with different shapes/molecular weights, we collected fractions around 400 kDa and around 700 kDa, respectively, and re-run these fractions. In both cases we obtained bands corresponding to either 400 kDa or 700 kDa, which shows that there is a distinct distribution of particles in the SEC that is not distinguishable in the EM analysis.
Next, we compared the molecular weight of the native nanodiscs identified in the SEC experiment to the size obtained from the EM analysis. Assuming an average size of 650 kDa (from the SEC experiment) and an average protein (including the lipids) density of 0.8 Da/Å3, the volume of the isolated particles is ~800 nm3. Assuming spherical particles they would have a diameter of ~12 nm, which approximately corresponds to the particle size as seen using EM.
As already mentioned above, the particles observed using EM correspond to native nanodiscs containing one CytcO per disc. Based on a structural model of the S. cerevisiae CytcO [28] or the structure of the bovine heart CytcO [29], the dimension of a CytcO monomer is approximately 7 nm × 9 nm when observed perpendicular to the membrane surface. As indicated above, the approximate dimensions of the isolated SMA-CytcO discs were 11 nm × 14 nm, where the smaller of these numbers, 11 nm, is presumably the “height” of the CytcO. The SMA ring around each native nanodisc is ~1 nm [12]. Consequently, assuming that the diameter in the plane of the membrane is ~14 nm, each CytcO is surrounded by a ring of lipids extending 1.5–2.5 nm from the enzyme. The surface area of one DOPC lipid is 70 Å2 [30], suggesting that ~100 lipids surround each CytcO molecule in the SMA native discs.
2.3. Spectral characteristics and activity
Fig. 2 shows absorption spectra of the reduced and oxidized (“as purified”) native nanodiscs (Fig. 2A), as well as a reduced minus oxidized difference spectrum (Fig. 2B). The difference spectrum displays features characteristic to CytcO; a peak at 603 nm, attributed primarily to heme a (with contribution from heme a3), a peak at 445 nm with contributions from both hemes in their reduced states and a negative peak at 418 nm, attributed to the oxidized hemes. The small peak at ~564 nm is presumably a superposition of local maxima at 560 nm and 568 nm originating from hemes a and a3, respectively [31]. We did not observe any peaks characteristic to the cyt. bc1 complex, which shows that this complex did not co-purify with the CytcO.
Fig. 2.
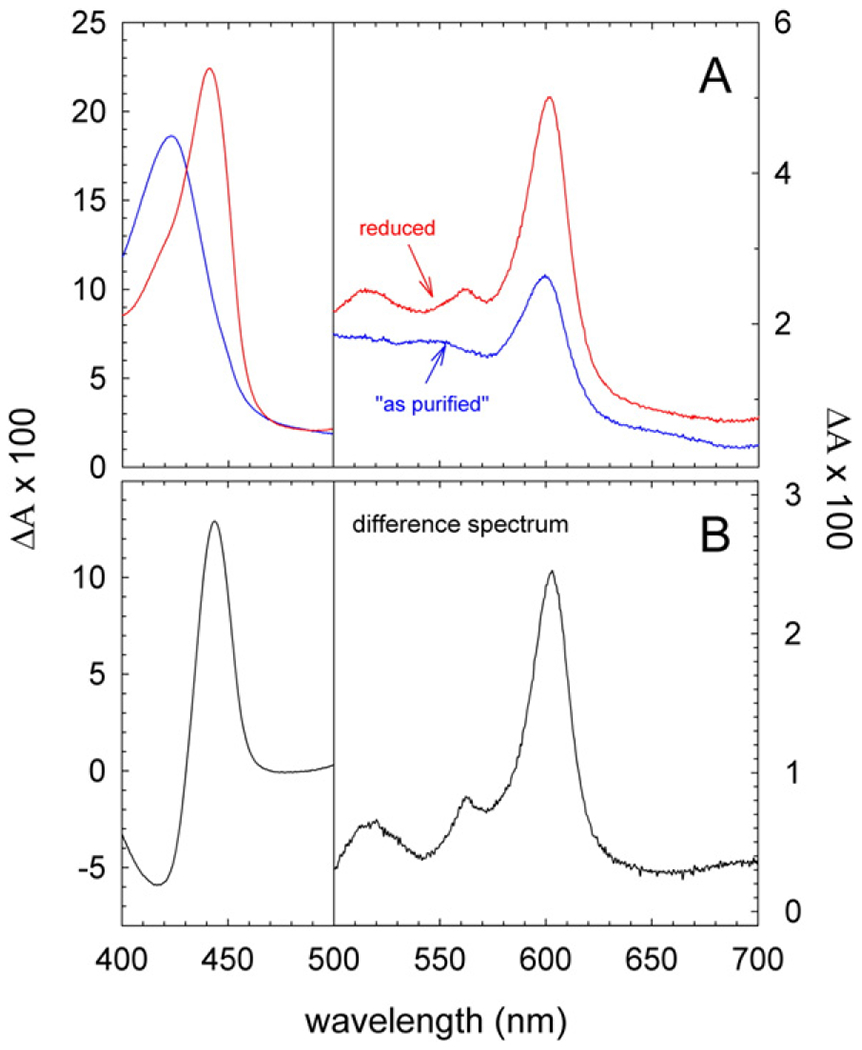
Optical absorption spectra of CytcO-SMA native nanodiscs. (A) Absolute absorption spectra of CytcO-SMA native nanodiscs “as purified” (blue) and after reduction with sodium dithionite (red). (B) a difference absorption spectrum of the reduced sample minus that of “as purified”. The concentration of CytcO was 0.95 μM in 0.2 M NaCl, 20 mM HEPES (pH 7.5).
The steady-state activity of the SMA-extracted CytcO was determined by measuring the O2-reduction rate in the presence of 10 mM ascorbate, 100 μM TMPD, and 50 μM cyt. c at pH 7.4. A value of 130 ± 20 e−/s (SD, n = 3) was obtained when normalized to the concentration of CytcO (cyt. aa3), based on an absorption coefficient for the reduced-minus-oxidized CytcO at 603 nm of 24 mM−1 cm−1 [31]. For comparison, the CytcO activity in intact S. cerevisiae mitochondria was 240 ± 20 e−/s (n = 6). The activity data are summarized in Table 1. Addition of low concentration of DDM (0.05%), which is a detergent typically used for CytcO purification, enhanced the activity of membrane-bound CytcO, both in the native nanodiscs and in mitochondria, by about a factor of 3 (Table 1). The similarity in the response to addition of detergent to both systems indicates that the CytcO structures were the same in both cases. Furthermore, the data demonstrate that a higher activity does not necessarily imply a “better” preparation. This is particularly evident from earlier studies of mammalian CytcO, which showed that addition of DDM at high concentration results in loss of regulatory subunits concomitantly with an increase in activity [32].
Table 1.
Steady state activity of CytcO-SMA native nanodiscs and of CytcO in the S. cerevisiae mitochondrial membrane*.
| Sample | Ratio heme aa3/protein, nmole/mg#) | CytcO activity, e−/s |
|
|---|---|---|---|
| No DDM | +0.05% DDM | ||
| CytcO-SMA | 0.9 ± 0.4 (n = 4) | 130 ± 20 (n = 3) | 300 ± 40 (n = 3) |
| 1.1 ± 0.3 (n = 5) | 90 ± 20 (n = 3) | 190 ± 40 (n = 3) | |
| Mitochondria | 0.08 ± 0.04 (n = 6) | 240 ± 20 (n = 6) | 700 ± 300 (n = 4) |
For CytcO in SMA native nanodiscs, the heme content was calculated either from the difference spectra “reduced-minus-as purified” or from absolute reduced spectrum (shown in italic, using ε = 39 mM−1 cm−1 [31] at 604 nm). Accordingly, two different values are given for the steady-state activity, which is normalized to the CytcO concentration. Because the isolated CytcO was slightly reduced in the “as purified” nanodiscs, the values obtained from the difference spectra yielded an underestimation of the heme concentration.
The ratio of heme aa3 concentration over the protein amount (see “Materials and Methods” section) is a measure of the CytcO purity. For 100% pure CytcO, with a molecular weight of 200 kDa, the ratio is . For detergent-purified CytcO we typically obtained a ratio of ~1. The data show that isolation of the CytcO native nanodiscs resulted in an enrichment of the CytcO by a factor of ~10.
2.4. Lipid analysis
Fig. 3 shows a thin-layer chromatography analysis of the native nanodiscs (lanes 4 and 5), which demonstrates the presence of the same major phospholipid components as in the original S. cerevisiae mitochondrial membranes (lane 3), i.e. PI + PS, PE, PC and CL (for full description, see sub-section “Analytical methods” in the Materials and Methods section). Similar results were observed earlier after preparation of other native nanodiscs from S. cerevisiae [8], from E. coli [4,9] or from Rhodobacter sphaeroides [18].
Fig. 3.
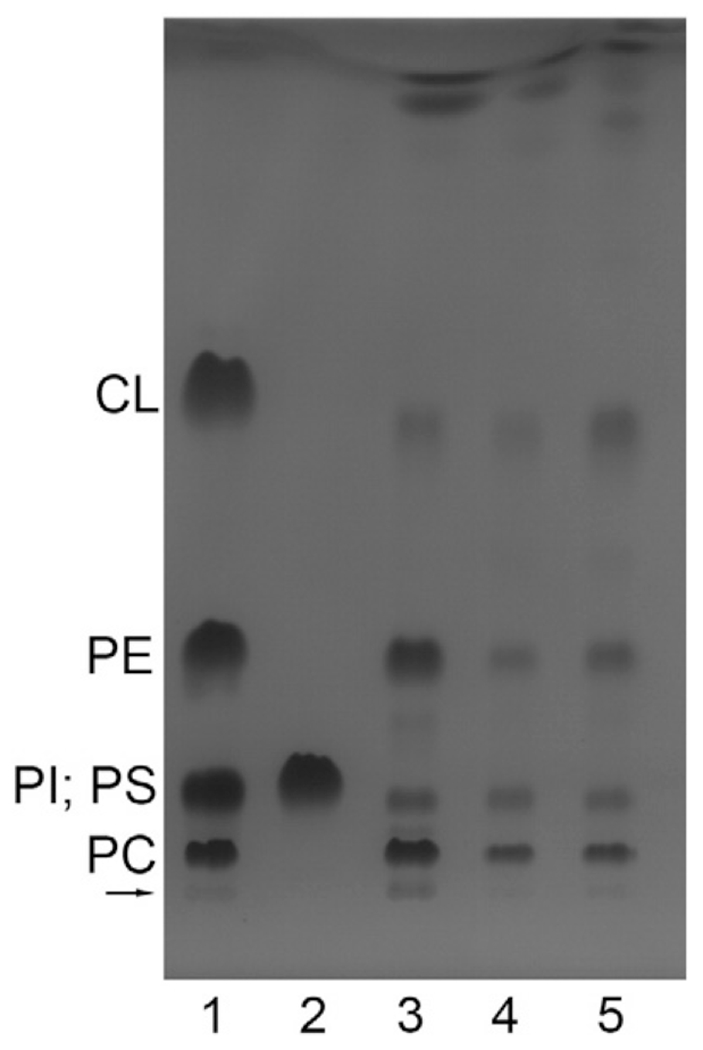
Thin-layer chromatogram of chloroform-methanol extracts of the yeast mitochondria and of CytcO-SMA native nanodiscs. From the left to the right: the 1st and the 2nd lanes are lipid standards (0.04 mg CL, 0.05 mg of each of DOPC, DOPE, PI and PS); the 3rd lane is the extract of the yeast mitochondria (the loaded lipids were extracted from a sample that originally contained ~0.6 mg protein); the 4th and 5th lanes are extracts of two preparations of CytcO-SMA. The loaded lipids (at the arrow) were extracted from a sample that originally contained ~0.2 mg protein. The bands in lane 4 are slightly weaker than those in lane 5, presumably because slightly less material was applied in the former. The bands were visualized by iodine staining. See Materials and methods for more details.
2.5. Ligand binding to the catalytic site
Upon addition of carbon monoxide to the reduced CytcO, the CO molecule binds to heme a3. The iron ligand bond is photolabile and can be fully dissociated using a short laser flash. The recombination rate of the heme a3-CO complex is sensitive to the structure around the catalytic site heme. Fig. 4A shows absorbance changes at 430 nm and 445 nm (minimum and maximum, respectively, in the CO difference spectrum) after flash-induced dissociation of the CO ligand from the CytcO in SMA native nanodiscs. The recombination reaction displayed components with time constants of 160 ± 20 ps and 6.0 ± 0.1 ms, respectively. The contribution of the faster of the two components was 30 ± 10% at 445 nm. As observed previously [26] and shown in Fig. 4A, also with the mitochondrial membranes we observed a small (~10% at 445 nm) rapid component with a time constant of ~100 μs (as well as a major kinetic component with a time constant of ~9 ms). This observation indicates that the altered structural state reflected by the faster CO recombination is present also in the native membranes, although at a lower relative concentration compared to that observed with the CytcO-SMA native nanodiscs. Fig. 4B shows a kinetic difference spectrum of the two components, i.e. the amplitudes of the absorbance changes as a function of wavelength in the ranges of 425–455 nm and 575–625 nm. In the Soret region, both phases displayed very similar difference spectra with coinciding isosbestic points, while in the alpha region the peak around 610 nm of the faster component was red-shifted by a few nanometers as compared to the slower component. These data indicate that the two components reflect CO re-binding to CytcO, however, the two CytcO forms display structural differences around the catalytic site, reflected by different recombination rates and slight spectral differences (c.f. the comparison to the reference kinetic difference spectrum obtained with the yeast mitochondria). In this context it should be noted that the rate of CO recombination may change significantly also upon minor structural changes. For example, the structures of this site are almost identical in both the Rhodobacter sphaeroides aa3 and Thermits thermophilus ba3 CytcOs, yet the CO recombination rate is almost a factor of 10 slower with the ba3 than with the aa3 CytcO [33–35].
Fig. 4.
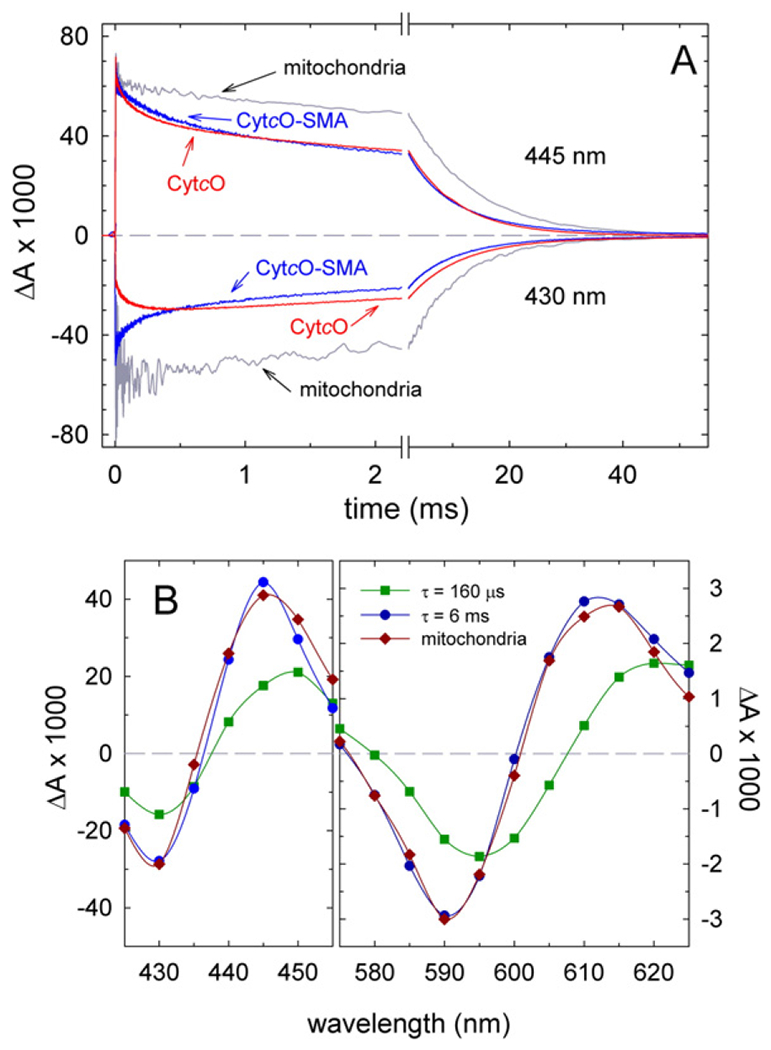
Absorbance changes after the flash-induced dissociation of CO from fully reduced samples. (A) Absorbance changes as a function of time at 445 nm and 430 nm (above and below the zero (dashed) line, respectively, as indicated in the graph) after flash-induced dissociation of CO (at t = 0). The traces represent: CytcO-SMA (blue), yeast mitochondria (grey) and yeast CytcO in DDM (red). Experimental conditions: 0.2 M NaCl, 20 mM HEPES (pH 8.0) for CytcO-SMA; 60 mM sorbitol, 20 mM HEPES (pH 7.4) for yeast mitochondria and 0.15 M KCl, 20 mM HEPES (pH 8.0), 0.05% DDM, 10% glycerol for detergent-solubilized CytcO. The temperature was ~22 °C. All amplitudes are normalized to 1 μM reactive enzyme. (B) Kinetic difference spectra for CytcO-SMA native nanodiscs. Amplitudes of the fast and the slow components of CO-recombination kinetic traces are plotted as a function of wavelength and shown in dark green (squares) and dark blue (circles), respectively. The sum of the amplitudes of these two components at 445 nm has been normalized to 1 μM reacting CytcO. A kinetic difference spectrum (CO-reduced – minus – reduced) of the major component (90%, τ ≅ 9 ms) in yeast CytcO in native mitochondria is shown for comparison (dark red diamonds). This difference spectrum is normalized to approximately overlap with the slow component (dark blue circles). Time constants and contributions of the kinetic components were determined using the software ProK (global fit).
With the detergent-purified CytcO, the main CO-recombination component displayed a time constant of 8 ms. In addition, we observed a rapid component with a time constant of ~140 μs and ~30% of the total absorbance change at 445 nm. In this case, however, the sign of the rapid component at 430 nm was opposite to that observed with the SMA-native nanodiscs and native mitochondrial membranes. Thus, with the detergent-purified CytcO, the rapid component represents CO recombination to heme a3 in a local structural environment that differs from that in the other two samples (SMA-extracted nanodiscs and native membranes, where the CytcO resides in a membrane). Results from an earlier study with mitochondria showed that a rapid component with a similar time constant of ~100 μs and the same spectral features as in detergent-purified CytcO (absorbance decrease at 430 nm) was observed upon removal of the Rcf1 protein [26]. Also this observation indicates that the 100-μs component observed with the detergent-purified CytcO reflects CO binding to a CytcO that is structurally altered. On the basis of this interpretation we conclude that isolation of CytcO using SMA yields CytcO, which displays CO-ligand binding dynamics that is similar to that observed in the native lipid environment.
2.6. Time-resolved kinetics of the oxidative part of the catalytic cycle
Fig. 5 shows absorbance changes associated with reaction of the reduced CytcO, in SMA native nanodiscs, with O2. In this experiment, the reduced CytcO with CO bound to heme a3 was mixed with an O2-containing solution. About 200 ms after mixing, the CO ligand was dissociated by means of a laser flash, which allowed O2 to bind to heme a3 and the absorbance changes at 445 nm were monitored over time. The unresolved increase in absorbance at t ≅ 0 is associated with dissociation of the CO ligand. The following decrease in absorbance is associated with binding of O2 and stepwise oxidation of the CytcO. The first step after binding of O2 reflects electron transfer from heme a to the catalytic site, with a time constant of ~20 μs [22], which results in breaking of the O-O bond and formation of the so-called “peroxy” state (PR). At 445 nm we also observed transfer of the fourth electron, originally residing at CuA, to the catalytic site resulting in formation of the oxidized CytcO (O state) with a main component with a time constants of ~0.8 ms (we also observed a small fraction absorbance decrease with a time constant of ~14 ms, see slope in Fig. 5). The main component was slightly slower than that observed with the detergent-purified CytcO (~0.4 ms) [22] or with mitochondria (~0.5 ms).
Fig. 5.
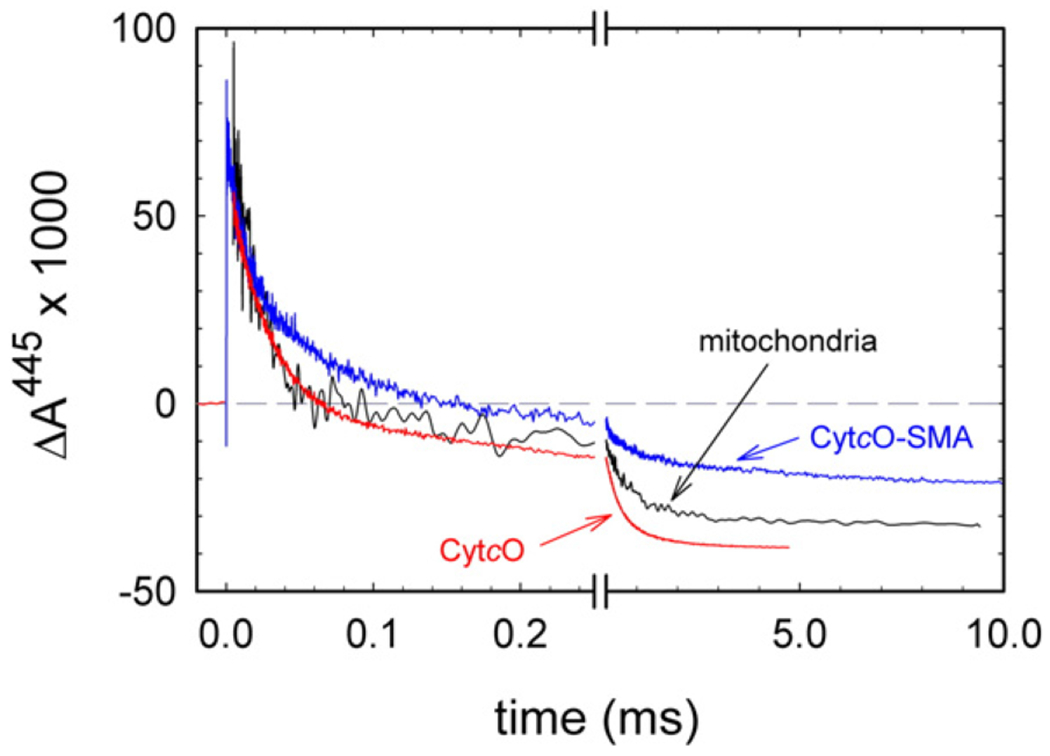
Absorbance changes associated with reaction of reduced CytcO with O2. The reaction was studied with three samples: CytcO in SMA native nanodiscs (blue), in native membranes (grey) and in the detergent DDM (red). The O2 concentrations after mixing were 0.9 mM, 0.6 mM and 0.6 mM, respectively. For experimental conditions, see Fig. 4. The CO ligand was dissociated by a laser flash at a time ~ 200 ms after mixing (the flash defines t = 0). All amplitudes were normalized to 1 μM reactive enzyme. The rate constants were determined using the software ProK.
The total absorbance-change amplitude, as shown in Fig. 5, associated with oxidation of the CytcO, was 83% (0.10) and 92% (0.11) (absorbance change from that at t = 0+ to the final level at ~10−2 s) with the SMA native nanodiscs and the inner mitochondrial membranes, respectively, of that in detergent-purified CytcO (0.12). The smaller extent of oxidation of the CytcO in SMA native nanodiscs may be an apparent effect that is explained by the method used to normalize the traces. A standard practice is to normalize all traces to the CO-dissociation absorbance change amplitude at t = 0 (which was done in this case). This procedure assumes that the CytcO population is uniform. However, as discussed above, we did observe two components in the CO-recombination reaction after addition of SMA, indicating that in the SMA native nanodiscs there is a small fraction CytcO with an altered catalytic site. If the absorbance changes in Fig. 5 would be normalized to the fraction “normal” CO recombination (i.e. equivalent to the amplitude of the 6-ms component) then the absorbance changes associated with oxidation of the CytcO would be more similar in all cases in Fig. 5. A more detailed discussion on the slightly larger amplitude observed with the detergent-purified CytcO than with the mitochondria is beyond the scope of this study and will be discussed elsewhere.
2.7. Inhibition of activity by SMA
A complication when using SMA for purification of CytcO (and probably also other membrane proteins) is that interaction of the co-polymer with the CytcO strongly inhibits the O2-reduction activity of the enzyme. The inhibition effect was reversible because after addition of SMA the activity was restored upon washing the sample with a buffer without SMA. These results indicate that addition of the SMA co-polymer does not result in altering the structure of the CytcO. The data in Fig. 6A shows the O2-reduction activity measured for different SMA concentrations added to S. cerevisiae mitochondria for two concentrations of cyt. c, 0.05 μM and 50 μM, respectively. For the lower cyt. c concentration, the O2-reduction activity dropped to 50% of the maximum value at ~0.002% SMA. For the higher cyt. c concentration, we first observed a stimulation of the activity, followed by a drop to 50% of the maximum activity at 0.03–0.04% SMA. This drop in activity at lower SMA concentrations for 0.05 μM than for 50 μM cyt. c indicates that SMA interferes with cyt. c binding to the CytcO. Because the effect was reversible, the excess SMA polymer could, for example, interfere with the electrostatically-controlled docking between cyt. c and CytcO on the p-side of the membrane. This conclusion is qualitatively consistent with the increase in activity at low SMA concentrations for the high cyt. c concentration, because optimum activity is observed when the electrostatic interactions are intermediate, i.e. upon increasing the ionic strength (at the lowest ionic strength product dissociation is rate limiting) [36].
Fig. 6.
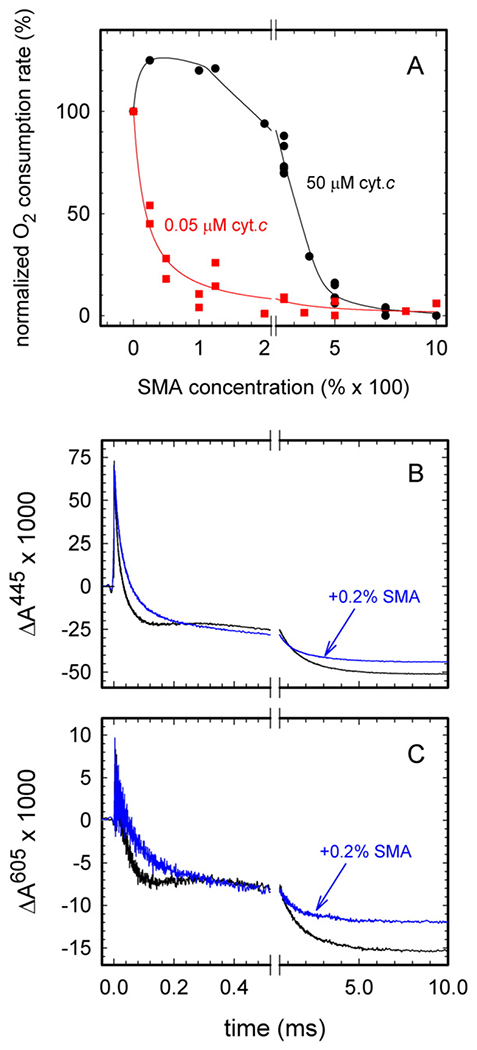
Effect of SMA on the activity of CytcO. (A) Steady-state O2-reduction activity of the S. cerevisiae mitochondria in the presence of SMA. The activity is defined by the oxygen-consumption rate measured with equine heart cyt. c at 50 μM (black dots) or 0.05 μM (red squares). The activity is presented as relative values compared to that in the absence of SMA (set as 100%). The 100% activities were 250 e−/s and 70 e−/s, respectively. The IC50 values for 50 μM and 0.05 μM cyt. c were 0.032 10−2% and 0.0019 10−2%, respectively. The concentration of yeast mitochondria protein was 0.2 mg/ml. For other experimental conditions, see the Materials and Methods section. (B,C) Reaction of fully reduced bovine CytcO with oxygen in the absence (black traces) and in the presence (blue traces) of 0.2% SMA, monitored at 445 nm (B) and 605 nm (C). The O2 concentration after mixing was 0.9 mM. The buffer contained 0.1 M KCl, 20 mM HEPES (pH 8.0), 0.1 mM EDTA and 0.05% DDM. For other experimental conditions, see Fig. 4. The CO ligand was dissociated by a laser flash at t = 0, ~200 ms after mixing. All amplitudes were normalized to 1 μM reactive enzyme. The rate constants were determined using the software ProK.
To test whether or not such an excess SMA co-polymer as used in the activity inhibition measurements interferes with proton uptake from the n-side of the membrane, we also investigated the reaction of the reduced mitochondrial CytcO from bovine heart and O2 (Fig. 6B). This enzyme is well characterized and the kinetic components associated with proton uptake have been identified in the past [37–39]. In the presence of SMA, the kinetic component having a time constant of ~100 μs (absorbance increase in the black traces in Fig. 6BC in the time range ~ 200–400 μs), associated with electron transfer from CuA to heme a during the PR → F reaction, was absent, which indicates that proton uptake was significantly slowed. However, the time constant of the next transition, F → O, (τ ≅ 1 ms) was about the same in the presence as in the absence of SMA, which indicates that the proton uptake was not slowed beyond ~1 ms. In other words, the slowed proton uptake during PR → F is not the (only) explanation for the observed SMA effect on the catalytic turnover. Furthermore, the total extent of oxidation was smaller with than without SMA added, which indicates that in the presence of SMA, a small CytcO fraction was not oxidized over the time scale of the experiment (c.f. also data with the SMA native nanodiscs in Fig. 5). Taken together, these data indicate that the dramatically slowed turnover activity seen upon addition of SMA most likely originates from interference with formation of the reduced cyt. c-oxidized CytcO complex. In addition, the presence of the SMA polymer presumably does slow proton uptake, however, this effect is not manifested in measurements of the turnover rate.
The activity measured with the isolated yeast SMA-CytcO in this work was 130 e−/s, i.e. about 50% of that measured with CytcO in mitochondria. The SMA concentration used for extraction of the CytcO native discs was 2% (note that the total protein content during protein extraction is orders of magnitude higher than that in the experiment shown in Fig. 6), but the sample was then washed in multiple steps; first in the affinity chromatography (without any quantitative estimation of the dilution) and then to further lower then SMA concentration by a factor of 5–10 × 103. Even though the SMA concentration after washing was negligible, we cannot exclude some remaining specific binding of the SMA co-polymer to the CytcO that would interfere with binding of cyt. c.
2.8. Comments on the extraction of CytcO
The use of the SMA co-polymer to extract CytcO offers a new method to isolate pure CytcO with negligible contamination from other heme proteins, yet containing the weakly bound and functionally important Rcf1 and Rcf2 proteins (see Introduction). The possibility to isolate only CytcO along with a significant amount of lipids and with the Rcf proteins is surprising given that in S. cerevisiae CytcO forms a supercomplex with cyt. bc1 and that the Rcf proteins are presumably weakly bound [23–25,40–49]. These complexes are typically isolated using weak detergents such as digitonin. Given our findings that the CytcO is extracted with a disc of surrounding lipids, one may expect that the CytcO would be found in the discs together with the cyt. bc1 complex. Indeed, we observed a subpopulation of cyt. bc1 in some preparations, but primarily when the cyt. bc1 to CytcO ratio was increased in some batches of the yeast mitochondrial membranes. It is possible that in the native membrane there is an equilibrium of supercomplexes and free CytcOs, as well as cyt. bc1 complexes. The suggested mechanism by which SMA extracts membrane proteins from the native membranes [4] indicates that the polymer enters the hydrophobic part before extraction. It is possible that this mechanism results in shifting the equilibrium of free complexes versus supercomplexes towards free protein components (i.e. weakening the cyt. bc1-CytcO interaction), before extraction of the CytcO native nanodiscs.
3. Conclusions
We present a detergent-free method to extract CytcO from the S. cerevisiae inner mitochondrial membranes into native nanodiscs using a SMA co-polymer. The CytcO native nanodiscs displayed significant O2-reduction activity. In addition, the ligand-binding kinetics and reaction of the reduced CytcO with O2 were similar to those measured in the native S. cerevisiae mitochondrial membranes. The size of the native nanodiscs indicates that ~100 lipid molecules surround each CytcO, which presumably yields a native lipid environment (see model in Fig. 7). The lipid composition in the native nanodiscs was similar to that found in the mitochondrial membrane. The preparation yields a relatively homogeneous size distribution. In future studies this method may prove valuable in functional and structural studies.
Fig. 7.
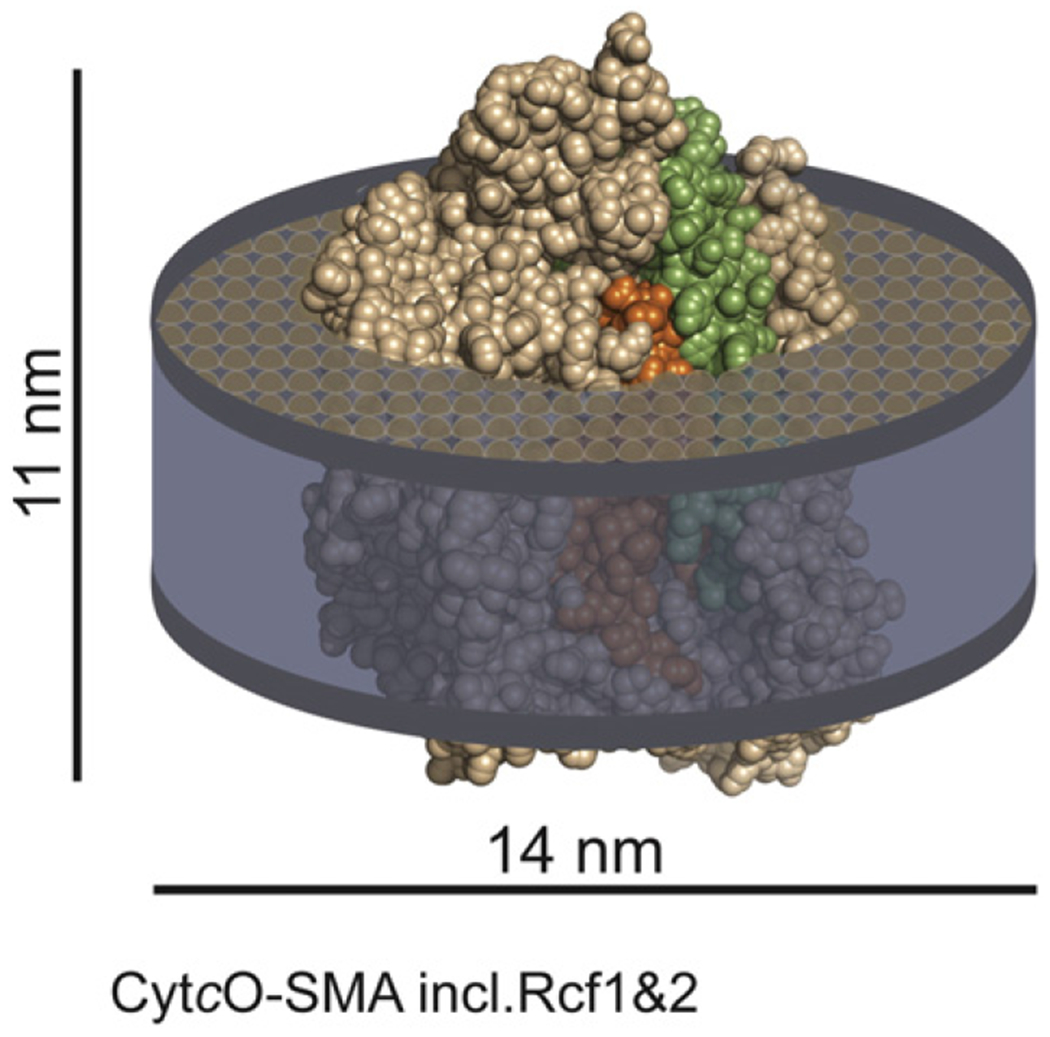
Schematic model of a CytcO-lipid SMA native nanodisc. The CytcO structure is based on a model of yeast CytcO [28] and prepared using the software PyMol [59]. Subunits I and II are shown in orange and green, respectively. The picture illustrates the approximate proportions of the lipid disc and of the protein.
4. Materials and methods
4.1. Preparation of the SMA co-polymer
Styrene maleic anhydride resin SMA EF30 (Cray Valley, the ratio of styrene to maleic anhydride was 3:1) was solubilized in water by alkali hydrolysis. The SMA powder (15 g) was stirred in 300 ml of 1 M NaOH overnight, the mixture was then heated to boiling with reflux and a stirred for another 4 h. After complete solubilization of the solid resin the mixture was allowed to cool to room temperature. The mixture was acidified with HCl to reach a pH value about 4. The pellet was collected by centrifugation and washed with water three times (about 400 ml each time). Finally, the pellet was dissolved in water by stepwise addition of 6 M NaOH under stirring; the final pH was about 8 and the final volume about 120 ml. Non-dissolved particles were removed by centrifugation. The obtained solution of hydrolyzed SMA was dialyzed against 50 mM Tris–HCl (pH 8.0). The SMA was quantified by drying a sample to constant weight and the volume of the stock solution was adjusted in order to obtain 5% SMA (w/v) concentration.
4.2. Cultivation of S. cerevisiae and preparation of mitochondria and SMA-CytcO native nanodiscs
For the construction of a yeast strain expressing Cox13 with a C-terminal His7Strep tag, the wild-type strain W303a (Mat A ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1) was transformed by homologous recombination with a PCR fragment coding for a HIS3 cassette and the sequences allowing to exchange the authentic stop codon of COX13 with a 6 amino acid linker followed by a His7/Strep tag and a new stop codon. The resultant strain (MOY764) was verified by PCR and showed normal respiratory growth. The yeast was grown on YPG medium with vigorous shaking at 30 °C according to Meisinger et al. [50]. Isolation of the crude mitochondrial fraction (for the SMA-treatment) and of highly purified mitochondria (for functional studies) from yeast was performed according to the procedure by Meisinger et al. [50] employing Zymolyase-20 T from Arthrobacter luteus (Nacalai Tesque). For time-resolved kinetic experiments (see below) the mitochondria were prepared devoid of their outer membrane according to [51]. Briefly, mitochondria after running the sucrose gradient [50] were treated with a hypotonic medium containing 60 mM sorbitol, 20 mM HEPES (pH 7.4) and mitoplasts were collected by centrifugation at 10,000 g.
For purification of CytcO-SMA native nanodiscs, crude yeast mitochondria containing CytcO with a 7-His-tag on subunit 13 were incubated with 2% SMA (3:1) at a SMA:protein ratio of 2.5 in 0.2 M NaCl, 50 mM Tris–HCl (pH 8.0) (buffer A) at 4 °C for 4 h with a slow rotation on a rotation wheel. Non-solubilized membranes were removed by centrifugation (17,000 g;20min; 4 °C) and the supernatant was collected. After this step, SMA was not present in the buffers and before the affinity chromatography the SMA concentration in the supernatant was lowered by a factor of about three by exchange of the buffer for 0.2 M NaCl, 20 mM HEPES (pH 8.0) (buffer B) using concentrator filters (Amicon Ultra, Ultracel-100 K, Merck Millipore). Then, the solution was loaded on a HisTrap HP column (5 ml, GE Healthcare) by overnight cycling. The column was washed with buffer B supplemented with 5 mM imidazole and the CytcO-SMA native discs were eluted with buffer B, supplemented with 20-50 mM imidazole. Finally, the eluted CytcO-SMA native discs were concentrated and imidazole was removed by exchange to buffer B (dilution by a factor of 5–10 × 103). The CytcO-SMA native discs were stored at 4 °C.
Pure CytcO from S. cerevisiae was prepared as described in [52], except that a TS Series Continuous Flow cell disruptor (Constant System Ltd) was used instead of glass beads. The bovine mitochondrial CytcO used for control experiments was prepared as described in [53].
4.3. Size-exclusion chromatography analysis
The SEC analysis of CytcO-SMA nanodiscs was performed on Superose 6 Increase 10/300 GL (GE Healthcare) linked to the Shimadzu Prominence HPLC system with SIL-20 A Autosampler and SPD-M20 A Detector. Buffer B (see above) was used as a liquid phase. A gel filtration calibration kit, HMW (GE Healthcare) was used for the calibration of the column. The data are shown in Fig. S2.
4.4. Analytical methods
The protein concentration was estimated using a Lowry assay in the presence of SDS.
Optical absorption spectra were recorded using Cary 100 UV–VIS or Cary 400 UV–VIS (Agilent Technologies) spectrophotometer. Sodium dithionite was used as a reducing agent. The concentration of hemes a and a3 was estimated from the difference “reduced-minus-oxidized” spectra at 603 nm using an absorption coefficient of 24 mM−1 cm−1, or from the reduced CytcO spectrum at 604 nm using an absorption coefficient of 39 mM−1 cm−1 [31].
SDS-PAGE was performed using pre-cast NuPAGE Bis-Tris 4–12% gels with MES-SDS running buffer. Samples were incubated in the NuPAGE SDS sample buffer with 50 mM DTT for 25 min at 50 °C. Western blotting was run with PVDF transfer membranes (0.45 μm; GE Healthcare) in NuPAGE Bicine transfer buffer. All buffers mentioned above in this paragraph were prepared according to recommendations provided by the manufacturer. The voltage was set according to the manufacturer’s recommendations. Page Ruler Plus (pre-stained protein ladder, Thermo Scientific) was used as a standard. Brilliant Blue R250 (Sigma) was used for gel staining. Immunostaining was done with anti-Rcf1 or anti-Rcf2 as primary antibodies (gift from H. Dawitz and M. Ott, Stockholm University) and anti-Rabbit IgG–Peroxidase antibody produced in goat (Sigma) as secondary antibodies. Chemiluminescence upon oxidation of luminol by hydrogen peroxide in the presence of peroxidase was used for visualization of blots (ECL Prime Western Blotting Detection Reagent, Amersham). Imaging of gels and blots was performed using a Luminescent Image Analyzer LAS-1000 plus (Fujifilm).
Lipid extraction and analysis were performed as follows. A volume of 0.25 ml of SMA-CytcO or of the yeast mitochondria suspension (the amount of starting material for CytcO-SMA and mitochondria was assayed as an amount of protein, which was approximately 2 mg or 4 mg, respectively) was mixed with three volumes (0.75 ml) of chloroform-methanol (1:2) and vortexed. Then, one volume of chloroform and one volume of water were added stepwise with vortexing of the samples after each addition. The samples were chilled on ice and spinned at 17,000 g at 4 °C for 7 min. The lipid-containing layer was washed three times with two volumes of cold water. If necessary, the lipid fraction was concentrated by evaporating with a stream of nitrogen. Thin-Layer Chromatography (TLC) of the chloroform-methanol extracts was performed on plates of Silica gel 60 (20 × 20 cm; Merck) with the mixture chloroform/methanol/acetic acid 85/25/10 (v/v/v) as a liquid phase. Iodine staining was used for band visualization. The used standards were 2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (DOPS); L-α-phosphatidylinositol from liver (PI) and 1,1′,2,2′-tetraoleoyl cardiolipin (sodium salt) (CL) (all from Avanti Polar Lipids). Imaging of TLC plates was performed using a Luminescent Image Analyzer LAS-1000 plus (Fujifilm).
The CytcO activity was monitored by measuring the oxygen consumption using an Oxygraph Plus System (Hansatech) at 25 °C with 10 mM ascorbate, 100 μM TMPD and 50 μM cytochrome c (from equine heart, Sigma). The reaction medium contained 10 mM KCl, 5 mM MgCl2, 10 mM KH2PO4 (pH 7.4), 0.1 mM EDTA and 0.3 M mannitol.
4.5. Flash photolysis and flow-flash measurements
Time-resolved kinetics experiments were performed as described e.g. in [33,54], using an LKS80 (Applied Photophysics) flash photolysis/flow-flash set-up. Briefly, the sample was transferred to an anaerobic Thunberg cuvette (path length 1.00 cm), the atmosphere was exchanged for N2 on a vacuum line after which the CytcO in the SMA native nanodiscs was reduced with 2 mM ascorbate and 3 μM PMS (4 mM and 1 μM, respectively, when using mitochondria). After the reduction was complete (based on the optical absorption spectrum), the atmosphere was exchanged for carbon monoxide. To monitor CO dissociation and recombination the sample was illuminated by a laser flash (Nd-YAG laser, pulse length 8 ns, 40 mJ at 532 nm) and absorbance changes were monitored. For the flow-flash measurements, the mixing with the oxygen-saturated buffer was 1:2.5 for CytcO-SMA and 1:1 for other samples, which gives the concentration of oxygen after mixing of ~0.9 mM and 0.6 mM, respectively. The reaction buffers used were: 0.2 M NaCl, 20 mM HEPES (pH 8.0) for CytcO-SMA; 0.15 M KCl, 20 mM HEPES (pH 8.0), 0.05% DDM, 10% glycerol for the purified CytcO; 60 mM sorbitol, 20 mM HEPES (pH 7.4) for the yeast mitochondria and 0.1 M KCl, 20 mM HEPES (pH 8.0), 0.1 mM EDTA, 0.05% DDM for CytcO from bovine heart. The concentration of reactive enzyme was estimated from the amplitude of the time-unresolved absorbance changes induced by the laser flash at 445 nm using an absorption coefficient of 67 mM−1 cm−1.
4.6. Negative stain electron microscopy
Aliquots of frozen SMA-CytcO native nanodiscs at 4 mg/ml were thawed slowly on ice before preparing the negative stain EM grid. The sample was diluted to 0.01 mg/ml in buffer B (see above). Prior to sample application, 400-mesh Cu-Rh maxtaformgrids (Electron Microscopy Sciences) that had been overlaid with a continuous carbon support were exposed to a 95% Ar/5% O2 plasma for 20 s using a Gatan Solarus (Gatan) to generate at hydrophilic surface. A volume of 4 μl of the sample was applied to the freshly cleaned grid. After 30 s of adsorption, excess protein was wicked away with a Whatman No. 1 filter paper (GE Healthcare Life Sciences), and 2% (w/v) uranyl formate solution (4 μl) was immediately applied to the grid. The grid was then inverted and placed on four 20 μl droplets of uranyl formate solution for 10 s each, in succession. Excess stain was then wicked away with filter paper and the grid set to dry on the edge of a fume hood to provide airflow.
Grids were inserted into a Tecnai Spirit (FEI Co) LaB6 electron microscope operating at 120 keV, and images collected using a random defocus range of −0.5 to −1.5 μm and an electron dose of 20 e−/Å2 using the Leginon automated image acquisition software (Suloway et al., 2005) at a nominal magnification of 52,000× (pixel size of 2.05 Å/pixel at the specimen level). 273 images were collected using a F416 CMOS 4 K× camera (T-VIPS, Oslo, Norway). Image analysis was performed using the Appion image processing pipeline [55]. Particles were selected from the micrographs using a difference-of-Gaussians-based automated particle picking program [56] and the contrast transfer function (CTF) of each micrograph was estimated using CTFFIND3 [57]. Using the estimated CTF, phases for each micrograph were corrected before extracting particles with a box size of 80 pixels. Particles were binned by a factor of two and normalized for analysis. The resulting stack of 79,343 particles was subjected to three rounds of reference-free 2D classification using iterative multivariate statistical analysis and multi-reference alignment [58], implemented in Appion. These rounds of 2D analysis were used to remove damaged, aggregated, or erroneously selected particles. The final 2D class averages result from 2D classification of the final stack of 46,763 particles. The particle size diameters of the class averages used to generate the histogram in Fig. 1 were calculated using SPIDER (PMID: 8742743). Class averages were rotationally averaged, and a 1D Radon transform was calculated in order to measure the width of the peak.
Supplementary Material
Acknowledgements
We would like to thank Susanna Seppälä (UCSB) for help with preparation of SMA in the initial phase of the project. This study was supported by a grant from the Knut and Alice Wallenberg Foundation (KAW 2013.0006) (to PB, CvB, PÄ, MH). Support was also obtained from the Pew Scholars program, the Searle Scholars program, and NIH grant DP2 EB020402-01 (to GCL), the Swedish Research Council (to PB, CvB, PÄ, MH), the Swedish Cancer Society (MH) and the Swedish Foundation for Strategic Research (MH).
Abbreviations:
- Bis-Tris
2,2-Bis(hydroxymethyl)-2,2′,2″-nitrilotriethanol
- cyt. c
cytochrome c
- CytcO
cytochrome c oxidase
- DDM
n-dodecyl β-D-maltoside
- EDTA
ethyl-enediaminetetraacetic acid
- HEPES
4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid
- MES
2-(N-Morpholino)ethanesulfonic acid
- PMS
phenazine methosulfate
- SDS
sodium dodecyl sulfate
- SMA
poly(styrene-alt-maleic acid)
- TMPD
N,N,N′,N′-tetramethyl-p-phenylenediamine
- Tris
Tris(hydroxymethyl)aminomethane
Transparency document
The Transparency document associated with this article can be found, in online version.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamem.2016.09.004.
References
- [1].Zoonens M, Popot JL, Amphipols for each season, J. Membr. Biol 247 (2014) 759–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rigaud JL, Lévy D, Reconstitution of membrane proteins into liposomes, METHODS IN ENZYMOLOGY, 372, 2003, pp. 65–86. [DOI] [PubMed] [Google Scholar]
- [3].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG, Chapter 11 reconstitution of membrane proteins in phospholipid bilayer nanodiscs, Methods in Enzymology, 464, 2009, pp. 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scheidelaar S, Koorengevel MC, Pardo JD, Meeldijk JD, Breukink E, Killian JA, Molecular model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers, Biophys. J 108 (2015) 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, Killian JA, The styrene–maleic acid copolymer: a versatile tool in membrane research, Eur. Biophys. J 45 (2016) 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lee SC, Pollock NL, Membrane proteins: is the future disc shaped? Biochem. Soc. Trans 44 (2016) 1011–1018. [DOI] [PubMed] [Google Scholar]
- [7].Logez C, Damian M, Legros C, Dupré C, Guéry M, Mary S, Wagner R, M’Kadmi C, Nosjean O, Fould B, Marie J, Fehrentz JA, Martinez J, Ferry G, Boutin JA, Baneìres JL, Detergent-free isolation of functional G protein-coupled receptors into nanometric lipid particles, Biochemistry 55 (2016) 38–48. [DOI] [PubMed] [Google Scholar]
- [8].Long AR, O’Brien CC, Malhotra K, Schwall CT, Albert AD, Watts A, Alder NN, A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs, BMC Biotechnol. 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, Van Der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M, Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer, J. Am. Chem. Soc 131 (2009) 7484–7485. [DOI] [PubMed] [Google Scholar]
- [11].Jamshad M, Charlton J, Lin YP, Routledge SJ, Bawa Z, Knowles TJ, Overduin M, Dekker N, Dafforn TR, Bill RM, Poyner DR, Wheatley M, G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent, Biosci. Rep 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jamshad M, Grimard V, Idini I, Knowles TJ, Dowle MR, Schofield N, Sridhar P, Lin Y, Finka R, Wheatley M, Thomas ORT, Palmer RE, Overduin M, Govaerts C, Ruysschaert JM, Edler KJ, Dafforn TR, Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins, Nano Res. 8 (2015) 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M, Poyner DR, Bill RM, Thomas ORT, Overduin M, Dafforn TR, Surfactant-free purification of membrane proteins with intact native membrane environment, Biochem. Soc. Trans 39 (2011) 813–818. [DOI] [PubMed] [Google Scholar]
- [14].Gulati S, Jamshad M, Knowles TJ, Morrison KA, Downing R, Cant N, Collins R, Koenderink JB, Ford RC, Overduin M, Kerr ID, Dafforn TR, Rothnie AJ, Detergent-free purification of ABC (ATP-binding-cassette) transporters, Biochem. J 461 (2014) 269–278. [DOI] [PubMed] [Google Scholar]
- [15].Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Gröbner G, Watts A, Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: lipodisq, Angew. Chem. Int. Ed 51 (2012) 4653–4657. [DOI] [PubMed] [Google Scholar]
- [16].Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A, Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer lipodisq particles for functional and biophysical studies, Nano Lett. 12 (2012) 4687–4692. [DOI] [PubMed] [Google Scholar]
- [17].Skaar K, Korza HJ, Tarry M, Sekyrova P, Högbom M, Expression and subcellular distribution of GFP-tagged human tetraspanin proteins in Saccharomyces cerevisiae, PLoS One 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Swainsbury DJK, Scheidelaar S, Van Grondelle R, Killian JA, Jones MR, Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability, Angew. Chem. Int. Ed 53 (2014) 11803–11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Postis V, Rawson S, Mitchell JK, Lee SC, Parslow RA, Dafforn TR, Baldwin SA, Muench SP, The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy, Biochim. Biophys. Acta Biomembr 1848 (2015) 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brzezinski P, Gennis RB, Cytochrome c oxidase: exciting progress and remaining mysteries, J. Bioenerg. Biomembr 40 (2008) 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brzezinski P, Johansson AL, Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase, Biochim. Biophys. Acta Bioenerg 1797 (2010) 710–723. [DOI] [PubMed] [Google Scholar]
- [22].Näsvik Öjemyr L, Maréchal A, Vestin H, Meunier B, Rich PR, Brzezinski P, Reaction of wild-type and Glu243Asp variant yeast cytochrome c oxidase with O2, Biochim. Biophys. Acta Bioenerg 1837 (2014) 1012–1018. [DOI] [PubMed] [Google Scholar]
- [23].Vukotic M, Oeljeklaus S, Wiese S, Vögtle FN, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M, Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex, Cell Metab. 15 (2012) 336–347. [DOI] [PubMed] [Google Scholar]
- [24].Strogolova V, Furness A, Micaela MR, Garlich J, Stuart RA, Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc 1-cytochrome c oxidase supercomplex, Mol. Cell. Biol 32 (2012) 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J, Identification of a protein mediating respiratory supercomplex stability, Cell Metab. 15 (2012) 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rydström Lundin C, Von Ballmoos C, Ott M, Ädelroth P, Brzezinski P, Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A 113 (2016) E4476–E4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Poyton RO, Schatz G, Cytochrome c oxidase from bakers’ yeast. III. Physical characterization of isolated subunits and chemical evidence for two different classes of polypeptides, J. Biol. Chem 250 (1975) 752–761. [PubMed] [Google Scholar]
- [28].Maréchal A, Meunier B, Lee D, Orengo C, Rich PR, Yeast cytochrome c oxidase: a model system to study mitochondrial forms of the haem-copper oxidase superfamily, Biochim. Biophys. Acta Bioenerg 1817 (2012) 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yano N, Muramoto K, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yoshikawa S, Tsukihara T, X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 Å resolution, Acta Crystallogr. F Struct. Biol. Commun 71 (2015) 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nagle JF, Tristram-Nagle S, Structure of lipid bilayers, Biochim. Biophys. Acta Rev. Biomembr 1469 (2000) 159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vanneste WH, The stoichiometry and absorption spectra of components a and a-3 in cytochrome c oxidase, Biochemistry 5 (1966) 838–848. [DOI] [PubMed] [Google Scholar]
- [32].Kadenbach B, Hüttemann M, The subunit composition and function of mammalian cytochrome c oxidase, Mitochondrion 24 (2015) 64–76. [DOI] [PubMed] [Google Scholar]
- [33].Von Ballmoos C, Gonska N, Lachmann P, Gennis RB, Ädelroth P, Brzezinski P, Mutation of a single residue in the ba3 oxidase specifically impairs protonation of the pump site, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 3397–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ädelroth P, Brzezinski P, Malmström BG, Internal electron transfer in cytochrome c oxidase from Rhodobacter sphaeroides, Biochemistry 34 (1995) 2844–2849. [DOI] [PubMed] [Google Scholar]
- [35].Woodruff WH, Coordination dynamics of heme-copper oxidases. The ligand shuttle and the control and coupling of electron transfer and proton translocation, J. Bioenerg. Biomembr 25 (1993) 177–188. [DOI] [PubMed] [Google Scholar]
- [36].Witt H, Malatesta F, Nicoletti F, Brunori M, Ludwig B, Cytochrome-c-binding site on cytochrome oxidase in Paracoccus denitrificans, Eur. J. Biochem 251 (1998) 367–373. [DOI] [PubMed] [Google Scholar]
- [37].Ädelroth P, Ek M, Brzezinski P, Factors determining electron-transfer rates in cytochrome c oxidase: investigation of the oxygen reaction in the R. sphaeroides and bovine enzymes, Biochim. Biophys. Acta 1367 (1998) 107–117. [DOI] [PubMed] [Google Scholar]
- [38].Karpefors M, Ädelroth P, Zhen Y, Ferguson-Miller S, Brzezinski P, Proton uptake controls electron transfer in cytochrome c oxidase, Proc. Natl. Acad. Sci. U. S. A 95 (1998) 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karpefors M, Ädelroth P, Aagaard A, Sigurdson H, Svensson Ek M, Brzezinski P, Electron-proton interactions in terminal oxidases, Biochim. Biophys. Acta 1365 (1998) 159–169. [DOI] [PubMed] [Google Scholar]
- [40].Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA, The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria, J. Biol. Chem 275 (2000) 18093–18098. [DOI] [PubMed] [Google Scholar]
- [41].Schägger H, Respiratory chain supercomplexes, IUBMB Life 52 (2002) 119–128. [DOI] [PubMed] [Google Scholar]
- [42].Stuart RA, Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria, J. Bioenerg. Biomembr 40 (2008) 411–417. [DOI] [PubMed] [Google Scholar]
- [43].Chaban Y, Boekema EJ, Dudkina NV, Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation, Biochim. Biophys. Acta Bioenerg 1837 (2014) 418–426. [DOI] [PubMed] [Google Scholar]
- [44].Heinemeyer J, Braun HP, Boekema EJ, Kouřil R, A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria, J. Biol. Chem 282 (2007) 12240–12248. [DOI] [PubMed] [Google Scholar]
- [45].Winge DR, Sealing the mitochondrial respirasome, Mol. Cell. Biol 32 (2012) 2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mileykovskaya E, Penczek PA, Fang J, Mallampalli VKPS, Sparagna GC, Dowhan W, Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy, J. Biol. Chem 287 (2012) 23095–23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Genova ML, Lenaz G, Functional role of mitochondrial respiratory supercomplexes, Biochim. Biophys. Acta Bioenerg 1837 (2014) 427–443. [DOI] [PubMed] [Google Scholar]
- [48].Acin-Perez R, Enriquez JA, The function of the respiratory supercomplexes: the plasticity model, Biochim. Biophys. Acta Bioenerg 1837 (2014) 444–450. [DOI] [PubMed] [Google Scholar]
- [49].Boumans H, Grivell LA, Berden JA, The respiratory chain in yeast behaves as a single functional unit, J. Biol. Chem 273 (1998) 4872–4877. [DOI] [PubMed] [Google Scholar]
- [50].Meisinger C, Pfanner N, Truscott KN, Isolation of yeast mitochondria, Methods Mol. Biol 313 (2006) 33–39. [DOI] [PubMed] [Google Scholar]
- [51].Björck M, Zhou S, Rydström Lundin C, Ott M, Ädelroth P, Brzezinski P, Kinetics of CO binding to flavohemoglobin in different cellular compartments of S. cerevisiae(submitted for publication) 2016. [DOI] [PubMed]
- [52].Meunier B, Maréchal A, Rich PR, Construction of histidine-tagged yeast mitochondrial cytochrome c oxidase for facile purification of mutant forms, Biochem. J 444 (2012) 199–204. [DOI] [PubMed] [Google Scholar]
- [53].Brandt U, Schägger H, von Jagow G, Purification of cytochrome-c oxidase retaining its pulsed form, Eur. J. Biochem 182 (1989) 705–711. [DOI] [PubMed] [Google Scholar]
- [54].Vilhjálmsdóttir J, Johansson AL, Brzezinski P, Structural changes and proton transfer in cytochrome c oxidase, Sci. Rep 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B, Appion: an integrated, database-driven pipeline to facilitate EM image processing, J. Struct. Biol 166 (2009) 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B, DoG picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy, J. Struct. Biol 166 (2009) 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mindell JA, Grigorieff N, Accurate determination of local defocus and specimen tilt in electron microscopy, J. Struct. Biol 142 (2003) 334–347. [DOI] [PubMed] [Google Scholar]
- [58].Ogura T, Iwasaki K, Sato C, Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking, J. Struct. Biol 143 (2003) 185–200. [DOI] [PubMed] [Google Scholar]
- [59].DeLano WL, The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA, USA, 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


