Abstract
Female eutherians transcriptionally silence one X chromosome to balance gene dosage between the sexes. X chromosome inactivation (XCI) is initiated by the lncRNA Xist, which assembles many proteins within the inactive X chromosome (Xi) to trigger gene silencing and heterochromatin formation. It is well established that gene silencing on the Xi is maintained through repressive epigenetic processes, including histone deacetylation and DNA methylation. Recent studies revealed a new mechanism where RNA-binding proteins that interact directly with the RNA contribute to the maintenance of Xist localization and gene silencing. In addition, a surprising plasticity of the Xi was uncovered with many genes becoming upregulated upon experimental deletion of Xist. Intriguingly, immune cells normally lose Xist from the Xi suggesting that this Xist-dependence is utilized in vivo to dynamically regulate gene expression from the Xi. These new studies expose fundamental regulatory mechanisms for the chromatin association of RNAs, highlight the need for studying the maintenance of XCI and Xist localization in a gene- and cell-type-specific manner, and are likely to have clinical impact.
Keywords: Xist, lncRNA, X-inactivation, PTBP1, Xi reactivation
Introduction
X-chromosome inactivation (XCI) is established in all cells early in embryonic development and is maintained for the whole lifespan of an individual [1-7]. To trigger the formation of the inactive X chromosome (Xi) in the embryo, the long noncoding RNA (lncRNA) Xist becomes expressed on either the maternally or paternally inherited X chromosome [1,3-7]. Without Xist, XCI cannot be initiated, and the increased expression of X-linked genes causes early embryonic lethality in mice [8-10]. Similarly, dysregulation of XCI in female human pluripotent stem cells (hPSCs) correlates with an altered differentiation potential [11-13]. Recent work in hPSCs showed that the absence of XCI severely alters the transcriptome and proteome across the entire genome, due to increased expression of X-linked transcription factors and regulators of translation [14,15]. Although the importance of XCI during development is well established, it is less clear how critical the process is for adult homeostasis. Addressing this challenge is not trivial as the Xi in somatic cells is not only controlled by Xist and its interacting proteins but also by multiple repressive chromatin layers that are difficult to disassemble, leading to the belief that the Xi is extremely stable throughout life and that Xist may not be critical for Xi maintenance [16,17]. However, recent studies have revealed that not all genes on the Xi are controlled by the same epigenetic layers, such that some are more malleable than previously thought and become upregulated upon loss of Xist [18-22]. Intriguingly, this plasticity particularly affects genes that normally escape complete silencing on the Xi [22,23]. Moreover, Xist, long thought to coat the Xi in all differentiated cells in vivo, loses its localization to the Xi in specific cell types of the immune system [20,21,24], opening the door to the tuning of X-linked gene expression. These new findings have critical implications for disease susceptibility as well as the development of new treatment strategies of X-linked disorders, which underscores the importance of understanding Xi maintenance.
In this review, after briefly considering the regulation of XCI initiation, we discuss how gene silencing on the Xi is maintained. We will focus on the recent discovery of a new maintenance mechanism that involves the RNA-binding proteins Polypyrimidine Tract Binding Protein 1 PTBP1, Matrin 3 MATR3, CUGBP Elav-like family member 1 CELF1, and TAR DNA-binding protein 43 TDP-43, which are typically known for their role in RNA processing, but are now shown to play a general role in controlling RNA localization and function in the nucleus. We then introduce the emerging principle that Xist is not only critical for the initiation of XCI but also for the maintenance of silencing for many genes on the Xi, and discuss how the Xist-dependence is exploited in vivo.
Initiation of X-chromosome inactivation by Xist
To achieve XCI, Xist recruits diverse proteins, including RNA-binding proteins, transcriptional repressors, and architectural and heterochromatin proteins (Figure 1a). These factors ensure that the RNA stays localized on the X chromosome it is transcribed from, induces gene silencing, and alters the chromatin environment and the three-dimensional organization of the X chromosome to form the Xi compartment, also known as the Barr body [25-36]. Most of the Xist-interacting proteins are recruited through one of the six repeat arrays in the RNA termed A – F [1,3-7,28] (Figure 1a), which are conserved across eutherians but can differ in copy number between species [37,38]. For instance, mouse Xist has a C-repeat expansion and the B-repeat in human XIST is split into two elements (B and Bh) [38]. The A repeat interacts with the transcriptional repressor SPEN (also called SHARP (silencing mediator for retinoid or thyroid-hormone receptors SMRT/histone deacetylase 1 HDAC1 Associated Repressor Protein)), which is critical for the silencing of virtually all genes on the Xi [25,26,30,36] (Figure 1a). The B and C repeats bind hnRNP-K, which recruits the polycomb repressive complexes (PRC) 1 and 2 that deposit the histone marks H3K27me3 and H2AK119Ub1 [32,39], as well as the architectural chromatin protein Structural Maintenance of Chromosomes flexible Hinge Domain Containing 1 SMCHD1 [27,40] (Figure 1a), to regulate chromatin reconfiguration and compaction [29,35]. After gene silencing has occurred, DNA methylation is established at CpG islands of many genes on the Xi through SMCHD1-dependent and independent action of the DNA methyltransferase DNMT3B [41,42]. DNA methylation in particular has long been recognized as a key factor in XCI maintenance [16]. While most genes on the Xi are completely silenced, 3-7% of mouse and 20-30% of human X-linked genes remain partially expressed, lack the repressive epigenetic marks described above, and are referred to as escape genes [23].
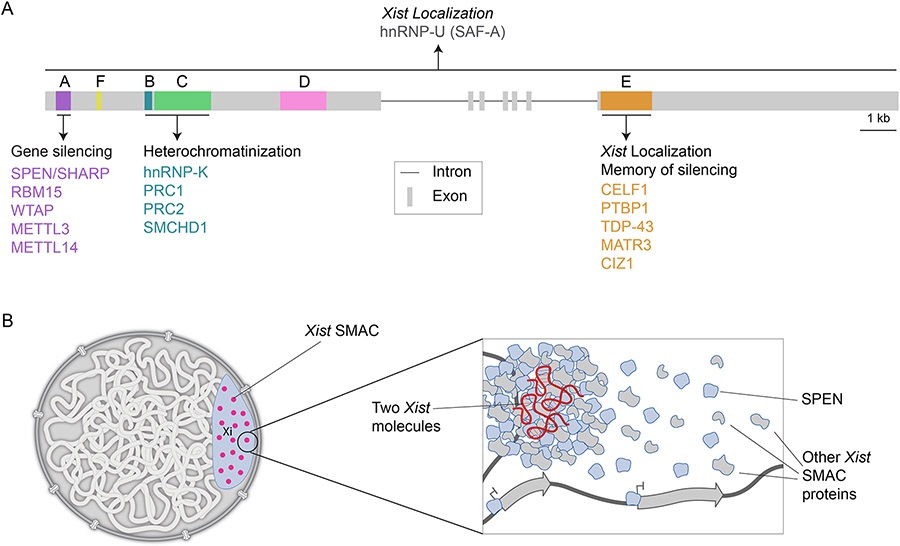
Figure 1. Xist structure and mechanisms of action.
A) Diagram of the mouse genomic Xist locus with its exons and introns and the location and key binding partners of the Xist repeat elements A-F. Some proteins such as scaffold attachment factor A (also called hnRNP-U : heterogeneous nuclear ribonucleoprotein U) SAF-A bind across the RNA. (kb=kilobase).
B) Xist silences X-linked genes by seeding supra-molecular complexes (SMACs). Left: Depiction of a nucleus with the Xi and its 50 Xist-SMACs that are locally constrained within the Xi. Right: Depiction of one Xist-SMAC highlighting that it is formed by two RNA molecules and a large number of proteins that bind to the RNA and undergo extensive protein-protein interactions. The dynamic behavior of most protein constituents of SMACs is thought to generate protein gradients, allowing free proteins to act on genes on the X chromosome to initiate and maintain their silencing and induce the heterochromatin state of the Xi.
A recently identified aspect of XCI initiation is that Xist distributes to only approximately 50 sites along the Xi [29,43-45]. At each of these 50 sites, two RNA molecules are tethered to chromatin with high affinity, locally confining their movement [29,34]. Where exactly the pairs of Xist molecules bind on chromatin is currently not defined. Specific Xist-interacting proteins such as the RNA and DNA binding protein SAF-A (also termed hnRNP-U) are involved in anchoring Xist on chromatin [46-50], whereas others are critical for the coupling of the two Xist molecules [34]. At each Xist hub, many protein molecules are recruited via RNA-protein and extensive protein-protein interactions to form supramolecular protein complexes (SMACs) [29] (Figure 1b). In contrast to Xist molecules, which persist for minutes to hours at these sites [29,34], most protein components of SMACs exhibit very short residence times in the range of seconds [29]. Therefore, SMACs are highly dynamic structures that allow most constituent proteins to rapidly bind and dissociate, which creates a local concentration gradient of these proteins around each Xist hub [29] (Figure 1b). Mechanistically, this is achieved by fleeting, low-affinity interactions between disordered regions found in Xist-interacting proteins as shown by the deletion of the intrinsically disordered domain of SPEN [29]. As a result, it has been posited that “free” SPEN proteins silence genes across the X without requiring continuous association with the RNA [29]. Importantly, the SMAC model of Xist action may provide a general mechanism for lncRNAs and other RNA species to achieve large regulatory effects even if they are very lowly expressed, via concentrating a highly dynamic pool of proteins [29].
A requirement for PTBP1, MATR3, CELF1, and TDP-43 for XCI maintenance
Experimental manipulations have identified a developmental switch where Xist-mediated silencing rapidly transitions from a fully Xist-dependent and reversible process to a largely Xist-independent and irreversible process [17]. Surprisingly, this switch occurs before the deposition of DNA methylation at CpG islands of genes on the Xi [41]. Consequently, additional mechanisms must contribute to the maintenance of gene silencing in the absence of Xist. Recent work has shed light on one such mechanism [31]. It was found that the proteins PTBP1, MATR3, TDP-43, and CELF1 proteins, which normally function in RNA processing, bind to the E-repeat of Xist [31] (Figures 1a, 2a). Since this sequence harbors a large number of putative binding sites for each of these proteins, many protein copies likely bind each Xist molecule [31]. The deletion of the E-repeat does not disrupt the initial Xist spread across the X or the initiation of gene silencing. Instead, the maintenance of gene silencing and Xist localization is impacted [31] (Figure 2a). These defects develop when XCI normally transitions to the Xist-independent stage, that is, when the experimentally induced loss of Xist from the Xi should have only minor consequences. Thus, E-repeat-dependent proteins shape the Xi-compartment in a way that enables it to maintain an epigenetic memory of gene silencing independently of Xist. This requires self-association of and interactions between these proteins, and is independent of their role in RNA processing [31] (Figure 2a). Together, these findings suggest that the composition of the Xi-compartment changes over time and that these changes are essential for the developmental switch to Xist-independence. Consistent with this idea, SPEN and CELF1 levels have been shown to increase over time in the Xi or Xist-SMACs [29,31], and proteomics studies have uncovered changes in the Xist-interactome with differentiation [22,25].
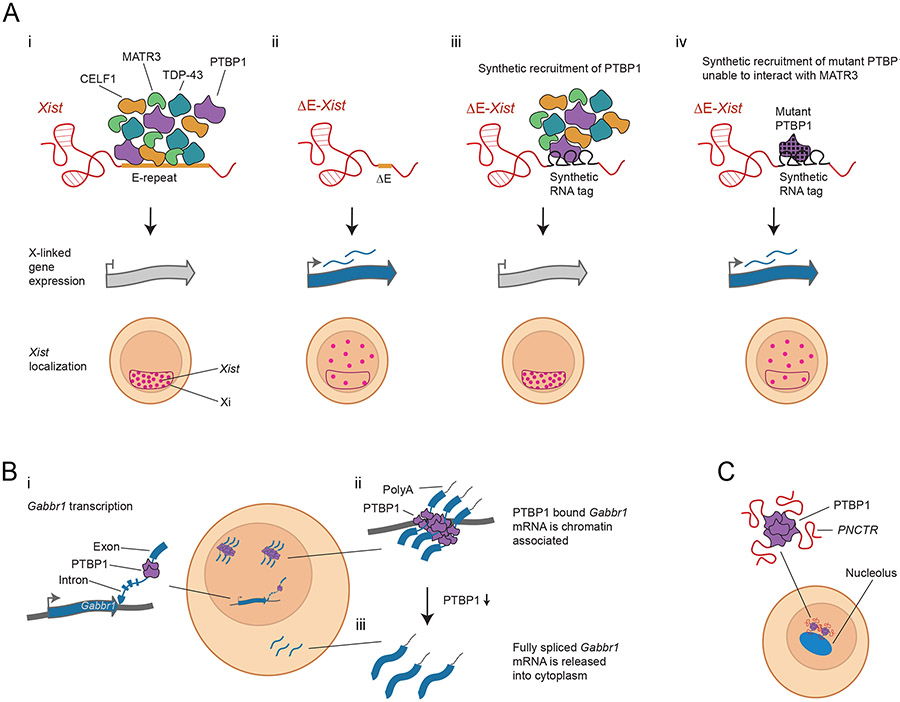
Figure 2. Stabilization of the Xi by the Xist E-repeat binding proteins PTBP1, MATR3, TDP-43 and CELF1.
A) i) The E-repeat binding proteins PTBP1, MATR3, TDP-43 and CELF1 bind to the E repeat in wildtype Xist, a 1.4kb region that contains a large number of sequence motifs for each of these proteins (top row). The proteins also interact with each other. In differentiating female mouse embryonic stem cells expressing wildtype Xist, the RNA exhibits coating of the Xi and gene silencing occurs (second and third row). ii) If the E repeat is deleted, PTBP1, MATR3, TDP-43 and CELF1 no longer bind to Xist. These cells initiate Xist coating and genes begin to silence, but after initiation of XCI, Xist becomes dispersed through the nucleus, and genes on the Xi reactivate. iii) If the E repeat is deleted but multiple PTBP1 molecules are artificially tethered to the mutant RNA, MATR3, TDP-43 and CELF1 can be indirectly recruited and silencing of X-linked genes and Xist coating of the Xi are retained. This result also holds when either MATR3, TDP-43, or CELF1 are artificially tethered to Xist [31]. iii) If a PTBP1 mutant that prevents the interaction with MATR3 is artificially tethered to Xist, the protein complex no longer forms and Xist fails to maintain its localization and silencing is not maintained. This result also holds if MATR3 is artificially tethered to Xist but contains a mutation preventing it from interacting with PTBP1 or blocking its self-interaction [31], supporting the importance of protein-protein interactions.
B) PTBP1 regulates the chromatin association of the Gabbr1 mRNA. i) The Gabbr1 transcript is transcribed from its locus. ii) The fully transcribed Gabbr1 mRNA is polyadenylated with most introns spliced out, but PTBP1 binding prevents splicing of one intron. The PTBP1-associated transcript is chromatin associated (although the specific location is currently unknown). iii) Following reduction of PTBP1 (by knockdown or neural differentiation), the intron is spliced and the transcript is released into the cytoplasm [85].
C) PTBP1 also maintains the localization of the lncRNA PNCTR to the peri-nucleolar compartment [86].
The mechanism of action of the XCI memory proteins described above remains unclear and will be an exciting problem to tackle in the future. One observation has been that CELF1 can remain enriched on the Xi in a PTBP1, MATR3, and TDP-43-dependent manner after Xist is deleted from the Xi in differentiated cells [31]. The CELF1 Xi enrichment upon Xist deletion appears to be stable only in a subset of cells and only for a short time (maybe for a few cell divisions) [31], suggesting that Xist is normally continuously needed to reinforce the protein interactions in the Xi-compartment. Regardless, it will be interesting to explore if the silencing protein SPEN can remain transiently localized together with the memory proteins in the Xi-compartment after Xist depletion in differentiated cells. This could explain the long-standing observation that silencing is maintained in the absence of Xist before the establishment of DNA methylation early in differentiation [17]. Intriguingly, in addition to the silencing protein SPEN, PTBP1 and MATR3 have been identified as essential for maintaining the repression of a candidate gene on the Xi in adult human cells, supporting a critical role of these E-repeat binding proteins in the maintenance of XCI [22] (Figure 3a). Intriguingly, another E-repeat binding protein, Cip1-interacting zinc finger protein CIZ1 (Figure 1a), is also important for Xist localization [34], However, it does not appear to function together with PTBP1, MATR3, TDP-43, and CELF1 in maintaining gene silencing [22,31] (Figure 3a).
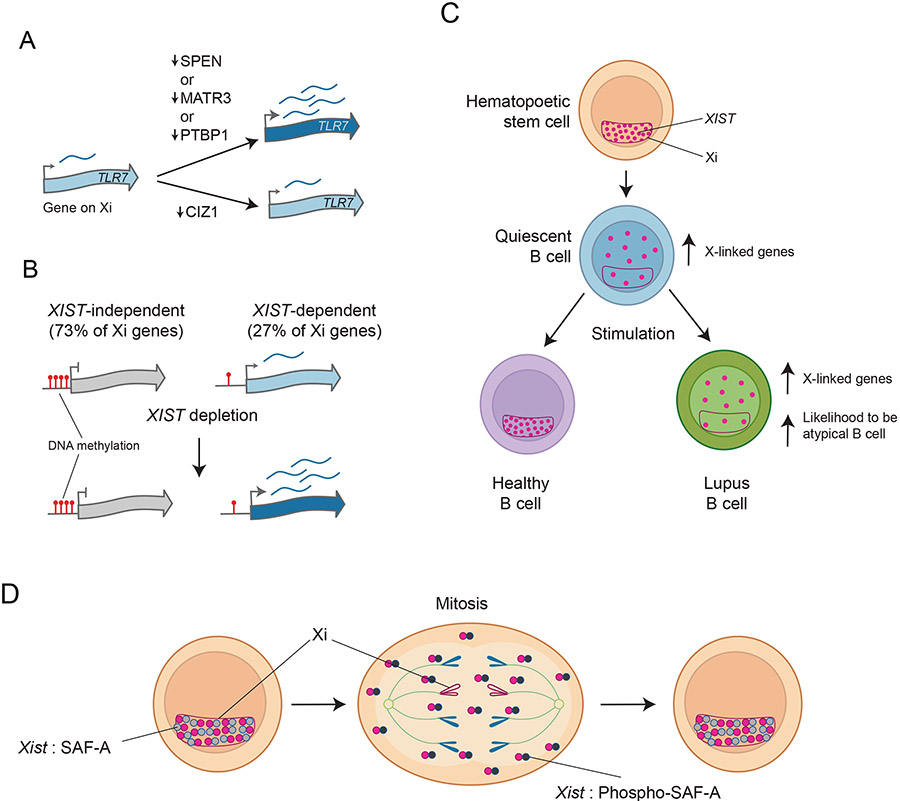
Figure 3. Loss of XIST/Xist impacts Xi maintenance and differentiation potential.
A) Similar to the transcriptional repressor SPEN, PTBP1, and MATR3 are required to maintain the repression of the inflammatory gene TLR7 on the Xi in the human B cell line GM12878 (which has an Xi-localized XIST). Like PTBP1 and MATR3, CIZ1 binds the E-repeat of XIST, but is not required to maintain silencing of Toll-like receptor 7 TLR7 [22,31], consistent with the E-repeat acting via two distinct pathways.
B) Requirement of XIST for Xi maintenance. The deletion of XIST in the human B cell line GM12878 showed that the majority of genes on the Xi are redundantly silenced by XIST and various layers of epigenetic regulation such as DNA methylation. However, the removal of XIST is sufficient to upregulate some genes on the Xi. These XIST-dependent genes often escape XCI and are lowly methylated in unperturbed B cells [22]. One example of an XIST dependent escape gene is TLR7.
C) Changes in XIST localization in lymphocyte development. XIST associates with the Xi in hematopoietic progenitor cells, forming its well-known territory over the Xi, but becomes delocalized from the Xi in quiescent B cells, distributing across the nucleus. Upon stimulation and re-entry into the cell cycle, B cells from healthy individuals regain the proper localization of XIST, while XIST remains delocalized in B cells from SLE (Lupus) patients [18]. SLE B cells display higher expression of XIST-dependent immune-regulatory genes on the X chromosome, including TLR7, and have an increased likelihood of forming atypical B cells (ABCs) [22].
D) During the normal cell cycle, XIST dissociates from the Xi in mitosis and one known regulatory mechanism is the aurora B kinase-mediated phosphorylation of SAF-A, which leads to the dissociation of XIST-SAF-A complexes [48]. In the new cell cycle round, newly transcribed XIST recoats the Xi. The dynamic localization of Xist is therefore a process that occurs in every dividing cell type, and therefore has the potential to be a regulatory or a disease mechanism in tissues beyond the hematopoietic system.
Overall, these studies identified an unanticipated role for a group of well-studied RNA-binding proteins in tethering RNAs to chromatin and controlling nuclear compartment functions, which does not require their RNA processing functions [31]. Excitingly, this emerging function of these proteins may not be limited to Xist (Box 1) (Figures 2b, c).
Box 1: A broader role for PTBP1 and MATR3 in the control of RNA localization, chromatin organization, and nuclear compartmentalization.
Xist studies have revealed numerous mechanisms that are also used by other RNAs. This now also extends to the PTBP1, MATR3, TDP-43 or CELF1-dependent control of RNA localization on chromatin. Various recent studies suggest that these E-repeat binding proteins, in particular PTBP1, mediate the chromatin localization of diverse RNAs such as Line L1 retrotransposon transcripts, lncRNAs and pre-mRNAs. Specifically, a recent publication showed that PTBP1, MATR3, TDP-43 and homologs of CELF1 densely bind antisense L1 sequences contained in mRNAs [82]. lncRNAs of the asynchronous replication and autosomal RNA (ASAR) family, which spread from their transcription locus in cis to control chromosome-wide replication timing, contain antisense L1 sequences that are required for the function and chromatin association of ASARs [83]. Together, these findings suggest that PTBP1, MATR3, TDP-43 and CELF proteins and their homologs may contribute to the chromatin association of ASARs and mRNAs containing antisense L1s, a function that may have been coopted by Xist during its evolution. Another example of Xist repurposing transposon-related mechanisms can be seen in the A-repeat of Xist, which is derived from an insertion of the endogenous retrovirus K (ERVK) [84]. Intriguingly, another study identified polyadenylated but incompletely spliced transcripts from protein-coding genes that are densely bound by PTBP1, associated with chromatin, and absent from the cytoplasm as mature mRNAs [85]. For one such gene, Gabbr1, it has been shown that its transcripts are released from chromatin upon depletion of PTBP1, suggesting that PTBP1 mediates its chromatin anchoring [85] (Figure 2b). Similarly, PTBP1 is also required for the localization of the lncRNA pyrimidine-rich noncoding transcript PNCTR in a peri-nucleolar compartment [86] (Figure 2c). The chromatin association of RNAs often directly impacts chromatin architecture [49]. Xist is a classic example for this type of regulation as the Xi becomes compacted and reorganized relative to the active X, through various architectural and heterochromatin regulators [29,35]. Similarly, thousands of other RNAs are maintained on chromatin through their interaction with RNA-binding proteins, such as SAF-A and MATR3, to prevent the compaction of active chromatin regions [49]. Moreover, MATR3 can bind the architectural proteins CCCTC-binding factor CTCF and ohesion [87] and thereby may directly link chromatin organization to chromatin-associated RNA molecules. Further studies of PTBP1, MATR3, TDP-43 and CELF1 and their homologs are therefore likely to reveal exciting new insights into the chromatin association of RNAs and the control of functional nuclear compartments.
Xist’s contribution to the maintenance of the inactive X chromosome
Classic experiments that focused on a few X-linked genes suggested that Xist is no longer required in somatic cells [16,17,51]. Interference with DNA methylation in Xist-deleted cells partially upregulated genes on the Xi, particularly when combined with histone deacetylase inhibition, consistent with the redundancy of Xi maintenance through various epigenetic layers [16]. Consequently, a common feature of Xi reactivation studies is that multiple inhibitors are combined [52-54]. However, an exciting recent study reassessed the role of XIST in the adult human B cell line GM12878 by applying various genomics approaches to define chromatin modifications and expression state across the Xi [22]. The authors found that XIST is critical for maintaining the silencing of a surprisingly large number of X-linked genes, as a quarter of genes on the Xi were upregulated upon experimental deletion of the RNA [22] (Figure 3b). These XIST-dependent genes carry lower methylation levels at their promoters on the normal Xi than genes resistant to XIST deletion [22] (Figure 3b). Genes known to escape XCI are enriched among these XIST-dependent genes [22]. Escape genes are normally expressed from the Xi, but often at lower levels than on the active X chromosome (Xa) [23,55]. Accordingly, escape genes have an active epigenetic state, unlike silenced genes on the Xi [56-59], which is consistent with them requiring continuous silencing from XIST [22]. The silencing environment created by XIST likely is explained by constant recruitment of the transcriptional repressor SPEN, as it has been shown to be required for the dampening of escape genes on the Xi [22,26] (Figure 3a). Since escape from XCI is more prevalent in humans than mice [60], genes on the human Xi may have a greater reliance on XIST than in mice. Nevertheless, recent studies using conditional Xist knockout approaches in mice have revealed a limited re-establishment of expression from the Xi [54,61,62]. Interestingly, the tissue-specific deletion of Xist in mice is typically well tolerated except for two tissues where induced Xist loss has dramatic consequences. These include blood, where loss of Xist in hematopoietic stem cells is oncogenic, and the gut, where Xist loss increases tumor burden upon exposure to chronic stress [61,62]. It is currently unknown why there are variable consequences of Xist deletion across different tissues, but tissue-specific escape from XCI may provide an explanation [61]. Additionally, recent studies suggest that gene repression on the Xi is balanced by upregulation of genes on the Xa in a tunable manner, via a process known as X chromosome upregulation [63]. Therefore, it remains possible that the upregulation of genes on the Xi results in dampening of their counterparts on the Xa, which could make most cell types robust to the consequences of XCI defects [63].
A link between escape genes and dependence on XIST may also exist in hPSCs. Normally, hPSCs carry an Xi with its classic epigenetic hallmarks, yet in most cell lines XIST eventually becomes repressed by de novo DNA methyltransferases [64]. XIST loss is typically followed by the re-expression of a subset of previously silenced genes on the Xi and loss of CpG island methylation at the affected genes [12,14,65]. This Xi-erosion occurs most often close to XCI escapees [65]. Thus, the degradation of the Xi-compartment appears to spread from genes already evading multiple layers of XCI repression, which are now known to be dynamically regulated by XIST. Intriguingly, genes proximal to escapees are also the earliest to become re-expressed during the reactivation of the Xi during mouse induced pluripotent stem cell iPSC reprogramming [66,67]. Since boundary elements surrounding escape genes are required to prevent activation of adjacent silenced genes [68], the erosion of boundaries could explain this consistent order of reactivation.
Xist localization is disrupted in immune cells
The aforementioned examples of XIST/Xist loss are either experimentally induced or represent culture-induced abnormalities. Although it was long thought that coating of the Xi by Xist is maintained in all somatic cells, recent studies have described an unusual distribution of the RNA in human and mouse quiescent lymphocytes, where it is dispersed throughout the interphase nucleus instead of being localized to the Xi [20,21,69] (Figure 3c). The mechanism underlying the redistribution of XIST/Xist in lymphocytes is currently unclear. However, XIST/Xist re-localizes to the Xi upon stimulation of lymphocytes and re-entry into the cell cycle (Figure 3c), requiring the RNA-interacting proteins Yin Yang 1 YY1 and SAF-A [20,21,69].
Curiously, in dividing cells, XIST/Xist is normally released from the Xi and disperses across the nucleocytoplasm in mitosis [70]. SAF-A is one of the proteins implicated in controlling the localization of the RNA in this process [47,48,50,71]. For instance, a recent paper demonstrated that the mitotic dispersal of XIST can be prevented by the inhibition of the cell cycle regulator aurora B kinase [48], which phosphorylates SAF-A to release the protein, together with XIST, from mitotic chromosomes [48] (Figure 3d). Although the question of how exactly XIST is anchored on chromatin is complex and might be highly cell type-specific [46-50,71], similar regulatory mechanisms may be at play to control SAF-A and/or other XIST/Xist chromatin anchors in quiescent lymphocytes to modify the localization of the RNA.
The deregulation of XIST/Xist localization in lymphocytes is particularly intriguing, given the recently discovered XIST-dependence of some genes on the Xi [22]. Indeed, a comparison of female and male lymphocytes showed that the dispersion of XIST/Xist is accompanied by an increase in the expression of X-linked immune regulators in female cells [21]. Allelic studies are required to systematically define which genes on the Xi become derepressed upon loss of the RNA from the Xi.
Together these findings show that Xist localization can be dynamically regulated in normal cells and that the cell type-specific dispersion of Xist results in XCI deregulation, which in lymphocytes increases the expression of X-linked immune regulators. Thus, Xist-dependence of some genes combined with the control of Xist localization on the Xi provides a mechanism to achieve the cell-type specific regulation of a subset of X-linked genes that would not be achievable if they were regulated via multiple repressive chromatin mechanisms.
Consequences of partial X chromosome reactivation due to Xist loss
It is well established that females mount stronger immune responses than males, which results in faster clearance of pathogens but also in increased susceptibility to autoimmune diseases [72]. This difference has been linked to XCI escape in females [72], and the aforementioned studies [20-22,69] suggest that the deregulation of XIST in lymphocytes potentiates the expression of escapees in females. These new studies of normally occurring dysregulation of XCI maintenance indeed support a link to the female-specific development of autoimmune diseases. For instance, it is well established that elevated levels of the X-linked escapee TLR7 enhance the formation of CD11c+ atypical B cells (ABCs), a cell type that greatly expands in a female-specific manner with age and in patients with autoimmune diseases [22]. ABCs lack proper XIST localization and express elevated levels of X-linked genes in female patients with autoimmune diseases, including systemic lupus erythematosus (SLE) [18-20,22] (Figure 3c). Intriguingly, there is also a causal link between XIST deregulation and cell fate specification, because the differentiation of stimulated naïve B cells into ABCs is enhanced when XIST is experimentally deleted in cultured cells [22]. Together, these findings open the door for a better understanding of the sexual dimorphism of human diseases, which might lead to new treatment opportunities, and suggest that the dispersal of XIST could be a useful biomarker for detecting autoimmune disorders. The latter idea is particularly important for diseases such as SLE, where diagnosis is often extensively delayed, resulting in worse patient outcomes [73].
The finding that the Xi is more plastic than previously thought may have implications for approaches that aim to reactivate the Xi to present a cure for heterozygous X-linked diseases such as the neurodevelopmental disorder Rett syndrome (RTT) caused by mutations in the X-linked gene methyl CpG binding protein 2 MECP2 [74]. When a deleterious copy of the X-linked gene MECP2 is expressed from the active X chromosome in RTT patients, reactivating the wildtype copy that is silenced on the Xi may rescue disease phenotypes [74]. Strengthening this idea, it was found that the induction of Mecp2 expression in mutant mice after the onset of symptoms can reverse disease symptoms [75]. Therefore, several studies have attempted to disrupt the Xi in mice, typically via deletion of Xist and/or inhibition of DNA methylation, and assess the activation of genes on the Xi as well as the physiological consequences [54,61,62]. Since escape from XCI (and therefore likely XIST-dependence) is more predominant in human than in mouse cells [23,55], similar studies need to be performed in human models of these X-linked diseases [76] to assess if Xi reactivation processes are well-tolerated in human tissues. The targeted demethylation and reactivation of specific genes on the Xi through programmable epigenetic editing represents an alternative strategy [77].
Conclusion
The study of XCI continues to provide fundamental insights into RNA biology, gene regulation and nuclear organization and is beginning to yield a better understanding of human diseases. Accordingly, elucidating the mechanisms that underlie the delocalization of XIST in lymphocytes and other immune cell types [20,24,69] may pave the way for the development of therapeutic approaches that prevent the loss of XIST from the Xi and upregulation of XIST-dependent genes. It is intriguing that XIST is also more dispersed in human pre-implantation embryos [78] and germ cells [79] where X chromosome dampening occurs [79,80], indicating that the modulation of XIST localization is exploited at different developmental stages to achieve a cell-type specific output of X-linked genes. In general, a deeper understanding of escape from the action of Xist-SMACs is an important focus, particularly as these genes appear to contribute disproportionately to sex-biased disorders [81]. Finally, the recent insights into the function of MATR3, PTBP1, SAF-A, CELF1 and TDP-43 in the control of RNA localization and chromatin organization suggest that further studies of these proteins will reveal tremendous insights into the extended molecular structures that surround genes and how RNA-protein interactions segregate nuclear compartments and ultimately gene function.
Acknowledgements
K.P. is supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, the David Geffen School of Medicine, the NIH (1R01MH109166, R01HD098387, P01GM099134), and a Faculty Scholar grant from the Howard Hughes Medical Institute.
Footnotes
Declaration of Interest
The authors declare no conflict of interest.
References
- 1.Deng X, Berletch JB, Nguyen DK, Disteche CM: X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet 2014, 15:367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disteche CM, Berletch JB: X-chromosome inactivation and escape. J Genet 2015, 94:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galupa R, Heard E: X-Chromosome inactivation: A crossroads between chromosome architecture and gene regulation. Annu Rev Genet 2016, 52:1–32. [DOI] [PubMed] [Google Scholar]
- 4.Jégu T, Aeby E, Lee JT: The X chromosome in space. Nat Rev Genet 2017, 18:377–389. [DOI] [PubMed] [Google Scholar]
- 5.Brockdorff N, Bowness JS, Wei G: Progress toward understanding chromosome silencing by Xist RNA. Genes Dev 2020, 34:733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.**. Dossin F, Heard E: The molecular and nuclear dynamics of X-Chromosome inactivation. Cold Spring Harb Perspect Biol 2021, doi: 10.1101/cshperspect.a040196. In this study the authors establish the importance of SPEN for the silencing of virtually all X-linked genes and define various mechanisms of action for the protein.
- 7.Raposo AC, Casanova M, Gendrel A-V, daRocha ST: The tandem repeat modules of Xist lncRNA: a swiss army knife for the control of X-chromosome inactivation. Biochem Soc Trans 2021, 49:2549–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N, et al. : Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 2017, 24:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R: Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 1997, 11:156–166. [DOI] [PubMed] [Google Scholar]
- 10.Takagi N, Abe K: Detrimental effects of two active X chromosomes on early mouse development. Dev Camb Engl 1990, 109:189–201. [DOI] [PubMed] [Google Scholar]
- 11.Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. : Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 2012, 11:75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel S, Bonora G, Sahakyan A, Kim R, Chronis C, Langerman J, Fitz-Gibbon S, Rubbi L, Skelton RJP, Ardehali R, et al. : Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep 2017, 18:54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokobayashi S, Yabuta Y, Nakagawa M, Okita K, Hu B, Murase Y, Nakamura T, Bourque G, Majewski J, Yamamoto T, et al. : Inherent genomic properties underlie the epigenomic heterogeneity of human induced pluripotent stem cells. Cell Rep 2021, 37:109909. [DOI] [PubMed] [Google Scholar]
- 14.*. Brenes AJ, Yoshikawa H, Bensaddek D, Mirauta B, Seaton D, Hukelmann JL, Jiang H, Stegle O, Lamond AI: Erosion of human X chromosome inactivation causes major remodeling of the iPSC proteome. Cell Rep 2021, 35:109032. Using proteomics approaches, the authors show that erosion of the Xi in hPSCs results in increased transcript and protein levels of X-linked genes including regulators of translation initiation, which in turn contributes to the upregulation of autosomally-encoded protein levels without changes in their RNA level.
- 15.Bruck T, Yanuka O, Benvenisty N: Human pluripotent stem cells with distinct X inactivation status show molecular and cellular differences controlled by the X-linked ELK-1 gene. Cell Rep 2013, 4:262–270. [DOI] [PubMed] [Google Scholar]
- 16.Csankovszki G, Nagy A, Jaenisch R: Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 2001, 153:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wutz A, Jaenisch R: A shift from reversible to irreversible X inactivation is triggered during ES Cell differentiation. Mol Cell 2000, 5:695–705. [DOI] [PubMed] [Google Scholar]
- 18.**. Pyfrom S, Paneru B, Knox JJ, Cancro MP, Posso S, Buckner JH, Anguera MC: The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc Natl Acad Sci U S A 2021, 118:2024624118. In this study, the authors show that upon stimulation and re-entry to the cell cycle, Xist relocalizes to the Xi in lymphocytes of healthy controls but not in SLE patients, providing evidence for Xist deregulation in auto-immune diseases.
- 19.Syrett CM, Sierra I, Beethem ZT, Dubin AH, Anguera MC: Loss of epigenetic modifications on the inactive X chromosome and sex-biased gene expression profiles in B cells from NZB/W F1 mice with lupus-like disease. J Autoimmun 2020, 107:102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syrett CM, Paneru B, Sandoval-Heglund D, Wang J, Banerjee S, Sindhava V, Behrens EM, Atchison M, Anguera MC: Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 2019, 4:e126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC: Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci U S A 2016, 113:E2029–E2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.**. Yu B, Qi Y, Li R, Shi Q, Satpathy AT, Chang HY: B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 2021, 184:1790–1803.e17. Exploring the role of XIST in human B cells, the authors show that many immune genes on the Xi are XIST-dependent, that the increased expression of these genes is observed in female patients with autoimmune diseases or COVID-19 infection, and that XIST deregulation is causal in specifying disease-associated cell types.
- 23.Fang H, Disteche CM, Berletch JB: X Inactivation and Escape: Epigenetic and Structural Features. Front Cell Dev Biol 2019, 7:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syrett CM, Sindhava V, Sierra I, Dubin AH, Atchison M, Anguera MC: Diversity of epigenetic features of the inactive X-chromosome in NK cells, dendritic cells, and macrophages. Front Immunol 2019, 10:3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY: Systematic discovery of Xist RNA binding proteins. Cell 2015, 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S, Saux AL, Chelmicki T, Attia M, Kapoor V, Zhan Y, et al. : SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 2020, 578:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansz N, Nesterova T, Keniry A, Iminitoff M, Hickey PF, Pintacuda G, Masui O, Kobelke S, Geoghegan N, Breslin KA, et al. : Smchd1 targeting to the inactive X Is dependent on the Xist-HnrnpK-PRC1 pathway. Cell Rep 2018, 25:1912–1923.e9. [DOI] [PubMed] [Google Scholar]
- 28.*. Lu Z, Guo JK, Wei Y, Dou DR, Zarnegar B, Ma Q, Li R, Zhao Y, Liu F, Choudhry H, et al. : Structural modularity of the XIST ribonucleoprotein complex. Nat Commun 2020, 11:6163. Employing several structure and interaction mapping approaches, the authors define the folding of Xist RNA and identify structural modules created by the 3D organization of the RNA.
- 29.**. Markaki Y, Chong JG, Wang Y, Jacobson EC, Luong C, Tan SYX, Jachowicz JW, Strehle M, Maestrini D, Banerjee AK, et al. : Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 2021, 184:6174–6192.e32. In this study, the authors use super-resolution microscopy to show that Xist seeds supra-molecular complexes (SMACs) at ~50 sites across the X chromosome and that the crowding of proteins SMACs is required for chromosome-wide silencing and compaction.
- 30.McHugh CA, Chen C-K, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. : The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.**. Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Leon WRM, Damianov A, Chronis C, Papp B, Chen C-K, McKee R, et al. : A protein assembly mediates Xist localization and gene silencing. Nature 2020, 587:145–151. In this study, the authors show that the Xist E-repeat binding proteins PTBP1, MATR3, TDP-43 and CELF1 (but not CIZ1) interact with each other to form a protein assembly that is dispensable for XCI initiation, but necessary for XCI maintenance.
- 32.Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. : hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell 2017, 68:955–969.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridings-Figueroa R, Stewart ER, Nesterova TB, Coker H, Pintacuda G, Godwin J, Wilson R, Haslam A, Lilley F, Ruigrok R, et al. : The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev 2017, 31:876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodermund L, Coker H, Oldenkamp R, Wei G, Bowness J, Rajkumar B, Nesterova T, Pinto DMS, Schermelleh L, Brockdorff N: Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 2021, 372:eabe7500. [DOI] [PubMed] [Google Scholar]
- 35.Wang CY, Jégu T, Chu HP, Oh HJ, Lee JT: SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 2018, 174:406–421.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Żylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, daRocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. : The implication of early chromatin changes in X chromosome inactivation. Cell 2019, 176:182–197.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nesterova TB, Slobodyanyuk SYa, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, SM Zakian: Characterization of the Genomic Xist Locus in Rodents Reveals Conservation of Overall Gene Structure and Tandem Repeats but Rapid Evolution of Unique Sequence. Genome Res 2001, 11:833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen ZC, Meyer IM, Karalic S, Brown CJ: A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 2007, 90:453–463. [DOI] [PubMed] [Google Scholar]
- 39.Bousard A, Raposo AC, Żylicz JJ, Picard C, Pires VB, Qi Y, Gil C, Syx L, Chang HY, Heard E, et al. : The role of Xist -mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep 2019, 20:e48019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C-Y, Colognori D, Sunwoo H, Wang D, Lee JT: PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat Commun 2019, 10:2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gendrel A-V, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, Tang YA, Huntley D, Montana G, Taylor S, et al. : Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell 2012, 23:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yagi M, Kabata M, Tanaka A, Ukai T, Ohta S, Nakabayashi K, Shimizu M, Hata K, Meissner A, Yamamoto T, et al. : Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat Commun 2020, 11:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, et al. : Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci 2014, 111:2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, Cerase A, Sterr M, Fiedler S, Demmerle J, Popken J, et al. : Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 2014, 7:8–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunwoo H, Colognori D, Froberg JE, Jeon Y, Lee JT: Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc Natl Acad Sci 2017, 114:10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S: The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 2010, 19:469–476. [DOI] [PubMed] [Google Scholar]
- 47.Sakaguchi T, Hasegawa Y, Brockdorff N, Tsutsui K, Tsutsui KM, Sado T, Nakagawa S: Control of Chromosomal Localization of Xist by hnRNP U Family Molecules. Dev Cell 2016, 39:11–12. [DOI] [PubMed] [Google Scholar]
- 48.*. Sharp JA, Perea-Resa C, Wang W, Blower MD: Cell division requires RNA eviction from condensing chromosomes. J Cell Biol 2020, 219:e201910148. In this study, the authors show that aurora kinase B phosphorylates SAF-A during mitosis to remove SAF-A-interacting RNAs from chromatin, including Xist.
- 49.**. Creamer KM, Kolpa HJ, Lawrence JB: Nascent RNA scaffolds contribute to chromosome territory architecture and counter chromatin compaction. Mol Cell 2021, 81:3509–3525.e5. In this study, authors propose that chromatin-associated nascent RNAs and lncRNAs perform important structural roles in the nucleus by binding to proteins like SAF-A and MATR3.
- 50.Kolpa HJ, Creamer KM, Hall LL, Lawrence JB: SAF-A mutants disrupt chromatin structure through dominant negative effects on RNAs associated with chromatin. Mamm Genome 2021, doi: 10.1007/s00335-021-09935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown CJ, Willard HF: The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 1994, 368:154–156. [DOI] [PubMed] [Google Scholar]
- 52.Minkovsky A, Sahakyan A, Bonora G, Damoiseaux R, Dimitrova E, Rubbi L, Pellegrini M, Radu CG, Plath K: A high-throughput screen of inactive X chromosome reactivation identifies the enhancement of DNA demethylation by 5-aza-2′-dC upon inhibition of ribonucleotide reductase. Epigenetics Chromatin 2015 81 2015, 8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lessing D, Dial TO, Wei C, Payer B, Carrette LLG, Kesner B, Szanto A, Jadhav A, Maloney DJ, Simeonov A, et al. : A high-throughput small molecule screen identifies synergism between DNA methylation and Aurora kinase pathways for X reactivation. Proc Natl Acad Sci U S A 2016, 113:14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrette LLG, Wang C-Y, Wei C, Press W, Ma W, Kelleher RJ, Lee JT: A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc Natl Acad Sci 2018, 115:E668–E675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, et al. : Landscape of X chromosome inactivation across human tissues. Nature 2017, 550:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ: Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 2015, 24:1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. : Structural organization of the inactive X chromosome in the mouse. Nature 2016, 535:575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duncan CG, Grimm SA, Morgan DL, Bushel PR, Bennett BD, Barnabas BB, Bouffard GG, Brooks SY, Coleman H, Dekhtyar L, et al. : Dosage compensation and DNA methylation landscape of the X chromosome in mouse liver. Sci Rep 2018, 8:10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sousa LBDAE, Jonkers I, Syx L, Dunkel I, Chaumeil J, Picard C, Foret B, Chen CJ, Lis JT, Heard E, et al. : Kinetics of Xist-induced gene silencing can be predicted from combinations of epigenetic and genomic features. Genome Res 2019, 29:1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balaton BP, Fornes O, Wasserman WW, Brown CJ: Cross-species examination of X-chromosome inactivation highlights domains of escape from silencing. Epigenetics Chromatin 2021, 14:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.**. Yang L, Yildirim E, Kirby JE, Press W, Lee JT: Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut. Proc Natl Acad Sci 2020, 117:4262–4272. In this study, the authors delete Xist in a variety of tissues in mice and find no phenotypic consequences except for an increased rate of gut tumors, but only under stress, suggesting that the loss of Xist and the upregulation of Xist-dependent genes is tolerated well in most adult tissues under normal conditions.
- 62.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT: Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013, 152:727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.**. Lentini A, Cheng H, Noble JC, Papanicolaou N, Coucoravas C, Andrews N, Deng Q, Enge M, Reinius B: Elastic dosage compensation by X-chromosome upregulation. Nat Commun 2022, 13:1854. In this study, the authors found that as the Xi is silenced during early development, the Xa becomes upregulated, such that gene dosage of the X is balanced in every cell, regardless of what phase of XCI the cell is at. This process is known as X chromosome upregulation (XCU).
- 64.Fukuda A, Hazelbaker DZ, Motosugi N, Hao J, Limone F, Beccard A, Mazzucato P, Messana A, Okada C, Juan IGS, et al. : De novo DNA methyltransferases DNMT3A and DNMT3B are essential for XIST silencing for erosion of dosage compensation in pluripotent stem cells. Stem Cell Rep 2021, 16:2138–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bansal P, Ahern DT, Kondaveeti Y, Qiu CW, Pinter SF: Contiguous erosion of the inactive X in human pluripotency concludes with global DNA hypomethylation. Cell Rep 2021, 35:109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bauer M, Vidal E, Zorita E, Üresin N, Pinter SF, Filion GJ, Payer B: Chromosome compartments on the inactive X guide TAD formation independently of transcription during X-reactivation. Nat Commun 2021, 12:3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Janiszewski A, Talon I, Chappell J, Collombet S, Song J, Geest ND, To SK, Bervoets G, Marin-Bejar O, Provenzano C, et al. : Dynamic reversal of random X-Chromosome inactivation during iPSC reprogramming. Genome Res 2019, 29:1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horvath LM, Li N, Carrel L: Deletion of an X-Inactivation Boundary Disrupts Adjacent Gene Silencing. PLoS Genet 2013, 9:e1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Syrett CM, Sindhava V, Hodawadekar S, Myles A, Liang G, Zhang Y, Nandi S, Cancro M, Atchison M, Anguera MC: Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet 2017, 13:e1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall LL, Byron M, Pageau G, Lawrence JB: AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J Cell Biol 2009, 186:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolpa HJ, Fackelmayer FO, Lawrence JB: SAF-A Requirement in Anchoring XIST RNA to Chromatin Varies in Transformed and Primary Cells. Dev Cell 2016, 39:9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klein SL, Flanagan KL: Sex differences in immune responses. Nat Rev Immunol 2016, 16:626–638. [DOI] [PubMed] [Google Scholar]
- 73.Kernder A, Richter JG, Fischer-Betz R, Winkler-Rohlfing B, Brinks R, Aringer M, Schneider M, Chehab G: Delayed diagnosis adversely affects outcome in systemic lupus erythematosus: Cross sectional analysis of the LuLa cohort. Lupus 2021, 30:431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carrette LLG, Blum R, Ma W, Kelleher RJ, Lee JT: Tsix–Mecp2 female mouse model for Rett syndrome reveals that low-level MECP2 expression extends life and improves neuromotor function. Proc Natl Acad Sci 2018, 115:201800931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guy J, Gan J, Selfridge J, Cobb S, Bird A: Reversal of neurological defects in a mouse model of Rett syndrome. Science 2007, 315:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samarasinghe RA, Miranda OA, Buth JE, Mitchell S, Ferando I, Watanabe M, Allison TF, Kurdian A, Fotion NN, Gandal MJ, et al. : Identification of neural oscillations and epileptiform changes in human brain organoids. Nat Neurosci 2021, 24:1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Halmai JANM, Deng P, Gonzalez CE, Coggins NB, Cameron D, Carter JL, Buchanan FKB, Waldo JJ, Lock SR, Anderson JD, et al. : Artificial escape from XCI by DNA methylation editing of the CDKL5 gene. Nucleic Acids Res 2020, 48:2372–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okamoto I, Patrat C, Thépot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf J-P, Renard J-P, Duranthon V, et al. : Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011, 472:370–374. [DOI] [PubMed] [Google Scholar]
- 79.Chitiashvili T, Dror I, Kim R, Hsu F-M, Chaudhari R, Pandolfi E, Chen D, Liebscher S, Schenke-Layland K, Plath K, et al. : Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nat Cell Biol 2020, 22:1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F: Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sauteraud R, Stahl JM, James J, Englebright M, Chen F, Zhan X, Carrel L, Liu DJ: Inferring genes that escape X-Chromosome inactivation reveals important contribution of variable escape genes to sex-biased diseases. Genome Res 2021, 31:1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.*. Attig J, Agostini F, Gooding C, Chakrabarti AM, Singh A, Haberman N, Zagalak JA, Emmett W, Smith CWJ, Luscombe NM, et al. : Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 2018, 174:1067–1081.e17. In this study, the authors show that Xist E-repeat binding proteins (PTBP1, MATR3, TDP-43 and homologs of CELF1) bind densely to antisense LINE1 elements in introns and prevent splicing at cryptic splice sites within the L1 sequence to maintain proper structure of the host gene mRNA.
- 83.*. Platt EJ, Smith L, Thayer MJ: L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J Cell Biol 2018, 217:541–553. This study shows that ASAR lncRNAs that control chromosome-wide replication timing in cis require the L1 antisense sequences contained within them for their chromatin association.
- 84.Carter AC, Xu J, Nakamoto MY, Wei Y, Zarnegar BJ, Shi Q, Broughton JP, Ransom RC, Salhotra A, Nagaraja SD, et al. : Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. eLife 2020, 9:e54508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeom K-H, Pan Z, Lin C-H, Lim HY, Xiao W, Xing Y, Black DL: Tracking pre-mRNA maturation across subcellular compartments identifies developmental gene regulation through intron retention and nuclear anchoring. Genome Res 2021, 31:1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yap K, Mukhina S, Zhang G, Tan JSC, Ong HS, Makeyev EV: A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol Cell 2018, 72:525–540.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.*. Cha HJ, Uyan Ö, Kai Y, Liu T, Zhu Q, Tothova Z, Botten GA, Xu J, Yuan G-C, Dekker J, et al. : Inner nuclear protein Matrin-3 coordinates cell differentiation by stabilizing chromatin architecture. Nat Commun 2021, 12:6241. In this study the authors show that MATR3 directly interacts with the chromatin organization proteins CTCF and cohesin to stabilize chromatin structure, which is required for the correct regulation of cell fate transitions.