ABSTRACT
SARS-CoV-2 has mutated frequently since its first emergence in 2019. Numerous variants, including the currently emerging Omicron variant, have demonstrated high transmissibility or increased disease severity, posing serious threats to global public health. This study describes the identification of an immunodominant non-neutralizing epitope on SARS-CoV-2 receptor-binding domain (RBD). A subunit vaccine against this mutant RBD, constructed by masking this epitope with a glycan probe, did not significantly affect RBD’s receptor-binding affinity or antibody-binding affinity, or its ability to induce antibody production. However, this vaccine enhanced the neutralizing activity of this RBD and its protective efficacy in immunized mice. Specifically, this vaccine elicited significantly higher-titer neutralizing antibodies than the prototypic RBD protein against Alpha (B.1.1.7 lineage), Beta (B.1.351 lineage), Gamma (P.1 lineage), and Epsilon (B.1.427 or B.1.429 lineage) variant pseudoviruses containing single or combined mutations in the spike (S) protein, albeit the neutralizing antibody titers against some variants were slightly lower than against original SARS-CoV-2. This vaccine also significantly improved the neutralizing activity of the prototypic RBD against pseudotyped and authentic Delta (B.1.617.2 lineage) and Omicron (B.1.1.529 lineage) variants, although the neutralizing antibody titers were lower than against original SARS-CoV-2. In contrast to the prototypic RBD, the mutant RBD completely protected human ACE2 (hACE2)-transgenic mice from lethal challenge with a prototype SARS-CoV-2 strain and a Delta variant without weight loss. Overall, these findings indicate that this RBD vaccine has broad-spectrum activity against multiple SARS-CoV-2 variants, as well as the potential to be effective and have improved efficacy against Omicron and other pandemic variants.
IMPORTANCE Several SARS-CoV-2 variants have shown increased transmissibility, calling for a need to develop effective vaccines with broadly neutralizing activity against multiple variants. This study identified a non-neutralizing epitope on the receptor-binding domain (RBD) of SARS-CoV-2 spike protein, and further shielded it with a glycan probe. A subunit vaccine based on this mutant RBD significantly enhanced the ability of prototypic RBD against multiple SARS-CoV-2 variants, including the Delta and Omicron strains, although the neutralizing antibody titers against some of these variants were lower than those against original SARS-CoV-2. This mutant vaccine also enhanced the protective efficacy of the prototypic RBD vaccine against SARS-CoV-2 infection in immunized animals. In conclusion, this study identified an engineered RBD vaccine against Omicron and other SARS-CoV-2 variants that induced stronger neutralizing antibodies and protection than the original RBD vaccine. It also highlights the need to improve the effectiveness of current COVID-19 vaccines to prevent pandemic SARS-CoV-2 variants.
KEYWORDS: coronavirus, COVID-19, SARS-CoV-2, Omicron and other variants, subunit vaccine, improved neutralizing activity, enhanced protection
INTRODUCTION
Coronavirus disease 2019 (COVID-19), first reported in 2019 (1), has led to a global pandemic with severe economic loss. As of July 19, 2022, more than 559 million COVID-19 cases and at least 6.3 million deaths have been reported worldwide. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of COVID-19, consists of four structural proteins, with the surface spike (S) protein playing a critical role in viral infection and pathogenesis (2, 3). The S protein consists of two subunits, S1 and S2. The receptor-binding domain (RBD) in the S1 subunit binds to a cellular receptor, angiotensin-converting enzyme 2 (ACE2), to initiate the viral entry process, whereas the S2 subunit mediates fusion between the virus and the cell membrane (4, 5). Therefore, the SARS-CoV-2 S protein and its RBD fragment are key targets for the development of COVID-19 vaccines.
Since its emergence in December 2019, SARS-CoV-2 has undergone continuous mutations, resulting in the occurrence of multiple variants, including Alpha (B.1.1.7 lineage), Beta (B.1.351 lineage), Gamma (P.1 lineage), Delta (B.1.617.2 lineage), and Epsilon (B.1.427 or B.1.429 lineage) (6–8). A new variant, Omicron (B.1.1.529 lineage), which was first identified in South Africa in late November 2021, has spread rapidly to other countries and currently accounts for the majority of cases in the United States and many other countries (9–11). These variants have been found to include multiple mutations in the S protein or RBD. Thus, it is crucial to understand and quickly determine whether current vaccines which target the prototype virus strain are effective against SARS-CoV-2 variants, especially Omicron.
Compared to other vaccine types, such as viral vectored vaccines, subunit vaccines generally have low immunogenicity due to their intrinsic limitations involving the inclusion of various immunodominant non-neutralizing epitopes (12, 13). Masking of such epitopes by glycan probes or other approaches may focus immune responses against the neutralizing epitopes, thereby potentially increasing the neutralizing capacity of subunit vaccines (12, 14). A neutralizing immunogenicity index (NII) approach has been developed to calculate the contribution of epitopes to the overall neutralizing immunogenicity of subunit vaccines. This led to the successful design of several mutant subunit vaccines with improved efficacy against the Middle East respiratory syndrome coronavirus (MERS-CoV) and Zika virus (12, 15).
This study describes the identification of an immunodominant non-neutralizing epitope containing residue Asn519 of the SARS-CoV-2 S protein, and its masking with a N-linked glycan probe. This enabled the design of a mutant RBD subunit vaccine containing a glycosylation site at residues 519 and 521. The NII of this epitope was determined, as were the receptor-binding affinity and antibody-binding affinity of the resultant mutant RBD and its ability to induce antibody production. This mutant protein was found to improve the neutralizing activity of the RBD, enabling it to neutralize multiple variants of SARS-CoV-2, including Omicron, and to enhance the ability of the RBD to protect animals against SARS-CoV-2 infection.
RESULTS
Introduction of a glycan probe onto an epitope and characterization of a glycosylated mutant SARS-CoV-2 RBD protein.
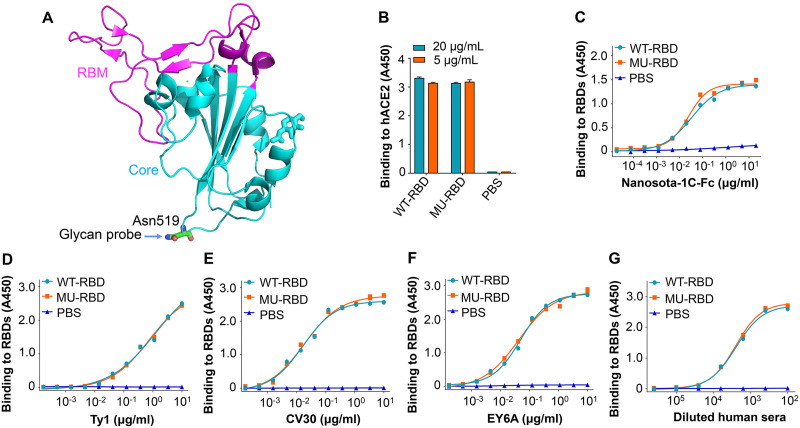
A glycan probe was attached to an epitope surrounding residue Asn519 of the RBD of SARS-CoV-2 S protein (Fig. 1A), and mutations were introduced at residues 519 and 521 to generate an N-linked glycosylation site. A recombinant mutant RBD protein was constructed based on this glycosylation site and purified from supernatants of 293T cells transfected with a recombinant plasmid expressing this protein. The binding of the glycosylated mutant RBD protein (MU-RBD) to human ACE2 (hACE2) receptor was similar to that of the prototypic wild-type (WT)-RBD did (Fig. 1B), indicating that masking of this epitope did not affect the ability of the RBD to bind to its receptor. Further analysis revealed that this mutant RBD protein also bound to SARS-CoV-2 RBD-specific neutralizing nanobodies (Nanosota-1C-Fc and Ty1) (Fig. 1C and D) and neutralizing monoclonal antibodies (MAbs) (CV30 and EY6A) (Fig. 1E and F), as well as serum neutralizing antibodies from COVID-19-vaccinated humans (Fig. 1G), similar to the binding of prototypic RBD, suggesting that the masked epitope did not affect the antigenicity of the RBD or its ability to bind to specific neutralizing antibodies.
FIG 1.
Introduction of glycan probe and characterization of glycosylated mutant SARS-CoV-2 RBD protein. (A) Crystal structure of SARS-CoV-2 RBD (PDB access code: 6M0J). The core structure is colored in cyan, and the receptor-binding motif (RBM) in magenta. Mutated residue (Asn519) is shown where an N-linked glycan probe was introduced. (B) Receptor-binding affinity of mutant receptor-binding domain (RBD) (MU-RBD) subunit vaccine. An enyzme-linked immunosorbent assay (ELISA) was carried out to assess the binding of MU-RBD protein to soluble human angiotensin-converting enzyme 2 (hACE2) protein. Prototypic wild-type (WT)-RBD protein was included as comparison. (C to G) Antibody-binding affinity of MU-RBD subunit vaccine. ELISA was carried out to detect the binding of MU-RBD protein to SARS-CoV-2 RBD-specific neutralizing nanobodies Nanosota-1C-Fc (C) and Ty1 (D), neutralizing MAbs CV30 (E) and EY6A (F), and neutralizing human sera (G). WT-RBD was used as comparison. Data (panels B to G) are presented as mean ± standard error of the mean (SEM) of quadruple wells. The experiments were repeated twice, resulting in similar results.
Glycosylated mutant SARS-CoV-2 RBD induced significantly higher-titer neutralizing antibodies than the prototypic RBD against SARS-CoV-2 Alpha, Beta, Gamma, and Epsilon variants.
To assess the neutralizing activity of glycosylated mutant RBD against multiple SARS-CoV-2 variants, hACE2-transgenic (Tg) mice were immunized with this protein (Fig. 2), and the titers of induced neutralizing antibodies against SARS-CoV-2 variants were compared with the titers of antibodies induced by the prototypic RBD. Neutralization analyses were performed using pseudoviruses expressing the S protein of the Alpha, Beta, Gamma, and Epsilon variants of SARS-CoV-2. Mice were immunized three times with prototypic or mutant RBD, and sera of these mice collected 10 days after both the second and the third immunizations were used for the following tests (Fig. 2).
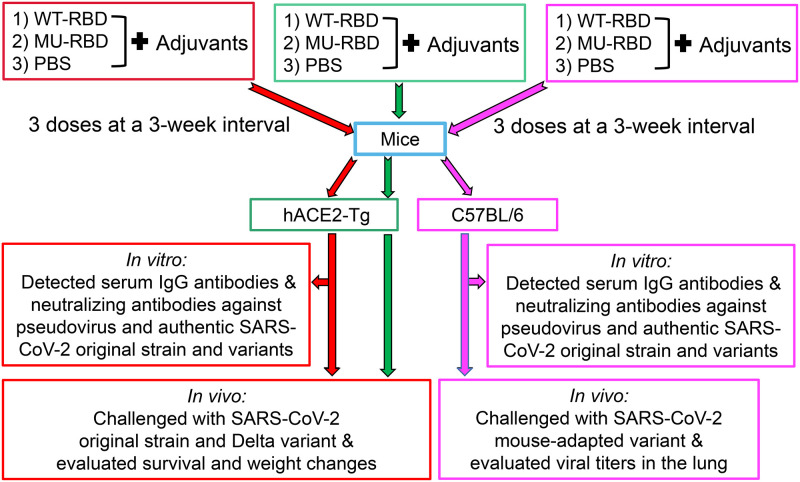
FIG 2.
Immunization and challenge schedules. C57BL/6 and hACE2-transgenic (Tg) mice were immunized with the prototypic WT-RBD, MU-RBD, or phosphate-buffered saline (PBS) control in the presence of adjuvants for three times at 3-week intervals. Sera were collected at 10 days after the second and third immunizations, and tested for IgG antibodies and neutralizing antibodies against pseudotyped and authentic SARS-CoV-2 original strain and variants. Immunized hACE2-Tg mice were challenged with SARS-CoV-2 original strain or Delta variant and observed for survival and weight changes for 14 days. Immunized C57BL/6 mice were challenged with a mouse-adapted SARS-CoV-2 variant and evaluated for viral titers in the lungs at day 2 after virus challenge.
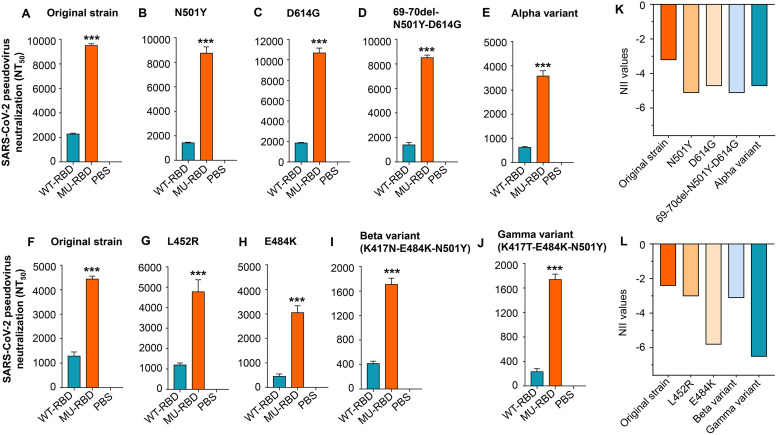
Sera collected from hACE2-Tg mice 10 days after the third immunization demonstrated that, compared with the prototypic RBD, the mutant RBD elicited significantly higher titers of neutralizing antibodies against B.1.1.7 (Alpha) variant pseudoviruses harboring key mutations, including N501Y, D614G, or 67-70del-N501Y-D614G, in the S protein. Moreover, the titers of neutralizing antibodies induced by mutant RBD against pseudovirus containing these mutations were similar to, or slightly different from, those against original SARS-CoV-2 (Fig. 3A to D). The titers of the neutralizing antibodies induced by mutant RBD were also significantly higher than those induced by the prototypic RBD protein against an Alpha variant pseudovirus containing all 10 amino acid changes in the S protein, although the neutralizing antibody titer induced by mutant RBD was about two- to three-fold less potent against this variant than against original SARS-CoV-2 (Fig. 3A and E).
FIG 3.
Glycosylated mutant SARS-CoV-2 RBD protein elicited improved neutralizing antibodies against SARS-CoV-2 Alpha, Beta, Gamma, and Epsilon variants. hACE2-Tg mice were immunized with the prototypic WT-RBD or MU-RBD protein and boosted twice at 3 weeks. Mice injected with PBS were included as control. Mouse sera collected 10 days after the third immunization (A to E) and 10 days after the second immunization (F to J) were assessed for neutralizing activity against infection of pseudoviruses expressing S protein of the SARS-CoV-2 original strain and each variant harboring mutation(s) at the indicated amino acid(s), respectively. Alpha variant (B.1.1.7 lineage) contains all 10 amino acid mutations (69 to 70 deletion, 145 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) in the S protein of SARS-CoV-2. Neutralizing activity was expressed as 50% neutralizing antibody titers (NT50) against pseudovirus infection in 293T cells expressing hACE2 receptor (hACE2/293T). Data are presented as mean ± SEM of quadruple wells from pooled sera of five mice in each group. ***, (P < 0.001) indicates significant differences between the MU-RBD and WT-RBD groups. The experiments were repeated twice, resulting in similar results. (K to L) Calculated neutralizing immunogenicity index (NII) values based on the neutralizing antibody titers (NT50) and the following formula: (NT50-WT – NT50-MU)/NT50-WT, where NT50-WT and NT50-MU represent NT50 induced by the WT-RBD and MU-RBD, respectively.
Sera collected from hACE2-Tg mice 10 days after the second immunization showed that the glycosylated mutant RBD protein induced high titers of neutralizing antibodies against pseudoviruses of Epsilon (B.1.427 or B.1.429 lineage) variants harboring a single L452R mutation, with titers similar to those against original SARS-CoV-2 (Fig. 3F and G). These antibodies also efficiently neutralized pseudoviruses bearing a single E484K mutation, or triple mutations of K417N-E484K-N501Y (Beta variant: B.1.351 lineage) and K417T-E484K-N501Y (Gamma variant: P.1 lineage), although the neutralizing antibody titers against the Beta and Gamma variants were lower than those against original SARS-CoV-2 (Fig. 3F, H to J).
The neutralizing immunogenicity index of an epitope has been defined as the contribution of that epitope to the overall neutralizing immunogenicity of the subunit vaccines (12, 15). Measurement of the NII of the glycan-shielded epitope on the RBD showed that its NIIs against original SARS-CoV-2 and its variants were all negative (Fig. 3K and L), indicating that this non-neutralizing epitope makes a negative contribution to the overall neutralizing immunogenicity of RBD. Thus, masking the epitope with a glycan probe significantly improved the overall neutralizing activity of the RBD subunit vaccine.
Glycosylated mutant SARS-CoV-2 RBD elicited higher neutralizing antibody titers than the prototypic RBD against SARS-CoV-2 Delta and Omicron variants.
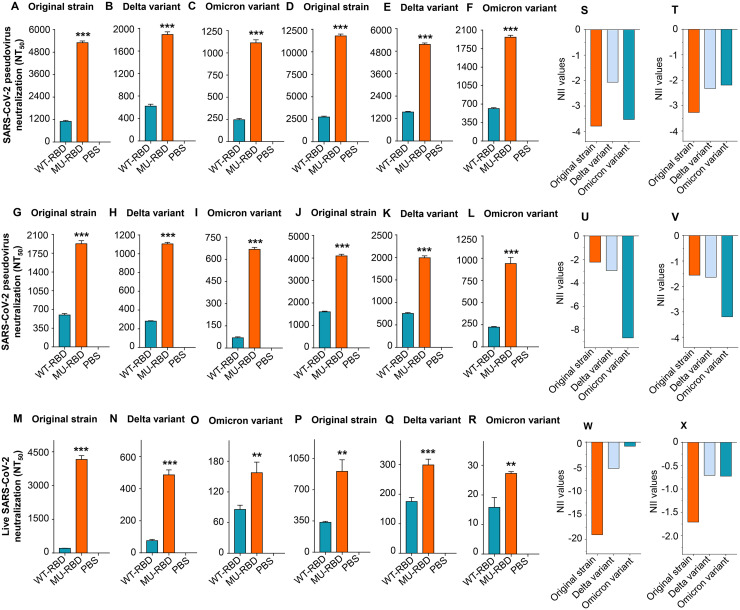
The SARS-CoV-2 Delta and Omicron variants have been identified as the variants of concern (VOCs) with high transmissibility and, in the case of Delta, disease severity (6, 10, 16). To investigate the neutralizing activities of glycosylated mutant SARS-CoV-2 RBD against these variants, sera of C57BL/6 and C57BL/6-background hACE2-Tg mice collected 10 days after the second and third immunizations were tested for neutralizing activity against pseudotyped Delta and Omicron variants which contained L452R-T478K-P681R and 38 amino acid mutations in their S protein, respectively, or against authentic Delta and Omicron variants (Fig. 2).
Compared with the prototypic RBD, the mutant RBD induced similar levels of IgG antibodies specific to the SARS-CoV-2 wild-type (WT)-RBD, Delta-RBD, Omicron BA.1-RBD, and Omicron BA.2-RBD proteins, respectively (Fig. 4A to P), but significantly higher-titer neutralizing antibodies against pseudotyped (Fig. 5A to L) and authentic (Fig. 5M to R) SARS-CoV-2 original strain, Delta, and Omicron variants (Fig. 5A to R) in both C57BL/6 and hACE2-Tg mice. In particular, the third dose of the mutant RBD vaccine further improved neutralizing antibody titers compared to the second dose (Fig. 5A to L). These results suggest that masking the non-neutralizing epitope Asn519 maintained the immunogenicity of the RBD in the induction of specific IgG antibodies but increased its ability to induce potent antiviral neutralizing antibodies against SARS-CoV-2 Delta and Omicron variants, in addition to the original virus strain. Similarly, the negative NIIs of the mutant RBD against the SARS-CoV-2 original strain, Delta, and Omicron variants confirm that the identified epitope makes a negative contribution to the overall neutralizing immunogenicity of the RBD (Fig. 5S to X). Notably, the neutralizing activity induced by the glycosylated mutant RBD was less effective against the Delta and Omicron variants than against original SARS-CoV-2 (Fig. 5A to R), and the numbers of IgG antibodies binding to the Omicron BA.1 and BA.2 RBD proteins were also lower than the numbers binding to the WT-RBD and Delta-RBD proteins (Fig. 4).
FIG 4.
Antibody responses induced by glycosylated mutant SARS-CoV-2 RBD protein. C57BL/6 and hACE2-Tg mice were immunized with the prototypic WT-RBD or MU-RBD protein, or with PBS control as described above, and collected for sera 10 days after the second and third immunizations. ELISA for detection of IgG antibodies specific to the SARS-CoV-2 WT-RBD (A, E, I, and M), Delta-RBD (B, F, J, and N), Omicron BA.1-RBD (C, G, K, and O), and Omicron BA.2-RBD (D, H, L, and P) from sera of C57BL/6 mice (A to H) and hACE2-Tg (I to P) mice after the second (A to D, I to L) and third (E to H, M to P) immunizations. Data are presented as mean ± SEM of quadruple wells from pooled sera of five mice in each group. The experiments were repeated twice, resulting in similar results.
FIG 5.
Glycosylated mutant SARS-CoV-2 RBD protein elicited improved neutralizing antibodies against SARS-CoV-2 Delta and Omicron variants. The same sera described in Fig. 4 were tested for neutralizing antibodies against infection of pseudoviruses expressing S protein of the SARS-CoV-2 original strain (A, D, G, and J), Delta variant (harboring L452R-T478K-P681R mutations in the S1 region) (B, E, H, and K), and Omicron variant (harboring 38 amino acid mutations in the S protein) (C, F, I, and L), respectively. These sera were collected from C57BL/6 (A to F) and hACE2-Tg (G to L) mice 10 days after the second (A to C, G to I) and third (D to F, J to L) immunizations. Sera from C57BL/6 (M to O) and hACE2-Tg (P to R) mice after the third immunization were also tested for neutralizing antibodies against infection of authentic SARS-CoV-2 original strain (M and P), Delta (N and Q), and Omicron (O and R) variants. Neutralizing activity was calculated as NT50 against infection of each pseudotyped or authentic SARS-CoV-2. **, (P < 0.01) and ***, (P < 0.001) indicate significant differences between the MU-RBD and WT-RBD groups. Data (panels A to R) are shown as mean ± SEM of quadruple wells from pooled sera of five mice in each group. The experiments were repeated twice, resulting in similar results. (S to X) Calculated neutralizing NII values based on the neutralizing antibody titers (NT50) and the formula described in Fig. 3.
These data indicate that glycosylated mutant RBD vaccine elicited antibodies that were able to effectively neutralize pseudotyped and authentic SARS-CoV-2 Delta and Omicron variants with multiple mutations in their S protein or RBD, albeit the neutralizing titers were relatively lower than those against the original virus strain.
Glycosylated mutant SARS-CoV-2 RBD showed greater efficacy than prototypic RBD in protecting mice against infection from SARS2-CoV-2 original strain and Delta variant.
To investigate the ability of glycosylated mutant RBD to induce protective immunity against SARS-CoV-2 infection, immunized hACE2-Tg mice were challenged with lethal doses of the original strain or Delta variant of SARS-CoV-2 25 to 50 days after the last immunization, and their survival and weight changes were monitored for 14 days. Immunized C57BL/6 mice were also challenged with a mouse-adapted SARS-CoV-2 variant (SARS2-N501YMA30), and viral titers were detected in the lung on day 2 after virus challenge (Fig. 2).
All of the hACE2-Tg mice immunized with the mutant RBD protein survived challenge with original SARS-CoV-2 infection and showed no weight loss, whereas only 80% of mice immunized with the prototypic RBD protein survived after this SARS-CoV-2 challenge with increased weight loss (Fig. 6A and B). Although all of the hACE2-Tg mice immunized with the mutant or prototypic RBD protein survived challenge with Delta SARS-CoV-2 variant, the mice immunized with the prototypic RBD protein had more weight loss than those immunized with the mutant RBD (Fig. 6C and D). In contrast, control hACE2-Tg mice receiving phosphate-buffered saline (PBS) and related adjuvants all lost weight after SARS-CoV-2 original or Delta variant challenge, and all died by 8 or 9 days after challenge (Fig. 6A to D). The immunized C57BL/6 mice potently inhibited replication of the mouse-adapted SARS2 (N501YMA30), with significantly lower viral titers in the lung than the PBS control mice after challenge (Fig. 6E). These findings indicate that glycosylation of the SARS-CoV-2 RBD of a single identified epitope enhanced neutralizing immunogenicity and protection of mice against SARS-CoV-2, leading to complete protection against infection from the original virus strain and Delta variant without obvious weight loss.
FIG 6.
Glycosylated mutant SARS-CoV-2 RBD protein induced enhanced protective efficacy against infection of the SARS-CoV-2 original strain and Delta variant. At 25 days after the third immunization, hACE2-Tg mice (5 mice/group) were challenged with prototype SARS-CoV-2 human strain (2019n-CoV/USA-WA1/2009, 5,000 PFU/mouse), and observed for survival (A) and body weight changes (B) for 14 days post-challenge. In addition, 50 days after the third immunization, hACE2-Tg mice (5 mice/group) were challenged with SARS-CoV-2 Delta variant (10,000 PFU/mouse) and observed for survival (C) and body weight changes (D) for 14 days post-challenge. At 50 days after the third immunization, C57BL/6 mice were challenged with mouse-adapted strain of SARS-CoV-2 (SARS2-N501YMA30, 5,000 PFU/mouse), and measured for lung viral titers on day 2 post-challenge. Data (in panels B, D, and E) are shown as mean ± SEM of 5 mice in each group. *, (P < 0.05); **, (P < 0.01); and ***, (P < 0.001) indicate significant differences between the WT-RBD and PBS (cyan), MU-RBD and PBS (red), or WT-RBD and MU-RBD (black) groups.
DISCUSSION
Subunit vaccines are generally safe but have relatively low immunogenicity. This study describes the design of a novel subunit vaccine with improved neutralizing immunogenicity and protection. Alignment of the SARS-CoV-2 and SARS-CoV RBD sequences indicates that SARS-CoV carries an asparagine (N) in its RBD corresponding to residue 519 (histidine, H) of SARS-CoV-2 RBD (17, 18). In addition, this residue is located on a protruding loop in the core region of SARS-CoV-2 RBD (Fig. 1A), a potential immunodominant non-neutralizing epitope. Therefore, a glycan probe was placed onto residue 519 of SARS-CoV-2 RBD to form an N-linked glycan probe (N-X-T, where X is any amino acid other than proline) with residue 521, on which the design of a mutant RBD subunit vaccine was based. This mutant RBD protein had the same receptor-binding affinity and antibody-binding activity as the prototypic RBD, presenting similar immunogenicity in the induction of effective IgG antibodies. Similar to the glycan-shielded subunit vaccines against MERS-CoV and Zika virus (12, 15), the mutant RBD induced significantly higher titers of neutralizing antibodies than the prototypic RBD. The hACE2-Tg mice express SARS-CoV-2 receptor human ACE2, and thus they are an effective lethal animal model for the evaluation of COVID-19 vaccines and therapeutic agents (19, 20). Here, we found that the mutant RBD demonstrated improved efficacy in protecting immunized hACE2-Tg mice against lethal challenge with the prototype SARS-CoV-2 strain and Delta variant. Our findings also confirm that the identified non-neutralizing epitope made a negative contribution to the overall neutralizing immunogenicity of the RBD subunit vaccine, enhancing its neutralizing activity and protective efficacy.
Several SARS-CoV-2 VOCs have been identified, all of which have mutations in the viral S protein, including the RBD (21–23). The Alpha, Beta, Gamma, and Delta VOCs induce more severe infections than the prototype virus strain, and the Delta variant may lead to more serious infection than other variants. In contrast, the Omicron variant appears to have increased transmissibility but cause attenuated infection and disease (8, 10, 11, 16, 24–27). Although at least three COVID-19 vaccines have been approved or authorized to prevent SARS-CoV-2 infection in humans (28–31), the emergence of these VOCs, particularly the Omicron variant, has limited current prevention strategies based on the prototype virus strain. Therefore, developing effective vaccines with broad-spectrum activity against different variants will be critical in preventing the spread of SARS-CoV-2 infection and ending the COVID-19 pandemic.
Interestingly, the glycosylated RBD described in this study was able to elicit neutralizing antibodies against pseudotyped and authentic prototype strains of SARS-CoV-2, as well as against all mutant VOC strains tested, including Alpha, Beta, Gamma, Delta, and Omicron. Moreover, the neutralizing antibody titers against these strains were significantly higher than those induced by the prototypic RBD. These results suggest that the mutant RBD subunit had the ability to induce broadly neutralizing antibodies against multiple SARS-CoV-2 variants, including Omicron.
There are some potential limitations of this study. For example, the antibodies induced by the mutant RBD had lower neutralizing activity against the SARS-CoV-2 variants, particularly the Omicron variant, than against the original virus strain, indicating that these variants, especially Omicron, which contains 38 mutations in the S protein, escape immune responses specific to the RBD of prototypic SARS-CoV-2 S protein. Thus, the mutant RBD protein with a single glycan probe might not induce neutralizing antibodies as potent as those induced by other proteins with several combined glycan probes against all SARS-CoV-2 variants, or cross-neutralizing antibodies against SARS-CoV and SARS-related coronaviruses with pandemic potential. Thus, future studies will be needed to identify more non-neutralizing epitopes within the RBD of SARS-CoV-2 or SARS-CoV and combine them to design vaccines with improved neutralizing immunogenicity and protection. In addition, future studies will also be needed to evaluate the structure of the mutant RBD complexed with the ACE2 receptor, the study of which will be important to understand whether the masked epitope affects the functionality of the RBD to bind its receptor in structure perspectives.
Overall, our results are consistent with those of other studies, which have demonstrated that antibodies induced by immunization with two doses of current COVID-19 vaccines had reduced neutralizing activity against Omicron and other variants than against prototype SARS-CoV-2 (32–34). Moreover, although neutralizing antibodies against the Omicron variant could be induced by three doses of the Moderna or Pfizer mRNA vaccines, their titers were relatively low (35). Therefore, effective vaccines specifically targeting variant S proteins or their RBDs, or universal vaccines targeting the conserved epitopes of SARS-CoV-2 S protein, are needed for further development. Improvements in the effectiveness of COVID-19 vaccines are urgently needed to prevent infection or at least disease caused by SARS-CoV-2 Omicron and other variants.
MATERIALS AND METHODS
Construction and purification of recombinant proteins.
The RBD sequence of prototypic SARS-CoV-2 was amplified by PCR using a plasmid encoding S protein of SARS-CoV-2 (GenBank accession no. QHR63250.2). The RBD sequence of SARS-CoV-2 Omicron BA.1 variant was amplified by PCR using a plasmid encoding RBD of SARS-CoV-2 Omicron BA.1 variant (GISAID accession no. EPI_ISL_6795835). The RBD sequence of SARS-CoV-2 Omicron BA.2 variant (GISAID accession no. EPI_ISL_9401700) was amplified using ClonExpress MultiS One Step Cloning kit (Cellagen Technology LLC) based on the prototypic SARS-CoV-2 RBD sequence described above. The amplified PCR fragments were fused with a C-terminal Fc fragment of human IgG. The mutant RBD and Delta RBD were constructed using a multi-site-directed mutagenesis kit (Agilent Technologies) based on the prototypic SARS-CoV-2 RBD sequence. The recombinant plasmids were transfected into HEK293T/293F cells, and the respective protein was purified from the culture supernatants using nProtein A Sepharose 4 Fast Flow (GE Healthcare).
ELISA.
The binding between each SARS-CoV-2 RBD protein and human ACE2 (hACE2) protein was detected by enzyme-linked immunosorbent assay (ELISA) (36). Briefly, the ELISA plates were coated with respective RBD protein (2 μg/mL) at 4°C overnight and blocked with 2% fat-free milk in PBS containing 0.05% Tween 20 (PBST) at 37°C for 2 h. The plates were then incubated with hACE2 protein (20 or 5 μg/mL, R&D Systems) at 37°C for 2 h. After washing 3 times using PBST, the binding was detected by goat anti-hACE2 IgG antibody (0.2 μg/mL, R&D Systems), followed by horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG antibody (1:5,000, R&D Systems). The plates were further washed and incubated with TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Sigma-Aldrich), and the reaction was stopped by 1 N H2SO4. The absorbance at 450 nm (A450) was measured using a Cytation 7 Microplate Multi-Mode Reader (BioTek Instruments).
The binding between SARS-CoV-2 RBD proteins and RBD-specific serum antibodies or neutralizing antibodies (nAbs) was detected by ELISA (36, 37). Briefly, the ELISA plates were coated with each RBD protein (1 μg/mL) at 4°C overnight. After the aforementioned blockage and washing steps, the plates were incubated with serially diluted mouse or human sera (38), or with nAbs (e.g., CV30, EY6A, and Ty1) (Absolute Antibody), at 37°C for 2 h, followed by incubation with HRP-conjugated anti-mouse IgG antibody (1:5,000, Abcam; for mouse sera), anti-human IgG-Fab antibody (1:4,000, Abcam; for CV30, EY6A, and human sera), or anti-His antibody (1:4,000; for Ty1) at 37°C for 1 h. Alternatively, the ELISA plates were coated with Nanosota-1C-Fc (20) as described above, followed by sequential addition of each RBD protein, mouse anti-SARS-CoV-2-S polyclonal antibody, and HRP-conjugated anti-mouse antibody (1:5,000, Abcam). The other procedures were performed as described above.
Construction of recombinant plasmids.
Recombinant plasmids expressing S protein of the SARS-CoV-2 original strain (GenBank accession no. QHR63250.2) or Alpha (B.1.1.7) variant strain (GISAID accession no. EPI_ISL_718813) were constructed by inserting the respective DNA sequence into pcDNA3.1/V5-His-TOPO vector (Thermo Fisher Scientific) (17, 36). Omicron BA.1 variant (B.1.1.529) (GISAID accession no. EPI_ISL_6795835) and recombinant plasmids expressing S protein of other variants containing single or multiple amino acid mutations were constructed using a multi-site-directed mutagenesis kit (Agilent Technologies) and confirmed by sequencing analysis. The constructed plasmids were used to generate pseudoviruses as described below.
Generation of SARS-CoV-2 pseudoviruses and neutralization assay.
This was performed as previously described with some modifications (17, 36, 37, 39). Briefly, SARS-CoV-2 pseudoviruses were produced by cotransfection of 293T cells with pLenti-CMV-Luciferase, PS-PAX2, and each of the plasmids encoding SARS-CoV-2 original or mutant S protein using a polyetherimide (PEI) transfection method. The pseudovirus-containing supernatants were harvested at 72 h post-transfection and used for neutralization assays. For pseudovirus neutralization, SARS-CoV-2 pseudoviruses expressing original or mutant S protein were incubated with serially diluted sera at 37°C for 2 h, and then added to 293T cells expressing hACE2 (hACE2/293T) cells in 96-well plates, followed by the addition of fresh medium 24 h later. At 72 h later, the cells were lysed using cell lysis buffer (Promega) and incubated with luciferase substrate (Promega). The relative luciferase activity was measured using the Cytation 7 Microplate Multi-Mode Reader described above. SARS-CoV-2 pseudovirus neutralization was calculated and expressed as 50% neutralizing antibody titer (NT50).
Plaque reduction neutralization assay.
Sera collected from the immunized mice described below were measured for neutralizing activity against prototype SARS-CoV-2 human strain (2019n-CoV/USA-WA1/2009), as well as against authentic Delta (B.1.617.2) and Omicron (B.1.1.529) variants, using a plaque reduction neutralization assay as previously described, with some modifications (40). Briefly, sera which had been serially diluted in Dulbecco’s modified Eagle medium (DMEM) were mixed with each virus (40 to 80 PFU/well) at 37°C for 1 h. The serum-virus mixture was incubated with Vero E6 (for prototype strain and Delta variant) or Vero E6 in the presence of ACE2 and TMPRSS2 (for Omicron variant) cells at 37°C for 45 min. After the inoculum was removed, the cells were overlaid with 0.6% agarose and cultured for 3 days. The overlays were then removed, and plaques were visualized by staining with 0.1% crystal violet. The neutralizing antibody titer was calculated as NT50 (i.e., the highest serum dilution able to reduce the number of virus plaques by 50%).
Ethics statement.
C57BL/6 and hACE2-Tg mice were used in the study. The animal welfare and experimental procedures were approved by the Committee on the Ethics of Animal Experiments of Georgia State University, New York Blood Center, and University of Iowa. All mouse-related experiments were performed in strict accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health.
Mouse immunization and challenge studies.
The following three immunization and challenge studies were performed as previously described with some modifications (20, 39). First, 4-month-old hACE2-Tg mice (mixed, 1 to 5 male or female) were intramuscularly (IM) immunized with SARS-CoV-2 prototypic-RBD protein, mutant RBD protein (10 μg/mouse), or PBS control in the presence of aluminum (500 μg/mouse) and monophosphoryl lipid A (MPL, 10 μg/mouse) adjuvants (InvivoGen). The immunized mice were boosted twice with the same immunogen and adjuvants at 3 weeks, and sera were collected at 10 days after the 2nd and 3rd immunizations to detect specific IgG antibodies or neutralizing antibodies (as described above). At 25 days after the last immunization, mice were transferred to an Animal Biosafety Level 3 facility, intranasally (IN) challenged with a prototype SARS-CoV-2 human strain (2019n-CoV/USA-WA1/2009, 5,000 PFU/mouse, 50 μL/mouse), and observed for survival and body weight changes for 14 days after the challenge. Second, 2- to 3-month-old hACE2-Tg mice (mixed, 1 to 5 male or female) were immunized with each protein or the control, collected for sera as described above, challenged with SARS-CoV-2 Delta (B.1.617.2) variant (10,000 PFU/mouse, 50 μL/mouse) at 50 days after the last immunization, and then observed for survival and body weight changes for 14 days after the challenge. Third, 2- to 4-month-old female C57BL/6 mice were immunized with each protein or the control, collected for sera as described above, and challenged with a virulent mouse-adapted strain of SARS-CoV-2 (SARS2-N501YMA30, 5,000 PFU/mouse, 50 μL/mouse) (40) at 50 days after the last immunization. Mouse lungs were collected on day 2 after virus challenge for detection of viral titers as described below. The immunization and challenge schedules are summarized in Fig. 2.
Detection of viral titers.
The lung tissues from challenged mice were homogenized and centrifuged for collection of supernatants. The supernatants were then serially diluted in DMEM cell culture medium and incubated with Vero E6 cells at 37°C for 1 h. This was followed by the same procedures as for the plaque reduction neutralization assay described above. Viral titers were quantified as PFU/mL of lung tissue.
Statistical analysis.
Statistical significance was analyzed using GraphPad Prism 9 statistical software. Statistically significant differences in neutralizing antibody or viral titers between the mutant RBD protein, prototypic RBD protein, and/or PBS control groups were calculated using a two-tailed independent Student’s t test. Statistically significant differences between survival curves were calculated by a Kruskal-Wallis test. Statistically significant differences between weight curves were calculated by an Ordinary one-way analysis of variance. P < 0.05 was considered significant. *, **, and *** in the figures represent P < 0.05, P < 0.01, and P < 0.001, respectively.
ACKNOWLEDGMENTS
This study was supported by NIH grants (R01AI139092, R01AI137472, and R01AI157975).
We declare no conflicts of interest.
Contributor Information
Stanley Perlman, Email: stanley-perlman@uiowa.edu.
Lanying Du, Email: ldu3@gsu.edu.
Stacey Schultz-Cherry, St. Jude Children’s Research Hospital.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang S, Zhang X, Yang Y, Hotez PJ, Du L. 2020. Neutralizing antibodies for the treatment of COVID-19. Nat Biomed Eng 4:1134–1139. 10.1038/s41551-020-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S, Hillyer C, Du L. 2020. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41:355–359. 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang S, Zhang X, Du L. 2021. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin Ther Targets 25:415–421. 10.1080/14728222.2020.1820482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Radhakrishnan VS, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, Piccoli L, Barclay WS, Rakshit P, CITIID-NIHR BioResource COVID-19 Collaboration ., et al. 2021. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thye AY, Law JW, Pusparajah P, Letchumanan V, Chan KG, Lee LH. 2021. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines 9:1303. 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin L, Liu Y, Tang X, He D. 2021. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front Public Health 9:775224. 10.3389/fpubh.2021.775224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker JM, Nakayama JY, O’Hegarty M, McGowan A, Teran RA, Bart SM, Mosack K, Roberts N, Campos B, Paegle A, McGee J, Herrera R, English K, Barrios C, Davis A, Roloff C, Sosa LE, Brockmeyer J, Page L, Bauer A, Weiner JJ, Khubbar M, Bhattacharyya S, Kirking HL, Tate JE. 2022. SARS-CoV-2 B.1.1.529 (Omicron) variant transmission within households: four U.S. jurisdictions, November 2021–February 2022. MMWR Morb Mortal Wkly Rep 71:341–346. 10.15585/mmwr.mm7109e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Shaman J. 2022. COVID-19 pandemic dynamics in South Africa and epidemiological characteristics of three variants of concern (Beta, Delta, and Omicron). medRxiv Preprint. 10.1101/2021.12.19.21268073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kannan S, Shaik Syed Ali P, Sheeza A. 2021. Omicron (B.1.1.529)—variant of concern—molecular profile and epidemiology: a mini review. Eur Rev Med Pharmacol Sci 25:8019–8022. 10.26355/eurrev_202112_27653. [DOI] [PubMed] [Google Scholar]
- 12.Du L, Tai W, Yang Y, Zhao G, Zhu Q, Sun S, Liu C, Tao X, Tseng CK, Perlman S, Jiang S, Zhou Y, Li F. 2016. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat Commun 7:13473. 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Shang J, Jiang S, Du L. 2020. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol 11:298. 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong PD, Mascola JR. 2018. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48:855–871. 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 15.Tai W, Chen J, Zhao G, Geng Q, He L, Chen Y, Zhou Y, Li F, Du L. 2019. Rational design of Zika virus subunit vaccine with enhanced efficacy. J Virol 93:e02187. 10.1128/JVI.02187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butt AA, Dargham SR, Chemaitelly H, Al Khal A, Tang P, Hasan MR, Coyle PV, Thomas AG, Borham AM, Concepcion EG, Kaleeckal AH, Latif AN, Bertollini R, Abou-Samra A-B, Abu-Raddad LJ. 2022. Severity of illness in persons infected with the SARS-CoV-2 Delta variant vs Beta variant in Qatar. JAMA Intern Med 182:197–205. 10.1001/jamainternmed.2021.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. 2020. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 17:613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv H, Mok CKP, Wilson IA. 2020. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368:630–633. 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Dick EJ, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye G, Gallant J, Zheng J, Massey C, Shi K, Tai W, Odle A, Vickers M, Shang J, Wan Y, Du L, Aihara H, Perlman S, LeBeau A, Li F. 2021. The development of Nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates. Elife 10:e64815. 10.7554/eLife.64815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papanikolaou V, Chrysovergis A, Ragos V, Tsiambas E, Katsinis S, Manoli A, Papouliakos S, Roukas D, Mastronikolis S, Peschos D, Batistatou A, Kyrodimos E, Mastronikolis N. 2022. From Delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene 814:146134. 10.1016/j.gene.2021.146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupala CS, Ye Y, Chen H, Su XD, Liu H. 2022. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun 590:34–41. 10.1016/j.bbrc.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Du L. 2022. Neutralizing antibodies and their cocktails against SARS-CoV-2 Omicron and other circulating variants. Cell Mol Immunol 19:962–964. 10.1038/s41423-022-00890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, Scheaffer SM, Darling TL, Joshi A, Loeber S, Singh G, Foster SL, Ying B, Case JB, Chong Z, Whitener B, Moliva J, Floyd K, Ujie M, Nakajima N, Ito M, Wright R, Uraki R, Warang P, Gagne M, Li R, Sakai-Tagawa Y, Liu Y, Larson D, Osorio JE, Hernandez-Ortiz JP, Henry AR, Ciuoderis K, Florek KR, Patel M, Odle A, Wong L-YR, Bateman AC, Wang Z, Edara V-V, Chong Z, Franks J, Jeevan T, Fabrizio T, DeBeauchamp J, Kercher L, Seiler P, Gonzalez-Reiche AS, Sordillo EM, Chang LA, van Bakel H, Consortium Mount Sinai Pathogen Surveillance (PSP) study group , et al. 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603:687–692. 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan S, Ye ZW, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Man Poon VK, Chan CC, Mok BW, Qin Z, Xie Y, Chu AW, Chan WM, Ip JD, Sun H, Tsang JO, Yuen TT, Chik KK, Chan CC, Cai JP, Luo C, Lu L, Yip CC, Chu H, To KK, Chen H, Jin DY, Yuen KY, Chan JF. 2022. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science 377:428–433. 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 26.Espenhain L, Funk T, Overvad M, Edslev SM, Fonager J, Ingham AC, Rasmussen M, Madsen SL, Espersen CH, Sieber RN, Stegger M, Gunalan V, Wilkowski B, Larsen NB, Legarth R, Cohen AS, Nielsen F, Lam JUH, Lavik KE, Karakis M, Spiess K, Marving E, Nielsen C, Wiid Svarrer C, Bybjerg-Grauholm J, Olsen SS, Jensen A, Krause TG, Müller L. 2021. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Euro Surveill 26:2101146. 10.2807/1560-7917.ES.2021.26.50.2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, Ip JD, Chan WM, Chu AW, Chan KH, Jin DY, Chen H, Yuen KY, To KK. 2022. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–283. 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb YN. 2021. BNT162b2 mRNA COVID-19 vaccine: first approval. Drugs 81:495–501. 10.1007/s40265-021-01480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodworth KR, Moulia D, Collins JP, Hadler SC, Jones JM, Reddy SC, Chamberland M, Campos-Outcalt D, Morgan RL, Brooks O, Talbot HK, Lee GM, Bell BP, Daley MF, Mbaeyi S, Dooling K, Oliver SE. 2021. The Advisory Committee on Immunization Practices’ Interim Recommendation for use of Pfizer-BioNTech COVID-19 vaccine in children aged 5–11 Years: United States, November 2021. MMWR Morb Mortal Wkly Rep 70:1579–1583. 10.15585/mmwr.mm7045e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, McClung N, Campos-Outcalt D, Morgan RL, Mbaeyi S, Romero JR, Talbot HK, Lee GM, Bell BP, Dooling K. 2021. The Advisory Committee on Immunization Practices’ Interim Recommendation for use of Moderna COVID-19 vaccine: United States, December 2020. MMWR Morb Mortal Wkly Rep 69:1653–1656. 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacNeil JR, Su JR, Broder KR, Guh AY, Gargano JW, Wallace M, Hadler SC, Scobie HM, Blain AE, Moulia D, Daley MF, McNally VV, Romero JR, Talbot HK, Lee GM, Bell BP, Oliver SE. 2021. Updated recommendations from the Advisory Committee on Immunization Practices for use of the Janssen (Johnson & Johnson) COVID-19 vaccine after reports of thrombosis with thrombocytopenia syndrome among vaccine recipients: United States, April 2021. MMWR Morb Mortal Wkly Rep 70:651–656. 10.15585/mmwr.mm7017e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edara VV, Manning KE, Ellis M, Lai L, Moore KM, Foster SL, Floyd K, Davis-Gardner ME, Mantus G, Nyhoff LE, Bechnak S, Alaaeddine G, Naji A, Samaha H, Lee M, Bristow L, Gagne M, Roberts-Torres J, Henry AR, Godbole S, Grakoui A, Saxton M, Piantadosi A, Waggoner JJ, Douek DC, Rouphael N, Wrammert J, Suthar MS. 2022. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep Med 3:100529. 10.1016/j.xcrm.2022.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, Nutalai R, Zhou D, Mentzer AJ, Zhao Y, Duyvesteyn HME, López-Camacho C, Slon-Campos J, Walter TS, Skelly D, Johnson SA, Ritter TG, Mason C, Costa Clemens SA, Gomes Naveca F, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Dold C, Temperton N, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert SC, Malik T, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Baillie V, Serafin N, Ditse Z, Da Silva K, Paterson NG, Williams MA, et al. 2021. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 184:4220.e13–4236.e13. 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi A, Koch M, Wu K, Dixon G, Oestreicher J, Legault H, Stewart-Jones GBE, Colpitts T, Pajon R, Bennett H, Carfi A, Edwards DK. 2021. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. J Virol 95:e0131321. 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, Berrios C, Ofoman O, Chang CC, Hauser BM, Feldman J, Roederer AL, Gregory DJ, Poznansky MC, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. 2022. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 185:457–466.e4. 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai W, Zhang X, Drelich A, Shi J, Hsu JC, Luchsinger L, Hillyer CD, Tseng CK, Jiang S, Du L. 2020. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res 30:932–935. 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai W, Zhang X, He Y, Jiang S, Du L. 2020. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res 179:104820. 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Huang L, Ye G, Geng Q, Ikeogu N, Harris M, Dileepan G, Burrack K, Du L, Frosch A, Li F. 2022. Vaccine booster efficiently inhibits entry of SARS-CoV-2 omicron variant. Cell Mol Immunol 19:445–446. 10.1038/s41423-022-00837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geng Q, Tai W, Baxter VK, Shi J, Wan Y, Zhang X, Montgomery SA, Taft-Benz SA, Anderson EJ, Knight AC, Dinnon KH, 3rd, Leist SR, Baric RS, Shang J, Hong SW, Drelich A, Tseng CK, Jenkins M, Heise M, Du L, Li F. 2021. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog 17:e1009897. 10.1371/journal.ppat.1009897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Zheng J, Zhang X, Tai W, Odle AE, Perlman S, Du L. 2022. RBD-mRNA vaccine induces broadly neutralizing antibodies against Omicron and multiple other variants and protects mice from SARS-CoV-2 challenge. Transl Res. 10.1016/j.trsl.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]