ABSTRACT
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a recently emerging bat-borne coronavirus responsible for high mortality rates in piglets. In vitro studies have indicated that SADS-CoV has a wide tissue tropism in different hosts, including humans. However, whether this virus potentially threatens other animals remains unclear. Here, we report the experimental infection of wild-type BALB/c and C57BL/6J suckling mice with SADS-CoV. We found that mice less than 7 days old are susceptible to the virus, which caused notable multitissue infections and damage. The mortality rate was the highest in 2-day-old mice and decreased in older mice. Moreover, a preliminary neuroinflammatory response was observed in 7-day-old SADS-CoV-infected mice. Thus, our results indicate that SADS-CoV has potential pathogenicity in young hosts.
IMPORTANCE SADS-CoV, which likely has originated from bat coronaviruses, is highly pathogenic to piglets and poses a threat to the swine industry. Little is known about its potential to disseminate to other animals. No efficient treatment is available, and the quarantine strategy is the only preventive measure. In this study, we demonstrated that SADS-CoV can efficiently replicate in suckling mice younger than 7 days. In contrast to infected piglets, in which intestinal tropism is shown, SADS-CoV caused infection and damage in all murine tissues evaluated in this study. In addition, neuroinflammatory responses were detected in some of the infected mice. Our work provides a preliminary cost-effective model for the screening of antiviral drugs against SADS-CoV infection.
KEYWORDS: coronavirus, SADS-CoV, suckling mice, infection, maternal protection
INTRODUCTION
Coronaviruses (CoVs) are a group of enveloped, single-stranded positive-sense RNA viruses within the subfamily Coronavirinae of the family Coronaviridae. CoVs infect diverse hosts, including birds, mammals, and humans, and cause respiratory and intestinal diseases (1, 2). In the 21st century, three novel human coronaviruses (HCoVs), namely, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome (MERS-CoV), and SARS-CoV-2 (causing an ongoing pandemic), have emerged and spread globally, posing severe threats to public health and the economy (3–6). Four other HCoVs, namely, HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1, cause cold-like symptoms in humans. The current data suggest that all HCoVs originate from wildlife, including bats, rodents, and probably other animals (7). CoVs have led to significant epidemics affecting animal husbandry globally (8). Six CoVs infect pigs, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine respiratory coronavirus (PRCV), swine acute diarrhea syndrome coronavirus (SADS-CoV), porcine hemagglutinating encephalomyelitis virus (PHEV), and porcine deltacoronavirus (PDCoV) (9, 10). Among them, TGEV, PEDV, SADS-CoV, and PDCoV are enteric viruses that cause acute diarrhea, vomiting, and high mortality in pigs. Pigs play an important role as an end host, which threatens animal health and economics, or as an intermediate host that enables virus transmission from natural hosts to humans (11, 12).
SADS-CoV, also named swine enteric alphacoronavirus or porcine enteric alphacoronavirus, is a novel swine enteric CoV belonging to the genus Alphacoronavirus that was identified during fatal swine disease outbreaks in Guangdong province, China, in October 2016 and led to more than 20,000 piglet deaths (13–15). A retrospective study showed that SADS-CoV emerged in Guangdong Province in August 2016 (16). Reemergence of SADS-CoV was reported in Fujian and Guangdong provinces in 2018 and 2019, respectively (17, 18). We previously reported that SADS-CoV is phylogenetically close to bat HKU2-CoV, and SADS-related CoVs (SADSr-CoV) were detected in four species of Rhinolophus bats (R. affinis, R. sinicus, R. rex, and R. pusillus) in several provinces in southern China (15, 19, 20). In addition, our and other groups have reported that SADS-CoV has broad cell tropism and a high replication efficiency, including in primary human cells in vitro, suggesting its potential for interspecies transmission (21–23).
Currently, pathogenicity studies of SADS-CoV in vivo rely on piglet infection, which is complicated by its high cost and requirement of manipulation in a biosafety laboratory, thus limiting further understanding. Studies on therapeutic treatments targeting SADS-CoV and related viruses are needed. In the present study, we experimentally infected suckling BALB/c and C57BL/6J mice and found efficient replication of SADS-CoV in suckling mice. In addition, we demonstrated that advanced maternal immunization partially protects suckling mice from SADS-CoV infection.
RESULTS
Suckling BALB/c and C57BL/6J mice are susceptible to SADS-CoV infection.
To investigate the pathogenicity of SADS-CoV toward mice, wild-type BALB/c and C57BL/6J mice of different ages, including 2-, 5-, and 7-day-old and 3- to 4-week-old mice, were infected with SADS-CoV. First, six 3- to 4-week-old mice in each group were intragastrically and intranasally inoculated with 8 × 105 50% tissue culture infective doses (TCID50) of virus per animal or an equal volume of Dulbecco’s modified Eagle’s medium (DMEM) as a control. The mice were weighed and observed for clinical signs daily for 14 days. None of the mice developed weight loss, diarrhea, or other clinical symptoms, and viral RNA was not detected in the tested tissues, anal swabs, oral swabs, or feces by reverse transcription-quantitative PCR (RT-qPCR) (data not shown).
A similar dose of SADS-CoV was intragastrically inoculated into 2-, 5-, and 7-day-old suckling BALB/c and C57BL/6J mice as described. Some of the suckling mice experienced weight loss, hyperspasmia, and death, and the remaining mice survived to the end point without evident signs of infection. All 2-day-old C57BL/6J mice (n = 7) died by 5 days postinfection (dpi), three out of five 5-day-old mice died by 5 dpi, two out of eight 7-day-old mice died by 7 dpi, and similar mortality was observed in BALB/c mice (Fig. 1B). Gross lesions were observed in some of the virus-infected suckling mice, including a transparent and thin-walled gastrointestinal tract; abdominal distension due to gas accumulation; yellow, watery contents in the intestine; and hemorrhage and edema in the brain. BALB/c mother mice ate infected pups, leading to difficulties in collecting tissue samples; therefore, we used C57BL/6J mice in the follow-up work.
FIG 1.
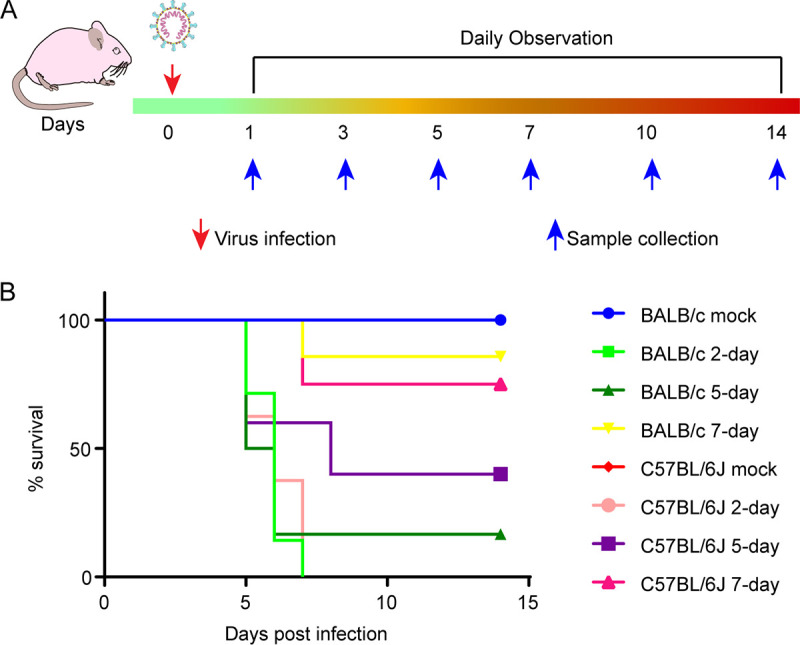
Experimental scheme and mortality of suckling mice after SADS-CoV infection. (A) Sixteen female C57BL/6J mice were randomly divided into four groups and used to produce suckling mice. Two-, 5-, and 7-day-old suckling mice were infected with SADS-CoV, and 2-day-old mock-infected mice were used as the control group. Mice were observed for clinical signs and survival daily for 14 days, and three mice from each group were sacrificed to collect tissue samples at 1, 3, 5, 7, 10, and 14 dpi. (B) Survival rates of 2 (n = 8)-, 5 (n = 5)-, and 7-day-old (n = 8) infected and 2-day-old (n = 6) mock-infected C57BL/6J mice, and 2 (n = 7)-, 5 (n = 6)-, and 7-day-old (n = 7) infected and 2-day-old (n = 6) mock-infected BALB/c mice.
Viral replication in multiple mouse tissues.
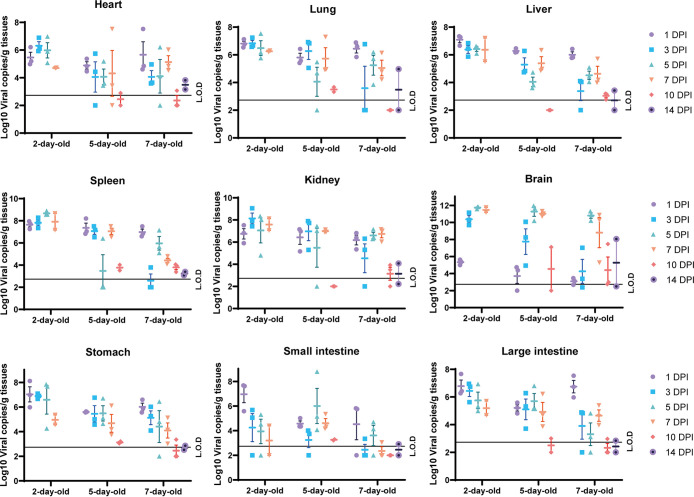
To better understand viral replication in suckling mouse tissues, viral RNA in different tissues from 2-, 5-, and 7-day-old mice euthanized at different time points was quantified by RT-qPCR (Fig. 2). All animals were successfully infected with SADS-CoV as indicated by the detection of viral nucleic acids in collected tissues, including the heart, lungs, liver, spleen, kidneys, stomach, small intestine, large intestine, and/or brain. Two-day-old mice were the most susceptible, followed by 5- and 7-day-old mice. In the 2-day-old group, the viral load peaked in all collected tissues, except the small intestine and brain, at 1 dpi until death. The viral load in the brain in this group increased 3 dpi and peaked 5 dpi. However, in the 5- and 7-day-old groups, we observed intrahost variation in viral replication in all tissues tested.
FIG 2.
Virus quantification in SADS-CoV-infected mice. Viral RNA in collected tissues from infected mice at 1, 3, 5, 7, 10, and 14 dpi was quantified using RT-qPCR. The data represent the means and standard errors.
Histopathological analysis of virus-infected mice.
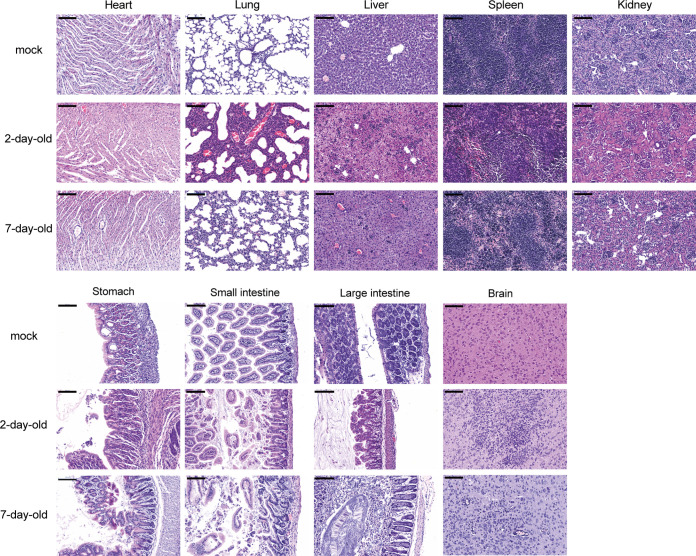
To further characterize the tissue damage in virus-infected suckling mice, all collected tissues of virus- and mock-infected 2- and 7-day-old mice at 5 dpi were histopathologically examined (Fig. 3). Histopathological lesions were more pronounced in 2-day-old mice than in 7-day-old mice. Different levels of pathological damage were observed throughout the course of infection. Some minor changes were observed in the heart and kidney tissues, including minor multifocal lesions, slight myocardial cell edema, and necrosis in the heart and slight edema in the kidneys. Severe damage appeared in the other tissues, including multifocal lesions, pulmonary fibrin exudation, and alveolar wall thickening with monocyte and lymphocyte infiltration in the lungs; edema and increased inflammatory cells in the liver; hemorrhage, white pulp basic structure destruction, and increased numbers of lymphocytes in the red pulp in the spleen; edema, necrosis, and disruption of gastric epithelial cells and intestinal villi in the gastrointestinal tract; and extensive neuronal cell necrosis and an increased number of lymphocytes in the brain.
FIG 3.
Pathological changes in suckling mice after SADS-CoV infection. Three 2- and 7-day-old SADS-CoV-infected and mock-infected mice were euthanized and used to examine the pathological changes in the heart, lungs, liver, spleen, kidneys, stomach, small intestine, large intestine, and brain. Images were acquired using a Pannoramic MIDI system. Scale bars: 100 μm.
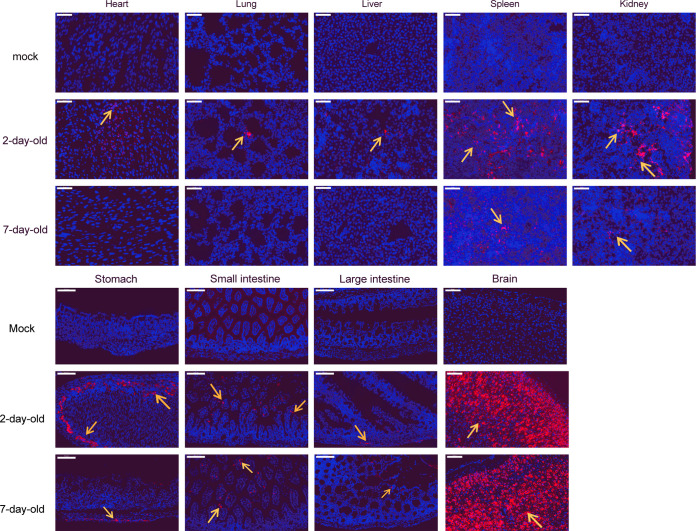
To analyze viral expression in suckling mouse tissues, viral antigens were detected by immunofluorescence assay (IFA) in all collected tissues of virus- and mock-infected 2- and 7-day-old mice at 5 dpi (Fig. 4). Viral antigens were detected in all tissues of 2-day-old infected mice, and in the spleen, kidneys, stomach, small intestine, and brain, but not the heart, lungs, and liver, of 7-day-old infected mice. Consistent with the viral genome detection results, the brain showed the highest viral nucleoprotein expression level in infected mice. Based on morphological observation, pulmonary epithelial cells, gastric basal cells, intestinal villous epithelial cells, and neurons were identified as major target cells, whereas viral-positive cells were not easily detected in the heart, liver, spleen, and kidneys.
FIG 4.
Viral antigen detection in mice after SADS-CoV infection. Viral antigens (orange arrows) in the heart, lungs, liver, spleen, kidneys, stomach, small intestine, large intestine, and brain were detected using a rabbit anti-SADSr-CoV N protein polyclonal antibody (red). Images were acquired using a Pannoramic MIDI system. Scale bars: 50 μm (heart, lungs, liver, spleen, and kidneys) and 100 μm (stomach, small intestine, large intestine, and brain).
Viral replication dynamics in the brain.
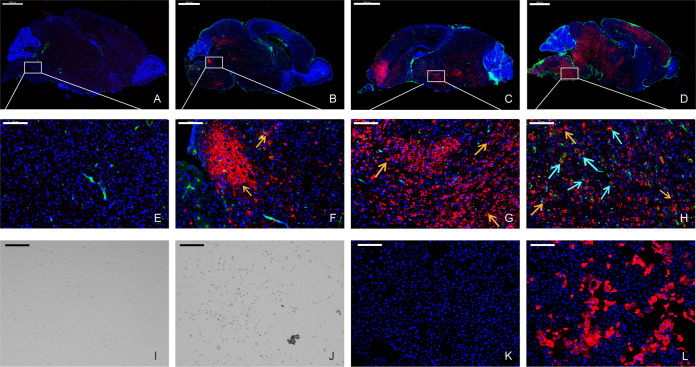
Notably, the brain was the most susceptible to SADS-CoV, and to better understand the dynamics of SADS-CoV infection in the brain, viral antigens were detected in 2-day-old mouse brains using IFA (Fig. 5A to H). Infection was first detected in the neuronal cells of the brain stem and the fourth ventricle near the spine on the ventral side and then spread into the entire brain through the circulation of cerebrospinal fluid. In addition, elevated expression of microglia markers was observed at 7 dpi. When we inoculated Vero cells with the homogenate supernatants of the brains of two infected 2-day-old mice sampled at 5 dpi, we observed a noticeable cytopathic effect (CPE) in the infected cells (Fig. 5J). The virus reisolated from the brain was confirmed by IFA (Fig. 5L) and next-generation sequencing (Table 1). The two brain isolates had three and five mutations, respectively, compared with the original viral genome from the supernatant before mouse infection, including A5498G (ORF1ab, E-G), A5996G (ORF1ab, E-G), and T14207G (ORF1ab, G-G) in one mouse and T357Y (ORF1ab, A-A/A), G14681T (ORF1ab, M-I), C21552T (spike, A-V), A22467R (spike, P-P/L), and A22496R (spike, E-E/K) in the other. These results suggest that SADS-CoV might have been adapted in infected individuals.
FIG 5.
Evaluation of the viral replication dynamics in the brain using IFA. Two-day-old mouse brains were stained for the viral antigen (SADS-CoV NP, red) and microglial cell antigen (CD68, green) at 1 (A and E), 3 (B and F), 5 (C and G), and 7 (D and H) dpi. The viral antigen (orange arrows) has been detected since 3 dpi, and a low level of microglial cell antigen (cyan arrows) was detected at 7 dpi. Images were acquired using a Pannoramic MIDI system. Infected mouse brain tissues were homogenized in DMEM and used to infect Vero cells. Compared to mock-infected cells (I and K), a cytopathic effect (CPE) (J) was observed and viral antigens (L) were detected in infected cells. Images were acquired using an Olympus FV1200 confocal microscope. Scale bars: 1000 μm (A to D), 100 μm (E to H), and 200 μm (I to L).
TABLE 1.
Genome comparison of the reisolated virus with the original viral genome from supernatanta
| Sample | Genome (start, end) | Gene mutation (nt, aa) |
|---|---|---|
| Reference | 1–27,163 | |
| Brain 1 | 21–27,163 | ORF1ab, A5498G, E-G ORF1ab, A5996G, E-G ORF1ab, T14207G, G-G ORF1ab, T357Y, A-A/A ORF1ab, G14681T, M-I |
| Brain 2 | 21–27,163 | Spike, C21552T, A-V Spike, A22467R, P-P/L Spike, A22496R, E-E/K |
nt, Nucleotide; aa, amino acid.
SADS-CoV infection induces inflammatory responses in the brain.
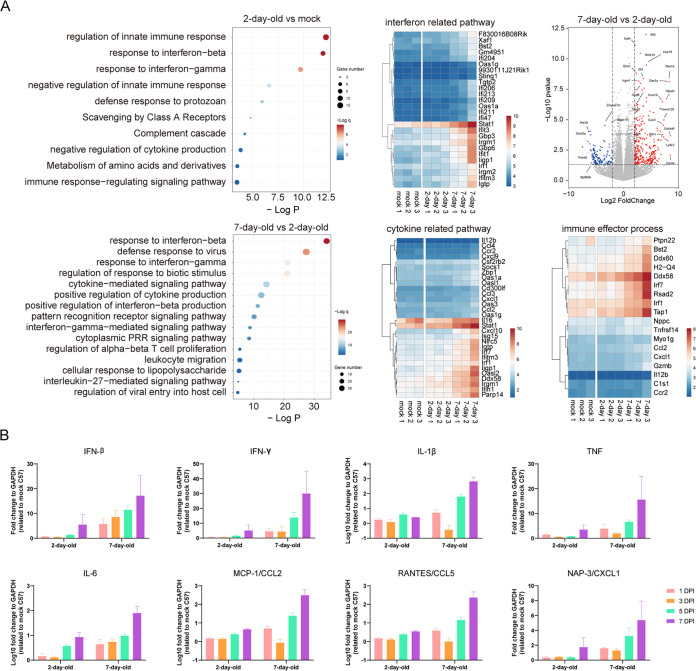
The rapid onset of morbidity and mortality in suckling mice in response to SADS-CoV challenge prompted us to explore the host cytokine/chemokine responses in the brains of the infected mice. First, we performed RNA-sequencing (RNA-seq) for pathway enrichment analysis using total RNA extracted from the brains of infected 2- and 7-day-old mice at 5 dpi. As expected, higher immune responses were observed in the brain tissues of 7-day-old mice than in those of 2-day-old mice (Fig. 6A).
FIG 6.
Immune responses in the brains of suckling mice. (A) Total RNA was extracted from the brains of 2- and 7-day-old virus- and mock-infected mice at 5 dpi and subjected to direct RNA-seq and pathway enrichment analysis. (B) Transcripts of genes encoding antiviral and proinflammatory cytokines and chemokines were detected by RT-qPCR. Relative transcript levels of were calculated using the comparative threshold cycle (ΔΔCT) method. The experiment was conducted in duplicate, using three mice. The data represent the means and standard errors.
Next, we determined the transcriptional activation of genes, including classic antiviral cytokines, proinflammatory cytokines, and chemokines in the brain tissues of infected 2- and 7-day-old mice using RT-qPCR. Interferon (IFN) and inflammatory cytokine/chemokine transcript production in the brain increased rapidly after infection in 7-day-old mice, whereas 2-day-old infected mice showed a weaker and later response. Proinflammatory cytokines (interleukin-1β [IL-1β] and interleukin-6 [IL-6]) and chemokines (MCP-1 and RANTES) were significantly induced in SADS-CoV-infected mice (Fig. 6B).
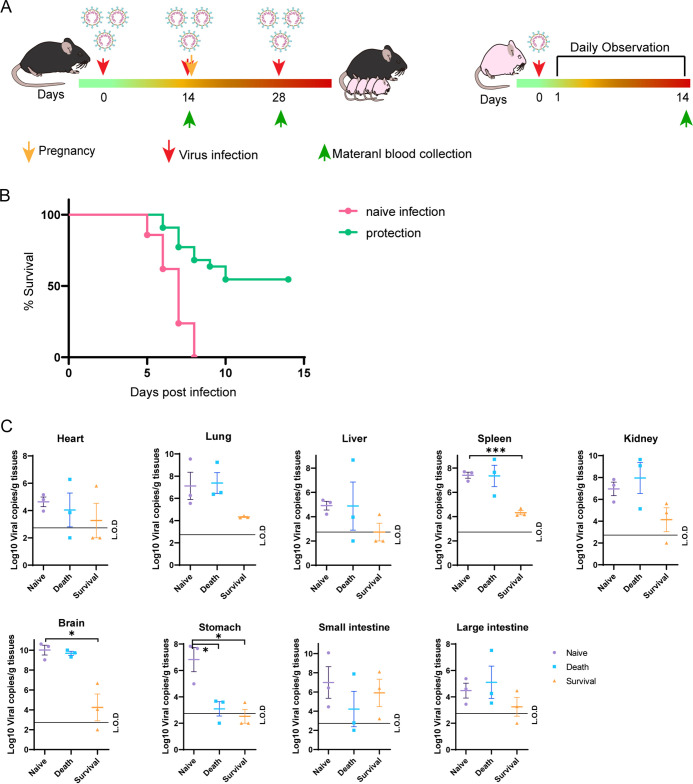
Maternal immunization through SADS-CoV infection partially protects suckling mice.
To investigate whether preimmunization of mother mice would protect suckling mice from viral infection, we infected female mice with 1.5 × 106 TCID50 SADS-CoV virus three times every 14 days, using DMEM as the control. After the second infection, the mice were mated, and their 2-day-old suckling mice were infected with 8 × 105 TCID50 SADS-CoV (Fig. 7A). In the group in which the mother mice were infected with the virus, 12 out of 23 suckling mice survived, whereas all animals in the control group died (Fig. 7B). Tissue viral loads were low, especially in neuronal tissues, in the surviving suckling mice in the protected group 14 dpi compared to those in the naive infected group or those mice that died in the protected group (Fig. 7C). No obvious differences in viral loads were observed between dead mice in the naive infected and protected groups. Twenty-one days after the third infection, the mother mice in the protected group generated antibodies that could neutralize 100 TCID50 SADS-CoV at dilutions of 1:20 to 1:80 (Table 2). We observed a correlation between the survival rate in the suckling mice and the neutralization antibody titers in the mother mice.
FIG 7.
Experimental scheme and mortality and virus quantification in suckling mice after SADS-CoV infection. (A) Female mice were intragastric inoculated with 1.5 × 106 TCID50 SADS-CoV or an equal volume of DMEM (mock control) three times (left) and mated. Two-day-old suckling mice were intragastrically inoculated with 8 × 105 TCID50 SADS-CoV and were observed for clinical signs daily for 14 days (right). Survival rate (B) and viral loads (C) in the heart, lungs, liver, spleen, kidneys, stomach, small intestine, large intestine, and brain, collected from 2-day-old suckling mice at the end point of the trial. *, P < 0.05; ***, P < 0.0001.
TABLE 2.
Neutralization antibody titer of female mouse seruma
| Group | Mice no. | Neutralization antibody titers (21 days after the 3rd infection) | Offspring survival rate |
|---|---|---|---|
| Naive infection | 4/4 | ND | 21/21 (0%) |
| Protection | 1 | 1:20 | 1/4 (25%) |
| 2 | 1:40 | 4/7 (57.1%) | |
| 3 | 1:20 | 2/5 (40%) | |
| 4 | 1:80 | 5/7 (71.5%) |
ND, not detected.
DISCUSSION
Many coronaviruses infect humans and domestic animals, posing a threat to public health and causing economic losses in the livestock industry (24, 25). PEDV, PDCoV, and SADS-CoV, which have emerged/reemerged in China and other countries, cause acute gastroenteritis in neonatal piglets (10, 26). Studies have shown that newborn piglets younger than 5 days infected with SADS-CoV develop typical clinical signs, such as rapid weight loss, watery diarrhea, acute vomiting, and even death, whereas infected sows suffer only mild diarrhea and most of them recover within 2 days (14, 15, 27, 28). Experimental infection in adult mice has demonstrated that SADS-CoV has a low level of replication in wild-type C57 and immunodeficient interferon receptor-knockout mice, without clinical manifestations and pathological damage in tissues (21, 22). In this study, we demonstrated that wild-type BALB/c and C57BL/6J suckling mice are highly susceptible to SADS-CoV infection through intragastric inoculation and exhibit severe morbidity and mortality. Suckling mice developed severe clinical signs such as weight loss, hyperspasmia, and death. Mortality was as high as 100% in 2-day-old mice and 25% in 7-day-old mice, which is consistent with findings in SADS-CoV-infected piglets younger than 5 days (12, 22). These results suggest that SADS-CoV is highly pathogenic to young animals.
Viral RNA was detected in all tissues collected from infected suckling mice and with different levels of damage, which indicates the wide tissue tropism of SADS-CoV and is consistent with the results of previous studies in vitro (21, 23). However, the key difference between murine and pig SADS-CoV infection is the high neuronal susceptibility and neurotropism in mice versus the intestinal infection in pigs. This illustrates that tissue tropism and clinical symptoms upon SADS-CoV infection vary among different hosts. Similarly, bovine coronavirus and feline infectious peritonitis coronavirus, which normally show intestinal tropism, showed brain invasion in infected suckling mice (29, 30). In our study, rather than injecting the virus directly into the brain as done in these previous studies, we administered the virus via intragastric inoculation, but we still observed efficient viral replication in the brain of tested suckling mice. Although this neonatal mouse model does not completely simulate the tissue tropism observed in SADS-CoV-infected piglets, it provides a preliminary cost-effective model to investigate the pathogenesis of SADS- and SADSr-CoVs and subsequently to screening for relevant antiviral drugs.
Strong cytokine and chemokine responses are associated with disease severity (31–33). To elucidate why SADS-CoV severity decreases with age, we performed RNA-seq for pathway enrichment analysis and detected the immune response in brain tissues that revealed elevated mRNA expression of early interferons, proinflammatory cytokines, and chemokines. These results indicated that the immature immune system in younger mice is responsible for their increased susceptibility to SADS-CoV. A recent study reported that placenta associated 8 (PLAC8) is highly expressed in piglets and it may account for the higher susceptibility of younger animals to SADS-CoV (34). However, according to the RNA-seq data, PLAC8 is barely expressed in the newborn mouse brain, which implies that PLAC8 is not involved in viral replication in suckling mice. A future assessment of host responses is needed to better understand the pathogenesis of this virus.
Reemergence of SADS-CoV in the Fujian and Guangdong provinces was reported in 2018 and 2019, respectively (18, 26). In a porcine diarrhea outbreak in 11 Chinese provinces, an 81.7% seroprevalence of SADS-CoV antibodies was detected by an enzyme-linked immunosorbent assay targeting its spike protein (35), suggesting that SADS-CoV has a substantially wider spread than expected. These results indicate that effective countermeasures should be in place to prevent the future spread of SADS-CoV. In our study, we mimicked a vaccine schedule and found that female mice infected with SADS-CoV generated 1:20 to 1:80 neutralizing antibody titers that partially protected their offspring from SADS-CoV infection. It is unclear whether the antibodies are present in the mother milk or in the suckling mice at birth. As the suckling mice were breastfed since birth, the mother’s milk likely plays a role in the protection against viral infection. These results show the possibility of vaccine development in sows to protect against SADS-CoV infection in piglets.
Bats play an important role in the emergence of highly lethal zoonotic viruses, such as Hendra, Nipah, Ebola, and Marburg viruses (36, 37). Moreover, they are the probable natural origin of many emerging CoVs, such as SARS-CoV, MERS-CoV, and SARS-CoV-2, which infect humans through intermediate hosts (4, 38–41). Our previous study suggested that SADS-CoV likely originated from bats, as closely related CoVs were detected in Rhinolophus bats in the same location as the SADS outbreak (15). Furthermore, SADS-CoV can replicate well in cells derived from humans and different animal species, as well as in primary human lung and intestinal cells, suggesting that this virus has a wide host range and a high potential for interspecies transmission (21–23). In addition, genetically diverse SADSr-CoV has been widely detected in at least four species of Rhinolophus bats (R. affinis, R. sinicus, R. rex, and R. pusillus) in the southern provinces of China (19, 20), highlighting the need for surveillance of this group of viruses in these regions. Further surveillance of bats, pigs, and other animal populations and the development of efficacious antiviral treatments or vaccines are needed to protect animals against potential SADS-CoV infection.
MATERIALS AND METHODS
Ethics statement.
Mouse infections were performed in an animal biosafety level 2 (ABSL2) facility in accordance with recommendations for the care and use of laboratory animals and the Institutional Review Board of the Wuhan Institute of Virology, Chinese Academy of Sciences (ethics no. WIVA05201905). All of the experiments complied with all relevant ethical regulations.
Cells lines and virus.
Vero cells lines (ATCC CRL-81) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% Anti-Anti (Invitrogen). The SADS-CoV virus was propagated and titrated in Vero cell lines with maintenance medium (MM) containing DMEM, 2% tryptose phosphate broth (TPB; Sigma), 5 μg/ml trypsin (Gibco), and 1% Anti-Anti (Invitrogen). Viral titrations were performed with 10-fold serial dilutions in Vero cells. The 50% tissue culture infective dose (TCID50) was expressed as the reciprocal of the highest dilution showing CPE by the Reed-Muench method.
Mouse infection.
Wild-type pregnant BALB/c and C57BL/6J mice were purchased from Hunan SJA Laboratory Animal Co., Ltd. and housed in an ABSL2 facility at the Wuhan Institute of Virology, Chinese Academy of Sciences, under specific pathogen-free conditions. The infection experiments included two independent sections, a survival monitoring experiment and a pathology progression experiment. For the survival monitoring experiment, 2-day-, 5-day-, 7-day-, and 3- to 4-week-old BALB/c and C57BL/6J mice were anesthetized with isoflurane and intragastric inoculated with 8 × 105 TCID50 SADS-CoV or an equal volume of DMEM (mock-infection control), and mice were observed for clinical signs daily for 14 days. For the pathology progression experiment, three C57BL/6J mice were dissected at 1, 3, 5, 7, 10, and 14 dpi, and tissues were harvested, including the heart, liver, spleen, lung, kidney, brain, stomach, small intestine, and large intestine.
For the protection test, the female C57BL/6J mice were infected with the virus three separate times every 14 days with 1.5 × 106 TCID50 SADS-CoV or an equal volume of DMEM (mock-infection control). After the second infection, they were mated and their 2-day-old suckling mice were infected with 8 × 105 TCID50 SADS-CoV, clinical signs were monitored daily for 14 days, and tissues were harvested at the end point.
RNA extraction and reverse-transcription quantitative PCR.
Mouse tissues were homogenated in RNALater, then centrifuged at 8,000 rpm for 10 min at 4°C. Viral RNA was extracted from tissues homogenate supernatant with the QIAamp 96 Virus QIAcube HT kit (Qiagen), RNA was used as a template for RT-qPCR for assessing SADS-CoV-specific genome targeting RdRp gene using HiSxript II One step RT-qPCR SYBR Green kit (Vazyme) as we previously described (15). Average values from duplicates of each gene were used to calculate the viral genomic copies. The 10-μl qPCR mix contained 5 μl 2×One Step SYBR green mix, 0.5 μl One Step SYBR green Enzyme mix, 0.2 μl (10 μM) of each primer, and 2 μl RNA. Amplification was performed as follows: 50°C for 3 min, 95°C for 30 s, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s, and a default melting curve step in a Bio-Rad Real-time PCR machine.
Histopathological analysis.
The tissue samples were fixed with 4% paraformaldehyde, paraffin embedded, and cut into 3.5-μm sections. Fixed tissue samples were used for hematoxylin-eosin (H&E) staining and indirect immunofluorescence (IFA) staining for the detection of the SADS-CoV antigen. For routine histology, tissue sections were stained with H&E. For IFA, slides were deparaffinized and rehydrated, followed by 15-min heat-induced antigen retrieval with EDTA pH 8.0 in a microwave oven. The slides were treated with PBS/0.025% Triton X-100 for 15 min for cell membrane perforation and then blocked with 5% BSA at room temperature for 1 h, followed by overnight incubation at 4°C with rabbit anti-SADS-CoV N protein polyclonal antibody (1:400; made in-house) and/or anti-CD68 antibody (1:100; Abcam; ab955), and then washed in PBS. Then, the slides were incubated with Cy3-conjugated goat-anti-rabbit IgG (1:150; Abcam; ab6939) and/or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:100; Abcam; ab150117). After being washed in PBS, slides were stained with DAPI (1:100; Beyotime). The image information was collected using a Pannoramic MIDI system (3DHISTECH, Budapest).
Virus isolation and sequencing.
Infected brain tissues from euthanized mice were homogenized with DMEM, and the supernatant was used for virus isolation. A total of 200 μl supernatant was inoculated onto monolayer Vero cells in a 24-well plate. After 1 h of inoculation, the supernatant was removed and replaced with fresh DMEM containing 5 μg/ml trypsin plus 1% Anti-Anti. CPE was monitored daily, and viral antigens were detected using IFA. Images were collected by EVOS M5000 (Thermo). The supernatants were subjected to viral RNA extraction for the next-generation sequencing (NGS). The NGS was performed by using Illumina MiSeq 3000 sequencers, and the sequences were analyzed by Geneious 10.2.6 for Windows.
Transcriptome analysis.
RNA was extracted from 2- and 7-day-old SADS-CoV infection and mock-infection brain tissues at 5 dpi (3 replicates each) and subjected for RNA-seq analysis. After mapping clean reads to GRCh38.p13 with HISAT2 v2.1.0 and format conversion with samtools v1.10-24, we used stringtie v2.1.0 to assemble and quantitate transcripts. Reads counts table of transcriptome generated by prepDE.py, a tool in stringtie, was used for gene differential expression analysis in R v4.1.0 with package DESeq2 v1.32.0. The gene with log2 fold change >2 and P value of <0.05 was selected to perform enrichment analysis using online tools Metascape.
Transcriptional profiling of cytokine and chemokine responses by RT-qPCR assays.
RNA was extracted from 2- and 7-day-old SADS-CoV infection and mock-infection spleen, small intestine, and brain tissues (3 replicates each) and subjected to RT-qPCR analyses of transcriptional expression of selected genes, including interferon-β, IL-1β, IFN-γ, IL-6, macrophage inflammatory protein 1-α (MIP1-α)/CCL3, IP-10/CXCL10, tumor necrosis factor (TNF), RANTES/CCL5, monocyte chemoattractant protein-1 (MCP-1)/CCL2, NAP-3/CXCL1, and MIP1-β/CCL4, using endogenous mouse glyceraldehyde-3-phosphate dehydrogenase (GADPH) gene as internal controls (primers provided upon request). The PCR systems and procedures were used as above. The relative abundance of transcripts of each gene was calculated according to the comparative ΔΔCT method.
Viral antibody testing and neutralization assay.
All serum samples were heat-inactivated by incubation at 56°C for 30 min before use. Mouse plasma (50 μl) was serially 2-fold diluted from 1:5 to 1:640, and then an equal volume of virus solution with 100 TCID50 virus was added and incubated at 37°C for 1 h in a 5% CO2 incubator. Diluted negative sera were mixed with an equal volume of virus solution and were used as a negative control. After incubation, 50-μl mixtures were inoculated onto monolayer Vero cells in a 96-well plate for 1 h. After the supernatant was removed, the plate was washed twice with DMEM, cells were incubated with DMEM supplemented with 2 μg/ml trypsin, 2% TPB, and 2% Anti-Anti for 5 days. Each serum concentration was duplicated. Cells were observed for CPE, and neutralizing titers were recorded as the final serum dilution that could neutralize all inoculated wells.
Statistical analysis.
Statistical analyses were performed using PRISM 8.0.2 for Windows (GraphPad). For contrasting two experimental groups, two-tailed Student’s t tests were performed to determine significant differences. Statistical significance was as follows: *, P < 0.05; ***, P < 0.001.
ACKNOWLEDGMENTS
This work was supported by the China Natural Science Foundation of China (31830096).
We thank Jing-Yun Ma (College of Animal Science, South China Agricultural University) for providing SADS-CoV isolate CN/GDWT/2017, Xue-Fang An and Fan Zhang from the Center for Animal Experiment of the Wuhan Institute of Virology for the animal facility support, and Juan Min from Center for Instrumental Analysis and Metrology of the Wuhan Institute of Virology for technical support.
Conceptualization, Z.-L.S.; Methodology, Y.C., R.-D.J., X.-L.Y., and P.Z.; Investigation, Y.C., Q.W., Y.L., M.-Q.L., Y.Z., X.L., and Y.-T.H.; Resources, Writing-Original Draft, Y.C.; Writing-Review & Editing, Z.-L.S.; Funding Acquisition, Z.-L.S.
We declare no conflict of interest.
Contributor Information
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity.
REFERENCES
- 1.Chan JF, To KK, Tse H, Jin DY, Yuen KY. 2013. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol 21:544–555. 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PC, Lau SK, Huang Y, Yuen KY. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood) 234:1117–1127. 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 3.Peiris JSM, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat Med 10:S88–S97. 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Corman VM, Muth D, Niemeyer D, Drosten C. 2018. Hosts and sources of endemic human coronaviruses. Adv Virus Res 100:163–188. 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson LJ, Tong S. 2010. Update on SARS research and other possibly zoonotic coronaviruses. Int J Antimicrob Agents 36(Suppl 1):S21–S25. 10.1016/j.ijantimicag.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y-L, Yu J-Q, Huang Y-W. 2020. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): an update three years after its discovery. Virus Res 285:198024. 10.1016/j.virusres.2020.198024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Vlasova AN, Kenney SP, Saif LJ. 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol 34:39–49. 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepin KM, Lass S, Pulliam JR, Read AF, Lloyd-Smith JO. 2010. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol 8:802–813. 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao D, Liu R, Zhang X, Li F, Wang J, Zhang J, Liu X, Wang L, Zhang J, Wu X, Guan Y, Chen W, Wang X, He X, Bu Z. 2019. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg Microbes Infect 8:438–447. 10.1080/22221751.2019.1590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis 23:1607–1609. 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. 2017. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol 211:15–21. 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Sun Y, Lan T, Wu R, Chen J, Wu Z, Xie Q, Zhang X, Ma J. 2019. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis 66:687–695. 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K, Li H, Bi Z, Gu J, Gong W, Luo S, Zhang F, Song D, Ye Y, Tang Y. 2018. Complete genome sequence of a novel swine acute diarrhea syndrome coronavirus, CH/FJWT/2018, isolated in Fujian, China, in 2018. Microbiol Resour Announc 7:e01259-18. 10.1128/MRA.01259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Li QN, Su JN, Chen GH, Wu ZX, Luo Y, Wu RT, Sun Y, Lan T, Ma JY. 2019. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis 66:2180–2183. 10.1111/tbed.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, Zhao K, Shi ZL, Zhou P. 2019. Bat coronaviruses in China. Viruses 11:210. 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, Chmura AA, Field HE, Zambrana-Torrelio C, Epstein JH, Li B, Zhang W, Wang LF, Shi ZL, Daszak P. 2020. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun 11:4235. 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. 2019. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol 93:e01448-19. 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KH, Sims AC, Swanstrom J, Gully K, Scobey TD, Cooley MR, Currie CG, Randell SH, Baric RS. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci USA 117:26915–26925. 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Y, Chen Y, Geng R, Li B, Chen J, Zhao K, Zheng XS, Zhang W, Zhou P, Yang XL, Shi ZL. 2021. Broad cell tropism of SADS-CoV in vitro implies its potential cross-species infection risk. Virol Sin 36:559–563. 10.1007/s12250-020-00321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson CB, Beaudette FR. 1932. Infection of the cloaca with the virus of infectious bronchitis. Science 76:34. 10.1126/science.76.1958.34-b. [DOI] [PubMed] [Google Scholar]
- 25.Islam A, Ferdous J, Islam S, Sayeed MA, Dutta Choudhury S, Saha O, Hassan MM, Shirin T. 2021. Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, domestic animals, and wildlife. Viruses 13:1908. 10.3390/v13101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Su S, Bi Y, Wong G, Gao GF. 2018. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol 26:466–470. 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Zhang Y, Gong L, Huang L, Lin Y, Qin J, Du Y, Zhou Q, Xue C, Cao Y. 2019. Isolation and characterization of a highly pathogenic strain of Porcine enteric alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound Emerg Dis 66:119–130. 10.1111/tbed.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Cheng J, Luo Y, Yan XL, Wu ZX, He LL, Tan YR, Zhou ZH, Li QN, Zhou L, Wu RT, Lan T, Ma JY. 2019. Attenuation of a virulent swine acute diarrhea syndrome coronavirus strain via cell culture passage. Virology 538:61–70. 10.1016/j.virol.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horzinek MC, Osterhaus AD, Wirahadiredja RM, de Kreek P. 1978. Feline infectious peritonitis (FIP) virus. III. Studies on the multiplication of FIP virus in the suckling mouse. Zentralbl Veterinarmed B 25:806–815. 10.1111/j.1439-0450.1978.tb01056.x. [DOI] [PubMed] [Google Scholar]
- 30.Akashi H, Inaba Y, Miura Y, Sato K, Tokuhisa S, Asagi M, Hayashi Y. 1981. Propagation of the Kakegawa strain of bovine coronavirus in suckling mice, rats and hamsters. Arch Virol 67:367–370. 10.1007/BF01314841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type i interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunders MJ, Altfeld M. 2020. Implications of sex differences in immunity for SARS-CoV-2 pathogenesis and design of therapeutic interventions. Immunity 53:487–495. 10.1016/j.immuni.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin CF, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:971–977.e3. 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tse LV, Meganck RM, Araba KC, Yount BL, Shaffer KM, Hou YJ, Munt JE, Adams LE, Wykoff JA, Morowitz JM, Dong S, Magness ST, Marzluff WF, Gonzalez LM, Ehre C, Baric RS. 2022. Genomewide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection. Proc Natl Acad Sci USA 119:e2118126119. 10.1073/pnas.2118126119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng P, Gao Y, Zhou Q, Jiang T, Zheng S, Huang M, Xue C, Cao Y, Xu Z. 2021. Development of an indirect ELISA for detecting swine acute diarrhoea syndrome coronavirus IgG antibodies based on a recombinant spike protein. Transbound Emerg Dis 69:2065–2075. 10.1111/tbed.14196. [DOI] [PubMed] [Google Scholar]
- 36.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545. 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irving AT, Ahn M, Goh G, Anderson DE, Wang LF. 2021. Lessons from the host defences of bats, a unique viral reservoir. Nature 589:363–370. 10.1038/s41586-020-03128-0. [DOI] [PubMed] [Google Scholar]
- 38.Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortazar-Schmidt C, Drosten C, Koopmans MP. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13:859–866. 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drosten C, Kellam P, Memish ZA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 371:1359–1360. 10.1056/NEJMc1409847. [DOI] [PubMed] [Google Scholar]
- 41.Tu C, Crameri G, Kong X, Chen J, Sun Y, Yu M, Xiang H, Xia X, Liu S, Ren T, Yu Y, Eaton BT, Xuan H, Wang LF. 2004. Antibodies to SARS coronavirus in civets. Emerg Infect Dis 10:2244–2248. 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]