Abstract
Purpose
Benefit from convalescent plasma therapy for coronavirus disease 2019 (COVID-19) has been inconsistent in randomized clinical trials (RCTs) involving critically ill patients. As COVID-19 patients are immunologically heterogeneous, we hypothesized that immunologically similar COVID-19 subphenotypes may differ in their treatment responses to convalescent plasma and explain inconsistent findings between RCTs .
Methods
We tested this hypothesis in a substudy involving 1239 patients, by measuring 26 biomarkers (cytokines, chemokines, endothelial biomarkers) within the randomized, embedded, multifactorial, adaptive platform trial for community-acquired pneumonia (REMAP-CAP) that assigned 2097 critically ill COVID-19 patients to either high-titer convalescent plasma or usual care. Primary outcome was organ support free days at 21 days (OSFD-21) .
Results
Unsupervised analyses identified three subphenotypes/endotypes. In contrast to the more homogeneous subphenotype-2 (N = 128 patients, 10.3%; with elevated type i and type ii effector immune responses) and subphenotype-3 (N = 241, 19.5%; with exaggerated inflammation), the subphenotype-1 had variable biomarker patterns (N = 870 patients, 70.2%). Subphenotypes-2, and -3 had worse outcomes, and subphenotype-1 had better outcomes with convalescent plasma therapy compared with usual care (median (IQR). OSFD-21 in convalescent plasma vs usual care was 0 (− 1, 21) vs 10 (− 1, to 21) in subphenotype-2; 1.5 (− 1, 21) vs 12 (− 1, to 21) in suphenotype-3, and 0 (− 1, 21) vs 0 (− 1, to 21) in subphenotype-1 (test for between-subphenotype differences in treatment effects p = 0.008).
Conclusions
We reported three COVID-19 subphenotypes, among critically ill adults, with differential treatment effects to ABO-compatible convalescent plasma therapy. Differences in subphenotype prevalence between RCT populations probably explain inconsistent results with COVID-19 immunotherapies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00134-022-06869-w.
Keywords: Precision medicine, Subphenotypes, Convalescent plasma
Take-home message
| We report three COVID-19 subphenotypes with differences in treatment response to ABO-compatible high-titer convalescent plasma therapy among critically ill adults, participating in a large international multi centre randomized clinical trial. Our findings support the hypothesis that immunotherapies in critically ill adults with COVID-19 could be enhanced with patient selection based on host immune response characteristics. |
Introduction
The coronavirus SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) causes COVID-19 (coronavirus disease 2019), an acute illness affecting pulmonary and extrapulmonary organs [1]. COVID-19 patients requiring hospitalization (moderate-to-severe disease) have significant viral load in the respiratory tract [2] and/or detectable viral ribonucleic acid (RNA) in blood [3]. Therefore, in moderate-to-severe COVID-19, antiviral therapies (either passive immunotherapy or antiviral medications) are considered potential treatment options [4]. The benefit of passive immunotherapy with convalescent plasma (blood product containing SARS-CoV-2–specific polyclonal antibodies) reported in cohort studies have not been observed in randomised clinical trials (RCTs), with guidelines recommending against the use of convalescent plasma outside of RCTs [5].
It is well recognized that in hospitalized patients, COVID-19 is an immunologically heterogenous illness [6–13]. It is also recognized that immunological heterogeneity in COVID-19 patients is observable at protein level, i.e., differences in cytokine, chemokines, and other biomarker profiles [8, 13]. Abnormal immune responses persist throughout critical illness in COVID-19 patients [13]. Thus, we hypothesized that differences in immune responses will manifest as subphenotypes and may be associated with subphenotype differences in treatment effect to convalescent plasma therapy (also known as heterogeneity of treatment effect (HTE) [14]) on OSFD-21 (primary outcome of pandemic appendix to REMAP-CAP) [15] and on hospital mortality (outcome of interest).
We tested these hypotheses in a biological sampling substudy conducted in the United Kingdom (UK) within the immunoglobulin domain of the REMAP-CAP, which randomized 2097 patients with severe COVID-19 into two units of high-titer convalescent plasma or usual care, and found no overall benefit with convalescent plasma therapy in critically ill COVID-19 patients [15]. Informed by previous work on protein biomarkers [6–13, 16], we explored whether differences in treatment effect (HTE [14] on OSFD-21 and mortality outcomes) was detectable between COVID-19 subphenotypes that were identified based on unsupervised analyses of changes in the CXC family of chemokines (CXCL1, CXCL5, CXCL8, CXCL9, CXCL10, CXCL11), the CC family of chemokines (CCL3, CCL4, CCL11, CCL17, CCL20), transforming growth factor-beta 1(TGF-β1), vascular endothelial growth factor (VEGF), interleukins (IL-6, IL-2, IL-4, IL-5, IL-10), interferons (IFN-α2, IFN-β, IFN-λ1, IFN-y), granulocyte monocyte colony stimulating factor (GM-CSF), soluble tumor necrosis factor receptor-1 (sTNF-R1), angiopoietin-2, intercellular adhesion molecule-1 (ICAM-1), A proliferation-inducing ligand (APRIL, TNFSF13) and B cell-activating factor (BAFF, TNFSF13B) [17]. We selected these biomarkers a priori, i.e., before our primary RCT results were available.
Methods
Study design
Briefly, REMAP-CAP is an international, multicentre, open-label adaptive platform designed to determine the best treatment strategies for patients with severe pneumonia in both settings during the pandemic and outside the pandemic [18]. This trial’s design, eligibility criteria, and results regarding glucocorticoids [19], anticoagulants [20, 21], antivirals [22], interleukin-6 (IL-6) receptor antagonists [23], antiplatelet therapy, and immunoglobulin domain convalescent plasma [15] for treatment of COVID-19 have been reported previously.
Study population
Our study population consisted of critically ill adult patients (> 18 years old) with microbiologically confirmed COVID-19, randomized to receive 2 units of high-titer, ABO-compatible convalescent plasma (total volume approximately 550 mL ± 150 mL) within 48 h of randomization or no convalescent plasma, between March 9, 2020 and January 18, 2021 [15]. These patients had a baseline blood sample collected after consent and before administration of the allocated intervention (convalescent plasma vs no convalescent plasma (usual care)).
Biomarker measurements
Serum was separated from whole blood by centrifugation (1300 g for 10 min at room temperature) and stored in 200 µl aliquots at − 80 °C until analyses. Two custom 14-plex Legendplex™ (BioLegend) bead-based multiplex assays were used to measure a priori selected biomarkers described in “Introduction”, as per manufacturer’s instructions (eMethods-1). SARS-CoV-2 immunoglobulin G (IgG) antibody against spike was measured using enzyme-linked immunosorbent assay (ELISA), as reported previously [2, 24]. Viral loads and strains in the respiratory tract were measured as described previously [2] (eMethods-2).
Data analyses
We described the study cohort characteristics (overall and by randomized allocation status) in terms of age, sex, body mass index (BMI), pre-existing chronic health conditions defined using the Acute Physiology and Chronic Health Evaluation II (APACHE II) score, SARS-CoV-2 antibody status, viral loads in respiratory tract, respiratory support status, concomitant COVID-19 therapy use, and allocation status.
Before biomarker analyses, two proteins (IL-2 and TGF-β1) with more than 20% missing data were excluded from the dataset (eFigure-1a, b). We then used Gibbs sampler-based left-censored missing value imputation approach (GSIMP) [25], which considers the lower limit of detection calculated in the LEGENDplex™ data analysis software suite. We checked data for batch effects and did not observe any batch effects (eFigure-1c, d). Thus, the final analytic dataset consisted of 26 biomarkers. The following analyses were performed in R statistical environment [26]. First, we assessed biomarker differences by SARS-CoV-2 antibody status and by hospital mortality, as differences in immune responses may be associated with antibody status and clinical outcomes. Second, we assessed biomarker differences by allocation status (convalescent plasma vs usual care), to check for baseline biomarker imbalances by randomization status, as any imbalances will need to be accounted for in the subsequent unsupervised analyses. Third, we used the agglomerative hierarchical clustering method with WARD2 linkage function on log10 transformed data. [27]. Additional details are reported in online supplement (eMethods-3).
Finally, we explored the associations between sub-populations, allocation status and outcomes, with regression models incorporating robust standard errors using Stata 15.1 [28]. We report the association between the outcome of OSFD-21 and subphenotypes and by allocation to convalescent plasma using ordered logistic regression models to relate our substudy results to the primary outcome in our original publication [15]. OSFD-21 is an ordinal scale outcome, where mortality is given a score of −1 and among survivors OSFD is calculated up to day 21, such that a higher number represents faster recovery [15]. We also report the associations between hospital mortality and SARS-CoV-2 antibody status in the overall cohort, by subphenotypes and by allocation to convalescent plasma using logistic regression models. We report unadjusted analyses, as testing associations between subphenotypes and treatment effects of convalescent plasma is equivalent to performing a subgroup analyses with moderate-sized RCT data, where the additional value of baseline prognostic covariates adjustment needs careful consideration, due to risk of alpha error [29]. After the regression models, we used post-estimation commands and test of heterogeneity for differences in treatment effects.
Results
Study cohort and clinical characteristics
Amongst the 2097 participants randomized to a COVID-19 immunoglobulin domain within the REMAP-CAP, 1023 were assigned to convalescent plasma and 868 to usual care in the UK [15]. Our report is based on data from 1239 participants (737/1023 (72%) assigned to convalescent plasma and 502/868 (57.8%) assigned to usual care) from the UK, who had baseline blood samples after consent but before intervention (Table 1). Clinical characteristics of our substudy cohort were similar to the overall trial population [15]. Importantly, clinical characteristics were similar between patients assigned to convalescent plasma and usual care, enabling us to analyse this UK sub-population as a cohort (eTable-1).
Table 1.
Clinical characteristics of subphenotypes
| Characteristic | Overall (n = 1,239) | Phenotype-1 (n = 870) | Phenotype-2 (n = 128; 10.3%) | Phenotype-3 (n = 241; 19.5%) |
|---|---|---|---|---|
| Allocation n (%) | ||||
|
Convalescent plasma Usual care |
737 (59.5%) 502 (40.5%) |
534 (61.4%) 336 (38.6%) |
67 (52.3%) 61 (47.7%) |
136 (56.4%) 105 (43.6%) |
| Age, median (IQR) y | 61 (52, 70) | 62 (53, 70) | 58 (48, 65.5) | 61 (52, 70) |
| Female n (%) | 408 (32.9%) | 286 (32.9%) | 39 (30.5%) | 83 (34.4%) |
| BMI (kg/BSA m2) | 30.9 (26.7, 36.3) [n = 1,111] | 30.5 (26.3, 36) [n = 776] | 33.4 (28.1, 37.1) [n = 116] | 30.9 (27.4, 36.1) [n = 219] |
| Pre-existing conditions | ||||
| Diabetes | 358 (28.9%) | 247 (28.4%) | 35 (27.3%) | 76 (31.5%) |
| Respiratory disease | 292 (23.6%) | 204 (23.5%) | 37 (28.9%) | 51 (21.2%) |
| Severe CVS disease | 103 (8.3%) | 73 (8.4%) [n = 848] | 14 (10.9%) [n = 124] | 16 (6.6%) [n = 236] |
| Immunosuppression treatment/disease | 83 (6.7%) [n = 1,237] | 61 (7%) | 6 (4.7%) [n = 127] | 16 (6.6%) [n = 240] |
| SARS-CoV-2 type | ||||
| Wild type | 424 (34.2%) | 301 (34.6%) | 45 (35.2%) | 78 (32.3%) |
| B.1.1.7 | 270 (21.8%) | 183 (21%) | 27 (21.1%) | 60 (24.9%) |
| Inconclusive | 391 (31.6%) | 265 (30.5%) | 39 (30.5%) | 87 (36.1%) |
| Not available | 154 (12.4%) | 121 (13.9%) | 17 (13.3%) | 16 (6.6%) |
| SARS-CoV-2 viral load, median (IQR) (105 IU /ml) | ||||
| Wild type | 7.88 (0.62—96.28) | 8.21 (0.63 – 115.68) | 14.27 (2.35 – 88) | 3.03 (0.34 – 47.28) |
| B.1.1.7 | 24.38 (1.39 – 248.65) | 19.82 (1.32 – 185.69) | 57.91 (0.38 – 420.74) | 88.01 (2.60 – 307.7) |
| Inconclusive | 0.01 (0.00001 – 0.047) | 0.013 (0.0016 – 0.050) | 0.0085 (0.00010 – 0.031) | 0.01 (0.00010 – 0.05) |
| SARS-CoV-2 antibody, n (%) | ||||
| Detected | 846 (68.3%) | 582 (66.9%) | 95 (74.2%) | 169 (70.1%) |
| Not detected | 348 (28.1%) | 259 (29.8%) | 29 (22.7%) | 60 (24.9%) |
| Not available | 45 (3.6%) | 29 (3.3%) | 4 (3.1%) | 12 (5%) |
| SARS-CoV-2 antibody positive n (%) | ||||
| Wild type | 262 (64.8%) | 184 (64.1%) | 29 (67.4%) | 49 (66.2%) |
| B.1.1.7 | 176 (66.7%) | 115 (64.2%) | 24 (88.9%) | 37 (63.8%) |
| Inconclusive | 318 (83.2%) | 212 (80.9%) | 32 (84.2%) | 74 (90.2%) |
| Not available | 90 (62.5%) | 71 (62.8%) | 10 (64.1%) | 0 (60%) |
| APACHE II score, median (IQR) | 13 (8, 19) [n = 1,196] | 13 (8, 19) [n = 841] | 11.5 (8, 17) [n = 122] | 13 (8, 18) [n = 233] |
| Use and type of acute respiratory support, n (%) | ||||
| Non-invasive mechanical ventilation | 562 (45.3%) | 397 (45.6%) | 61 (47.7%) | 104 (43.2%) |
| Invasive mechanical ventilation | 418 (33.7%) | 290 (33.3%) | 43 (33.6%) | 85 (35.3%) |
| High-flow nasal cannula | 243 (19.6%) | 172 (19.8%) | 23 (18%) | 48 (19.9%) |
| None or supplemental oxygen only | 16 (1.3%) | |||
| COVID-19 therapy use | ||||
| Glucocorticoids | 1,166 (94.1%) | 820 (94.3%) | 121 (94.5%) | 225 (93.4%) |
| Remdesivir | 442 (35.7%) | 300 (34.5%) | 48 (37.5%) | 94 (39%) |
| Il-6 receptor antagonists | 41 (3.3%) | |||
| Outcomes | ||||
| Overall | ||||
| Number of OSFD at D21* (median (IQR)) | 1 (− 1, 21) | 0 (− 1, 16) | 6 (− 1, 17) | 8 (− 1, 17) |
| Hospital mortality | ||||
| Overall | 444 (35.9%) [N = 1214] | 331 (38.7%) [n = 855] | 37 (30.1%) [N = 123] | 76 (32.2%) [n = 236] |
| Seropositive | 260/827 (31.4%) | 195/571 (34.2%) | 21/91 (23.1%) | 44/165 (26.7%) |
| Seronegative | 166/343 (48.4%) | 123/255 (48.2%) | 14/28 (50%) | 29/60 (48.3%) |
APACHE II score measures the severity of illness based on age, medical history, and physiological variables. Scores range from 0 to 71; higher numbers represent greater risk of death. The median score of 12 is typical for critically ill COVID-19 patients [15]. Immunosuppression treatment refers to recent chemotherapy, radiation, high dose, or long-term glucocorticoid treatment
APACHE II, Acute Physiology and Chronic Health Evaluation II score; BMI, body mass index; BSA, body surface area; COVID-19, coronavirus disease 2019; CVS, cardiovascular; IQR, interquartile range; OSFD, organ support free days
The study cohort had a median (interquartile range, IQR) age of 61 (52, 70) years, 408 (32.9%) were females and median (IQR) APACHE II score was 13 (8, 19). Typing of the specific SARS-CoV-2 variant was successful in 56% of cases, of which wild type and B.1.1.7 variants represented 61.1% and 38.9%, respectively. SARS-CoV-2 antibodies (seropositive) were detected at baseline in 846 (68.3%) patients. Nearly all (98.7%) patients required either invasive or non-invasive respiratory support; 94.1% received low dose corticosteroids and 35.7% received remdesivir. The overall cohort had a hospital mortality of 35.9%. Seronegative patients had higher hospital mortality compared to seropositive patients (odds ratio (OR) (95% confidence interval (CI)) of 2.05 (1.58–2.65; p < 0.001), which is consistent with the literature and explained by the deficient or delayed humoral immunity in severe COVID-19 [30].
Unsupervised clustering identified three subphenotypes
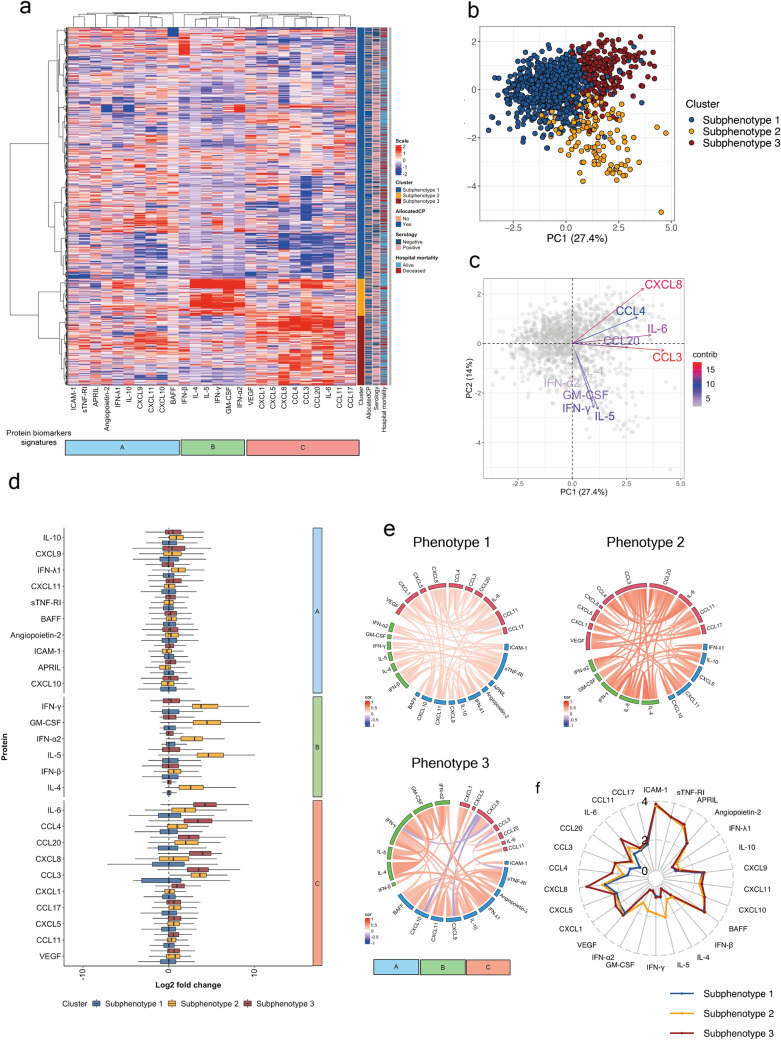
We found that a three subphenotype model (Fig. 1a, b; eFigure-2) optimally explained our biomarker data (eFigure-3). Subphenotype-1 was most common (70.2%; n = 870/1239), followed by subphenotype-3 (19.5%; 241/1239) and subphenotype-2 (10.3%;128/1239) (Table 1). The top ten contributing proteins to principal component-1 (PC1) and PC2 were biomarkers determining subphenotype-2 (IL-4, IFN-α2, GM-CSF, IFN-γ, IL-5) and subphenotype-3 (CXCL8, CCL4, IL-6, CCL20, CCL3) allocations (Fig. 1c). We observed striking differences between these subphenotypes on biomarker changes (Fig. 1d), with biologically plausible correlations between biomarkers (Fig. 1e) and differences in average biomarker concentrations (Fig. 1f).
Fig. 1.
Unsupervised clustering of 26 protein biomarkers identified three sub-subphenotypes of critically ill COVID-19 patients. a Heatmap displaying the agglomerative hierarchal clustering identified three subphenotypes. Each row is a patient (N = 1239) and each column a biomarker. Each cell is coloured by the scaled log10-transformed protein levels (high = red, low = blue). Rows are annotated by subphenotype (subphenotype-1 = blue, subphenotype-2 = orange, subphenotype-3 = red); allocation of convalescent plasma (yes = dark blue and no = orange); serology (positive = pink and negative = navy) and hospital mortality (alive = blue and deceased = red). b Principal component analysis (PCA) of the same 26 protein biomarkers coloured by subphenotype. Subphenotype-1 = blue, subphenotype-2 = orange and subphenotype-3 = red. Columns are annotated by protein biomarker signature. A = sky blue, B = light green, and C = light red. c Top ten contributing variables to principal component (PC) PC1 and PC2. Arrows are coloured based on their respective protein contribution to variation from low (blue) to high (red). d Box and whisker plots of Log2 fold change of protein biomarkers normalized to median of subphenotype-1 and grouped by protein signature (A–B). Boxes are coloured by subphenotype. The bottom border of the box represents the 25th percentile; line bisecting the box represents the median; upper border of the box is the 75th percentile. The whiskers represent extremes, 1.5 times the 75th (highest) and 25th (lowest) values. e Circos plots of each patient subphenotype represent Spearman correlations between each protein biomarker. Only correlations of an adjusted p value < 0.001 are shown. Positive and negative correlations are coloured by red and blue, respectively. The strength of the correlation is depicted by the strength of the colour. Proteins are grouped into three signatures: A = sky blue (representing biomarkers associated with dysregulated COVID-19 immune responses), B = light green (representing Type ii, Type i and altered interferon responses), C = light red (co-regulated innate immune responses with chemokines and cytokines associated with leukocyte migration and activation). Subphenotype-1 had the weakest positive correlations between the biomarkers evaluated. In subphenotype-2, all 26 biomarkers were positively correlated, consistent with the mixed immune response pattern. In subphenotype-3, CXCL8 was negatively correlated with CXCL9, CXCL10, IFN-γ, and IFN-α2, as previously reported in COVID-19. f Summary radar plot of the 26 protein biomarkers. Medians of the log10-transformed values of each protein by subphenotype are plotted. Lines are coloured by subphenotype: subphenotype-1 = blue, subphenotype-2 = orange, subphenotype-3 = red
Immunologically, subphenotype-1 patients had variable levels of proinflammatory chemokines involved in leukocyte trafficking (CXCL9, CXCL10, CXCL11) [31], immune activating cytokines (IL-10 [32]), interferons [33] (IFN- λ1, IFN-β), TNF family biomarkers (APRIL, BAFF, sTNF-R1), and endothelial biomarkers of COVID-19 severity (ICAM-1 [34], angiopoietin-2 [35]). Taken together, subphenotype-1 represents a dysregulated immune state with biomarkers strongly associated with severe COVID-19 [6, 13], without a dominant effector or co-regulated immune responses.
In contrast, subphenotype-2 appears homogeneous on biomarker patterns, with elevated levels of IL-4 and IL-5 (known to polarize naïve T cells into T-helper 2 (Th2), enable selective B cell class switch, and macrophage activation) [36], elevated levels of interferons (IFN-γ, IFN-α2) dysregulated in severe COVID-19 [33], and high GM-CSF levels with a central role in endothelial injury, Th17 T cell response [37], neutrophil recruitment, and thrombosis seen in COVID-19 [38, 39] (Fig. 1a). Taken together, subphenotype-2 resembles the mixed immune response pattern reported previously in COVID-19 [13].
Subphenotype-3 also appears more homogeneous compared to subphenotype-1 on biomarker patterns, with elevated proinflammatory cytokines such as IL-6 (with prognostic [6, 8, 13]/therapeutic relevance [40]), and elevated chemokines indicating leukocyte recruitment/activation [41] (Fig. 1a). Taken together, subphenotype-3 represents a heightened early innate immune response state [42].
Clinical features of subphenotypes
These subphenotypes were broadly similar in terms of age, sex, prevalence of comorbidities, illness severity, types of respiratory support, and prevalence of immunosuppression. SARS-CoV-2 wild-type infections were detected in a third of patients within each of the three subphenotypes. Compared to other subphenotypes, subphenotype-3 had the highest proportion of SARS-CoV-2 B.1.1.7 variant and the highest B.1.1.7 variant viral load. Compared to seronegative patients, seropositive patients had lower viral loads for all variants, in all three subphenotypes (eFigure-4). There were no differences between subphenotypes in terms of glucocorticoid and remdesivir therapy (Table 1).
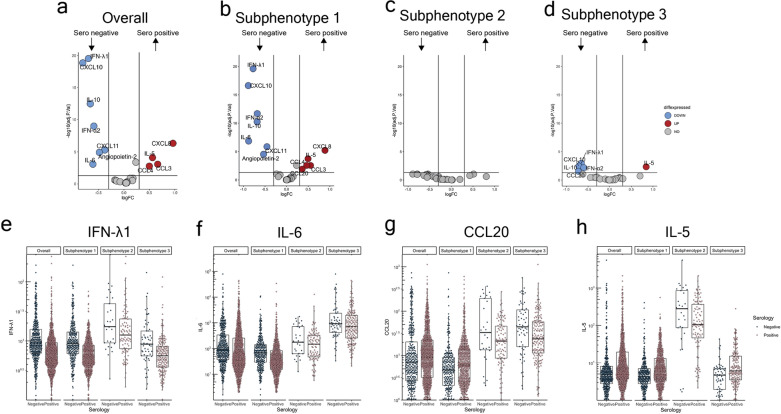
Biomarker associations between subphenotypes and serology status
Subphenotype-2 had the highest proportion of seropositive patients (74.2%) (Table 1). In the overall cohort, compared to seronegative, seropositive patients had significantly higher CXCL8, IL-5, CCL3 and CCL4, and lower levels og IFN-λ1, CXCL10, IL-10, IL-6, IFN-α2, CXCL11, and angiopoietin-2 (Fig. 2a; eFigure-5). Serology status did not segregate patients on principal component analyses (PCA; eFigure-6a). In subphenotype-1, the seropositive–seronegative comparison highlighted a pattern similar to the overall cohort (Fig. 2b). There were no significant biomarker differences between seropositive and seronegative patients within subphenotype-2 (Fig. 2c). In addition, seropositive subphenotype-1 patients had higher levels of CCL20, while seropositive subphenotype-3 patients had lower levels of CCL20 (Fig. 2d). Individual biomarker differences by serology status are shown in Fig. 2e–h.
Fig. 2.
Biomarker associations between subphenotypes and serology status. Comparison of the overall cohort and subphenotypes by serology status. a Volcano plot of the overall cohort. b Volcano plot of subphenotype-1. c Volcano plot of subphenotype-2. d Volcano plot of subphenotype-3. e–h Box and violin plot of (e) IFN- λ1, (f) IL-6, (g) CCL20 a chemokine increased during microbial insult and required for effective humoral responses [54], and (h) IL-5 by overall and subphenotypes by serology status. For volcano plots, upregulated proteins (higher in serology positive compared to serology negative) are coloured red and defined as log2 fold change > 0.3 and P ≤ 0.05. Downregulated proteins (lower in serology negative compared to serology positive) are coloured blue and defined as log2 fold change < − 0.3 and P ≤ 0.05. For box and whisker plots, the bottom border of the box represents the 25th percentile; line bisecting the box represents the median; upper border of the box is the 75th percentile. The whiskers represent 1.5 times the 75th (highest) and 25th (lowest) values
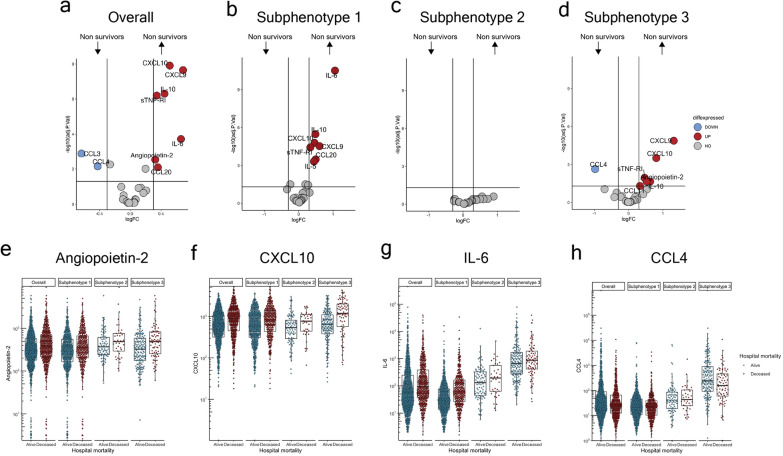
Biomarker associations between subphenotypes and hospital mortality
Hospital mortality differed between subphenotypes (p = 0.03, Chi-square test), with the highest mortality observed in subphenotype-1 (38.7%), and the lowest hospital mortality in subphenotype-2 (30.1%). In all three subphenotypes, seronegative patients had a higher (and importantly similar) hospital mortality compared to seropositive patients (Table 1).
In the overall cohort, compared to survivors, non-survivors had significantly higher levels of CXCL10, CXCL9, IL-10, sTNF-RI, IL-6, angiopoietin-2, CCL20 and lower levels of CCL3 and CCL4 (Fig. 3a; eFigure-7). Mortality did not segregate patients on PCA (eFigure-6b). In subphenotype-1, and subphenotype-3, the comparison of survivors versus non-survivors had a biomarker pattern similar to the overall cohort (Fig. 3b-d). Although the volcano plot appears to show no biomarker differences between survivors versus non-survivors in subphenotype-2 (Fig. 3c), non-survivors in this cohort had higher IL-6, CXCL-10, and angiopoietin-2 (Fig. 3e–h), which is consistent with the pattern seen in the overall cohort.
Fig. 3.
Biomarker associations between subphenotypes and hospital mortality. Comparison of the overall cohort and subphenotypes by hospital mortality. a Volcano plot of the overall cohort. b Volcano plot of subphenotype-1. c Volcano plot of subphenotype-2. d Volcano plot of subphenotype-3. e–h Box and violin plot of (e) angiopoietin-2, (f) CXCL10, (g) IL-6, and (h) CCL4 by overall and subphenotypes by mortality status. For volcano plots, upregulated proteins (higher in deceased patients compared to survivors) are coloured red and defined as log2 fold change > 0.3 and P ≤ 0.05. Downregulated proteins (lower in deceased patients compared to survivors) are coloured blue and defined as log2 fold change < − 0.3 and P ≤ 0.05. For box and whisker plots, the bottom border of the box represents the 25th percentile; the line bisecting the box represents the median; the upper border of the box is the 75th percentile. The whiskers represent 1.5 times the 75th (highest) and 25th (lowest) values
Association between subphenotypes and treatment effect of convalescent plasma
Within each subphenotype, there was no difference in baseline biomarkers between subjects allocated to convalescent plasma or usual care (eFigure-8), and allocation status did not segregate patients based on PCA (eFigure-6c).
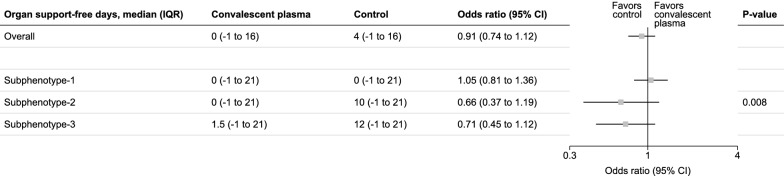
The overall treatment effect of convalescent plasma compared to usual care for OSFD-21 was OR (95%CI) of 0.91 (0.74–1.11), which is consistent with our original RCT result [15] (OR (95%CI) 0.97 (0.82 to 1.14)). The overall treatment effect of convalescent plasma compared to usual care for mortality was OR (95%CI) 1.01 (0.80–1.29), which was also consistent with our original RCT result [15] with OR (95%CI) 1.04 (0.85 to 1.27) [15].
There were no major differences in the main baseline prognostic clinical characteristics [43] (age, sex, BMI, comorbidities, and need for mechanical ventilation) by allocation status, within each subphenotype (eTable-2). In subphenotype-1, the median (IQR) OSFD-21 was 0 (−1, 21) in convalescent plasma and 0 (−1, to 21) in the usual care arm. In subphenotype-2 and -3, the usual care had higher OSFD-21 compared to the convalescent plasma arm (subphenotype-2 (OSFD-21 median (IQR) 0 (−1, 21) in convalescent plasma vs 10 (−1, to 21) in usual care) and subphenotype-3 (OSFD-21 median (IQR) 1.5 (−1, 21) in convalescent plasma vs 12 (−1, to 21) in usual care). The corresponding odds ratio differed by subphenotype (test of heterogeneity; p = 0.008 (Fig. 4).
Fig. 4.
Treatment effect of convalescent plasma compared to usual care for organ support-free days by subphenotypes. Forest plot comparing organ support-free days at day 21 (OSFD-21) of the overall cohort and by subphenotypes when treated with convalescent plasma, compared to usual care population. Median and inter-quartile ranges (IQR) for OFSD are displayed. Odds ratio was calculated using ordered logistic regression, and 95% confidence intervals are reported. Square dots represent odds ratio of the respective row, and the black line denotes 95% confidence intervals. Odds ratio < 1 favours control. The P value is reported based on the test of heterogeneity estimated post-ordered logistic regression. The odds ratio represents the average odds ratio for each possible cut points of the outcome variable. Proportional odds assumption means that the odds ratios are about the same regardless of the cut point of the ordinal outcome variable
In subphenotype-1, the hospital mortality in the convalescent plasma group was lower than that in the usual care group (37.6% vs 40.5%). In contrast, in subphenotype-2 and subphenotype-3, the hospital mortality in the convalescent plasma group was higher than that in usual care (subphenotype-2 = 35.4% vs 24.1% and subphenotype-3 = 34.1% vs 29.7%). The corresponding odds ratio differed by subphenotype (test of heterogeneity; p = 0.02) (eFigure-9).
Association between serology status of subphenotypes and treatment effect of convalescent plasma
In our main trial publication [15], the treatment effect of convalescent plasma did not meaningfully vary in the prespecified serology status subgroup. Consistent with the main trial result, the treatment effect on mortality did not vary by serology status of the subphenotypes (p = 0.69, for the three-way interaction test between allocation to convalescent plasma, serology status, and subphenotype). It could be that our substudy is underpowered to assess this subgroup within a subgroup effect (i.e., serology status within subphenotypes). Of note, only in subphenotype-1, seronegative patients who received convalescent plasma had lower mortality, compared to seronegative patients who received usual care (eFigure-10). As serology is a prognostic covariate, our sensitivity analyses including serology status as a covariate in two additional regression models (for OSFD-21 and mortality) was consistent with the main findings.
Discussion
We report the largest biomarker study conducted within an RCT in critically ill COVID-19 patients. We highlight three distinct subphenotypes based on biomarker profiles within critically ill COVID-19 patients who had similar clinical features, but with differences in clinical outcomes and treatment effect estimates for OSFD-21 and hospital mortality. Compared to subphenotype-1, mortality was lower despite higher inflammation in suphenotype-2 and subphenotype-3. Our observations, if validated, favour avoiding convalescent plasma therapy in subphenotype-2 and -3.
Our subphenotypes have biological plausibility. The median (IQR) IL-6 levels in our cohort was 62.2 (23.8–290.6) pg/mL, which is consistent with literature [44]. Negative association between IL-10, CXCL-10, and IL-6 with seropositive status in COVID-19 has been reported previously [6], explaining the prognostic utility of this biomarker triad. Biomarker differences between seropositive and seronegative patients in our study represent altered interferon responses [33], and compromised humoral immunity [12, 13, 30] in critically ill COVID-19 patients. Prognostic associations with many of these biomarkers have been reported previously in acute respiratory distress syndrome [45].
As we are unable to assess potential molecular mechanisms, we propose the following hypotheses as to why convalescent plasma therapy could theoretically worsen outcomes in the more proinflammatory subphenotypes [46–49]. High-affinity antibodies present in convalescent plasma elicit SARS-CoV-2 neutralization [46–48]. However, the low-affinity antibodies present either in donor plasma or formed in recipients following convalescent plasma administration could activate proinflammatory pathways [49], worsening the outcomes. The presence of autoantibodies reported in COVID-19 patients [50, 51] may be present in convalescent plasma, which could worsen outcomes in the more proinflammatory subphenotypes. Although a rare event in our primary trial [15], convalescent plasma is a blood product that can cause transfusion-related adverse events.
Our sampled population appears representative of the overall RCT publication [15]. Our findings also highlight the value of enriching trial populations [52]. Although our findings are likely to be widely applicable to moderate or severe COVID-19 patients, our primary RCT was not powered to detect subgroup effects. We neither have non-COVID controls nor validation cohorts. Research blood sampling was not possible outside the UK, and not every patient enrolled in the UK had sampling. As our RCT was conducted early on in the pandemic, SARS-CoV-2 vaccination may alter the prevalence of reported subphenotypes.
Our results have clinical utility for the following reasons. Our findings support the hypothesis that immunotherapy in COVID-19 could be useful with better patient selection based on host immune response characteristics. It is feasible to determine subphenotype-2 and -3, where we observed a harm signal by measuring a limited biomarker set based on discriminant value (such as IL-6, CCL3, and IL-8 based our data in Fig. 1). Lower overall mortality in the more inflammatory subphenotypes in our cohort supports the notion that strong prognostic association to cytokines such as IL-6 in mild COVID-19 [8] may be masked by complex cytokine networks or hubs in severe inflammatory illnesses [53] observed with severe COVID-19, highlighting the futility of measuring single cytokines as value-added biomarkers for informing treatment decisions.
Our novel findings highlight future research questions. A key next step is to study the molecular mechanisms underpinning these subphenotypes, to consider them as COVID-19 endotypes [52]. A related research question is whether these subphenotypes are associated with HTE to other immunomodulators. It is important to determine whether these subphenotypes are identifiable in non-critically ill COVID-19 patients and whether they have HTE to immunotherapies, as our study focused on critically ill COVID-19. In any viral pandemic, as convalescent plasma will be a potential treatment, understanding the mechanisms for harm may lead to better selection of donor plasma in the future.
Conclusions
We report three COVID-19 subphenotypes with differences in treatment response to ABO-compatible high-titer convalescent plasma therapy among critically ill adults, participating in an RCT. Given the distinct immunological mechanisms, these subphenotypes could be termed endotypes. These findings support the hypothesis that the benefits of immunotherapy in COVID-19 could be enhanced with patient selection based on host immune response characteristics.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the NIHR Clinical Research Network (UK) for their support in participant recruitment. We are grateful to the UK Blood Services (NHS Blood and Transplant, Northern Ireland Blood Transfusion Service, Scottish National Blood Transfusion Service and Welsh Blood Service) for the supply of convalescent plasma in the UK. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health and Social Care. This research was supported by the National Institute for Health and Care Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London and/or the NIHR Clinical Research Facility.
Author contributions
MS-H, LE, JR, MF had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. MF and JR contributed equally to this article. Concept and design: LE, HH, DKM, DR, MS-H. Acquisition, analysis, or interpretation of data: JR, MF, AJ, AL, JR, LE, HH, DKM, DR, MS-H Drafting of the manuscript: JR, MF, MS-H. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: JR, MF, MS-H. Obtained funding: LE, HH, DKM, DR, MS-H Supervision: MS-H.
Funding
This study was supported by National Institute for Health (UKRIDHSC COVID-19 Rapid Response Rolling Call, “The use of convalescent plasma to treat hospitalized and critically ill patients with COVID-19 disease”, Grant Reference Number COV19-RECPLAS). MF was supported by a National Institute of Academic Anesthesia BJA/RCoA fellowship (WKRO-2018–0047). JR was supported by the Marshall Scholarship and Clarendon Fund. ACG was funded by an NIHR Research Professorship (RP-2015-–06-18). DKM was supported by the NIHR through the Cambridge NIHR BRC and the Addenbrooke’s Charitable Trust. MS-H is funded by a clinician scientist fellowship 2016-16-011 from the National Institute for Health Research. The REMAP-CAP has four regional nonprofit sponsors: Monash University, Melbourne, Australia (Australasian sponsor); Utrecht Medical Center, Utrecht, the Netherlands (European sponsor); St Michael’s Hospital, Toronto, Ontario, Canada (Canadian sponsor); and the Global Coalition for Adaptive Research, San Francisco, California (US sponsor). REMAP-CAP was additionally funded by FP7-health-2013-innovation-1 (#602,525) from the European Union Platform for European Preparedness Against Reemerging Epidemics grant, the Rapid European COVID-19 Emergency Research response (RECOVER) consortium by the European Union’s Horizon 2020 research and innovation programme (#101,003,589), the UK National Institute for Health Research (NIHR) and the NIHR Imperial Biomedical Research Centre, Collection of UK plasma was funded by the DHSC through core funding under COVID-19 and EU SoHo Grants. The funders/sponsors had no role in the design and conduct of the trial; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health and Social Care.
Declarations
Conflicts of interest
LE reported receiving grants from the National Institute for Health Research and European Union Horizon 2020. ACG reported receiving grants from the National Institute for Health Research and receiving personal fees from 30 Respiratory, GlaxoSmithKline, and Bristol Myers Squibb. DKM reports consultancy agreements, educational support, or research collaborations with NeuroTrauma Sciences LLC, GlaxoSmithKline Ltd, Calico Ltd, Lantmannen AB, Cortirio Ltd, and Gryphon Inc., all outside the research in this manuscript. MSH reports receiving grant from the Chief Scientists Office, Scotland, for time-critical precision medicine in adult critically ill patients (TRAITS Programme) and highlights industry support for TRAITS research programme (https://www.ed.ac.uk/inflammation-research/clinical-trials/traits-ci-trial).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh SA, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Bilezikian JP, Landry DW. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 2.Ratcliff J, Nguyen D, Fish M, Rynne J, Jennings A, Williams S, Al-Beidh F, Bonsall D, Evans A, Golubchik T, Gordon AC, Lamikanra A, Tsang P, Ciccone NA, Leuscher U, Slack W, Laing E, Mouncey PR, Ziyenge S, Oliveira M, Ploeg R, Rowan KM, Shankar-Hari M, Roberts DJ, Menon DK, Estcourt L, Simmonds P, Harvala H, Investigators R-CIDU. Virological Characterization of critically Ill patients with COVID-19 in the United Kingdom: interactions of viral load, antibody status, and B.1.1.7 infection. J Infect Dis. 2021;224:595–605. doi: 10.1093/infdis/jiab283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutmann C, Takov K, Burnap SA, Singh B, Ali H, Theofilatos K, Reed E, Hasman M, Nabeebaccus A, Fish M, McPhail MJ, O'Gallagher K, Schmidt LE, Cassel C, Rienks M, Yin X, Auzinger G, Napoli S, Mujib SF, Trovato F, Sanderson B, Merrick B, Niazi U, Saqi M, Dimitrakopoulou K, Fernandez-Leiro R, Braun S, Kronstein-Wiedemann R, Doores KJ, Edgeworth JD, Shah AM, Bornstein SR, Tonn T, Hayday AC, Giacca M, Shankar-Hari M, Mayr M. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat Commun. 2021;12:3406. doi: 10.1038/s41467-021-23494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Veerdonk FL, Giamarellos-Bourboulis E, Pickkers P, Derde L, Leavis H, van Crevel R, Engel JJ, Wiersinga WJ, Vlaar APJ, Shankar-Hari M, van der Poll T, Bonten M, Angus DC, van der Meer JWM, Netea MG. A guide to immunotherapy for COVID-19. Nat Med. 2022;28:39–50. doi: 10.1038/s41591-021-01643-9. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal A, Rochwerg B, Lamontagne F, Siemieniuk RA, Agoritsas T, Askie L, Lytvyn L, Leo YS, Macdonald H, Zeng L, Amin W, Barragan FAJ, Bausch FJ, Burhan E, Calfee CS, Cecconi M, Chanda D, Dat VQ, De Sutter A, Du B, Freedman S, Geduld H, Gee P, Gotte M, Harley N, Hashimi M, Hunt B, Jehan F, Kabra SK, Kanda S, Kim YJ, Kissoon N, Krishna S, Kuppalli K, Kwizera A, Lado Castro-Rial M, Lisboa T, Lodha R, Mahaka I, Manai H, Mino G, Nsutebu E, Preller J, Pshenichnaya N, Qadir N, Relan P, Sabzwari S, Sarin R, Shankar-Hari M, Sharland M, Shen Y, Ranganathan SS, Souza JP, Stegemann M, Swanstrom R, Ugarte S, Uyeki T, Venkatapuram S, Vuyiseka D, Wijewickrama A, Tran L, Zeraatkar D, Bartoszko JJ, Ge L, Brignardello-Petersen R, Owen A, Guyatt G, Diaz J, Kawano-Dourado L, Jacobs M, Vandvik PO. A living WHO guideline on drugs for covid-19. BMJ. 2020;370:m3379. doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- 6.Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, Munoz-Ruiz M, McKenzie DR, Hayday TS, Francos-Quijorna I, Kamdar S, Joseph M, Davies D, Davis R, Jennings A, Zlatareva I, Vantourout P, Wu Y, Sofra V, Cano F, Greco M, Theodoridis E, Freedman JD, Gee S, Chan JNE, Ryan S, Bugallo-Blanco E, Peterson P, Kisand K, Haljasmagi L, Chadli L, Moingeon P, Martinez L, Merrick B, Bisnauthsing K, Brooks K, Ibrahim MAA, Mason J, Lopez Gomez F, Babalola K, Abdul-Jawad S, Cason J, Mant C, Seow J, Graham C, Doores KJ, Di Rosa F, Edgeworth J, Shankar-Hari M, Hayday AC. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 7.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D'Andrea K, Manne S, Chen Z, Huang YJ, Reilly JP, Weisman AR, Ittner CAG, Kuthuru O, Dougherty J, Nzingha K, Han N, Kim J, Pattekar A, Goodwin EC, Anderson EM, Weirick ME, Gouma S, Arevalo CP, Bolton MJ, Chen F, Lacey SF, Ramage H, Cherry S, Hensley SE, Apostolidis SA, Huang AC, Vella LA, Unit UPCP, Betts MR, Meyer NJ, Wherry EJ. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hojyo S, Uchida M, Tanaka K, Hasebe R, Tanaka Y, Murakami M, Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflamm Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodriguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, Jia X, Wu M, Shi B, Xu S, Chen J, Wang W, Chen B, Jiang L, Yu S, Lu J, Wang J, Xu M, Yuan Z, Zhang Q, Zhang X, Zhao G, Wang S, Chen S, Lu H. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 12.julian.knight@well.ox.ac.uk CO-M-oBACEa, Consortium CO-M-oBA A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185(916–938):e958. doi: 10.1016/j.cell.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IT, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kravitz RL, Duan N, Braslow J. Evidence-based medicine, heterogeneity of treatment effects, and the trouble with averages. Milbank Q. 2004;82:661–687. doi: 10.1111/j.0887-378X.2004.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Writing Committee for the R-CAPI. Estcourt LJ, Turgeon AF, McQuilten ZK, McVerry BJ, Al-Beidh F, Annane D, Arabi YM, Arnold DM, Beane A, Begin P, van Bentum-Puijk W, Berry LR, Bhimani Z, Birchall JE, Bonten MJM, Bradbury CA, Brunkhorst FM, Buxton M, Callum JL, Chasse M, Cheng AC, Cove ME, Daly J, Derde L, Detry MA, De Jong M, Evans A, Fergusson DA, Fish M, Fitzgerald M, Foley C, Goossens H, Gordon AC, Gosbell IB, Green C, Haniffa R, Harvala H, Higgins AM, Hills TE, Hoad VC, Horvat C, Huang DT, Hudson CL, Ichihara N, Laing E, Lamikanra AA, Lamontagne F, Lawler PR, Linstrum K, Litton E, Lorenzi E, MacLennan S, Marshall J, McAuley DF, McDyer JF, McGlothlin A, McGuinness S, Miflin G, Montgomery S, Mouncey PR, Murthy S, Nichol A, Parke R, Parker JC, Priddee N, Purcell DFJ, Reyes LF, Richardson P, Robitaille N, Rowan KM, Rynne J, Saito H, Santos M, Saunders CT, Serpa Neto A, Seymour CW, Silversides JA, Tinmouth AA, Triulzi DJ, Turner AM, van de Veerdonk F, Walsh TS, Wood EM, Berry S, Lewis RJ, Menon DK, McArthur C, Zarychanski R, Angus DC, Webb SA, Roberts DJ, Shankar-Hari M. Effect of convalescent plasma on organ support-free days in critically ill patients with COVID-19: a randomized clinical trial. JAMA. 2021;326:1690–1702. doi: 10.1001/jama.2021.18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buszko M, Nita-Lazar A, Park JH, Schwartzberg PL, Verthelyi D, Young HA, Rosenberg AS. Lessons learned: new insights on the role of cytokines in COVID-19. Nat Immunol. 2021;22:404–411. doi: 10.1038/s41590-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent FB, Saulep-Easton D, Figgett WA, Fairfax KA, Mackay F. The BAFF/APRIL system: emerging functions beyond B cell biology and autoimmunity. Cytokine Growth Factor Rev. 2013;24:203–215. doi: 10.1016/j.cytogfr.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angus DC, Berry S, Lewis RJ, Al-Beidh F, Arabi Y, van Bentum-Puijk W, Bhimani Z, Bonten M, Broglio K, Brunkhorst F, Cheng AC, Chiche JD, De Jong M, Detry M, Goossens H, Gordon A, Green C, Higgins AM, Hullegie SJ, Kruger P, Lamontagne F, Litton E, Marshall J, McGlothlin A, McGuinness S, Mouncey P, Murthy S, Nichol A, O'Neill GK, Parke R, Parker J, Rohde G, Rowan K, Turner A, Young P, Derde L, McArthur C, Webb SA. The REMAP-CAP (randomized Embedded multifactorial adaptive platform for community-acquired pneumonia) study. Rationale and design. Ann Am Thorac Soc. 2020;17:879–891. doi: 10.1513/AnnalsATS.202003-192SD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angus DC, Derde L, Al-Beidh F, Annane D, Arabi Y, Beane A, van Bentum-Puijk W, Berry L, Bhimani Z, Bonten M, Bradbury C, Brunkhorst F, Buxton M, Buzgau A, Cheng AC, de Jong M, Detry M, Estcourt L, Fitzgerald M, Goossens H, Green C, Haniffa R, Higgins AM, Horvat C, Hullegie SJ, Kruger P, Lamontagne F, Lawler PR, Linstrum K, Litton E, Lorenzi E, Marshall J, McAuley D, McGlothin A, McGuinness S, McVerry B, Montgomery S, Mouncey P, Murthy S, Nichol A, Parke R, Parker J, Rowan K, Sanil A, Santos M, Saunders C, Seymour C, Turner A, van de Veerdonk F, Venkatesh B, Zarychanski R, Berry S, Lewis RJ, McArthur C, Webb SA, Gordon AC, Writing Committee for the R-CAPI, Al-Beidh F, Angus D, Annane D, Arabi Y, van Bentum-Puijk W, Berry S, Beane A, Bhimani Z, Bonten M, Bradbury C, Brunkhorst F, Buxton M, Cheng A, De Jong M, Derde L, Estcourt L, Goossens H, Gordon A, Green C, Haniffa R, Lamontagne F, Lawler P, Litton E, Marshall J, McArthur C, McAuley D, McGuinness S, McVerry B, Montgomery S, Mouncey P, Murthy S, Nichol A, Parke R, Rowan K, Seymour C, Turner A, van de Veerdonk F, Webb S, Zarychanski R, Campbell L, Forbes A, Gattas D, Heritier S, Higgins L, Kruger P, Peake S, Presneill J, Seppelt I, Trapani T, Young P, Bagshaw S, Daneman N, Ferguson N, Misak C, Santos M, Hullegie S, Pletz M, Rohde G, Rowan K, Alexander B, Basile K, Girard T, Horvat C, Huang D, Linstrum K, Vates J, Beasley R, Fowler R, McGloughlin S, Morpeth S, Paterson D, Venkatesh B, Uyeki T, Baillie K, Duffy E, Fowler R, Hills T, Orr K, Patanwala A, Tong S, Netea M, Bihari S, Carrier M, Fergusson D, Goligher E, Haidar G, Hunt B, Kumar A, Laffan M, Lawless P, Lother S, McCallum P, Middeldopr S, McQuilten Z, Neal M, Pasi J, Schutgens R, Stanworth S, Turgeon A, Weissman A, Adhikari N, Anstey M, Brant E, de Man A, Lamonagne F, Masse MH, Udy A, Arnold D, Begin P, Charlewood R, Chasse M, Coyne M, Cooper J, Daly J, Gosbell I, Harvala-Simmonds H, Hills T, MacLennan S, Menon D, McDyer J, Pridee N, Roberts D, Shankar-Hari M, Thomas H, Tinmouth A, Triulzi D, Walsh T, Wood E, Calfee C, O'Kane C, Shyamsundar M, Sinha P, Thompson T, Young I, Bihari S, Hodgson C, Laffey J, McAuley D, Orford N, Neto A, Detry M, Fitzgerald M, Lewis R, McGlothlin A, Sanil A, Saunders C, Berry L, Lorenzi E, Miller E, Singh V, Zammit C, van Bentum Puijk W, Bouwman W, Mangindaan Y, Parker L, Peters S, Rietveld I, Raymakers K, Ganpat R, Brillinger N, Markgraf R, Ainscough K, Brickell K, Anjum A, Lane JB, Richards-Belle A, Saull M, Wiley D, Bion J, Connor J, Gates S, Manax V, van der Poll T, Reynolds J, van Beurden M, Effelaar E, Schotsman J, Boyd C, Harland C, Shearer A, Wren J, Clermont G, Garrard W, Kalchthaler K, King A, Ricketts D, Malakoutis S, Marroquin O, Music E, Quinn K, Cate H, Pearson K, Collins J, Hanson J, Williams P, Jackson S, Asghar A, Dyas S, Sutu M, Murphy S, Williamson D, Mguni N, Potter A, Porter D, Goodwin J, Rook C, Harrison S, Williams H, Campbell H, Lomme K, Williamson J, Sheffield J, van't Hoff W, McCracken P, Young M, Board J, Mart E, Knott C, Smith J, Boschert C, Affleck J, Ramanan M, D'Souza R, Pateman K, Shakih A, Cheung W, Kol M, Wong H, Shah A, Wagh A, Simpson J, Duke G, Chan P, Cartner B, Hunter S, Laver R, Shrestha T, Regli A, Pellicano A, McCullough J, Tallott M, Kumar N, Panwar R, Brinkerhoff G, Koppen C, Cazzola F, Brain M, Mineall S, Fischer R, Biradar V, Soar N, White H, Estensen K, Morrison L, Smith J, Cooper M, Health M, Shehabi Y, Al-Bassam W, Hulley A, Whitehead C, Lowrey J, Gresha R, Walsham J, Meyer J, Harward M, Venz E, Williams P, Kurenda C, Smith K, Smith M, Garcia R, Barge D, Byrne D, Byrne K, Driscoll A, Fortune L, Janin P, Yarad E, Hammond N, Bass F, Ashelford A, Waterson S, Wedd S, McNamara R, Buhr H, Coles J, Schweikert S, Wibrow B, Rauniyar R, Myers E, Fysh E, Dawda A, Mevavala B, Litton E, Ferrier J, Nair P, Buscher H, Reynolds C, Santamaria J, Barbazza L, Homes J, Smith R, Murray L, Brailsford J, Forbes L, Maguire T, Mariappa V, Smith J, Simpson S, Maiden M, Bone A, Horton M, Salerno T, Sterba M, Geng W, Depuydt P, De Waele J, De Bus L, Fierens J, Bracke S, Reeve B, Dechert W, Chasse M, Carrier FM, Boumahni D, Benettaib F, Ghamraoui A, Bellemare D, Cloutier E, Francoeur C, Lamontagne F, D'Aragon F, Carbonneau E, Leblond J, Vazquez-Grande G, Marten N, Wilson M, Albert M, Serri K, Cavayas A, Duplaix M, Williams V, Rochwerg B, Karachi T, Oczkowski S, Centofanti J, Millen T, Duan E, Tsang J, Patterson L, English S, Watpool I, Porteous R, Miezitis S, McIntyre L, Brochard L, Burns K, Sandhu G, Khalid I, Binnie A, Powell E, McMillan A, Luk T, Aref N, Andric Z, Cviljevic S, Dimoti R, Zapalac M, Mirkovic G, Barsic B, Kutlesa M, Kotarski V, Vujaklija Brajkovic A, Babel J, Sever H, Dragija L, Kusan I, Vaara S, Pettila L, Heinonen J, Kuitunen A, Karlsson S, Vahtera A, Kiiski H, Ristimaki S, Azaiz A, Charron C, Godement M, Geri G, Vieillard-Baron A, Pourcine F, Monchi M, Luis D, Mercier R, Sagnier A, Verrier N, Caplin C, Siami S, Aparicio C, Vautier S, Jeblaoui A, Fartoukh M, Courtin L, Labbe V, Leparco C, Muller G, Nay MA, Kamel T, Benzekri D, Jacquier S, Mercier E, Chartier D, Salmon C, Dequin P, Schneider F, Morel G, L'Hotellier S, Badie J, Berdaguer FD, Malfroy S, Mezher C, Bourgoin C, Megarbane B, Voicu S, Deye N, Malissin I, Sutterlin L, Guitton C, Darreau C, Landais M, Chudeau N, Robert A, Moine P, Heming N, Maxime V, Bossard I, Nicholier TB, Colin G, Zinzoni V, Maquigneau N, Finn A, Kress G, Hoff U, Friedrich Hinrichs C, Nee J, Pletz M, Hagel S, Ankert J, Kolanos S, Bloos F, Petros S, Pasieka B, Kunz K, Appelt P, Schutze B, Kluge S, Nierhaus A, Jarczak D, Roedl K, Weismann D, Frey A, Klinikum Neukolln V, Reill L, Distler M, Maselli A, Belteczki J, Magyar I, Fazekas A, Kovacs S, Szoke V, Szigligeti G, Leszkoven J, Collins D, Breen P, Frohlich S, Whelan R, McNicholas B, Scully M, Casey S, Kernan M, Doran P, O'Dywer M, Smyth M, Hayes L, Hoiting O, Peters M, Rengers E, Evers M, Prinssen A, Bosch Ziekenhuis J, Simons K, Rozendaal W, Polderman F, de Jager P, Moviat M, Paling A, Salet A, Rademaker E, Peters AL, de Jonge E, Wigbers J, Guilder E, Butler M, Cowdrey KA, Newby L, Chen Y, Simmonds C, McConnochie R, Ritzema Carter J, Henderson S, Van Der Heyden K, Mehrtens J, Williams T, Kazemi A, Song R, Lai V, Girijadevi D, Everitt R, Russell R, Hacking D, Buehner U, Williams E, Browne T, Grimwade K, Goodson J, Keet O, Callender O, Martynoga R, Trask K, Butler A, Schischka L, Young C, Lesona E, Olatunji S, Robertson Y, Jose N, Amaro dos Santos Catorze T, de Lima Pereira TNA, Neves Pessoa LM, Castro Ferreira RM, Pereira Sousa Bastos JM, Aysel Florescu S, Stanciu D, Zaharia MF, Kosa AG, Codreanu D, Marabi Y, Al Qasim E, Moneer Hagazy M, Al Swaidan L, Arishi H, Munoz-Bermudez R, Marin-Corral J, Salazar Degracia A, Parrilla Gomez F, Mateo Lopez MI, Rodriguez Fernandez J, Carcel Fernandez S, Carmona Flores R, Leon Lopez R, de la Fuente Martos C, Allan A, Polgarova P, Farahi N, McWilliam S, Hawcutt D, Rad L, O'Malley L, Whitbread J, Kelsall O, Wild L, Thrush J, Wood H, Austin K, Donnelly A, Kelly M, O'Kane S, McClintock D, Warnock M, Johnston P, Gallagher LJ, Mc Goldrick C, Mc Master M, Strzelecka A, Jha R, Kalogirou M, Ellis C, Krishnamurthy V, Deelchand V, Silversides J, McGuigan P, Ward K, O'Neill A, Finn S, Phillips B, Mullan D, Oritz-Ruiz de Gordoa L, Thomas M, Sweet K, Grimmer L, Johnson R, Pinnell J, Robinson M, Gledhill L, Wood T, Morgan M, Cole J, Hill H, Davies M, Antcliffe D, Templeton M, Rojo R, Coghlan P, Smee J, Mackay E, Cort J, Whileman A, Spencer T, Spittle N, Kasipandian V, Patel A, Allibone S, Genetu RM, Ramali M, Ghosh A, Bamford P, London E, Cawley K, Faulkner M, Jeffrey H, Smith T, Brewer C, Gregory J, Limb J, Cowton A, O'Brien J, Nikitas N, Wells C, Lankester L, Pulletz M, Williams P, Birch J, Wiseman S, Horton S, Alegria A, Turki S, Elsefi T, Crisp N, Allen L, McCullagh I, Robinson P, Hays C, Babio-Galan M, Stevenson H, Khare D, Pinder M, Selvamoni S, Gopinath A, Pugh R, Menzies D, Mackay C, Allan E, Davies G, Puxty K, McCue C, Cathcart S, Hickey N, Ireland J, Yusuff H, Isgro G, Brightling C, Bourne M, Craner M, Watters M, Prout R, Davies L, Pegler S, Kyeremeh L, Arbane G, Wilson K, Gomm L, Francia F, Brett S, Sousa Arias S, Elin Hall R, Budd J, Small C, Birch J, Collins E, Henning J, Bonner S, Hugill K, Cirstea E, Wilkinson D, Karlikowski M, Sutherland H, Wilhelmsen E, Woods J, North J, Sundaran D, Hollos L, Coburn S, Walsh J, Turns M, Hopkins P, Smith J, Noble H, Depante MT, Clarey E, Laha S, Verlander M, Williams A, Huckle A, Hall A, Cooke J, Gardiner-Hill C, Maloney C, Qureshi H, Flint N, Nicholson S, Southin S, Nicholson A, Borgatta B, Turner-Bone I, Reddy A, Wilding L, Chamara Warnapura L, Agno Sathianathan R, Golden D, Hart C, Jones J, Bannard-Smith J, Henry J, Birchall K, Pomeroy F, Quayle R, Makowski A, Misztal B, Ahmed I, KyereDiabour T, Naiker K, Stewart R, Mwaura E, Mew L, Wren L, Willams F, Innes R, Doble P, Hutter J, Shovelton C, Plumb B, Szakmany T, Hamlyn V, Hawkins N, Lewis S, Dell A, Gopal S, Ganguly S, Smallwood A, Harris N, Metherell S, Lazaro JM, Newman T, Fletcher S, Nortje J, Fottrell-Gould D, Randell G, Zaman M, Elmahi E, Jones A, Hall K, Mills G, Ryalls K, Bowler H, Sall J, Bourne R, Borrill Z, Duncan T, Lamb T, Shaw J, Fox C, Moreno Cuesta J, Xavier K, Purohit D, Elhassan M, Bakthavatsalam D, Rowland M, Hutton P, Bashyal A, Davidson N, Hird C, Chhablani M, Phalod G, Kirkby A, Archer S, Netherton K, Reschreiter H, Camsooksai J, Patch S, Jenkins S, Pogson D, Rose S, Daly Z, Brimfield L, Claridge H, Parekh D, Bergin C, Bates M, Dasgin J, McGhee C, Sim M, Hay SK, Henderson S, Phull MK, Zaidi A, Pogreban T, Rosaroso LP, Harvey D, Lowe B, Meredith M, Ryan L, Hormis A, Walker R, Collier D, Kimpton S, Oakley S, Rooney K, Rodden N, Hughes E, Thomson N, McGlynn D, Walden A, Jacques N, Coles H, Tilney E, Vowell E, Schuster-Bruce M, Pitts S, Miln R, Purandare L, Vamplew L, Spivey M, Bean S, Burt K, Moore L, Day C, Gibson C, Gordon E, Zitter L, Keenan S, Baker E, Cherian S, Cutler S, Roynon-Reed A, Harrington K, Raithatha A, Bauchmuller K, Ahmad N, Grecu I, Trodd D, Martin J, Wrey Brown C, Arias AM, Craven T, Hope D, Singleton J, Clark S, Rae N, Welters I, Hamilton DO, Williams K, Waugh V, Shaw D, Puthucheary Z, Martin T, Santos F, Uddin R, Somerville A, Tatham KC, Jhanji S, Black E, Dela Rosa A, Howle R, Tully R, Drummond A, Dearden J, Philbin J, Munt S, Vuylsteke A, Chan C, Victor S, Matsa R, Gellamucho M, Creagh-Brown B, Tooley J, Montague L, De Beaux F, Bullman L, Kersiake I, Demetriou C, Mitchard S, Ramos L, White K, Donnison P, Johns M, Casey R, Mattocks L, Salisbury S, Dark P, Claxton A, McLachlan D, Slevin K, Lee S, Hulme J, Joseph S, Kinney F, Senya HJ, Oborska A, Kayani A, Hadebe B, Orath Prabakaran R, Nichols L, Thomas M, Worner R, Faulkner B, Gendall E, Hayes K, Hamilton-Davies C, Chan C, Mfuko C, Abbass H, Mandadapu V, Leaver S, Forton D, Patel K, Paramasivam E, Powell M, Gould R, Wilby E, Howcroft C, Banach D, Fernandez de Pinedo Artaraz Z, Cabreros L, White I, Croft M, Holland N, Pereira R, Zaki A, Johnson D, Jackson M, Garrard H, Juhaz V, Roy A, Rostron A, Woods L, Cornell S, Pillai S, Harford R, Rees T, Ivatt H, Sundara Raman A, Davey M, Lee K, Barber R, Chablani M, Brohi F, Jagannathan V, Clark M, Purvis S, Wetherill B, Dushianthan A, Cusack R, de Courcy-Golder K, Smith S, Jackson S, Attwood B, Parsons P, Page V, Zhao XB, Oza D, Rhodes J, Anderson T, Morris S, Xia Le Tai C, Thomas A, Keen A, Digby S, Cowley N, Wild L, Southern D, Reddy H, Campbell A, Watkins C, Smuts S, Touma O, Barnes N, Alexander P, Felton T, Ferguson S, Sellers K, Bradley-Potts J, Yates D, Birkinshaw I, Kell K, Marshall N, Carr-Knott L, Summers C. (2020) Effect of Hydrocortisone on Mortality and Organ Support in Patients With Severe COVID-19: The REMAP-CAP COVID-19 corticosteroid domain randomized clinical trial. JAMA. 324: 1317–1329 [DOI] [PMC free article] [PubMed]
- 20.Investigators A, Investigators AC-a, Investigators R-C. Lawler PR, Goligher EC, Berger JS, Neal MD, McVerry BJ, Nicolau JC, Gong MN, Carrier M, Rosenson RS, Reynolds HR, Turgeon AF, Escobedo J, Huang DT, Bradbury CA, Houston BL, Kornblith LZ, Kumar A, Kahn SR, Cushman M, McQuilten Z, Slutsky AS, Kim KS, Gordon AC, Kirwan BA, Brooks MM, Higgins AM, Lewis RJ, Lorenzi E, Berry SM, Berry LR, Aday AW, Al-Beidh F, Annane D, Arabi YM, Aryal D, Baumann Kreuziger L, Beane A, Bhimani Z, Bihari S, Billett HH, Bond L, Bonten M, Brunkhorst F, Buxton M, Buzgau A, Castellucci LA, Chekuri S, Chen JT, Cheng AC, Chkhikvadze T, Coiffard B, Costantini TW, de Brouwer S, Derde LPG, Detry MA, Duggal A, Dzavik V, Effron MB, Estcourt LJ, Everett BM, Fergusson DA, Fitzgerald M, Fowler RA, Galanaud JP, Galen BT, Gandotra S, Garcia-Madrona S, Girard TD, Godoy LC, Goodman AL, Goossens H, Green C, Greenstein YY, Gross PL, Hamburg NM, Haniffa R, Hanna G, Hanna N, Hegde SM, Hendrickson CM, Hite RD, Hindenburg AA, Hope AA, Horowitz JM, Horvat CM, Hudock K, Hunt BJ, Husain M, Hyzy RC, Iyer VN, Jacobson JR, Jayakumar D, Keller NM, Khan A, Kim Y, Kindzelski AL, King AJ, Knudson MM, Kornblith AE, Krishnan V, Kutcher ME, Laffan MA, Lamontagne F, Le Gal G, Leeper CM, Leifer ES, Lim G, Lima FG, Linstrum K, Litton E, Lopez-Sendon J, Lopez-Sendon Moreno JL, Lother SA, Malhotra S, Marcos M, Saud Marinez A, Marshall JC, Marten N, Matthay MA, McAuley DF, McDonald EG, McGlothlin A, McGuinness SP, Middeldorp S, Montgomery SK, Moore SC, Morillo Guerrero R, Mouncey PR, Murthy S, Nair GB, Nair R, Nichol AD, Nunez-Garcia B, Pandey A, Park PK, Parke RL, Parker JC, Parnia S, Paul JD, Perez Gonzalez YS, Pompilio M, Prekker ME, Quigley JG, Rost NS, Rowan K, Santos FO, Santos M, Olombrada Santos M, Satterwhite L, Saunders CT, Schutgens REG, Seymour CW, Siegal DM, Silva DG, Jr, Shankar-Hari M, Sheehan JP, Singhal AB, Solvason D, Stanworth SJ, Tritschler T, Turner AM, van Bentum-Puijk W, van de Veerdonk FL, van Diepen S, Vazquez-Grande G, Wahid L, Wareham V, Wells BJ, Widmer RJ, Wilson JG, Yuriditsky E, Zampieri FG, Angus DC, McArthur CJ, Webb SA, Farkouh ME, Hochman JS, Zarychanski R. Therapeutic anticoagulation with heparin in noncritically ill patients with Covid-19. N Engl J Med. 2021;385:790–802. doi: 10.1056/NEJMoa2105911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goligher EC, Bradbury CA, McVerry BJ, Lawler PR, Berger JS, Gong MN, Carrier M, Reynolds HR, Kumar A, Turgeon AF, Kornblith LZ, Kahn SR, Marshall JC, Kim KS, Houston BL, Derde LPG, Cushman M, Tritschler T, Angus DC, Godoy LC, McQuilten Z, Kirwan BA, Farkouh ME, Brooks MM, Lewis RJ, Berry LR, Lorenzi E, Gordon AC, Ahuja T, Al-Beidh F, Annane D, Arabi YM, Aryal D, Baumann Kreuziger L, Beane A, Bhimani Z, Bihari S, Billett HH, Bond L, Bonten M, Brunkhorst F, Buxton M, Buzgau A, Castellucci LA, Chekuri S, Chen JT, Cheng AC, Chkhikvadze T, Coiffard B, Contreras A, Costantini TW, de Brouwer S, Detry MA, Duggal A, Dzavik V, Effron MB, Eng HF, Escobedo J, Estcourt LJ, Everett BM, Fergusson DA, Fitzgerald M, Fowler RA, Froess JD, Fu Z, Galanaud JP, Galen BT, Gandotra S, Girard TD, Goodman AL, Goossens H, Green C, Greenstein YY, Gross PL, Haniffa R, Hegde SM, Hendrickson CM, Higgins AM, Hindenburg AA, Hope AA, Horowitz JM, Horvat CM, Huang DT, Hudock K, Hunt BJ, Husain M, Hyzy RC, Jacobson JR, Jayakumar D, Keller NM, Khan A, Kim Y, Kindzelski A, King AJ, Knudson MM, Kornblith AE, Kutcher ME, Laffan MA, Lamontagne F, Le Gal G, Leeper CM, Leifer ES, Lim G, Gallego Lima F, Linstrum K, Litton E, Lopez-Sendon J, Lother SA, Marten N, Saud Marinez A, Martinez M, Mateos Garcia E, Mavromichalis S, McAuley DF, McDonald EG, McGlothlin A, McGuinness SP, Middeldorp S, Montgomery SK, Mouncey PR, Murthy S, Nair GB, Nair R, Nichol AD, Nicolau JC, Nunez-Garcia B, Park JJ, Park PK, Parke RL, Parker JC, Parnia S, Paul JD, Pompilio M, Quigley JG, Rosenson RS, Rost NS, Rowan K, Santos FO, Santos M, Santos MO, Satterwhite L, Saunders CT, Schreiber J, Schutgens REG, Seymour CW, Siegal DM, Silva DG, Jr, Singhal AB, Slutsky AS, Solvason D, Stanworth SJ, Turner AM, van Bentum-Puijk W, van de Veerdonk FL, van Diepen S, Vazquez-Grande G, Wahid L, Wareham V, Widmer RJ, Wilson JG, Yuriditsky E, Zhong Y, Berry SM, McArthur CJ, Neal MD, Hochman JS, Webb SA, Zarychanski R. Therapeutic Anticoagulation with Heparin in Critically Ill Patients with Covid-19. N Engl J Med. 2021;385:777–789. doi: 10.1056/NEJMoa2103417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arabi YM, Gordon AC, Derde LPG, Nichol AD, Murthy S, Beidh FA, Annane D, Swaidan LA, Beane A, Beasley R, Berry LR, Bhimani Z, Bonten MJM, Bradbury CA, Brunkhorst FM, Buxton M, Buzgau A, Cheng A, De Jong M, Detry MA, Duffy EJ, Estcourt LJ, Fitzgerald M, Fowler R, Girard TD, Goligher EC, Goossens H, Haniffa R, Higgins AM, Hills TE, Horvat CM, Huang DT, King AJ, Lamontagne F, Lawler PR, Lewis R, Linstrum K, Litton E, Lorenzi E, Malakouti S, McAuley DF, McGlothlin A, McGuinness S, McVerry BJ, Montgomery SK, Morpeth SC, Mouncey PR, Orr K, Parke R, Parker JC, Patanwala AE, Rowan KM, Santos MS, Saunders CT, Seymour CW, Shankar-Hari M, Tong SYC, Turgeon AF, Turner AM, Van de Veerdonk FL, Zarychanski R, Green C, Berry S, Marshall JC, McArthur C, Angus DC, Webb SA, Investigators R-C. Lopinavir-ritonavir and hydroxychloroquine for critically ill patients with COVID-19: REMAP-CAP randomized controlled trial. Intensive Care Med. 2021;47:867–886. doi: 10.1007/s00134-021-06448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Investigators R-C, Gordon AC, Mouncey PR, Al-Beidh F, Rowan KM, Nichol AD, Arabi YM, Annane D, Beane A, van Bentum-Puijk W, Berry LR, Bhimani Z, Bonten MJM, Bradbury CA, Brunkhorst FM, Buzgau A, Cheng AC, Detry MA, Duffy EJ, Estcourt LJ, Fitzgerald M, Goossens H, Haniffa R, Higgins AM, Hills TE, Horvat CM, Lamontagne F, Lawler PR, Leavis HL, Linstrum KM, Litton E, Lorenzi E, Marshall JC, Mayr FB, McAuley DF, McGlothlin A, McGuinness SP, McVerry BJ, Montgomery SK, Morpeth SC, Murthy S, Orr K, Parke RL, Parker JC, Patanwala AE, Pettila V, Rademaker E, Santos MS, Saunders CT, Seymour CW, Shankar-Hari M, Sligl WI, Turgeon AF, Turner AM, van de Veerdonk FL, Zarychanski R, Green C, Lewis RJ, Angus DC, McArthur CJ, Berry S, Webb SA, Derde LPG. Interleukin-6 receptor antagonists in critically Ill patients with Covid-19. N Engl J Med. 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ainsworth M, Andersson M, Auckland K, Baillie JK, Barnes E, Beer S, Beveridge A, Bibi S, Blackwell L, Borak M, Bown A, Brooks T, Burgess-Brown NA, Camara S, Catton M, Chau KK, Christott T, Clutterbuck E, Coker J, Cornall RJ, Cox S, Crawford-Jones D, Crook DW, D'Arcangelo S, Dejnirattsai W, Dequaire JMM, Dimitriadis S, Dingle KE, Doherty G, Dold C, Dong T, Dunachie SJ, Ebner D, Emmenegger M, Espinosa A, Eyre DW, Fairhead R, Fassih S, Feehily C, Felle S, Fernandez-Cid A, Fernandez Mendoza M, Foord TH, Fordwoh T, Fox McKee D, Frater J, Gallardo Sanchez V, Gent N, Georgiou D, Groves CJ, Hallis B, Hammond PM, Hatch SB, Harvala HJ, Hill J, Hoosdally SJ, Horsington B, Howarth A, James T, Jeffery K, Jones E, Justice A, Karpe F, Kavanagh J, Kim DS, Kirton R, Klenerman P, Knight JC, Koukouflis L, Kwok A, Leuschner U, Levin R, Linder A, Lockett T, Lumley SF, Marinou S, Marsden BD, Martinez J, Martins Ferreira L, Mason L, Matthews PC, Mentzer AJ, Mobbs A, Mongkolsapaya J, Morrow J, Mukhopadhyay SMM, Neville MJ, Oakley S, Oliveira M, Otter A, Paddon K, Pascoe J, Peng Y, Perez E, Perumal PK, Peto TEA, Pickford H, Ploeg RJ, Pollard AJ, Richardson A, Ritter TG, Roberts DJ, Rodger G, Rollier CS, Rowe C, Rudkin JK, Screaton G, Semple MG, Sienkiewicz A, Silva-Reyes L, Skelly DT, Sobrino Diaz A, Stafford L, Stockdale L, Stoesser N, Street T, Stuart DI, Sweed A, Taylor A, Thraves H, Tsang HP, Verheul MK, Vipond R, Walker TM, Wareing S, Warren Y, Wells C, Wilson C, Withycombe K, Young RK. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. 2020;20:1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei R, Wang J, Jia E, Chen T, Ni Y, Jia W. GSimp: A gibbs sampler based left-censored missing value imputation approach for metabolomics studies. PLoS Comput Biol. 2018;14:e1005973. doi: 10.1371/journal.pcbi.1005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing. In: Editor (eds) Book R: A language and environment for statistical computing. R Foundation for Statistical Computing. City
- 27.Murtagh F, Legendre P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 28.StataCorp. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC; 2017. [Google Scholar]
- 29.Kahan BC, Jairath V, Dore CJ, Morris TP. The risks and rewards of covariate adjustment in randomized trials: an assessment of 12 outcomes from 8 studies. Trials. 2014;15:139. doi: 10.1186/1745-6215-15-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zohar T, Loos C, Fischinger S, Atyeo C, Wang C, Slein MD, Burke J, Yu J, Feldman J, Hauser BM, Caradonna T, Schmidt AG, Cai Y, Streeck H, Ryan ET, Barouch DH, Charles RC, Lauffenburger DA, Alter G. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183(1508–1519):e1512. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalil BA, Elemam NM, Maghazachi AA. Chemokines and chemokine receptors during COVID-19 infection. Comput Struct Biotechnol J. 2021;19:976–988. doi: 10.1016/j.csbj.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu L, Zhang H, Dauphars DJ, He YW. A Potential role of interleukin 10 in COVID-19 pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sposito B, Broggi A, Pandolfi L, Crotta S, Clementi N, Ferrarese R, Sisti S, Criscuolo E, Spreafico R, Long JM, Ambrosi A, Liu E, Frangipane V, Saracino L, Bozzini S, Marongiu L, Facchini FA, Bottazzi A, Fossali T, Colombo R, Clementi M, Tagliabue E, Chou J, Pontiroli AE, Meloni F, Wack A, Mancini N, Zanoni I. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184(4953–4968):e4916. doi: 10.1016/j.cell.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagashima S, Mendes MC, Camargo Martins AP, Borges NH, Godoy TM, Miggiolaro A, da Silva DF, Machado-Souza C, de Noronha L. Endothelial dysfunction and thrombosis in patients with COVID-19-brief report. Arterioscler Thromb Vasc Biol. 2020;40:2404–2407. doi: 10.1161/ATVBAHA.120.314860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smadja DM, Guerin CL, Chocron R, Yatim N, Boussier J, Gendron N, Khider L, Hadjadj J, Goudot G, Debuc B, Juvin P, Hauw-Berlemont C, Augy JL, Peron N, Messas E, Planquette B, Sanchez O, Charbit B, Gaussem P, Duffy D, Terrier B, Mirault T, Diehl JL. Angiopoietin-2 as a marker of endothelial activation is a good predictor factor for intensive care unit admission of COVID-19 patients. Angiogenesis. 2020;23:611–620. doi: 10.1007/s10456-020-09730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junttila IS. Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front Immunol. 2018;9:888. doi: 10.3389/fimmu.2018.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeachy MJ. GM-CSF: the secret weapon in the T(H)17 arsenal. Nat Immunol. 2011;12:521–522. doi: 10.1038/ni.2044. [DOI] [PubMed] [Google Scholar]
- 38.Lang FM, Lee KM, Teijaro JR, Becher B, Hamilton JA. GM-CSF-based treatments in COVID-19: reconciling opposing therapeutic approaches. Nat Rev Immunol. 2020;20:507–514. doi: 10.1038/s41577-020-0357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thwaites RS, Sanchez Sevilla Uruchurtu A, Siggins MK, Liew F, Russell CD, Moore SC, Fairfield C, Carter E, Abrams S, Short CE, Thaventhiran T, Bergstrom E, Gardener Z, Ascough S, Chiu C, Docherty AB, Hunt D, Crow YJ, Solomon T, Taylor GP, Turtle L, Harrison EM, Dunning J, Semple MG, Baillie JK, Openshaw PJ, investigators IC, Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci Immunol. 2021;6:9873. doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Group WHOREAfC-TW. Shankar-Hari M, Vale CL, Godolphin PJ, Fisher D, Higgins JPT, Spiga F, Savovic J, Tierney J, Baron G, Benbenishty JS, Berry LR, Broman N, Cavalcanti AB, Colman R, De Buyser SL, Derde LPG, Domingo P, Omar SF, Fernandez-Cruz A, Feuth T, Garcia F, Garcia-Vicuna R, Gonzalez-Alvaro I, Gordon AC, Haynes R, Hermine O, Horby PW, Horick NK, Kumar K, Lambrecht BN, Landray MJ, Leal L, Lederer DJ, Lorenzi E, Mariette X, Merchante N, Misnan NA, Mohan SV, Nivens MC, Oksi J, Perez-Molina JA, Pizov R, Porcher R, Postma S, Rajasuriar R, Ramanan AV, Ravaud P, Reid PD, Rutgers A, Sancho-Lopez A, Seto TB, Sivapalasingam S, Soin AS, Staplin N, Stone JH, Strohbehn GW, Sunden-Cullberg J, Torre-Cisneros J, Tsai LW, van Hoogstraten H, van Meerten T, Veiga VC, Westerweel PE, Murthy S, Diaz JV, Marshall JC, Sterne JAC. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: a meta-analysis. JAMA. 2021;326:499–518. doi: 10.1001/jama.2021.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 43.Ferrando-Vivas P, Doidge J, Thomas K, Gould DW, Mouncey P, Shankar-Hari M, Young JD, Rowan KM, Harrison DA, Team IC Prognostic factors for 30-day mortality in critically ill patients with coronavirus disease 2019: an observational cohort study. Crit Care Med. 2021;49:102–111. doi: 10.1097/CCM.0000000000004740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, Hirayama AV, Mastroiani F, Turtle CJ, Harhay MO, Legrand M, Deutschman CS. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway Morris A, Kohler K, Shankar-Hari M. ARDS subphenotypes: searching for Rorschach among the roentgenograms? Thorax. 2022;77:2–4. doi: 10.1136/thoraxjnl-2021-217428. [DOI] [PubMed] [Google Scholar]
- 46.Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, Zhou Y, Du L, Li F. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015–e2019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu G, Zhu C, Zhu Y, Sun F. Minireview of progress in the structural study of SARS-CoV-2 proteins. Curr Res Microb Sci. 2020;1:53–61. doi: 10.1016/j.crmicr.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abraham J. Passive antibody therapy in COVID-19. Nat Rev Immunol. 2020;20:401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty S, Gonzalez J, Edwards K, Mallajosyula V, Buzzanco AS, Sherwood R, Buffone C, Kathale N, Providenza S, Xie MM, Andrews JR, Blish CA, Singh U, Dugan H, Wilson PC, Pham TD, Boyd SD, Nadeau KC, Pinsky BA, Zhang S, Memoli MJ, Taubenberger JK, Morales T, Schapiro JM, Tan GS, Jagannathan P, Wang TT. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Wijst MGP, Vazquez SE, Hartoularos GC, Bastard P, Grant T, Bueno R, Lee DS, Greenland JR, Sun Y, Perez R, Ogorodnikov A, Ward A, Mann SA, Lynch KL, Yun C, Havlir DV, Chamie G, Marquez C, Greenhouse B, Lionakis MS, Norris PJ, Dumont LJ, Kelly K, Zhang P, Zhang Q, Gervais A, Le Voyer T, Whatley A, Si Y, Byrne A, Combes AJ, Rao AA, Song YS, Fragiadakis GK, Kangelaris K, Calfee CS, Erle DJ, Hendrickson C, Krummel MF, Woodruff PG, Langelier CR, Casanova JL, Derisi JL, Anderson MS, Ye CJ. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Beziat V, Manry J, Shaw E, Haljasmagi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Le Pen J, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, Haerynck F, Dalmau D, Martinez-Picado J, Brodin P, Nussenzweig MC, Boisson-Dupuis S, Rodriguez-Gallego C, Vogt G, Mogensen TH, Oler AJ, Gu J, Burbelo PD, Cohen JI, Biondi A, Bettini LR, D'Angio M, Bonfanti P, Rossignol P, Mayaux J, Rieux-Laucat F, Husebye ES, Fusco F, Ursini MV, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Castagnoli R, Montagna D, Licari A, Marseglia GL, Duval X, Ghosn J, Lab H, Group N-UIRtC, Clinicians C, Clinicians C-S, Imagine CG, French CCSG, Milieu Interieur C, Co VCC, Amsterdam UMCC-B, Effort CHG, Tsang JS, Goldbach-Mansky R, Kisand K, Lionakis MS, Puel A, Zhang SY, Holland SM, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova JL, (2020) Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370: eabd4585
- 52.Shankar-Hari M, Rubenfeld GD. Population enrichment for critical care trials: phenotypes and differential outcomes. Curr Opin Crit Care. 2019;25:489–497. doi: 10.1097/MCC.0000000000000641. [DOI] [PubMed] [Google Scholar]
- 53.Schett G, McInnes IB, Neurath MF. Reframing immune-mediated inflammatory diseases through signature cytokine hubs. N Engl J Med. 2021;385:628–639. doi: 10.1056/NEJMra1909094. [DOI] [PubMed] [Google Scholar]
- 54.Lee AYS, Korner H. The CCR6-CCL20 axis in humoral immunity and T-B cell immunobiology. Immunobiology. 2019;224:449–454. doi: 10.1016/j.imbio.2019.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.