ABSTRACT
The geographical spread and inter-host transmission of the subgroup J avian leukosis virus (ALV-J) may be the most important issues for epidemiology. An integrated analysis, including phylogenetic trees, homology modeling, evolutionary dynamics, selection analysis and viral transmission, based on the gp85 gene sequences of the 665 worldwide ALV-J isolates during 1988–2020, was performed. A new Clade 3 has been emerging and was evolved from the dominating Clade 1.3 of the Chinese Yellow-chicken, and the loss of a α-helix or β-sheet of the gp85 protein monomer was found by the homology modeling. The rapid evolution found in Clades 1.3 and 3 may be closely associated with the adaption and endemicity of viruses to the Yellow-chickens. The early U.S. strains from Clade 1.1 acted as an important source for the global spread of ALV-J and the earliest introduction into China was closely associated with the imported chicken breeders in the 1990s. The dominant outward migrations of Clades 1.1 and 1.2, respectively, from the Chinese northern White-chickens and layers to the Chinese southern Yellow-chickens, and the dominating migration of Clade 1.3 from the Chinese southern Yellow-chickens to other regions and hosts, indicated that the long-distance movement of these viruses between regions in China was associated with the live chicken trade. Furthermore, Yellow-chickens have been facing the risk of infections of the emerging Clades 2 and 3. Our findings provide new insights for the epidemiology and help to understand the critical factors involved in ALV-J dissemination.
IMPORTANCE Although the general epidemiology of ALV-J is well studied, the ongoing evolutionary and transmission dynamics of the virus remain poorly investigated. The phylogenetic differences and relationship of the clades and subclades were characterized, and the epidemics and factors driving the geographical spread and inter-host transmission of different ALV-J clades were explored for the first time. The results indicated that the earliest ALV-J (Clade 1.1) from the United States, acted as the source for global spreads, and Clades 1.2, 1.3 and 3 were all subsequently evolved. Also the epidemiological investigation showed that the early imported breeders and the inter-region movements of live chickens facilitated the ALV-J dispersal throughout China and highlighted the needs to implement more effective containment measures.
KEYWORDS: Avian leukosis virus subgroup J, clades/subclades, diversification, emergence, geographical spread, gp85 gene, inter-host transmission, live chicken trade
INTRODUCTION
Avian leukosis (AL), caused by avian leukosis virus (ALV), is an important infectious tumor disease affecting poultry. In major developed countries and regions like the United States and Europe, the disease has been well under control by management measures; however, in developing countries like China, the eradication of ALV still remains a difficult task. ALV belongs to the genus Alpharetrovirus and the family Retroviridae (1). ALV contains two identical sense-strand RNA monomers (2), and the arrangement of the proviral DNA of the ALV genome is 5′ LTR-gag/pro-pol-env-LTR 3′, which consists of approximately 7.8 kb nucleotides (nt) (3). Major stages in the virus replication are: synthesis of the minus-strand of viral DNA through reverse transcription of viral RNA by reverse transcriptase; formation on the template of minus-strand DNA of the plus-strands of viral DNA, giving rise to linear DNA duplexes; migration of linear DNA to the cell nucleus; and the linear viral DNA becomes linearly integrated into the host DNA under the influence of the enzyme integrase. Subsequently, the proviral genes are transcribed into viral RNAs, which are translated to produce precursor and mature proteins that constitute the virion (3). ALV could be transmitted horizontally and vertically, as well as through contaminated commercial live vaccines route (4–7). No effective commercial vaccines or drugs are available so far to prevent and control ALV infection. Eradication of the virus in the breeders is the only way to eliminate the virus from chicken flocks. The nationwide eradication program begun to implement in China since 2008 has greatly reduced the disease burdens in the White-chicken and layers that were mainly imported from foreign breeding companies known as the grandparent breeders, but it is still one of the most important infectious diseases in the Yellow-chickens due to the non-eradication and/or incomplete-eradication practice on farms (8–10).
ALV is currently divided into 11 subgroups (A–K) (3, 11, 12). As it is well known, the subgroup J ALV (ALV-J) causes severe economic loss to the poultry industry worldwide (11) as it is the most contagious and pathogenic virus among all the subgroups. ALV-J is responsible for more than 90% of the AL cases (8), which accounts for one of the major clinical tumors in chickens (5, 8). ALV-J, as an oncogenic exogenous retrovirus, is a recombinant virus between the endogenous and exogenous viruses and induces malignant tumors including myelocytoma, hemangioma, and nephroma (3, 13, 14). ALV-J infection also causes severe immunosuppression (5, 15). The pathogenicity of ALV-J is closely related to its env gene (16) and its env gene is distinct from the other ALV subgroups (3). The ENV protein, coded by the env gene is divided into gp85 and gp37 subunits. The gp85 constitutes the surface (SU) portion of the structural protein and is closely associated with the process of viral binding to the cell receptor and the determination of the specificity of subgroup (3, 17). Zhang et al. (18) reported that the bipartite sequence motif, spanning throughout the regions of the 38–131 and 159–283 amino acid (aa) locations of gp85, plays a crucial role in receptor binding and viral entry. The gp85 sequence is usually used for the analysis of the molecular epidemiology (19–21), and it is also the most available sequenced gene of ALV-J in GenBank. Also, the variation of the viruses was more likely to be better found in gp85 gene (9, 22, 23).
ALV-J was first isolated from commercial meat-type chickens in the United Kingdom in 1988 and the isolate HPRS103 is considered as the prototype strain (24). The disease was reported in the United States in the early 1990s (14). In China, ALV-J was first officially recognized in the white-feathered broilers (White-chickens) in 1999 (17). In the first 2–3 years, ALV-J was only found in the offspring of imported White-chicken breeders, which were isolated in Shandong, Henan, Jiangsu, etc. (17, 20). Since 2004, ALV-J has been found in layers (25), which have a high death rate caused by superficial hemangioma and vascular rupture. The layers mainly in the northern China experienced severe outbreaks of ALV-J during 2007–2010 (20, 26, 27). And then the clinical outbreaks in the White-chickens and the layers, especially in the northern China, seemed being disappeared until the reemerging outbreaks of ALV-J in the White-chickens have been taking place in some lines of chickens that seemed to have a direct rink with the imported grandparent breeders since 2018 (28, 29). The Yellow-chickens mainly in the southern China were found to be about 10% positive infection but no obvious clinical disease symptoms during the 2005–2010 surveillance and began to experience significant clinical diseases since 2011 and till now (8, 30–32). Besides, ALV-J has been reported to be isolated in wild birds during 2010–2012 (33) and in gamecocks during 2013–2014 (21), and it has a wide host range (34).
Certain studies have proved the live poultry trade as an important source of viral spread like avian influenza virus (AIV) (35, 36). In this study, live chicken trades include the introduced breeder chickens used for breeding and the sale of slaughter chickens and yang chicks, which were involved in the inter-region movement of live chickens. Those practice may have resulted in the spread of viruses especially favor the viruses like ALV, and subclinically infected chickens with less/no significant signs and symptoms, which could be transmitted vertically to their offspring and/or horizontally to other chickens. This situation could happen frequently, especially in the chicken flocks that have not been completely eradicated and/or quarantined to clean out of the ALV. This situation may also make it difficult for veterinarians to detect the existence of ALV during routine inspections. Here, the slaughter chickens refer to birds that are sold to the market as a commodity, including White-chickens, Yellow-chickens, 817 broilers (a crossbred meat-type chicken derived from commercial layer hens and meat-type cocks), and used layers.
In the past 20 years in China, production of White-chickens and layers has mainly been practiced by import of breeders from the United States and Europe countries, whereas the Yellow-chickens have been produced by independent breeding farms in the country. Since 2000, Shandong province in East China has rapidly become a major producer of White-chickens and its broilers’ slaughter volume has always ranked number one, with approximately 2.5 billion in 2020. According to poultry industry statistics in 2020, the slaughter volumes of White-chickens and Yellow-chickens were about 5 billion for each. The development and change of the chicken industry driven mainly by human consumption of commercial chicken meat and eggs, and the economic profits of the industry. The chicken sector has already been significantly expanded in the last 50 years in China, as a result of the short supply of chicken meat due to the outbreaks of the H5N6 and H7N9 highly pathogenic avian influenza viruses (HPAIVs) and the short supply of pork due to the outbreaks of African swine fever (ASF) during 2016–2018 (37–39) (Fig. S1A in the supplemental material). According to the industry statistics in 2020, the top regions of White-chickens’ slaughter volume are distributed in Eastern China (provinces Shandong, Jiangsu and Fujian), Northern China (province Hebei), and Northeastern China (province Liaoning), which accounted for about 80% of the output; the top regions of Yellow-chickens’ slaughter volume are distributed in Southern China (provinces Guangxi and Guangdong), Southwestern China (provinces Sichuan and Yunnan), Central China (provinces Hunan and Hubei), and Eastern China (provinces Jiangsu and Jiangxi), which accounted for about 90% of the output (Fig. S1B and 1C). Guangxi and Guangdong are the major provinces in the production of Yellow-chickens in China (10), accounted for about 40% of the total 5 billion in 2020. And Guangxi also produces about 1.6 billion Yellow-chicken commercial chicks each year and ranks number one in China. About 20% of White-chickens and 85% of Yellow-chickens were sold as “live birds.” It is worth noting that the slaughtering of the used breeder hens and layer hens, which were sold as live birds, are transported from the northern to the southern regions (as much as hundreds of millions per year). This flow of live chickens likely caused the viruses to transmit in the northern regions and entered the southern chicken flocks.
In this study, we employed a dynamics analysis using the most comprehensive data sets publicly available to characterize the evolutionary history and the relationship of viruses of different clades and subclades classified, and to explore the risk factors associated with the inter-region spatial diffusion and the inter-host transmission of the endemic field isolates of ALV-J. We aimed to evaluate the important aspects of the evolution and epidemiology of ALV-J, such as the viral transmission in association with the live bird trading events that may have played an important role in the adaption and endemicity of ALV-J to the Yellow-chickens in China.
RESULTS
Geographic and host characteristics.
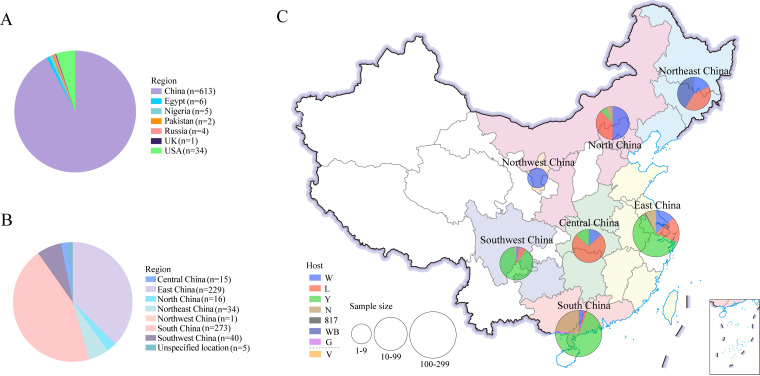
The 665 viruses were categorized by the patterns of sampled geographic sources and hosts. The 665 viruses isolated from samples collected during 1988–2020 were from seven countries: United Kingdom (1), United States (34), China (613), Russia (4), Nigeria (5), Pakistan (2), and Egypt (6) (Fig. 1A). Of the 613 viruses isolated from samples collected in China during 1999–2020, 15 were originated from Central China, 229 were from East China, 16 were from North China, 34 were from Northeast China, 1 was from Northwest China, 273 were from South China, 40 were from Southwest China, and 5 were from unspecified locations (Fig. 1B). The 15 viruses collected in Central China were originated from White-chickens (2), layers (11), and Yellow-chickens (2). The 229 viruses collected in Central China were originated from White-chickens (30), layers (42), Yellow-chickens (137), gamecock (1), 817 broilers (2), and unrecorded breeds of chickens (17). The 16 viruses collected in North China were originated from White-chickens (8), layers (6), Yellow-chickens (1), and unrecorded breeds (1). The 34 viruses collected in Northeast China were originated from White-chickens (6), layers (14), and wild birds (14). The one virus collected in Northwest China was originated from White-chickens (1). The 273 viruses collected in South China were originated from White-chickens (11), layers (5), Yellow-chickens (189), gamecock (5), and unrecorded breeds of chickens (62). The 40 viruses collected in Southwest China were originated from White-chickens (1), layers (3), Yellow-chickens (35), and unrecorded breeds (1) (Fig. 1C). These sequences are globally distributed and represent the most comprehensive ALV-J data currently available.
FIG 1.
Geographic region and host distribution of ALV-J viruses during 1988–2020. Geographic region distribution of ALV-J virus in the world (A) and in China (B). Host distribution in 7 different regions of China (C). W: White-chickens; L: layers; Y: Yellow-chickens; N: unrecorded breeds; 817: a crossbred meat-type chicken; WB: wild birds; G: gamecock; V: contaminated live vaccines.
Phylogenetic clusters.
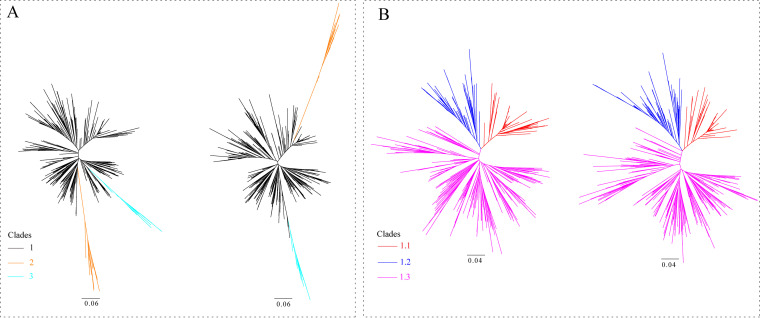
Clades 1 and 2 had 617 and 25 isolates, respectively (Fig. 2 and Table S1 in the supplemental material). The phylogenetic trees of Clade 1 were constructed under the IQ-TREE and RAxML methods, respectively. The former includes 183 isolates of Clade 1.1, 95 isolates of Clade 1.2, and 339 isolates of Clade 1.3; the latter includes 180 isolates of Clade 1.1, 106 isolates of Clade 1.2, and 331 isolates of Clade 1.3 (Fig. 2 and Table S1). Clade 1.3 is the dominant subclade of Clade 1. A new clade, Clade 3, has been defined for the first time (Fig. 2 and Table S2) based on the genetic distance that is >10% between the clades of viruses and has a common ancestor with the strain GD18HZ11 of Clade1.3. The phylogenetic trees illustrated that the Clade 3 was derived from the Clade 1.3. The Clade 3 viruses that were just newly emerging and were collected from Yellow-chickens during the years 2017–2019 in China. The strains GD17ZQ08 and JX19DX4 were selected to construct the Pilot tree. Fifty new isolates from our group all fell into the Clade 1, including 22 isolates to Clade 1.1, one isolate to Clade 1.2, and 27 isolates to Clade 1.3.
FIG 2.
Phylogenetic analysis was performed based on the nucleotide sequences of ALV-J gp85 gene. (A) Two radial trees were constructed based on the gp85 sequences (n = 572) to identify the first-order clades with IQ-TREE (right) and RAxML (left) software, respectively. (B) Two radial trees were constructed based on the gp85 sequences (n = 530) to identify the second-order clades of Clade 1 with IQ-TREE (right) and RAxML (left) software, respectively. The trees were drawn to scale, with branch lengths measured in the number of substitutions per site.
Three-dimensional structure and aa-site mutations in gp85.
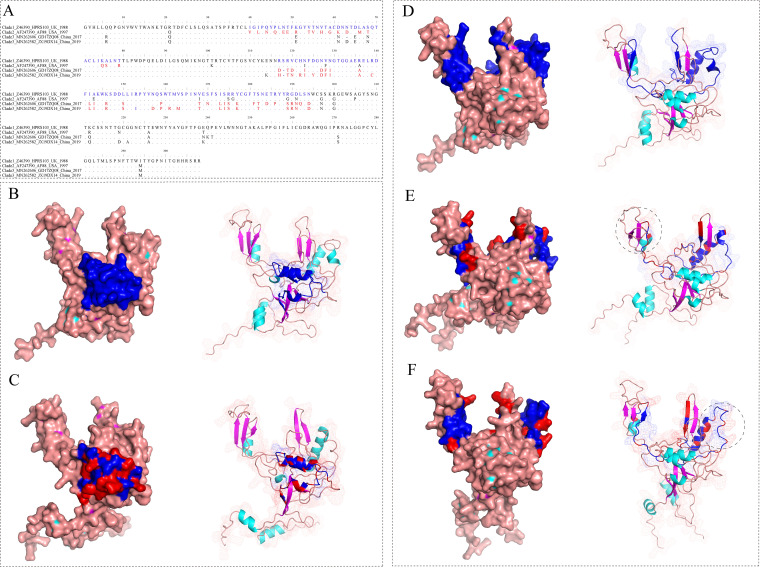
Compared with the gp85 of Clade 1, Clade 2 mainly differed at the aa-residuals 40–79 (calculated based on the aa-sequence of HPRS103 reference strain). Clade 1 and Clade 2 did share 3 similar α-helix structures at residuals 40–79. We found that although some aa-residues were substituted, the number of the secondary structure elements remains unchanged. The domain showed considerable conservative (Fig. 3A–C). Compared with the gp85 of Clade 1, Clade 3 mainly differed at residuals 117–194 and exhibited the loss of the secondary structure element, a α-helix or β-sheet, due to the aa substitutions (Fig. 3A, D-F). In addition, Clade 1.1 is mainly characterized by the residual 61D in a novel hypervariable region; Clade 1.2 is mainly characterized by the residual 61Q in a novel hypervariable region, and an aa-deletion among the residuals 112YKENNRSRV120 in the hr1 region; Clade 1.3 is mainly characterized by an aa-deletion both at residual 61 in a novel hypervariable region and among the residuals 112YKENNRSRV120 in the hr1 region, respectively. The different aa sites of the prevalent and dominant Clade 1.3 from Yellow-chickens in 2010–2018 and newly generated Clade 3 from Yellow-chickens in 2017–2019 were also summarized (Fig. S2 in the supplemental material).
FIG 3.
Display the differences on the amino acid (aa) residuals of the gp85 protein monomer in different Clades by three-dimensional structure. (A) Display of aa-mutation residuals at position 40–79 of Clades 1 and 2, and at position 117–194 of Clades 1 and 3, respectively; (B) Three-dimensional structure of strain HPRS103 gp85 protein from Clade 1; (C) Three-dimensional structure of gp85 protein of the strain AF88 from Clade 2. (D) Three-dimensional structure of strain HPRS103 from Clade 1; (E) Three-dimensional structure of strain GD17ZQ08 from Clade 3; (F) Three-dimensional structure of strain JX19DX14 from Clade 3.
Estimations of evolutionary rate, time origin and population dynamics.
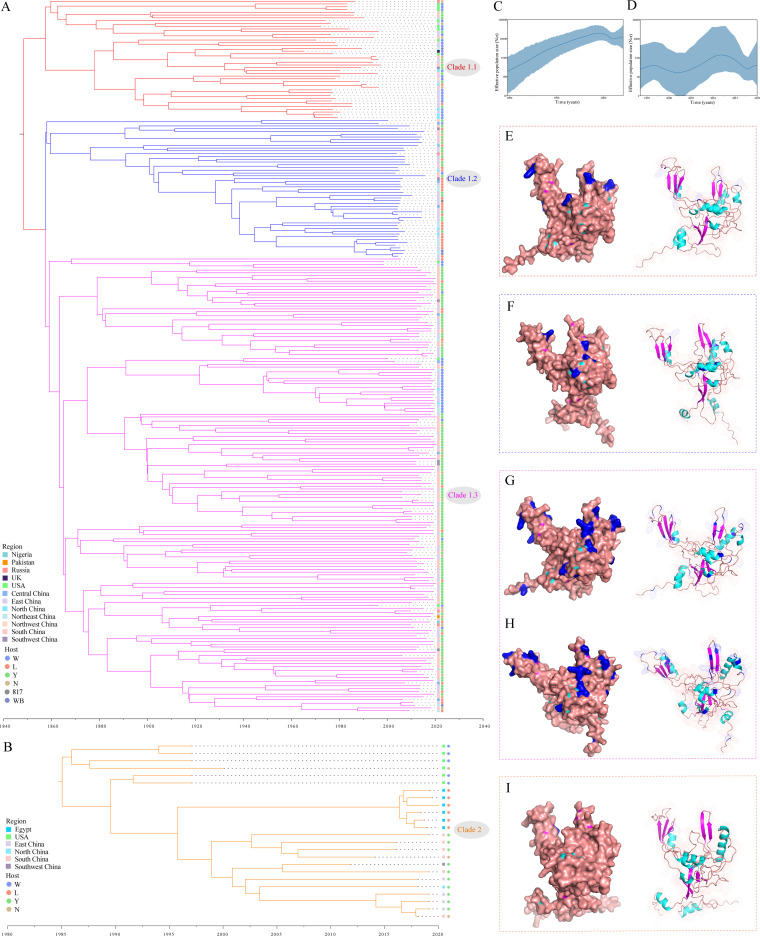
A root-to-tip regression analysis of genetic divergence and sampling date using the best-fitting root showed that the 2 data sets (257 isolates of Clade 1 and 24 isolates of Clade 2) exhibited a positive correlation between the genetic divergences and the sampling times, which ranged from slightly strong to relatively strong (Fig. S3 in the supplemental material). The fitting of the uncorrected lognormal relaxed molecular clock model and Bayesian Skygrid coalescent model were finally selected. For Clade 1, the estimated evolutionary rate and the time to the most recent common ancestor (tMRCA) of the gp85 gene sequences (n = 257) were 7.57 × 10−4 substitutions/site/year (95% highest posterior density [HPD]: 5.95 × 10−4 – 9.54 × 10−4) and 1,859.8 (95% HPD interval: 1818.1–1897.3), respectively. Upon estimation of the MCC tree (Fig. 4A), the Clade 1 isolates were divided into three subclades, which was in agreement with a report on the previously identified second-order clades (19). For Clade 2, the estimated evolutionary rate and tMRCA of the gp85 gene sequences (n = 24) were 2.87 × 10−3 substitutions/site/year (95% HPD: 1.79 × 10−3 – 4.12 × 10−3) and 1985 (95% HPD: 1,974.5–1,992.4), respectively. A Bayesian Skygrid model was used to further explore the changes in genetic diversity of the populations of these data sets mentioned above (that reflects the changes in effective population size (Ne) over time). Ne of Clades 1 (n = 257) showed an obvious period, that is, a relatively high level since 2005 (Fig. 4A and C). This also revealed that the population dynamics actually reflected the increased genetic diversity of the Yellow-chickens derived isolates (127/257, 49.4%). The estimated Ne of Clade 2 showed that the phase from the beginning to present has remained relatively stable (Fig. 4B and D).
FIG 4.
Evolution of ALV-J viruses and structural mapping of the positively selected sites on the gp85 molecule. (A) Bayesian phylogenetic analysis of the 257 sequences of gp85 in Clade 1 during 1988–2020; (B) Bayesian phylogenetic analysis of the 24 sequences of gp85 in Clade 2 during 1997–2020; Bayesian Skygrid demographic reconstruction of Clades 1 (C) and 2 (D), respectively. The vertical axis shows the effective number of infections (Ne) multiplied by mean viral generation time (τ). The solid line and shaded region represent the median and 95% credibility interval, respectively, of the inferred Neτ through time. Three-dimensional structures of the gp85 of strains HPRS103 (E), JS09GY2 (F), GD14J2 (G), GX10GL08 (H), and AF88 (I). Mapping of positively selected aa-sites identified onto the three-dimensional structure of the gp85, and their locations in the three-dimensional structure are indicated with blue (E-I).
High numbers of positively selected sites detected in Clade 1 viruses.
Some positively selected sites in different clades were detected by all the four methods (mixed effects model of evolution (MEME), fixed effects likelihood (FEL), fast, unconstrained Bayesian approximation (FUBAR), and single-likelihood ancestor counting (SLAC)) used (Table 1 and Fig. 4E–I). Clade 1.1 had 5 positive selection sites; Clade 1.2 had 7 positive selection sites; Clade 1.3 had 27 positive selection sites. A rapid increase in diversity was confirmed in Clade 1, with Clade 1.3 having the highest diversity, whereas only one selection site was detected in Clade 2.
TABLE 1.
Detecting positively selected sites (amino acid residuals) by codon substitution models for gp85 of ALV-J using four methodsa
| Clades | No. of positive sites | Locations of positive sites |
|---|---|---|
| 1.1 | 5 | 64, 150, 200, 212, 239 |
| 1.2 | 7 | 61, 75, 143, 168, 189, 197, 241 |
| 1.3 | 27 | 5, 21, 44, 49, 75, 76, 113, 114, 120, 143, 148, 150, 156, 168, 189, 190, 192, 197, 202, 212, 216, 219, 238, 239, 240, 271, 304 |
| 2 | 1 | 192 |
Only visualize P-values <0.05 and pp > 0.95 and their close values. Strain HPRS103 was used as the reference strain for aa-sequence alignment and to determine the position of the positive selection sites.
Region and host ancestral reconstruction of Clades 1 and 2.
Phylogeographic analysis indicated there were two strongly supported migration routes with Bayes factor (BF) >10 found for the Clade 1.1 viruses that primarily spread from the United States to the United Kingdom and to China (Fig. 5A). This was further supported by the numbers of the observed region state changes (that is, the number of geographical locations transition/year). The Bayes factor of migration from the United States to East China (mainly Shandong) was BF > 10 and those to other regions of China were BF < 3 (Fig. 5B). State counts showed that high numbers of outward and inward migrations took place in East China, indicating that East China acted as a primary source of migrations within China. Host analysis indicated that there were three migration pathways supported the transmission from White-chickens to Yellow-chickens, wild birds, and unrecorded breeds with BF > 3 (Fig. 5C). State counts showed migration out of White-chickens to all other hosts and the high numbers of inward migration for Yellow-chickens (Fig. 5D). The MCC tree indicated that White-chickens from the USA and East China played a key role in seeding the early virus epidemics (Fig. S4A in the supplemental material). Notably, the MCC tree showed that the early isolates of the United States were at the root of the tree and certain early U.S. strains (i.e., ADOL-Hcl) appeared earlier than the prototype strain HPRS103 from the U.K. and the Chinese strains. The MRCA estimates that the first U.K.-reported isolated ALV-J prototype strain HPRS103 is branched off from a fully U.S.-located backbone of the phylogenetic tree (pp = 0.87).
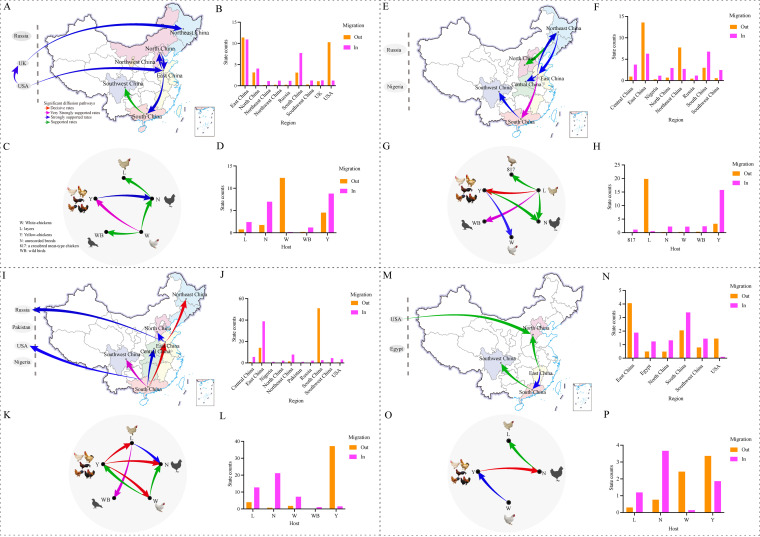
FIG 5.
Spatial diffusion and host transmission of the ALV-J Clades 1.1, 1.2, 1.3 and 2 viruses. Spatial diffusion pathways and histograms of total number of state transitions (A, B, E, F, I, J, M and N); Host transmission pathways and histograms of total number of state transitions (C, D, G, H, K, L, O and P). Green arrows, supported rates with 3 ≤ BF < 10; blue arrows, strongly supported rates with 10 ≤ BF < 100; purple arrows, very strongly supported rates with 100 ≤ BF < 1,000 and red arrows, decisive rates with BF ≥ 1,000. W: White-chickens; L: layers; Y: Yellow-chickens; WB: wild bird; 817: a crossbred meat-type chicken; N: unrecorded breeds.
Phylogeographic analysis of the Clade 1.2 indicated that there were 5 strongly supported migration pathways with BF > 10 in China, of which, at least 3 were outward from East China (Fig. 5E). East China acted as a major source of the diffusion and an infection center. State counts showed the migration out from East China to other regions, which supports the conclusion that East China may have acted as the transmission reservoir (Fig. 5F). Host analysis indicated that there were four migration pathways supported the transmission from layers to the other hosts (BF > 3). The transmission route to the Yellow-chickens was a decisive support (BF > 1,000) (Fig. 5G). State counts reflected this dynamic with dominating outward migration from layers and with dominating inward migration to Yellow-chickens (Fig. 5H). The MCC tree also supported that the layers of East China had acted as the transmission reservoir (Fig. S4B in the supplemental material).
Three very strong supported migration pathways (BF > 100) suggested there were two regions (South and East China) that served as the important sources for Clade 1.3 in China (Fig. 5I). This was further supported by the numbers of the observed state changes with outward migrations from South and East China, whereas four regions (East China, Northeast China, Central China and Southwest China) showed high numbers of inward migrations (Fig. 5J). Host analysis indicates three migration pathways with decisive support (BF > 1,000) that have mediated transmission from the Yellow-chickens to the other breeds (Fig. 5K). State counts also reflected this dynamic with dominating outward migration from the Yellow-chickens (Fig. 5L). The MCC tree supported that the Yellow-chickens in South China have become the epicenter for the dissemination of Clade 1.3 (Fig. S4C in the supplemental material).
The Clade 2 virus spread from the United States to other parts of the world was confirmed, and the Chinese isolates further spread from East China to other regions (Fig. 5M). State counts reflected this dynamic with dominating outward migrations from the USA and East China (Fig. 5N). Host dynamics analysis found 2 migration pathways with strong support (BF > 10) from White-chickens to Yellow-chickens, and from Yellow-chickens to chickens of unrecorded breeds (Fig. 5O). States counts also reflected the dynamic with the outward migration from White-chickens, the more or less equal numbers of the outward and inward migrations from the Yellow-chicken, and the inward migrations from layers (Fig. 5P). The MCC tree also supported that White-chickens from the United States acted as the source of viral dissemination for the Clade 2 (Fig. S4D).
DISCUSSION
Phylodynamic methods have not yet been used for the epidemiological investigation of ALV-J. However, it has been widely used to analyze the evolution and spatial spreading of other viruses such as SARS-CoV-2 (40), HIV (41), and influenza virus (42). It is well known that China has the largest free-range Yellow-chicken production (5 billion birds each year) and the most frequent inter-provincial live chicken trade in the world, which might be in favor of the transmission and the evolution of viruses including ALV. Through comprehensive analysis of the gp85 gene sequences from a total of 665 ALV-J global isolates collected during the past 3 decades, the geographical spread and inter-host transmission of the ALV-J field strains were tracked and the findings would be useful for advancing the understanding of the epidemiology of this important virus, and further for improving the prevention and better control of the diseases caused by ALV-J in China.
The phylogenetic trees showed the ALV-J strains fell into five clades/subclades (Clades 1.1, 1.2, 1.3 of Clade 1, 2 and 3). Only 8.5% (45/530) of the strains in the phylogenetic reconstruction were found to be inconsistent in classification under the IQ-TREE and RAxML methods (Table S1 in the supplemental material), so the phylogenetic analysis is largely reliable, and the results were also consistent with the previously reported results (19). Clade 1.1 includes the isolates originated from various hosts sources; Clade 1.2 was mainly a group of strains originated from the layers; Clade 1.3 dominated by the Chinese strains was originated from the Yellow-chickens. The present study found that isolates were closely related to different host backgrounds, which was consistent with the results of previous studies (9, 21). The major key sites (61 and 112–120) of the dominating Clade 1.3 of the Chinese Yellow-chicken are summarized, of which 61 is located on the α-helix at position 57VTACD61, and the other is located on the two β-sheet at 110VCYKE114 and 117RSRVCH122, so the deletion of these two sites may play an important role in the adaption and endemicity of Clade 1.3 viruses to the Yellow-chickens. Also, the Bayesian inference of Clade 1 suggested that the earliest ALV-J (Clade 1.1) from the USA acted as the source for global spreads, and the Clades 1.2 and 1.3 were all subsequently evolved. As a matter of fact, although the inter-regional commercial live poultry trades are commonly practices in the White-chickens, layers, and Yellow-chickens, they belong to different breeds and are often produced by different companies. It is almost impossible to raise all those on a single farm. This made the tracking and analysis of correlation almost impossible (except the 817 breed). Thus, it is speculated that after the spread of Clade 1.1 virus from the White-chickens to the layers, virus was prevalent in layers for years, resulting in some site mutations and evolved into Clade 1.2, and then better adapted and led to endemicity in layers. By the same token, it is speculated that after the spread of Clade 1.1 virus from the White-chickens to the Yellow-chickens, the virus was prevalent in Yellow-chickens for years, resulting in some site mutations and evolved into Clade 1.3, and then better adapted and led to endemicity in Yellow-chickens.
Chinese Yellow-chickens have been facing the emerging Clades 2 and 3 viruses. In the homology modeling on the gp85 protein, the differences between Clades 2 or 3 and Clade 1 were explained. Although some aa-mutations (aa 40–79) occurred in Clade 2, the secondary structure of the domain showed considerable conservative, those aa-mutations do not significantly affect the receptor binding ability and the entry activity of the viruses. However, some aa-mutations (aa 117–194) found in the new Clade 3 isolates from the Yellow-chickens (Fig. 3D to G) caused differences in the secondary structure elements. The emerging Clade 3, first defined in this study, was evolved from the Clade 1.3 strains of the Yellow-chicken. It is speculated that Clade 1.3 was prevalent in Yellow-chickens for years, resulting in some site mutations and got better adapted and led to endemicity in Yellow-chickens and finally evolved to Clade 3. Over the past years, the live bird trade and the trade of related chicken products have been subjected to drastic changes. The Yellow-chickens industry has grown rapidly, and the slaughter volume of Yellow-chickens has exceeded that of White-chickens in 2019–2020, which fueled the trade of live Yellow-chickens in China. Specifically, after the outbreaks of H5N6 and H7N9 HPAIVs during 2016–2017 (37, 38), the meat price of Yellow-chickens was increased from a low CNY 10/kg to a much higher CNY 17.6/kg between July to November in 2017 and the meat price of Yellow-chickens had always been stable at around CNY 14–16/kg in the following 2 years (Fig. S1A in the supplemental material). Furthermore, the outbreaks of ASF began at the end of 2018 (39), resulting in a severe short supply of pork. These events boosted the growing of the Yellow-chicken production from 3.6 billion in 2017 to 4.3 billion birds in 2018, and to 5 billion in 2019–2020. Also, the chicken trading, including the live bird trading and especially the Yellow-chicken trading, was subsequently expanded from southern China to other regions, which might have favored the spread of the viruses through the inter-regional live bird movement. This reveals that the spreads of Clade 1.3 and 3 viruses from the Yellow-chickens were closely associated with the live chicken trade. In addition, population dynamics also reflected the high genetic diversity of Clade 1.3 viruses. Selection pressure in the Clade 1.3 viruses involved in 27 positive selection sites (aa substitutions) in the gp85 gene were statistically significant (P < 0.05 and pp > 0.95). It may result in the viral evasion from host immunity response through the continuous and rapid mutations to better adapt to and efficiently transmit in a new environment. The current Clade 1.3 isolates mainly contain one aa-deletion at each of the residue 61 and at the residues 112–120 positions. It is speculated that there was a co-evolution event took place in the Clade 1.3 strains at the two positions, which was different from the layer isolates of Clade 1.2 and the White-chickens isolates of Clade 1.1. These results revealed that the rapid evolution in Clades 1.3 and 3 may be in favor of better adaption and endemicity of viruses into the Yellow-chickens.
The tMRCA of Clade 1 indicated that this clade’s isolates originated in the 1800s. The U.S. breeder chickens, advanced in technology, have been exported to other countries especially to Europe and Asia since the mid-1900s. The live bird trade serves as an ideal way for the circulation of different clades/subclades viruses between different regions or breeds. The viral transmission could have happened along the trading routes. Our study reconstructed the transmission dynamics of Clade 1.1 that strongly supported the migration routes with BF > 10 found that the viruses of this clade were primarily transmitted from the United States to the United Kingdom and to China. The MRCA demonstrated that the early strains from the United States were positioned at the root of the tree, and certain early U.S. strains (i.e., ADOL-Hcl appeared earlier than the prototype strain HPRS103 from the United Kingdom). The prototype strain HPRS103 identified in the United Kingdom is a branch of the fully U.S.-located backbone of the phylogenetic tree. This evidence showed that the Clade 1.1 viruses infected breeders in the United States acted as an important source for the global spreads. This suggests that the virus may have been circulated, but undetected, in the United States before the pandemic took place. By the 1990s, these viruses had been spread to China (20). At that time, an early case of ALV-J infection in China was found in the imported White-chicken breeders and believed that early Chinese strains might be related to the imported breeders from the United States (17). The Chinese Clade 1.1 viruses appeared in the White-chicken in East China first, and then the White-chicken of East China acted as the source of diffusion in China.
Clade 1.2 was a subclade that most of strains originated from layers and a small group of strains that newly emerged in the Yellow-chickens (in Guangxi, Guangdong, and Jiangxi). The transmission dynamics of Clade 1.2 reconstructed in the study indicated that the layers of East China have acted as a hub for dispersal to the provinces that are relatively close geographically and also to the southern China. A strong correlation was found between the ALV-J outbreaks in layers in 2007–2009 and the trade activity during these years. From 2007 to 2009, a conservative estimate of the layer losses due to ALV-J outbreaks was around 50 to 60 million (20, 27). However, there were more than 555,800 sets of imported egg-type grandparent breeder and self-bred grandparent breeder nationwide in 2008, a net increase of 181,900 sets over 2007 with a growth rate of 48.65% (43). Among them, the number of egg-type grandparent breeder in the northern regions of China accounted for more than 80% of the total number of egg-type grandparent breeder. The layers in the northern regions accounted for the majority of the total 1.2 billion/year in China and some of the used layers were sold to southern China as slaughtered chickens (44). In addition, one of our isolates, GX20LZ03J, was identified on a farm where the layer hens were used in the crossbreeding with the broiler males for the commercial broilers, and it belongs to the Clade 1.2. At least 2.5 billion chickens were produced by using the commercial layer hens crossbred with the cocks of the meat-type chicken in 2020. This type of breeding practice that quickly provided meat production has resulted in a relatively high risk of virus transmission in this sector. These evidenced data revealed that likely there were more viral movements from northern layers to the southern Yellow-chickens.
Most importantly, our study reconstructed the transmission dynamics of Clade 1.3 (Fig. 5I–L), which suggested that there was a viral spread with very strong support from South China to 3 other regions in China (BF > 100) and a highly likely viral movement from Yellow-chicken to other breeds. The Yellow-chicken in South China was a major source of the diffusion and infection center of this clade of viruses. Interestingly, these virus spreads were closely related to the live chicken trade. Guangxi and Guangdong provinces in southern China have been the top producers of the Yellow-chicken, producing about 40% of total Yellow-chickens in China (10). A historical event, after the outbreak of H5N6 and H7N9 HPAIVs during 2016–2017, the meat prices of Yellow-chickens were nearly doubled in the second half of 2017. Yellow-chickens industry has grown rapidly, and the volume of slaughtered Yellow-chickens had exceeded that of White-chickens during 2018–2020. Yellow-chickens are mainly sold as live birds, accounting for more than 85% of the total. South China (e.g., Guangxi and Guangdong) is also the main region for Yellow-chickens breeding, production and consumption. For decades, more than 60 new varieties of Yellow-chickens (matching lines) have been approved and produced by a large number of breeding companies of different scales in China (10). According to reports, in 2018–2020, a large number of the dominant Clade 1.3 strains were isolated from the Yellow-chickens in South China (19, 45). This evidence suggests that live Yellow-chickens trade in South China has driven the transmission and endemicity of Clade 1.3 ALV-J. At the same time, the production of Yellow-chickens in East China (e.g., Jiangxi) are also expanding rapidly, where many Clade 1.3 viruses have been isolated (46). The prevention and control of ALV during this period is also complicated. The closure of live chicken markets alone was unlikely to eliminate the virus threat. Therefore, the ALV eradication program should be further strengthened in the Yellow-chicken breeding companies. In the perspective of live chicken trading, spatially structured spread of ALV-J presented. Therefore, additional interventions should be jointly implemented to restrict viral expansion along trade paths. A strict supervision by government departments is essential to establish a safer trade management system for live chicken trading. Thus, the risk of ALV-J transmission from the Yellow-chicken flocks in South China to other flocks in the other regions might be effectively mitigated. In addition, most breeder companies also conduct detection to eliminate possible exogenous viruses contaminated in the live vaccines. Notably, the emergence of the new Chinese ALV-J strains that caused the outbreaks in the White-chicken during 2018–2020 was estimated at 2014 (95% HPD interval: 2013–2015). Those ALV-J strains share the closest genetic relationship with a Guangdong isolate, GD14J2, which originated from the White-chickens imported from abroad (29, 47). A broiler breeder strain, WA1112, identified in Guangdong in 2012 also belongs to this same source, indicating that the viruses might have been circulated in Guangdong for years before the outbreak in 2018–2020 took place in the northern and these viruses most likely favored the adaptation into Yellow-chickens in Guangdong between 2013 and 2015. However, certain limitations of the data and analysis should be considered when interpreting our findings, such as the number of samples with its inhomogeneous in different years. Therefore, follow-up studies should focus on the phylodynamic analysis of ALV-J using the available whole genome or other genes besides gp85, which could contribute to a better understanding of the spread of ALV-J.
In conclusion, phylogenetic analysis showed that there is an emerging Clade 3, which is defined for the first time. The viruses of this Clade 3 were specifically evolved from the dominating Clade 1.3 strains of the Yellow-chickens after the epidemic of these isolates for years. Compared with the gp85 proteins of the Clade 1, the main differences of Clade 3 were found at residuals 117–194 with deletion of a α-helix or β-sheet resulted from aa substitutions. The rapid evolution found in Clades 1.3 and 3 might be closely associated with the adaption and endemicity of viruses to the Yellow-chickens. The early U.S. strains from Clade 1.1 acted as an important source for the global spread of ALV-J, and its earliest introduction into China was closely associated with the imported chicken breeders in the 1990s. The dominant outward migrations of the Clades 1.1 and the Clades 1.2 from the northern White-Chickens and the northern layers to the southern Yellow-chickens, and the dominating migration of the Clade 1.3 from the southern Yellow-chickens to other regions and hosts, respectively, indicated that the long-distance movement of these viruses between regions in China was associated with the live chicken trade. In addition, Yellow-chickens have been facing the risk of infections of the emerging Clades 2 and 3. Strengthening of the epidemiological surveillance and eradication measurements against ALV-J in the Yellow-chicken in South and East China should be crucial and necessary for better ALV-J control in the poultry industry from now on.
MATERIALS AND METHODS
Virus sequences.
The gp85 gene sequences from a total of 665 ALV-J isolates were initially used in this study. Those sequences were downloaded from the GenBank submitted before May 2021 and from our laboratory (Table S1 in the supplemental material). From the GenBank, 506 sequences with known sampling information (i.e., time, geographic source and host) were retrieved, which include 454 sequences from China sampled during 1999–2020; 52 sequences from other countries sampled during 1988–2019; another 159 sequences from our laboratory including 109 sequences sampled during 2010–2020 (6, 8, 9, 19, 21, 48); and 50 newly generated sequences collected during 2016–2020 (GenBank accession numbers: MZ393151-MZ393200).
Classification analysis.
The 665 sequences were aligned using MAFFT v7.475 (49) and MEGA 7 (50) and adjusted manually in BioEdit v7.2.5 (51). Duplicated gene sequences were automatically identified and excluded (93/665) by BioEdit v7.2.5 (Table S3 in the supplemental material). Low-quality sequences were removed by filtering (i.e., at least 900 nt in length). Following the classification method previously described by our group (19), Maximum likelihood (ML) trees of the first-order clades (n = 572) and second-order clades of Clade 1 (n = 530) were constructed using IQ-TREE (52) and RAxML software through the CIPRES Science Gateway V.3.3 (53). All trees were visualized in FigTree v.l.4.3 (https://github.com/rambaut/figtree/releases). In addition, the different clades were used to estimate the mean inter-clade distances using MEGA7.
Predictive structural modeling.
To study the specific aa-mutation sites of gp85 in different clades (Clades 1, 2, and 3) that took place during years 1988–2020, the representative gp85 of ALV-J were selected according to the results of phylogenetic classification. We used machine learning to display a three-dimensional model of the virus protein monomer to gain a better understanding of this gp85 subunit. AlphaFold2 was used to predict the three-dimensional structure of the protein monomer (54) and the model with the highest predicted local distance difference test value among five models was selected. To visualize the locations of aa-mutation sites identified, the identified aa-residue was mapped onto a three-dimensional structure of gp85 using PyMOL v 2.5 (https://pymol.org/2/).
Evolutionary dynamics inferences.
Based on the classification results, Clades 1 and 2 data sets were obtained independently. To determine the topological structure for Clades 1 and 2, the clock signal using the ML trees was investigated with Tempest v1.5.3 (55). Careful filtering was conducted to remove identical/near-identical sequences from the same source and/or case and the sequences that were highly consistent with the endogenous ALV ev/J, as well as the sequences showing evidence of recombination events based on recombination analysis using RDP4 (56). The numbers of sequences were appropriately standardized to minimize the sampling bias and to increase the interpretability of the results after the analyses. The remaining sequences were used as the final two data sets (Clade 1, n = 257; Clade 2, n = 24) for the temporal dynamics analysis, respectively.
To explore the evolutionary history of ALV-J Clades 1 and 2, a Bayesian phylogenetic approach was taken in estimating the rate of evolution, the tMRCA, and the population dynamics history. Bayesian inference through a Markov chain Monte Carlo (MCMC) framework was implemented in BEAST v1.10.4 (57, 58) using the CIPRES Science Gateway V.3.3. The fitting nucleotide-substitution model for two data sets was selected using the IQ-TREE software under Bayesian information criterion (BIC). The combination of two molecular clock models (the strict molecular clock model and the uncorrected lognormal relaxed molecular clock model) and four population models (constant size coalescent, exponential growth coalescent, Bayesian skyline coalescent and Bayesian Skygrid coalescent) were compared (59–61). Two independent MCMC runs were performed for each combination, using a random starting tree for each run. Each MCMC ranged from 50 to 600 million steps, sampling every 10,000 steps. Convergence was evaluated by calculating the effective sample sizes (ESS) (>200) of all parameters using Tracer v1.7.1 (62). Trees were summarized as maximum-clade credibility (MCC) trees using TreeAnnotator v1.10.4 after discarding the first 10% as burn-in, and then visualized in FigTree v1.4.3.
Positively selected site analysis.
The Datamonkey (http://www.datamonkey.org) (63) was used to find whether site/sites in the gp85 gene were under positive selection in November 2021. Based on the results of the evolutionary dynamics, positive selection sites were compared for 3 subclades of Clade 1 and the Clade 2. Four different methods (MEME, FEL, FUBAR, and SLAC) were used to identify positively selected sites of gp85, and then visualized in PyMOL v 2.5. The significance level of MEME, FEL and SLAC was set to P < 0.05. The significance level of FUBAR was based on a posterior probability (pp) of >0.95. Sites under positive selection were identified based on statistical significance (P < 0.05 or pp < 0.95) using the four methods.
Ancestral reconstructions of discrete traits.
A Bayesian phylogenetic method was further employed to infer the ancestral discrete traits for 3 subclades of Clade 1 and the Clade 2. Two types of traits (i.e., geographic regions and hosts) were modeled as a diffusion process among discrete states in BEAST v1.10.4 (42, 64). The isolates from each of the countries (except China) were treated in an independent category, while the isolates from China are currently divided into 7 categories combined with the distribution of the chicken production and the Chinese regional geography. The Chinese isolates were grouped into 7 geographic categories: Central China (provinces Henan, Hubei and Hunan), East China (provinces Shandong, Jiangsu, Shanghai, Jiangxi, Anhui, Fujian and Taiwan), North China (provinces Beijing, Tianjin, Hebei, Inner Mongolia and Shanxi), Northeast China (provinces Heilongjiang, Jilin and Liaoning), Northwest China (province Ningxia), South China (provinces Guangxi and Guangdong), and Southwest China (provinces Sichuan and Guizhou). The host category refers to the category standard reported by Deng et al. (19). The asymmetric substitution model with the Bayesian stochastic search variable selection (BSSVS) was used to infer asymmetric diffusion rates between each pair of traits and allowing the BF calculations to test the diffusion rates. The expected number of regions or hosts state transitions (that is, Markov jump counts) was also estimated following the reported procedures (65). The convergence of the MCMC chain was evaluated based on the ESS (>200) of the parameters. Significant diffusions pathways were summarized based on the combination of both BF > 3 and the posterior probability (pp) of >0.5. The degrees of rate support were as followings: 3 ≤ BF < 10 indicating supported, 10 ≤ BF < 100 indicating strong support, 100 ≤ BF < 1,000 indicating very strong support, and BF ≥ 1,000 indicating decisive support (42). The significant diffusion pathways were visualized via maps from SpreaD3 v0.9.7.1 (66).
Data availability.
All data has been included in the main tables, figures, and supplementary information file. Fifty sequences generated and used in this study have been deposited in GenBank under the accession numbers: MZ393151-MZ393200.
ACKNOWLEDGMENTS
The manuscript was kindly reviewed by Richard Roberts, Aurora, CO, USA. This work was supported by the Guangxi Program for Modern Agricultural Industry Technical System Construction-Chicken Industry [nycytxgxcxtd-19-03], the Guangxi Special Funding on Science and Technology Research [AA17204057] and the Shandong Provincial Natural Science Foundation [ZR2019BC047].
Conceptualization, P. Wei and Q. Deng; Methodology, P. Wei and Q. Deng; Analysis, Q. Deng, Q. Li, M. Li, S. Zhang, P. Wang, F. Fu and W. Zhu; Writing-original draft, Q. Deng and P. Wei; Writing–reviewing and editing, P. Wei and H. Zhang; Supervision, T. Wei, M. Mo, T. Huang and H. Zhang; Funding acquisition, P. Wei and P. Wang.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Ping Wei, Email: pingwei8@126.com.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.ICTV. 2020. International committee on taxonomy of viruses. Virus taxonomy: 2019 release. https://talk.ictvonline.org/taxonomy/.
- 2.Ortiz-Conde BA, Hughes SH. 1999. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J Virol 73:7165–7174. 10.1128/JVI.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair V, Fadly AM, et al. 2013. Leukosis/sarcoma group, p 553–592. In Swayne DE (ed) Diseases of poultry, 13th ed. Wiley, Blackwell, Ames, IA. [Google Scholar]
- 4.Li Y, Cui S, Li W, Wang Y, Cui Z, Zhao P, Chang S. 2017. Vertical transmission of avian leukosis virus subgroup J (ALV-J) from hens infected through artificial insemination with ALV-J infected semen. BMC Vet Res 13:204. 10.1186/s12917-017-1122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Lin L, Shi M, Li H, Gu Z, Li M, Gao Y, Teng H, Mo M, Wei T, Wei P. 2020. Vertical transmission of ALV from ALV-J positive parents caused severe immunosuppression and significantly reduced Marek's disease vaccine efficacy in three-Yellow chickens. Vet Microbiol 244:108683. 10.1016/j.vetmic.2020.108683. [DOI] [PubMed] [Google Scholar]
- 6.Wang P, Lin L, Li H, Shi M, Gu Z, Wei P. 2018. Full-length genome sequence analysis of an avian leukosis virus subgroup J (ALV-J) as contaminant in live poultry vaccine: The commercial live vaccines might be a potential route for ALV-J transmission. Transbound Emerg Dis 65:1103–1106. 10.1111/tbed.12841. [DOI] [PubMed] [Google Scholar]
- 7.Witter RL, Bacon LD, Hunt HD, Silva RE, Fadly AM. 2000. Avian leukosis virus subgroup J infection profiles in broiler breeder chickens: association with virus transmission to progeny. Avian Dis 44:913–931. [PubMed] [Google Scholar]
- 8.Li H, Wang P, Lin L, Shi M, Gu Z, Huang T, Mo ML, Wei T, Zhang H, Wei P. 2019. The emergence of the infection of subgroup J avian leucosis virus escalated the tumour incidence in commercial Yellow chickens in Southern China in recent years. Transbound Emerg Dis 66:312–316. 10.1111/tbed.13023. [DOI] [PubMed] [Google Scholar]
- 9.Wang P, Li M, Li H, Lin L, Shi M, Gu Z, Gao Y, Huang T, Mo M, Wei T, Wei P. 2020. Full-length cDNA sequence analysis of 85 avian leukosis virus subgroup J strains isolated from chickens in China during the years 1988–2018: coexistence of 2 extremely different clusters that are highly dependent upon either the host genetic background or the geographic location. Poult Sci 99:3469–3480. 10.1016/j.psj.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei P. 2019. Challenge and development opportunity of quality chicken industry in china: a review about 9 issues on the topic. China Poultry 41:1–6. (In Chinese). [Google Scholar]
- 11.Payne LN, Nair V. 2012. The long view: 40 years of avian leukosis research. Avian Pathol 41:11–19. 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Lin W, Chang S, Zhao P, Zhang X, Liu Y, Chen W, Li B, Shu D, Zhang H, Chen F, Xie Q. 2016. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese Yellow broiler in South China. Arch Virol 161:2717–2725. 10.1007/s00705-016-2965-x. [DOI] [PubMed] [Google Scholar]
- 13.Payne LN, Gillespie AM, Howes K. 1993. Recovery of acutely transforming viruses from myeloid leukosis induced by the HPRS-103 strain of avian leukosis virus. Avian Dis 37:438–450. [PubMed] [Google Scholar]
- 14.Silva RF, Fadly AM, Hunt HD. 2000. Hypervariability in the envelope genes of subgroup J avian leukosis viruses obtained from different farms in the United States. Virology 272:106–111. 10.1006/viro.2000.0352. [DOI] [PubMed] [Google Scholar]
- 15.Ye F, Wang Y, He Q, Cui C, Yu H, Lu Y, Zhu S, Xu H, Zhao X, Yin H, Li D, Li H, Zhu Q. 2020. Exosomes transmit viral genetic information and immune signals may cause immunosuppression and immune tolerance in ALV-J i nfected HD11 cells. Int J Biol Sci 16:904–920. 10.7150/ijbs.35839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Wang P, Li M, Lin L, Shi M, Li H, Deng Q, Teng H, Mo M, Wei T, Wei P. 2020. Recombinant subgroup B avian leukosis virus combined with the subgroup J env gene significantly increases its pathogenicity. Vet Microbiol 250:108862. 10.1016/j.vetmic.2020.108862. [DOI] [PubMed] [Google Scholar]
- 17.Cui Z, Du Y, Zhang Z, Silva RF. 2003. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis 47:1321–1330. 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Yu M, Xing L, Liu P, Chen Y, Chang F, Wang S, Bao Y, Farooque M, Li X, Guan X, Liu Y, Liu A, Qi X, Pan Q, Zhang Y, Gao L, Li K, Liu C, Cui H, Wang X, Gao Y. 2020. The bipartite sequence motif in the N and C termini of gp85 of subgroup J avian leukosis virus plays a crucial role in receptor binding and viral entry. J Virol 94:4187–4192. 10.1128/JVI.01232-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Q, Li M, He C, Lu Q, Gao Y, Li Q, Shi M, Wang P, Wei P. 2021. Genetic diversity of avian leukosis virus subgroup J (ALV-J): toward a unified phylogenetic classification and nomenclature system. Virus Evol 7:veab037. 10.1093/ve/veab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Meng F, Li W, Wang Y, Chang S, Zhao P, Cui Z. 2018. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult Sci 97:3532–3539. 10.3382/ps/pey241. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Lin L, Li H, Yang Y, Huang T, Wei P. 2018. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989–2016: Coexistence of five extremely different clusters. Arch Virol 163:377–389. 10.1007/s00705-017-3601-0. [DOI] [PubMed] [Google Scholar]
- 22.Ma M, Yu M, Chang F, Xing L, Bao Y, Wang S, Farooque M, Li X, Liu P, Chen Y, Qi X, Pan Q, Gao L, Li K, Liu C, Zhang Y, Cui H, Wang X, Sun Y, Gao Y. 2020. Molecular characterization of avian leukosis virus subgroup J in Chinese local chickens between 2013 and 2018. Poult Sci 99:5286–5296. 10.1016/j.psj.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao Y, Li W, Dong X, Liu J, Zhao P. 2013. Different quasispecies with great mutations hide in the same subgroup J field strain of avian leukosis virus. Sci China Life Sci 56:414–420. 10.1007/s11427-013-4479-z. [DOI] [PubMed] [Google Scholar]
- 24.Payne LN, Brown SR, Bumstead N, Howes K, Frazier JA, Thouless ME. 1991. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol 72:801–807. 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 25.Xu B, Dong W, Yu C, He Z, Lv Y, Sun Y, Feng X, Li N, Lee LF, Li M. 2004. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol 33:13–17. 10.1080/03079450310001636237a. [DOI] [PubMed] [Google Scholar]
- 26.Pan W, Gao Y, Qin L, Ni W, Liu Z, Yun B, Wang Y, Qi X, Gao H, Wang X. 2012. Genetic diversity and phylogenetic analysis of glycoprotein GP85 of ALV-J isolates from mainland China between 1999 and 2010: coexistence of two extremely different subgroups in layers. Vet Microbiol 156:205–212. 10.1016/j.vetmic.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Z, Liu J, Cui Z, Zhang L. 2010. Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J Vet Med Sci 72:1027–1033. 10.1292/jvms.09-0564. [DOI] [PubMed] [Google Scholar]
- 28.Zhou D, Xue J, Zhang Y, Wang G, Feng Y, Hu L, Shang Y, Cheng Z. 2019. Outbreak of myelocytomatosis caused by mutational avian leukosis virus subgroup J in China, 2018. Transbound Emerg Dis 66:622–626. 10.1111/tbed.13096. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Su Q, Zhang Z, Cui Z, Chang S, Zhao P. 2020. Molecular characteristics of the re-emerged avian leukosis virus in China, 2018–2019. Transbound Emerg Dis 67:1141–1151. 10.1111/tbed.13440. [DOI] [PubMed] [Google Scholar]
- 30.Sun S, Cui Z. 2007. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local “yellow” chickens. Avian Pathol 36:221–226. 10.1080/03079450701332345. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Lu S, Zhang H, Chen P, Lai H, Liao M, Cao W. 2009. Isolation and identification of SCAU-0901 strain of subgroup J avian leukosis virus. Chinese Veterinary Science 39(08):674–678. (In Chinese). [Google Scholar]
- 32.Li X, Yan Y, Li G, Liu Y, Dai Z, Chen W, Chen F, Xie Q. 2016. Isolation, identification and full-genome sequence analysis of subgroup J avian leukosis virus from Yellow broilers. Progress in Veterinary Medicine 37:27–31. (In Chinese). [Google Scholar]
- 33.Jiang L, Zeng X, Hua Y, Gao Q, Fan Z, Chai H, Wang Q, Qi X, Wang Y, Gao H, Gao Y, Wang X. 2014. Genetic diversity and phylogenetic analysis of glycoprotein gp85 of avian leukosis virus subgroup J wild-bird isolates from Northeast China. Arch Virol 159:1821–1826. 10.1007/s00705-014-2004-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Guan X, Chen Z, Cao D, Kang Z, Shen Q, Lei Q, Li F, Li H, Leghari MF, Wang Y, Qi X, Wang X, Gao Y. 2018. The high conserved cellular receptors of avian leukosis virus subgroup J in Chinese local chickens contributes to its wide host range. Poult Sci 97:4187–4192. 10.3382/ps/pey331. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q, Zhao X, Lemey P, Suchard MA, Bi Y, Shi W, Liu D, Qi W, Zhang G, Stenseth NC, Pybus OG, Tian H. 2020. Assessing the role of live poultry trade in community-structured transmission of avian influenza in China. Proc Natl Acad Sci USA 117:5949–5954. 10.1073/pnas.1906954117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Zhang T, Bai Y, Li H, Wang Y, Bi Y, Chang J, Xu B. 2018. Live poultry trading drives China's H7N9 viral evolution and geographical network propagation. Front Public Health 6:210. 10.3389/fpubh.2018.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi Y, Chen Q, Wang Q, Chen J, Jin T, Wong G, Quan C, Liu J, Wu J, Yin R, Zhao L, Li M, Ding Z, Zou R, Xu W, Li H, Wang H, Tian K, Fu G, Huang Y, Shestopalov A, Li S, Xu B, Yu H, Luo T, Lu L, Xu X, Luo Y, Liu Y, Shi W, Liu D, Gao GF. 2016. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20:810–821. 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Shi J, Deng G, Ma S, Zeng X, Yin X, Li M, Zhang B, Cui P, Chen Y, Yang H, Wan X, Liu L, Chen P, Jiang Y, Guan Y, Liu J, Gu W, Han S, Song Y, Liang L, Qu Z, Hou Y, Wang X, Bao H, Tian G, Li Y, Jiang L, Li C, Chen H. 2018. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe 24:558–568. 10.1016/j.chom.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, Zhang S, Cao P, Li X, Tian K, Qiu HJ, Hu R. 2018. Emergence of African swine fever in China, 2018. Transbound Emerg Dis 65:1482–1484. 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- 40.Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, Goodfellow I, Loman NJ, Pybus OG, Robertson DL, Thomson EC, Rambaut A, Connor TR, COG-UK Consortium. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184:64–75.e11. 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vrancken B, Zhao B, Li X, Han X, Liu H, Zhao J, Zhong P, Lin Y, Zai J, Liu M, Smith DM, Dellicour S, Chaillon A. 2020. Comparative circulation dynamics of the five main HIV types in China. J Virol 94:e00683-20. 10.1128/JVI.00683-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su YCF, Bahl J, Joseph U, Butt KM, Peck HA, Koay ESC, Oon LLE, Barr IG, Vijaykrishna D, Smith GJD. 2015. Phylodynamics of H1N1/2009 influenza reveals the transition from host adaptation to immune-driven selection. Nat Commun 6:7952. 10.1038/ncomms8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong G. 2009. Production status of layers in 2008 and development trend in 2009 in China. Guide to Chinese Poultry 26:4–8. (In Chinese). [Google Scholar]
- 44.Deng Q, Shi M, Li Q, Wang P, Li M, Wang W, Gao Y, Li H, Lin L, Huang T, Wei P. 2021. Analysis of the evolution and transmission dynamics of the field MDV in China during the years 1995–2020, indicating the emergence of a unique cluster with the molecular characteristics of vv+MDV that has become endemic in southern China. Transbound Emerg Dis 68:3574–3587. 10.1111/tbed.13965. [DOI] [PubMed] [Google Scholar]
- 45.Liu P, Li L, Jiang Z, Yu Y, Chen X, Xiang Y, Chen J, Li Y, Cao W. 2021. Molecular characteristics of subgroup J avian leukosis virus isolated from Yellow breeder chickens in Guangdong, China, during 2016–2019. Infect Genet Evol 89:104721. 10.1016/j.meegid.2021.104721. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Tan M, Zhang F, Ji H, Zeng Y, Yang Q, Tan J, Huang J, Su Q, Huang Y, Kang Z. 2021. Diversity of avian leukosis virus subgroup J in local chickens, Jiangxi, China. Sci Rep 11:4797. 10.1038/s41598-021-84189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma M, Yu M, Xu C, Huang Q, Meng Z, Wang S, Xing L, Chang F, Liu C, Qi X, Wang Y, Sun Y, Wang X, Gao Y. 2019. Full-length genome sequence analysis of subgroup J avian leukosis viruses isolated from imported white feather broiler breeders. Chinese J Preventive Veterinary Medicine 41:750–754. (In Chinese). [Google Scholar]
- 48.Lin L, Wang P, Yang Y, Li H, Huang T, Wei P. 2017. Full-length genome sequence analysis of four subgroup J avian leukosis virus strains isolated from chickens with clinical hemangioma. Virus Genes 53:868–875. 10.1007/s11262-017-1490-7. [DOI] [PubMed] [Google Scholar]
- 49.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf): 95–98. [Google Scholar]
- 52.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE): 1–8. [Google Scholar]
- 54.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2021. ColabFold: making protein folding accessible to all. bioRxiv. [DOI] [PMC free article] [PubMed]
- 55.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pépin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. HIV epidemiology: the early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nie Q, Li X, Chen W, Liu D, Chen Y, Li H, Li D, Tian M, Tan W, Zai J. 2020. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res 287:198098. 10.1016/j.virusres.2020.198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol 35:773–777. 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hicks JT, Dimitrov KM, Afonso CL, Ramey AM, Bahl J. 2019. Global phylodynamic analysis of avian paramyxovirus-1 provides evidence of inter-host transmission and intercontinental spatial diffusion. BMC Evol Biol 19:108. 10.1186/s12862-019-1431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Brien JD, Minin VN, Suchard MA. 2009. Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol 26:801–814. 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. 2016. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol 33:2167–2169. 10.1093/molbev/msw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download jvi.00717-22-s0001.xlsx, XLSX file, 0.1 MB (53.9KB, xlsx)
Fig. S1 to S4. Download jvi.00717-22-s0002.pdf, PDF file, 3.1 MB (3.1MB, pdf)
Data Availability Statement
All data has been included in the main tables, figures, and supplementary information file. Fifty sequences generated and used in this study have been deposited in GenBank under the accession numbers: MZ393151-MZ393200.