Abstract
The large majority of proteins of alkaliphilic Bacillus pseudofirmus OF4 grown at pH 7.5 and 10.5, as studied by two-dimensional gel electrophoresis analyses, did not exhibit significant pH-dependent variation. A new surface layer protein (SlpA) was identified in these studies. Although the prominence of some apparent breakdown products of SlpA in gels from pH 10.5-grown cells led to discovery of the alkaliphile S-layer, the largest and major SlpA forms were present in large amounts in gels from pH 7.5-grown cells as well. slpA RNA abundance was, moreover, unchanged by growth pH. SlpA was similar in size to homologues from nonalkaliphiles but contained fewer Arg and Lys residues. An slpA mutant strain (RG21) lacked an exterior S-layer that was identified in the wild type by electron microscopy. Electrophoretic analysis of whole-cell extracts further indicated the absence of a 90-kDa band in the mutant. This band was prominent in wild-type extracts from both pH 7.5- and 10.5-grown cells. The wild type grew with a shorter lag phase than RG21 at either pH 10.5 or 11 and under either Na+-replete or suboptimal Na+ concentrations. The extent of the adaptation deficit increased with pH elevation and suboptimal Na+. By contrast, the mutant grew with a shorter lag and faster growth rate than the wild type at pH 7.5 under Na+-replete and suboptimal Na+ conditions, respectively. Logarithmically growing cells of the two strains exhibited no significant differences in growth rate, cytoplasmic pH regulation, starch utilization, motility, Na+-dependent transport of α-aminoisobutyric acid, or H+-dependent synthesis of ATP. However, the capacity for Na+-dependent pH homeostasis was diminished in RG21 upon a sudden upward shift of external pH from 8.5 to 10.5. The energy cost of retaining the SlpA layer at near-neutral pH is apparently adverse, but the constitutive presence of SlpA enhances the capacity of the extremophile to adjust to high pH.
Bacillus species have been a major component of the extremely alkaliphilic bacterial flora isolated both from highly selective environments such as alkaline lakes and from ostensibly unselective environments such as conventional soils (21, 24, 26). While many studies have focused on useful products of alkaliphilic bacilli (21), others have focused on the basis for alkaliphily itself (19, 26, 28). Among the questions that immediately arise are how can those membranous and protein structures that are exposed to the alkaline medium function, and how can cells growing above pH 10 maintain a cytoplasmic pH that is well below the external pH? With respect to the first question, recent structural studies of extracellular enzymes from extreme alkaliphiles and numerous deduced protein sequences of alkaliphile proteins have begun to indicate properties that may correlate with the ability to function at extremely high pH (26). The adaptations, moreover, appear to depend upon whether a high net charge is important to the function of the molecule or molecular segment. When that is the case, there is a general paucity of arginine and lysine residues and/or an increase in acidic residues that would remain charged at high external pH (26, 46). What has not yet been examined, however, is whether facultative alkaliphiles that grow from pH 7.5 to 11 or higher possess alternate neutral-pH and high-pH versions of a significant proportion of extracellular and membrane-associated proteins. In the current studies, two-dimensional electrophoresis was used as an analytical tool to develop an initial estimate of the fraction of alkaliphile Bacillus pseudofirmus OF4 proteins that differ in pH 7.5- and 10.5-grown cells. This approach to proteome characterization under diverse conditions has been broadly and productively applied to prokaryotes (2, 7, 35, 45) since its initial development (36).
With respect to cytoplasmic pH regulation, molecular biological and physiological studies have especially made use of two alkaliphilic strains, Bacillus halodurans C-125 (19, 26) and B. pseudofirmus OF4 (26, 28). In both strains, there is strong evidence for a key role for an Na+ cycle. This cycle comprises Na+ efflux via electrogenic Na+/H+ antiporters and Na+ reentry via Na+/solute symporters and, probably, via the Na+ channel associated with Na+-dependent alkaliphile motility (28, 43). Net proton as well as solute uptake is coupled to this cycle. Work on B. halodurans C-125 has shown that in addition to the critical role of the active transporters of the Na+ cycle, acidic cell surface polymers contribute to pH homeostasis and alkaliphily, in particular a teichuronopeptide that is more highly expressed at high pH then at low pH (3–5). By contrast, B. pseudofirmus OF4 did not appear to possess these acidic polymers (18). Moreover, H+-coupled ATP synthesis occurred robustly in membrane preparations of B. pseudofirmus OF4 that lacked detectable cell wall polymer (18).
ATP synthesis is the second physiologically important process that, in addition to Na+-dependent pH homeostasis, requires inward proton translocation from the highly alkaline milieu in alkaliphilic Bacillus species (26, 29). It was thus of interest to determine whether B. pseudofirmus OF4 possesses proteins whose levels increase at high pH that might modulate the properties of the membranes themselves (e.g., determinants of phospholipid content) or comprise external layers different from those in B. halodurans C-125. That is, B. pseudofirmus OF4 might have alternate functional counterparts to the uronic acid-containing polymers of B. halodurans C-125.
The current studies focus on a newly discovered surface layer (S-layer) protein, designated SlpA, that was identified in the follow-up analyses of the two-dimensional gel electrophoresis studies; while shown to have a role in alkaliphily, its expression is not significantly pH dependent. Several other more tentatively identified proteins raise the possibility of a pH 10.5-associated increase in enzymes that could be involved in remodeling of membrane lipids and/or lipid catabolism.
MATERIALS AND METHODS
Bacterial strains and cell growth.
B. pseudofirmus OF4811M, a methionine auxotroph (11), was used as the wild type in all studies. It was grown in malate medium at pH 7.5 or 10.5 as described previously (16). The cells were grown at 30°C with shaking at 200 rpm. Escherichia coli strain DH5α (Gibco-BRL, Gaithersburg, Md.) was used for cloning purposes and grown in Luria broth (LB). To maintain plasmids, ampicillin was present at 100 μg/ml. For two-dimensional gel analysis of steady-state growth conditions, B. pseudofirmus OF4811M was grown to the mid-logarithmic phase at either pH 7.5 or 10.5 in malate-containing medium. After harvesting, the cells were washed in 50 mM Tris-Cl (pH 8)–1 mM EDTA and finally resuspended in 50 mM Tris-Cl (pH 8)–1 mM EDTA–1 mM phenylmethylsulfonyl fluoride (PMSF)–5 mM p-aminobenzamidine.
Two-dimensional gel electrophoresis.
Cells were broken, and membranes and cytoplasm were isolated as described previously (17), except that membranes were washed only once in 50 mM Tris-Cl (pH 8)–1 mM EDTA–0.1 mM PMSF. Two-dimensional gel electrophoresis was performed according to the method of O'Farrell (36) by Kendrick Labs, Inc. (Madison, Wis.) as follows. Isoelectric focusing (IEF) was carried out in glass tubes (inner diameter, 2.0 mm), using 2.0% Resolytes pH 4–8 ampholines (BDH from Hoefer Scientific Instruments, San Francisco, Calif.) for 9600 V-h. One microgram of an IEF internal standard, tropomyosin (Mr, 33,000, pI 5.2) was added to the samples. This standard is indicated by an arrow on the stained gel. After IEF, the tube gels were equilibrated for 10 min in buffer O (10% glycerol, 50 mM dithiothreitol, 2.3% sodium dodecyl sulfate [SDS], 0.0625 M Tris [pH 6.8]) and sealed at the top of the stacking gels which were on top of 10% acrylamide slab gels (0.75-mm thick). The SDS slab gel electrophoresis was carried out for 4 h at 12.5 mA/gel. Following electrophoresis, the gels were stained with Coomassie brilliant blue R-250 and dried between two sheets of cellophane. For computer analysis of protein spot densities, the gels were scanned using a laser densitometer and analyzed using Phoretix software (Phoretix, Newcastle-upon-Tyne, England, U.K.) by Kendrick Labs Inc. Two independent samples were prepared for each analysis, and two identical gels were run for each sample.
Protein sequence determinations.
Spots chosen for protein sequence were transferred to polyvinylidene difluoride (PVDF) membranes in 0.025 M Tris–0.2 M glycine–10% methanol (pH 8.8). N-terminal sequence was determined using a Perkin Elmer model 494 amino acid sequencer. For internal sequence, the protein was digested with endoproteinase LysC, and peptides were separated using high-pressure liquid chromatography.
DNA manipulations.
E. coli cells were made competent by RbCl treatment and transformed as described by Hanahan (20). For PCR, chromosomal DNA from B. pseudofirmus OF4811M was isolated by the method of Ausubel et al. (6). Radioactive probes were prepared using a random priming kit (Boehringer Mannheim). Restriction enzymes were purchased from New England Biolabs.
Isolation and sequencing of the slpA gene.
Degenerate primers were designed based on the sequence of the N terminus and two internal peptides of spot 17. The forward primer NTERF (GCICCIGCIGAYGCIAARTTYWSIGAYGT) was designed based on the N-terminal sequence, and the two reverse primers (R1 [RTTIACDATIGGIGCIGTIGTRTCRTC] and R2 [YTTRAARAAYTGICCIGTIGG]) were based on the sequence of two internal peptides. R, D, Y, W, and S are equal mixtures of A/G, G/A/T, C/T, A/T, and G/C, respectively. Primers were synthesized by Gibco-BRL. PCRs were performed in a 100-μl volume containing Amplitaq DNA polymerase (2.5 U), 2.5 mM MgCl2, 2 mM each of the four deoxynucleoside triphosphates, and 0.5 μM primers. A Perkin Elmer Cetus thermocycler was used to perform 32 cycles, each of which had a denaturation step at 94°C for 1 min, an annealing step at 37°C for 2 min, and an elongation step at 72°C for 3 min. PCR products were electrophoresed on 0.8% agarose gels, purified by a gel extraction kit (Qiagen, Chatsworth, Calif.), and ligated to the pGEM T vector (Promega, Madison, Wis.). Recombinant plasmids were selected by blue-white screening on LB-ampicillin plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). Plasmid inserts were sequenced at the DNA core of Utah State University using an ABI50 automated DNA sequencer. The remaining downstream region of the gene was isolated by using the PCR product to screen pBK36 plasmid libraries of alkaliphilic genomic DNA, as previously described (22). The upstream region of the gene was isolated by inverse PCR using Taq1-cleaved chromosomal DNA which had been religated (M. Ito, unpublished results).
Northern analyses.
Northern analyses were conducted on RNA isolated from pH 7.5- and pH 10.5-grown cells as described by others (13) and probed with the same PCR product used in the gene isolation above.
Construction of S-layer deletion strain RG1.
A recombinant pGEM3Zf(+) plasmid was constructed containing the N-terminal fragment of slpA that was originally isolated by PCR with degenerate primers, as described above. This plasmid was digested with XbaI and SalI. The resultant 3.6-kb fragment contained the plasmid and the first 650 nucleotides of slpA. This fragment was ligated to an XbaI-SalI fragment from plasmid pBK36 isolated after colony hybridization, which contained the remainder of the downstream slpA sequence. The resulting recombinant plasmid contained the entire slpA sequence. A 1.95-kb fragment was deleted from this gene by XbaI-HpaI digestion. Following mung bean nuclease treatment to make blunt ends, the fragment for the deletion construct was ligated to a spectinomycin resistance cassette. After selection for spectinomycin resistance in E. coli, the deletion construct was excised from pGEM3Zf(+) by ApaI-HincII treatment and ligated to pG+Host4 digested with ApaI and SmaI. After selection in E. coli for erythromycin resistance at 30°C, the construct was isolated and used to transform protoplasts of B. pseudofirmus OF4811M. Cell walls were regenerated at 30°C on modified DM-3 medium containing erythromycin (0.1 μg/ml). Transformants were confirmed by streaking on complex medium plates with spectinomycin (100 μg/ml). Generation of deletion strains followed the procedure previously described for this plasmid in B. pseudofirmus OF4 (23). Single-crossover mutants were isolated by growing the transformants to the mid-logarithmic phase at 30°C in complex medium at pH 7.5 with erythromycin (0.6 μg/ml) and then plating at 40°C on the same medium. After isolation of the single-crossover mutant, the double-crossover strains were isolated by growing the single-crossover strain to mid-logarithmic phase at 30°C in complex medium at pH 7.5 with spectinomycin (100 μg/ml) followed by plating on the same medium at 40°C. The double-crossover strains were resistant to spectinomycin and sensitive to erythromycin. The deletion was confirmed by PCR and Southern analyses. Two individual isolates, designated RG21 and RG22, were characterized and showed essentially identical properties, as shown for RG1 in this report.
Characterization of the wild type and S-layer mutant RG21.
For SDS-polyacrylamide gel electrophoresis (PAGE) and electron microscopy, the wild type and mutant were grown at either pH 7.5 or 10.5 as described above. SDS-PAGE was conducted by the method of Laemmli (31) on 10% gels. For electron microscopy, ultrathin sectioning and freeze-etching were performed as described previously (33, 40). The specimens were investigated using a Philips CM100 transmission electron microscope at 80 kV acceleration voltage. For growth experiments, the wild type and mutant were grown overnight in malate medium containing 200 mM Na+ at pH 9.5. An inoculum of 0.5 ml of this culture was added to 50 ml of malate-containing medium buffered with various combinations of sodium or potassium carbonate salts (pH 10.5 or 11.0) or medium buffered with various combinations of sodium or potassium phosphate salts (pH 7.5). The final concentration of Na+ was 10 or 200 mM at pH 10.5 or 11.0 or 25 or 200 mM at pH 7.5. Cultures were grown with shaking at 30°C, and the A600 was recorded at intervals.
For pH shift experiments, cells grown at pH 9.5 to the mid-logarithmic phase were harvested, washed and equilibrated at pH 8.5, and subjected to a shift in external pH from 8.5 to 10.5 exactly as described previously (23). Cells for the assay of α-aminoisobutyric acid (AIB) were also grown to mid-logarithmic phase at pH 9.5. The assay for AIB uptake at either pH 7.5 or 10.5 was performed as described elsewhere (17). For measurements of ATP synthesis, mid-logarithmic-phase cells were starved for 8 h and reenergized by the addition of 10 mM potassium malate. The amount of ATP synthesized was determined by the luciferin-luciferase system as described previously (18).
RESULTS
Two-dimensional gel electrophoresis analyses.
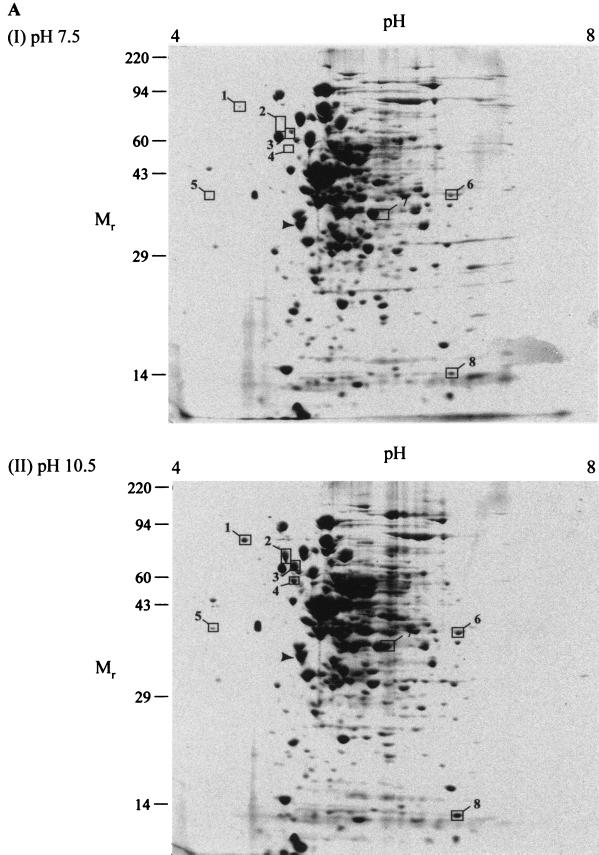
Cytoplasmic and membrane fractions of B. pseudofirmus OF4 grown to the mid-logarithmic phase at pH 7.5 and 10.5 were subjected to two-dimensional gel electrophoresis. No attempts were made to exclude membrane-associated proteins as opposed to directly anchored proteins from the membrane fraction, inasmuch as these were of interest in connection with possible cell surface layers. In fact, since the analyses were not expected to resolve highly hydrophobic and generally low-copy polytopic proteins, the more peripheral membrane-associated proteins were the likely target of study in this fraction. A pH gradient of 4 to 8 was chosen for the IEF due to the stability and reproducibility of the gradient in this pH range. An SDS–10% PAGE gel was considered most useful for the second dimension, since most proteins of interest were expected to be between 10 and 100 kDa. The results of the two-dimensional electrophoresis are shown in Fig. 1. The gel patterns of the cytoplasmic fractions from pH 7.5- and pH 10.5-grown cells were very similar (Fig. 1A). Eight protein spots were, however, clearly elevated in the pH 10.5 sample and are boxed in Fig. 1A.
FIG. 1.
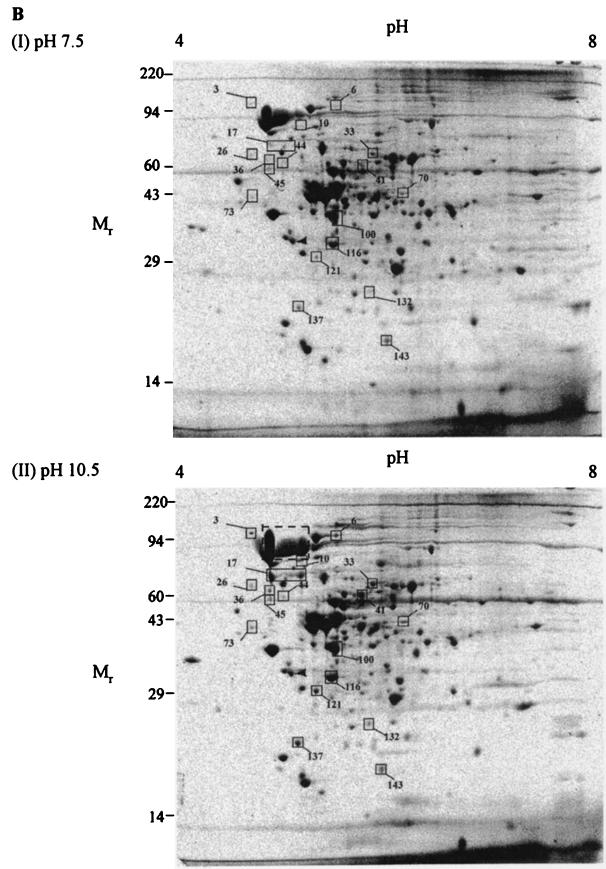
Two-dimensional gel electrophoresis of B. pseudofirmus OF4811M under steady-state growth conditions. Cytoplasmic (A) and membrane (B) fractions were subjected to two-dimensional electrophoresis as described under Materials and Methods. A total of 250 μg of protein was loaded for each gel. Prior to electrophoresis, samples were incubated in SDS buffer (5% SDS, 5% β-mercaptoethanol, 10% glycerol, 60 mM Tris [pH 6.8]) at room temperature for 15 min. After electrophoresis, gels were stained with Coomassie blue R-250 and dried between sheets of cellophane. Spots whose density was increased in the pH 10.5 gels are boxed. The region in the bottom of panel B that is indicated by a dashed box is the region of a prominent band from which three samples were analyzed (described in text). The following molecular weight standards were used in SDS-PAGE: myosin (220,000), phosphorylase A (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000), and lysozyme (14,000).
Because the challenge of pH homeostasis is central to alkaliphiles and appears to determine the upper pH limit for their growth (42), our interest was especially focused on the membrane-associated proteins. In order to quantify differences between the spots visualized on gels of membrane samples from cells grown at the different pHs (Fig. 1B), the gels were scanned by a laser densitometer and analyzed by Pherotix software. This software calculates the spot densities and estimates isoelectric point (pI) and relative molecular weight (Mr) for each spot on the gel. One hundred and forty-eight spots were resolved in this analysis. Of these 148, the densities of 19 spots were increased more than twofold at pH 10.5, whereas the densities of 18 were decreased more than twofold. Spots whose density increased at the higher pH were considered potentially important for growth in an alkaline environment, and so their analysis was pursued. The 19 spots with >2-fold-increased density in cells grown at pH 10.5 are boxed in Fig. 1B. The corresponding spots are also boxed in the pH 7.5 gel figure for easy comparison. A summary of the spots, including estimated pI and Mr, is shown in Table 1. Most of the spots are increased in density between two- and eightfold, with two being increased over 20-fold. In contrast, of the 18 proteins that are decreased in density at pH 10.5, only 1 was down more than fourfold, with the majority decreased by two- to threefold (data not shown).
TABLE 1.
Summary of membrane-associated protein spots whose levels increase at least twofold at pH 10.5a
| Protein spot no. | pI | Mr | Spot density
|
Ratio, pH 10.5/pH 7.5 | |
|---|---|---|---|---|---|
| pH 7.5 | pH 10.5 | ||||
| 3 | 4.87 | 136,782 | 0.01 | 0.07 | 7.44 |
| 6 | 5.48 | 125,257 | 0.29 | 0.58 | 2.00 |
| 10 | 5.23 | 86,756 | 0.05 | 0.11 | 2.34 |
| 17 | 5.07 | 78,185 | 0.08 | 1.84 | 24.30 |
| 26 | 4.81 | 71,074 | 0.01 | 0.04 | 3.67 |
| 33 | 6 | 73,346 | 0.02 | 0.09 | 4.55 |
| 36 | 4.95 | 67,688 | 0.02 | 0.08 | 4.33 |
| 41 | 5.87 | 65,532 | 0.04 | 0.13 | 3.49 |
| 44 | 5.09 | 64,302 | 0.00 | 0.03 | 29.42 |
| 45 | 4.95 | 62,270 | 0.03 | 0.07 | 2.08 |
| 70 | 6.29 | 51,395 | 0.07 | 0.16 | 2.15 |
| 73 | 4.8 | 48,073 | 0.00 | 0.04 | 7.59 |
| 100 | 5.67 | 41,704 | 0.41 | 1.04 | 2.53 |
| 116 | 5.64 | 35,448 | 0.79 | 1.86 | 2.36 |
| 121 | 5.47 | 32,504 | 0.07 | 0.34 | 4.63 |
| 132 | 6.03 | 26,686 | 0.02 | 0.10 | 4.64 |
| 137 | 5.64 | 24,753 | 0.13 | 0.39 | 2.91 |
| 143 | 6.17 | 20,415 | 0.10 | 0.23 | 2.29 |
Spot density values are expressed as spot percentages (individual spot volumes as a percentage of total volume in all spots) and are the averages of analyses for two separate gels.
In order to characterize a selection of the individual proteins expressed more highly under alkaline relative to near-neutral growth conditions, gels containing the protein spot of interest were transferred to PVDF membranes. The proteins were then subjected to N-terminal amino acid sequencing. The results for spots from the membrane fraction that gave satisfactory sequence data are summarized in Table 2. Three of the spots, 17, 36, and 73, were found to have the same N-terminal sequence. Of these, spot 17 was the largest and most abundant. Spot 17 appeared as a horizontal smear on the gel (Fig. 1B). This suggests possible charge heterogeneity leading to a diffuse band in the IEF. In addition, the N-terminal sequences of spot 2 from the cytoplasm and spot 17 were identical. The observation of the same protein in the membrane and cytoplasm, with most being in the membrane, suggested that spot 17 was membrane associated but not an integral membrane protein. A database search using the N-terminal sequence of spot 17 failed to yield any significant matches, although subsequent analyses revealed direct homologues and showed that a higher-molecular-weight species that is even more prominent than spot 17 in gels of both fractions from pH 7.5- and 10.5-grown cells is the major form (see below).
TABLE 2.
N-terminal sequence of membrane-associated proteins showing increased levels at pH 10.5
| Spot no. | N-terminal sequencea | Database match | Function or comments |
|---|---|---|---|
| 17 | APADAKFSDVSSxH(V)xID | None | Subsequent cloning of a gene fragment identified this spot as a surface layer protein |
| 33 | SNTAERVVKGGSFLVEDIE | B. subtilis YusJ | Similar to butyryl-CoA dehydrogenase |
| 36 | APADAKFSDVSS(S)H(W)SI | Spot 17 | Degradation product of spot 17? |
| 41 | ATEITMPQLGESVTEG(T)I | B. subtilis BfmBB | Branched-chain α-keto acid dehydrogenase |
| 73 | APADAKFSDVxx(N)(H)(E) | Spot 17 | Degradation product of spot 17? |
| 100 | MNFDLTKEQKMIRDMVRD | B. subtilis YngJ | Similar to butyryl-CoA dehydrogenase |
| 121 | AVISYIEAVTLALKEEM | B. subtilis BfmBAB | Branched-chain α-keto acid dehydrogenase |
x, unknown; probable residues are shown in parentheses.
Of the four remaining membrane spots, 41 and 121 appeared to be a significant match with two subunits of the branched-chain α-keto acid dehydrogenase from Bacillus subtilis (47). Spot 121 was 65% identical to the N terminus of the E1β subunit, and spot 41 was 84% identical to the E2 subunit. The branched-chain α-keto acid dehydrogenase is an enzyme complex with three components. The E1 component consists of two subunits, E1α and E1β, and is a branched-chain α-keto acid decarboxylase (37). The two remaining spots, 33 and 100, matched two proteins of unknown function in B. subtilis. Spot 33 was 50% identical to YusJ (30), and spot 100 was 75% identical to YngJ (44). Although no biochemical data are available for these proteins, they both show strong sequence similarity to known butyryl and other short-chain acylcoenzyme A (acyl-CoA) dehydrogenases. BLAST analysis (1) indicated that YngJ (380 amino acids) was 50% identical to the butyryl-CoA dehydrogenase from Clostridium acetobutylicum (10) over the entire length of the protein. YusJ (594 amino acids) and YngJ showed 38% identity to each other over 400 amino acids, with some gaps.
Cloning, analysis, and deletion of an S-layer gene (spot 17).
In order to further characterize spot 17, internal sequence was obtained from two peptides isolated after treatment with endopeptidase LysC. The sequence of the two peptides did not reveal any significant database matches. Using the sequence of the N terminus and the two internal peptides, degenerate primers were designed, and a fragment of the gene was amplified by PCR. Sequence analysis indicated a single, incomplete open reading frame coding for 370 amino acids. The remaining downstream sequence was obtained from plasmids isolated after colony hybridization of E. coli cells carrying a B. pseudofirmus OF4 plasmid library. The remaining upstream region was isolated by inverse PCR.
Analysis of the DNA sequence identified a single open reading frame of 931 amino acids (Fig. 2). A BLAST search revealed homology to a number of surface layer proteins from various Bacillus species, including Bacillus sphaericus, Bacillus thuringiensis, Bacillus stearothermophilus, and Bacillus anthracis (38, 41). The identity and similarity of the alkaliphile protein to each of these homologues were approximately 25 and 40%, respectively. Three S-layer homology domains could be identified in the N-terminal region of the protein (residues 7 to 67, 69 to 128, and 132 to 189). The domains are represented by three sequence repeat regions of approximately 60 amino acids and are involved in binding of S-layer proteins to the underlying cell surface (32, 38). The overall charge associated with the alkaliphile S-layer is much more acidic than that of its homologues. This is due to a paucity of lysine and arginine residues in the protein. The sequence of SlpA has been deposited in GenBank (accession number AF242295). Since some bacteria have multiple, sometimes related S-layer-encoding genes, the slpA gene was used for Southern analyses of B. pseudofirmus OF4 DNA at low stringency. Although not shown, there was no indication of a second gene that was related to slpA.
FIG. 2.
Nucleotide and deduced amino acid sequence of the slpA gene from B. pseudofirmus OF4. The sequence of the chromosomal region including the slpA gene is shown together with the deduced amino acid sequence of SlpA. Candidates for promoter (−35 and −10) and ribosome-binding site (RBS) sequences are in bold and underlined. The amino acid residues that were identified from N-terminal and internal peptide sequencing are also shown in bold.
Characterization of RG21, the slpA mutant of B. pseudofirmus OF4811M, and reexamination of the gels for the major SlpA form.
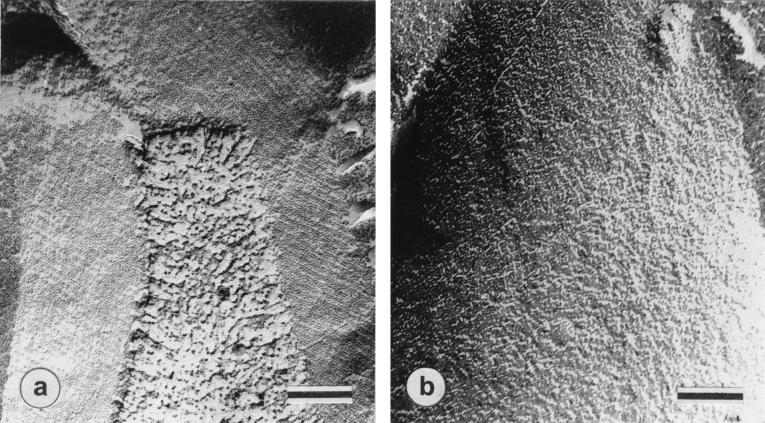
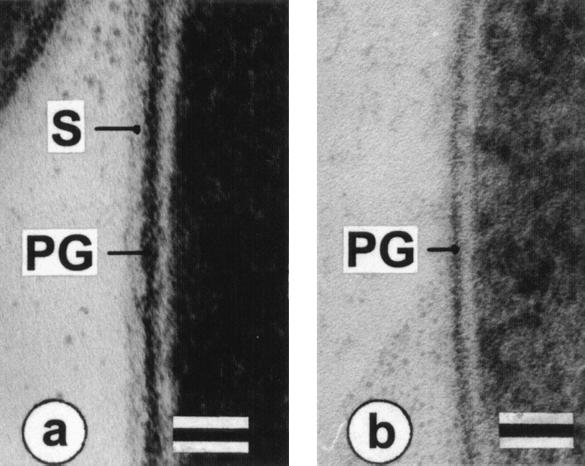
In freeze-fracture electron micrographs of intact cells of wild-type B. pseudofirmus OF4811M grown at pH 7.5 (Fig. 3A), a smooth, oblique S-layer lattice with center-to-center spacings of a = 9.2 nm, b = 12.8 nm, and γ ∼ 77° was observed. Wild-type cells grown at pH 10.5 revealed identical S-layers, while no regularly arrayed S-layer was found in RG21 at either pH (shown for pH 7.5-grown cells in Fig. 3B). Thin sections of the wild type showed the S-layer as the outermost cell envelope component (Fig. 4A) that was absent in ultrathin sections of RG21 (as shown for pH 10.5-grown cells in Fig. 4B).
FIG. 3.
Freeze-etched and metal-shadowed electron micrographs. Images from freeze-etching electron microscopic examination of pH 7.5-grown cells of B. pseudofirmus OF4811M (a) and mutant RG21 (b) are shown. Bars, 100 nm.
FIG. 4.
Electron micrographs of ultrathin sections of cell surface region of B. pseudofirmus OF4811M wild type and mutant strain RG21. Wild-type B. pseudofirmus OF4811M (a) and mutant RG21 (b) were grown at pH 10.5. S, S-layer; PG, peptidoglycan. Bars, 50 nm.
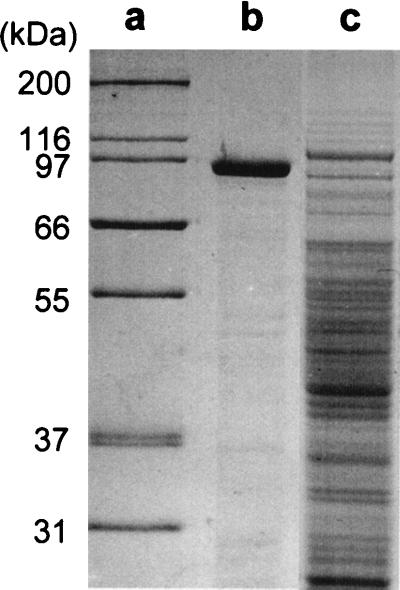
SDS-PAGE analyses of soluble whole-cell extracts of the wild-type strain and mutant RG21 confirmed the results of the ultrastructural examination. In extracts from both pH 7.5- and pH 10.5-grown cells of the wild type, a very prominent band of approximately 90 to 95 kDa was observed; it dominated the protein profile of wild-type cells (shown for pH 7.5-grown cells in Fig. 5B). In gels loaded with the same amount of total cell extract protein to facilitate detection of any SlpA, a comparable band was lacking from extracts of the mutant RG21 (shown for pH 7.5-grown cells in Fig. 5C). In preliminary experiments, periodic acid-Schiff staining of SDS-PAGE gels (as described in reference 25) did not reveal the presence of carbohydrates with unsubstituted vicinal OH groups, as would be found in most glycoprotein S-layers (25) (data not shown). Although two-dimensional gel electrophoresis experiments suggested that at least some breakdown products of SlpA were present in greater amounts at pH 10.5, the level of the 90- to 95-kDa species was not consistently higher in extracts from pH 10.5-grown cells than in extracts from pH 7.5-grown cells. In some preparations, this was the case, but in others there was no difference or even a higher intensity of the 90- to 95-kDa band in the preparations from pH 7.5-grown cells. Although not shown, Northern analyses clearly showed a band corresponding in size to that anticipated for slpA alone (i.e., with the potential to encode a protein of no more than 98.8 kDa), but there was no difference in the abundance of that RNA between preparations from pH 7.5- and 10.5-grown cells.
FIG. 5.
SDS-PAGE analyses of whole-cell extracts of B. pseudofirmus OF4811M and mutant strain RG21. Lanes: a, molecular mass standards; b, wild type grown at pH 7.5; c, RG21 grown at pH 7.5.
The SDS-PAGE analyses and Northern results suggested that the high level of SlpA in B. pseudofirmus OF4811M might be largely, if not entirely, pH independent, even though attention had been drawn to this molecule by the particular prominence of some breakdown products in the two-dimensional gels from pH 10.5-grown cells. Gels of both the cytoplasmic and membrane-associated proteins (Fig. 1A and B) at both pHs exhibited pronounced, somewhat smeared bands in the region corresponding to a size of about 94 kDa. This was an apparent candidate for a major SlpA form correlating better with the size of the band observed in the SDS-PAGE gels and the anticipated molecular size of SlpA. Three distinct samples from that region, taken in the area shown by the dashed box in the bottom panel of Fig. 1B, were therefore analyzed. All three samples had the same N-terminal sequence as spot 17, APADAKFSDV.
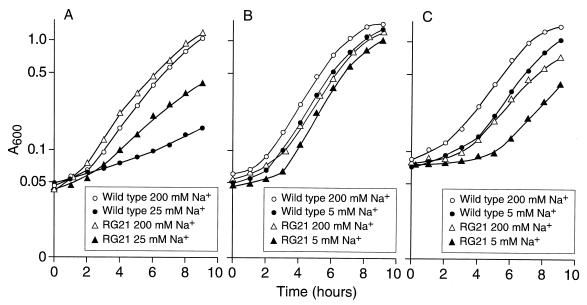
Growth studies of the wild type and RG21 were conducted in highly buffered batch cultures at starting pHs of 7.5, 10.5, and 11. B. pseudofirmus OF4 generally exhibits slightly faster growth on malate at pH 10.5 than at either of the other two pHs (42). We also sought to examine both an Na+-replete condition and one in which the Na+ concentration was expected to be suboptimal for the function of the Na+ cycle. Since B. pseudofirmus OF4 requires a higher concentration of Na+ during growth at pH 7.5 (23), the suboptimal Na+ condition for pH 7.5 was 25 mM added Na+, whereas that chosen for pH 10.5 and 11 was 5 mM. For the Na+-replete condition, 200 mM added Na+ was used at all three pHs. As shown in Fig. 6, the most striking differences between the mutant and wild-type strains were observed at suboptimal pH and/or Na+ concentration, but general trends were evident. (i) At pH 7.5, the slpA deletion mutant RG21 grew better than the wild type, i.e., with a slightly but reproducibly shorter lag under Na+-replete conditions and with a significantly faster growth rate at suboptimal Na+ levels. (ii) At pH 10.5 and 11, the wild type grew with a shorter lag than was observed in the mutant RG21, and the extent of the “adjustment defect” in the mutant was exacerbated by both the higher pH and suboptimal Na+ conditions. (iii) After the more extended lag of the mutant at pH 10.5 or 11, logarithmic growth proceeded at rates that differed little if at all from those of the wild type. Phase microscopic examination indicated that the mutant and wild type exhibited comparable motility at pH 10.5. Several other variables were examined in growth experiments because of the possibility that a highly charged S-layer might contribute to iron and/or magnesium acquisition, which is particularly challenging at very high pH, and might be involved in binding extracellular enzymes such as amylase to facilitate the retrieval of the enzymatic products by the alkaliphile. Although not shown, the wild-type and mutant strains did not exhibit differences in their growth patterns on limiting iron or magnesium concentrations at high or low pH or in formation of a zone of digestion on starch plates.
FIG. 6.
Growth of B. pseudofirmus OF4811M and RG21 at different external pHs. Cultures were grown at pH 7.5 (A) with either 25 or 200 mM added Na+ or at pH 10.5 (B) or 11 (C) with either 5 or 200 mM added Na+. The A600 was monitored at intervals.
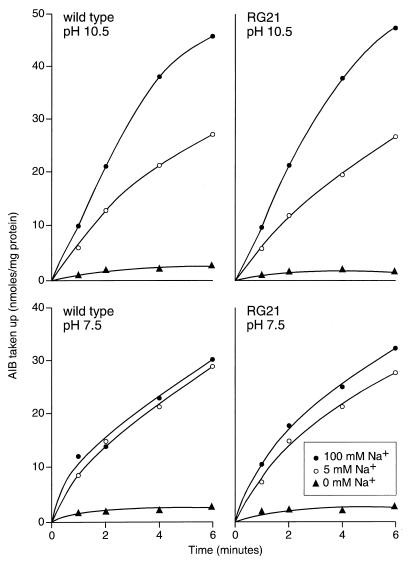
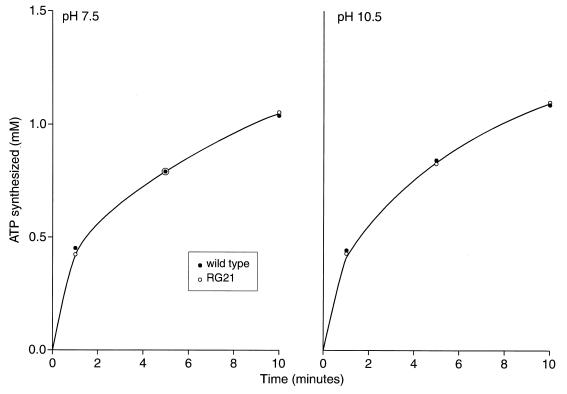
Experiments were then focused on cation-coupled energetic processes. The capacity for cytoplasmic pH regulation was examined under conditions of a sudden shift to external pH and was also assessed in “steady-state” logarithmically growing cells. In the logarithmic-phase cells, two other ion-coupled processes, apart from motility, were also examined. When the external pH was suddenly shifted from 8.5 to 10.5, the mutant consistently exhibited a relatively high internal pH (9.12 ± 0.12 and 9.10 ± 0.11 at 5 and 100 mM Na+, respectively), while the wild type maintained an internal pH of 8.53 ± 0.04 and 8.3 ± 0.12 at added Na+ of 5 and 100 mM, respectively. By contrast, the cytoplasmic pH determined for cells of the wild type and RG21 in the middle of logarithmic growth at pH 10.5 at either suboptimal or replete Na+ conditions (as in Fig. 6B) was between 8.3 and 8.4 (data not shown). Other ion-coupled functions in logarithmic-phase cells were similarly unaffected by the slpA status. The Na+-dependent uptake of AIB was the same in wild-type and mutant RG21 at either pH 7.5 or 10.5, in the presence or absence of a high or low concentration of Na+ (Fig. 7). The rate of ATP synthesis, which is H+ coupled in B. pseudofirmus OF4, was also comparable in the wild type and mutant at both pHs (Fig. 8).
FIG. 7.
Transport of AIB by cells of wild-type B. pseudofirmus OF4811M and mutant RG21. Cells grown and washed as described in Materials and Methods were assayed for the uptake of AIB at either pH 10.5 or 7.5 in the presence of no added Na+ or 5 or 100 mM Na+.
FIG. 8.
Synthesis of ATP upon addition of malate to starved cells of wild-type B. pseudofirmus OF4811M or RG21. The cells were starved for 8 h as described in Materials and Methods and reenergized at either pH 7.5 or 10.5 by the addition of 10 mM potassium malate. Samples were taken and assayed for ATP content at various times.
DISCUSSION
The general findings that emerge from two-dimensional gel electrophoresis analyses of steady state pH 7.5- and pH 10.5-grown cells of B. pseudofirmus OF4 are that only a very few cytoplasmic proteins and even a minority of the resolved membrane-associated proteins show significant changes in level as a function of growth pH. The observation is not surprising for the cytoplasmic proteins, inasmuch as the facultative alkaliphile maintains a cytoplasmic pH that is below 8.5 during growth in the entire range from pH 7.5 to 10.5 (16, 42). The cytoplasmic pH is only above pH 7.5 at external pHs above 9.5 and is only about pH 8.3 at an external pH of 10.5 (42). A cytoplasmic pH of 7.5 is normal for nonalkaliphilic prokaryotes under near-neutral external pH conditions (27). Adaptation to optimal growth at a cytoplasmic pH as high as 8.3 during growth of B. pseudofirmus OF4 at pH 10.5 may require subtle accommodations. However, it seems unlikely that these accommodations would be so drastic as to preclude robust growth at near-neutral pH and therefore force major changes in the cytoplasmic proteome. On the other hand, the features of the cell surface polymers and proteins that are entirely or partially exposed to the external pH might have more radical adaptations than the cytoplasmic complement. Such major differences might mandate production of different versions of a substantial number of these proteins during steady-state growth at pH 10.5 from those that are present at pH 7.5.
The findings here suggest, by contrast, that B. pseudofirmus OF4 is a well-adapted extremophile that appears to constitutively express much of the protein complement that supports extremely alkaliphilic growth even though that is not optimal for growth at near-neutral pH. For example, the failure of B. pseudofirmus OF4 to grow on malate below pH 7.5 is associated with properties of the cell membrane phospholipid composition that are apparently important for growth at very alkaline pH (12, 26). The requirement of the alkaliphile for a higher Na+ concentration to support its growth at pH 7.5 than at much more alkaline pHs probably also arises from the alkaliphile's use of a largely constant panoply of Na+-coupled transporters over a broad pH range. The transporters may be poised, by features of their primary sequence and structure, for maximal activity at very high pH, at which the competition by a low H+ concentration is relatively small. At lower pHs, the much more abundant protons, which cannot serve as coupling ions, may be effective competitive inhibitors. This could account for the requirement for higher Na+ levels for operation of the Na+ cycle at pH 7.5 than at pH 10.5.
In making generalizations about the protein complement based on the two-dimensional gel electrophoresis analyses, it is important to note a caveat. This study has not focused on proteins specially expressed during an alkaline shift, nor has it identified all the membrane-associated proteins with an important role and increased expression during growth at high pH. Two major reasons can be cited for the failure to identify such steady-state proteins. First, standard two-dimensional gels, as used in this work, only resolve proteins within the pI range from 4 to 8. Many proteins have pI values above and below this range and so cannot be analyzed under the standard conditions used here. For example, the pI for the flagellin protein from Bacillus firmus RAB, an obligate alkaliphile related to B. pseudofirmus OF4, is 3.3 (15). The B. pseudofirmus OF4 homologue of this protein may therefore not be resolved in this study despite the fact that its production probably is increased at the alkaline pH: motility is only observed at an external growth pH above 8.5 (42). Second, many membrane proteins, such as an inducible antiporter inferred to be a contributor to pH homeostasis in B. pseudofirmus OF4 (23) or even more readily extracted proteins, exist in such low amounts that even large increases in expression may not be observed, especially if the protein migrates in a region that contains a large number of polypeptides.
In spite of this caveat, the findings here for SlpA strongly support the general thesis that proteins in alkaliphilic B. pseudofirmus OF4 that have roles in alkaliphily are likely to be expressed significantly at pH 7.5 to facilitate adaptation to rapid upward changes in pH. For SlpA, this is the case even though its expression is apparently somewhat detrimental at “low” pH and even though SlpA is not essential for either adaptation to or growth under the high-pH condition. B. pseudofirmus OF4 SlpA is the first S-layer protein of which we are aware to be described in an extreme alkaliphile. Across a wide spectrum of other prokaryotes and archaea, S-layers have been documented and noted to be found in amounts that represent a significant investment of cellular energy (28). Some organisms have multiple S-layer-encoding genes whose relative expression varies with particular conditions, allowing an inference with respect to a role of the S-layer in responding to a particular stress (39). A wide variety of such functions have been proposed, including roles in pathogenicity, divalent cation sequestration and acquisition, adaptation to different oxygen tensions, anchoring of extracellular enzymes, and others, but no single function has emerged as being a common basis for the prevalence of S-layers in microorganisms (8, 9, 38). Rather, as suggested recently by Sára and Sleytr (38), there appears to have been so much functional evolution that the primary, current functions of these widely occurring and “expensive” macromolecular layers have diverged to reflect very specific needs of specific organisms. The findings here support that concept. SlpA plays a role in alkaliphily, a new role for an S-layer. Moreover, other roles that have been suggested in connection with other S-layers and which might have provided a direct rationale for SlpA production at pH 7.5 were not supported, i.e., divalent cation sequestration or growth on substrates dependent upon extracellular amylase activity.
The role of the B. pseudofirmus OF4 SlpA in alkaliphily is somewhat analogous to that proposed for the cell wall teichuronopeptide in B. halodurans C-125 (3–5, 21). Neither of these cell wall polymers in the two different alkaliphiles is essential for alkaliphily, but both are involved in supporting the Na+-dependent pH homeostasis that principally relies upon active transport mechanisms. The role of SlpA in B. pseudofirmus OF4 seems largely restricted to the period of adaptation to a more alkaline exterior. The presence or absence of SlpA did not affect growth rate, pH homeostasis, motility, or two additional ion-coupled processes in logarithmic-phase cells. It is likely that the acidic surface polymers sequester Na+ and/or H+ in some manner that is useful in connection with the antiport of cytoplasmic Na+ for external H+, but only before full induction of the crucial, active Na+ cycle occurs (19, 28). The finding that SlpA is more acidic than homologues from other bacteria is consistent with SlpA's being among those alkaliphile proteins that are exposed to the external medium and that must maximize their content of amino acids that will retain charge at very alkaline pH.
The SlpA is likely to be the outermost layer of the B. pseudofirmus envelope, although it is possible that, like B. anthracis (34), this alkaliphile also possesses a capsule that is external to the S-layer. An operon has been identified in B. pseudofirmus OF4 that is likely to encode a polyglutamate capsule (22), but attempts to visualize such a capsule in living cells have thus far been negative (A. Guffanti, unpublished results). The SlpA-dependent layer of B. pseudofirmus OF4 appears as a smooth oblique S-layer lattice that probably consists, as is generally the case, of identical protein monomers that interact with each other by hydrophobic and electrostatic interactions to cover the cell. Based on the size of the species observed in the SDS-PAGE gels in Fig. 5, and the largest form of SlpA identified on the two-dimensional gels, the monomer size in B. pseudofirmus OF4 is likely to be 90 to 95 kDa. While variable glycosylation products could account for a heterogeneous smear of various pIs, as seen on the two-dimensional gels, more work is needed to rigorously show whether there may be a periodate-Schiff-unreactive carbohydrate component. We conclude that there is little or no increase in SlpA production in steady-state cells at pH 10.5 versus pH 7.5. Rather, there is some instability of SlpA in extracts, and perhaps natural breakdown in cells themselves, such that some high-molecular-weight SlpA degradation products are especially pronounced in the preparations from pH 10.5-grown cells. If there is, in fact, also somewhat greater expression of SlpA at pH 10.5, it is probably achieved at the level of translation, since the slpA RNA abundance was the same in pH 10.5- and 7.5-grown cells. Translational controls have been implicated in expression of an S-layer in Thermus thermophilus HB8 (14).
The other set of proteins elevated at pH 10.5, tentatively identified as spots 33, 41, 100, and 121, appear to be involved in catabolism or synthesis of the derivatives of branched-chain fatty acids and branched-chain amino acids. It is possible that catabolism of branched-chain fatty acids or fatty acids in general is of particular importance in meeting the energy needs of the alkaliphile at high pH. Another interesting possibility is that significant turnover and remodeling of the fatty acid complement are required for adaptation, or even in steady-state growth, at pH 10.5 versus pH 7.5. Another possible synthetic fate of branched-chain fatty acids is their incorporation into lipopeptide antibiotics. There is no current basis for anticipating such a use in B. pseudofirmus OF4. However, in B. subtilis, a number of antibacterial and antifungal agents are known to be lipopeptides containing branched-chain fatty acids (48). The synthetase genes for the antifungal fengycin have been identified in B. subtilis (44). Immediately downstream of the peptide synthetase genes is a gene cluster involved in fatty acid metabolism that has been proposed to be important for the production of branched-chain fatty acid attachment to fengycin. One of the genes in this cluster, yngJ (originally called yotC), is a homologue of spot 100 in the current study (Table 2). For this group of B. pseudofirmus OF4 membrane-associated proteins with elevated levels at pH 10.5, i.e., those apart from SlpA that have tentative identifications, further work will be needed to confirm or modify the identifications and to probe the actual functions in relation to alkaliphily.
ACKNOWLEDGMENTS
The work described was supported by research grants GM 28454 from the National Institutes of Health and DE-FG02-86ER13559 from the Department of Energy to T.A.K. and from the Austrian Science Fund, project P12966-MOB, to P.M.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 3.Aono R. Isolation and partial characterization of structural components of the walls of alkalophilic Bacillus strain C-125. J Gen Microbiol. 1985;131:105–111. [Google Scholar]
- 4.Aono R, Ito M, Joblin K N, Horikoshi K. A high cell wall negative charge is necessary for the growth of the alkaliphile Bacillus lentus C-125 at elevated pH. Microbiology. 1995;141:2955–2964. [Google Scholar]
- 5.Aono R, Ito M, Machida T. Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphilic Bacillus lentus C-125. J Bacteriol. 1999;181:6600–6606. doi: 10.1128/jb.181.21.6600-6606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7.Bernhardt J, Volker U, Volker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 8.Beveridge T J. Role of cellular design in bacterial metal accumulation and mineralization. Annu Rev Microbiol. 1989;43:147–171. doi: 10.1146/annurev.mi.43.100189.001051. [DOI] [PubMed] [Google Scholar]
- 9.Beveridge T J, Pouwels P H, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer E M, Schocher I, Sleytr U B, Morelli L, Callegari M L, Nomellini J F, Bingle W H, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Beguin P, Ohayon H, Gounon P, Matuschek M, Koval S F. Functions of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 10.Boynton Z L, Bennet G N, Rudolph F B. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clejan S, Guffanti A A, Cohen M A, Krulwich T A. Mutation of Bacillus firmus OF4 to duramycin resistance results in substantial replacement of membrane lipid phosphatidylethanolamine by its plasmalogen form. J Bacteriol. 1989;171:1722–1746. doi: 10.1128/jb.171.3.1744-1746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clejan S, Krulwich T A, Mondrus K R, Seto-Young D. Membrane lipid composition of obligately and facultatively alkalophilic strains of Bacillus spp. J Bacteriol. 1986;168:334–340. doi: 10.1128/jb.168.1.334-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimari J F, Bechhofer D. Initiation of mRNA decay in Bacillus subtilis. Mol Microbiol. 1991;7:705–717. doi: 10.1111/j.1365-2958.1993.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Herrero L A, Olabarria G, Berenguer J. Surface proteins and a novel transcription factor regulate the expression of the S-layer gene in Thermus thermophilus HB8. Mol Microbiol. 1997;24:61–72. doi: 10.1046/j.1365-2958.1997.3191683.x. [DOI] [PubMed] [Google Scholar]
- 15.Guffanti A A, Eisenstein H C. Purification and characterization of flagella from the alkalophile Bacillus firmus RAB. J Gen Microbiol. 1983;129:3239–3242. [Google Scholar]
- 16.Guffanti A A, Hicks D B. Molar growth yields and bioenergetic parameters of extremely alkaliphilic Bacillus species in batch cultures, and growth in a chemostat at pH 10.5. J Gen Microbiol. 1991;137:2375–2379. doi: 10.1099/00221287-137-10-2375. [DOI] [PubMed] [Google Scholar]
- 17.Guffanti A A, Krulwich T A. Features of apparent nonchemiosmotic energization of oxidative phosphorylation by alkaliphilic Bacillus firmus OF4. J Biol Chem. 1992;267:9580–9588. [PubMed] [Google Scholar]
- 18.Guffanti A A, Krulwich T A. Oxidative phosphorylation by ADP + Pi-loaded membrane vesicles of alkaliphilic Bacillus firmus OF4. J Biol Chem. 1994;269:21576–21582. [PubMed] [Google Scholar]
- 19.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. I. New York, N.Y: IRL Press; 1985. pp. 109–136. [Google Scholar]
- 21.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito M, Cooperberg B, Krulwich T A. Diverse genes of alkaliphilic Bacillus firmus OF4 that complement K+-uptake-deficient Escherichia coli include an ftsH homologue. Extremophiles. 1997;1:22–28. doi: 10.1007/s007920050011. [DOI] [PubMed] [Google Scholar]
- 23.Ito M, Guffanti A A, Zemsky J, Ivey D M, Krulwich T A. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J Bacteriol. 1997;179:3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones B E, Grant W D, Duckworth A W, Owenson G G. Microbial diversity of soda lakes. Extremophiles. 1998;2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- 25.Kosma P, Neuninger C, Christian R, Schulz G, Messner P. Glycan structure of the S-layer glycoprotein of Bacillus sp. L420-91. Glycoconjugate J. 1995;12:99–107. doi: 10.1007/BF00731875. [DOI] [PubMed] [Google Scholar]
- 26.Krulwich T A. Alkaliphilic prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes: an evolving database for the microbiological community. 3rd ed. Springer-Verlag; 1999. www.prokaryotes.com , version 3.1. [Online.] www.prokaryotes.com. . [Google Scholar]
- 27.Krulwich T A, Guffanti A A, Ito M. Novartis Found. Sym. 1999. pH tolerance in Bacillus: alkaliphile vs non-alkaliphile; pp. 167–182. p. 221: Mechanisms by which bacterial cells respond to pH. Wiley, Chichester, England. [DOI] [PubMed] [Google Scholar]
- 28.Krulwich T A, Ito M, Gilmour R, Hicks D B, Guffanti A A. Energetics of alkaliphilic Bacillus species: physiology and molecules. Adv Microb Physiol. 1998;40:410–438. doi: 10.1016/s0065-2911(08)60136-8. [DOI] [PubMed] [Google Scholar]
- 29.Krulwich T A, Ito M, Hicks D B, Gilmour R, Guffanti A A. pH homeostasis and ATP synthesis: studies of two processes that necessitate inward proton translocation in extremely alkaliphilic Bacillus species. Extremophiles. 1998;2:217–222. doi: 10.1007/s007920050063. [DOI] [PubMed] [Google Scholar]
- 30.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M-F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinios S, Lauber J, Lazarevic V, Lee S-M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O'Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S-H, Parro V, Pohl T M, Portelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Lupas A, Engelhardt H, Peters J, Santarius U, Volker S, Baumeister W. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Stauffer K, Busse H-J, Rainey F A, Burghardt J, Scheberl A, Hollaus F, Kuen B, Makristhatis A, Sleytr U B, Messner P. Description of Bacillus thermoaerophilus sp. nov., to include sugar beet isolates and Bacillus brevis ATCC 12990. Int J Syst Bacteriol. 1996;46:532–541. [Google Scholar]
- 34.Mesnage S, Tosi-Couture E, Gounon P, Mock M, Gouet A. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J Bacteriol. 1998;180:52–58. doi: 10.1128/jb.180.1.52-58.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor C D, Farris M, Fowler R, Qi S Y. The proteome of Salmonella enterica serovar Typhimurium: current progress on its determination and some applications. Electrophoresis. 1997;18:1483–1490. doi: 10.1002/elps.1150180823. [DOI] [PubMed] [Google Scholar]
- 36.O'Farrel P H. High resolution two dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 37.Oku H, Kaneda T. Biosynthesis of branched-chain fatty acids in Bacillus subtilis: a decarboxylase is essential for branched-chain fatty acid synthetase. J Biol Chem. 1988;263:18386–18396. [PubMed] [Google Scholar]
- 38.Sára M, Sleytr U B. S-layer proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sára M, Kuen B, Mayer H F, Mandl F, Schuster K C, Sleytr U B. Dynamics in oxygen-induced changes in S-layer protein synthesis from Bacillus stearothermophilus PV72 and the S-layer-deficient variant T5 in continuous culture and studies of the cell wall composition. J Bacteriol. 1996;178:2108–2117. doi: 10.1128/jb.178.7.2108-2117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleytr U B, Messner P, Pum D. Analysis of crystalline bacterial cell surface layers by freeze-etching, metal shadowing, negative staining, and ultrathin sectioning. Methods Microbiol. 1988;20:29–60. [Google Scholar]
- 41.Sleytr U B, Messner P, Pum D, Sára M. Crystalline bacterial cell surface layers. Mol Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 42.Sturr M G, Guffanti A A, Krulwich T A. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J Bacteriol. 1994;176:3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiyama S. Na+-driven flagellar motors as a likely Na+ re-entry pathway in alkaliphilic bacteria. Mol Microbiol. 1995;15:592. doi: 10.1111/j.1365-2958.1995.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 44.Tosato V, Albertini A M, Zotti M, Sonda S, Bruschi C V. Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology. 1997;143:3443–3450. doi: 10.1099/00221287-143-11-3443. [DOI] [PubMed] [Google Scholar]
- 45.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene protein database of E. coli K-12, edition 6. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2067–2117. [Google Scholar]
- 46.Van der Laan J M, Gerritse G, Mulleners L J S M, Van der Hock R A C, Quax W J. Cloning, characterization, and multiple chromosomal integration of a Bacillus alkaline protease gene. Appl Environ Microbiol. 1991;57:901–909. doi: 10.1128/aem.57.4.901-909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G F, Kuriki T, Roy K L, Kaneda T. The primary structure of branched-chain alpha-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other alpha-oxo acid dehydrogenases. Eur J Biochem. 1993;213:1091–1099. doi: 10.1111/j.1432-1033.1993.tb17858.x. [DOI] [PubMed] [Google Scholar]
- 48.Zuber P, Nakano M M, Marahiel M A. Peptide antibiotics. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: ASM Press; 1993. pp. 897–916. [Google Scholar]