Abstract
Streptococcus thermophilus, unlike many other gram-positive bacteria, prefers lactose over glucose as the primary carbon and energy source. Moreover, lactose is not taken up by a phosphoenolpyruvate-dependent phosphotransferase system (PTS) but by the dedicated transporter LacS. In this paper we show that CcpA plays a crucial role in the fine-tuning of lactose transport, β-galactosidase (LacZ) activity, and glycolysis to yield optimal glycolytic flux and growth rate. A catabolite-responsive element (cre) was identified in the promoter of the lacSZ operon, indicating a possible role for regulation by CcpA. Transcriptional analysis showed a sevenfold relief of repression in the absence of a functional CcpA when cells were grown on lactose. This CcpA-mediated repression of lacSZ transcription did not occur in wild-type cells during growth on galactose, taken up by the same LacS transport system. Lactose transport during fermentation was increased significantly in strains carrying a disrupted ccpA gene. Moreover, a ccpA disruption strain was found to release substantial amounts of glucose into the medium when grown on lactose. Transcriptional analysis of the ldh gene showed that expression was induced twofold during growth on lactose compared to glucose or galactose, in a CcpA-dependent manner. A reduced rate of glycolysis concomitant with an increased lactose transport rate could explain the observed expulsion of glucose in a ccpA disruption mutant. We propose that CcpA in S. thermophilus acts as a catabolic regulator during growth on the preferred non-PTS sugar lactose. In contrast to other bacteria, S. thermophilus possesses an overcapacity for lactose uptake that is repressed by CcpA to match the rate-limiting glycolytic flux.
Carbon catabolite repression (CR) in bacteria is the phenomenon of using a rapidly metabolizable carbon source in the growth medium by inhibiting utilization of other substrates. The mechanism underlying CR is best understood in enteric bacteria, where the glucose-specific enzyme IIA of the phosphoenolpyruvate-dependent phosphotransferase system (PTS) modulates adenylate cyclase activity. Controlled by the level of cyclic AMP, the cyclic AMP receptor protein is a transcriptional regulator modulating expression of target genes (36, 38). In low-G+C gram-positive bacteria, the mechanism of CR is distinctly different. The catabolite control protein A (CcpA) is the central regulator of CR, as was shown first for Bacillus subtilis, in which it mediates glucose repression of the α-amylase gene (9). CcpA is a member of the LacI-GalR family of bacterial regulator proteins and appears to be widespread among low-G+C gram-positive bacteria (4, 12, 21, 29). Genes affected by CR typically contain a catabolite-responsive element (cre) near their promoter regions (44). CcpA has been shown to bind to these cre sites in vitro in a way that can be enhanced by indicators of a high energy state in the cell, e.g., glucose 6-phosphate (6, 27). Another important factor in this catabolite control mechanism is the PTS phosphocarrier HPr. In B. subtilis, high concentrations of the glycolytic intermediate fructose-1,6-diphosphate (FBP) trigger an ATP-dependent protein kinase that phosphorylates HPr at residue Ser-46. P-Ser-HPr subsequently enhances the binding of CcpA to cre and hence links glycolytic activity to CR (2, 5, 15). Catabolite control by CcpA involves not only repression of genes and operons but also activation. In B. subtilis, transcription of the alsS and ackA genes (encoding α-acetolactate synthase and acetate kinase, respectively) is activated by CcpA when glucose is present in the medium (7, 37). More direct evidence for a link between catabolite control and glycolytic activity was reported recently for Lactococcus lactis. In the presence of glucose in the medium, CcpA was found to be a transcriptional activator of the las operon, thus modulating glycolytic flux rates by controlling the production of the three key glycolytic enzymes, phosphofructokinase, pyruvate kinase, and lactate dehydrogenase (22).
Although the mechanism of CR differs between gram-negative and low-G+C gram-positive bacteria, they have in common that a rapidly metabolizable PTS sugar reduces the expression of genes involved in the utilization of other PTS or non-PTS carbon sources. Glucose is the classical example of such a rapidly metabolizable PTS sugar in most bacteria. However, glucose is a non-PTS carbon source for Streptococcus thermophilus and is a poor substrate for growth (34). Lactose is also a non-PTS sugar for this organism but is a very good growth substrate on which growth is even more rapid than on a PTS sugar like sucrose. This indicates that S. thermophilus, a homofermentative thermophilic lactic acid bacterium, is highly adapted to growth on lactose as the primary carbon and energy source. Together with other lactic acid bacteria, this organism is used as a starter culture for the production of yogurt and certain cheeses, where it mainly contributes to the rapid acidification of milk by conversion of lactose to lactic acid.
The S. thermophilus lac operon contains the genes encoding a lactose permease (lacS) and a β-galactosidase (lacZ) for the transport and hydrolysis of lactose, and its transcription is induced during growth on lactose (32, 40). Studies of the lac operon revealed a cre site located in the lacSZ promoter, suggesting a possible involvement of CcpA in the regulation of this operon (34). The S. thermophilus galM and galE genes, encoding enzymes of the Leloir pathway for galactose fermentation, were found upstream of this lac operon (33). The complete galKTE operon was recently identified in strain CNRZ302, which is unable to grow on galactose, like most S. thermophilus strains (E. E. Vaughan, P. T. C. van den Bogaard, P. Catzeddu, O. P. Kuipers, and W. M. de Vos, submitted for publication). From this strain, galactose-fermenting mutants were isolated, and their molecular characterization showed that these mutants were all galK promoter-up mutants. One of these mutants, used in this study, was designated NZ302G. Insertional mutagenesis studies of the galR gene located upstream of the galKTE operon, encoding a regulator protein of the LacI-GalR family of transcriptional regulators, showed that the GalR protein was an activator of the galK promoter (Vaughan et al., submitted). Transcription of this promoter was induced when cells were grown on medium containing lactose or galactose. Furthermore, GalR was also found to be a transcriptional activator of the lac operon, which is expressed at a basal level when cells are grown on glucose, while it is expressed at least twice as high in lactose- or galactose-grown cells.
In this study we show that in S. thermophilus, CcpA is acting as a transcriptional repressor of the lac operon and an activator of genes encoding key glycolytic enzymes, induced by the non-PTS sugar lactose. This catabolite control is probably regulated by the glycolytic intermediates that are derived from the glucose moiety of lactose rather than from a PTS sugar in the growth medium. We provide evidence that CcpA is involved in fine-tuning the rate of lactose transport with glycolytic activity, enabling rapid fermentation and high growth rate of S. thermophilus.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
S. thermophilus was routinely grown at 42°C in M17 broth (Difco, Surrey, U.K.) containing 1% of the chosen carbon source unless stated otherwise. Escherichia coli strains were grown in TY broth (39) with aeration at 37°C. The antibiotics used for selection in growth media were chloramphenicol (Cm, 4 μg/ml) and erythromycin (Em, 2.5 μg/ml) for S. thermophilus and ampicillin (Ap, 50 μg/ml), Cm (10 μg/ml), and Em (150 μg/ml) for E. coli.
DNA manipulations and transformations.
Transfer to and isolation of plasmid DNA from E. coli was performed using established protocols (39). Plasmid and chromosomal DNA from S. thermophilus was isolated as described previously for L. lactis (43). Electroporation of S. thermophilus was performed by the procedure described by Mollet et al. (28) with the modification that the harvested cells were incubated in the electroporation buffer at 4°C for at least 4 h prior to electroporation. Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were used as recommended by the suppliers (Gibco-BRL or Boehringer Mannheim). DNA fragments were recovered from agarose gels using the glass matrix DNA isolation system (Gibco-BRL).
Cloning and disruption of the S. thermophilus ccpA gene.
Total genomic DNA from S. thermophilus CNRZ302 was digested with HindIII and KpnI, and the DNA fragments were separated by agarose gel (0.7%) electrophoresis. The DNA was transferred to a Gene Screen Plus (Dupont, Boston, Mass.) membrane by standard procedures (39). A 3.3-kb hybridizing fragment was identified using a 1-kb fragment of the B. subtilis 1G33 ccpA gene (kindly provided by E. Luesink) that was gel purified and labeled by nick translation using [α-32P]dATP (Amersham International plc, London, U.K.). S. thermophilus chromosomal DNA was digested with HindIII and KpnI, and fragments with sizes of between 3.0 and 3.5 kb were recovered, ligated with HindIII- and KpnI-digested pUC19 (45), and transformed into E. coli MC1061. Clones carrying the ccpA gene were selected by colony blotting of the Ap-resistant colonies (39) onto Gene Screen Plus membranes and probing the transferred minibank with the radiolabeled B. subtilis ccpA gene. Sequence analysis of the positive clones confirmed that a 3.3-kb insert in pUC19 contained a ccpA-like gene. This construct was designated pNZ6100 and used in further experiments. A 799-bp internal PCR fragment of the S. thermophilus ccpA gene was generated from pNZ6100 as a template using primers CCPAKF (5′-GCTCGAAGTCATTGATCG-3′) and CCPAKR (5′-AGTCAACATACGCATGCT-3′) and ligated into pGEM-T (Promega), yielding pNZ6101. Plasmid pNZ6102 was generated by cloning the insert with ApaI and SalI from pNZ6101 into the thermosensitive pGh9 vector (23, 24) digested with the same restriction enzymes. S. thermophilus strains CNRZ302 and NZ302G transformed with pNZ6102 were selected at 28°C on M17 sucrose medium supplemented with Em. To facilitate integration of pNZ6102, cultures grown overnight in M17 glucose broth with Em at 28°C were diluted 100-fold into fresh medium and reincubated at 28°C to allow the exponential phase of growth to resume. The cultures were then shifted to 42°C and grown until stationary phase. Dilutions of the cultures were plated at 42°C, and integrants appeared as Em-resistant colonies after 24 to 48 h of incubation. Integration of pNZ6102 into the ccpA locus of CNRZ302 and NZ302G should result in two truncated and inactive copies of the ccpA gene. Correct integration was confirmed by PCR and Southern analysis and yielded strains NZ6150 and NZ6151, which were handled further at 42°C with Em to maintain the integrated plasmid. For complementation studies of the ccpA disruption mutant, plasmid pNZ6100 was digested with AccI, and the ends were filled in using Klenow polymerase followed by a second digestion with HindIII. The 1.3-kb fragment containing the ccpA gene was ligated in pNZ273 (31), from which the gusA gene was removed by digestion with ScaI and HindIII. The resulting plasmid (pNZ6103) harbors the S. thermophilus ccpA gene under the control of its own promoter. The S. thermophilus CNRZ302 ldh promoter was obtained by PCR using Pwo DNA polymerase (Boehringer Mannheim) and primers LDH1 (5′-ACACTCATGGCATAATCGATA-3′) and LDH2 (5′-AGTTCTTGAGCGATACCTTG-3′) based on the sequence of the ldh locus of strain M-192 (14). The promoter fragment was adenylated using Taq polymerase and ligated into pGEM-T (Promega), yielding pNZ6104.
RNA isolation, Northern blot, and primer extension analysis.
S. thermophilus strains were grown in M17 broth (30 ml) containing 1% glucose or lactose to an optical density at 600 nm (OD600) of 1.0. Total RNA was isolated from the harvested cells using the Macaloid method as described by Kuipers et al. (18) with the following adaptation. Prior to bead beating, the resuspended cells were incubated with lysozyme for 5 min on ice to increase RNA yield. Per sample, 4.5 μg of RNA was size separated on a 1.0% formaldehyde gel (39) and transferred to Gene Screen Plus membranes (Dupont) according to the protocols provided by the manufacturers. RNA size markers were obtained from Bethesda Research Laboratories. Hybridizations were performed at 65°C in a 0.5 M sodium phosphate buffer (pH 7.2) containing 1.0% bovine serum albumin (fractionV), 1.0 mM EDTA, and 7.0% sodium dodecyl sulfate (SDS), and subsequently, blots were washed at 55 to 65°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Internal PCR fragments were generated using S. thermophilus CNRZ302 chromosomal DNA as the template and primers based on published sequence data: lacS, LACSF (5′-TAACACAGGTGATCCAAAGCA-3′) and LACSR (5′-GGTGACCAGAACTCAAGAAG-3′) (30); and ldh, STLDH-F (5′-GTCATCCTTGTTGGTGACGG-3′) and STLDH-R (5′-TGCTTCATCAATGATAGCTTGC-3′) (12). These fragments were glass matrix purified, labeled by nick translation with [α-32P]dATP (Amersham International plc), and subsequently used as hybridization probes (39). The images of the Northern blots were exposed to a Storage Phosphor Screen (Molecular Dynamics) and scanned using a STORM 840 PhosphorImager (Molecular Dynamics). The Northern signals were quantified using the ImageQuant 1.2 program (Molecular Dynamics). Per Northern blot, a final 16S rDNA probe, created by PCR with 16S-specific primers (NR7 [5′-GAAGCAGCGTGG-3′] and NR19 [5′-GTCGTTATGCGGTA-3′]) with S. thermophilus CNRZ302 chromosomal DNA as the template, was used to correct the gene-specific signals for the total amount of RNA loaded per sample, which never differed by more than 20%. Primer extension was performed as previously described (39) by annealing 20 ng of oligonucleotide PECCPA (5′-CGATACTCCAGCTTCACGCGC-3′; ccpA) or PELDH (5′-CGGCACCGTCACCAACAAGG-3′; ldh) to 15 μg of total S. thermophilus CNRZ302 mRNA. The primer extension reaction was loaded on a 5% polyacrylamide gel together with a sequencing reaction obtained using the same oligonucleotide primer and an appropriate template.
β-Galactosidase and protein assays.
The S. thermophilus strains were grown to an OD600 of 1.0 in M17 broth containing 1% lactose, galactose, or glucose. For the preparation of cell extracts, cells were disrupted with zirconium glass beads in a Bead Beater (Biospec Products, Bartlesville, Okla.) by 3-min treatments, with intervals of 1 min in between on ice; cellular debris was removed by centrifugation. The extracts were kept on ice, and enzyme assays were performed within 2 h using 1 to 6 μg of protein per reaction. β-Galactosidase was assayed at 42°C by the method of Miller (26). Lactate dehydrogenase was assayed at 25°C by the method of Hillier and Jago (11). All enzyme activity measurements presented are the means of at least two independent experiments. Protein concentrations were estimated by a dye-binding assay (1) using bovine serum albumin as the standard.
Small-scale sugar fermentations.
The S. thermophilus strains were grown to an OD600 of 1.0 in M17 broth containing 1% lactose or galactose, washed, and resuspended in a 4% β-glycerophosphate solution at a final OD600 of 10.0. The cells were preincubated for 2 min at 42°C, and fermentation was started by addition of 20 mM lactose. Consecutive samples were taken at regular time intervals from the primary fermentation suspension and immediately transferred to −5°C in salted ice water to prevent further uptake and metabolic conversion. The samples were centrifuged, and supernatants were analyzed by high-performance liquid chromatography (HPLC). Sugars were separated on a Polyspher CHPb18 column (Merck) with water as the eluent. Organic acids were separated on a Rezex organic acid column (Phenomenics) using 5 mM sulfuric acid as the eluent. The separations were carried out on an isocratic pumping system (M6000; Perkin-Elmer) in combination with an automatic sample injector (717+; Waters) and a refractive index detector (M410; Waters).
Western blot analysis.
For CcpA detection, cells were grown to an OD600 of 1.0 in M17 broth containing 1% lactose or glucose. For the preparation of cell extracts, cells were disrupted with zirconium glass beads in a Bead Beater (Biospec Products) by three treatments of 1 min, interspaced by 1 min of cooling of the samples on ice; cellular debris was removed by centrifugation. For LacS analysis, portions of the cells that were used for the small-scale sugar fermentations were protoplasted by extensive treatment with a combination of lysozyme (2 mg/ml) and mutanolysin (25 U/ml) in THMS buffer (30 mM Tris [pH 8.0], 3 mM MgCl2 in 25% sucrose). The protoplasts were washed once in THMS buffer and dissolved in a buffer containing 50 mM potassium phosphate (pH 8.0), 100 mM NaCl, 20% (vol/vol) glycerol, and 0.5% (wt/vol) Triton X-100 to solubilize the LacS protein (16). The suspensions were mixed, and after 20 min of incubation at 4°C, the insoluble material was removed by centrifugation.
Protein concentrations were estimated by a dye-binding assay (1). Samples were equalized and separated by SDS–12.5% polyacrylamide gel electrophoresis. The separated proteins were transferred to Gene Screen Plus membranes (Dupont) using electroblot equipment (LKB 2051 Midget Multiblot). CcpA proteins were detected using antibodies raised against Bacillus megaterium CcpA that were shown to cross-react with CcpAs from various organisms (19). LacS proteins were detected using antibodies raised against the COOH terminus of LacS (35). These antibodies were detected using goat anti-rabbit immunoglobulin peroxidase-conjugated antibodies (Gibco-BRL) as described by the manufacturer.
Nucleotide sequence accession number.
These sequence data have been submitted to the GenBank database under accession number AF231985.
RESULTS
Cloning, characterization, and disruption of the S. thermophilus ccpA gene.
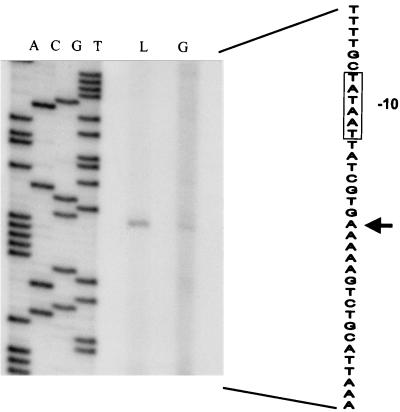
To determine the mechanism of CR in S. thermophilus CNRZ302, its ccpA gene was identified on a 3.3-kb chromosomal fragment on basis of its hybridization with the B. subtilis ccpA gene and subsequently cloned, resulting in plasmid pNZ6100. Nucleotide sequence analysis showed the fragment to contain two open reading frames (ORFs). Translation of one ORF predicted a protein of 333 amino acids, corresponding to a calculated molecular mass of 36.7 kDa, which is referred to as the S. thermophilus CcpA, since it shared 49% amino acid identity with B. subtilis CcpA (9). The greatest identity, however, was shared with the CcpA of Streptococcus mutans (80% identical amino acid residues) (41). An inverted repeat structure and a stretch of five T residues, which could function as a rho-independent transcriptional terminator, followed the coding ccpA sequence. Primer extension experiments using total RNA from S. thermophilus CNRZ302 grown on glucose or lactose revealed an identical transcriptional start site located 38 bp upstream of the ccpA coding region (Fig. 1). Northern analysis revealed a single transcript of approximately 1.2 kb, supporting the functional role of the terminator (data not shown). The second ORF could encode a product with a high level of sequence similarity to proline peptidases (S. mutans, 52% identical amino acid residues; Lactobacillus delbrueckii, 49% identical amino acid residues) (30, 41) and was designated pepQ. The ccpA and pepQ genes were found in a back-to-back organization, as has been reported for several other lactic acid bacteria (Lactobacillus pentosus, Lactobacillus casei, L. delbrükii, S. mutans, and L. lactis) (25).
FIG. 1.
Primer extension analysis of the ccpA promoter. The transcriptional start site is indicated with an arrow. The −10 region in the coding strand is boxed. RNA was isolated from S. thermophilus CNRZ302 grown on glucose (G) or lactose (L), and primer extension products were run parallel to a sequence ladder (lanes A, C, G, and T) obtained with the same primer. Approximately 15 μg of RNA was used per primer extension reaction.
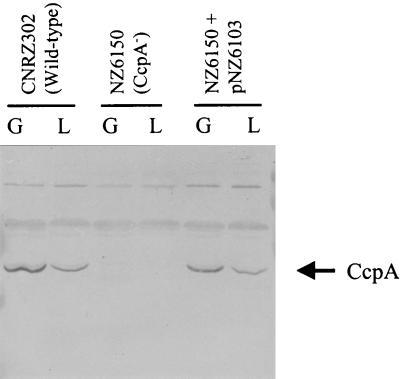
The ccpA gene was disrupted in strain CNRZ302 and its galactose-fermenting derivative NZ302G by a single crossing-over event of the integration vector pNZ6102, resulting in strains NZ6150 and NZ6151, respectively. Both ccpA disruption strains contained two truncated ccpA gene copies, as verified by Southern blot and PCR analysis (data not shown). The ccpA gene copy that is still under the control of the ccpA promoter encodes a CcpA that lacks the last 26 amino acids and should be nonfunctional, as was shown for similar C-terminal deletions of B. megaterium CcpA and its E. coli structural homologue, the LacI repressor (17, 20). The second, promoterless ccpA gene (the ribosome-binding site is also missing; no expression expected) copy would encode a CcpA that lacked the first 41 amino acids, including the DNA-binding region. Neither of these truncated forms of CcpA could be detected by Western blot analysis using antibodies raised against B. megaterium CcpA, while the intact S. thermophilus CcpA, expressed in wild-type and complemented ccpA disruption cells, could be detected (19) (Fig. 2). These results confirm that the C-terminal truncation of CcpA leads to a highly unstable form of this protein, as was shown for its structural homologue in E. coli, the LacI repressor (20). In wild-type cells, CcpA was identified as a stained band of approximately 37 kDa, and the amount of CcpA protein was at least twofold higher in cells grown on glucose relative to cells grown on lactose, indicating a form of regulation on the CcpA production. Interestingly, complementation of the ccpA disruption strain with pNZ6103 restored not only CcpA production but also sugar-dependent regulation of its production.
FIG. 2.
Western blot analysis of total protein extracts of S. thermophilus strains CNRZ302 (wild type), NZ6510 (CcpA−), and NZ6510 plus pNZ6103 grown on glucose (G) or lactose (L). Per sample, 10 μg of total protein was loaded, and CcpA proteins were detected using antibodies raised against B. megaterium CcpA. S. thermophilus CcpA was identified as a stained band of approximately 37 kDa. Next to the CcpA protein, several a-specific bands with a higher molecular weight were detected in every sample.
Effect of ccpA disruption on growth.
To analyze the physiological effects of ccpA disruption, S. thermophilus wild-type strain and NZ6150 (CcpA−) were grown on M17 medium supplemented with glucose, lactose, or sucrose, while the galactose-fermenting variant NZ302G and its CcpA− derivative NZ6151 were grown on M17 medium containing galactose (Fig. 3). Wild-type cells showed the highest maximal growth rate on lactose (2.48 h−1) relative to glucose (1.01 h−1), sucrose (1.72 h−1), and NZ302G on galactose (0.67 h−1). Moreover, the growth kinetics of wild-type cells grown on a combination of glucose and lactose were similar to those of lactose-grown cells (data not shown), indicating a preference for lactose as a carbon and energy source. The primary effects of ccpA disruption were a prolonged lag time and reduced growth rate on all sugars tested. In addition, lactose-grown NZ6150 cells reached a significantly lower OD600 than wild-type cells, which was not observed for growth on the other sugars (data not shown). To rule out pleiotropic effects of the insertion of the integration vector, NZ6150 was complemented with plasmid pNZ6103, expressing the S. thermophilus ccpA gene. Wild-type growth characteristics could be restored to NZ6150, showing that disruption of the ccpA gene was responsible for the observed impaired growth (data not shown).
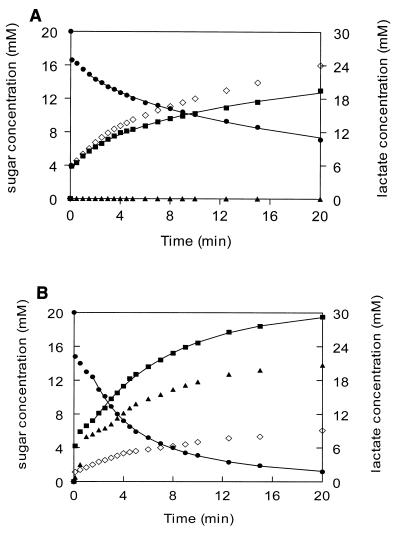
FIG. 3.
Small-scale fermentation of lactose of CNRZ302 (wild type) (A) and NZ6150 (ccpA disruption mutant) (B). Strains were grown to an OD600 of 1.0 in lactose-containing medium and resuspended in a 4% β-glycerophosphate buffer. Fermentation was started by addition of 20 mM lactose. Medium components were analyzed by HPLC. ●, lactose; ■, galactose; ▴, glucose; ◊, lactate.
Sugar uptake and utilization of lactose in ccpA disruption strains.
To obtain insight on the kinetics of lactose fermentation in the ccpA disruption mutant compared to the wild-type strain, small-scale fermentations were performed using resting cells in a strongly buffered system (Fig. 3). The wild-type cells show a very rapid initial uptake of lactose accompanied by the appearance of an equimolar amount of galactose in the buffer, as was observed in previous S. thermophilus studies (13, 32) (Fig. 3A). The values for lactose internalization and galactose expulsion are probably somewhat overestimated in the first 1.5 min due to the high initial transport and hydrolysis that continued for several seconds on ice during sampling. Hence, the later samples were used to calculate the rates of lactose internalization and galactose expulsion, as the overestimation decreases greatly with the decrease in transport rate. For these samples, the rates agree well with the nonlinear kinetic model for lactose uptake by the S. thermophilus LacS transporter (32). The wild-type strain consumed half of the added amount of lactose in 10 min, whereas the ccpA disruption mutant achieved this in 2.5 min, consuming almost all added lactose within 20 min (Fig. 3B). Remarkably, after a very short lag period (30 s), glucose appeared in the fermentation medium of the ccpA disruption strain and amounted after 20 min to two-thirds of the internalized lactose. This indicates that the ccpA disruption strain ferments only one-third of the glucose derived from the internalized lactose, whereas the wild-type strain ferments this completely. Moreover, no detectable β-galactosidase activity was found in the fermentation buffer of either strain, which rules out the possibility that the difference in lactose consumption and appearance of galactose or glucose in the fermentation buffer was caused by release of β-galactosidase due to differential lysis of the ccpA disruption strain. The rate and amount of lactate production agreed with the influx of lactose-derived glucose that was strongly reduced in the ccpA disruption strain. No end products other than lactate could be detected in the fermentation buffer, in contrast to what was found in an L. lactis ccpA disruption mutant, which showed a mixed acid fermentation (22). In a similar experiment with NZ6151 cells fermenting galactose, no apparent differences were observed in the sugar consumption rate compared to NZ302G cells, indicating that the CcpA effect on galactoside uptake is lactose specific (data not shown).
Regulation of the lac operon by CcpA.
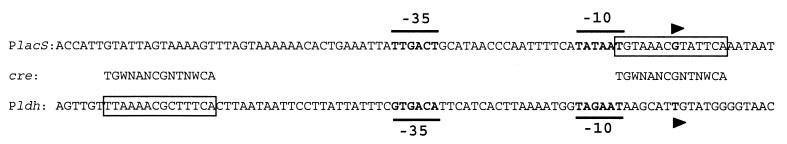
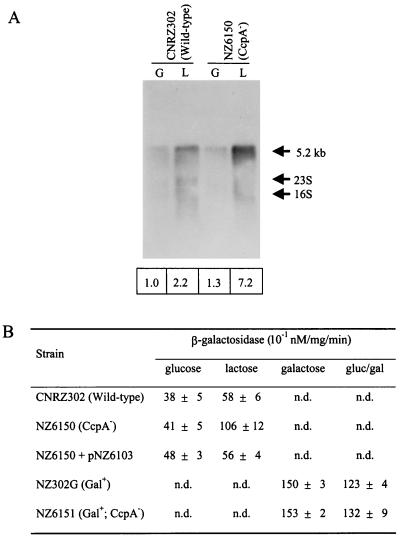
The efficiency of the S. thermophilus lacS promoter in CNRZ302 is induced during growth on lactose and galactose as a consequence of GalR activity (Vaughan et al., submitted). This lac promoter contains a cre site overlapping the −10 box and the transcriptional start site, identical in sequence and location to that previously published for strain A147 (Fig. 4) (34). To study the effect of CcpA as an additional transcriptional regulator of the lac operon, total RNA was isolated from the ccpA disruption and wild-type strains grown on various carbon sources. An internal fragment of the lacS gene was used in Northern blots to detect the single 5.2-kb lacSZ messenger (Fig. 5A). Lactose-grown wild-type cells showed a twofold increase in lacSZ expression relative to glucose-grown cells, due to GalR activity. In contrast, this increase was sevenfold in the ccpA disruption strain (NZ6150). Galactose-grown NZ302G cells showed a high amount of lacSZ transcript, even relative to lactose-grown NZ6150 cells. This did not differ significantly in strain NZ6151, indicating that CcpA-mediated repression of the lacSZ promoter during growth on galactose did not occur (data not shown). The basal level of lacSZ transcription in cells grown on glucose was not significantly affected by the loss of a functional CcpA. To further substantiate the effect of the ccpA gene disruption on the expression of the lac operon, the lacZ gene of this operon was used as a reporter (Fig. 5B). Lactose-grown wild-type cells showed 1.5-fold-higher β-galactosidase activity than cells grown on glucose. This induction was approximately threefold in the ccpA disruption mutant grown on lactose, while the β-galactosidase activity of glucose-grown cells was not significantly affected by the loss of a functional CcpA. The absolute induction values were lower relative to those of the transcriptional analysis but showed the same tendencies. Galactose-grown NZ302G cells showed a twofold-increased β-galactosidase activity relative to CNRZ302 cells grown on lactose, which was not increased further by the disruption of ccpA in this strain. NZ302G cells grown on a combination of galactose and glucose showed only a slightly lower β-galactosidase activity than galactose-grown NZ302G. Introduction of pNZ6103 in the ccpA disruption strain (NZ6150) was found to restore the wild-type β-galactosidase activity levels.
FIG. 4.
Alignment of the lacS and ldh promoter regions of S. thermophilus CNRZ302. The −35 and −10 boxes are in bold, and the determined transcriptional start sites are indicated by arrows. The putative cre sites are boxed and aligned with the consensus sequence.
FIG. 5.
CcpA regulation of the S. thermophilus lacSZ operon. (A) Northern blot analysis of lacSZ expression of strains CNRZ302 (wild type) and NZ6510 (CcpA−) grown on glucose (G) or lactose (L). Below each lane, the relative amounts of the lacSZ-specific transcripts are given, which were obtained by phosphor image analysis of the Northern blot. These values were corrected for the total amount of RNA loaded; the lacSZ transcript amount of glucose-grown CNRZ302 was set at 1.0. (B) β-Galactosidase activities of the strains used in this study. Average values are presented of at least two independent experiments. n.d., not determined.
Since the ccpA disruption strain was able to transport lactose with a significantly higher rate than the wild-type strain, the amount of LacS protein in these strains was compared. Total protein was isolated from parent (CNRZ302 and NZ302G) and ccpA disruption (NZ6150 and NZ6151) strains grown on lactose or galactose. Using antibodies raised against LacS, protein bands of the expected molecular weights were detected (16). Significantly higher amounts of LacS protein were detected for both ccpA disruption strains NZ6150 and NZ6151 grown on lactose compared to their parent strains (data not shown), indicating relief of a repressing effect by CcpA on LacS production. In analogy with the β-galactosidase results, the galactose-grown NZ302G cells contained high amounts of LacS protein compared to lactose-grown wild-type cells, which was not significantly affected by the ccpA disruption. These results indicate that the repression of the lacS promoter during growth on lactose is relieved by the loss of a functional CcpA and is not occurring during growth on galactose. This strongly suggests that the glucose moiety of lactose is responsible for this CcpA-mediated repression.
Expression of the ldh gene.
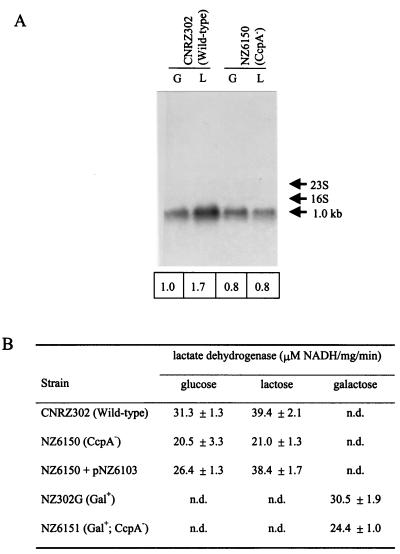
The observation that the ccpA disruption strain grown on lactose produced less lactate than the wild-type strain (Fig. 3) could indicate that glycolysis was affected by the disruption of the ccpA gene. The production of lactate from pyruvate by lactate dehydrogenase is an essential step in homofermentative lactic acid bacteria to reoxidate NADH that is generated during glycolysis. The las operon of L. lactis, comprising the pfk, pyk, and ldh genes, was found to be transcriptionally activated by CcpA on glucose (22). In S. thermophilus, the ldh gene (14) and the pfk/pyk operon (F. Crispie, J. Anba, P. Renault, S. D. Ehrlich, G. F. Fitzgerald, and D. van Sinderen, unpublished data) are located at distinct chromosomal locations. Sequence analysis of the ldh promoter region revealed a cre site upstream of the −35 region (Fig. 4). This promoter was isolated from CNRZ302, and sequence analysis confirmed the presence of this cre site. Therefore, the involvement of CcpA in the regulation of transcription of the ldh gene was analyzed by Northern blot analysis using an internal fragment of the ldh gene as a probe (Fig. 6A). The ldh gene showed a single transcript of 1.0 kb, of which the amount was twofold higher in lactose-grown compared to glucose-grown wild-type cells. Galactose-grown NZ302G cells showed an even lower amount of ldh transcript than glucose-grown wild-type cells (data not shown). This sugar-dependent regulation of ldh expression was completely lost in the ccpA disruption strains. In analogy, lactate dehydrogenase activity was highest in wild-type cells grown on lactose but significantly lower in the ccpA disruption strains in a sugar-independent way (Fig. 6B). Introduction of pNZ6103 in the ccpA disruption strain could restore lactate dehydrogenase activities towards wild-type levels. These results show that in S. thermophilus, CcpA is a positive regulator of the ldh gene and that this activation is stronger in lactose-grown cells than in glucose- or galactose-grown cells.
FIG. 6.
CcpA regulation of the S. thermophilus ldh gene. (A) Northern blot analysis of ldh gene expression of strains CNRZ302 (wild type) and NZ6510 (CcpA−) grown on glucose (G) or lactose (L). Below each lane, the relative amounts of ldh-specific transcripts are given, which were obtained by phosphor image analysis of the Northern blot. These values were corrected for the total amount of RNA loaded; the ldh transcript amount of glucose-grown CNRZ302 was set at 1.0. (B) Lactate dehydrogenase activities of the strains used in this study. Average values are presented of at least two independent experiments. n.d., not determined.
DISCUSSION
The role of the S. thermophilus catabolite control protein CcpA in fine-tuning of transport and hydrolysis of the non-PTS sugar lactose and glycolytic flux was established by the cloning, characterization, and disruption of the ccpA gene. CcpA was found to repress the lacSZ operon, encoding lactose permease and β-galactosidase, that is under positive control of the GalR activator (Vaughan et al., submitted). In contrast, the gene encoding lactate dehydrogenase was found to be transcriptionally activated by CcpA. Western blot analysis showed that CcpA production was sugar source dependent, with more than a twofold-higher amount found in glucose-grown cells than in lactose-grown cells. The observed regulation of S. thermophilus CcpA production could be regulated at the transcriptional level as negative autoregulation, as has been found for some but not all other ccpA genes (4, 25, 29). Inspection of the ccpA sequence showed the presence of two cre-like elements. One putative cre (5′-TGTAAGCGATGAAT-3′) is located at −150 relative to the transcription start site of the ccpA promoter and is thus more closely linked to the divergently transcribed pepQ gene (located at −100 relative to its putative promoter), suggesting its involvement in regulation of pepQ expression rather than ccpA autoregulation. The second putative cre (5′-ATTCACCGGTTACA-3′) is located within the coding sequence of the ccpA gene (+873 bp), which could suggest a role in transcriptional control. An internal cre site has also been found in the coding region of the L. lactis ccpA gene, but autoregulation could not be established in this organism (22). An alternative mechanism for the observed autoregulation involving the two cre sites could be that CcpA binds to these sites to form a DNA loop, thereby inhibiting ccpA promoter activity. The large distance found between the two sites could then explain the small magnitude of the observed regulatory effect. The restoration of sugar source-dependent regulation of CcpA production observed in the ccpA disruption strain when complemented with the complete ccpA gene in trans indicates that the regulation mechanism is not impaired by the presence of multiple ccpA gene copies, supporting the suggested autoregulation of CcpA. Nevertheless, some form of posttranslational regulation of CcpA production cannot be excluded on the basis of our experiments. Disruption of the ccpA gene seriously impaired the growth of S. thermophilus, as has also been observed for other gram-positive bacteria (4, 12, 29). On both PTS (sucrose) and non-PTS (glucose, lactose, and galactose) sugars, growth of the ccpA disruption mutants showed a prolonged lag phase and a reduced maximum growth rate. In contrast to the other sugars tested, the ccpA disruption mutant NZ6150 grown on lactose reached a significantly lower final cell density in the stationary phase compared to the wild-type cells. It is likely that growth ceased because all lactose in the medium was depleted by the high-lactose transport and hydrolysis capacity. Apparently, growth of NZ6150 does not continue on the expelled glucose.
Until now, CR by CcpA was only found for PTS substrates. In this paper, we present evidence of CcpA-mediated CR by the non-PTS substrate lactose. Northern analysis of the lacS promoter revealed that the negative regulation by CcpA when cells were grown on lactose occurred at the transcriptional level. The cre site in the lacS promoter is overlapping the −10 box and the transcriptional start site, in accordance with negative regulation by CcpA (10). This repression was not present in cells grown on galactose which is transported by the same LacS permease. This was further substantiated by the results from β-galactosidase activity and LacS Western blot analyses. The lactose-mediated lacSZ repression could not be achieved by growing strain NZ302G on a combination of glucose and galactose. These results suggest that the glucose moiety derived from lactose induces CR of the lacS promoter. Glucose that is internalized from the growth medium is not metabolized as fast as the glucose moiety from lactose, giving virtually no CR. This indicates that glucose is not a preferred carbon source for S. thermophilus compared to lactose or sucrose and that uptake is probably the limiting factor for efficient glucose metabolism.
CcpA-mediated CR in low-G+C gram-positive bacteria is dependent on the intracellular amounts of FBP, as relatively high concentrations of this glycolytic intermediate stimulate the HPr kinase in B. subtilis to convert HPr to P-Ser-HPr (2). The LacS permease of S. thermophilus constitutes a very fast and efficient system for lactose uptake that facilitates high influx of glucose into glycolysis. At the maximal growth rate of S. thermophilus, P-Ser-HPr appears to be the dominant phosphorylated species, whereas P-His-HPr dominates in the stationary phase (8). This reflects a relatively high intracellular FBP concentration that subsequently induces CR of the lacS promoter. Galactose metabolism by the relatively slow Leloir pathway probably yields insufficient intracellular FBP concentrations for induction of CcpA-mediated repression. CR of the lac operon in S. thermophilus may not be so much carbon source dependent as determined by the rate of glycolysis relative to sugar uptake, in which the FBP concentration may act as the intracellular indicator of this glycolytic flux. Small-scale fermentation experiments substantiated the negative regulation of CcpA on the uptake and utilization of lactose, but also showed involvement of this regulator in the central metabolism of S. thermophilus. In the absence of a functional CcpA, the cells not only take up lactose and expel galactose at least four times faster than the wild-type cells but also show a significant reduction in the amount of lactate produced. The increased lactose uptake by the ccpA disruption strain does not result in an increased growth rate. Moreover, glucose was expelled into the fermentation medium by the ccpA disruption mutant, and its amount correlates with the amount of internalized lactose and that of lactate produced, closing the carbon balance. Obviously, this glucose is derived from lactose, since not only the amount of LacS transporter, and hence its transport capacity, was increased in the ccpA disruption mutant, but also the β-galactosidase activity. Since this glucose is expelled with a short lag time, whereas galactose is expelled instantaneously and in equimolar amounts with lactose uptake, it is likely that the additional amount of glucose entering glycolysis (from the increased uptake of lactose) in the ccpA disruption mutant cannot be processed by glycolysis and is expelled.
S. thermophilus ldh expression is sugar regulated and mediated by CcpA. The cre site found in the ldh promoter region is situated upstream of the −35 box, agreeing with positive control by CcpA (10). ldh induction is highest during growth on lactose and decreased during growth on glucose and galactose, the order of which correlates with the growth rates observed. The activating effect of CcpA is presumably also mediated by P-Ser-HPr, explaining the high induction of ldh transcription on lactose and lower induction on glucose and galactose. However, as lactate dehydrogenase catalyzes the last step in homolactic fermentation, it is unlikely that this is the sole glycolytic step regulated by CcpA, causing the massive glucose expulsion. The ccpA disruption mutant of strain CNRZ302 still produces only lactate as its end product, although several strains of S. thermophilus have been reported to produce other end products, like acetoin, α-acetolactate, and diacetyl (42). Apparently, no massive accumulation of the pyruvate pool occurs in this mutant, indicating that glycolysis indeed is failing at additional steps, similar to what has been reported for L. lactis (22; E. Jamet, C. Delorme, S. D. Ehrlich, A. Bolotine, A. Sorokine, and P. Renault, Proc. 6th Symp. Lactic Acid Bacteria, abstr. H58, 1999). In the small-scale lactose fermentations, the internalization of lactose was four times faster in the ccpA disruption strain compared to the wild-type strain, while lactate expulsion was reduced twofold. This indicates that during exponential growth, S. thermophilus has a lactose transport capacity that exceeds the maximal glycolysis rate by at least twofold, suggesting that glycolysis tunes down the total lactose transport capacity to meet maximal glycolytic flux. This is in contrast to the situation in various other bacteria, where uptake of a PTS substrate is the principal rate-limiting factor in sugar metabolism (36). During late exponential and stationary growth, P-His-HPr becomes the predominant phosphorylated form of HPr, which indicates that lactose transport probably becomes rate limiting (8).
CcpA has been studied in many low-G+C gram-positive bacteria, where it mediates CR when cells are grown on PTS carbon sources, of which glucose is the most preferred. To the best of our knowledge, no other catabolic systems have been reported in which non-PTS carbon sources induce CR at the transcriptional level. Lactose, a non-PTS sugar to which S. thermophilus is highly adapted for growth, causes not only repression of the lac operon but also activation of glycolysis, both events being mediated by CcpA. Glucose, also a non-PTS sugar for S. thermophilus, is not able to repress the lac operon, and the activation of glycolysis is not as strong as that induced by lactose. In conclusion, CcpA simultaneously tunes the uptake of lactose and the capacity for glycolysis to yield optimal glycolytic flux and growth rate of S. thermophilus.
ACKNOWLEDGMENTS
We thank Roelie Holleman for HPLC analysis and Jeroen Hugenholz, Roland Siezen, and Elaine Vaughan for critically reading the manuscript.
This work was partially supported by the Biotech Programme of the European Community (contracts ERBBI04-CT96-0439 and ERBBI04-CT96-0498).
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 3.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In Genetics and biotechnology of lactic acid bacteria. Gasson, M. G. and de Vos, W. M. (eds). Chapman and Hall, U.K. 1994. pp. 52–105. [Google Scholar]
- 4.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 5.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 6.Gösseringer R, Küster E, Galinier E, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 7.Grundy F J, Waters D A, Allen S H, Henkin T M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunnewijk, M. G. W., and B. Poolman. 2000. Phosphorylation state of HPr determines the level of expression and the extent of phosphorylation of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem., in press. [DOI] [PubMed]
- 9.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 10.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 11.Hillier A J, Jago G R. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 1982;89:362–367. doi: 10.1016/s0076-6879(82)89065-4. [DOI] [PubMed] [Google Scholar]
- 12.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyzes of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 13.Hutkins R, Morris H A, McKay L L. Galactose transport in Streptococcus thermophilus. Appl Environ Microbiol. 1985;50:772–776. doi: 10.1128/aem.50.4.772-776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito Y, Sasaki T. Cloning and nucleotide sequencing of l-lactate dehydrogenase gene from Streptococcus thermophilus M-192. Biosci Biotechnol Biochem. 1994;58:1569–1573. doi: 10.1271/bbb.58.1569. [DOI] [PubMed] [Google Scholar]
- 15.Jones B E, Dossonnet V, Küster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 16.Knol J, Veenhoff L, Liang W, Henderson P J H, Leblanc G, Poolman B. Unidirectional reconstitution into detergent-destabilized liposomes of the purified lactose transport system of Streptococcus thermophilus. J Biol Chem. 1996;271:15358–15366. doi: 10.1074/jbc.271.26.15358. [DOI] [PubMed] [Google Scholar]
- 17.Kraus A, Hillen W. Analysis of CcpA mutations defective in carbon catabolite repression in Bacillus subtilis. FEMS Microbiol Lett. 1997;153:221–226. doi: 10.1111/j.1574-6968.1997.tb10485.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the Tn5276-located nisin gene cluster nisABTCIPR of Lactococcus lactis and evidence for the involvement of expression of nisI and nisA in producer immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 19.Küster E, Luesink E J, de Vos W M, Hillen W. Immunological crossreactivity to the catabolite control protein CcpA from Bacillus megaterium is found in many Gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Matthews K S. Characterization of mutants affecting the KRK sequence in the carboxyl-terminal domain of lac repressor. J Biol Chem. 1995;270:10640–10649. doi: 10.1074/jbc.270.18.10640. [DOI] [PubMed] [Google Scholar]
- 21.Lokman B C, Heerikhuisen M, Leer R J, van den Broek A, Borsboom Y, Chaillou S, Postma P W, Pouwels P H. Regulation of expression of the Lactobacillus pentosus xylAB operon. J Bacteriol. 1997;179:5391–5397. doi: 10.1128/jb.179.17.5391-5397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luesink E J, van Herpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 23.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahr K, Hillen W, Titgemeyer F. Carbon catabolite repression in lactobacillus pentosus: analysis of the ccpA region. Appl Environ Microbiol. 2000;66:277–283. doi: 10.1128/aem.66.1.277-283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Miwa Y, Nagura K, Eguchi S, Kukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 28.Mollet B, Knol J, Poolman B, Marciset O, Delley M. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J Bacteriol. 1993;175:4315–4324. doi: 10.1128/jb.175.14.4315-4324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monedero V, Gosalbes M J, Perez-Martinez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morel F, Frot-Coutaz J, Aubel D, Portalier R, Atlan D. Characterization of a prolidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397 with an unusual regulation of biosynthesis. Microbiology. 1999;145:437–446. doi: 10.1099/13500872-145-2-437. [DOI] [PubMed] [Google Scholar]
- 31.Platteeuw C, Simons G, De Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990;172:4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 35.Poolman B, Knol J, Mollet B, Nieuwhuis B, Sulter G J. Regulation of the bacterial sugar-H+ symport by phosphoenolpyruvate-dependent enzyme I/HPr-mediated phosphorylation. Proc Natl Acad Sci USA. 1995;92:778–782. doi: 10.1073/pnas.92.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in the post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier M H., Jr Regulatory interactions involving the proteins of the phosphotransferase system in enteric bacteria. J Cell Biochem. 1993;51:62–68. doi: 10.1002/jcb.240510112. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schroeder C J, Robert C, Lenzen G, McKay L L, Mercenier A. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus β-galactosidase sequences. J Gen Microbiol. 1991;137:369–380. doi: 10.1099/00221287-137-2-369. [DOI] [PubMed] [Google Scholar]
- 41.Simpson C L, Russell R R. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teraguchi S, Ono J, Kiyosawa I, Okonogi S. Oxygen uptake activity and aerobic metabolism of Streptococcus thermophilus STH450. J Dairy Sci. 1987;70:514–523. doi: 10.3168/jds.S0022-0302(87)80036-X. [DOI] [PubMed] [Google Scholar]
- 43.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repressor operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]