Abstract
We describe swarming in Pseudomonas aeruginosa as a third mode of surface translocation in addition to the previously described swimming and twitching motilities. Swarming in P. aeruginosa is induced on semisolid surfaces (0.5 to 0.7% agar) under conditions of nitrogen limitation and in response to certain amino acids. Glutamate, aspartate, histidine, or proline, when provided as the sole source of nitrogen, induced swarming, while arginine, asparagine, and glutamine, among other amino acids, did not sustain swarming. Cells from the edge of the swarm were about twice as long as cells from the swarm center. In both instances, bacteria possessing two polar flagella were observed by light and electron microscopy. While a fliC mutant of P. aeruginosa displayed slightly diminished swarming, a pilR and a pilA mutant, both deficient in type IV pili, were unable to swarm. Furthermore, cells with mutations in the las cell-to-cell signaling system showed diminished swarming behavior, while rhl mutants were completely unable to swarm. Evidence is presented for rhamnolipids being the actual surfactant involved in swarming motility, which explains the involvement of the cell-to-cell signaling circuitry of P. aeruginosa in this type of surface motility.
Pseudomonas aeruginosa is a gram-negative bacterium living in soil and aqueous environments, where it survives due to its extraordinary metabolic abilities. P. aeruginosa is also a typical opportunistic pathogen which colonizes the lungs of cystic fibrosis patients and causes severe infections in immunocompromised hosts. Due to its notorious elevated intrinsic resistance to antimicrobial agents and its ability to attach to and to form biofilms on medical devices (9), P. aeruginosa is difficult to eradicate in the hospital environment.
P. aeruginosa has a single polar flagellum which enables the cell to swim in aqueous environments and in low-agar (<0.4%) medium. The flagellum and the chemotaxis system, consisting of chemoreceptors (11, 49) and a signal relay system similar to that of Escherichia coli (25, 31), allow the bacterium to respond to attractants and repellents. In addition, P. aeruginosa is able to propagate at surface interfaces by twitching motility, which is mediated by type IV pili (5, 12, 53). Twitching motility is believed to result from the extension and retraction of the pilus filament, which propels the cells across a surface. Pilus synthesis and assembly require at least 40 genes which are located in several unlinked regions on the chromosome (22). The nature of the environmental signal that triggers the expression of pili is not known. Pili are important for attachment to epithelial cells (8, 17) and contribute to the virulence of P. aeruginosa in animal models (19, 50, 51). Furthermore, twitching motility and, hence, type IV pili are required for the formation of biofilms on abiotic surfaces (38).
Besides swimming and twitching, several gram-negative bacteria are able to propagate on semisolid surfaces (i.e., 0.4 to 1.0% agar) in a coordinated manner by swarming motility. Swarmer cells, which are usually elongated and hyperflagellated, differentiate from vegetative cells probably by sensing the viscosity of the surface or in response to nutritional signals (20).
In the present study, we demonstrate swarming of the normally polar, monotrichously flagellated bacterium P. aeruginosa. The swarming process is induced on 0.5 to 0.7% agar when certain amino acids are provided as the sole source of nitrogen. We further show that swarmer cells of P. aeruginosa are elongated and can possess two polar flagella. Unlike all other swarming bacteria, P. aeruginosa also requires type IV pili for this type of motility. Our results suggest that rhamnolipids are the biosurfactant involved in swarming motility, which indicates that this type of surface propagation is dependent on the las and rhl cell-to-cell signaling circuitry.
MATERIALS AND METHODS
Bacteria and media.
The strains used in this study are listed in Table 1. Swarm agar was based on M9 salts (30) without NH4Cl (termed here M8 medium, for convenience), supplemented with 0.2% glucose, 2 mM MgSO4, and trace elements (composition available upon request) and solidified with 0.5% agar. Amino acids as a sole nitrogen source were added at a final concentration of 0.05%, unless otherwise indicated. After solidification, plates were briefly dried and then inoculated by toothpick with individual colonies from a fresh Luria-Bertani (LB) agar plate. Incubation was done at 37°C or as otherwise stated. Rhamnolipid plates were prepared according to a previously described protocol (46). The medium composition was modified, however: it was based on M8 salts supplemented with 0.2% glucose (instead of 2% glycerol), 2 mM MgSO4, trace elements, 0.0005% methylene blue, 0.02% cetyltrimethylammonium bromide, and a nitrogen source and was solidified with agar (1.6% final concentration). Plates were incubated at 37°C for 24 h and then at room temperature until the appearance of a blue halo, indicating the production of rhamnolipids (usually requiring a further 24 h for the wild-type strain).
TABLE 1.
Bacterial strains and plasmids
| Strain, plasmid, or phage | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PT5 | PAO1, wild type | Laboratory collection |
| PT454 | PT5ΔrhlI::Tn501, transduced from PDO100 | 6, this study |
| PT462 | PT5rhlR::Tn501, transduced from PAO-JP3 | 6, this study |
| PT466 | PT5ΔlasI::Tc, transduced from PAO-JP1 | 42, this study |
| PT498 | PT5ΔlasR::Tc, transduced from PAO-JP3 | 42, this study |
| PT610 | PAK | S. Lory |
| PT611 | PAK-R1, pilR::Gm | S. Lory |
| PT612 | PT5pilR::Gm, transduced from PAK-R1 | 24, this study |
| PT613 | PAK-NP, pilA::Tc | 45 |
| PT623 | PT5pilA::Tc, transduced from PAK-NP | 45, this study |
| PT688 | PAO1fliC::Gm | R. Ramphal |
| PT690 | PT5fliC::Gm | This study |
| PT711 | PAO1ΔrhlA::Gm | U. Ochsner |
| PT712 | PT5ΔrhlA::Gm, transduced from PT711 | This study |
| Plasmids | ||
| pMMB207 | Broad-host-range vector, Cmr | 32 |
| pKI23 | ptac::pilR in pMMB67EH, Apr | 4 |
| p207R1 | ptac::pilR in pMMB207, Cmr | This study |
| pJPP6 | rhlABRI in pBSIISK+, Apr | J. P. Pearson |
| pJMC30 | lasRI-carrying pUC derivative, Apr | 39 |
| Bacteriophage E79tv2 | LPS-specific transducing phage | 33 |
Tc, tetracycline; Cm, chloramphenicol; Ap, ampicillin; Gm, gentamicin; LPS, lipopolysaccharide.
Strain and plasmid constructions.
Mutations in the cell-to-cell-signaling regulator genes were transferred from previously described strains into wild-type strain PT5, using the transducing phage E79tv2 (33) in order to obtain isogenic strains. Single colonies obtained by transduction were checked by Southern hybridization using digoxigenin (Roche Diagnostics)-labeled DNA probes. The lasR and lasI mutants were analyzed using a 2.1-kbp BamHI-SphI DNA fragment from pJMC30 containing lasI and the 3′ end of lasR (39), while rhlR and rhlI mutants were hybridized with a 3.5-kbp BglII fragment from pJPP6 containing the rhlAB and rhlR genes. Genomic DNA from each strain was digested with either PstI or XhoI. Southern hybridizations were carried out according to the manufacturer's protocols (digoxigenin system user's guide; Boehringer Mannheim). Banding patterns of the transduced strains obtained after hybridization were always identical to those of the original donor strains.
Plasmid p207R1 was constructed by ligating a 1.6-kbp EcoRI-BamHI DNA fragment carrying the pilR gene on plasmid pKI21 into EcoRI-BamHI-cleaved pMMB207.
Electron microscopy.
Cells from the swarm edge and from the swarm center were deposited with a toothpick on a drop of water. Formvar (0.5%)-coated 75-mesh grids were placed on top of the drop for 15 to 20 s to allow the adhesion of bacterial cells. Grids were then stained for 20 to 30 s with a freshly prepared 1% solution of potassium phosphotungstate (pH 7.0) and washed twice for 10 s in a drop of water. The grid was air dried and examined on a Zeiss EM10 electron microscope at 60 to 80 keV. At least 10 fields of view were analyzed for each sample from either the swarm edge or the swarm center.
Autoinducer assay.
The presence of N-butyryl-homoserine lactone (C4-HSL) in filtered culture supernatants was determined. Two milliliters of the supernatant was extracted twice with 2 ml of ethyl acetate (containing 0.01% acetic acid) and analyzed using the previously described bioassay (42).
RESULTS
P. aeruginosa swarming is induced by amino acids.
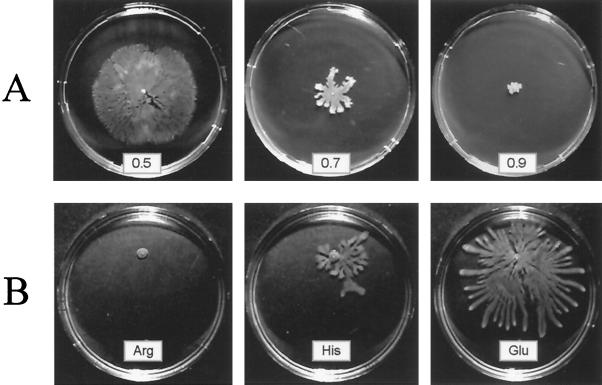
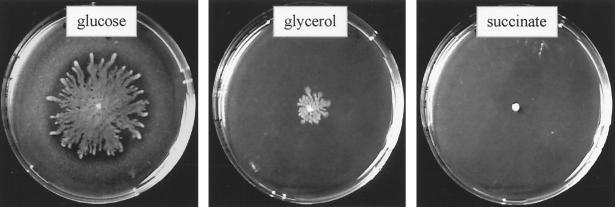
P. aeruginosa is able to swim in a low-percentage agar (<0.4%) using its single polar flagellum, and it propagates between the agar and an artificial interface (usually a petri dish) by type IV pilus-mediated twitching motility (5). After decreasing the agar concentration of the twitching motility plates to 0.7%, we noticed that wild-type P. aeruginosa strain PT5 was able to propagate on the surface of the agar in a manner similar to the typical swarming behavior described for several gram-negative bacteria (20). When PT5 was inoculated in the middle of a swarm agar plate (M8-glucose-glutamate–0.5% agar), the strain began to produce a fluid at the point of inoculation after about 6 h of incubation at 37°C. After a further 6 h, cells had propagated on the plate, forming dendritic structures which covered the whole surface of an 8.5-cm petri dish by 24 h. The same swarming phenotype was also observed with PAO1 strains from other laboratories. However, the P. aeruginosa PAK strain showed only very weak surface propagation (data not shown). Swarming usually requires the presence of Casamino Acids or peptone in the swarm agar plates. Only Proteus mirabilis has been shown to respond to a single amino acid, namely glutamine, as an inducer of swarming motility (1). We therefore tested all 20 amino acids, provided as the sole source of nitrogen at a final concentration of 0.05%, for their ability to induce swarming in P. aeruginosa. As shown in Table 2, a majority of amino acids did not induce swarming, although they sustained growth on these plates. The strongest response was obtained with glutamate (Fig. 1) and aspartate. Swarming was dependent on the amino acid concentration used. For both aspartate and glutamate, swarming was observed at final concentrations between 0.01 and 0.1%. When ammonium chloride was provided as the sole nitrogen source (≥5 mM), no swarming was observed. However, when the ammonium chloride concentration was ≤1 mM, some swarming could be observed after prolonged incubation (>48 h). Swarming was also dependent on the carbon source. For instance, when aspartate served as a nitrogen source, glucose permitted optimal cell propagation, while glycerol was less efficient and succinate did not sustain swarming at all (Fig. 2). However, when aspartate or glutamate served as both carbon and nitrogen sources, no swarming was observed.
TABLE 2.
Rhamnolipid production and swarming as a function of the amino acid provided as the sole nitrogen sourcea
| Amino acid | Swarming | Rhamnolipid |
|---|---|---|
| Alanine | ± | ± |
| Arginine | − | − |
| Asparagine | − | − |
| Aspartate | ++ | ++ |
| Cysteine | ± | + |
| Glutamine | − | ± |
| Glutamate | ++ | ++ |
| Glycine | + | ± |
| Histidine | ++ | ++ |
| Isoleucine | + | ++ |
| Leucine | − | + |
| Lysine | ± | ++ |
| Methionine | − | − |
| Phenylalanine | − | + |
| Proline | + | + |
| Serine | ± | + |
| Threonine | − | + |
| Tryptophan | − | + |
| Tyrosine | − | + |
| Valine | + | + |
Swarming and rhamnolipid production were determined as described in Materials and Methods on M8-based minimal medium plates with glucose as the carbon source and supplemented with the indicated amino acid at a final concentration of 0.05%. Swarm plates were incubated for 24 h at 37°C, while rhamnolipid production was scored after incubation for 24 h at 37°C and 24 h at room temperature. −, not detected; ±, weak; +, intermediate; ++, strong.
FIG. 1.
Swarming in P. aeruginosa is induced by agar concentrations below 0.7% (A) and by specific amino acids (B). Colonies of wild-type strain PT5 from a fresh LB agar plate were inoculated by toothpick into the middle of the swarm plates. In panel A the medium contained 0.2% glucose as the carbon source and 0.05% (wt/vol) glutamate as the nitrogen source. Plates in panel B contained 0.02% glucose, a 0.05% concentration of the indicated amino acid as the nitrogen source, and 0.6% agar. Plates were incubated for 24 h at 37°C.
FIG. 2.
Swarming is dependent on the carbon source. M8 swarm plates with aspartate as the nitrogen source were supplemented with either glucose, glycerol, or succinate at a final concentration of 100 mM each. PT5 was inoculated in the center, followed by 24 h of incubation at 37°C.
Swarming in P. aeruginosa requires both flagella and pili.
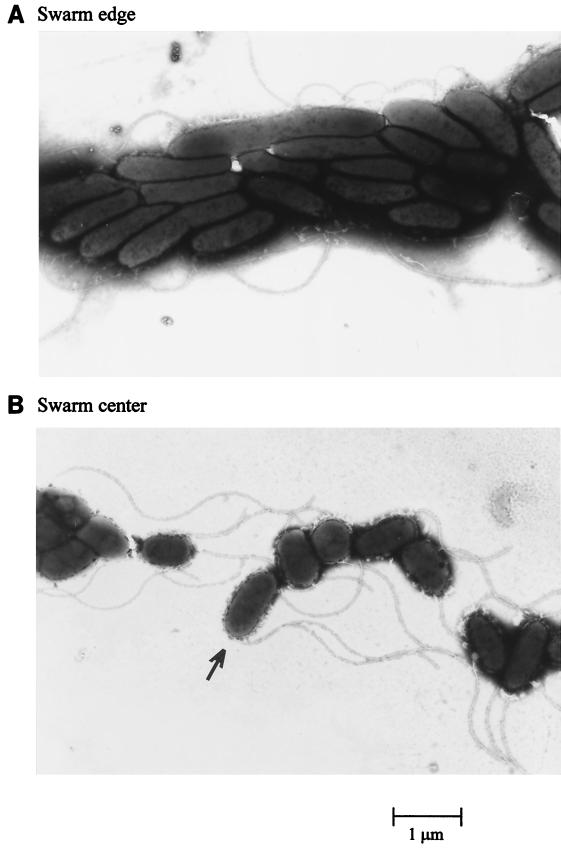
So far, swarming has been described as a phenomenon exclusively requiring propulsion by flagella, which in E. coli (21), Serratia marcescens, and P. mirabilis is linked to the differentiation into swarmer cells, which is characterized by cell elongation and hyperflagellation (20). Light microscopic analysis of P. aeruginosa cells taken from the edge of a swarming colony showed a significant proportion of highly motile cells which were approximately twice as long as cells taken from the center of the swarm. Further examination by electron microscopy confirmed that a majority of cells from the swarm edge were elongated (Fig. 3A). Surprisingly, some bacteria from both the center and the swarm edge presented two flagella which were located at one pole of the cell (Fig. 3B).
FIG. 3.
Electron microscopy of PT5 cells taken from the swarm edge and the swarm center. (A) Elongated cells of approximately 3 to 4 μm were observed at the periphery of the swarming colony. (B) Smaller cells, of about 2 μm, were observed at the swarm center. A cell expressing two polar flagella is indicated by the arrow. A few elongated cells from the swarm edge were also found to possess two flagella. Magnification is ×8,500.
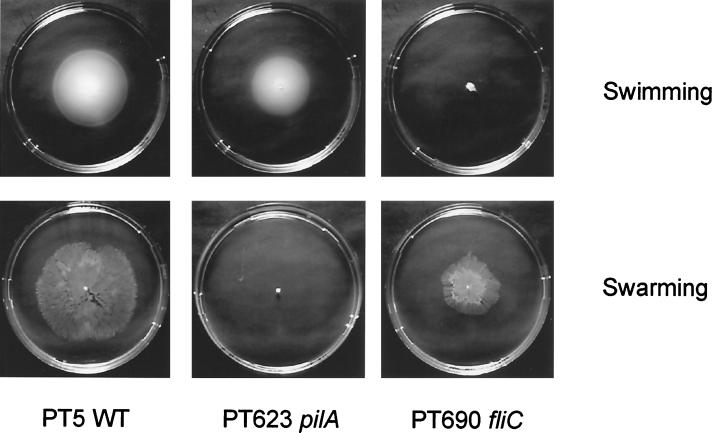
All previously described swarming bacteria are peritrichous, while P. aeruginosa possesses only a single polar flagellum. We therefore tested the swarming motility of a fliC mutant of P. aeruginosa PAO1 (kindly provided by R. Ramphal). This mutant does not synthesize any flagella, as judged by flagellum staining and observation under the light microscope. Interestingly, the fliC transductant PT690 and the original fliC mutant were unable to swim in 0.3% agar (Fig. 4) but were still able to propagate on swarm plates, albeit to a lesser extent than the wild type (Fig. 4). This suggests that swarming of P. aeruginosa is not dependent exclusively on flagella. Besides flagella, P. aeruginosa produces additional surface structures of which the type IV pili are the best characterized. Since pili are responsible for twitching motility (5, 12, 53), we analyzed their involvement in swarming. To our surprise, we observed a complete lack of swarming for the pilA mutant PT623 (Fig. 4) as well as for the pilR mutant PT612 (data not shown), both of which are completely deficient for the production of type IV pili. Swarming, albeit at a lower level, could be restored in the pilR mutant by introduction of the pilR-expressing plasmid p207R1. This is the first report demonstrating the involvement of pili in swarming motility.
FIG. 4.
Swimming and swarming motilities in P. aeruginosa wild-type (WT) PT5 and its fliC and pilA mutant derivatives. Swimming plates were made of LB agar with 0.3% agar. After inoculation, plates were incubated at room temperature for 24 h. Swarm plates were M8-glucose-glutamate plates containing 0.5% agar. Incubation was done at 37°C for 24 h.
Swarming is controlled by the las and rhl cell-to-cell signaling system.
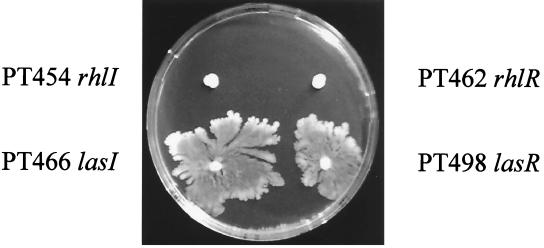
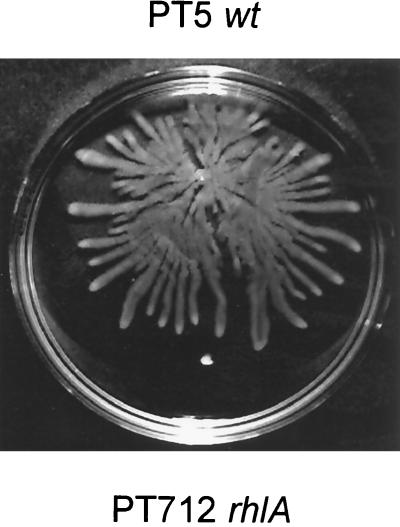
In Serratia liquefaciens, Eberl et al. (14) have identified a cell-to-cell signaling system, called swr, that is responsible for the initiation of swarming. The swrI gene belongs to the luxI class of homoserine lactone synthases directing the production in S. liquefaciens of two autoinducers, N-butanoyl-l-homoserine lactone and N-hexanoyl-l-homoserine lactone. P. aeruginosa possesses two well-characterized cell-to-cell signaling systems, las (36, 39–41) and rhl (6, 27), which contain the LasR and RhlR transcriptional regulators, their cognate autoinducer synthases, LasI and RhlI, and the corresponding signaling molecules, N-(3-oxo-dodecanoyl)-l-homoserine lactone and C4-HSL, respectively. These two regulatory networks control the expression of a number of extracellular virulence factors, including elastase, alkaline protease, rhamnolipids, and pyocyanin (52). We tested whether the las and rhl systems were also required for swarming in P. aeruginosa. Isogenic derivatives of strain PT5 that had been inactivated in either lasI, lasR, rhlI, or rhlR were inoculated on a swarm plate. While swarming by the lasR and lasI mutants was reduced and occurred only after prolonged incubation (>48 h), it was completely abolished in the rhlR and rhlI mutants (Fig. 5). We concluded that a factor under the control of the las and rhl systems is required for swarming in P. aeruginosa. Rhamnolipid production is mainly controlled by the rhl cell-to-cell signaling system (6, 34, 35), which regulates the transcription of the rhlAB operon, encoding rhamnosyltransferase. In order to test whether rhamnolipids are required for swarming, we inoculated the rhlA-deficient strain PT712 and its parental wild-type strain, PT5, on a swarm plate (Fig. 6). The rhlA mutant was completely deficient in swarming, although it produced wild-type levels of the C4-HSL autoinducer (data not shown). Swarming of PT712 could be rescued by coinoculation with the wild-type strain, since about 50% of the colonies from the swarm edge were rhlA mutant colonies, as identified by their gentamicin resistance. This observation clearly designates rhamnolipids as the actual biosurfactant required for swarming in P. aeruginosa and explains the absence of swarming in the rhl mutants.
FIG. 5.
Swarming requires the las and rhl cell-to-cell signaling systems. The lasI, lasR, rhlI, and rhlR mutants were inoculated on a swarm plate which was incubated at 37°C for 24 and 48 h at room temperature. As a comparison, the wild-type strain, PT5, would have covered the whole plate by that time.
FIG. 6.
Rhamnolipids are the biosurfactant required for swarming in P. aeruginosa. The wild-type (wt) strain, PT5, and rhlA mutant PT712 were inoculated on a swarm plate and incubated at 37°C for 24 h.
Furthermore, we noticed a good correlation between rhamnolipid production and swarming for the wild-type strain, PT5. Glutamate, aspartate, proline, and histidine not only were excellent inducers of swarming motility but also yielded large amounts of rhamnolipids in the plate assay when they were provided as the sole nitrogen source (Table 2). In contrast, asparagine, glutamine, and arginine, which do not induce swarming, also completely repressed the synthesis of rhamnolipids. Our results suggest a novel function for rhamnolipids in P. aeruginosa, namely as a biosurfactant promoting surface translocation on semisolid surfaces.
DISCUSSION
We show in this study that P. aeruginosa, already known for its swimming and twitching motility, is also able to propagate on semisolid surfaces by swarming. This makes P. aeruginosa one of the rare bacteria to possess three types of motility. Swarming, described so far only for peritrichously flagellated organisms, requires in P. aeruginosa the interplay of several features, namely amino acids as a nitrogen source, the presence of both flagella and type IV pili, and the secretion of rhamnolipids as a surface-active compound.
The surprising finding that P. aeruginosa swarmer cells can express two polar flagella is in agreement with a recent report of a fleN mutant of P. aeruginosa (13) which was found to express between three and six polar flagella. Although the fleN mutant was nonmotile, this observation suggests that flagellum number is indeed regulated in this organism. Swarming could be a natural situation where flagellum upregulation could provide a more efficient propagation on semisolid surfaces. Indeed, all swarming bacteria described so far are peritrichous or are able to synthesize additional lateral flagella, as in the case of Vibrio parahaemolyticus (3). That pili are required for swarming is a completely novel observation among flagellated bacteria. The polar type IV pili have been reported, so far, to be involved only in twitching motility. In Myxococcus xanthus, a nonflagellated organism, type IV pili are involved in gliding, a type of surface propagation comparable to the twitching motility described for P. aeruginosa and Neisseria gonorrhoeae. It seems likely that the rectractable type IV pili of P. aeruginosa assist the flagellum in surface propagation. Alternatively, the pili could be involved in sensing the viscocity of the surface and sending a signal for initiation of swarming.
During preparation of the manuscript, an article by Rashid and Kornberg (44) also reported on swarming motility in P. aeruginosa. These authors also demonstrated the presence of two flagella and the elongation of swarmer cells. In contrast to our findings, these investigators reported that a pilA mutant was not affected in swarming, while a fliC mutant was completely unable to swarm. Whether these results are due to strain differences or to the medium used in the swarm assay is unclear. One should also keep in mind that P. aeruginosa strains can vary in the composition of the pilin (47) and flagellum proteins (48), which could affect these types of motility.
In this study, we further demonstrate that swarming is regulated by the availability of nitrogen. Glutamate, aspartate, histidine, and proline provided as the sole nitrogen source were the best inducers of swarming, while high NH4+ concentrations (≥5 mM) and the amino acids glutamine, asparagine, and arginine, among others, completely prevented swarming of P. aeruginosa. Rhamnolipid production is also subject to nitrogen regulation and requires the RpoN sigma factor (34). Although some rhamnolipid production was detectable in our plate assay at NH4+ concentrations below 2 mM (unpublished results), this level of production was not sufficient to promote swarming motility, which thus requires the presence of specific amino acids. The response to these amino acids could be mediated by the chemotaxis system of P. aeruginosa, which is sensitive to several amino acids and small peptides and is also subject to nitrogen regulation, probably at the level of chemoreceptors and transducers (10). The chemotaxis system, but not chemotaxis per se, previously has been demonstrated to be required for swarming by E. coli (7).
It is tempting to speculate that nitrogen limitation might affect pilus synthesis. Indeed, transcription of the pilin operon pilABCD is controlled by the two-component regulatory system pilS-pilR (22) and by the RpoN sigma factor (23), which is involved in transcription of nitrogen-regulated genes. Furthermore, the pilE gene has been isolated, in a mutant unable to assimilate or dissimilate nitrate (16). Thus, under N excess conditions, pilus transcription could be reduced to levels preventing swarming of P. aeruginosa. Recently, the global carbon metabolism regulator Crc was also shown to be involved in type IV pilus synthesis (37).
The inability of the rhlI and rhlR mutants to sustain swarming is unlikely to be caused by effects on flagellum synthesis, since both rhl mutants are still able to swim (our unpublished observation). However, an rhlI mutant was reported to be deficient in type IV-pilus-mediated twitching motility (15). Although the synthesis of pili per se was not affected in the rhlI mutant, final surface piliation was decreased compared to that in the wild type, suggesting an involvement of the rhl cell-to-cell signaling system in pilus assembly (15). The inability of the rhl mutants to swarm could therefore be the result of both reduced rhamnolipid production and decreased surface piliation.
Swarming is associated in several bacterial species with the secretion of a surfactant which reduces friction between bacterial cells and surfaces. Examples of such biosurfactants are a cell surface-attached polysaccharide in P. mirabilis (18) and a secreted lipopeptide in S. liquefaciens (28). P. aeruginosa rhamnolipid is a biosurfactant involved in solubilization and degradation of hydrocarbons (2). In conjunction with phospholipase C, rhamnolipids also act as a hemolysin (29).
The fact that the rhlA mutant, which produces normal amounts of C4-HSL, is also unable to swarm suggests that rhamnolipids per se are crucial to swarming motility in P. aeruginosa. However, rhamnolipid production is regulated predominantly by the rhl system and partly also by the las system, based on the cell-to-cell signaling hierarchical circuitry (26, 43). Indeed, production of rhamnolipids is also reduced in the lasI and lasR mutants, which would explain the delayed swarming behavior observed in the las mutants. The cell-to-cell signaling circuitry could therefore play a role in sensing nutrient deficiency and inducing rhamnolipid production, which would allow the bacteria to migrate towards nutrient-replete environments.
The fact that P. aeruginosa retains three types of motility probably reflects the variety of its habitats. Swarming is certainly one possible mode for colonizing its natural environments, but swarming could also play a role in colonization in vivo, where nitrogen availability might be a limiting factor.
ACKNOWLEDGMENTS
We are grateful to B. Iglewski, R. Ramphal, S. Lory, J. P. Pearson, U. Ochsner, and D. Haas for providing strains, plasmids, and phage. We thank M. Michéa-Hamzehpour for critical reading of the manuscript.
T.K. was supported by a grant from the Swiss National Science Foundation.
REFERENCES
- 1.Allison C, Lai H C, Gygi D, Hughes C. Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol Microbiol. 1993;8:53–60. doi: 10.1111/j.1365-2958.1993.tb01202.x. [DOI] [PubMed] [Google Scholar]
- 2.Arino S, Marchal R, Vandecasteele J P. Involvement of a rhamnolipid-producing strain of Pseudomonas aeruginosa in the degradation of polycyclic aromatic hydrocarbons by a bacterial community. J Appl Microbiol. 1998;84:769–776. doi: 10.1046/j.1365-2672.1998.00412.x. [DOI] [PubMed] [Google Scholar]
- 3.Belas M R, Colwell R R. Scanning electron microscope observation of the swarming phenomenon of Vibrio parahaemolyticus. J Bacteriol. 1982;150:956–959. doi: 10.1128/jb.150.2.956-959.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd J M, Lory S. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J Bacteriol. 1996;178:831–839. doi: 10.1128/jb.178.3.831-839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 6.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkart M, Toguchi A, Harshey R M. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comolli J C, Waite L L, Mostov K E, Engel J N. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect Immun. 1999;67:3207–3214. doi: 10.1128/iai.67.7.3207-3214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 10.Craven R, Montie T C. Regulation of Pseudomonas aeruginosa chemotaxis by the nitrogen source. J Bacteriol. 1985;164:544–549. doi: 10.1128/jb.164.2.544-549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven R C, Montie T C. Chemotaxis of Pseudomonas aeruginosa: involvement of methylation. J Bacteriol. 1983;154:780–786. doi: 10.1128/jb.154.2.780-786.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta N, Arora S K, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberl L, Winson M K, Sternberg C, Stewart G S, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-hormoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 15.Glessner A, Smith R S, Iglewski B H, Robinson J B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J Bacteriol. 1999;181:1623–1629. doi: 10.1128/jb.181.5.1623-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldflam M, Rowe J J. Evidence for gene sharing in the nitrate reduction systems of Pseudomonas aeruginosa. J Bacteriol. 1983;155:1446–1449. doi: 10.1128/jb.155.3.1446-1449.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S K, Berk R S, Masinick S, Hazlett L D. Pili and lipopolysaccharide of Pseudomonas aeruginosa bind to the glycolipid asialo GM1. Infect Immun. 1994;62:4572–4579. doi: 10.1128/iai.62.10.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gygi D, Rahman M M, Lai H C, Carlson R, Guard-Petter J, Hughes C. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol Microbiol. 1995;17:1167–1175. doi: 10.1111/j.1365-2958.1995.mmi_17061167.x. [DOI] [PubMed] [Google Scholar]
- 19.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 20.Harshey R M. Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 21.Harshey R M, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbs M, Collie E S, Free P D, Livingston S P, Mattick J S. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishimoto K S, Lory S. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol. 1992;174:3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- 26.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 27.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 28.Lindum P W, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P V. Extracellular toxins of Pseudomonas aeruginosa. J Infect Dis. 1974;130(Suppl.):S94–S99. doi: 10.1093/infdis/130.supplement.s94. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales V M, Bäckman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 33.Morgan A F. Transduction of Pseudomonas aeruginosa with a mutant of bacteriophage E79. J Bacteriol. 1979;139:137–140. doi: 10.1128/jb.139.1.137-140.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochsner U A, Fiechter A, Reiser J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem. 1994;269:19787–19795. [PubMed] [Google Scholar]
- 35.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Toole G A, Gibbs K A, Hager P W, Phibbs P V, Jr, Kolter R. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2000;182:425–431. doi: 10.1128/jb.182.2.425-431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 39.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 40.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson J P, Pesci E C, Iglewski B H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashid M H, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saiman L, Ishimoto K, Lory S, Prince A. The effect of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J Infect Dis. 1990;161:541–548. doi: 10.1093/infdis/161.3.541. [DOI] [PubMed] [Google Scholar]
- 46.Siegmund I, Wagner F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth in mineral agar. Biotechnol Tech. 1991;5:265–268. [Google Scholar]
- 47.Spangenberg C, Fislage R, Sierralta W, Tummler B, Romling U. Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiol Lett. 1995;125:265–273. doi: 10.1111/j.1574-6968.1995.tb07367.x. [DOI] [PubMed] [Google Scholar]
- 48.Spangenberg C, Heuer T, Burger C, Tummler B. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 1996;396:213–217. doi: 10.1016/0014-5793(96)01099-x. [DOI] [PubMed] [Google Scholar]
- 49.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 50.Tang H, Kays M, Prince A. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Delden C, Iglewski B H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]