Abstract
Background
The role of fomites in the transmission of SARS-CoV-2 is unclear.
Aim
To assess whether SARS-CoV-2 can be transmitted through fomites, using evidence from viral culture studies.
Methods
Searches were conducted in the World Health Organization COVID-19 Database, PubMed, LitCovid, medRxiv, and Google Scholar to December 31st, 2021. Studies that investigated fomite transmission and performed viral culture to assess the cytopathic effect (CPE) of positive fomite samples and confirmation of SARS-CoV-2 as the cause of the CPE were included. The risk of bias using a checklist modified from the modified Quality Assessment of Diagnostic Accuracy Studies – 2 (QUADAS-2) criteria was assessed.
Findings
Twenty-three studies were included. The overall risk of bias was moderate. Five studies demonstrated replication-competent virus from fomite cultures and three used genome sequencing to match fomite samples with human clinical specimens. The mean cycle threshold (CT) of samples with positive viral culture was significantly lower compared with cultured samples that returned negative results (standardized mean difference: –1.45; 95% confidence interval (CI): –2.00 to –0.90; I2 = 0%; P < 0.00001). The likelihood of isolating replication-competent virus was significantly greater when CT was <30 (relative risk: 3.10; 95% CI: 1.32 to 7.31; I2 = 71%; P = 0.01). Infectious specimens were mostly detected within seven days of symptom onset. One study showed possible transmission of SARS-CoV-2 from fomites to humans.
Conclusion
The evidence from published studies suggests that replication-competent SARS-CoV-2 is present on fomites. Replication-competent SARS-CoV-2 is significantly more likely when the PCR CT for clinical specimens and fomite samples is <30. Further studies should investigate the duration of infectiousness of SARS-CoV-2 and the frequency of transmission from fomites.
Keywords: COVID-19, SARS-CoV-2, Viral culture, Transmission, Fomite, Systematic review
Introduction
The SARS-CoV-2 (COVID-19) pandemic continues to be a major public health concern. Based on World Health Organization (WHO) statistics, there have been more than 566 million confirmed cases and more than six million deaths globally as of July 25th, 2022 [1]. Although several vaccines have been developed and programmes have been implemented globally, mutations in the virus have resulted in the occurrence of variants that reduce vaccine effectiveness [2]. In addition, the duration of protection appears to be limited [3] and booster doses are considered necessary [4].
In addition to the use of vaccines, a better understanding of the transmission dynamics of the virus will help with developing interventions that can interrupt the chain of transmission and reduce the spread of infection. In the early stages of the pandemic, we previously found that the evidence from published studies assessing the risk of transmission of SARS-CoV-2 via fomites was limited [5].
Furthermore, the quality of included primary studies was low to very low, probably due to limited understanding of the dynamics of SARS-CoV-2 shedding at the onset of the pandemic, the types of studies done, the timing of collection, and sampling issues. In addition, less than a fifth of included studies at that time examined the cytopathic effect of SARS-CoV-2 and relied on detecting RNA from SARS-CoV-2. Since we published our previous review, several studies examining the potential transmission of SARS-CoV-2 via fomites have been published. Accordingly, we considered it timely to update the evidence on fomite transmission using the highest quality evidence from all the published studies to date.
To assess the transmission potential of fomites of SARS-CoV-2, we aimed to address the following questions:
-
(1)
Are fomite samples infectious (i.e. replication-competent virus is present on the samples)?
-
(2)
If so, what proportions are infectious, and what is the duration of infectiousness?
-
(3)
What is the relationship between fomite presence of SARS-CoV-2, infectiousness, and polymerase chain reaction (PCR) cycle threshold (CT)?
-
(4)
Is there evidence of a chain of transmission that establishes instances of fomite transmission of SARS-CoV-2?
The aim was to identify, appraise, and summarize the evidence relating to the role of fomite presence of SARS-CoV-2, its relationship with infectiousness (viral culture and/or serial quantitative reverse transcriptase (qRT)–PCR with or without gene sequencing), and the factors influencing transmissibility.
Methods
Search strategy
The WHO COVID-19 Database, PubMed, LitCovid, medRxiv, and Google Scholar for SARS-CoV-2 were searched using keywords and associated synonyms (see Appendix 1). The searches were conducted up to December 31st, 2021. No language restrictions were imposed. An information specialist (J.B.) conducted the searches. For relevant papers, forward citation was undertaken to identify relevant studies. The protocol is available at https://www.medrxiv.org/content/10.1101/2022.01.26.22269917v1.
Studies of any design (and in any setting) were included that investigated fomites as a potential source for transmission of SARS-CoV-2, defining a fomite as any inanimate object that, when contaminated with or exposed to an infectious agent, can transfer the infectious agent to a new host and be capable of inducing disease in the new host [6]. Studies that performed viral culture from fomite samples to assess the cytopathic effect (CPE) with verification techniques to ensure the cultured virus was SARS-CoV-2 were included.
Viral culture has previously been defined as encompassing several methods that can uniquely identify the replicating agent as SARS-CoV-2 [7]. Most commonly, this would be a plaque assay combined with a specific PCR confirmation, or immunological staining or gene sequencing of viral RNA. Viral genome sequencing is a process that helps to determine the order, or sequence, of the nucleotides in each of the genes present in a virus's genome (see https://www.cdc.gov/flu/about/professionals/genetic-characterization.htm). Therefore, studies that performed serial RT–PCR (with or without genomic sequencing) in addition to viral culture were included. Predictive or modelling studies were excluded. Laboratory studies that solely aimed to investigate viral stability and/or infectiousness through inoculation of fomites with already isolated SARS-CoV-2 virus were also excluded. Two reviewers (I.J.O., E.A.S.) independently screened study abstracts to determine eligibility. Any disagreements were resolved through discussion and consensus decisions.
Quality assessment
The risk of bias in included studies was assessed using modified Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria [8]. Using the patient selection, index test and flow and timing domains, we formulated the following questions to assess the risk of bias: (i) Was the source population clearly described? (ii) Were the study methods detailed enough to allow for replication of the study? (iii) Were sample sources clearly described? (iv) Were the analysis and reporting appropriate? (v) Was the pattern and number of fomite samples sufficient to demonstrate fomite transmission? Each domain was rated as low, unclear, or high risk of bias. One reviewer (I.J.O.) assessed the risk of bias, while a second reviewer (E.A.S.) independently cross-checked the ratings. Disagreements were resolved via consensus.
For studies that reported positive viral cultures, the level of reporting of the methods used to obtain replicable and appropriate viral culture results was assessed using the following four items: (1) adequate description of specimen sampling and management; (2) adequate exclusion of contamination or co-infection (use of good controls and appropriate antibacterials and antimycotics; (3) adequate reporting of results of inspection/culture; and (4) definition of CPE and identification of the agent causing the CPE was verified as SARS-CoV-2. Adapted SARS-CoV-2 case causality criteria were also used to assess transmission [7]. The causality categories include: certain, probable/likely, possible, unlikely, or unclear (see Appendix 2).
Data extraction
For each included study, the following data were extracted on to customized Excel spreadsheets: study ID, setting, types of participants, sources of fomites, methods used to detect SARS-CoV-2, timing of sample collection and number of samples, PCR CT, internal controls for CT, hygiene procedures, methods and thresholds for viral culture, methods for genomic sequencing, and study results. One reviewer (I.J.O.) extracted the data, and these data were independently verified by a second reviewer (C.J.H.). Disagreements were resolved through discussion.
Data analysis
Summary tables were used to present the frequency of SARS-CoV-2-positive tests from fomites, CT, CPE results, information on viral cultures, timing of collection from patients with respect to symptom onset, including the methods (timing, media), and verification techniques [7]. For studies attempting viral cultures, mean and standard deviation (SD) and median with interquartile range (IQR) were used as measures of central dispersion. Mean differences (MD) with SD were used to compute effect estimates for individual studies. For two studies that did not provide suitable data to compute SD, the average measure of SD for positive samples across the other studies was used to compute their SD [9]. Using the random-effects model of the standard meta-analysis software (RevMan 5.4) [10], the CT values for positive versus negative viral cultures were compared. Because of lack of standardization in CT values across studies, standardized mean differences (SMD) with 95% confidence intervals (CI) were used to compute overall effect estimates. Post hoc, the pooled relative effect of a binary cycle threshold of <30 was calculated. I 2 statistics were used to assess heterogeneity; values of 25%, 50%, and 75% represented mild, moderate, and substantial heterogeneity, respectively. Sensitivity analyses were performed by excluding two studies for which SDs were imputed, and through exclusion of one study in which fomite samples were not collected serially. One reviewer (I.J.O.) conducted the meta-analyses; these were independently cross-checked by a second reviewer (C.J.H.).
Results
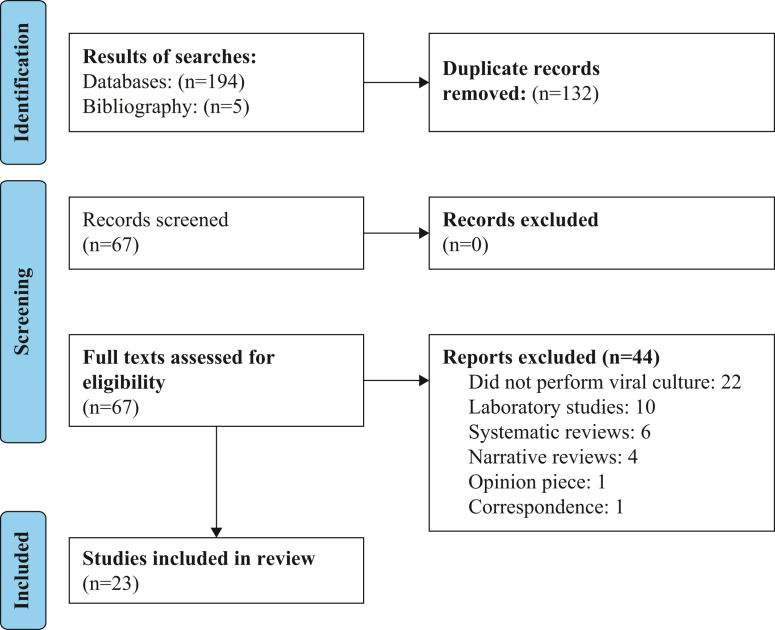
Sixty-two citations were identified (see Figure 1 ), and a further five through bibliography searches resulting in a total number of 67 eligible studies. Twenty-two studies that did not perform viral cultures were excluded, 10 that were laboratory-based, six systematic reviews, and four because they were narrative reviews. One study was excluded because it was an opinion piece and another study which was correspondence. (See Appendix 3 for reference list of excluded studies.) Finally, 23 studies [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33]] were included in the review. The main characteristics of included studies are detailed in Table I, Table II, Table III.
Figure 1.
PRISMA flow chart showing the process for the inclusion of studies that performed viral culture to assess transmission of SARS-CoV-2 through fomites.
Table I.
Main characteristics of included studies that assessed fomite transmission of SARS-CoV-2
| Study ID | Country | Setting dates | Types of participants | Sources of fomites | Test used to detect SARS-CoV-2 | Notes |
|---|---|---|---|---|---|---|
| Adenaiye 2021 [11] | USA | University campus May 2020 to Apr 2021 |
Participants with active SARS-CoV-2 infection (N = 49) | Mobile phones. | qRT–PCR | Used data from seronegative cases. The limit of detection (LOD) was 75 copies/sample. |
| Ahn 2020 [12] | South Korea | Tertiary care hospital Mar 2020 |
Three lab-confirmed COVID-19 patients who required high-flow oxygen therapy or mechanical ventilation. | Bedside tables, blood pressure cuffs, pillows, bedsheets, nasal prongs, outside surface of the ventilator circuit, tubing, masks, telephones, thermometers, keyboards, and fixed structures in the room (such as the doorknob, bedrails, floor, walls, window, and faucet handles), and grills of the ventilation exits in the ceiling. | rRT–PCR | Patient 1 was a 71-year-old man who presented with severe pneumonia. He was started on mechanical ventilation on hospital admission, 15 days after the onset of symptoms. Patient 2 was a 67-year-old woman with rapidly progressing pneumonia who was started on mechanical ventilation on hospital day 2, five days after the onset of symptoms. Patient 3 was a 44-year-old man with underlying terminal lung cancer and progressive pneumonia caused by SARS-CoV-2; high-flow oxygen therapy via high-flow nasal cannula at 60 L/min was started from hospital day 3, five days from the onset of symptoms. A positive test result was defined as CT ≤35 for the RdRp and E genes. |
| Alvis-Chirinos 2021 [13] | Peru | Public spaces Nov and Dec 2020 |
Food surfaces and inert surfaces in three districts. | Foods: bulk rice, avocado, banana, mango, lemon, tomato, lettuce, potato, cheese, chicken. Inert surfaces: handrails, seats, counter, touch screen, shopping cart handle, vending stand, and ATMs. |
Nucleic acid detection kit | Three districts with the highest number of COVID-19 cases were selected. Public spaces: market, supermarket, public transport bus, train, and bank agency. CT >39 for the ORF1ab gene considered negative. |
| Ang 2021 [14] | Singapore | Quaternary care university teaching hospital Feb–May 2020 |
Active COVID-19 patients. | Patient care and staff area: floor, table, bed handrail, nurse call button, cell phone, the sink, and door handles. Toilet area: door handle, the sink, toilet ledge, toilet bowl, and the flush button. |
RT–qPCR | The presence of SARS-CoV-2 was detected by RT–qPCR of the E-gene and N-gene. Positive control CT values ranged between 30.0 and 30.8 for the 1 pfu-positive control and 33.8–35.1 for the 0.1 pfu-positive control. All patients involved during the study; however, could be considered as mildly symptomatic without significant hypoxia or need for oxygen. |
| Bartlett 2021 [15] | USA | University hospital Apr 16th–30th, 2020 |
HCPs (N = 42) directly caring for non-ICU patients infected with SARS-CoV-2. | 1. High-touch surface areas outside the rooms of COVID-19 patients: donning/doffing stations, doorknobs, door thresholds, and shared workstations (mouse and keyboard). 2. High-touch surface areas in the emergency room and other COVID-19 wards: door handles and shared workstations). |
RT–qPCR | Spike assay was evaluated using 10-fold serial dilutions of SARS-CoV-2 RNA. |
| Ben-Shmuel 2020 [16] | Israel | COVID-19 isolation units in two hospitals and one quarantine facility | Patients with COVID-19. | ‘Mild COVID-19: floor, bedrails, bedside table, faucet handle, mobile phones, eyeglasses, patient's walker, air sampling filter Severe COVID-19: bedrails, faucet handle, ventilator, staff computer mouse, staff mobile phone, bedside table, trash bin top, bench top, air sampling filter Patient's toilets: toilet seat, handle grip, door handle Nurse station: floor, bench top, computer mouse, staff mobile phone, glucometer, electric thermometer, blood pressure cuff, air sampling filter Doffing area: floor, door handle, trash bin top, air sampling filter’ |
rRT–PCR | Mild COVID-19 patients required no ventilation. Patients stayed in private rooms either alone or as a family but were free to move around the hotel and socialize in public spaces. |
| Binder 2020 [17] | USA | University hospital Apr–May 2020 |
Hospitalized COVID-19 patients (N = 20) and their close contacts (N = 6). | 1. Toilet seat and interior of toilet bowl, TV remote control, cell phone, bedrailings and bed tray. 2. Hospital ward break room and head nurse and physician workstation. |
RT–PCR | For quality control, all RT–PCR assays were run twice; positive and negative controls used. |
| Coil 2021 [18] | USA | Academic medical centre Apr–Aug 2020 |
Active COVID-19 hospitalized in three ICU and two medical wards. | Doffing station, hand sanitizers, nightstand, computer station, door handle, ventilator, endotracheal tube straps, floor, intravenous pump, arterial line plunger, window, soiled linen, telemetry screen, heating, ventilation and air-conditioning (HVAC) system. | qRT–PCR | Air pressure in the HVAC system was temporarily reduced during sampling. Samples were collected both from the floor with some COVID-19 patients, as well as from another floor with no known COVID-19 patients. |
| Colaneri 2020 [19] | Italy | Emergency unit and the sub-intensive care ward | Area where febrile patients with respiratory symptoms were evaluated, and an infectious disease sub-intensive care ward that allows advanced respiratory care. |
Pre-intensive care ward Ward and buffer zone: exit buffer zone (1); computer keyboards in staff lounge (1). Rooms of patients A and B with CPAP helmet: bedrails (1), infusion pump (1), multi-parameter monitor (1), nurse buzzer (1), CPAP helmet, exterior surface (1), table (1) room of patient C in high-flow oxygen therapy: bedrails (1), multi-parameter monitor (1), infusion pump (1), table (1). Staff PPE: liquid-repelling gowns after 1 h of use (1), face shield and eye goggles (1), woven gown worn over liquid-repelling gown after 1 h of use (1), staff gloves, internal pair (1). Infectious disease emergency unit Ward: computer keyboards in triage and examination rooms (1), telephones (1), doorknobs and water taps in patient toilets (1). Staff equipment: portable X-ray machine (1), ECG machine (1), Medication cart (1). Patient rooms: beds (1), bed of patient with CPAP helmet (1). Staff equipment: liquid-repelling gowns, after 7 h of use (1), face shields (1), nurse gloves, internal pair gloves (1), staff mobile phone (1). |
rRT–PCR | Real-time RT–PCR targeting RNA-dependent RNA polymerase and E genes. |
| da Silva 2021 [20] | Brazil | Public areas Feb 2021 |
N/A | Transport terminals: (i) toilets; (ii) benches; (iii) public bike station; (iv) outdoor gym; (v) fresh green coconut; (vi) handrails; (vii) faucet; (viii) traffic light button; (ix) bus stop; (x) resting area. Healthcare units: (i) playground; (ii) recreation area; (iii) outdoor gym; (iv) toilet; (v) handrail; (vi) bus stop; (vii) public bike station; (viii) traffic light button; (ix) coffee shop; (x) faucet. Beach areas: (i) toilet; (ii) restaurant; (iii) handrail; (iv) resting area. Public parks: (i) playground; (ii) recreation area; (iii) outdoor gym; (iv) toilet; (v) handrail; (vi) bus stop; (vii) public bike station; (viii) traffic light button; (ix) coffee shop; (x) faucet. Supply centre: (i) toilet; (ii) restaurant; (iii) handrail; (iv) resting area. Public markets: (i) principal entrance; (ii) side entrance; (iii) public market access; (iv) toilet; (v) kiosk; (vi) store; (vii) food hall; (viii) traffic light button; (ix) faucet; (x) resting area; (xi) outside area. |
RT–qPCR | Samples collected from highly frequented areas. Samples were considered positive when they presented amplification for N1 target, CT value of <40. |
| Dumont-Leblond 2021 [21] | Canada | Long-term care facilities Spring 2020 |
Residents and HCWs. | Shelving units, door frames. | RT–qPCR | Only RT–qPCR results under 40 CT were considered positive. |
| Espinoza 2021 [22] | Brazil | COVID-19 ICU of a teaching hospital | HCWs in close contact with adults infected with COVID-19. | Mobile phones of HCWs. | RT–PCR | Amplification was carried out using the Roche Light Cycler® 96 System. A sample was considered positive when at least one of the target genes (S and E) was detected. |
| Kowta 2021 [23] | Canada | Six acute care hospitals March–May 2020 |
Consecutive patients (N = 78) hospitalized on any ward with lab-confirmed COVID-19. | (1) Bathroom doorknob, (2) phone (all surfaces of the patient's phone and room phone), (3) overbed table and chair (pooled), (4) bed (bedrail and pillow) and light switch or pullcord in patient's bedspace (pooled), and (5) toilet and sink faucet handles. | RT–PCR | Two separate gene targets were used for detection of SARS-CoV-2, the 5ʹ untranslated region (UTR) and the envelope (E) gene. Samples with CT <40 in both UTR and E genes were considered positive. In logistic regression analysis, the likelihood of positive viral culture was significantly more if the fomite sample was collected within 7 days of symptom onset (P = 0.02). |
| Lin 2021 [24] | Canada | Four hospitals Apr 2020 to Mar 2021 |
Patients with COVID-19 (N = 75). | Facial tissues, nasal prongs, call bells/cell phones, dentures, and sputum deposits | rRT–PCR | Samples were considered positive when E gene CT value was <35. For community participants, the responsible Medical Officer of Health Alberta Health Services (AHS) Public Health (PH) in Calgary provided a list of people who had tested positive for COVID-19 confirmed by RT–PCR and were approached by telephone to participate in the study in their community setting. Hand samples were collected post handshake from each hand in a patient who was 1 day from symptom onset and had a cough. |
| Ma 2021 [25] | China | Dock, Shandong Province Sep 2020 |
Two asymptomatic infected dock workers. | Cold chain products (frozen cods): outer packaging. | rRT–PCR | The two infected dock workers only unloaded frozen cod from abroad. Both had no history of living in high-risk areas in China and had no contact with patients from high-risk areas or patients with unexplained fever. There were no imported cases in their communities for 55 days, and neither dock worker had contact history with people returning from overseas. rRT–PCR CT value of ORF1ab and N gene target ≤40 was positive. |
| Ma 2021a [26] | China | Dock, Liaoning Province Jun–Jul 2020 |
Dock workers (N = 63) in contact with cold-chain products. | Operating table, floor, tools, sinks, sewers, and other environmental locations. | RT–qPCR Nucleic acid |
Samples were taken from the food processing area of the dock company. A TaqMan probe-based kit was designed to detect the ORF1ab and N genes of COVID-19 virus in one reaction. |
| Marcenac 2021 [27] | USA | Households Mar 30th to Apr 25th, 2020 |
Index patient in a household living with ≥1 person, and positive for SARS-CoV-2 on a nasopharyngeal swab collected ≤10 days prior to enrolment. | Light switches (2 samples/household), toilet handles (1/household), bathroom sink handles (1/household), pillows or nightstands of index cases (1/household), pillows or nightstands of contacts (2/household), and refrigerator handles (1/household). | RT–PCR | Remaining surfaces were selected by requesting that household members identify frequently touched surfaces in their respective household, which at times were additional surfaces of pre-assigned types (e.g. bathroom sink handles). |
| Nannu Shankar 2021 [28] | USA | Residential rooms of two adults with COVID-19 Sep 2020 |
Two adults with COVID-19. | High-touch surface areas: Volunteer A: mobile phone, a laptop computer touch pad, and the left elbow. Volunteer B: mobile phone, room doorknob. |
rRT–PCR | rRT–PCR tests were performed in a BioRad CFX96 Touch Real-Time PCR detection system using the SARS-CoV-2 N-gene detection primers and probe and rRT–qPCR parameters. |
| Newey 2021 [29] | USA | Near the campus of a university | N/A | 1. Cash from university vault: comprises currency from university-based stores, restaurants, dormitories and vending machines 2. Fresh samples of cash and/or coins were obtained from local restaurants. |
LAMP assays | |
| Oksanen 2021 [30] | Finland | COVID-19 ward, home July 2020–Mar 2021 |
Unvaccinated participants with RT–PCR-confirmed symptomatic COVID-19 infection (N = 31). | High-touch surfaces: bed remote, cellphone, drinking glass, computer, door handle. Low-touch surfaces: hospital equipment, floor, table, bedrail, airvent. Toilet surfaces: toilet seat, toilet flush button, toilet tap, toilet bowl. Other surfaces: staff PPE. |
RT–PCR | 31 collections were performed, 24 of them on the cohort COVID-19 ward and 7 in patients' homes in normal rooms where the patients spent time during illness. PCR was positive if CT value <38. RNA extracted from the Fin/20 strain culture was used as a positive control and nuclease-free water as a negative control. |
| Rajendiran 2021 [31] | Malaysia | ICU and general wards Mar 25th to Apr 17th, 2020 |
COVID-19 patients hospitalized in ICU and general wards. | High-touch surface areas: 1. Patient's cubicle or room (doorknob, bedrail, pillowcase, side table, cardiac table, floor at 1 m from patient's bed, ventilation outlet or window, blood pressure cuff, oximeter). 2. Areas in toilet (sink and toilet bowl) of general wards. 3. ICU: patient's cubicle (doorknob, bedrail, bedsheet, side table, floor 1 m from patient's bed, ventilation outlet, intravenous drip stand) and staff area (oximeter, monitor, phone, keyboard, mouse, soles of medical staff). |
RT–PCR | |
| Winslow 2021 [32] | UK | University hospital | 30 hospitalized patients with COVID-19 requiring supplemental oxygen. | Floor, table, high-object surface. | RT–qPCR | A sample was defined as positive for viral RNA if both E and ORF1a RT–qPCR assays gave CT <45. |
| Zhou 2020 [33] | UK | Teaching hospital Apr 2020 |
Adults with COVID-19. | High-touch surfaces: bedrails, blood pressure monitors, ward telephones, computer keyboards, clinical equipment (syringe pumps, urinary catheters), hand-cleaning facilities (hand-washing basins, alcohol gel dispensers). | RT–qPCR | SARS-CoV-2 viral RNA was detected using AgPath-ID. One-step RT–PCR reagents with specific primers and probes targeting the envelope (E) gene. Samples were defined as positive if both duplicates had CT <40.4 |
ATM, automated teller machine; rRT–PCR, real-time reverse transcription–polymerase chain reaction; q, quantitative; LAMP; loop-mediated isothermal amplification; ICU, intensive care unit.
Table II.
Sampling, hygiene procedures and CT test results
| Study ID | Timing of sample collection | Hygiene procedures | No. of fomite samples | No. of SARS-CoV-2-positive samples | Sites of positive samples | Comments |
|---|---|---|---|---|---|---|
| Adenaiye 2021 [11] | Phone/tablet swab done at each visit | Surgical masks | 74 | 42/80 (52.5%) | Mobile phone | 6/6 of alpha variant and 36/74 of other variant positive. |
| Ahn 2020 [12] | Patient 1: hospital day 7 when patient was febrile with poor oxygenation, and on that day, chest imaging demonstrated severe ARDS. Patient 2: hospital day 4 when patient had a sustained fever with rapid deterioration to severe ARDS. Patient 3: hospital day 13 when patient had a persistent cough with sputum and shortness of breath and spat out sputum frequently. |
Nurses performed daily routine cleaning, but disinfection was performed only after the patients were discharged. | 76 Patients 1 and 2: 48 Patient 3: 28 |
15/76 (19.7%) Patients 1 and 2: 2/48 (4.2%) Patient 3: 13/28 (46.4%) |
Patients 1 and 2: outside surface of the endotracheal tubes in the area connected to the ventilator circuit. Patient 3: thermometer, restraints, bedsheets, cup, nasal prongs, NIV mask, high-flow oxygen generator, telephone, remote control, and fixed structures including bedrails, floor, and the grill of an air outlet fan in the ceiling. |
All patients were symptomatic, and immunocompromised by virtue of their critical care stay or underlying cancer. Respiratory specimens persistently tested positive for SARS-CoV-2 by rRT–PCR up to the time of environmental sampling, which varied between 9 and 22 days post symptom onset. CT of sputum samples: 13.32 to 40.0 between hospital days 1–14 and CT of environmental samples 28.85 to 31.78 for positive PCR samples that were culture positive. CT of positive samples: 22.31 to 29.65. |
| Alvis-Chirinos 2021 [13] | The frequency of sample collection was once a week and four times during a month from Nov–Dec 2020 | Not specified | 2055 960 food surfaces 1095 inert surfaces |
1/2055 (0.1%) Food surfaces: 0/960 Inert surfaces: 1/1095 (0.1%) |
ATM | Non-probabilistic convenience sampling was applied to select the samples. Prior to sample collection, authorization for the entry of field personnel was coordinated with those responsible authorities. CT of positive samples: 35.01–37.08. |
| Ang 2021 [14] | Samples were collected once during the last campaign of air sampling in the same wards. The chosen swab sites were not cleaned for at least 8 h prior to swabbing. The timing for sample collection and analysis were subject to the availability of the trained medical staff, consent of patients, and the capacity of the BSL-3 processing laboratory. |
All patients were masked with regular surgical mask whenever possible. | 73 | 7/73 (9.6%) | Toilets, patient bedside table | Surface samples were collected from one isolation ward and two open-cohort wards. A portion of some swab sites, e.g. the bedside table in the patient care areas and ledge in the toilet, were covered with a sieve one day before swabbing to avoid direct contact from the patient. The LOD of the RT–qPCR assay was 3.9 copies per reaction. |
| Bartlett 2021 [15] | Before and after encounters between the patients and HCP | In all clinical encounters, the HCP wore hair bouffant, surgical mask, contact gown, and gloves. Pre-exposure samples were collected prior to donning of PPE, while post-exposure samples were collected following PPE doffing and upon exiting the patient's room. |
82 | 2/82 (2.4%) | Patient's room on the door threshold following the encounter with the HCP. Door handle leading to a physician's workroom in the emergency department. |
70 samples from clinical encounters; 12 samples from high-touch surface areas from heavy traffic COVID-19 hospital care areas. Samples were collected from skin around the nose and mouth. Samples were also collected from the HCP's exposed skin at the temples, cheeks, and neck. Additional samples were taken from the sides of the HCP's footwear. CT for positive samples: 30.6, 37.2. |
| Ben-Shmuel 2020 [16] | Not specified | Routine cleaning and decontamination were done once daily at best only in communal areas. Staff routinely used gowns, masks, shoe covers, face shields, and disposable surgical caps. |
55 | 29/55 (51%) | Outside the patient's room on the door threshold following the encounter with the HCP door handle leading to a physician's workroom in the emergency department. | Patients had mild to moderate disease. No patients on ventilation. CT for positive samples: 34 to 37.9. |
| Binder 2020 [17] | Not specified | No scheduled cleaning of rooms while COVID-19 patients were occupying them. Deep cleaning of the rooms and sanitation procedures such as disinfecting floors and surfaces with bleach solution and UV light emitter treatment for 45 min were performed between patients. | 112 | 7/112 (6.3%) | TV remote, bedrails, tray, toilet bowl, cell phone. | Fomite sampling was conducted in an empty hospital room (no patient contact for four days) in the Duke University Hospital COVID-19 ward which had been disinfected by bleach solution wipe downs and UV light treatment for 45 min. CT for positive samples: 36.4–39.8. |
| Coil 2021 [18] | Not specified | Improved cleaning protocols with a change in the frequency/duration/composition of cleaning material in the hospital between April and August 2020. | 224 56 in April 2020 168 in Jul/Aug 2020 |
11/224 (4.9%) 6/56 (11%) for Apr 2020 5/168 (3%) for Jul/Aug 20 |
Telemetry screen, soiled linen, endotracheal tube strap, computer station. | Changes to hygiene procedures were instituted after collection of the first samples in Apr 2020. CT for positive samples: 32–44. |
| Colaneri 2020 [19] | Swabs were performed around 12 noon, ∼4 h after cleaning. | Ward surfaces were routinely cleaned twice daily (morning and early afternoon). HCWs wore PPE comprising liquid-repelling gowns, double gloves, class 2 filtering face-piece respirators and eye protection. | 26 | 2/26 (7.7%) | External surface of the CPAP helmet. | In the sub-intensive care ward, swabs were performed in a double room where two patients with CPAP helmets were allocated. In the emergency room, samples were collected from two different rooms. Each room accommodated three patients, one of them with a CPAP helmet. CT for two patients in sampling area was 23 and 26 respectively. |
| da Silva 2021 [20] | Between 09:00 and 13:00 | Qualified technicians wore PPE. | 400 Transport terminals 84 Healthcare units 84 Beach areas 21 Public parks 105 Supply centre 21 Public markets 81 |
97/400 (24.2%) | Toilets, ATMs, handrails, playground and outdoor gym. | SARS-CoV-2 RNA was found most frequently on rock (10/22, 45.4%), followed by plastic (18/50, 36%), wood (12/47, 25.5%), metal (45/179, 25.1%), glass (2/10, 20%), concrete (8/55, 14.5%) and others (ceramic and rubber) (2/37, 5.4%). CT for positive samples: 31.0 to 38.7. |
| Dumont-Leblond 2021 [21] | Rooms were sampled from 8 to 30 days after the patient was diagnosed and from 9 to 48 days since of the first confirmed case in the corresponding long-term care facility. | Standard infection control practices instituted. The swabbed surfaces were out of reach and unfrequently cleaned. | 62 | 20/62 (32%) | Shelving units, door frames. | Positive tests: Shelving units: median 761.5 genome equivalents/surface (1st quartile = 74.6, 3rd quartile = 2615.6). Door frames: median 830.8 genome equivalents/surface (1st quartile = 391, 3rd quartile = 2772). |
| Espinoza 2021 [22] | Not specified | HCWs wore lab coats, N95 face masks and surgical caps inside the unit, and a surgical gown, face shield and gloves when entering the patient room. | 51 | 2/51 (3.9%) | Mobile phones | An educational campaign on SARS-CoV-2 cross-transmission, its permanence on fomites, and the proper use and disinfection of mobile phones was performed. CT of positive samples: 34,36. |
| Kowta 2021 [23] | At enrolment and every three days | Not specified | 474 | 125/474 (25%) | Bathroom door, bed and switch, phone, table and chair, toilet and sink. | Median CT value of surface samples testing positive for SARS-CoV-2 was 35.1 (IQR: 32.8–36.9). Factors significantly associated with positive environmental samples: hypoxia on admission, PCR-positive nasopharyngeal swab with CT ≤30 on or after the environmental sampling date, higher Charlson Comorbidity Index, and shorter time from onset of illness to environmental sample date. |
| Lin 2021 [24] | Not specified | No data were collected on the frequency of cleaning of fomites. | 21 Used tissues 5 Nasal prongs 4 Dentures 2 Phone/call bell 10 |
10/21 (47.6%) Used tissues 4/5 (80%) Nasal prongs 3/4 (75%) Dentures 1/2 (50%) Phone/call bell 2/10 (20%) |
Used tissues, nasal prongs, face cloth, dentures, phone/call bell, bedrail. | They found the ability to detect culturable virus was within the first 7 days after symptom onset and then declined dramatically with the exception of immunocompromised patients. Positive results also identified in specimens also collected from a patient's two hands after one hand had been coughed on, kiss samples inside of the polyethylene bag touched by a patient's lip, and dried saliva on a Petri dish (post 2 h). Duration of infectiousness on fomites was assessed by placing a respiratory clinical sample from an infected patient on fomites such as a call bell, computer keyboard, stethoscope diaphragm and an N95 respirator and demonstrating minimal reduction in quantitative burden of SARS-C0V-2 from baseline after 4 h and in dried saliva on a plastic surface for 2 h with no reduction. |
| Ma 2021 [25] | Not specified | Not specified | 919 Stage 1: 420 Stage 2: 499 |
102/919 (11.1%) Stage 1: 51/420 (12.1%) Stage 2: 51/499 (10.2%) |
Outer surface of cold-chain containers. | |
| Ma 2021 [26] | Not specified | Not specified | 5370 Processing workplace: 39 Cold-chain seafood: 4963 Cold-chain pollock packaging: 368 |
148/5370 (2.8%) Processing workplace: 14/39 (35.9%) Cold-chain seafood: 0/4963 Cold-chain pollock packaging: 134/368 (36.4%) |
Cold-chain containers | Surface swab samples of the inner and outer packaging of the cold-chain imported seafood that were suspected to be contaminated by COVID-19 virus were carefully smeared and collected. Imported cold-chain seafood by Company K; all other imported cold-chain seafood products from the two cargo ships that were temporarily stored at Company K were sampled and tested. |
| Marcenac 2021 [27] | Four households on enrolment visit (day 0). Five households on median 7 days (range: 4–9). One household on day 20. |
Not specified. Two households reported using disinfecting wipes and sprays on high-touch surfaces after someone became ill with COVID-19. | 150 | 23/150 (15%) | Nightstands (4/6 samples, 67%), pillows (4/23, 17%), and light switches (3/21, 14%). Also, on doorknobs (2/17, 12%); kitchen surfaces and appliances, including a sink handle (1/5, 20%), countertop (1/9, 11%), table (1/4, 25%), refrigerator handle (1/11, 9%), microwave (1/7, 14%), and trash can lid (1/1, 100%); and electronic items, including a phone (1/3, 33%), computer (1/4, 25%), and TV remote control (1/7, 14%). |
15 surfaces were sampled in each household. CT for positive samples: Median 33.8 (26.4–37.2). Positive culture from a nightstand sample (CT = 26.4) was found in a 35-year-old man) with respiratory symptoms whose nasopharyngeal swab was positive for SARS-CoV-2 (CT = 15.5) on the environmental sampling date. This man first tested positive for SARS-CoV-2 two days prior to environmental sampling. |
| Nannu Shankar 2021 [28] | Surface samples collected from volunteer A's room on days 5 and 6 after positive test. Surface samples collected from volunteer B's room on days 2 and 6 after positive test. |
Self-isolation | 7 Volunteer A: 3 Volunteer B: 4 |
2/7 (28.6%) Volunteer A: 0/3 Volunteer B: 2/4 (50%) |
Mobile phone (front screen and back cover). | Both positive samples for Volunteer B were collected Oct 2nd, 2020; negative samples collected Oct 6th, 2020. CT for positive samples: 35.91 and 36.4 |
| Newey 2021 [29] | Fresh environmental samples were obtained from surfaces of bank notes and coins from local restaurants and assayed within 1 h | Not specified | 813 Money cards and ID cards 279 Bank notes 429 Coins 105 |
17/813 (2.1%) Money cards and ID cards: 17/279 (6.1%) Bank notes: 0/429 Coins: 0/105 |
Bank notes | |
| Oksanen 2021 [30] | Not specified | Hand hygiene, universal masking for staff (FFP2/3 for ICU and surgical masks for the COVID ward), guidance on social distancing (2 m), and PPE following droplet precautions. The room was cleaned twice a day. | 252 Hospital 182 Homes 70 |
25/252 (9.9%) | High touch surfaces: bed remote, cell phone, drinking glass, computer, door handle. Low touch surfaces: hospital equipment, floor, table, bedrail. Toilet surfaces: toilet seat, toilet flush button. |
Positive surface samples were detected more often when there were multiple COVID-19 patients in the ward/house during the sampling (P = 0.02). |
| Rajendiran 2021 [31] | In the morning before cleaning | Investigators wore PPE and complied with the respective hospital protocol on donning and doffing procedures. | 124 Stage 1: 104 samples from high-touch surfaces and areas used by patients and HCWs. Stage 2: 20 samples from general wards. |
6/124 (4.8%) Stage 1: 3/104 (2.9%) Stage 2: 3/20 (15%) |
Cardiac table, sink in an isolation room and open ward in stage 1. Floor, sink and toilet bowl in stage 2. |
2 stage sample collection: In stage 1, sampling was performed on random days in the intensive care unit (ICU) and general wards. In stage 2, samples were collected serially on alternate days for 7 days in two selected general wards. CT for positive samples: 33.49 to 37.52. |
| Winslow 2021 [32] | 0–15 days in hospital with illness | Not specified | 90 | 6/90 (7%) | Floor (5), high-object surface (1) | Environmental samples varied according to clinical and operational needs. Participants in the HFNO group were sampled significantly later in their illness compared to those receiving only supplemental oxygen: mean 16 days (95% CI: 13 to 19) vs mean 9 days (95% CI: 5 to 13), from symptom onset, respectively. Clinical surfaces were more contaminated with viral RNA than the air samples. CT for positive samples: 35–40. |
| Zhou 2020 [33] | Not specified | All areas were disinfected daily with an additional twice-daily disinfection of high-touch surfaces. | 218 | 23/218 (10.6%) | Computer keyboards/mice, alcohol gel dispensers, and chairs, and >50% of toilet seats, sink taps, and patient bedrails. |
ARDS, acute respiratory distress syndrome; CT, cycle threshold; CPAP, continuous positive airway pressure; HCP, healthcare professional; HCW, healthcare worker; HFNO, high-flow nasal oxygen; ID, identity cards; PPE, personal protective equipment.
Table III.
Methods and results of viral culture in studies assessing fomite transmission of SARS-CoV-2
| Study ID | Method used for viral culture | Threshold for viral culture | Duration of incubation | Results of viral culture | Comments |
|---|---|---|---|---|---|
| Adenaiye 2021 [11] | Virus propagated on Vero E6 cells stably expressing TMPRSS2, then transferring the media to A549 cells stably expressing human ACE2. Infected A549-ACE2 cells were quantified using immunofluorescence staining with anti-SARS-CoV-2 nucleocapsid antibody and Hoechst 33342 and imaging with a Celigo Imaging Cytometer. | CT values: max. 1.7 × 104 and 1.2 × 106 RNA copies for alpha and other variants respectively. | Unclear | No CPE observed. | |
| Ahn 2020 [12] | Confluent monolayers of Vero E6 cells in 96-well plates were infected by 10-fold dilutions of the SARS-CoV-2 supernatants from the environmental samples. The inoculated cultures were grown in a humidified 37 °C incubator with 5% CO2. After 72 h, areas of cell clearance with Crystal Violet staining were used to demonstrate the CPE. When the CPE was observed, detection of SARS-CoV-2 nucleic acid by rRT–PCR in the supernatant was performed to confirm a successful culture. | All positive samples (CT 22.31 to 31.78). | 3 days | Viable viruses were detected in seven samples (patient 3) from a nasal prong, bedside table, floor near the patient, remote control, bedrails, bedsheets, and NIV mask. One sample from patient 1 (nasal prong). | |
| Alvis-Chirinos 2021 [13] | Vero-E6 cells were cultured in DMEM culture medium supplemented with streptomycin 100 mg/L, ampicillin 25 mg/L, 20% inactivated FBS and kept at 37 °C in a humid atmosphere of 5% CO2. The filtered sample was inoculated into the cells, and incubated for 7–10 days for the isolation of the SARS-CoV-2 virus. The presence of CPE was observed and the supernatant and cells were collected at the end of incubation. The diagnosis of isolation was determined by RT–PCR values. | All positive SARS-CoV-2 samples (N = 1; CT 37.08) | 7–10 days | No CPE after 10 days. | |
| Ang 2021 [14] | Culture was performed using Vero E6 cells grown in DMEM supplemented with 2% heat-inactivated FBS and buffered with 2 g sodium hydrogen carbonate. 400 μL of each qPCR-positive UTM filtrate was added to Vero E6 cells and incubated at 37 °C with 5% CO2. After 4–7 days of incubation, CPE was monitored using light microscopy. The supernatant was passaged twice more following a 4–7-day incubation period at 37 °C with 5% CO2. RNA was then extracted from these cultures after the third passage using the QIAamp Viral RNA Mini Kit and qPCR performed. | Any qPCR-positive samples were processed for culturing. | 4–7 days | No CPE after 7 days. | |
| Bartlett 2021 [15] | Conducted in a BSL 3. Vero E6 cells were grown to 90% confluency in a 24-well plate with DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Selected VTM samples that tested positive with the RT–qPCR, along with a selection of samples that tested negative, were thawed and filtered to ensure sterility. Each well of Vero E6 cells was inoculated with 100 μL of filtered VTM. Cells were incubated at 37 °C in a humidified incubator for 8 days. Cultures were monitored every 48 h for CPE by microscopy. Control conditions were performed with either the SARS-CoV-2 isolate or with media only. Infectious SARS-CoV-2 was confirmed when CPE was detected in the inoculated wells. | Two SARS-CoV-2 samples met threshold: one from door handle leading to a physician's workroom in the ED (CT 37.8) and one from outside the patient's room on the door threshold. following the encounter with the HCP (CT 30.6). | 8 days | No CPE detected after 8 days in culture or media alone. | |
| Ben-Shmuel 2020 [16] | Virus infectivity was tested by seeding quadruplets of 200 μL on Vero E6 cells for CPE assay. Applied 200 μL from 10-fold serial sample dilutions upon Vero E6 cell cultures in 24-well plates. After 1 h, wells were overlaid with 1 mL of MEM medium supplemented with 2% FBS, MEM non-essential amino acids, 2 mM l-glutamine, 100 units/mL penicillin, 0.1% streptomycin, 12.5 units/mL nystatin, and 0.15% sodium bicarbonate. Cells were incubated for 5 days (37 °C, 5% CO2), and CPEs were observed after fixation with Crystal Violet solution. | Not specified. CT was 34–37.9. | 5 days | None of the surface and air samples from the three sites (0/97) were found to contain infectious titres of SARS-COV-2 on tissue culture assay. | Unlikely to have positive viral culture because of high CT. |
| Binder 2020 [17] | BSL-3 lab. Specimens were inoculated on to Vero E6 cells in two passages by transferring 250 μL of supernatant at 7 days post inoculation for a total 14 days of incubation. Cells were monitored for CPE every 48 h. The cells and supernatant harvested 14 days post inoculation were screened for SARS-CoV-2 by molecular assay. Infectious SARS-CoV-2 was confirmed when (i) CPE was detected in inoculated wells and (ii) SARS-CoV-2 was detected in inoculated wells by real-time RT–PCR, at least two CT values below the original sample. | All seven positive samples included for culture: CT 36.4–39.8. | 14 days | No CPE detected after 14 days. | Two aerosol samples with the lowest CT (19.4, 20.9) showed CPE. |
| Coil 2021 [18] | Vero E6 cells were maintained in DMEM supplemented with 10% FBS and 100 IU/mL of penicillin–streptomycin. The mNeonGreen SARS-CoV-2 was propagated and titrated in Vero E6 cells. All swab samples and positive controls were diluted in D10-CoV medium consisting of DMEM supplemented with 10% FBS, 100 IU/mL penicillin–streptomycin, 250 μg/mL amphotericin B and 250 μg/mL gentamicin. Six-well plates of Vero E6 cells were infected with either 300 μL of the viral transport medium from qRT–PCR positive environmental swab samples diluted 1:1 in D10-CoV medium, or 300 μL of mNeonGreen SARS-CoV-2 serially diluted 10-fold in D10-CoV medium. Following 1 h incubation at 37 °C, the cells were replenished with fresh D10-CoV medium and incubated at 37 °C + 5% CO2 for 5 days. A mock-treated control consisting of cells only maintained in D10-CoV medium was included in the assay and treated identically. All samples were tested in duplicate. Two and five days post infection, the cells were assessed microscopically for any visible CPE. Five days post infection, 2 mL of cell culture supernatant was collected from each well and mixed with 6 mL of Trizol LS reagent. All Trizol-treated samples were used for RNA extraction and qRT–PCR. | Five swabs (identified as positive by qRT–PCR) were tested (CT unspecified). | Five days | No signs of CPE 5 days post infection. | CT for the 11 positive samples ranged from 32 to 44. Environmental samples especially may have been degraded or diluted, affecting the genomic RNA available for reverse transcription. |
| Colaneri 2020 [19] | Culture attempted with Vero E6 cell line. A 200 μL sample was inoculated on to a Vero E6 confluent 24-well microplate for virus isolation. After 1 h of incubation at 33 °C in 5% CO2 in air, the inoculum was discarded and 1 mL of medium for respiratory viruses was added to each well. Cells were incubated at 33 °C in 5% CO2 in air and observed by light microscopy every day for CPE. After a 7-day incubation, 200 μL of supernatant was used for molecular assays. To further confirm negative data, supernatants collected on day 7 were tested by real-time RT–PCR. | All 26 samples were inoculated on to susceptible Vero E6 cells. | 7 days | No CPE observed after 7 days. | |
| da Silva 2021 [20] | BSL-3. Vero CCL-81 was used for virus isolation from positive environmental samples. Cells were cultured in DMEM, supplemented with 10% heat-inactivated foetal bovine serum, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin; and maintained in a humidified atmosphere, at 37 °C and 5% CO2. Vero CCL-81 cells were cultured in 12-well plates at a density of 2 × 105 cells/well. After 24 h, the culture media was removed and cells were incubated with 300 μL of undiluted and filtered surface samples at 37 °C, 5% CO2, for 1 h. Fresh media supplemented with 2% FBS (700 μL) was added to the cells and they were maintained at 37 °C, 5% CO2. Cells were monitored daily for the visualization of virus-induced CPE. CPE images were acquired in Carl Zeiss Axio Observer 5 microscope coupled to a photographic camera. After 3 days post-infection supernatants were collected and 300 μL were transferred to a new 12-well plate. This procedure was repeated until completing three passages (P1, P2 and P3). Following this, cell culture supernatants were collected on t = 0 h and t = 72 h in each passage for viral RNA extraction and possible SARS-CoV-2 detection by RT–qPCR. | Nine samples with Cq value <34 (CT ranging from 31.0 to 33.7). | 3 days | No CPE after 3 days. | |
| Dumont-Leblond 2021 [21] | Vero E6 cells were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 100 U/mL penicillin and 100 μg/mL streptomycin. Vero E6 cells were seeded at a concentration of 3 × 105 cells/well in a 6-well plate. The next day, 400 μL of PCR-positive sample was centrifuged at 10,000 rpm to remove debris and the supernatant supplemented with 16 μg/mL TPCK-treated trypsin and 2× antibiotic–antimycotic was added to the cells. The plate was incubated for 1 h in a humidified 37 °C incubator with 5% CO2 and rocked every 15 min. After 1 h, the inoculum was removed and replaced with 2 mL of DMEM supplemented with 2% FBS, 2× antibiotic–antimycotic, 100 U/mL/100 μg/mL P/S and 6 μg/mL TPCK-treated trypsin. The plate was returned to the incubator and observed for CPE for 5 days. If no CPE was observed, the supernatant (500 μL) was passaged on to fresh Vero E6 cells and observed for another 5 days. | Viral culture on swab samples with the seven positive RT–qPCR signals was attempted. | 5 days | Viral culture was negative for all samples. | Method for viral culture based on previous publication by the authors: https://pubmed.ncbi.nlm.nih.gov/33206022/ |
| Espinoza 2021 [22] | Performed by inoculating an aliquot of the sample collected from the mobile phones into Vero cells in DMEM supplemented with FBS (5%), antibiotics and antimycotics at 37 °C in an atmosphere with 5% CO2. Vero cells were examined for CPE daily and new RT–PCRs were performed from the culture supernatant on the 3rd, 7th, and 14th days of culture. | Two positive samples with CT 34 and 36 included for culture. | 14 days | 1/2 (50%) was positive. The sample with CT 34 showed CPE on the 3rd day, but subsequent RT–PCR of this isolate was negative. | The supernatant from both cultures was monitored for 14 days without observing any other CPE. The swabs that had a positive SARS-CoV-2 RT–PCR corresponded to HCWs with high exposure to patients with COVID-19. |
| Kowta 2021 [23] | Vero E6 cells were seeded at a concentration of 3 × 105 cells/well in a six well-plate. The next day, 500 μL of sample containing 16 μg/mL TPCK-treated trypsin, 2× Pen/Strep and 2× antibiotic-antimycotic were used to inoculate cells. Plates were returned to a 37 °C, 5% CO2 incubator for 1 h and rocked every 15 min. After 1 h, the inoculum was removed and replaced with DMEM containing 2% FBS, 6 μg/mL TPCK-treated trypsin, 2× Pen/Strep, and 2× antibiotic-antimycotic. Cells were observed daily under a light microscope for CPE for 5 days post infection. Cell cultures not showing any CPE were blind-passaged on to fresh Vero cells and observed for a further 5 days. The RT–PCR assay described above was used to confirm SARS-CoV-2 isolation from supernatant. | Attempted if CT <34.0 (36 samples). | 5 days | 6/36 (16.7%) had CPE. Bathroom door (1/4), bed and switch (2/13), phone (1/5), table and chair (1/10), toilet and sink (1/4). The highest CT of a sample yielding virus by culture was 29.1. |
The median time between onset of illness and surface sample date was 10 days (IQR 6–12) for all environmental surface swabs, nine days (IQR 5–12; range 3–20 days) for PCR-positive surface swabs, and four days (range 4–5) for culture positive surface swabs. |
| Lin 2021 [24] | BSL-3 lab. Vero (ATCC #CCL-81) and Vero E6/TMPRSS2 (JCRB cell bank 1819) were cultured with DMEM supplemented with 100 units/mL of penicillin, 100 μg/mL of streptomycin, 0.25 μg/mL of amphotericin B (Gibco), and 10% FBS. For virus culture, 2×105 cells were seeded into each well of the 12-well plates one day before titration. Ten-fold serial dilutions of the virus were plated in duplicate on Vero CCL-81 cells and cultured for 3 days at 37 °C in MEM supplemented with 100 units/mL of penicillin, 100 μg/mL of streptomycin, 0.25 μg/mL of amphotericin B, and 1% carboxymethyl cellulose (CMC) (Sigma C4888). In a few cases, detected late in the study, some slow-growing viruses that produced small plaques on Vero CCL-81 cells were also plated on Vero E6/TMPRSS2 cells. The cells were then fixed and stained with a solution containing 0.13% (w/v) Crystal Violet, 11% formaldehyde (v/v), and 5% ethanol (v/v) to permit plaque counts. | All samples were cultured. | 3 days | Frequency of infectious virus: used tissues 4/5 (80%) nasal prongs 3/4 (75%) dentures 1/2 (50%) phone/call bell 2/10 (20%) |
Quantity of SARS-CoV-2: used tissues 40 to 2.0×103 pfu/mL; nasal prongs 5 to 1.1×102 pfu/mL; dentures 45 pfu/mL; phone/call bell 2.0×102 to 1.9×103 pfu/mL. Virus was also cultured in samples from a patient's hand, dried saliva and ‘kiss’ bag. Samples that contained infectious material had significantly lower CT values compared with plaque negative specimens (P < 0.0001). Infectious samples had a mean CT of 19.2, whereas the mean CT in non-infectious samples was 29.5. |
| Ma 2021 [25] | Not described. | Six samples with high viral loads (CT values 25, 28, 30, 31, 31, and 32) collected from Fish Cluster Pallet 1 were obtained for further viral isolation. | Unclear | 1/6 sample (CT 25) showed CPE. | Methods used for viral culture not well described. Unclear which samples were used for viral culture. The sequencing match also suggested that the positive virus culture was not a result of contamination. |
| Ma 2021a [26] | Each of the selected RT–qPCR-positive samples was seeded separately in Vero cells. The cells were monitored daily with light microscopy for cytopathic effects. | Nucleic acid positive samples from the cold-chain pollock from cargo ship. | Unclear | No virus was isolated. | |
| Marcenac 2021 [27] | The archived aliquots of positive swab samples were further tested by cell culture on Vero cells (ATCC CCL-81) to check the infectivity of SARS-CoV-2, with minor modifications from the original protocol. Swab eluates were filtered and cultured in 96-well plates (200 μL) and T-25 cm2 flasks (1 mL) in a humidified 37 °C incubator at 5% CO2 for one week, with daily monitoring of virus-induced CPE. CPE-positive samples were confirmed for SARS-CoV-2 by RT–PCR. Positive and negative controls were included in all RT–PCR and culture assays. | Median CT for positivity was 33.8 (26.4–37.2). All 23 RT–PCR-positive samples used to inoculate Vero cells. | 7 days | One out of 23 samples (4%) was positive after 7 days. Sample came from a nightstand swab (CT = 26.4) belonging to an index case whose nasopharyngeal swab was positive for SARS-CoV-2 (CT = 15.5) on the environmental sampling date. | |
| Nannu Shankar 2021 [28] | BSL-3 Lab. Vero E6 cells procured from the ATCC, were used as the host cells. The cells were grown and maintained as monolayers and were inoculated with aliquots of material collected at volunteer B's room once they attained 80% confluence in T-25 flasks. The spent cell culture medium was removed from the T-25 flasks with Vero E6 cells and replaced with 1 mL of sterile cell culture medium and 50 μL of the filtered samples, and the flasks with inoculated cells incubated for 1 h at 37 °C. Following that, 2 mL of complete media was added to the flasks, which were reincubated at 37 °C. The cells were periodically observed for CPE. An aliquot of the nucleic acids was also tested using a GenMarkDx multiplex PCR eSensor XT-8 respiratory viral panel. | Two positive samples with CT 35.91 and 36.4 included for culture. | 14 days | No CPE observed after 14 days. | CPE were observed in Vero E6 cells inoculated with surface samples collected from volunteer B's room within 4 days of their inoculation. The resulting amplicon was sequenced and informed that volunteer B was co-infected with HAdV species B type 3. Genomic sequencing was performed for an air sample with lowest CT. |
| Newey 2021 [29] | BSL-3 lab. Viral cultivation and plaque assays were performed using VERO 76, C1008 cells obtained from ATCC. VERO cells were maintained in T-75 flasks containing DMEM supplemented with 10% FBS; Corning, 35-010-CV) at 37 °C, and 5% CO2. For 24-well plate preparation, cells were seeded at 200,000 cells per well in DMEM with 10% FBS and used for assays 18–24 h later. Currency was sampled at the following four post-inoculation time-points: 30 min (used as time zero) then 4, 24, and 48 h. | 14 samples were interrogated for live virus. | 3 days | No plaques were seen for any of the samples. | In vitro, viable SARS-CoV-2 appeared to be most stable on plastic money cards, with banknotes providing the least stability of all four surfaces tested in this study. |
| Oksanen 2021 [30] | BSL-3 lab. Vero E6 cells (VE6) and their TMPRSS2-expressing clone VE6-TMPRSS2-10 (VE6T) were grown as previously described. To inhibit fungal growth, 0.205 μg/mL of amphotericin B (Fungizone, Thermo Scientific) was added to the medium of the cells that were taken to the hospital for aerosol collections. Samples were cultured at 37 °C for 10–14 days and checked for CPE. | Culturing was considered positive if CPE was detected and the CT value of PCR performed from the culture media was <20. | 10–14 days | Viable virus was not detected in any of the 212 cultured surface samples. | |
| Rajendiran 2021 [31] | BSL-3 lab. Vero E6 cells were grown overnight in HMEM supplemented with 10% FBS in 15 mL Corning culture tubes. Then 200 μL of each environmental specimen in VTM was added to the Vero E6 cells and incubated for an hour at room temperature. The infected cells were then maintained with 1 mL of HMEM supplemented with 2% FBS and monitored for CPE. Next, the infected culture tubes were frozen at –80 °C overnight and then thawed. The culture supernatants were filtered and tested for the growth of SARS-CoV-2 by determining the CT value using real-time RT–PCR. | Stage 1: 3 positive samples with CT 34.43, 37.01, and 37.52, respectively. Stage 2: 3 positive samples with CT 33.49, 35.59, and 37.01, respectively. |
Unclear | No CPE detected. | |
| Winslow 2021 [32] | Vero E6 cells expressing ACE2 and TMPRSS2 were used to culture virus from any positive/suspected-positive viral RNA sample. Vero cells were maintained in DMEM, supplemented with heat-inactivated FBS (10%) and penicillin/streptomycin (10,000 IU/mL and 10,000 μg/mL). For virus isolation, 200 μL of samples were added to 24-well plates. On day 0 and after 5–7 days the cell supernatants were collected, and RT–qPCR used to detect SARS-CoV-2 RNA as described above. Samples with ≥1 log10 increase in copy numbers for the E gene (reduced CT values relative to the original samples) after 5–7 days of propagation in cells compared with the starting value were considered positive by viral culture. | All positive SARS-CoV-2 samples (N = 6); CT 35–40. | 5–7 days | No CPE after 7 days. | |
| Zhou 2020 [33] | Vero E6 cells were used to culture virus from samples. The cells were maintained in DMEM supplemented with heat-inactivated FBS (10%) and penicillin/streptomycin (10,000 IU/mL and 10,000 μg/mL). For virus isolation, 200 μL of samples were added to 24-well plates. On day 0 and after 5–7 days, cell supernatants were collected, and RT–qPCR to detect SARS-CoV-2 performed. Samples with at least one log10 increase in copy numbers for the E gene (reduced CT values relative to the original samples) after 5–7 days propagation in cells compared with the starting value were considered positive by viral culture. | Positive SARS-CoV-2 samples. | 5–7 days | No CPE after 7 days. |
ACE2, angiotensin converting enzyme 2; CPE, cytopathic effect; CT, cycle threshold; DMEM, Dulbecco's modified Eagle's medium; FBS, foetal bovine serum; HMEM, Hanks' minimum essential medium; RT–PCR, reverse transcriptase–polymerase chain reaction; RT–qPCR, quantitative reverse transcription–polymerase chain reaction; UTM, universal transport medium; TMPRSS2, transmembrane serine protease 2; VTM, viral transport medium.
Seven studies were conducted in the USA, three in Canada, two each in Brazil, China and the UK, and one each in Finland, Israel, Italy, Malaysia, Peru, Singapore, and South Korea (Table I) (Table 4). Eleven studies were conducted exclusively in hospitals [12,14,15,[17], [18], [19],22,23,[31], [32], [33]], one of which was exclusively in an ICU [22]. Three studies were conducted in both hospital and quarantine or home/community settings [16,24,30]. One study was performed in a university setting (college campus and community) [11], two in public and/or open spaces [13,20], four were exclusively in residential settings [21,[27], [28], [29]] and two at dockyards [25,26].
Table IV.
Sources of fomites in included studies
| Source | High-frequency touch surfaces | Low-frequency touch surfaces |
|---|---|---|
| Patients' rooms/wards | Call bells, bedside tables, medical equipment, cell phones, bedrails, tissues, washcloths. Healthcare workers' (HCWs) stations. |
Hospital equipment: ventilator, endotracheal tube straps, floor, intravenous pump, arterial line plunger, X-ray machine, medication cart, etc.). |
| Outside patients' rooms/wards | Ward telephones, computer keyboards, clinical equipment (syringe pumps, urinary catheters), hand-cleaning facilities (hand-washing basins, alcohol gel dispensers). | Air vent, floors, tables. |
| In the community | Cold-chain containers at dockyards, and food surfaces (bulk rice, chicken, cheese, fruits). | – |
Sixteen studies collected fomite samples from the environment of patients with confirmed SARS-CoV-2 infection (see Table I ). In two studies [12,32] all patients required high-flow oxygen therapy or mechanical ventilation. Two studies [25,26] involved workers at dockyards, and three [13,20,29] did not involve any human subjects.
The fomite sources included objects/areas within patients' rooms/wards, objects/areas outside patients' rooms, and objects/areas in the community which were classified as high and low touch (see Table I and Table 4 ).
Quality assessment
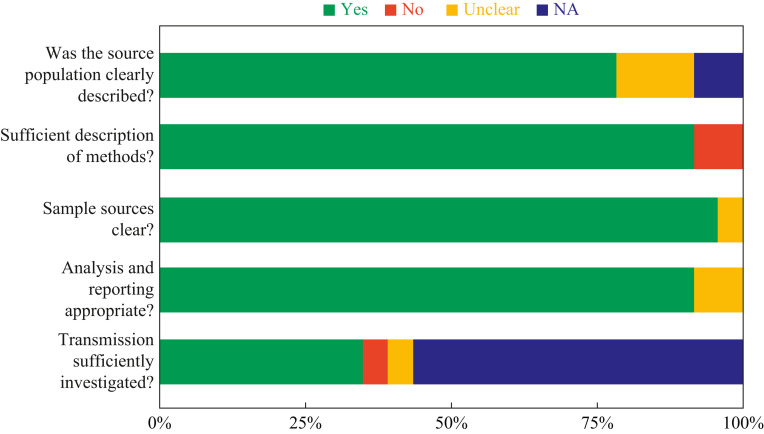
The reporting quality of included studies is shown in Figure 2 (see also Appendix Table 1). Eighteen studies (78%) adequately described the study population, and 21 (91%) described their study methods in sufficient detail. Twenty studies (87%) clearly specified the fomite sources, and 20 (87%) sufficiently reported the analysis of their results. Seven studies (30%) sufficiently investigated fomite transmission. None of the included studies published a protocol. The overall reporting quality was rated as moderate.
Figure 2.
Risk of bias in studies assessing fomite transmission of SARS-CoV-2.
Methods used to detect SARS-CoV-2 in fomites
Twenty-one studies (91%) used RT–PCR to detect the presence of SARS-CoV-2 RNA in fomites (Table I); the methods generally involved targeting the nucleocapsid (N) and/or envelope (E) genes of SARS-CoV-2. One study [24] used immunostaining and another [26] used nucleic acid, in addition to RT–PCR to confirm that the cultured virus was SARS-CoV-2. The remaining two studies used a nucleic acid detection kit [13] and loop-mediated isothermal amplification (LAMP) assays [29].
SARS-CoV-2 PCR cycle threshold values for positive tests from fomite samples
A total of 11 studies (48%) pre-specified the CT for positive tests (Table I); studies using commercially available RT–PCR assays used the manufacturer-specified CT threshold as indicated in the product insert for the assay. Four studies [20,21,23,25] used a cut-off of <40, and two studies [12,24] used <35. Four studies used the following cut-offs: <45 [32], <40.4 [33], <39 [13], and <38 [30]. One study [14] used two CT range values (30.0–30.8 and 33.8–35.1) as positive controls depending on the concentration of plaque-forming units (pfu) (1 pfu and 0.1 pfu, respectively); negative controls were reagent blanks without any sample. Another study [11] used the limit of detection (LOD) of 75 SARS-CoV-2 RNA copies/sample as cut-off point for positivity, and a third used LOD of 3.9 copies/reaction. Twelve studies (52%) did not report pre-specified CT values for positivity.
SARS-CoV-2 PCR results in fomite samples
The number of swab samples across the studies ranged from seven to 4370. All 23 studies reported positive SARS-CoV-2 PCR tests from the samples taken, ranging from 0.1% to 52.5% of the samples obtained across all the studies (Table II). The CT values were lowest in fomite samples from two studies [12,19] that included participants receiving either high-flow oxygen or mechanical ventilation: 22.3 to 29.7, and 23 to 26, respectively. A third study [32], including participants on oxygen, reported higher CT values (35–40); however, the investigators noted that participants in the high-flow nasal oxygen (HFNO) group were sampled significantly later in their illness compared to those receiving only supplemental oxygen: mean 16 days vs mean 9 days, from symptom onset, respectively. The authors of one study of SARS-CoV-2 patients in acute care [23] reported that hypoxia on admission (P = 0.003), CT ≤30 on or after the sampling date (P = 0.006), higher Charlson Comorbidity Index score (P = 0.002), and shorter time from illness onset (≤7 days) to sampling date (P = 0.02) were significantly associated independent risk factors for positive SARS-CoV-2 PCR tests from fomite samples.
In one study that included 31 SARS-CoV-2-infected participants [30], the frequency of positive tests on fomite samples was significantly higher when multiple patients were in the same ward or home. Findings from one study [13] reported that SARS-CoV-2 RNA was not detected on food surfaces. One study [29] showed that SARS-CoV-2 RNA is more likely to be detected on bank cards and ID cards compared to bank notes and coins.
SARS-CoV-2 viral culture methods in fomite samples
A summary of the key details for viral culture is shown in Table III. All the studies described the methods used to isolate SARS-CoV-2 except one [25]; in another study [26], the methods used to culture virus from fomite surfaces were not sufficiently described. All the studies that described their methods used Vero cells for testing viral growth. In three studies [11,24,32], culture media was supplemented with recombinant Vero cells expressing transmembrane serine protease 2 (TMPRSS2). In 12 studies (52.2%), the Vero cells were cultured in Dulbecco's modified Eagle's medium (DMEM), and one study [31] used Hanks' minimum essential medium (HMEM). The criteria used for including fomite samples for viral culture varied across the studies (see Table III). In 15 studies (65.2%), all positive SARS-CoV-2 samples were included for viral culture (Table III). In another three studies (13%) [16,19,24], all surface samples were included for viral culture. The duration of incubation ranged from three to 14 days across 19 studies (see Table III). In four studies (17.4%) [11,25,26,31], the duration of incubation was not specified.
SARS-CoV-2 viral culture results in fomite samples
Five studies (22%) [12,[23], [24], [25],27] reported recoverable infectious virus from culture (Table III). Four of these five studies [12,23,24,27] adequately reported the methods used to isolate infectious virus, whereas the reporting was unclear in one study (Appendix Table 2). The highest CT value across these studies from which culturable SARS-CoV-2 was detected was 31.78. A sixth study [22] that reported a positive viral culture in one sample had a CT value of 34; however, the sample tested negative for SARS-CoV-2 on RT–PCR on the third day.
The highest proportion of positive cultures (10/21, 48%) involved adult patients with SARS-CoV-2 both in hospital and in the community [24]. The authors reported that the likelihood of infectious SARS-CoV-2 was significantly higher with lower CT values (mean CT 19.6 ± 5.1 vs 29.2 ± 4.2, P < 0.0001). With the exception of patients with immunodeficiency or critically ill patients, all the infectious specimens were detected within seven days after symptom onset. In Ahn et al., of three patients (all critically ill and/or cancer patients) with SARS-CoV-2 and on oxygen supplementation, nearly half (47%) of the 15 positive SARS-CoV-2 fomite samples were positive on viral culture [12]. In Kowta et al., the likelihood of positive viral culture was significantly more likely if the fomite sample was collected within seven days of symptom onset (P = 0.02); this likelihood was an independent risk factor on logistic regression [23]. In Lin et al., environmental samples were assessed by time post symptom onset and the actual quantity of virus was determined by quantitative culture [24]. They found the ability to detect culturable virus occurred most readily within the first seven days after symptom onset and then declined rapidly with the exception of immunocompromised patients who had positive cultures in both clinical and fomite samples for prolonged periods. Facial tissues (80% of all samples, 4.0 × 101 to 2.0 × 103 pfu/mL), nasal prongs (75% of all samples, 5 × 100 to 1.1 × 102 pfu/mL), and dentures found lying on the bedside table for ≥4 h, (4.5 × 101 pfu/mL) were some of the types of fomites found to be positive and had very large burdens of SARS-CoV-2. Some phone and call bell samples were also found to contain infectious virus ranging from 2.0 × 102 to 1.9 × 103 pfu/mL. The positive culture from a nightstand swab in the only other study providing symptom onset and/or test date positivity was at two days and from a household case in a healthy person [27]. The median CT for positive samples was lower compared with negative samples: 26.1 (IQR: 6.2) vs 31.6 (IQR: 4.2) (see Appendix Figure 1).
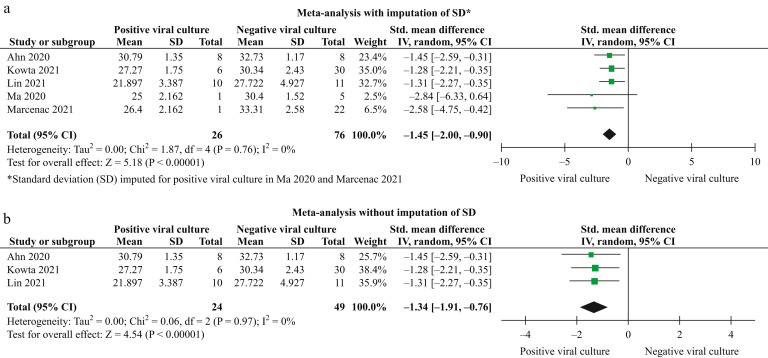
A meta-analysis of five studies after imputing SDs for the two studies with only one positive culture [12,[23], [24], [25],27] showed that a lower CT was significantly associated with positive viral cultures (SMD: –1.45; 95% CI: –2.00 to –0.90; I 2 = 0%; P < 0.00001; N = 102) (Figure 3 a). A meta-analysis of only three studies without imputation of SDs [12,23,24] showed similar results (SMD: –1.34; 95% CI: –1.91 to –0.76; I 2 = 0%; P < 0.00001; N = 73) (Figure 3b).
Figure 3.
Relationship between cycle threshold and viral culture results in fomites.
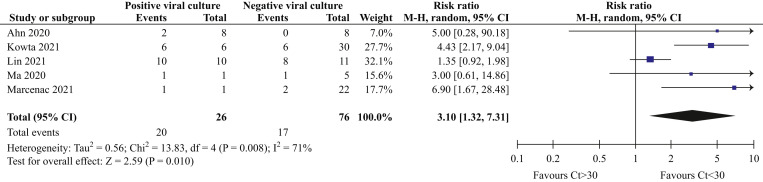
A post-hoc analysis showed that the relative risk (RR) of a positive viral culture result was three times more likely if the CT was <30 (RR: 3.10; 95% CI: 1.32 to 7.31; I 2 = 71%; P = 0.01; N = 102 samples) (Figure 4 ). Sensitivity analyses after removing one study that did not collect fomite samples serially [24] showed similar results, but with significant reduction in heterogeneity (RR: 4.55; 95% CI: 2.55 to 8.14; I 2 = 0%; P < 0.00001; N = 81 samples).
Figure 4.
Forest plot showing the relationship between CT and viral culture results.
One of the included studies [24] reporting positive viral cultures from fomites assessed the duration of infectiousness by demonstrating that a clinical sample with infectious virus retained its infectiousness on fomites such as a call bell, computer keyboard, stethoscope diaphragm, and a N95 respirator for up to 4 h with minimal reduction in quantitative burden and in dried saliva for 2 h with no reduction. One study [25] demonstrated evidence of possible SARS-CoV-2 transmission from fomites (infected cold-chain containers) to humans (dock workers); however, the study authors did not describe the methods used for viral culture.
SARS-CoV-2 gene sequencing
Five studies [18,[23], [24], [25], [26]] performed gene sequencing (Appendix Table 3); however, only three of these [[23], [24], [25]] reported positive viral cultures (see Table III). All three studies with positive viral cultures from fomite samples that used gene sequencing showed associations with each other and/or with genetic material from human respiratory samples. In Kowta et al., 23 out of 152 samples passed quality control and were submitted to Global Initiative on Sharing Avian Influenza Data (GISAID) (https://www.gisaid.org/) [23]. All 23 samples clustered with the nasopharyngeal swab from SARS-CoV-2 patients who were admitted to the same room (Appendix Table 3). Lin et al. used a SARS-CoV-2 strain from GISAID as positive control and plaque reference [24]. Genomic sequences were submitted to the Pangolin lineage assigner (https://pangolin.cog-uk.io/). Subsets of virus isolates were also confirmed through de-staining and immunostaining. In the plaque assays, characteristic halos were observed in samples from facial tissues (with one sampled about 9 h after use), a face cloth lying on a patient's bed, a patient gown, a bedrail, dentures, nasal prongs, a cell phone, and a call bell used by patients infected with SARS-CoV-2. In the Ma et al. study of samples from dockyards, two case (dock workers) samples and 11 frozen cod samples with positive nucleic acid results shared 12 nucleotide mutation sites with the Wuhan reference strain [25]. Five outer packaging-related strains shared one unique nucleotide mutation with the strain from one of the case samples, demonstrating that the strains were identical. The sequences for the 11 samples were also deposited on the GISAID database.
Discussion
Summary of main findings
Our results show that infectious SARS-CoV-2 can be readily detected on fomites in specific settings. Genome sequencing following positive culture results in three studies also provided supportive evidence for the SARS-CoV-2 strains being identical on these fomites.
The likelihood of positive SARS-CoV-2 PCR tests on fomites is significantly greater if the CT from nasopharyngeal swabs in SARS-CoV-2-infected individuals is <30, and if multiple SARS-CoV-2-infected patients share the same space. The number of samples with positive PCR test results was extensive (see Table II), and CT values were lowest in fomite samples from participants receiving either high-flow oxygen or mechanical ventilation based on the studies analysed.
The evidence from the included studies suggests that positive SARS-CoV-2 culture is at least three times more likely when the positive RT–PCR CT value is <30. The significant heterogeneity observed in the overall results is largely due to one study [24] that used different methods (and varied intervals) to collect the fomite samples; the removal of this study resulted in homogeneity in the results. Four of the infectious viral culture samples were in a hospital setting with virus samples found on various high-touch surfaces and one was in a fish cluster pallet at the port dock (see Box 1 ).
Box 1. Sites of replication-competent samples.
-
–
Ahn 2020 [12]: nasal prong, bedside table, floor near the patient, remote control, bedrails, bedsheets, and non-invasive ventilation mask.
-
–
Kowta 2021 [23]: bathroom door, bed and switch, phone, table and chair, toilet and sink (1/4).
-
–
Lin 2021 [24]: used tissues, face cloth, gown deposit, nasal prongs, dentures, phone/call bells, bedrail, dentures on bedside tables.
-
–
Ma 2021 [25]: fish cluster pallet.
-
–
Marcenac 2021 [27]: nightstand swab.
Alt-text: Box 1
The findings in the four studies providing data on timing of collection and symptom onset [12,23,24,27] demonstrate that collection of fomite specimens in the environment of infected persons within the first few days post symptom onset in normal hosts is most likely, which is not only biologically probable but also consistent with the recent human challenge experiments with SARS-CoV-2 demonstrating the kinetics of viral culturability by time, post symptom onset [34].
That higher Charlson Comorbidity Index scores were independently associated with increased risk of transmission in one study [23] may be related to more severely ill persons having higher levels of infectious virus due to their underlying illness, which then more readily transmit into the environment.
Evidence for a chain of transmission of SARS-CoV-2 from fomites to humans was not reported in the studies reviewed. Nonetheless, a chain of transmission was demonstrated by SARS-CoV-2 from respiratory tract specimens which can deposit on ‘fomites’; these can survive for many hours and may readily cause invasive infection in the Syrian hamster model [24] at very low infecting doses [37], fulfilling both the Gwaltney–Hendley postulates of viral causation and Koch's postulates [38]. These very low infectious doses are consistent with the recently published human challenge experiments demonstrating infection in 53% using an inoculum of only 10 median tissue culture infectious dose (TCID50: ∼7 pfu) in 0.1 mL of a wild-type virus (SARS-CoV-2/human/GBR/484861/2020) [39]. The overall quality of the evidence was moderate, but the results are strengthened by study replication, the homogeneity of the pooled results, and the genomic sequencing demonstrating identical strains on the fomite and from the infected source person.
Although three studies [[23], [24], [25]] showed evidence of transmission using genome sequencing, the results should be interpreted with caution because the default settings of the databases currently used for phylogenetic analysis are not robust enough to conclusively demonstrate transmission of SARS-CoV-2 [35,36].
Comparison with existing reviews
The present review has identified several reviews in the published literature that investigate the role of fomite transmission in SARS-CoV-2. In a previous review on fomite transmission [5], 11 studies that attempted culture were found, but none was able to demonstrate replication-competent virus. Similar results were found in another systematic review that investigated fomite transmission [40] – no virus was isolated after culture of 242 fomite samples across six included studies. The findings from the present review differ because we identified 23 studies and included five studies that demonstrated replication-competence on culture and looked at more appropriate timing of collection.
Another review of 39 studies [41] concluded that the duration of exposure of fomites to infected patients was largely responsible for the variation in the prevalence of fomite contamination. However, our results indicate that inconsistency in CT cut-offs and timing of sample collection from symptom onset also markedly influence the probability of obtaining positive test results. A fourth systematic review of 51 cross-sectional studies examining fomite transmission in hospitals [42] showed that culturable SARS-CoV-2 was demonstrated in two studies. Our review includes three additional studies that were not available in that review, plus pooled results along with reporting the results of genome sequencing. The latter finding is important as it demonstrates the identical strain found on the fomite and the infected source person.
Replication-competent virus was recovered from several pieces of commonly used medical equipment and had little loss of infectiousness after 4 h on several items in one study [24]. Several laboratory studies have demonstrated survival of SARS-CoV-2 on multiple commonly encountered fomites for several hours [43,44], strengthening the plausibility of a chain of transmission from fomites.
Strengths and limitations
To our knowledge, this is the most comprehensive review to date that investigates the potential for fomite transmission of SARS-CoV-2. Extensive literature searches were conducted to identify relevant studies (including those awaiting peer review) and the reporting quality of included studies was accounted for. Our review is the first meta-analysis of CT results for viral culture studies on fomites. All results (except for one) showed no heterogeneity, suggesting that the direction of effects is likely to be the same irrespective of the methods used for viral culture. In addition, we investigated whether the results of genome sequencing were consistent with viral culture results. However, there are some limitations. The present review may not have identified all available studies investigating viral culture in fomites. In addition, the failure in many studies to sufficiently demonstrate replication-competent SARS-CoV-2, coupled with variation in CT cut-off values and differences in sampling methods, limits the robustness of our conclusions. However, studies that have patients with high CT values that lack infectiousness or that sample at a late stage in the infection will not detect replication-competent virus in environmental samples; this is a major shortcoming in many studies and represents a methodologic flaw that would underestimate the true nature of positive fomite samples, leading to erroneous conclusions. Furthermore, the small number of studies included in our meta-analyses limits the applicability of the study findings.
Implications for research and policy
Our findings suggest that SARS-CoV-2 transmission through fomites exists and may have been under-appreciated due to methodologic shortcomings in many early studies during the pandemic. Indeed, results of laboratory studies have demonstrated the infectivity of SARS-CoV-2 on fomites and especially with the omicron variant [45,46]. However, there is still a paucity in the number of well-conducted studies demonstrating SARS-CoV-2 transmission through fomites.
One study [24] found replication-competent virus on patients' hands, a kiss sample, and dried saliva (see Table III). These samples were not included in our review because it is unclear whether they could be defined as fomites. Some authors have suggested that all surfaces and objects should be considered as fomites [6]. The definitions of what might be considered a fomite requires further clarification.
Our findings suggest that hand hygiene and surface decontamination procedures should be encouraged to limit the risk of fomite transmission and should remain a cornerstone of public health prevention policies. Although some authors suggested that fomite transmission of SARS-CoV-2 is exaggerated, and surface decontamination represents excesses that are counterproductive [47,48], the relevance of hand hygiene and surface decontamination procedures should not be underestimated. The likelihood of detecting replication-competent SARS-CoV-2 on fomite samples is greater in high-risk patient settings. Consistent with our review findings, Liu and colleagues have shown that fomite contamination of surfaces is significantly associated with CT values and the interval to collection of samples [49]. Future studies should consider using lower CT values as cut-off to detect replication-competent SARS-CoV-2. In addition, investigators in future studies should ensure that fomite samples are collected within the first few days of symptom onset in patients to maximize yield. We have recently suggested a framework for studies investigating the transmission of viral respiratory pathogens [50].
Although one study [25] provided evidence of transmission of SARS-CoV-2 from cold-chain containers to humans, the methodology used for viral isolation and cross-identification was unclear. Whereas some groups of authors have suggested that SARS-CoV-2 can be transmitted through fomites in relation to cold-chain food products [[51], [52], [53]], others have argued that the risk of transmission through this source is low [54,55]. Further well-designed studies are required to improve our understanding in this area.
In conclusion, the evidence from published studies suggests that replication-competent SARS-CoV-2 may be found in fomites and that factors of major importance – including the timing of sampling from symptom onset, severity of the patients' illness, and the CT <30 (high viral titre) – are associated with replication-competent virus in fomite samples. Replication-competent SARS-CoV-2 can be readily detected on fomites in specific settings. The results suggest a plausible chain of transmission that is strengthened by study replication, the homogeneity of the pooled results and genomic sequencing demonstrating identical strain on the fomite and the source person. However, more rigorous studies are required to demonstrate the extent and settings in which fomites are responsible for transmission with subsequent invasive infection from SARS-CoV-2. Future studies should also investigate how long replication-competent virus remains infectious on various fomites in real-world scenarios and should obtain specimens that are appropriately timed to avoid critical sampling bias.
Author contributions
I.J.O.: conceptualization, methodology, screening of abstracts, data extraction, quality assessment, data analysis, data interpretation, co-drafting; C.J.H.: conceptualization, methodology, data analysis, data interpretation, co-drafting; E.A.S.: conceptualization, methodology, screening of abstracts, data extraction, quality assessment, data analysis, data interpretation, co-drafting; J.B.: conceptualization, electronic searches, methodology, data interpretation, co-drafting; E.C.R.: conceptualization, methodology, data interpretation, co-drafting; S.M.: conceptualization, methodology, data interpretation, co-drafting; A.P.: conceptualization, methodology, data extraction, data interpretation, co-drafting; D.H.E: conceptualization, methodology, data interpretation, co-drafting; J.M.C.: conceptualization, methodology, data extraction, data interpretation, co-drafting; T.J.: conceptualization, methodology, data extraction, data interpretation, co-drafting.
Conflict of interest statement
T.J. received a Cochrane Methods Innovations Fund grant to develop guidance on using regulatory data in Cochrane reviews (2015–18). From 2014 to 2016, he was a member of three advisory boards for Boehringer Ingelheim. T.J. was a member of an independent data monitoring committee for a Sanofi Pasteur clinical trial on an influenza vaccine. Market research companies occasionally interview T.J. about phase I or II pharmaceutical products for which he receives fees (current). T.J. was a member of three advisory boards for Boehringer Ingelheim (2014–16). T.J. was a member of an independent data monitoring committee for a Sanofi Pasteur clinical trial on an influenza vaccine (2015–17). T.J. is a relator in a False Claims Act lawsuit on behalf of the United States that involves sales of Tamiflu for pandemic stockpiling. If resolved in the United States favour, he would be entitled to a percentage of the recovery. T.J. is coholder of a Laura and John Arnold Foundation grant for the development of a RIAT support centre (2017–20) and Jean Monnet Network Grant, 2017–20 for The Jean Monnet Health Law and Policy Network. T.J. is an unpaid collaborator to the Beyond Transparency in Pharmaceutical Research and Regulation led by Dalhousie University and funded by the Canadian Institutes of Health Research (2018–22). T.J. consulted for Illumina LLC on next-generation gene sequencing (2019–20). T.J. was the consultant scientific coordinator for the HTA Medical Technology programme of the Agenzia per I Servizi Sanitari Nazionali (AGENAS) of the Italian MoH (2007–19). T.J. is Director Medical Affairs for BC Solutions, a market access company for medical devices in Europe. T.J. was funded by NIHR UK and the World Health Organization (WHO) to update Cochrane review A122, Physical Interventions to interrupt the spread of respiratory viruses. Oxford University funds T.J. to carry out a living review on the transmission epidemiology of COVID-19. Since 2020, T.J. receives fees for articles published by The Spectator and other media outlets. T.J. is part of a review group carrying out a Living rapid literature review on the modes of transmission of SARS-CoV-2 (WHO Registration 2020/1077093 0). He is a member of the WHO COVID-19 Infection Prevention and Control Research Working Group, for which he receives no funds. T.J. is funded to co-author rapid reviews on the impact of COVID restrictions by the Collateral Global Organisation.
C.J.H. holds grant funding from the NIHR, the NIHR School of Primary Care Research, the NIHR BRC Oxford and the World Health Organization for a series of Living rapid reviews on the modes of transmission of SARS-CoV-2, reference WHO registration No2020/1077093, and to carry out a scoping review of systematic reviews of interventions to improve vaccination uptake, reference WHO Registration 2021/1138353–0. He has received financial remuneration from an asbestos case and given legal advice on mesh and hormone pregnancy tests cases. He has received expenses and fees for his media work, including occasional payments from BBC Radio 4 Inside Health and The Spectator. He receives expenses for teaching evidence-based medicine and is also paid for his GP work in NHS out of hours (contract Oxford Health NHS Foundation Trust). He has also received income from the publication of a series of toolkit books and appraising treatment recommendations in non-NHS settings. He is the Director of CEBM, an NIHR Senior Investigator and an advisor to Collateral Global.
D.H.E. holds grant funding from the Canadian Institutes for Health Research and Li Ka Shing Institute of Virology relating to the development of COVID-19 vaccines and the Canadian Natural Science and Engineering Research Council concerning COVID-19 aerosol transmission. He is a recipient of WHO and Province of Alberta funding which supports the provision of BSL3 based SARS-CoV-2 culture services to regional investigators. He also holds public and private sector contract funding relating to the development of poxvirus-based COVID-19 vaccines, SARS-CoV-2 inactivation technologies, and serum neutralization testing.
J.M.C. holds grants from the Canadian Institutes for Health Research on acute and primary care preparedness for COVID-19 in Alberta, Canada, and was the primary local Investigator for a Staphylococcus aureus vaccine study funded by Pfizer, for which all funding was provided only to the University of Calgary. He is a co-investigator on a WHO-funded study using integrated human factors and ethnography approaches to identify and scale innovative IPC guidance implementation supports in primary care with a focus on low-resource settings and using drone aerial systems to deliver medical supplies and PPE to remote First Nations communities during the COVID-19 pandemic. He also received support from the Centers for Disease Control and Prevention (CDC) to attend an Infection Control Think Tank Meeting. He is a member and Chair of the WHO Infection Prevention and Control Research and Development Expert Group for COVID-19 and a member of the WHO Health Emergencies Programme (WHE) Ad hoc COVID-19 IPC Guidance Development Group, both of which provide multidisciplinary advice to the WHO, for which no funding is received and from which no funding recommendations are made for any WHO contracts or grants. He is also a member of the Cochrane Acute Respiratory Infections Group.
J.B. is a major shareholder in the Trip Database search engine (www.tripdatabase.com) as well as being an employee. In relation to this work, Trip has worked with many organizations over the years; none has any links with this work. The main current projects are with AXA and Collateral Global.
E.C.R. was a member of the European Federation of Neurological Societies (EFNS)/European Academy of Neurology (EAN) Scientist Panel, Subcommittee of Infectious Diseases (2013–2017). Since 2021, she has been a member of the International Parkinson and Movement Disorder Society (MDS) Multiple System Atrophy Study Group, the Mild Cognitive Impairment in Parkinson Disease Study Group, and the Infection Related Movement Disorders Study Group. She was an External Expert and sometimes Rapporteur for COST proposals (2013, 2016, 2017, 2018, 2019) for Neurology projects. She is a Scientific Officer for the Romanian National Council for Scientific Research.
S.M. is a pharmacist working for the Italian National Health System since 2002 and a member of one of the three Institutional Review Boards of Emilia-Romagna Region (Comitato Etico Area Vasta Emilia Centro) since 2018.
A.P. holds grant funding from the NIHR School for Primary Care Research.
I.J.O. and E.A.S. have no interests to disclose.
Funding sources
This work is part-funded by the NIHR School for Primary Care Research [project 569]. The work also received funding from the University of Calgary. WHO funded the first iteration of this review: Living rapid review on the modes of transmission of SARS-CoV-2, reference WHO registration No. 2020/1077093. C.J.H., A.P. and E.A.S. also receive funding support from the National Institute of Health Research School of Primary Care Research Evidence Synthesis Working Group project 390. (https://www.spcr.nihr.ac.uk/eswg). The funders had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhin.2022.09.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available at: https://covid19.who.int/[last accessed May 2022].
- 2.Moore J.P., Offit P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA. 2021;325:821–822. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 3.Pouwels K.B., Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Karina-Doris Vihta K.-D., et al. Impact of Delta on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. medRxiv. 2021 doi: 10.1101/2021.08.18.21262237. 08.18.21262237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shekhar R., Garg I., Pal S., Kottewar S., Sheikh A.B. COVID-19 vaccine booster: to boost or not to boost. Infect Dis Rep. 2021;13:924–929. doi: 10.3390/idr13040084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onakpoya I.J., Heneghan C.J., Spencer E.A., Brassey J., Plüddemann A., Evans D.H., et al. SARS-CoV-2 and the role of fomite transmission: a systematic review. F1000Res. 2021;10:233. doi: 10.12688/f1000research.51590.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castaño N., Cordts S.C., Kurosu Jalil M., Zhang K.S., Koppaka S., Bick A.D., et al. Fomite transmission, physicochemical origin of virus-surface interactions, and disinfection strategies for enveloped viruses with applications to SARS-CoV-2. ACS Omega. 2021;6:6509–6527. doi: 10.1021/acsomega.0c06335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jefferson T., Spencer E.A., Brassey J., Onakpoya I.J., Rosca E.C., Plüddemann A., et al. Transmission of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from pre and asymptomatic infected individuals: a systematic review. Clin Microbiol Infect. 2022;28:178–189. doi: 10.1016/j.cmi.2021.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 9.Higgins JPT, Li T, Deeks JJ. Chapter 6: Choosing effect measures and computing estimates of effect. In: Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al, eds. Cochrane handbook for systematic reviews of interventions, version 6.3 (updated February 2022). Cochrane; 2022. Available from: www.training.cochrane.org/handbook.
- 10.Review Manager (RevMan) Cochrane Collaboration; 2020. [Google Scholar]
- 11.Adenaiye O.O., Lai J., de Mesquita PJB, Hong F., Youssefi S., German J., et al. Infectious SARS-CoV-2 in exhaled aerosols and efficacy of masks during early mild infection. Clin Infect Dis. 2021:ciab797. doi: 10.1093/cid/ciab797. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn J.Y., An S., Sohn Y., Cho Y., Hyun J.H., Baek Y.J., et al. Environmental contamination in the isolation rooms of COVID-19 patients with severe pneumonia requiring mechanical ventilation or high-flow oxygen therapy. J Hosp Infect. 2020;106:570–576. doi: 10.1016/j.jhin.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvis-Chirinos K., Angulo-Bazán Y., Escalante-Maldonado O., Fuentes D., Rodriguez M.G.P., Gonzales-Achuy E., et al. Presence of SARS-CoV-2 in food surfaces and public space surfaces in three districts of Lima, Peru. medRxiv. 2021 doi: 10.1101/2021.11.15.21265879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ang A.X., Luhung I., Ahidjo B.A., Drautz-Moses D.I., Tambyah P.A., Mok C.K., et al. Airborne SARS-CoV-2 surveillance in hospital environment using high-flowrate air samplers and its comparison to surface sampling. Indoor Air. 2022;32 doi: 10.1111/ina.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett C., Langsjoen J., Cheng Q., Yingling A.V., Weiss M., Bradfute S., et al. Presence of SARS-CoV-2 fomites in a university hospital setting. Exp Biol Med (Maywood) 2021;246:2039–2045. doi: 10.1177/15353702211024597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Shmuel A., Brosh-Nissimov T., Glinert I., Bar-David E., Sittner A., Poni R., et al. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clin Microbiol Infect. 2020;26:1658–1662. doi: 10.1016/j.cmi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binder R.A., Alarja N.A., Robie E.R., Kochek K.E., Xiu L., Rocha-Melogno L., et al. Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J Infect Dis. 2020;222:1798–1806. doi: 10.1093/infdis/jiaa575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coil D.A., Albertson T., Banerjee S., Brennan G., Campbell A.J., Cohen S.H., et al. SARS-CoV-2 detection and genomic sequencing from hospital surface samples collected at UC Davis. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colaneri M., Seminari E., Novati S., Asperges E., Biscarini S., Piralla A., et al. Severe acute respiratory syndrome coronavirus 2 RNA contamination of inanimate surfaces and virus viability in a health care emergency unit. Clin Microbiol Infect. 2020;26 doi: 10.1016/j.cmi.2020.05.009. 1094.e1–1094.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silva S.J.R., do Nascimento J.C.F., Dos Santos Reis W.P.M., da Silva C.T.A., da Silva P.G., Mendes R.P.G., et al. Widespread contamination of SARS-CoV-2 on highly touched surfaces in Brazil during the second wave of the COVID-19 pandemic. Environ Microbiol. 2021;23:7382–7395. doi: 10.1111/1462-2920.15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumont-Leblond N., Veillette M., Bhérer L., Boissoneault K., Mubareka S., Yip L., et al. Positive no-touch surfaces and undetectable SARS-CoV-2 aerosols in long-term care facilities: an attempt to understand the contributing factors and the importance of timing in air sampling campaigns. Am J Infect Control. 2021;49:701–706. doi: 10.1016/j.ajic.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espinoza E.P.S., Cortes M.F., Noguera S.V., Paula A.V., Guimarães T., Boas L.S.V., et al. Are mobile phones part of the chain of transmission of SARS-CoV-2 in hospital settings? Rev Inst Med Trop Sao Paulo. 2021;63:e74. doi: 10.1590/S1678-9946202163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotwa J.D., Jamal A.J., Mbareche H., Yip L., Aftanas P., Barati S., et al. Surface and air contamination with SARS-CoV-2 from hospitalized COVID-19 patients in Toronto, Canada, March-May 2020. J Infect Dis. 2021:jiab578. doi: 10.1093/infdis/jiab578. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y., Malott R.J., Ward L., Kiplagat L., Pabbaraju K., Gill K., et al. Detection and quantification of infectious severe acute respiratory coronavirus-2 in diverse clinical and environmental samples from infected patients: evidence to support respiratory droplet, and direct and indirect contact as significant modes of transmission. medRxiv. 2021 doi: 10.1101/2021.07.08.21259744. 07.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma H., Wang Z., Zhao X., Han J., Zhang Y., Wang H., et al. Long distance transmission of SARS-CoV-2 from contaminated cold chain products to humans – Qingdao City, Shandong Province, China. China CDC Wkly. 2021;3:637–644. doi: 10.46234/ccdcw2021.164. September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H., Zhang J., Wang J., Qin Y., Chen C., Song Y., et al. COVID-19 outbreak caused by contaminated packaging of imported cold-chain products – Liaoning Province, China. China CDC Wkly. July 2020;3:441–447. doi: 10.46234/ccdcw2021.114. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcenac P., Park G.W., Duca L.M., Lewis N.M., Dietrich E.A., Barclay L., et al. Detection of SARS-CoV-2 on surfaces in households of persons with COVID-19. Int J Environ Res Public Health. 2021;18:8184. doi: 10.3390/ijerph18158184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nannu Shankar S., Witanachchi C.T., Morea A.F., Lednicky J.A., Loeb J.C., Alam M.M., et al. SARS-CoV-2 in residential rooms of two self-isolating persons with COVID-19. J Aerosol Sci. 2022;159 doi: 10.1016/j.jaerosci.2021.105870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newey C.R., Olausson A.T., Applegate A., Reid A.A., Robison R.A., Grose J.H. Presence and stability of SARS-CoV-2 on environmental currency and money cards in Utah reveals a lack of live virus. PLoS One. 2022;17 doi: 10.1371/journal.pone.0263025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oksanen LAH, Virtanen J, Sanmark E, Venkat V, Sofieva S, Aaltonen K, et al. SARS-CoV-2 air and surface contamination on a COVID-19 ward and at home, December 6th, 2021, Preprint (Version 2). Available at: Research Square: 10.21203/rs.3.rs-1002547/v2. [DOI]
- 31.Rajendiran S., Thahir S.S.A., Veloo Y., Suppiah J., Pahrol M.A., Shakor A.S.A., et al. Environmental surface sampling of SARS-CoV-2 in selected hospitals in Malaysia. Trop Biomed. 2021;38:462–468. doi: 10.47665/tb.38.3.089. [DOI] [PubMed] [Google Scholar]
- 32.Winslow R.L., Zhou J., Windle E.F., Nur I., Lall R., Ji C., et al. SARS-CoV-2 environmental contamination from hospitalised patients with COVID-19 receiving aerosol-generating procedures. Thorax. 2021 Nov 4 doi: 10.1136/thoraxjnl-2021-218035. thoraxjnl-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J., Otter J.A., Price J.R., Cimpeanu C., Meno Garcia D., Kinross J., et al. Investigating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surface and air contamination in an acute healthcare setting during the peak of the coronavirus disease 2019 (COVID-19) pandemic in London. Clin Infect Dis. 2021;73 doi: 10.1093/cid/ciaa905. e1870–e1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killingley B., Mann A.J., Kalinova M., Boyers A., Goonawardane N., Zhou J., et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 35.Morel B., Barbera P., Czech L., Bettisworth B., Hübner L., Lutteropp S., et al. Phylogenetic analysis of SARS-CoV-2 data is difficult. Mol Biol Evol. 2021;38:1777–1791. doi: 10.1093/molbev/msaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pipes L., Wang H., Huelsenbeck J.P., Nielsen R. Assessing uncertainty in the rooting of the SARS-CoV-2 phylogeny. Mol Biol Evol. 2021;38:1537–1543. doi: 10.1093/molbev/msaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferson T., Heneghan C., Spencer E., Brassey J., Plüddemann A., Onakpoya I., et al. A hierarchical framework for assessing transmission causality of respiratory viruses. Preprints. 2021 doi: 10.20944/preprints202104.0633.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Killingley B., Mann A.J., Kalinova M., Boyers A., Goonawardane N., Zhou J., et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022 Mar 31 doi: 10.1038/s41591-022-01780-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Gonçalves J., da Silva P.G., Reis L., Nascimento M.S.J., Koritnik T., Paragi M., et al. Surface contamination with SARS-CoV-2: a systematic review. Sci Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belluco S., Mancin M., Marzoli F., Bortolami A., Mazzetto E., Pezzuto A., et al. Prevalence of SARS-CoV-2 RNA on inanimate surfaces: a systematic review and meta-analysis. Eur J Epidemiol. 2021;36:685–707. doi: 10.1007/s10654-021-00784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribaric N.L., Vincent C., Jonitz G., Hellinger A., Ribaric G. Hidden hazards of SARS-CoV-2 transmission in hospitals: a systematic review. Indoor Air. 2022;32 doi: 10.1111/ina.12968. [DOI] [PubMed] [Google Scholar]
- 43.Riddell S., Goldie S., Hill A., Eagles D., Drew T.W. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol J. 2020;17:145. doi: 10.1186/s12985-020-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.L., Chan M.C.W., et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virtanen J., Aaltonen K., Kivistö I., Sironen T. Survival of SARS-CoV-2 on clothing materials. Adv Virol. 2021;2021 doi: 10.1155/2021/6623409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastorino B., Touret F., Gilles M., de Lamballerie X., Charrel R.N. Prolonged infectivity of SARS-CoV-2 in fomites. Emerg Infect Dis. 2020;26:2256–2257. doi: 10.3201/eid2609.201788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldman E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet Infect Dis. 2020;20:892–893. doi: 10.1016/S1473-3099(20)30561-2. Epub 2020 Jul 3. [Erratum in: Lancet Infect Dis 2020;20:e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis D. COVID-19 rarely spreads through surfaces. So why are we still deep cleaning? Nature. 2021;590:26–28. doi: 10.1038/d41586-021-00251-4. [DOI] [PubMed] [Google Scholar]
- 49.Liu J., Liu J., He Z., Yang Z., Yuan J., Wu H., et al. Duration of SARS-CoV-2 positive in quarantine room environments: a perspective analysis. Int J Infect Dis. 2021;105:68–74. doi: 10.1016/j.ijid.2021.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jefferson T., Heneghan C.J., Spencer E., Brassey J., Plüddemann A., Onakpoya I., et al. A hierarchical framework for assessing transmission causality of respiratory viruses. Viruses. 2022;14:1605. doi: 10.3390/v14081605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi Y., Zheng S., Liu C., Wang Q. Transmission of SARS-CoV-2 on cold-chain food overpacks: a new challenge. J Glob Health. 2021;11 doi: 10.7189/jogh.11.03071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen G.S., Hu S., Zheng S.L., Liu C.D., Wang M. How to deal with the transmission of SARS-COV-2 on the surface of cold-chain foods to people: a review. Eur Rev Med Pharmacol Sci. 2021;25:6378–6385. doi: 10.26355/eurrev_202110_27011. [DOI] [PubMed] [Google Scholar]
- 53.Ji W., Li X., Chen S., Ren L. Transmission of SARS-CoV-2 via fomite, especially cold chain, should not be ignored. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2026093118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobolik J.S., Sajewski E.T., Jaykus L.A., Cooper D.K., Lopman B.A., Kraay A.N., et al. Low risk of SARS-CoV-2 transmission via fomite, even in cold-chain. medRxiv. 2021 Aug 26 doi: 10.1101/2021.08.23.21262477. 2021.08.23. [DOI] [Google Scholar]
- 55.Sobolik J.S., Sajewski E.T., Jaykus L.A., Cooper D.K., Lopman B.A., Kraay A.N.M., et al. Decontamination of SARS-CoV-2 from coldchain food packaging provides no marginal benefit in risk reduction to food workers. Food Control. 2022;136 doi: 10.1016/j.foodcont.2022.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.