Abstract
Nontypeable Haemophilus influenzae is a gram-negative commensal organism that is commonly associated with localized respiratory tract disease. The pathogenesis of disease begins with colonization of the nasopharynx, a process that likely depends on bacterial adherence to respiratory epithelial cells. Hia is the major adhesin expressed by a subset of nontypeable H. influenzae strains and promotes efficient adherence to a variety of human epithelial cell lines. Based on previous work, Hia is transported to the surface of Escherichia coli transformants and is capable of mediating E. coli adherence without the assistance of other H. influenzae proteins. In the present study, we examined the mechanism of Hia secretion. PhoA fusions, deletional mutagenesis, and N-terminal amino acid sequencing established that the signal for Hia export from the cytoplasm resides in the first 49 amino acids, including a 24-amino-acid stretch with striking similarity to the N terminus of a number of proteins belonging to the autotransporter family. Immunoelectron microscopy demonstrated that the Hia internal region defined by amino acids 221 to 779 is exposed on the bacterial surface. Secondary-structure analysis predicted that the C terminus of Hia forms a β-barrel with a central hydrophilic channel, and site-specific mutagenesis and fusion protein analysis demonstrated that the C terminus targets Hia to the outer membrane and functions as an outer membrane translocator, analogous to observations with autotransporter proteins. In contrast to typical autotransporter proteins, Hia undergoes no cleavage between the internal and C-terminal domains and remains fully cell associated. Together, these results suggest that Hia is the prototype of an important subfamily of autotransporter proteins.
The interaction between pathogenic bacteria and host cells is largely determined by microbial proteins presented on the bacterial surface or released extracellularly. In gram-negative bacteria, the process of protein secretion requires translocation across the inner membrane, the periplasm, and the outer membrane. Several pathways for gram-negative bacterial protein secretion are known to exist, including some that require the Sec proteins located in the cytoplasm and inner membrane and others that are independent of the Sec machinery (23, 44).
The autotransporter proteins are gram-negative bacterial extracellular proteins that are characterized by a distinctive mechanism of translocation across the outer membrane (15, 24). The Neisseria gonorrhoeae immunoglobulin A1 (IgA1) protease is the prototype member of the autotransporter family, and studies of this protein provide the basis for much of what we know about the family (25–27, 37). Autotransporter proteins are synthesized as precursor proteins with three functional domains: an N-terminal signal peptide, an internal passenger domain, and a C-terminal β domain. Following translation, these proteins are exported from the cytoplasm to the periplasm, presumably by the N-terminal signal peptide and the Sec translocase. Subsequently the signal peptide is cleaved, and the C-terminal domain inserts into the outer membrane. For all known autotransporter proteins, analysis of secondary structure of the C terminus predicts formation of a β-barrel with a central hydrophilic pore (24, 25). Studies of IgA1 protease suggest that the passenger domain may be extruded through the C-terminal β domain in an unfolded or partially unfolded state (26, 27, 57).
Once on the surface of the organism, IgA1 protease and all of the other well-characterized autotransporter proteins undergo processing, in most cases resulting in extracellular release of the passenger domain (15, 37). IgA1 protease and many members of the autotransporter family contain a serine protease motif, characterized by the sequence GDSGSP. In the case of IgA1 protease and the Haemophilus influenzae Hap adhesin, the serine protease motif mediates autoprocessing (17). In other cases, including Escherichia coli Tsh, E. coli EspC, Shigella flexneri SepA, and the E. coli and S. flexneri Pic toxin, mutation of the serine protease motif has no effect on processing, suggesting the presence of another protease (4, 14, 45, 46). The S. flexneri IcsA (VirG) protein lacks a protease motif and instead undergoes processing mediated by SopA (also called IcsP), an outer membrane protease with homology to OmpT (11). Whether OmpT, OmpU, or DegP may be responsible for cleaving some of the autotransporter proteins remains unclear. The E. coli Ag43 protein contains an aspartyl protease motif that may mediate the cleavage event responsible for release of the 60-kDa mature derivative of this protein (α43) (16).
Nontypeable H. influenzae is a common commensal organism in the human upper respiratory tract and an important cause of localized respiratory tract infection, including otitis media, sinusitis, bronchitis, and pneumonia (56). The pathogenesis of disease begins with colonization of the nasopharynx, followed by contiguous spread within the respiratory tract (34), usually facilitated by viral respiratory tract infection or allergic disease. Colonization requires multiple bacterial factors, including adhesins that promote direct bacterial interaction with the epithelial surface (47). Only a minority of nontypeable H. influenzae strains express adhesive pili (28), but virtually all nontypeable strains produce one or more nonpilus adhesins (52). In approximately 20 to 25% of strains, the predominant adhesin is a nonpilus protein referred to as Hia (3, 52).
In earlier work, we found that expression of the hia gene by nonadherent laboratory strains of E. coli was associated with a capacity for adherence to cultured epithelial cells (3). Of note, even clones containing very little DNA flanking hia conferred a capacity for in vitro adherence, suggesting that Hia may be an autotransporter protein. However, Hia is distinguished from other proteins that clearly belong to the autotransporter family. In particular, Hia undergoes no cleavage between the internal and C-terminal domains and thus remains full length and completely cell associated.
In the present study, we examined the hypothesis that Hia is an autotransporter protein. We found that the Hia N terminus contains the signal for export from the cytoplasm, that the mature protein is inserted into the outer membrane via the C-terminal domain, and that the C-terminal domain functions as an outer membrane translocator, transporting the internal passenger domain to the bacterial surface. Together these observations provide strong evidence that Hia represents a variant form of the autotransporter family. Based on homology analysis and secondary-structure predictions, it appears that several other proteins may belong to the same subfamily.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. E. coli DH5α is a nonadherent laboratory strain that has been described previously (40). E. coli CC118 contains a deletion of the phoA gene and lacks alkaline phosphatase activity (32). This strain was provided by V. Miller (Washington University, St. Louis) and was used to examine PhoA fusions.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | φ80dlacZΔM15 ΔlacU169 deoR recA endA1 | Life Technologies |
| CC118 | Δlac ΔphoA | 32 |
| BL21(DE3) | hsdS gal (λ cI ts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 53 |
| W3110 | F−mcrA mcrB IN(rrnD-rrnE)1 λ− | 40 |
| H. influenzae | ||
| 11 | Clinical isolate, nontypeable | 3 |
| 11hia | hia::kan | 3 |
| DB117 | Nonadherent laboratory strain, rec-1, cap-deficient serotype d | 42 |
| Plasmids | ||
| pT7-7 | Cloning vector, Ampr | 54 |
| pUC19 | Cloning vector, Ampr | New England Biolabs |
| pACYC184 | Cloning vector, Tetr Camr | 8 |
| pGJB103 | E. coli-H. influenzae shuttle vector, Tetr Ampr | 55 |
| pUC-Hia | pUC19 containing hia | This study |
| pUC-HiaΔ2-49 | Derivative of pUC-Hia with deletion of sequence encoding amino acids 2 to 49 | This study |
| pUC-HiaW1098Stop | Derivative of pUC-Hia with mutation of codon for C-terminal tryptophan, inserting a stop codon | This study |
| pACYC-Hia | 4.4-kb XbaI-BglII fragment from pUC-Hia cloned into pACYC184 digested with XbaI and BamHI | This study |
| pACYC-HiaΔ2-49 | 4.4-kb XbaI-BglII fragment from pUC-HiaΔ2-49 cloned into pACYC184 digested with XbaI and BamHI | This study |
| pACYC-HiaW1098Stop | 4.4-kb XbaI-BglII fragment from pUC-HiaW1098Stop cloned into pACYC184 digested with XbaI and BamHI | This study |
| pPHO7 | Derivative of pTZ18R containing phoA flanked on both sides by multiple cloning sites | 13 |
| pCH39 | pKT279 containing β-lactamase signal sequence fused to PhoA | 22 |
| pHiaN49 | pT7-7 derivative containing coding sequence for N-terminal 49 amino acids of Hia | This study |
| pHiaN49-PhoA | pT7-7 derivative containing coding sequence for N-terminal 49 amino acids of Hia fused to coding sequence for mature PhoA | This study |
| pUC-PhoA | pUC19 containing phoA gene from E. coli strain W3110 | This study |
| pPhoA-Hia(780-1098) | pUC19 containing phoA gene fused to hia fragment encoding Hia amino acids 780 to 1098 | This study |
| pHapS243A | pGJB103 containing hap gene with point mutation at active-site serine (S243) | 17 |
| pDH106 | pUC19 derivative containing hap fragment encoding Hap signal sequence and Haps, with SalI site at 3′ end of hap fragment | 18 |
| pJS106 | pGJB103 containing 6.7-kb PstI fragment with wild-type hap gene | 48 |
| pHaps-Hia(780-1098) | pGJB103 containing hap fragment encoding Hap signal sequence and Haps fused to hia fragment encoding Hia amino acids 780 to 1098 | This study |
| pHia(780-1098) | pGJB103 containing hia fragment encoding Hia signal sequence fused to hia fragment encoding Hia amino acids 780 to 1098 | This study |
E. coli strains were grown on Luria-Bertani (LB) agar or in LB broth and were stored at −80°C in LB broth with 50% glycerol. Antibiotics used to select for plasmids included ampicillin (100 μg/ml) and tetracycline (12.5 μg/ml). H. influenzae strains were grown on chocolate agar, on brain heart infusion agar supplemented with hemin and NAD, or in brain heart infusion broth supplemented with hemin and NAD, as previously described (49). Antibiotics used to select for plasmids included tetracycline (5 μg/ml) and chloramphenicol (2 μg/ml).
Plasmid construction.
The plasmid pUC-Hia contains the hia gene and was constructed by excising the 5.5-kb XbaI-EcoRI fragment from pHMW8-3 (3) and ligating it into XbaI- and EcoRI-digested pUC19. pUC-HiaΔ2-49 contains the hia gene with a deletion of coding sequence for amino acids 2 to 49 in the full-length Hia protein. This plasmid was constructed using recombinant PCR to generate a 0.9-kb XmnI-StyI fragment harboring the relevant deletion (21). The resulting fragment was digested with XmnI and StyI and then ligated in place of the wild-type XmnI-StyI fragment in pUC-Hia. pUC-HiaW1098Stop contains the hia gene with a stop codon in place of the codon for the C-terminal tryptophan residue. It was constructed using recombinant PCR to generate a 1.5-kb DsaI-BglII fragment harboring the desired mutations. The resulting fragments were digested with DsaI and BglII and then ligated in place of the wild-type DsaI-BglII fragment in pUC-Hia. The inserts in pUC-Hia, pUC-HiaΔ2-49, and pUC-HiaW1098Stop were excised by digestion with XbaI and BglII and cloned into pACYC184 to generate pACYC-Hia, pACYC-HiaΔ2-49, and pACYC-HiaW1098Stop, respectively. pHiaN49 contains the N terminus of full-length Hia. This plasmid was constructed using PCR to amplify a 0.5-kb EcoRI-BamHI fragment corresponding to hia upstream sequence and coding sequence for Hia amino acids 1 to 49, then digesting this fragment with EcoRI and BamHI and ligating into EcoRI and BamHI-digested pT7-7 (54). pHiaN49-PhoA contains coding sequence for Hia amino acids 1 to 49 fused in frame to coding sequence for mature PhoA (lacking the PhoA signal sequence). This plasmid was constructed by digesting pHiaN49 with BamHI and ligating in the BamHI fragment containing the phoA gene from pPHO7 (13), then performing restriction analysis to confirm ligation in the proper orientation. pUC-PhoA-Hia(780-1098) contains coding sequence for full-length PhoA fused in frame to coding sequence for the C-terminal end of Hia. To construct this plasmid, the phoA gene was amplified from E. coli strain W3110 and ligated into BanII- and BamHI-digested pUC19. The resulting plasmid was digested with BamHI and PstI, and a BamHI-PstI fragment containing coding sequence for Hia amino acids 780 to 1098 and downstream sequence was inserted. Because of the engineered BamHI sites at the 3′ end of phoA fragment and the 5′ end of the hia fragment, the resulting chimera contains a glycine and a serine joining PhoA to the Hia C teminus. The plasmid pHaps-Hia(780-1098) contains the upstream region of hap and coding sequence for the Hap signal sequence and Haps fused in frame to coding sequence for the C-terminal end of Hia. To construct this plasmid, coding sequence for Hia amino acids 780 to 1098 along with downstream sequence was amplified by PCR, introducing a SalI site at the 5′ end and a BamHI site at the 3′ end. The amplified fragment was digested with SalI and BamHI and then ligated into SalI- and BamHI-digested pDH106 (18). The resulting plasmid was digested with PstI and BamHI to liberate a 6.1-kb fragment containing the gene fusion, which was then ligated into PstI- and BglII-digested pGJB103 (55), producing the final plasmid. Because of the engineered SalI sites at the 3′ end of the hap fragment and the 5′ end of the hia fragment, the resulting chimeric protein contains a valine and an aspartic acid in place of the usual C-terminal three amino acids of Haps.
Recombinant DNA methods.
DNA ligations, restriction endonuclease digestions, and gel electrophoresis were performed according to standard techniques (40). Plasmids were introduced into E. coli by electroporation, while plasmids were transformed into H. influenzae by the MIV method (20).
Cell fractionation and protein analysis.
Whole-cell sonicates were prepared by resuspending bacterial pellets in 10 mM HEPES (pH 7.4) and sonicating to clarity. Outer membrane proteins were recovered on the basis of sarcosyl insolubility, using the protocol described by Carlone et al. (7), or by sucrose density centrifugation, as previously described (35, 51). Outer membrane fractions were confirmed to be virtually free of NADH oxidase activity, a marker for inner membrane proteins, by the method of Osborn et al. (35). Proteins released from the surface of the bacterium were recovered by precipitating culture supernatants with 10% trichloroacetic acid (2). To collect culture supernatants, bacteria were pelleted by ultracentrifugation at 100,000 × g for 60 min. Because Hia has a tendency to form aggregates, prior to loading onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, samples were denatured with formic acid, as described previously (52).
Proteins were resolved by SDS-PAGE using 7.5 to 10% polyacrylamide gels (29). Western blots were performed with rabbit polyclonal antiserum 36B raised against recombinant Hia, guinea pig polyclonal antiserum raised against Hia residues 221 to 658, guinea pig polyclonal antiserum raised against Hia residues 659 to 1098, guinea pig polyclonal antiserum raised against Haps, or a rabbit polyclonal antiserum raised against PhoA (3-Prime, 5-Prime, Boulder, Colo.).
In examining whole-cell sonicates, volumes containing 30 μg of total protein were loaded in each lane. Similarly, in assessing outer membrane proteins, volumes corresponding to 5 μg of total protein were compared.
Whole-cell (dot) immunoblots.
For whole-cell immunoblots, bacteria were grown to an optical density at 600 nm of 0.8, washed twice with phosphate-buffered saline (PBS), fixed for 30 min at room temperature with 4% paraformaldehyde in PBS, and resuspended in PBS to an optical density at 600 nm of 0.8. Aliquots of 100 μl were applied to nitrocellulose filters using a dotblot manifold apparatus (Schleicher and Schuell, Keene, N.H.). Samples were incubated for 30 min and then pulled through the filter by vacuum suction. Following blocking for 1 h with 5% skim milk in Tris-buffered saline, surface-exposed protein was detected using an appropriate primary antibody and then a horseradish peroxidase-conjugated secondary antibody and chemiluminscence (Pierce, Rockford, Ill.).
Alkaline phosphatase assays.
Alkaline phosphatase activity associated with PhoA fusion constructs was assessed by permeabilizing the relevant strains and measuring hydrolysis of para-nitrophenyl phosphate (Sigma 104), as described by Manoil (31). Production of PhoA fusion proteins was confirmed by Western analysis using anti-PhoA antiserum.
Determination of N-terminal sequence.
Outer membrane fractions were resolved on an SDS–7.5% polyacrylamide gel, and proteins were electrotransferred to a polyvinylidene membrane. After staining with Coomassie brilliant blue R-250, the ∼115-kDa Hia protein was excised from the membrane and submitted to Midwest Analytical, Inc. (St. Louis, Mo.). Amino-terminal sequence determination was performed by automated Edman degradation using a Perkin-Elmer Biosystems model 477A sequencing system.
Adherence assays.
Adherence assays were performed with Chang epithelial cells (Wong-Kilbourne derivative, clone 1-5c-4 [human conjunctiva]; ATCC CCL20.2) or with A549 respiratory epithelial cells (ATCC CCL185) as described previously (18, 50). Percent adherence was calculated by dividing the number of adherent CFU per monolayer by the number of CFU inoculated.
Immunoelectron microscopy.
For immunoelectron microscopy, bacteria were incubated on agar overnight and then resuspended in PBS. Glow-discharged, carbon-coated, Formvar-strengthened grids were overturned on a drop of bacterial suspension and incubated for 2 min. Subsequently, samples were blocked for 1 h with PBS containing 23% goat serum and 0.1% gelatin and then incubated for 2 h with a 1:25 dilution of guinea pig polyclonal antiserum raised against Hia amino acids 221 to 779. Next, samples were washed with PBS and incubated for 1 h with a 1:25 dilution of goat anti-guinea pig IgG conjugated to 12-nm colloidal gold beads (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Samples were then washed again with PBS, fixed with 1% glutaraldehyde in PBS, and negatively stained with 0.5% uranyl acetate.
RESULTS
Hia is an integral outer membrane protein and remains cell associated.
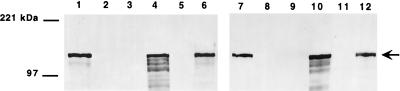
In considering the possibility that Hia is an autotransporter protein, we began by examining whether this protein is anchored in the outer membrane. Following disruption of organisms by sonication, membranes were isolated, and outer membrane proteins were then recovered on the basis of sarcosyl insolubility. As shown in Fig. 1, Western analysis of outer membrane fractions from H. influenzae strain 11, H. influenzae strain DB117/pACYC-Hia (wild-type hia), and E. coli DH5α/pUC19-Hia (wild-type hia) revealed the full-length 115-kDa Hia protein. As expected, outer membrane fractions from strains 11hia, DB117/pACYC184 (cloning vector alone), and DH5α/pUC19 (cloning vector alone) were devoid of Hia. Examination of outer membrane proteins recovered by sucrose density centrifugation confirmed these results (not shown).
FIG. 1.
Outer membrane localization of Hia. Lanes 1 to 6 show sonicates, and lanes 7 to 12 show outer membrane proteins detected by Western analysis with guinea pig antiserum against Hia residues 659 to 1098. Lanes 1 and 7, H. influenzae strain 11; lanes 2 and 8, H. influenzae strain 11hia; lanes 3 and 9, DH5α/pUC19; lanes 4 and 10, DH5α/pUC-Hia; lanes 5 and 11, DB117/pACYC184; lanes 6 and 12, DB117/pACYC-Hia. Arrow indicates full-length Hia.
Given that most autotransporter proteins undergo cleavage on the surface of the organism and then are released extracellularly, we examined culture supernatants from strains 11, DB117/pACYC-Hia, and DH5α/pUC19-Hia for the presence of a processed form of Hia. However, Western analysis using polyclonal antiserum against either full-length Hia, the Hia fragment corresponding to amino acids 221 to 658, or the Hia fragment corresponding to amino acids 659 to 1098 revealed no evidence of either full-length or processed Hia (not shown).
Together these results suggest that Hia is an integral outer membrane protein that undergoes no major proteolytic processing and remains cell associated in the native H. influenzae strain, a recombinant H. influenzae strain, and a recombinant E. coli strain.
Hia N terminus is the secretion signal for export from the cytoplasm.
According to the prevailing model for autotransporter proteins, secretion involves an N-terminal signal peptide and interaction with the Sec machinery. In this context, it is noteworthy that Hia has an unusual N terminus characterized by a 24-amino-acid sequence with three positively charged residues and then a 25-amino-acid sequence with hydrophobic residues and a predicted leader peptidase cleavage site between residues 49 and 50. To explore the possibility that the Hia N terminus functions as a signal sequence, we fused the N-terminal 49 amino acids to a truncated PhoA fragment lacking a signal sequence, then introduced this construct into E. coli strain CC118 and measured alkaline phosphatase activity. As a positive control, we examined CC118/pCH39, which contains the β-lactamase signal sequence fused upstream of the mature PhoA protein. As negative controls, we examined CC118/pPHO7 (which produces PhoA without a signal sequence) and CC118/pHiaN49 (which produces the Hia N-terminal 49 amino acids by themselves). Of note, both CC118/pHiaN49-PhoA and CC118/pCH39 were associated with significant levels of alkaline phosphatase activity (537 and 1,493 U, respectively). In contrast, CC118/pPHO7 and CC118/pHiaN49 were completely devoid of alkaline phosphatase activity (<2 U). Western analysis of periplasmic extracts from CC118/pHiaN49-PhoA and CC118/pCH39 revealed similar quantities of HiaN49-PhoA and wild-type PhoA, respectively (not shown).
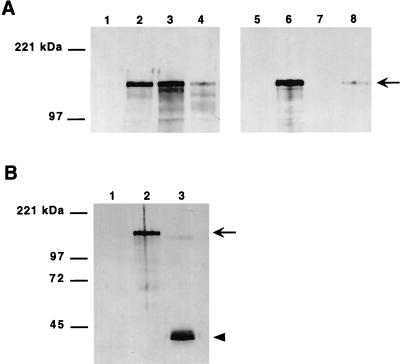
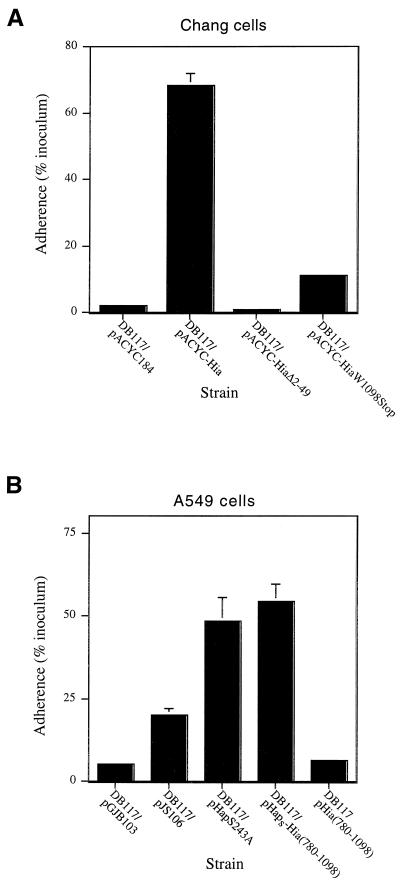
To extend these results, we sought to determine the N-terminal amino acid sequence of mature Hia. Outer membrane proteins were recovered from both H. influenzae strain DB117/pACYC-Hia and E. coli strain BL21(DE3)/pHMW8-7, then separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. (We used H. influenzae strain DB117/pACYC-Hia rather than H. influenzae strain 11 because the DB117 derivative produces slightly higher quantities of Hia.) Subsequently, in both cases the Hia band was excised and subjected to Edman degradation, revealing the sequence NNNTPVT, which corresponds to amino acids 50 to 56, indicating cleavage between residues 49 and 50. Consistent with these findings, in experiments with H. influenzae DB117, deletion of amino acids 2 to 49 eliminated secretion of Hia (Fig. 2) and abolished Hia-mediated adherence (Fig. 3A).
FIG. 2.
Deletion of the Hia N terminus and the Hia C-terminal tryptophan and expression of the C terminus. (A and B) H. influenzae derivatives detected by Western analysis with guinea pig antiserum against Hia residues 659 to 1098. (A) Lanes 1 to 4 show sonicates, and lanes 5 to 8 show outer membrane proteins. Lanes 1 and 5, DB117/pACYC184; lanes 2 and 6, DB117/pACYC-Hia; lanes 3 and 7, DB117/pACYC-HiaΔ2-49; lanes 4 and 8, DB117/pACYC-HiaW1098Stop. Arrow indicates full-length Hia. (B) Outer membrane proteins. Lane 1, DB117/pGJB103; lane 2, DB117/pACYC-Hia; lane 3, DB117/pHia(780-1098).
FIG. 3.
Adherence to epithelial cells by H. influenzae DB117 expressing Hia derivatives. (A) Adherence to Chang epithelial cells. (B) Adherence to A549 epithelial cells. Adherence was determined in 30-min assays and calculated by dividing the number of adherent bacteria by the number of inoculated bacteria. For all strains, inocula were approximately 2 × 107 CFU/ml. Bars represent the mean ± standard error of the mean of measurements made in triplicate from representative experiments.
These observations suggest that the N-terminal 49 amino acids of Hia serve as a secretion signal for export from the cytoplasm.
Hia C terminus inserts into the outer membrane and is predicted to form a β-barrel.
Among proteins belonging to the autotransporter family, the C-terminal portion inserts into the outer membrane and is predicted to form a β-barrel with a central hydrophilic pore, allowing translocation of the passenger domain from the periplasm to the bacterial surface. With this information in mind, we examined the structure of the C-terminal end of Hia. Based on analysis by algorithms developed to predict secondary structure and membrane topology (9, 39, 41), the final 319 amino acids of Hia contain a membrane-spanning α-helix followed by 14 antiparallel amphipathic membrane-spanning β-sheets, suggesting formation of a β-barrel structure (not shown).
To obtain more direct evidence for the conclusion that the C terminus of Hia inserts into the outer membrane and forms a β-barrel, we examined the effect of mutation of the terminal tryptophan. In the case of β-barrel outer membrane proteins, the C-terminal aromatic amino acid is proposed to target the protein to the outer membrane (19). As shown in Fig. 2A, deletion of the terminal tryptophan resulted in a marked reduction in the quantity of Hia in the outer membrane. Of note, the quantity of Hia in whole-cell sonicates was also reduced, suggesting rapid degradation of the mutant protein, perhaps related to abnormal targeting to the outer membrane or to instability in the outer membrane. Consistent with these findings, this deletion resulted in a ∼85% decrease in Hia-mediated adherence (Fig. 3A).
To extend these observations, we generated a fusion protein with the Hia signal peptide linked directly to the C-terminal 319 amino acids (residues 780 to 1098), thereby deleting residues 52 to 779. Following introduction of the resulting construct into DB117, fractionation studies were performed. As shown in Fig. 2B, the Hia C terminus was inserted into the outer membrane, similar to the situation with wild-type Hia.
Together these results indicate that the Hia C terminus likely targets Hia to the outer membrane, directing insertion and formation of a β-barrel.
Hia C terminus functions as a translocator.
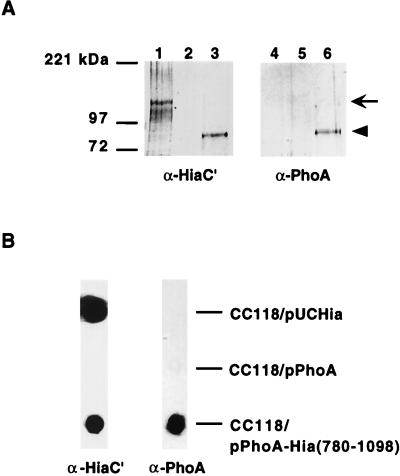
To examine the possibility that the C-terminal portion of Hia functions as a translocator, we fused the intact PhoA protein (including signal sequence) upstream of the C-terminal 319 amino acids of Hia, then assessed the fate of this chimera in E. coli DH5α. As shown in Fig. 4, Western analysis with anti-PhoA antiserum and anti-Hia antiserum demonstrated that the chimeric protein was targeted to the outer membrane. Furthermore, as assessed by whole-cell dot immunoblot, when organisms were grown in the presence of 10 mM β-mercaptoethanol (with the intent to prevent formation of disulfide bonds and inhibit folding of PhoA), PhoA was translocated across the outer membrane and presented on the bacterial surface (Fig. 4B). Of note, in the absence of β-mercaptoethanol, only minimal amounts of PhoA were present on the surface of the organism (not shown), suggesting that folding of PhoA impedes passage across the outer membrane. In control constructs grown in medium supplemented with β-mercaptoethanol, wild-type Hia was detectable in the outer membrane and on the bacterial surface, while wild-type PhoA by itself was not (Fig. 4).
FIG. 4.
Immunoblot analysis of PhoA-Hia(780-1098) chimera. (A) Outer membrane proteins detected by Western blot with guinea pig antiserum against Hia residues 659 to 1098 in lanes 1 to 3 and with antiserum against PhoA in lanes 4 to 6. Lanes 1 and 4, E. coli CC118/pUC-Hia; lanes 2 and 5, E. coli CC118/pUC-PhoA; lanes 3 and 6, E. coli CC118/pPhoA-Hia(780-1098). Arrow indicates full-length Hia, and arrowhead indicates PhoA-Hia(780-1098). (B) Whole-cell immunoblots performed with either guinea pig antiserum against Hia residues 659 to 1098 (left lane) or antiserum against PhoA (right lane).
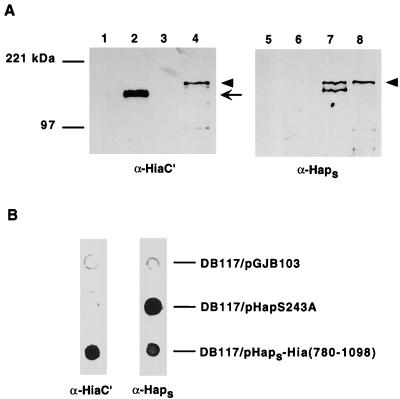
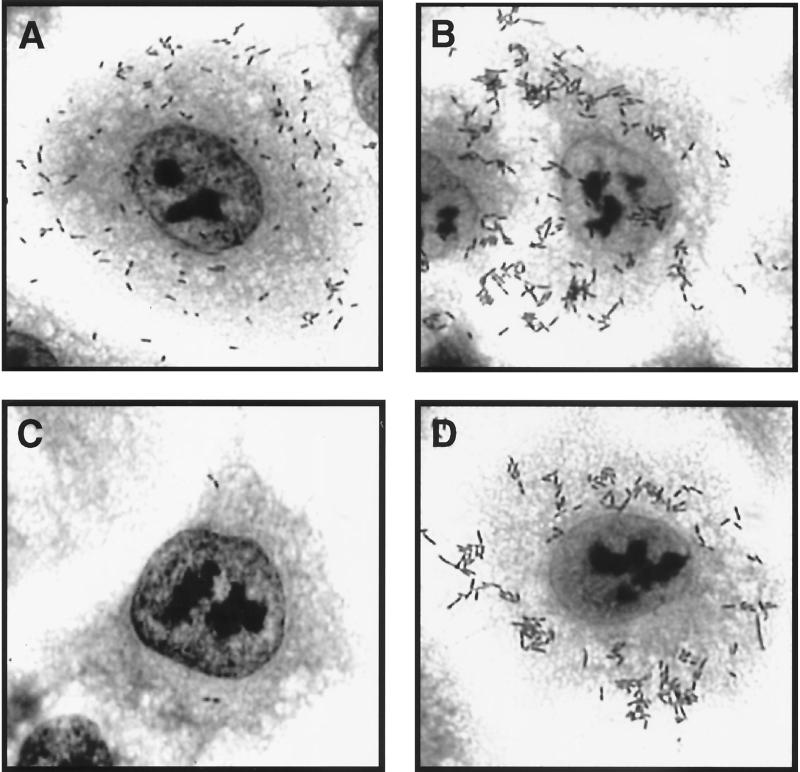
In related experiments, we fused the passenger domain of the H. influenzae Hap protein upstream of the C-terminal 319 amino acids of Hia and then expressed the resulting chimera in H. influenzae strain DB117. Hap is a member of the autotransporter family and contains a passenger domain called Haps, which mediates bacterial adherence to epithelial cells and bacterial aggregation on the epithelial surface (18). These properties are maximal when Hap is locked in the full-length, surface-associated state, which occurs when the active-site serine is mutated (HapS243A) (17, 18). As shown in Fig. 5, Western analysis confirmed that the Haps-Hia(780-1098) chimera was inserted into the outer membrane, and whole-cell dot immunoblotting established that the Haps domain was present on the bacterial surface. To address whether surface-associated Haps was properly folded, we examined DB117/pHaps-Hia(780-1098) for in vitro adherence. As shown in Fig. 3B, adherence to A549 respiratory epithelial cells was appreciable, similar to that observed with DB117 expressing full-length Hap (HapS243A). As controls, we examined DB117 harboring vector alone and DB117/pHia(780-1098), both of which were nonadherent with these cells (Fig. 3B). As an additional indication of proper folding, we assessed DB117/pHaps-Hia(780-1098) for bacterial aggregation on the epithelial surface. Samples were stained with Giemsa and then examined by light microscopy, which revealed significant bacterial aggregation, again comparable to the aggregation observed with DB117/pHapS243A (Fig. 6).
FIG. 5.
Immunoblot analysis of Haps-Hia(780-1098) chimera. (A) Outer membrane proteins detected by Western blot with guinea pig antiserum against Hia residues 659 to 1098 in lanes 1 to 4 and with antiserum against Haps in lanes 5 to 8. Lanes 1 and 5, DB117/pGJB103; lanes 2 and 6, DB117/pACYC-Hia; lanes 3 and 7, DB117/pHapS243A; lanes 4 and 8, DB117/pHaps-Hia(780-1098). Arrow indicates full-length Hia, and arrowhead indicates HapS243A and the Haps-Hia(780-1098) chimera. A degradation product of HapS243A is present in lane 7. (B) Whole-cell immunoblots performed with either guinea pig antiserum against Hia residues 659 to 1098 (left lane) or antiserum against Haps (right lane).
FIG. 6.
Bacterial adherence and aggregation associated with Haps-Hia(780-1098) chimera. Bacteria were inoculated onto monolayers of A549 epithelial cells, and samples were incubated for 30 min, rinsed, and stained with Giemsa. (A) DB117/pJS106 (wild-type Hap); (B) DB117/pHapS243A; (C) DB117/pHia(780-1098); (D) DB117/pHaps-Hia(780-1098).
Together these observations demonstrate that the C terminus of Hia functions as an outer membrane translocator and allows proper folding of a heterologous passenger domain.
Hia internal domain is surface localized.
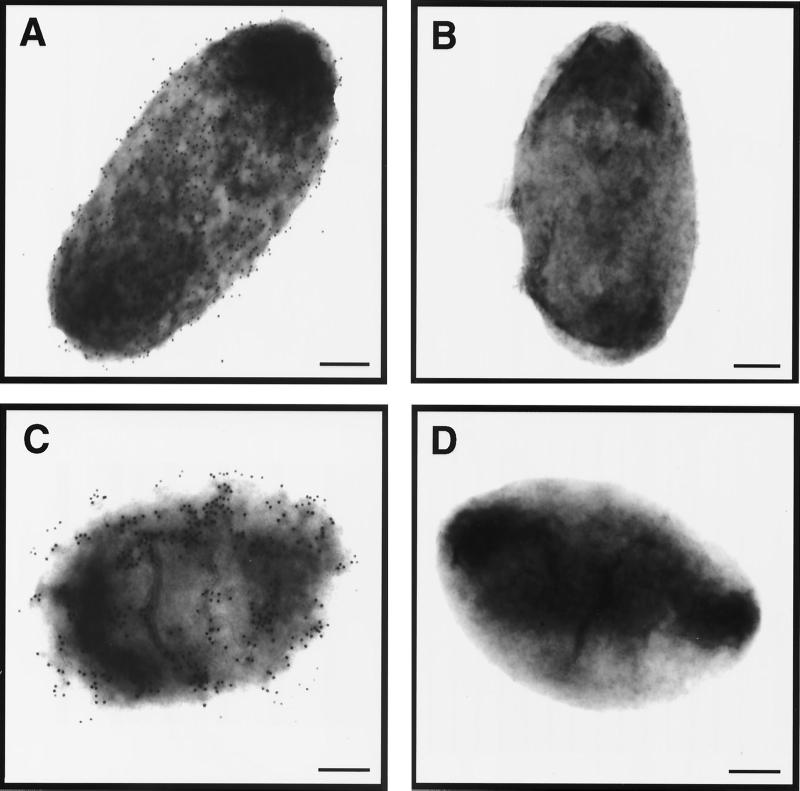
Given the resemblance between Hia and autotransporter proteins, we hypothesized that the internal region of Hia represents a passenger domain and is translocated across the outer membrane and localized on the surface of the organism. To confirm this conclusion, we generated a polyclonal antiserum against the polypeptide corresponding to Hia amino acids 221 to 779 and then performed immunoelectron microscopy. As shown in Fig. 7, examination of strain 11 and strain DB117/pACYC-Hia revealed abundant surface labeling, as assessed by transmission electron microscopy. In contrast, strains 11hia and DB117/pACYC184 were virtually devoid of gold labeling.
FIG. 7.
Immunoelectron microscopy showing surface localization of internal region of Hia. Samples were prepared using guinea pig antiserum against Hia residues 221 to 779 and a goat anti-guinea pig IgG antiserum conjugated to 12-nm gold beads. (A) DB117/pACYC-Hia; (B) DB117/pACYC184; (C) H. influenzae strain 11; (D) H. influenzae strain 11hia. Bars, 200 nm.
DISCUSSION
In the present study we examined the mechanism of secretion of the H. influenzae Hia adhesin and demonstrated that the N terminus directs export out of the cytoplasm, while the C terminus targets the protein to the outer membrane and translocates the passenger domain to the surface of the organism. Together these observations argue that Hia belongs to the autotransporter family of proteins. In this context, it is particularly noteworthy that Hia undergoes no processing event at the C terminus, distinguishing it from all other well-characterized autotransporter proteins and expanding the scope of the autotransporter family. The absence of a protease motif and the lack of extracellular release are other features of Hia that are unusual among known autotransporter proteins.
In the course of our studies, we found that the N-terminal 49 amino acids of Hia are cleaved, with cleavage occurring at a predicted leader peptidase recognition site. Additional experiments demonstrated that the N-terminal 49 amino acids are essential for export of Hia and are capable of directing export of PhoA as well, providing strong evidence that these residues constitute a signal sequence. It is noteworthy that the Hia N terminus is atypical for a signal sequence, with a long N-terminal extension. Interestingly, a similar extension is present in a number of other high-molecular-weight, extracellular gram-negative bacterial proteins, including the E. coli AIDA-I adhesin, EspP protease, Pet protease, and Ag43 protein, the S. flexneri SepA protease, the Bordetella pertussis FHA protein, and the H. influenzae HMW1 and HMW2 proteins, among others. Of these proteins, all but FHA and the HMW proteins belong to the autotransporter family, and all are presumed to be secreted by a Sec-dependent mechanism. One possibility is that this domain functions as an intramolecular cytoplasmic chaperone, preventing premature folding and facilitating export across the cytoplasmic membrane. Alternatively, the N-terminal extension may interact with an independent cytoplasmic chaperone and thereby facilitate secretion. Both of these possibilities are presently under investigation.
Consistent with studies of other autotransporter proteins, analysis of the secondary structure of the Hia C terminus predicted formation a β-barrel with a central hydrophilic channel. In early studies of Neisseria gonorrhoeae IgA1 protease, Pohlner and colleagues speculated that the IgA1 protease C terminus (Igaβ) forms a pore in the outer membrane, allowing extrusion of the covalently linked passenger domain (37). More recently a number of investigators have suggested the same possibility with other autotransporter proteins. Of note in this context, Shannon and Fernandez recently demonstrated that the B. pertussis BrkA C terminus is indeed capable of forming a pore (43). Using black lipid bilayers, these workers showed that purified recombinant protein was associated with a conductance of 3.0 nS in 1 M KCl, indicating a pore size similar to that described for PapC, an outer membrane usher through which P pilus subunits are exported in uropathogenic E. coli.
In autotransporter proteins, the sequence that joins the passenger domain to the C-terminal β-barrel is referred to as the linker region. In the case of the E. coli AIDA-I protein, cleavage occurs between residues 839 and 840, generating a 447-amino-acid C-terminal fragment. Based on secondary-structure predictions, the AIDA-I linker region is 160 amino acids, and the complete AIDA-I β-barrel is 287 amino acids with 14 transmembrane β-sheets. Recently Maurer et al. constructed a series of fusions between the cholera toxin B (CtxB) subunit and various portions of the C terminus of AIDA-I and then examined whether the CtxB moiety was displayed on the surface of the organism (33). Using this approach, these investigators found that a part of the linker region was absolutely necessary for translocation of the CtxB passenger. This portion of the linker region in AIDA-I corresponds to the membrane-spanning α-helical segment in Hia, which presumably allows the Hia passenger domain to swing across the outer membrane and gain surface accessibility.
In characterizing the chimeric protein containing PhoA fused to the Hia C terminus, we found that growth in the presence of β-mercaptoethanol was required for efficient surface localization of the PhoA moiety. This observation is reminiscent of previous studies of N. gonorrhoeae IgA1 protease. In particular, Klauser et al. examined the ability of the IgA1 protease C-terminal domain (Igaβ) to translocate CtxB across the outer membrane in Salmonella typhimurium and E. coli and observed that efficient translocation occurred only when folding of CtxB was prevented by either β-mercaptoethanol or a mutation in the dsbA gene, thus inhibiting periplasmic disulfide bond formation (26, 27). Based on these results, Klauser and coworkers proposed that the IgA1 protease passenger domain passes across the outer membrane in a linear or partially unfolded form (26, 27). More recently Veiga et al. analyzed the relationship between formation of disulfide bonds, folding, and secretion by constructing a chimera with a single-chain antibody (scFv) fused to Igaβ, exploiting the fact that scFv binding to its target antigen is entirely dependent on formation of disulfide bonds (57). Interestingly, these workers found that scFv was able to pass through the outer membrane in an active conformation with its disulfide bonds preformed in the periplasm, although the efficiency of passage was reduced approximately threefold. Thus, whether the passenger domain of IgA1 protease and other autotransporter proteins is unfolded or folded during translocation from the periplasm to the bacterial remains unclear.
It is interesting that H. influenzae elaborates at least three different proteins that belong to the autotransporter family, including IgA1 protease, the Hap adhesin, and now Hia. One possibility is that Hia evolved by acquiring and maintaining the activities of multiple distinct proteins, thus conserving space on the H. influenzae genome, which is relatively small compared to those of other pathogenic bacteria. Alternatively, Hia may have originated from IgA1 protease or Hap or perhaps another H. influenzae autotransporter protein and acquired mutations over time, resulting in a new function.
To summarize, the Hia adhesin is a novel member of the autotransporter family. In contrast to other well-characterized members of the family, this protein undergoes no processing between the internal and C-terminal domains and remains completely cell associated, allowing maximal adhesive activity. Based on homology analysis and secondary-structure predictions, the Moraxella catarrhalis UspA1 and UspA2 proteins, the enterotoxigenic E. coli TibA adhesin/invasin, the Helicobacter mustelae Hsr ring-forming protein, and the Pseudomonas aeruginosa EstA esterase may belong to the same subfamily (10, 15, 30, 36, 58).
ACKNOWLEDGMENTS
We thank M. Levy for assistance with cryoimmunoelectron microscopy and C. Stathopoulos for assistance with the secondary-structure predictions of the Hia C terminus and critical reading of the manuscript.
This work was supported by Public Health Service grants 1RO1 AI-44167 and 1RO1 DC-02873 to J.W.S.
REFERENCES
- 1.Barenkamp S J, Leininger E. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect Immun. 1992;60:1302–1313. doi: 10.1128/iai.60.4.1302-1313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barenkamp S J, St. Geme J W., III Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect Immun. 1994;62:3320–3328. doi: 10.1128/iai.62.8.3320-3328.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp S J, St. Geme J W., III Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- 4.Benjelloun-Touimi Z, Sansonetti P J, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 5.Benz I, Schmidt M A. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect Immun. 1989;57:1506–1511. doi: 10.1128/iai.57.5.1506-1511.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 7.Carlone G M, Thomas M L, Rumschlag H S, Sottnek F O. Rapid procedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:330–332. doi: 10.1128/jcm.24.3.330-332.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou P Y, Fasman G D. Prediction of β-turns. Biophys J. 1979;26:367–384. doi: 10.1016/S0006-3495(79)85259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cope L D, LaFontaine E R, Slaughter C A, Hasemann C A, Jr, Aebi C, Henderson F W, McCracken G H, Jr, Hansen E J. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J Bacteriol. 1999;181:4026–4034. doi: 10.1128/jb.181.13.4026-4034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egile C, d'Hauteville H, Parsot C, Sansonetti P J. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997;23:1063–1073. doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- 12.Eslava C, Navarro-Garcia F, Czeczulin J R, Henderson I R, Cravioto A, Nataro J P. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3155–3163. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson I R, Czeczulin J, Eslava C, Noriega F, Nataro J P. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–5596. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–377. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 16.Henderson I R, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrixson D R, de la Morena M L, Stathopoulos C, St. Geme J W., III Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Mol Microbiol. 1997;26:505–518. doi: 10.1046/j.1365-2958.1997.5921965.x. [DOI] [PubMed] [Google Scholar]
- 18.Hendrixson D R, St. Geme J W., III The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol Cell. 1998;2:841–850. doi: 10.1016/s1097-2765(00)80298-1. [DOI] [PubMed] [Google Scholar]
- 19.Henning U, Koebnik R. Outer membrane proteins of Escherichia coli: mechanism of sorting and regulation of synthesis. In: Ghuysen J-M, Hukenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 381–395. [Google Scholar]
- 20.Herriott R M, Meyer E M, Vogt M. Defined non-growth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 22.Hoffman C S, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jose J, Jähnig F, Meyer T F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol. 1995;18:378–380. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 25.Klauser T, Kramer J, Otzelberger K, Pohlner J, Meyer T F. Characterization of the Neisseria Iga beta-core: the essential unit for outer membrane targeting and extracellular protein secretion. J Mol Biol. 1993;234:579–593. doi: 10.1006/jmbi.1993.1613. [DOI] [PubMed] [Google Scholar]
- 26.Klauser T, Pohlner J, Meyer T F. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease beta-domain: conformation-dependent outer membrane translocation. EMBO J. 1990;9:1991–1999. doi: 10.1002/j.1460-2075.1990.tb08327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klauser T, Pohlner J, Meyer T F. Selective extracellular release of cholera toxin B subunit by Escherichia coli: dissection of Neisseria Igaβ-mediated outer membrane transport. EMBO J. 1992;11:2327–2335. doi: 10.1002/j.1460-2075.1992.tb05292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasan G P, Cutter D, Block S L, St. Geme J W., III Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect Immun. 1999;67:449–454. doi: 10.1128/iai.67.1.449-454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lindenthal C, Elsinghorst E A. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect Immun. 1999;67:4084–4091. doi: 10.1128/iai.67.8.4084-4091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 32.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer J, Jose J, Meyer T F. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J Bacteriol. 1999;181:7014–7020. doi: 10.1128/jb.181.22.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy T F, Bernstein J M, Dryja D M, Campagnari A A, Apicella M A. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to nontypeable Haemophilus influenzae: pathogenic and epidemiologic observations. J Infect Dis. 1987;5:723–731. doi: 10.1093/infdis/156.5.723. [DOI] [PubMed] [Google Scholar]
- 35.Osborn M J, Bander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium: isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;12:3962–3972. [PubMed] [Google Scholar]
- 36.O'Toole P W, Austin J W, Trust T J. Identification and molecular characterization of a major ring-forming surface protein from the gastric pathogen Helicobacter mustelae. Mol Microbiol. 1994;11:349–361. doi: 10.1111/j.1365-2958.1994.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 37.Pohlner J, Halter R, Beyreuther K, Meyer T F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 38.Relman D A, Domenighini M, Tuomanen E, Rappuoli R, Falkow S. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc Natl Acad Sci USA. 1989;86:2637–2641. doi: 10.1073/pnas.86.8.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Schirmer T, Cowan S W. Prediction of membrane-spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Setlow J K, Brown D C, Boling M E, Mattingly A, Gordon M P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968;95:546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon J L, Fernandez R C. The C-terminal domain of the Bordetella pertussis autotransporter BrkA forms a pore in lipid bilayer membranes. J Bacteriol. 1999;181:5838–5842. doi: 10.1128/jb.181.18.5838-5842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stathopoulos C, Hendrixson D R, Thanassi D G, Hultgren S J, St. Geme III J W, Curtiss R., III Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2000;2:1061–1072. doi: 10.1016/s1286-4579(00)01260-0. [DOI] [PubMed] [Google Scholar]
- 45.Stathopoulos C, Provence D L, Curtiss R., III Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect Immun. 1999;67:772–781. doi: 10.1128/iai.67.2.772-781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein M, Kenny B, Stein M A, Finlay B B. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178:6546–6554. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St. Geme J W., III Insights into the mechanism of respiratory tract colonization by nontypable Haemophilus influenzae. Pediatr Infect Dis J. 1997;16:931–935. doi: 10.1097/00006454-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 48.St. Geme J W, III, de la Morena M L, Falkow S. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol. 1994;14:217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 49.St. Geme J W, III, Falkow S. Haemophilus influenzae adheres to and enters cultured human epithelial cells. Infect Immun. 1990;58:4036–4044. doi: 10.1128/iai.58.12.4036-4044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St. Geme J W, III, Falkow S, Barenkamp S J. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci USA. 1993;90:2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.St. Geme J W, III, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- 52.St. Geme J W, III, Kumar V V, Cutter D, Barenkamp S J. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 54.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomb J-F, Barcak G J, Chandler M S, Redfield R J, Smith H O. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae Rd. J Bacteriol. 1989;171:3796–3802. doi: 10.1128/jb.171.7.3796-3802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turk D C. The pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 57.Veiga E, de Lorenzo V, Fernandez L A. Probing secretion and translocation of a β-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol Microbiol. 1999;33:1232–1243. doi: 10.1046/j.1365-2958.1999.01571.x. [DOI] [PubMed] [Google Scholar]
- 58.Wilhelm S, Tommassen J, Jaeger K-E. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6977–6986. doi: 10.1128/jb.181.22.6977-6986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]