Abstract
Extracellular vesicle (EV) secretion enables cell–cell communication in multicellular organisms. During development, EV secretion and the specific loading of signalling factors in EVs contributes to organ development and tissue differentiation. Here, we present an in vivo model to study EV secretion using the fat body and the haemolymph of the fruit fly, Drosophila melanogaster. The system makes use of tissue‐specific EV labelling and is amenable to genetic modification by RNAi. This allows the unique combination of microscopic visualisation of EVs in different organs and quantitative biochemical purification to study how EVs are generated within the cells and which factors regulate their secretion in vivo. Characterisation of the system revealed that secretion of EVs from the fat body is mainly regulated by Rab11 and Rab35, highlighting the importance of recycling Rab GTPase family members for EV secretion. We furthermore discovered a so far unknown function of Rab14 along with the kinesin Klp98A in EV biogenesis and secretion.
Keywords: Drosophila, exosomes, fat body, haemolymph, intercellular communication, Rab11, Rab35, Tsp96F
1. INTRODUCTION
Cells communicate through different forms of extracellular vesicles (EVs) that are released by discrete mechanisms and fulfil various functions depending on their cargo. They transport unique combinations of proteins, RNA/DNA and lipids to receiving cells. This capacity is frequently exploited by tumour cells to interact with each other and the surrounding stromal tissue and to modulate the tumour environment. Due to their pathogenic role in promoting tumour growth, survival and metastasis, EVs are an emerging field of research. They also mediate physiological processes such as immune responses, metabolism and reproduction. During development, EV secretion and the specific loading of signalling factors into EVs contributes to organ development and tissue differentiation (Yáñez‐Mó et al., 2015).
Investigating cell–cell communication has been predominantly done in mammalian two‐dimensional cell culture conditions. To gather three‐dimensional information, simpler model systems have to be employed, but whether this form of EV‐mediated communication also prevails in other metazoans remains unclear. EV formation was detected in several vertebrate and non‐vertebrate model organisms, including fish (Danio rerio) (Hyenne et al., 2019; Verweij et al., 2019), nematodes (Caenorhabditis elegans) (Liégeois et al., 2006), Leishmania parasites (Silverman et al., 2010), amoebae (Dictyostelium discoideum) (Kriebel et al., 2018) and fungi like the pathogenic yeasts Cryptococcus neoformans (Rodrigues et al., 2008) and Candida albicans (Silverman et al., 2010).
To understand the role of EVs in cell–cell communication in a multicellular organism, we study their biogenesis and secretion in the fruit fly Drosophila melanogaster. Many molecular mechanisms are conserved from fly to humans, and about 75% of the human genes known to cause diseases have an equivalent in the Drosophila genome. In addition, Drosophila provides a fast and easy means to manipulate any given gene to understand how it contributes to cell–cell communication in development, and always gives rise to the same structure in the adult animal. To date, Drosophila EVs were isolated from cultured cell lines to perform proteomic (Beckett et al., 2013; Koppen et al., 2011) or transcriptomic (Lefebvre et al., 2016) analyses. In the polarised Drosophila wing imaginal disc epithelium, the EV‐mediated secretion of the morphogens Wingless (Chaudhary & Boutros, 2019; Gross et al., 2012; Huang & Kornberg, 2015; Linnemannstöns et al., 2020; Witte et al., 2020) and Hedgehog (Bischoff et al., 2013; Gradilla et al., 2014; Matusek et al., 2014; Parchure et al., 2015) in the extracellular space as well as in association with cytonemes have been extensively described. EV‐mediated mechanisms have also been postulated for the presynaptic release of vesicles in the Drosophila larval neuromuscular junction (Ashley et al., 2018; Koles et al., 2012; Korkut et al., 2009 , 2013). The Drosophila male accessory gland/paragonium can be considered the homologue of the human prostate. Its secretory cells contain large intracellular membrane‐bound compartments and secrete EVs in the gland lumen (Corrigan et al., 2014; Fan et al., 2020; Redhai et al., 2016). These analyses of distinct Drosophila organs are based on immunofluorescent detection of EVs, as the experimental settings did not allow biochemical characterisation so far. Since EVs function as intercellular communication vehicles, in this study we tested if the Drosophila ‘blood’ called haemolymph contains EVs at sufficient levels for biochemical purification. This hypothesis was motivated firstly by the fact that virtually all mammalian body fluids contain EVs (Colombo et al., 2014). Secondly, previous proteomic studies identified orthologues of EV markers in the Drosophila haemolymph (Handke et al., 2013). Thirdly, flies have an open circulatory system and the haemolymph is in touch with all organs. We therefore envisioned to expand our analysis in order to establish a model for the systemic analysis of communication between haemolymph, as well as secreting and receiving organs that communicate via the haemolymph. The fat body is unique to insects and the equivalent to both vertebrate adipose tissue and liver. It is the central metabolic tissue responsible for energy/nutrient‐storage, immune response and endocrine secretion and along this line also functions as the main secreting organ (Arrese & Soulages, 2010). Dietary nutrients are absorbed in the gut and ultimately stored in fat body adipocytes as glycogen or triacylglyceride incorporated into lipid droplets (Ugrankar et al., 2019). The storage function of the fat body is vital for holometabolous insects since accumulated energy reserves will be used during metamorphosis (Arrese & Soulages, 2010).
While the larval fat body is an established model to study starvation‐induced autophagy (Lőrincz et al., 2017; Mauvezin et al., 2014), which puts the organism under extreme stress conditions, much less is known about the physiological steady state and the endosomal machinery upstream of autophagy. The endosomal system of membrane compartments includes early, recycling and late endosomes/multivesicular bodies (MVBs) and lysosomes and thereby provides a platform for sorting, recycling and degradation of internalised cargo (Klumperman & Raposo, 2014). Upon progressive maturation of early endosomes, intraluminal vesicles (ILVs) form by inward budding of the endosomal membrane, giving rise to MVBs. EVs can be shed as exosomes originating from ILVs of MVBs or as microvesicles directly from the plasma membrane. Even though the different EV subtypes originate from distinct sites within the cell, they are generated by common intracellular mechanisms and sorting machineries. This renders the analysis of their biogenesis very complex and requires standardised guidelines regarding nomenclature, separation methods and characterisation of vesicle types (Théry et al., 2018). The general term ‘extracellular vesicle’, or EV covers vesicles of both types of cellular origin and diameter without specifying origin and will be used from now on.
Multiple mechanisms can generate EVs. Both ESCRT (endosomal sorting complex required for transport)‐dependent (Baietti et al., 2012) and ‐independent (Stuffers et al., 2009; Trajkovic et al., 2008; Vaccari et al., 2009) mechanisms drive the incorporation of specific cargos into ILVs, leading to distinct MVB sub‐populations. The ESCRT family comprises four complexes (ESCRT‐0, ‐I, ‐II and ‐III) plus accessory proteins that are sequentially recruited on MVBs, induce inward membrane budding and subsequent ILV abscission (reviewed in van Niel et al. (2018)). In contrast, ESCRT‐independent mechanisms require the formation of ceramide which induces a negative curvature and thereby promotes ILV formation (Trajkovic et al., 2008). After ILV formation, MVBs follow one of two major trafficking pathways: towards the lysosome for degradation or towards the plasma membrane for fusion and release of ILVs as EVs (Palmulli & Van Niel, 2018). The final release of EVs requires the concerted action of Rab GTPases, SNAREs and other coat/tether proteins for transport, tethering and fusion of MVBs with the plasma membrane (Colombo et al., 2014). In this context, the role of Rab27 and Rab35 in EV biogenesis has been extensively characterised. Knockdown of Rab27A/B and Rab35 impairs MVB fusion with the membrane and thereby decreases exosome release in mammalian cells without affecting MVB morphology (Hsu et al., 2010; Ostrowski et al., 2010). Another Rab family member implicated in EV biogenesis is Rab11, classically recognised for its role in recycling endosome trafficking (Takahashi et al., 2012). Knockdown studies in human cells and Drosophila indicated a role for Rab11 family members in exosome secretion (Ashley et al., 2018; Beckett et al., 2013; Corrigan et al., 2014; Koles et al., 2012; Messenger et al., 2018; Savina et al., 2002) and one report even identified Rab11a‐positive MVBs in human cells (Savina et al., 2005). Rab11‐dependent EV secretion was furthermore suggest to occur specifically under metabolic stress conditions (Fan et al., 2020).
Tetraspanins are a class of transmembrane proteins, also found on neuronal surfaces, that regulate a diverse array of cellular processes (Hemler, 2003). The transmembrane proteins CD9, CD63 and CD81 of the tetraspanin family are mainly used to identify EVs (Kowal et al., 2016). Depending on the cell type they can derive both from the plasma membrane and MVBs (Mathieu et al., 2021). We generated several reporter fly lines expressing fluorescently labelled orthologues of these tetraspanins and of Rab11 (Fan et al., 2020) to allow for visualisation of EV biogenesis in different organs. Moreover, we established a technique that allows biochemical quantification of EVs from the haemolymph of larvae. Combining these approaches, we characterised haemolymph‐derived EVs in detail. Here we report our approach to establish EV secretion into haemolymph as a model system that allows both biochemical and microscopic in‐depth analysis of EV secretion in vivo. Comparing the knockdown of known factors of EV biogenesis in intracellular colocalisation studies, we confirmed a role for recycling Rab GTPases Rab11/Rab35 and identified Rab14 and Klp98A as two novel regulators of EV secretion from fat body cells into the haemolymph.
2. RESULTS
2.1. Purification and characterisation of EVs from D. melanogaster haemolymph
Drosophila is a holometabolous insect and its life cycle is divided into the four stages: embryo, larva, pupa and adult. The larvae feed and consequently grow, while molting twice into the so‐called second and third instar until they encapsulate at the end of the third instar stage to become a pre‐pupa. During pupal stage the animal undergoes a full metamorphosis leading to a complete remodelling of the body: most larval tissues are degraded and adult structures reform, finally followed by adult hatching (Figure 1A). We decided to purify EVs from the third instar larval stage for several reasons: (1) Many important developmental and physiological processes occur first in larval stage so that many gene functions are not needed in the embryo, (2) in contrast to pupa, developmental autophagy does not take place at larval stage yet (Rusten et al., 2004), (3) the third instar is still developmentally active, in contrast to adult flies and finally (4) we hypothesised that the level of communication should be high before the onset of metamorphosis.
FIGURE 1.

Purification and characterisation of EVs from D. melanogaster haemolymph. (A) Overview of the Drosophila life cycle. The third instar larval stage relevant for the following results is indicated. (B) Scheme of EV purification from L3 larval haemolymph by differential centrifugation. The P14 and P100 pellets representing large and small EVs were used for further studies. (C) Protein content of P14 and P100 is represented for five independent samples. (D) Total particle concentration per μl of resuspended P14 and P100 pellets as determined by Nanoparticle Tracking Analysis (NTA) from three independent samples. Three videos were recorded per sample and each point corresponds to one video. The line represents the mean. Size distribution of P14 and P100 were studied by electron microscopy (EM) (E–F) and NTA (G–I). (E) Representative EM image of P14 and P100 pellets. Exosome‐like vesicles with characteristic cup‐shape were discovered in both fractions. Scale bars represent 500 nm. (F) Size distribution of P14 and P100 as determined by EM of three independent samples. (G) Mean size of P14 and P100 as determined by NTA from three independent samples. Three videos were recorded per sample and each point corresponds to one video. The line represents the mean. (H) Size distribution of P14 as determined by NTA from three videos of three independent samples. (I) Size distribution of P100 as determined by NTA from three videos of three independent samples. Data are mean ± SD
We isolated haemolymph by gently opening the larvae to release undiluted haemolymph, careful not to destroy any internal organs. The haemolymph also contains blood cells called haemocytes that are partially released as well (Petraki et al., 2015). Subsequently, we performed a standard differential centrifugation‐based purification protocol (Figure 1B) to purify Drosophila EVs and characterised them according to published procedures (Kowal et al., 2016; Théry et al., 2006, 2018). For experimental standardisation, the same amount of starting material (10 μl haemolymph) was processed. Cells in the haemolymph pellet at 750 g. We use the terms P14 and P100 to refer to material pelleting at 14,000 g and subsequently at 100,000 g. While some small EVs may already pellet at 14,000 g, P100 is classically considered to contain exosomes (Théry et al., 2006). Total protein quantification was performed on both fractions and revealed that the protein content was higher in the P100 fraction (Figure 1C), yet the number of particles in P100 was significantly lower compared to P14 as determined by Nanoparticle measurement (NTA) (Figure 1D). The isolated EVs were analysed by transmission electron microscopy (TEM) to analyse if their morphology and size are comparable to mammalian EVs. Interestingly, we discovered typically shaped exosome‐like vesicles both in the P14 and P100 fraction, which were otherwise free of debris and other contaminations (Figure 1E). The ultrastructural studies revealed that the P14 fraction contains vesicles in a size range of 50–200 nm (mean diameter 116 ± 47 nm, median 108 nm), while the P100 contained vesicles in a broader range from 50 to 300 nm (mean diameter 95 ± 50 nm, median 86 nm) (Figure 1F). Nanoparticle tracking analysis (NTA) showed a mean size of about 100 nm for the P14 and above 150 nm for the P100 fraction (Figure 1G). The size distribution profile of the P14 pellet seems more similar to mammalian exosomes with a peak at 100 nm (Figure 1H), than the EVs in the P100, distributed over a broader size range and with more experimental fluctuations, as expected from less concentrated samples (Figure 1I). Strikingly, EVs isolated from cell supernatants from Kc167(Figure S1A–C) and S2 cells (Koppen et al., 2011) were found be similar to EVs released from mammalian cells, but we confirmed the peculiar size distribution of in vivo EVs also in other developmental stages (Figure S1D–O).
In summary, haemolymph from living animals contains a broad range of EVs that can be isolated by standard methods. Both TEM and NTA studies reveal more particles resembling small EVs in the P14 pellet compared to P100, suggesting in vivo Drosophila EVs pellet already at lower g‐forces compared to mammalian EVs.
2.2. Analysis of the haemolymph EV proteome
To investigate the protein composition of the secreted EVs of Drosophila haemolymph and to determine which fraction contains known EVs markers, we analysed the haemolymph‐derived EV P14 and P100 by label‐free mass spectrometry. As in vivo EV proteomics approaches are prone to contaminations, we decided to define the proteomics background by including the supernatant after P100 (SN100) into the analysis (Figure 2A). Using an annotated MS/MS spectral library derived from all three fractions, we obtained full quantitative profiles for a total of 2466 proteins by data‐independent acquisition mass spectrometry (DIA‐MS). To control for background, we filtered for P14 and P100 proteins exhibiting an at least twofold enrichment over SN100 and detection in two or more replicates. Applying these criteria, we found 1127 proteins enriched in the P14 pellet (Figure 2B) and 1114 proteins in the P100 pellet over background SN100 (Figure 2C, Table S1), respectively. Next, we performed a functional enrichment analysis of these proteins based on PANTHER classifications (Mi et al., 2013). Comparison of the respective Gene Ontology (GO) cellular compartment enrichments revealed that P14 proteins are associated with ER‐Golgi origin components, membranes and vesicles (Figure 2D), whereas P100 proteins are mostly ribosomal and non‐membrane‐bound components (Figure 2E, Table S2). Consistently, P14 revealed enrichment in GO biological processes related to vesicle trafficking, cell migration, vacuolar acidification, signalling and transport (Figure S2A). P14 enriched GO Molecular Function terms included receptor and transporter activity (Figure S2B). In contrast, proteins identified in P100 were highly enriched for terms related to ribosome assembly, cytoplasmic translation and splicing (Figure S2C) as well as proteasome activity, RNA and ribosome binding (Figure S2D). As control we also analysed the proteins enriched in the supernatant. The SN100 is enriched for GO Cellular Components of various soluble protein complexes (Figure S2E). Enriched GO Biological Processes and Molecular Function are involved in metabolism (Figure S2F,G).
FIGURE 2.

Analysis of the haemolymph EV proteome. (A) EVs were isolated from wildtype L3 larval haemolymph of three independent replicates by differential centrifugation. The composition of the resulting 14,000 g (P14) and 100,000 g (P100) pellets and remaining supernatant (SN100) was analysed by label‐free data independent acquisition mass spectrometry (DIA‐MS). (B and C) Volcano plots of all 2466 proteins quantified by DIA‐MS. Significance of enrichment was established using FDR‐based calculation with p ≤ 0.05 and S0 = 0.5, resulting in an at least 2‐fold enrichment as a requirement for significance. Proteins significantly enriched appear in blue (P14) (B) or green (P100) (C). Data points are annotated by gene name. (D and E) Enriched GO cellular components of P14 (D) and P100 (E) with a p‐value < 0.05 as determined by Fisher Exact Test with the Benjamini–Hochberg False Discovery Rate < 0.05. (F) Overlap of P14 and P100 with Drosophila orthologues of Top100 EV markers and FunRich identified markers for Drosophila. The Top100 EV markers were retrieved from EVpedia and ExoCarta and the corresponding Drosophila orthologues identified with DIOPT. FunRich database provides another Drosophila EV marker dataset based on Vesiclepedia. (G) Common proteins (annotated by gene name) identified in EVs and their closest human orthologues. Proteins found in P14 only are indicated in blue. Proteins found in P100 are indicated in green
Because the GO terms extracellular vesicle/exosome/organelle are not annotated in Drosophila yet, we next determined the prevalence of published EV markers in the haemolymph samples. For this we compared the proteins detected in our preparations to the published EV datasets EVpedia (Kim et al., 2013, 2015) and ExoCarta (Keerthikumar et al., 2016; Mathivanan et al., 2012) as well as the FunRich database (Pathan et al., 2015, 2017), which is based on Vesiclepedia (Kalra et al., 2012). The Drosophila orthologues for the Top100 human proteins published by EVpedia and ExoCarta were identified with DIOPT (Hu et al., 2011) and after removal of duplicates this returned 251 potential Drosophila EV markers. FunRich/Vesiclepedia comprises additional 540 potential marker proteins (Table S3). Comparing these published proteomes to our own dataset we discovered that the P100 proteins overlap with merely seven Drosophila orthologues of the Top100 EV markers. These include G protein subunit alphai, a proteasome subunit and chaperonins. There are 17 common proteins between P100 and FunRich, but these are mostly ribosomal protein, translation initiation factors and a proteasome subunit. Similarly, the 52 common proteins between P14, P100 and FunRich are all ribosomal proteins except for the RNA‐binding protein Arc1, which was shown to be secreted on EVs before (Ashley et al., 2018) (Figure 2F,G, Table S4). In contrast, much more proteins from P14 overlap with Top100 EV and/or FunRich (Figure 2F,G). Among these we found a number of interesting potential Drosophila EV markers, including orthologues of the intensively studied CD81 (Dm Tsp96F, common to P14 and TopEV), and two other tetraspanins, Dm Tsp42Ed and Tsp42Ee (common to P14, Top100 EV and FunRich. The closest human orthologue of the latter two proteins is the exosomal marker CD63 (DIOPT v8.0). We also retrieved the small GTPase ARF6 (Dm Arf51F, common to P14 and FunRich), RalA (Dm RalA, common to all) and the ESCRT component Tsg101 (common to P14, Top100 EV and FunRich). Other promising markers identified comprise members of the Annexin and solute carrier (SLC) families, as well as CK1γ kinase (Dm Gish), an ATPase (ATP6V0D1, Dm VhaAC39‐1) and one integrin family member (Integrin beta 1, Dm Myospheroid) (Figure 2F). 200/388 proteins enriched in the supernatant did not overlap with Top100EV and/or FunRich listed proteins and the overlapping 77 are proteins of low abundance on EVs (Figure S2 H, Table S4) (Pathan et al., 2019). Taken together, in agreement with TEM and NTA data, proteomic analyses confirmed that the haemolymph particles isolated at 14,000 g contain many more markers, which are typically associated with mammalian P100‐EVs/exosomes, then those particles isolated at 100,000 g.
2.3. Identification and characterisation of novel Drosophila EV markers
Combining our proteomic analysis and previous studies performed in cell culture (Beckett et al., 2013; Gradilla et al., 2014; Parchure et al., 2015), we assembled a list of potential in vivo EV markers (Figure S3A). Candidates (with available antibodies) were validated by immunoblot analysis of P14, P100 pellets and their respective cell lysates (P0.75) obtained from wildtype haemolymph. The cis‐Golgi marker GM130 served as negative control (Figure 3A, left column). We were able to detect the membrane trafficking protein Cystein string protein (Csp‐1 and ‐2), the tetraspanin Tsp42Em/Late bloomer (Lbm), IntegrinβPS/Myospheroid as well as Syntaxin 1A (Syx1A) in the P14 and Csp‐1 and ‐2 as well as Lbm also weakly in the P100. Weak signals for the ESCRT component Hrs (Hepatocyte growth factor regulated tyrosine kinase substrate) and the lysosomal protein Lamp1 (Lysosome‐associated membrane glycoprotein) came up in the P100. Antibodies against both human and Drosophila Alix, Flotillin‐2 and Tsg101 failed to detect the respective proteins (data not shown). We did not have an antibody against the transmembrane v‐ATPase component VhaAC39‐1/ATP6V0D1, but an antibody against human transmembrane ATP6V0A1 gave signals in P14 and P100. For the plasma membrane protein Basigin (Bsg), a mammalian marker for large EVs (Menck et al., 2015), and another Tetraspanin Tsp42Ee no antibodies were available either, but we were able to detect respective bands in P14, but not in P100 by using endogenous GFP insertion lines. Furthermore, we could also retrieve bands in both P14 and P100 for Hsp70 (heat shock protein 70), α‐Tubulin and the small GTPase Rab11 (Figure 3A, right column).
FIGURE 3.

Identification and characterisation of novel Drosophila EV markers. (A) EVs were isolated from wildtype L3 larval haemolymph by differential centrifugation and the resulting pellets analysed by immunoblot for previously published markers as indicated. (B–F) Overexpression of different tagged EV markers. (B) Transgenes encoding tagged EV markers were expressed with Collagen IV‐GAL4 (Cg‐GAL4), which is active on both the fat body and haemocytes. Haemolymph EVs were isolated from the corresponding progeny and analysed for both endogenous and overexpressed EV markers. (C) Overexpression of mouse UAS‐CD63‐pHluorin using CgGAL4. CD63‐pHluorin could be detected in the P0.75 and P14 of respective haemolymph EVs. (D) Close up of a phylogenetic tree of human CD9, CD63 and CD81 against all 36 Drosophila tetraspanin sequences. The full tree can be found in the Supplementary Material. (E) Overexpression of UAS‐mCherry‐Tsp96F using CgGAL4. mCherry‐Tsp96F could be detected in the P0.75 and P14 of respective haemolymph EVs. (F) CgGAL4 > UAS‐mCherry‐Tsp96F derived P14 EVs were treated with proteinase K in the presence or absence of TritonX‐100. The internal N‐terminal tag is protected and the extracellular part of Tsp96F is digested by proteinase K alone (lane 2). In the presence of detergent, the whole protein is degraded (lane 4) (G) CgGAL4, GFP‐Rab11 derived haemolymph was diluted with increasing amounts of wildtype haemolymph prior to EV isolation. (H) Quantification of (G). The dotted black line represents the haemolymph ratio. The solid‐colored lines represent the Rab11‐GFP signal normalised to endogenous controls as indicated. Data are mean ± SD. All immunoblots depicted are representative blots for at least three independent experiments
Since the expression of these markers is rather weak and/or variable and also with the perspective to label EVs in vivo, we decided to develop several fluorescently labelled markers in addition to the endogenous markers mentioned. This approach allows tissue‐ and time‐controlled specific expression of EV markers and thereby tracking the fate of labelled EVs using the versatile UAS/GAL4 system (Brand & Perrimon, 1993). Previously, a GFP‐tagged human CD63 construct was successfully expressed in flies to label exosomes (Corrigan et al., 2014; Gradilla et al., 2014; Gross et al., 2012; Panáková et al., 2005) and overexpression of mouse CD63‐GFP was shown to have the same effect on lysosomes as overexpression of the Drosophila orthologue Tsp29Fa (Ma et al., 2020).
Here, we generated transgenic flies expressing pHluorin‐tagged mouse CD63. Using a Collagen IV‐GAL4 (Cg‐GAL4) driver, which is active in both the fat body and haemocytes (Asha et al., 2003) (Figure 3B), we purified EVs from haemolymph. CD63‐pHLuorin could be detected in the P14 of haemolymph EVs (Figure 3C). The tetraspanins CD9 (Théry et al., 1999), CD63 and CD81(Escola et al., 1998) have been used as exosomal markers for the last two decades, with CD63 enriched in MVBs and CD9 and CD81 at the plasma membrane. To be able to rely on endogenous markers other than mammalian CD63, we aligned the protein sequences of human CD9, CD63 and CD81 against all 36 Drosophila tetraspanin sequences. This phylogenetic analysis revealed that Tsp96F is the closest homolog to both CD9 and CD81 (Figure 3D and S3B). We generated transgenic flies for N‐terminally mCherry‐tagged Tsp96F and expressed it with CgGAL4. mCherry‐Tsp96F is also secreted on EVs, since we could detect it in the P14 (Figure 3E) and in rare cases the P100 (Figure S3C). As expected for an N‐terminally tagged tetraspanin, mCherry‐Tsp96F retains its label upon proteinase K treatment, but is completely degraded following detergent solubilisation (Figure 3F). Similarly, endogenous Lbm, which is detected by an antibody against the extracellular domain, is trimmed by proteinase K alone and completely degraded in the presence of detergent (Figure S3D) This underlines that P14 EVs indeed originate from the endosomal system and carry tetraspanins in agreement with expected EV topology.
Finally, Rab11‐GFP positive exosomes were shown to be secreted from male accessory gland cells (Fan et al., 2020; Marie et al., 2020) and increasing evidence points at EV secretion from recycling endosomes (Beckett et al., 2013; Fan et al., 2020; Koles et al., 2012; Messenger et al., 2018; Savina et al., 2005, 2002). In line with these results, we identified CK1γ kinase by DIA Mass Spec, which is involved in Rab11‐mediated vesicle trafficking (Gault et al., 2012). We tested the expression of Rab11‐GFP with CgGAL4 and found Rab11‐GFP to be secreted in P14 and P100 (Figure S3C). We attempted to estimate the amount of fatbody‐derived EV in comparison to total haemolymph EV by two different approaches: (1) We compared CgGAL4‐expressed Rab11‐GFP with an endogenous Myc‐YFP‐Rab11 insertion (Dunst et al., 2015) as EV marker in haemolymph‐isolated EVs from both fly lines and (2) we diluted CgGAL4,‐expressed Rab11‐GFP haemolymph with increasing amounts of wild‐type haemolymph.
Regarding (1), the amount of UAS‐Rab11‐GFP (50 kD) can only originate from the fat body, while endogenous Rab11 (20 kD) and endogenous Rab11‐YFP (62 kD) supposedly come from all the cells that secrete EVs into the haemolymph. Indeed, we found both endogenous and overexpressed Rab11 to be secreted in P14 and P100 (Figure S3E). No strong differences between three biological replicates indicated a quantitative purification of EVs in P14 and P100. Similar levels of endogenous Rab11 (20 kD) in samples from the endogenous trap line and the UAS‐mediated overexpression line were detected with an anti‐rab11 antibody (Figure S3E, left panel P0.75, P14 and P100), indicating that the amount of endogenous Rab11 are similar in cells (P0.75), but also on EVs (P14, P100). In contrast, levels of Rab11‐GFP (50 kD) overexpressed from the fat body exceeded endogenous Rab11 (20 kD) in P14 and P100 several fold, showing that the amount of overexpressed Rab11 originating from the fat body significantly contributes to overall EVs in the haemolymph (Figure S3E). Consistently, diluting haemolymph from CgGAL4 > Rab11‐GFP larvae with wildtype haemolymph results in a decay of the Rab11‐GFP signal, while endogenously expressed proteins remain constant (Figure 3G,H). From these two experimental comparisons, we speculate that the fat body is a major contributor of EVs to the overall EV content in the haemolymph. Taken together, we confirmed the secretion of several overexpressed EV markers from fat body into the haemolymph that we identified by proteomics or that were previously detected on cell culture‐derived EVs. Our data therefore indicate that our model can reflect general aspects of EV secretion in vivo with the special feature of P14 EVs originating from the endosomal pathway.
2.4. Novel EV markers localise to distinct compartments in fat body cells
Next, we were interested in the subcellular localisation of these proteins. For this, we expressed the markers via UAS/Cg‐GAL4 in the fat body and analysed the tissue by immunofluorescence microscopy (Figure 4A,B). The larval fat body is composed of single‐cell layers of adipocytes. In the wandering L3 stage, the cells are flat and polygonal and tightly associated with each other. Along the entire anterior–posterior axis they organise in thin sheets that are suspended in the haemolymph (Nelliot et al., 2006). With a diameter of roughly 50 μm fat body cells are comparatively large and easy to microscope. They are filled with lipid droplets of variable sizes ranging from 0.5 to 10 μm in diameter (Beller et al., 2006). Lipid droplets appear as black holes when cytoplasmic eGFP is overexpressed (Figure 4B). We co‐overexpressed cytoplasmic eGFP and mCherry‐Tsp96F to distinguish lipid droplets, cytoplasm and luminal structures. Overexpressed Tsp96F does not co‐localise with lipid droplets, but labels vacuolar structures in the cytoplasm with a discernable lumen (Figure 4C,D). Tsp96F localises mainly to the limiting membrane of these vacuoles but can also be detected in internal puncta and ILVs (Figure 4D). To better understand the identity of these vacuolar structures we stained mCherry‐Tsp96F‐expresssing fat bodies with different organelle markers. mCherry‐Tsp96F‐positive structures did neither co‐localise with the ER‐marker Calnexin (Cnx) (Figure S4A), nor the Golgi marker Golgin (Figure S4B). We did not observe any co‐localisation with the early endosome marker Rab5 (Figure 4E), the ESCRT‐0 component Hrs (Figure 4F), and the late endosome marker Rab7 (Figure 4G) either. Strikingly, a subset of Tsp96F‐mCherry‐positive structures co‐localises with the tetraspanin Lbm. Lbm furthermore appears to label intraluminal puncta within the large vacuolar structures (Figure 4H,I). Next, we co‐expressed mCherry‐Tsp96F with Lamp1‐GFP and CD63‐pHluorin (Figure 4J–L). This revealed that the mCherry‐Tsp96F positive compartment also co‐localises with overexpressed eGFP‐tagged Lamp1 (Figure 4J). The Lamp1 construct contains the transmembrane domain and cytoplasmic tail, which is sufficient to direct proteins to lysosomes, from human Lamp1 (Pulipparacharuvil et al., 2005) and has been successfully used in Drosophila to label lysosomes (Jacomin et al., 2016). Consistently, mCherry‐Tsp96F positive compartments are positive for LysoTracker and LysoSensor, indicating acidification (Figure S4 C, D). Tsp96F‐mCherry and CD63‐pHluorin localise to the same intracellular compartment (Figure 4K,L). The localisation of overexpressed CD63‐pHluorin reflects the expression of endogenously tagged Tsp42Ee, which is most closely related to human CD63 (Figure S4E,F), demonstrating that the formation of these large structures is not caused by overexpression.
FIGURE 4.

Novel EV markers localise to distinct compartments in fat body cells. (A) Transgenes encoding tagged EV markers were expressed with Collagen IV‐GAL4 (Cg‐GAL4), which is active in both the fat body and haemocytes. Fat bodies were dissected from the corresponding progeny and analysed for both endogenous and overexpressed EV markers by immunofluorescence. (B) Overview of the fat body composed of flat polygonal adipocytes visualised by expression of cytoplasmic eGFP and co‐staining with Phalloidin to label F‐actin. (C) Co‐expression of eGFP and Tsp96F‐mCherry. Tsp96F localises to large vacuolar structures containing a lumen. Lipid droplets appear as GFP‐negative holes. (D) Magnified view of the boxed region in (C). The filled arrows point at the membrane of Tsp96F labelled vacuoles, the filled arrow heads point at internal puncta, the empty arrow heads point at intraluminal vesicles, the asterisks label lipid droplets. (E‐G) mCherry‐Tsp96F was expressed and co‐stained with endogenous endosomal markers Rab5 (E), Hrs (F), Rab7 (G). (H) The tetraspanin Lbm labels a subset of Tsp96F‐mCherry positive structures. (I) Magnified view of the boxed region in (H). mCherry‐Tsp96F co‐localises to the same compartment with eGFP‐Lamp1 (J) and CD63‐pHluorin (K). (L) Magnified view of the boxed region in (K). (M–P) Rab11‐GFP was expressed alone (M) and co‐stained with endogenous endosomal markers Rab5 (N), Hrs (O), Rab7 (P). (Q) Endogenous Myc‐YFP‐Rab11 reflects the localisation of overexpressed Rab11‐GFP in (M‐P). (R) Co‐expression of Rab11‐GFP and mCherry‐Tsp96F. Rab11‐GFP and Tsp96F‐mCherry mark different compartments. (S) Magnified view of the boxed region in (R). All images show representative confocal images. Scale bar represents 50 μm in (B) and 10 μm in all other images
Overexpressed UAS‐Rab11‐GFP localises to small puncta or larger endosomal structures as was described before (Figure 4L) (Shandala et al., 2011; Sorvina et al., 2016, 2020). Next, we also co‐stained Rab11‐GFP‐expressing fat bodies with a panel of organelle markers. Rab11‐GFP‐positive structures did neither co‐localise with the ER‐marker Cnx (Figure S4G), nor the Golgi marker Golgin (Figure S4H). We observed a mostly distinct localisation with partial co‐localisation with the early endosome marker Rab5 (Figure 4N), the ESCRT‐0 component Hrs (Figure 4O) and the late endosome markers Rab7 (Figure 4P). The same was observed for endogenously eYFP‐Myc‐tagged Rab11, indicating that overexpression of tagged Rab11 does not perturb its localisation (Figure 4Q). Next, we co‐expressed mCherry‐Tsp96F with Rab11‐GFP (Figure 4R). Rab11‐GFP positive puncta are in close proximity to the larger mCherry‐Tsp96F expressing luminal structures (Figure 4R,S). Rab11‐GFP also localises proximal to large acidic compartments in the absence of mCherry‐Tsp96F (Figure S4I). This suggests that Rab11 and Tsp96F mark partially overlapping endosomal compartments and contribute to subpopulations of EVs as previously shown (Fan et al., 2020).
These immunofluorescence studies confirm that prior to secretion, EV markers can be detected intracellularly and localise to late endosomal compartments, marking diverging sorting routes towards the extracellular space.
2.5. Fat‐body specific knockdown of ESCRT and RAB components can alter the amount of EVs secreted in the haemolymph
So far, EV‐focused biochemical studies have only been performed with Drosophila cell culture supernatants and in vivo analyses were based on immunofluorescence analyses. Encouraged by the overexpressed and endogenous rab11 comparison, we asked whether RNAi‐induced changes in EV secretion from the fat body, as the biggest organ in contact with haemolymph, can be detected biochemically by Western Blot analysis of haemolymph EV. Flies were generated that co‐expressed Rab11‐GFP or mCherry‐Tsp96F together with RNAi targeting the respective genes or a control RNAi (targeting the gene yellow) using the UAS/GAL4 system and Cg‐GAL4 as fat body specific driver (Figure 5A). No single EV marker is known to distribute evenly across all EV subtypes, so it is advisable to use several different antibodies to reliably normalise signals on EV Western Blots. To standardise our analysis in different knockdown conditions, we isolated EVs from an equivalent volume of haemolymph (10 μl), loaded the resulting EV preparations (P14 and P100) and directly compared the Western Blot signals of candidates and control (Figure 5A,B). We probed for the overexpressed EV proteins, either Rab11‐GFP or Tsp96F‐mCherry, and the endogenous EV markers Lbm and Syx1A (Figure 5B, representative blots for each candidate are depicted in Figure S5).
FIGURE 5.

Fat‐body mediated knockdown of ESCRT and RAB components can alter the amount of EVs secreted in the haemolymph. (A) Tagged EV markers were co‐expressed with RNAi using Cg‐GAL4. EVs were isolated from the corresponding L3 larval haemolymph and the resulting P14 and P100 pellets analysed by immunoblot. (B) Representative immunoblot probing for Rab11‐GFP and endogenous Syx1A and Lbm. RNAi against yellow was used as control. The intensity of the endogenous markers in P100 was too low to be quantified. (C) List of all knockdown candidates. (D) Quantification of Rab11‐GFP, Lbm and Syx1A signal intensity upon knockdown of ESCRT components Hrs, Tsg101, Shrb and Alix. One‐way ANOVA: *p = 0.00238. (E) Quantification of Rab11‐GFP, Lbm and Syx1A signal intensity upon knockdown of Rab7, Rab27 and Rab35. One‐way ANOVA: *p = 0.0451. (F) Quantification of Tsp96F‐mCherry, Lbm and Syx1A signal intensity upon knockdown of Rab11. Student's unpaired two‐tailed t‐test: **p = 0.0048 (P14 Tsp96F), **p = 0.0058 (P14 Lbm), *p = 0.0192, ****p < 0.0001. All quantifications were performed on at least three independent experiments. Data are mean ± SD
To test our system, we chose several candidates for knockdown based on their previously described functions in EV biogenesis and secretion: Rab GTPases Rab27 and Rab35, which are well characterised to mediate MVB fusion with the plasma membrane, the late endosome marker Rab7 and Rab11, associated with recycling endosomes. Furthermore, we chose members of the different ESCRT complexes, namely Hrs (ESCRT‐0), Tsg101 (ESCRT‐I), Shrb, homolog of human Chmp4B, (ESCRT‐III) and Alix (ESCRT accessory) (Figure 5C).
We quantified the signal intensities of Rab11‐GFP or Tsp96F‐mCherry, Syx1A and Lbm in P14 and those of Rab11‐GFP or Tsp96F‐mCherry in P100. The intensity of the endogenous markers was too low to be quantified in P100. As could be expected from an in vivo approach, variation between biological replicates was high and most of the multiple candidates and different RNAi lines per candidate did not alter EV secretion significantly. For example, the Rab11‐GFP and Syx1A signal in P14 were slightly increased upon knockdown of Tsg101, however this effect could not be reproduced with a second RNAi line (Figure 5D). In general, knockdown of different ESCRT components showed high data variation and did not allow clear conclusions (Figure 5D), although the lines used showed effects in other systems (Matusek et al., 2014). However, there was a 50% reduction of Lbm and Syx1A along with increase of Rab11‐GFP in P14 after Rab35 knockdown using two different Rab35 RNAi lines (Figure 5E). This is in line with studies from mouse oligodendroglial cells, where Rab35 was shown to mediate EV secretion (Frühbeis et al., 2013; Hsu et al., 2010). In the case of Rab11 RNAi, only Tsp96F‐mCherry was co‐expressed (Figure 5F). Knockdown of Rab11 led to a significant reduction of Tsp96F, but to an increase of Lbm and Syx1A in P14 and a concomitant increase of the detectable Tsp96F marker in P100 (Figure 5F). It has been observed before that perturbing EV biogenesis by neutral sphingomyelinase inhibition can shift EV markers from P14 to P100 (Menck et al., 2017).
Taken together, fat‐body mediated knockdown of a number of previously described mediators of EV biogenesis, including Rab7, Rab27 and several ESCRT components did not lead to a reduction of EV marker intensity as detected biochemically. Albeit the observed data variability of this in vivo approach, knockdown of Rab35 and Rab11 strongly reduced secretion of some EV markers. This supports previous findings that different EVs populations exist, and that they are secreted by multiple trafficking routes and in a cell‐type specific manner into the extracellular space.
2.6. ESCRT and RAB knockdown regulates EV trafficking in fat body cells
Since capturing changes in haemolymph EV levels by Western Blot was not trivial, we wanted to test in parallel if knockdown of the respective candidates affected endosomal compartments and trafficking of EV markers on a subcellular level. For this, we co‐expressed Rab11‐GFP together with RNAi targeting the respective genes or a control RNAi (yellow) using the UAS/GAL4 system and dissected the fat bodies from wandering L3 larvae (Figure 6A). None of the knockdown conditions analysed was lethal. The larvae developed, pupated and hatched normally, indicating that overall fat body function was not impaired.
FIGURE 6.
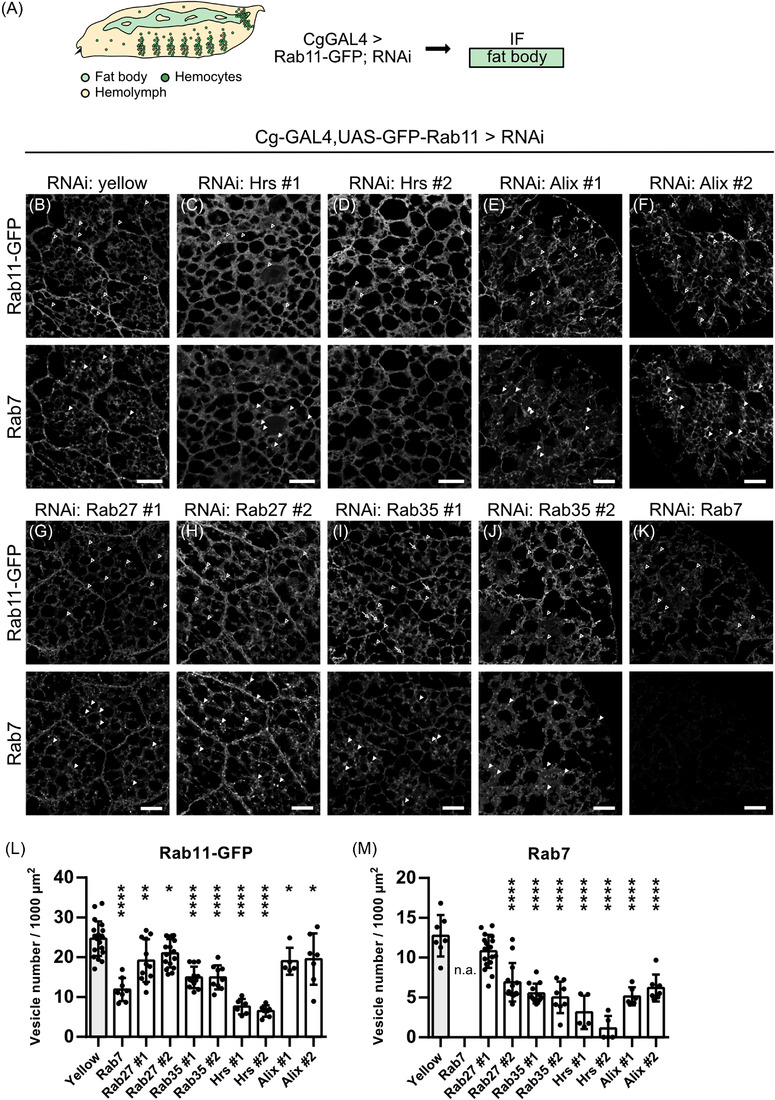
ESCRT and RAB knockdown regulates EV trafficking in fat body cells. (A) Rab11‐GFP was co‐expressed with RNAi using Cg‐GAL4. Fat bodies were dissected from the corresponding progeny and analysed by immunofluorescence. (B) RNAi against yellow is used as control. Fat bodies expressing RNAi against Hrs (C, D), Alix (E, F), Rab27 (G, H), Rab35 (I and J) and Rab7 (K) were stained for Rab11‐GFP (upper panel) and endogenous Rab7 (lower panel). Empty arrowheads in (B, upper panel) point at Rab11‐GFP positive puncta. Arrowheads in (B, lower panel) point at Rab7‐positive ring‐like structures. Arrows in (I, J) point at Rab11‐GFP mislocalising to Rab7‐positive ring‐like structures (L) Quantification of number of Rab11‐GFP positive vesicles (upper panel in B‐K). One‐way ANOVA: ****p < 0.0001, **p = 0.0017 (Rab27 #1), *p = 0.0424 (Rab27 #2), *p = 0.0318 (Alix #1), *p = 0.0256 (Alix #2). (M) Quantification of number of Rab7 positive vesicles (lower panel in B‐K). One‐way ANOVA: ****p < 0.0001. Data are mean ± SD. All images show representative confocal images and maximum intensity projection of two sections (distance 1 μm) are depicted for visualisation. Scale bar 20 μm
We stained against a panel of organelle markers, including Cnx (ER), Golgin84 (Golgi), Rab5 (early endosome), Hrs (ECSRT‐0/MVBs) and Rab7 (late endosome) and quantified the number of the respective puncta. We assumed that the number of exosome‐forming compartments could be affected by both upstream trafficking pathways or by secretion rate. The ER marker Cnx is diffusely distributed all over the cell and this localisation was not affected by any of the knockdown conditions tested (Figure S6A). Knockdown of any candidate did neither show an effect on Golgin‐positive puncta number or distribution, indicating that Golgi function was not impaired either (Figure S6B,E).
The control knockdown of yellow revealed a vesicular staining for the EV marker Rab11‐GFP (Figure 6B) as was observed before (Figure 4L–Q). Upon knockdown of the ESCRT‐0 component Hrs the Rab11‐GFP signal was rather diffuse and puncta were almost not discernable (Figure 6C,D), therefore the number of Rab11‐GFP puncta was strongly reduced (Figure 6L). Knockdown of the ESCRT accessory member Alix (Figure 6E,F) and the Rab GTPase Rab27 (Figure 6G,H) led to slight reductions of Rab11‐GFP positive puncta (Figure 6L). However, knockdown of the recycling endosome component Rab35 (Figure 6I,J) and the late endosomal marker Rab7 (Figure 6K) led to significant reductions of intracellular Rab11‐GFP positive puncta (Figure 6L). Interestingly, Rab35 knockdown leads to mislocalisation of the signal to the cortex of luminal structures resembling Rab7 endosomes, indicating that sorting or fusion is disturbed (Figure 6I,J). Since knockdown of Rab7, Rab35 and Hrs had strong effects on the Rab11‐GFP signal distribution, we wanted to see how other endosomal compartments behave upon knockdown. Rab5‐labelled early endosomes were distributed over the cell and also located at the cell membrane in the control (Figure S6C, top). Hrs‐positive endosomes/MVBs are mostly found intracellularly and not at the cortex (Figure S6D, top). Knockdown of Rab27 and Hrs did not change the number of Rab5‐ and Hrs‐positive puncta (Figure S6 C,D,F,G). The late endosome marker Rab7 localises to ring‐like structures in the control knockdown of yellow (Figure 6B, bottom) as was seen before (Figure 4G,N). The number of Rab7‐positive late endosomes was significantly reduced by knockdown of all candidates analysed (Figure 6C–K,M).
In summary, different endosomal maturation steps, among them steps necessary for EV biogenesis, can be monitored in the Drosophila fat body. Knockdown of several known factors involved in endosomal trafficking including Rab GTPases and the ESCRT‐0 member Hrs led to significant perturbations in the number and distribution of the EV marker Rab11‐GFP as well as the late endosomal marker Rab7, complementing and expanding our biochemical analysis.
2.7. Fat‐body mediated knockdown of Klp98A/Rab14 affects EV secretion in the haemolymph
Based on these findings with established EV biogenesis components, we next investigated EV secretion upon knockdown of so far undescribed candidates to gain insights into the regulation of EV secretion in fat bodies. We analysed the role of Rab14 and the kinesin Klp98A, which recently were described in Drosophila in lysosomal positioning during autophagy in fat bodies and Wingless secretion from wing imaginal disc cells (Mauvezin et al., 2016; Witte et al., 2020).
In our DIA‐MS analysis we detected Rab14 secreted on P14 EVs (Figure 2B,G). Rab14 knockdown in fat body cells did not alter levels of secreted EV markers isolated from haemolymph, but Klp98A knockdown reduced Lbm levels in P14, whereas all other markers were not affected in immunoblots (Figures 7A, S7A). The same trend was observed in Kc167 cells (Figure S7B,C). Immunofluorescence analysis of fat bodies revealed an opposite effect, namely that Klp98A knockdown did not affect the number of Rab11‐GFP, Rab5, Hrs‐ and Rab7‐positive puncta (Figure 7B,C,E–H). In contrast, Rab14 knockdown significantly reduced the number of Rab11‐GFP puncta (Figure 7B,D,E), as well as the number of Hrs‐ and Rab7‐positive puncta (Figure 7B,D,G,H). The number of Rab5‐positive endosomes was not affected by Rab14 knockdown (Figure 7B,D,F). Golgin‐positive puncta number or distribution was neither affected by Klp98A nor Rab14 knockdown, indicating that Golgi function was not impaired (Figure S7D). The trafficking pathways at the level of endosomes are diverse. Specificity and directionality of sorting depend on the Rab GTPases involved as well as overlapping effectors present on endosomal subpopulations (Nag et al., 2018; Sorvina et al., 2016; Yamamoto et al., 2010). To check whether knockdown affects organisation of these subpopulations, we analysed the colocalisation of Rab11‐GFP, Rab5, Rab7 and Hrs on endosomes upon RNAi. Since endosomal maturation occurs by a Rab5 to Rab7 conversion (Chavrier et al., 1990; Feng et al., 1995; Rink et al., 2005), changes in colocalisation could indicate maturation defects as well as shifting from one trafficking pathway to another (Chavrier et al., 1990). Rab14 knockdown significantly reduced Rab11‐GFP/Rab5 colocalisation (Figure 7I), whereas Rab11‐GFP/Hrs and Rab11‐GFP/Rab7 colocalisation were not affected (Figure 7J,K). As the number of Rab5‐positive puncta is not changed (Figure S7B,D,F), the decreased Rab11‐GFP/Rab5 colocalisation is likely caused by the reduced number of Rab11‐GFP puncta. This suggests that Rab14 knockdown specifically affects the recycling of Rab11 towards early endosomes at the dispense of late endosomal compartments. In contrast, Klp98A knockdown only affects late Rab7‐positive endosomes.
FIGURE 7.
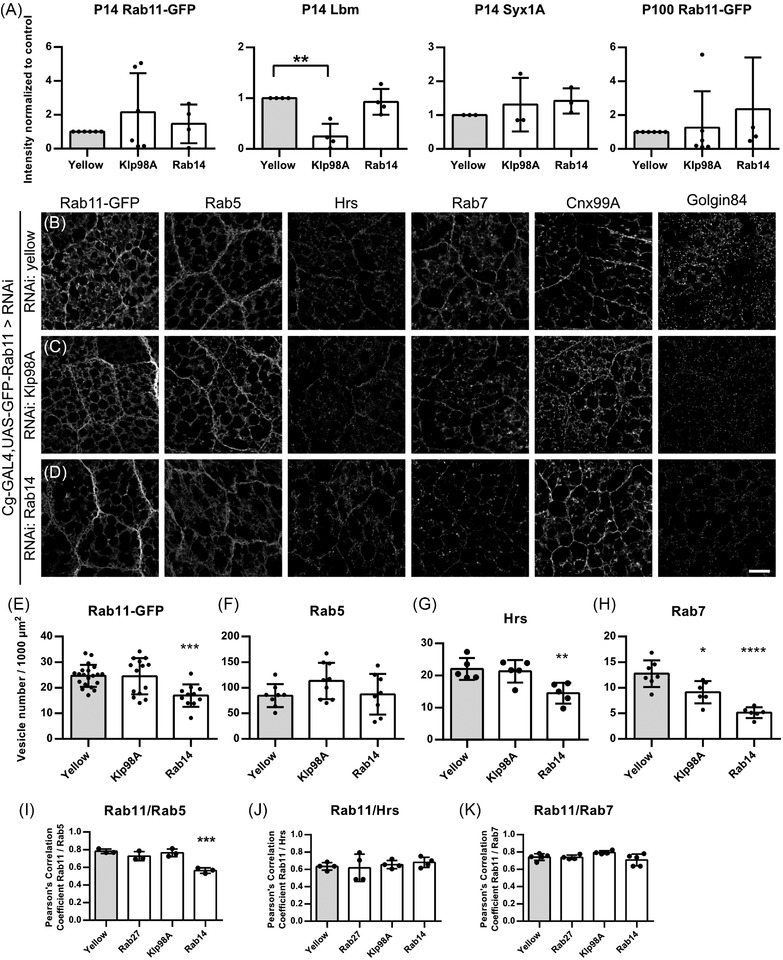
Fat‐body mediated knockdown of Klp98A/Rab14 affects EV secretion in the haemolymph. (A) Quantification of Rab11‐GFP, Lbm and Syx1A signal intensity upon knock down of Klp98A and Rab14. Quantifications were performed on at least three independent experiments. Data are mean ± SD. One‐way ANOVA: **p = 0.0011. (B‐D) Rab11‐GFP was co‐expressed with RNAi using Cg‐GAL4. Fat bodies were dissected from the corresponding progeny and analysed by immunofluorescence. RNAi against yellow was used as control. Fat bodies expressing RNAi against yellow (B), Klp98A (C) and Rab14 (D) were stained for Rab11‐GFP as well as endogenous Rab5, Hrs, Rab7, Cnx99A and Golgin as indicated. (E) Quantification of number of Rab11‐GFP positive vesicles in (B–D). One‐way ANOVA: ***p = 0.0008. (F) Quantification of number of Rab5‐positive vesicles in (B–D). (G) Quantification of number of Hrs‐positive vesicles in (B–D). One‐way ANOVA: **p = 0.0078. (H) Quantification of number of Rab7 positive vesicles in (B–D). One‐way ANOVA: *p = 0.0124, ****p < 0.0001. (I) Pearson's correlation coefficient of Rab11‐GFP/Rab5 is decreased upon Rab14 knockdown. One‐way ANOVA: ***p = 0.0004. Pearson's correlation coefficient of Rab11‐GFP/Hrs (J) and Rab11‐GFP/Rab7 (K) is not affected by the knockdown conditions tested. Data are mean ± SD. All images show representative confocal images and maximum intensity projection of two sections (distance 1 μm) are depicted for visualisation. Scale bar 20 μm
Taken together, we could show a novel role of Klp98A and Rab14 in regulating EV secretion. However, they do not appear to function at the same level of EV compartment trafficking in contrast to their previously described interaction in autophagosome positioning (Mauvezin et al., 2016), but affect discrete sorting steps in the endosomal system.
3. DISCUSSION
Propagation of endogenous exosomes in vivo is largely unknown, and data on their biogenesis and role in normal developing tissue and adult tissue homeostasis lags behind. Drosophila is one of the best organisms to study intercellular and ‐organ communication. The same genes and principles underlie fly and human development, so human diseases can be modelled in the fly and analysed on the molecular and cellular level (Droujinine & Perrimon, 2016). Drosophila has functionally corresponding organ systems to mammals. The work presented here establishes the fat body, equivalent to fat and liver, as a genetic model to study EV biogenesis in vivo. Our system allows both biochemical quantification of EVs from the haemolymph of larvae and expression of fluorescently tagged EV proteins to understand principles of interorgan communication by EVs.
We purified EVs from haemolymph by differential centrifugation and analysed the resulting pellets by Western Blot, NTA, EM and DIA mass Spectrometry. Both TEM and NTA studies suggest that fly EVs have a different pelleting behaviour compared to mammalian EVs. The majority of EVs, with the size and morphology of exosomes, can be pelleted at 14,000 g. So far, particle characterisation in Drosophila relied on purification of EVs from cell culture supernatants. For this, Kc167 and S2 cells (Koppen et al., 2011), derived from embryonic haemocytes, as well as Dm‐D17‐c3 (D17) and S2R+ cells (Lefebvre et al., 2016) were used. Drosophila cells are cultured in the same serum (FBS)‐containing medium as mammalian cells and generally switched to serum (FBS)‐free conditions only shortly before the start of isolation. We suspect that serum‐derived lipids are incorporated into membranes while cultured in the presence of serum (FBS). This might interfere with EV composition leading to the fact that cell culture studies do not reflect the in vivo situation. Drosophila membranes contain sphingolipids, but their composition is different compared to mammalian membranes. They lack sphingomyelin, and instead have phosphoethanolamine ceramide (PE‐ceramide) as a major membrane constituent. In PE‐ceramide, a phosphoethanolamine replaces the phosphocholine in sphingomyelin (Hildenbrandt et al., 1971). Interestingly, basic components for the synthesis of sphingolipids exist in Drosophila, including several sphingomyelinase homologs (Acharya & Acharya, 2005). In mammals, ceramide is generated via breaking down sphingomyelin by neutral and acid sphingomyelinases. Moreover, Drosophila lack all enzymes required for the synthesis of cholesterol and the sterol content in fly membranes is less compared to mammalian membranes (18% vs. 30%) (Rietveld et al., 1999). Nevertheless, they are capable of utilising ingested cholesterol for post‐translational modification of substrates like Hedgehog (Acharya & Acharya, 2005; Callejo et al., 2006; Dawber et al., 2005; Gallet et al., 2006). We suspect that Kc167 cells can incorporate and metabolise sphingomyelin and cholesterol from the cell culture medium supplemented with bovine serum and that this may influence EV lipid composition and result in the observed differences in pelleting behaviour.
Out of the described human EV markers we identified > 100 in Drosophila, including common exosome markers such as different Tetraspanins and Rab GTPases. Out of these, we decided to focus on overexpressed Rab11‐GFP, CD63‐pHluorin and mCherry‐Tsp96F as markers for immunofluorescence and ‐blot. Rab11‐GFP and tagged tetraspanins label distinct EV subpopulations within fat body cells. Rab11‐GFP positive puncta do not overlap with any of the other endosomal markers tested, underlining their unique identity as recycling endosomal MVBs (Fan et al., 2020). The expression of tagged Tsp96F and CD63 overlap and mCherry‐Tsp96F positive structures colocalised with endogenous Lbm and GFP‐Lamp1. Previously, it was postulated that the GFP‐LAMP1 fusion protein identified large perinuclear structures corresponding to lysosomes as well as smaller structures evenly distributed in the cytoplasm (Jacomin et al., 2016). A recent report, however, describes the secretion of Lamp1/2 on EVs from HeLa cells, indicating that Lamp1/2 does not only mark endosomes destined for lysosomal degradation (Mathieu et al., 2021). The tetraspanin Lbm/Tsp42Em was originally described to facilitate neuromuscular junction formation in larvae (Kopczynski et al., 1996) and localises to a cluster of 18 Tsp on the second chromosome (Fradkin et al., 2002; Todres et al., 2000).
Regarding immunoblot analysis, no single EV marker is known so far to distribute evenly across all EV subtypes that could be used for normalisation. We could not load complete haemolymph for normalisation because it is very protein‐rich and therefore only produces a smear so that no clear bands can be detected. It is neither possible to normalise against the P0.75 cell pellet containing the Drosophila blood cells, because the number of haemocytes varies between preparations and cannot be standardised since the larvae are punctured manually. Therefore, as described, we always isolated EVs from an equivalent volume of haemolymph, loaded the whole EV preparation and directly compared the Western Blot signals to standardise different knockdown conditions.
As proof of principle, we started by analysing the knockdown of members of the Rab and ESCRT families that have been shown to regulate EV biogenesis before. Rab proteins regulate EV biogenesis in a cell‐type specific manner. Originally, knockdown of Rab27A/B and Rab35 was shown to impair MVB fusion with the plasma membrane and thereby decrease exosome release in oligodendroglia and HeLa cells (Hsu et al., 2010; Ostrowski et al., 2010). In addition to Rab27 and Rab35, we also included Rab7 in our analysis. It is involved both in trafficking between early endosomes and lysosomes and is also associated with late endosomes and MVBs like Rab27. In Drosophila, Rab7 knockdown or expression of a dominant‐negative version of Rab7 (Rab7‐DN) in accessory gland cells disturbs exosome release (Corrigan et al., 2014). There is also evidence for its role in EV secretion from C. elegans (Hyenne et al., 2015) and mammalian cultured cells (Jaé et al., 2015). The recycling endosome component Rab11 was implied in EV secretion from human and Drosophila cells and also in vivo (Ashley et al., 2018; Beckett et al., 2013; Corrigan et al., 2014; Fan et al., 2020; Koles et al., 2012; Messenger et al., 2018; Savina et al., 2002) and one report even identified Rab11a‐positive MVBs in human cells (Savina et al., 2005).
The ESCRT‐0 component Hrs is recruited to endosomes via its FYVE domain and recognises ubiquitylated cargos. Tsg101 is one of the four ESCRT‐I components and bridges between ESCRT‐0 and ‐II. Shrb is the homolog of human Chmp4B and a filament forming subunit of ESCRT‐III, which drives inward membrane deformation (Henne et al., 2013; Raiborg & Stenmark, 2009). Alix (ALG‐2 interacting protein X) is an ESCRT accessory protein that can directly bind to Shrb/Chmp4B, thereby bridging cargo into MVBs in a ubiquitylation‐independent way (Baietti et al., 2012). For many of these candidates, data variability hampered the interpretation of immunoblot results, even though the lines had effects on intracellular EV marker trafficking as shown by immunofluorescence. This biochemical variability might be introduced by technical imprecision. Considering the low amount of starting material, it cannot be ruled out that insufficient resuspension led to the observed fluctuation in protein expression of P14/P100, however similar amounts of EV were purified from wildtype haemolymph in a regular manner. What is more likely, though, is that endosomal trafficking is adaptable and knockdown of single ESCRT members might not be sufficient to block EV secretion completely due to redundancy and despite subcellular perturbations detectable by IF. We decided against a ubiquitous GAL driver like tubulin‐GAL4 in the beginning of the study because complete knockdown is lethal for many of the candidates analysed. To circumvent this problem in the future, one could combine a ubiquitous driver with a temperature‐sensitive GAL80 element to restrict the knockdown to a short developmental time window.
Generally, a mixed population of EVs of different cellular origin are co‐purified as revealed by proteomic profiling of EV subtypes (Kowal et al., 2016). By manipulating ESCRT‐III and Alix we anticipated to perturb the formation of all ESCRT‐dependent EVs. However, EV biogenesis via Syndecan and Alix requires Syntenin (Baietti et al., 2012) and there is no Syntenin homolog in the Drosophila genome (FlyBASE version FB2021_01), indicating that other mechanisms must exist in flies. Furthermore, variability could also result from competing EV biogenesis mechanisms, where knockdown of one machinery favours the other as was shown for the generation of Hrs‐ and CD63‐dependent subpopulations (Edgar et al., 2014). Interestingly, we observed a significant reduction in EV secretion upon knockdown of Rab11 and Rab35, suggesting that recycling Rab family members strongly contribute to EV secretion from the fat body. In mammalian cells, Rab11 orchestrates slow recycling from MVBs to the plasma membrane, whereas Rab4 directs fast recycling from early endosomes towards the plasma membrane (De Renzis et al., 2002). Rab4 plays a role in apical secretion of the morphogens Hedgehog and Wingless in wing imaginal discs (D'Angelo et al., 2015; Linnemannstöns et al., 2020). In Drosophila fat body cells, Rab4 and Rab11 function in anti‐microbial peptide secretion. Here, UAS‐Rab11‐GFP localises to small puncta and to larger endosomal structures which are also positive for Rab4 (Shandala et al., 2011; Sorvina et al., 2016, 2020). Studies in human cells and Drosophila indicated a role for Rab11 in EV secretion (Ashley et al., 2018; Beckett et al., 2013; Corrigan et al., 2014; Koles et al., 2012; Messenger et al., 2018; Redhai et al., 2016; Savina et al., 2002) and a recent study highlighted the emerging importance of recycling MVBs in response to glutamine starvation (Fan et al., 2020). Rab11‐ and Rab35‐regulated exosomal secretion does not seem to be ESCRT‐dependent (reviewed in Blanc and Vidal (2018)), which supports our findings that EV secretion from the fat body is not impaired by knockdown of ESCRT components, but controlled by Rab11/Rab35. However, the detailed mechanism of Rab11‐ and Rab35‐mediated EV secretion remain to be elucidated. None of the various Rab35 effectors identified so far was shown to be involved in EV secretion (Chaineau et al., 2013). The exocyst complex is a direct target of Rab11 (Zhang et al., 2004) and its members were shown to regulate apical secretion of the morphogen Wingless in wing imaginal discs (Chaudhary & Boutros, 2019). Noteworthy, the suppression of any individual Rab family member was not sufficient to completely block exosome secretion. This can be due to redundancy or alternatively each Rab might regulate the secretion of a subset of EVs (McGough & Vincent, 2016).
For this reason, we decided to further study the role of Rab14, which was shown to regulate autophagosome localisation in the Drosophila fat body together with the kinesin 3 family member Klp98A, the orthologue of mammalian KIF16B, as upstream effector (Mauvezin et al., 2016). The larval fat body uses autophagy to mobilise nutrients throughout the animal in response to starvation. In the present work, Klp98A significantly affects secretion of Lbm‐ positive EV in the haemolymph as detected by immunoblot, whereas Rab14 knockdown affects the late endosome populations (as detected by IF). This could indicate that the two proteins collaborate under stressed autophagic conditions, but perform distinct functions in the physiological state, provided there are enough nutrients. We found Rab14 in the P14 pellet by DIA‐MS and work in human cell culture showed that components of functional protein complexes can be sorted differentially into EVs to modulate endosomal trafficking by the autophagic component ATG5 (Guo et al., 2017). As mentioned above, specificity and directionality of EV sorting depend on Rab GTPases as well as overlapping effectors present on the subpopulations. Rab14 and the fast recycling Rab4 share the effector RUFY1 and engage in recycling of TfR and Glut4 to the plasma membrane (Yamamoto et al., 2010). Rab14 was also shown to act prior to Rab11 and after Rab4/5 in an intermediate recycling compartment (Linford et al., 2012). Rab4 and KIF3A regulate melanosome/lysosome‐related organelle trafficking (Nag et al., 2018), which has overlapping functional components with EV biogenesis (Delevoye et al., 2019). Klp98A is also involved in apical Wingless recycling via Rab4‐endosomes to the plasma membrane (Witte et al., 2020) prior to Wingless secretion on EVs (Gross et al., 2012). Within the fat body Rab4/Rab11 are regulating exocytosis of innate immune cargo like antimicrobial peptides (Shandala et al., 2011; Sorvina et al., 2016). The interconnection of these components in targeting endosomal subpopulations and EV cargo proteins has to be elucidated in further studies.
Taken together, we introduce a novel in vivo model that allows to study the secretion of different EVs populations via multiple trafficking routes into the extracellular space. The larval fat body provides nutrients used by developing organs like imaginal discs and other tissues for growth. It remains to be identified whether fat body‐derived EVs act as communication device or provide mainly trophic support and if they are preferentially taken up by certain organs or tissues.
4. MATERIAL AND METHODS
4.1. Drosophila stocks and genetics
The following Drosophila stocks were used in this study: wildtype OregonR (a gift from J. Grosshans, Philipps University Marburg, Germany), UAS‐eGFP (chr. II, a gift from Martin Zeidler, University of Sheffield, UK), UAS‐Lamp1‐GFP (a gift from T. Vaccari, University of Milan, Italy), bsgCA (GFP trap Bsg, a gift from Ulrich Thomas, University of Magdeburg, Germany (Besse et al., 2007)). These stocks were obtained from Bloomington Drosophila stock centre: Cg‐Gal4(7011), eYFP‐Myc‐Rab11(62549), UAS‐Rab11‐GFP (8506), vas‐PhiC31; attP.ZH‐86Fb (24749), nos‐PhiC31; attP40.25C6(25709), AliX TRiP (33417(#1)), Hrs TRiP (28026(#1), 33900(#2)), Rab14 TRiP (28708), Rab35 TRiP (#2), Tsg101 TRiP (35710(#2)), Tsp42Ee‐GFP (Flytrap, BL51558), Tsp42Ee EGFP‐FlAsH‐StrepII‐TEV‐3xFlag (RMCE MiMIC, 63197). The following UAS‐RNAi stocks were obtained from Vienna Drosophila RNAi Center: Yellow (KK 106068), ALiX (GD 32047(#2)), Klp98A (GD 40603), Rab7 (KK 27051), Rab11 (KK 42709), Rab27 (KK 31887(#1), KK 35774 (#2)), Rab35 (KK 101361(#1), Shrb (KK 106823), Tsg101 (GD 23944(#2)). UAS‐mCherry‐Tsp96F and UAS‐CD63‐pHluorin transgenic lines were generated according to standard protocols by φC31 integrase‐mediated site‐specific insertion in the attP landing sites at 86Fb and 25C6, respectively (Bischof et al., 2007). Fly stocks were kept on standard medium containing agar, yeast and corn flour. Crosses were performed at 25°C.
4.2. Plasmids
The coding region of Drosophila Tsp96F was amplified from DGRC clone LD19727 and the resulting PCR product was recombined into pDONRTM221 vector using the Gateway BP Clonase II Enzyme mix (Life Technologies, Carlsbad, CA, USA). For generation of transgenic flies, the construct was subcloned into the expression vector pUASt‐attB‐mCherry‐rfA (kind gift from Sven Bogdan) by LR recombination (Life Technologies, Carlsbad, CA, USA). CD63‐pHLuorin was amplified from Addgene clone #70113(pCMV‐lyso‐pHluorin) and subcloned into pUASTattB (kind gift from Jörg Großhans) via XhoI/XbaI.
4.3. Antibodies
Antibodies were used against human Alix (1:100 WB; goat, Santa Cruz and mouse, Santa Cruz), ɑ‐Tubulin (1:100 WB; mouse, DSHB 12G10), human ATP6V0A1, (1:1000 WB; rabbit, Novus Bio), Cnx (1:10 IF; mouse, DSHB Cnx99A 6‐2‐1), Csp‐1(1:50 WB; mouse, DSHB ab49), Csp‐2(1:50 WB; mouse, DSHB 6D6), Flo2(1:1000 WB; BD Transduction Laboratories), GFP (1:1000 WB and IF; mouse (A11120) and rabbit (A11122), Molecular Probes), GM130 1:1000(WB; rabbit, Abcam ab30637), Golgin 1:10(IF; mouse DSHB Golgin84), Hrs (1:50 WB; 1:10 IF; mouse, DSHB Hrs 27‐4 and Hrs 8‐2), Hsp70(1:1000 WB; mouse Santa Cruz BRM22), (IntegrinβPS (1:50 WB; mouse, DSHB CF.6G11), Lamp1(1:500 WB; 1:100 IF; rabbit, Abcam ab30687), Lbm (1:50 WB; 1:10 IF; mouse, DSHB 10C9), mCherry (1:1000 IF and WB; rabbit, Abcam ab167453), Rab5(1:500 IF; rabbit, Abcam ab31261), Rab7(1:10 IF; mouse, DSHB), human Rab11(1:1000 WB; mouse, BD Transduction), Syx1A (1:50 WB; mouse, DSHB 8C3), Tsg101(1:500 WB; mouse, Abcam 4A10). Secondary antibodies directed against the species of interest were coupled to Alexa Fluor 488, 568 and 647(IF, 1:400, Invitrogen) and 680RD and 800CW (WB, 1:20,000, LiCor).
4.4. Haemolymph collection
Wandering L3 larvae were collected and cleaned in PBS. After drying them briefly on a paper towel to remove the remaining buffer the larvae were transferred into perforated 0.5 ml tubes. While transferring, the larvae are pierced with forceps near the mouth hooks to open the larval body wall and release haemolymph. The tubes were then inserted into 1.5 ml tubes and centrifuged for 20–30 s in a picofuge. The 1.5 ml tube containing the haemolymph was immediately snap‐frozen in liquid nitrogen and stored at –80°C. Ten larvae yield approximately 10 μl haemolymph.
4.5. Isolation of EVs from haemolymph
Ten microlitres haemolymph were collected and resuspended in 500 μl filtered PBS+PTU (Phenylthiourea 0.1 μg/ml) to prevent melanisation and centrifuged at 750 g for 15 min at 4°C. The supernatant (SN0.75) was transferred into a fresh 1.5 ml reaction tube and centrifuged at 1500 g for 10 min at 4°C to remove cell debris. The supernatant kept after 1500 g centrifugation (SN1.5) was transferred into a fresh 1.5 ml reaction tube and centrifuged at 14,000 g for 35 min at 4°C. The resulting P14 pellet was washed with 200 μl PBS and again centrifuged for 35 min at 4°C. The washed P14 pellet was resuspended in 20 μl filtered PBS and 5 μl RIPA + PI. The supernatant kept after the first 14,000 g centrifugation was centrifuged at 100,000 g for 2 h at 4°C (rotor: Beckmann Coulter TLA 120.1). The supernatant was discarded and the P100 pellet was resuspended with 25 μl filtered PBS (EM and NTA) or 20 μl filtered PBS and 5 μl RIPA + PI (Western Blot). Samples for NTA were stored short‐term at –20°C until further analysis. Samples for Western Blot analysis were boiled in reducing or non‐reducing 6X SDS sample buffer and stored at –20°C until further use.
4.6. Proteinase K protection assay
EVs were isolated from 40 μl haemolymph. The resulting P14 and P100 pellets were resuspended in 80 μl filtered PBS and split into four identical aliquots. Proteinase K digestion was then performed with or without 0.01 mg/ml proteinase K (QIAGEN) in presence or absence of 1% Triton X‐100 for 30 min at 37°C. The samples were then boiled in or non‐reducing 6X SDS sample buffer and subjected to Western Blot analysis.
4.7. Protein lysis
The P0.75 pellets, containing haemocytes and other cells present in the haemolymph, were lysed in 25 μl RIPA buffer + Proteinase Inhibitor (Roche) on ice for 20–30 min and then centrifuged 15 min at 14,000 g and 4°C. The supernatant was boiled in reducing or non‐reducing 6X SDS sample buffer and stored at –20°C until further use.
4.8. Immunoblot
Proteins were separated on 4%–12% gradient gels (Bolt Bis‐Tris Plus Gels, Thermo Scientific) and transferred to PVDF membrane (Merck). After blocking with 5% (wt/vol) milk‐TBST, membranes were incubated with corresponding primary antibodies in TBST at 4°C. After washing, membranes were incubated with fluorescently labelled secondary antibodies at room temperature in the dark. Detection was performed using Odyssey from Li‐COR. Quantitative measurements were done with LiCOR Image Studio software.
4.9. Electron microscopy
Purified EVs were left to settle on carbon‐coated grids. After washing with water, samples were stained with neutral uranyl acetate. After incubation with a 1:1 mixture of 4% uranylacetate and 2% methycellulose, grids were air‐dried and visualised at 30,000× magnification using a transmission electron microscope (Zeiss EM900). Representative images from three biological replicates of P14 and P100 from wild‐type haemolymph were obtained. Particle size was determined compared to a size marker in Fiji.
4.10. Nanoparticle tracking analysis
Nanoparticle tracking experiments were performed using a Nanosight LM10 (Malvern) instrument. P14 and P100 samples were diluted 1:200 in PBS and 400 μl diluted sample were measured. We used consistent settings with 30 s of measurement time at medium polydispersity and reproducibility under a level of illumination of 15 (LM10 software) and measured three consecutive rounds of 30 s. For P100 and P14 particles up to 500 nm were used in calculations and the rest of the registered particles (which corresponded to <1% in all cases) were taken out. The measured EV concentrations were corrected for the dilution factor and depicted as number of particles per μl haemolymph pellet (starting material was 10 μl haemolymph).
4.11. Mass spectrometry
Three independent biological replicates of haemolymph samples (P14, P100, SN100) were boiled 5 min in non‐reducing SDS sample buffer (300 mM Tris‐HCl pH 6.8, 12% SDS, 0.05% Bromphenolblue, 60% Glycerol, 12 mM EDTA) and run short‐distance (1.5 cm) into a 4%–12 % NuPAGE Novex Bis‐Tris Minigel (Invitrogen). Gels were stained with Coomassie Blue for visualisation purposes. Stained areas were cut out as a single fraction and diced. After washing, gel slices were reduced with dithiothreitol (DTT), alkylated with 2‐iodoacetamide and digested with trypsin overnight. The resulting peptide mixtures were then extracted, dried in a SpeedVac (Atanassov & Urlaub, 2013). For generation of an annotated MS/MS spectral library for DIA‐MS, equal amount aliquots from each sample were pooled to a total amount of 80 μg and separated into twelve fractions using a reversed phase spin column (Pierce High pH Reversed‐Phase Peptide Fractionation Kit, Thermo Fisher Scientific).
For mass spectrometric analysis, samples and reverse phase fractions were taken up in 20 μl loading buffer (0.1% aqueous formic acid, 2% acetonitrile) and separated by nanoflow chromatography. Injections of 1 μg protein equivalent enriched on a self‐packed precolumn (0.15 mm ID x 20 mm, Reprosil‐Pur120 C18‐AQ 5 μm, Dr. Maisch, Ammerbuch‐Entringen, Germany) and separated on an analytical RP‐C18 column (0.075 mm ID × 250 mm, Reprosil‐Pur 120 C18‐AQ, 3 μm, Dr. Maisch) using a 30 to 90 min linear gradient of 5%–35% acetonitrile/0.1% formic acid (v:v) at 300 nl/min. The eluents were analysed on a hybrid triple quadrupole‐TOF mass spectrometer (TripleTOF 5600+) equipped with a Nanospray III ion source (Ionspray Voltage 2400 V, Interface Heater Temperature 150°C, Sheath Gas Setting 12) and controlled by Analyst TF 1.7.1 software build 1163(all AB Sciex).
For generation of an MS/MS spectral library, reverse phase fractions as well as the individual samples were analysed by data‐dependent acquisition (DDA) using a Top30 data‐dependent acquisition method, with a survey scan of m/z 380‐1250 accumulated for 250 ms at a resolution of 35,000 full width at half maximum (FWHM). Precursors above a threshold MS intensity of 200 cps with charge states 2+, 3+ and 4+ were selected for MS/MS at 0.7 FWHM resolution, and fragmented using nitrogen as a collision gas using manufacturer's default rolling collision energy settings. MS/MS scans of m/z 180–1500 were accumulated for 100 ms at a resolution of 17,500 FWHM, resulting in a total cycle time of 3.4 s. Two replicate injections per reversed phase fraction were analysed to construct a spectral library.
For quantitative SWATH analysis, MS/MS data were acquired using 100 variable size windows (Zhang et al., 2015) across the 400‐1200 m/z range. Fragments were produced using rolling collision energy settings for charge state 2+, and fragments acquired over an m/z range of 180–1500 for 40 ms per segment. Including a 250 ms survey scan this resulted in an overall cycle time of 4.3 s. Two replicate injections per biological sample were acquired.
4.12. Mass spectrometry data processing
Data were analysed using Spectronaut SW v14.2.200619.47784 (Biognosys). For generation of a spectral library, both DDA and DIA data were searched against the UniProtKB D. melanogaster reference proteome (revision 02‐2017, 23,306 entries) augmented with a set of 51 known common laboratory contaminants and the sequence of the mCherry. Default setting including dynamic mass and retention time calibration, carbamidomethylation as fixed modification, and methionine oxidation and protein N‐terminal acetylation as variable modifications were used. The search resulted in a hybrid DDA/DIA spectral library of 31,104 precursors representing 23,940 peptide sequences and 2871 protein groups at an FDR of 1%, respectively, established by combined forward‐and‐reverse decoy searches.
Quantitative analysis of DIA data was achieved by peak extraction on the precursor/fragment level using the above spectral library and combining six fragments to the precursor and up to 10 peptides to the protein for obtaining protein area values, each at estimated FDR levels of 1%. Other than this, default software settings were used, including dynamic mass and retention time calibration. The resulting peak areas were summed to peptide, protein and protein group area values, which were used for further statistical analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al., 2019) partner repository with the dataset identifier PXD027290.
Statistical analysis was performed in Perseus 1.5.6.0 software (Max Planck Institute for Biochemistry, Martinsried, Germany). Following log 2 transformation, data were filtered for proteins with valid values in at least 60% of injections in at least one sample group (P14/P100/SN100). Missing values were imputed from a normal distribution (width 0.3, downshift 1.8) to avoid division by zero, resulting in a data matrix containing 2466 protein group areas. Significance of enrichment was established using FDR‐based calculation with p ≤ 0.05 and S0 = 0.5, resulting in an at least 2‐fold enrichment as a requirement for significance.
GO term enrichment analysis was performed with PANTHER (Mi et al., 2013) with a cut‐off p‐value < 0.05 as determined by Fisher Exact Test with the Benjamini–Hochberg False Discovery Rate < 0.05.
4.13. Immunofluorescent stainings and microscopy
For fat body isolation L3 larvae were inverted and collected in PBS on ice. Fixation and immunostainings were performed as per standard procedures. After transferring the tissue on a slide, FBs were separated from the rest of the larval tissue and the excess liquid was removed. Specimen were mounted in Mowiol and images were acquired using a Zeiss LSM780 confocal microscope. Staining with LysoTracker Red DND‐99, LysoTracker Deep Red and LysoSensor Green DND‐189 (1 μM each) was done as described (Juhasz & Neufeld, 2008). Staining and microscopy conditions were kept identical for comparisons. Z stacks were generated with 0.5‐1 μm intervals using a Plan Neofluar 63X/oil NA 1.4 objective.
4.14. Microscopy and image analysis
Confocal images were processed with Zen lite (Zeiss), Fiji/ImageJ (NIH) (Rueden et al., 2017; Schindelin et al., 2012; Schneider et al., 2012) and Affinity Designer (Affinity). Microscopy conditions were kept identical for colocalisation analyses. The number of endosomes were quantified using Fiji. Rab11‐GFP‐, Rab7‐ and Hrs‐positive endosomes were counted manually per cell (11‐20 fat body cells per RNAi) and normalised to 10 μm2. Rab5‐positive puncta > 4 px were counted automatically in Fiji using ComDet v.0.5.3 plugin for ImageJ (https://github.com/ekatrukha/ComDet). Quantification of colocalisation was performed by calculating Pearson's correlation coefficients of sections using the Fiji plug‐in JaCoP (Bolte & Cordelières, 2006).
4.15. Phylogenetic tree analysis
Protein sequences for all tetraspanins analysed were retrieved from UniProt. Phylogenetic tree was generated with the EMBL‐EBI Simple Phylogeny tool (Saitou & Nei, 1987) with default settings (no gap exclusion, no distance correction) using the neighbour‐joining method.
4.16. Statistics
All experiments were carried out at least in biological triplicates. All graphs depict the mean and error bars indicate standard deviation. Statistical significance was calculated by carrying out one‐way ANOVA with Dunnett's multiple comparison test to compare a control mean with the other means or Student's t‐test where appropriate. The data that support the findings of this study are available from the corresponding author upon reasonable request.
AUTHOR CONTRIBUTIONS
Karen Linnemannstöns and Julia Christina Gross designed the study, analysed data and wrote the manuscript. Kristin Steinmetz carried out experiments with the help of Leonie Witte, Dolma Choezom, Alex Simon Lagurin, Chantal Vanessa Schmidt, Shreya Shrikhande and Lara‐Kristin Steinmetz. Mona Honemann‐Capito generated fly lines and constructs. Christof Lenz designed and performed DIA‐MS analysis. Christof Lenz, Karen Linnemannstöns and Pradhipa Karuna M analysed MS data. Wiebke Möbius performed EM.
CONFLICT OF INTEREST
The authors have no competing financial interests.
Supporting information
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.
ACKNOWLEDGEMENTS
The authors thank the Core Facility Proteomics at the Institute of Clinical Chemistry, UMG, Sven Bogdan, Jörg Großhans and Andrew Peden for plasmids and fly reagents. We also thank Boguslawa Sadowski for excellent technical support in EM image acquisition. Research in the lab of JCG was supported by the DFG‐funded Research Center SFB1324/1 ‐ project number 331351713 and GR4810/2‐1, the Research program of the University Medical Center, University of Göttingen and a postdoctoral fellowship to KL by the Dorothea Schlözer Program, University of Göttingen.
Linnemannstöns, K. , Karuna M, P. , Witte, L. , Choezom, D. , Honemann‐Capito, M. , Lagurin, A. S. , Schmidt, C. V. , Shrikhande, S. , Steinmetz, L.‐K. , Wiebke, M. , Lenz, C. , & Gross, J. C. (2022). Microscopic and biochemical monitoring of endosomal trafficking and extracellular vesicle secretion in an endogenous in vivo model. Journal of Extracellular Vesicles, 11, e12263. 10.1002/jev2.12263
REFERENCES
- Acharya, U. , & Acharya, J. K. (2005). Enzymes of sphingolipid metabolism in Drosophila melanogaster . Cellular and Molecular Life Sciences, 62, 128–142. 10.1007/s00018-004-4254-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese, E. L. , & Soulages, J. L. (2010). Insect fat body: Energy, metabolism, and regulation. Annual Review of Entomology, 55, 207–225. 10.1146/annurev-ento-112408-085356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha, H. , Nagy, I. , Kovacs, G. , Stetson, D. , Ando, I. , & Dearolf, C. R. (2003). Analysis of Ras‐induced overproliferation in Drosophila hemocytes. Genetics, 163, 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, J. , Cordy, B. , Lucia, D. , Fradkin, L. G. , Budnik, V. , & Thomson, T. (2018). Retrovirus‐like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell, 172, 262–270.e11. 10.1016/j.cell.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassov, I. , & Urlaub, H. (2013). Increased proteome coverage by combining PAGE and peptide isoelectric focusing: Comparative study of gel‐based separation approaches. Proteomics, 13, 2947–2955. 10.1002/pmic.201300035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti, M. F. , Zhang, Z. , Mortier, E. , Melchior, A. , Degeest, G. , Geeraerts, A. , Ivarsson, Y. , Depoortere, F. , Coomans, C. , Vermeiren, E. , Zimmermann, P. , & David, G. (2012). Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nature Cell Biology, 14, 677–685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Beckett, K. , Monier, S. , Palmer, L. , Alexandre, C. , Green, H. , Raposo, G. , Thibault, P. , Borgne, R. L. , & Vincent, J. (2013). Drosophila Wingless is loaded on exosome‐like vesicles but forms a gradient in an exosome‐independent manner. Traffic (Copenhagen, Denmark), 14, 82–96. 10.1111/tra.12016.Drosophila [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller, M. , Riedel, D. , Jänsch, L. , Dieterich, G. , Wehland, J. , Jäckle, H. , & Kühnlein, R. P. (2006). Characterization of the Drosophila lipid droplet subproteome. Molecular & cellular proteomics : MCP, 5, 1082–1094. 10.1074/mcp.M600011-MCP200 [DOI] [PubMed] [Google Scholar]
- Besse, F. , Mertel, S. , Kittel, R. J. , Wichmann, C. , Rasse, T. M. , Sigrist, S. J. , & Ephrussi, A. (2007). The Ig cell adhesion molecule Basigin controls compartmentalization and vesicle release at Drosophila melanogaster synapses. Journal of Cell Biology, 177, 843–855. 10.1083/jcb.200701111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof, J. , Maeda, R. K. , Hediger, M. , Karch, F. , & Basler, K. (2007). An optimized transgenesis system for Drosophila using germ‐line‐specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America, 104, 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, M. , Gradilla, A.‐C. , Seijo, I. , Andrés, G. , Rodríguez‐Navas, C. , González‐Méndez, L. , & Guerrero, I. (2013). Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nature Cell Biology, 15, 1269–1281. 10.1038/ncb2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, L. , & Vidal, M. (2018). New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases, 9, 95–106. 10.1080/21541248.2016.1264352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte, S. , & Cordelières, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. Journal of Microscopy, 224, 213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Brand, A. H. , & Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England), 415, 401–415. [DOI] [PubMed] [Google Scholar]
- Callejo, A. , Torroja, C. , Quijada, L. , & Guerrero, I. (2006). Hedgehog lipid modifications are required for Hedgehog stabilization in the extracellular matrix. Development (Cambridge, England), 133, 471–483. 10.1242/dev.02217 [DOI] [PubMed] [Google Scholar]
- Chaineau, M. , Ioannou, M. S. , & Mcpherson, P. S. (2013). Rab35: GEFs, GAPs and effectors. Traffic (Copenhagen, Denmark), 14, 1109–1117. 10.1111/tra.12096 [DOI] [PubMed] [Google Scholar]
- Chaudhary, V. , & Boutros, M. (2019). Exocyst‐mediated apical Wg secretion activates signaling in the Drosophila wing epithelium. Plos Genetics, 15, e1008351. 10.1371/journal.pgen.1008351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier, P. , Parton, R. G. , Hauri, H. P. , Simons, K. , & Zerial, M. (1990). Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell, 62, 317–329. 10.1016/0092-8674(90)90369-P [DOI] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Théry, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Corrigan, L. , Redhai, S. , Leiblich, a. , Fan, S.‐J. , Perera, S. M. W. , Patel, R. , Gandy, C. , Wainwright, S. M. , Morris, J. F. , Hamdy, F. , Goberdhan, D. C. I. , & Wilson, C. (2014). BMP‐regulated exosomes from Drosophila male reproductive glands reprogram female behavior. Journal of Cell Biology, 206, 671–688. 10.1083/jcb.201401072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo, G. , Matusek, T. , Pizette, S. , & Thérond, P. P. (2015). Endocytosis of hedgehog through dispatched regulates long‐range signaling. Developmental cell, 32, 290–303. 10.1016/j.devcel.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Dawber, R. J. , Hebbes, S. , Herpers, B. , Docquier, F. , & van den Heuvel, M. (2005). Differential range and activity of various forms of the Hedgehog protein. BMC Developmental Biology [Electronic Resource], 5, 21. 10.1186/1471-213X-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye, C. , Marks, M. S. , & Raposo, G. (2019). Lysosome‐related organelles as functional adaptations of the endolysosomal system. Current Opinion in Cell Biology, 59, 147–158. 10.1016/j.ceb.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine, I. A. , & Perrimon, N. (2016). Interorgan communication pathways in physiology: Focus on Drosophila. Annual Review of Genetics, 10.1146/annurev-genet-121415-122024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunst, S. , Kazimiers, T. , von Zadow, F. , Jambor, H. , Sagner, A. , Brankatschk, B. , Mahmoud, A. , Spannl, S. , Tomancak, P. , Eaton, S. , Brankatschk, M. , von Zadow, F. , Jambor, H. , Sagner, A. , Brankatschk, B. , Mahmoud, A. , Spannl, S. , Tomancak, P. , Eaton, S. , & Brankatschk, M. (2015). Endogenously tagged Rab proteins: A resource to study membrane trafficking in Drosophila. Developmental cell, 33, 351–365. 10.1016/j.devcel.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, J. R. , Eden, E. R. , & Futter, C. E. (2014). Hrs‐ and CD63‐dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic (Copenhagen, Denmark), 15, 197–211. 10.1111/tra.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escola, J.‐M. , Kleijmeer, M. J. , Stoorvogel, W. , Griffith, J. M. , Yoshie, O. , & Geuze, H. J. (1998). Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B‐lymphocytes. Journal of Biological Chemistry, 273, 20121–20127. 10.1074/jbc.273.32.20121 [DOI] [PubMed] [Google Scholar]
- Fan, S. , Kroeger, B. , Marie, P. P. , Bridges, E. M. , Mason, J. D. , McCormick, K. , Zois, C. E. , Sheldon, H. , Khalid Alham, N. , Johnson, E. , Ellis, M. , Stefana, M. I. , Mendes, C. C. , Wainwright, S. M. , Cunningham, C. , Hamdy, F. C. , Morris, J. F. , Harris, A. L. , Wilson, C. , & Goberdhan, D. C. (2020). Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO Journal, 1–27. 10.15252/embj.2019103009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , Press, B. , & Wandinger‐Ness, A. (1995). Rab 7: An important regulator of late endocytic membrane traffic. Journal of Cell Biology, 131, 1435–1452. 10.1083/jcb.131.6.1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin, L. G. , Kamphorst, J. T. , DiAntonio, A. , Goodman, C. S. , & Noordermeer, J. N. (2002). Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proceedings of the National Academy of Sciences of the United States of America, 99, 13663–13668. 10.1073/pnas.212511099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeis, C. , Fröhlich, D. , Kuo, W. P. , Amphornrat, J. , Thilemann, S. , Saab, A. S. , Kirchhoff, F. , Möbius, W. , Goebbels, S. , Nave, K. A. , Schneider, A. , Simons, M. , Klugmann, M. , Trotter, J. , & Krämer‐Albers, E. M. (2013). Neurotransmitter‐triggered transfer of exosomes mediates oligodendrocyte‐neuron communication. Plos Biology, 11, e1001604. 10.1371/journal.pbio.1001604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet, A. , Ruel, L. , Staccini‐Lavenant, L. , & Thérond, P. P. (2006). Cholesterol modification is necessary for controlled planar long‐range activity of Hedgehog in Drosophila epithelia. Development (Cambridge, England), 133, 407–418. 10.1242/dev.02212 [DOI] [PubMed] [Google Scholar]
- Gault, W. J. , Olguin, P. , Weber, U. , & Mlodzik, M. (2012). Drosophila CK1‐γ, gilgamesh, controls PCP‐mediated morphogenesis through regulation of vesicle trafficking. Journal of Cell Biology, 196, 605–621. 10.1083/jcb.201107137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradilla, A.‐C. , González, E. , Seijo, I. , Andrés, G. , Bischoff, M. , González‐Mendez, L. , Sánchez, V. , Callejo, A. , Ibáñez, C. , Guerra, M. , Ortigão‐Farias, J. R. , Sutherland, J. D. , González, M. , Barrio, R. , Falcón‐Pérez, J. M. , & Guerrero, I. (2014). Exosomes as Hedgehog carriers in cytoneme‐mediated transport and secretion. Nature Communication, 5, 5649. 10.1038/ncomms6649 [DOI] [PubMed] [Google Scholar]
- Gross, J. C. , Chaudhary, V. , Bartscherer, K. , & Boutros, M. (2012). Active Wnt proteins are secreted on exosomes. Nature Cell Biology, 14, 1036–1045. 10.1038/ncb2574 [DOI] [PubMed] [Google Scholar]
- Guo, H. , Chitiprolu, M. , Roncevic, L. , Javalet, C. , Hemming, F. J. , Trung, M. T. , Meng, L. , Latreille, E. , Tanese de Souza, C. , McCulloch, D. , Baldwin, R. M. , Auer, R. , Côté, J. , Russell, R. C. , Sadoul, R. , & Gibbings, D. (2017). Atg5 disassociates the V1V0‐ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Developmental Cell, 43, 716–730.e7. 10.1016/j.devcel.2017.11.018 [DOI] [PubMed] [Google Scholar]
- Handke, B. , Poernbacher, I. , Goetze, S. , Ahrens, C. H. , Omasits, U. , Marty, F. , Simigdala, N. , Meyer, I. , Wollscheid, B. , Brunner, E. , Hafen, E. , & Lehner, C. F. (2013). The hemolymph proteome of fed and starved Drosophila larvae. Plos One, 8, 1–10. 10.1371/journal.pone.0067208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annual Review of Cell and Developmental Biology, 19, 397–422. 10.1146/annurev.cellbio.19.111301.153609 [DOI] [PubMed] [Google Scholar]
- Henne, W. M. , Stenmark, H. , & Emr, S. D. (2013). Sculpting ESCRT pathway. Cold Spring Harb. Perpectives Biol, 5, a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbrandt, G. R. , Abraham, T. , & Bieber, L. L. (1971). Metabolism of ceramide phosphorylethanolamine, phosphatidylinositol, phosphatidylserine and phosphatidylglycerol by housefly larvae. Lipids, 6, 508–516. 10.1007/BF02531237 [DOI] [PubMed] [Google Scholar]
- Hsu, C. , Morohashi, Y. , Yoshimura, S. I. , Manrique‐Hoyos, N. , Jung, S. Y. , Lauterbach, M. A. , Bakhti, M. , Grønborg, M. , Möbius, W. , Rhee, J. S. , Barr, F. A. , & Simons, M. (2010). Regulation of exosome secretion by Rab35 and its GTPase‐activating proteins TBC1D10A‐C. Journal of Cell Biology, 189, 223–232. 10.1083/jcb.200911018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Flockhart, I. , Vinayagam, A. , Bergwitz, C. , Berger, B. , Perrimon, N. , & Mohr, S. E. (2011). An integrative approach to ortholog prediction for disease‐focused and other functional studies. BMC Bioinformatics [Electronic Resource], 12, 357. 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , & Kornberg, T. B. (2015). Myoblast cytonemes mediate Wg signaling from the wing imaginal disc and Delta‐Notch signaling to the air sac primordium. Elife, 4, 1–22. 10.7554/eLife.06114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne, V. , Apaydin, a. , Rodriguez, D. , Spiegelhalter, C. , Hoff‐Yoessle, S. , Diem, M. , Tak, S. , Lefebvre, O. , Schwab, Y. , Goetz, J. G. , & Labouesse, M. (2015). RAL‐1 controls multivesicular body biogenesis and exosome secretion. Journal of Cell Biology, 211, 27–37. 10.1083/jcb.201504136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne, V. , Ghoroghi, S. , Collot, M. , Harlepp, S. , Mercier, L. , Busnelli, I. , Lefebvre, O. , Fekonja, N. , Machado, P. , Bons, J. , Delalande, F. , Amor, A. I. , Silva, G. , Verweij, F. J. , van Niel, G. , Schwab, Y. , Peinado, H. , Carapito, C. , Klymchenko, A. S. , … Goetz, J. G. (2019). Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Developmental Cell, 0, 1–19. 10.1016/J.DEVCEL.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Jacomin, A. C. , Fauvarque, M. O. , & Taillebourg, E. (2016). A functional endosomal pathway is necessary for lysosome biogenesis in Drosophila. Bmc Cell Biology [Electronic Resource], 17, 1–9. 10.1186/s12860-016-0115-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaé, N. , McEwan, D. G. , Manavski, Y. , Boon, R. A. , & Dimmeler, S. (2015). Rab7a and Rab27b control secretion of endothelial microRNA through extracellular vesicles. FEBS Letters, 589, 3182–3188. 10.1016/j.febslet.2015.08.040 [DOI] [PubMed] [Google Scholar]
- Juhasz, G. , & Neufeld, T. P. (2008). Experimental control and characterization of autophagy in drosophila. Methods in Molecular Biology, 445, 125–133. 10.1007/978-1-59745-157-4_8 [DOI] [PubMed] [Google Scholar]
- Kalra, H. , Simpson, R. J. , Ji, H. , Aikawa, E. , Altevogt, P. , Askenase, P. , Bond, V. C. , Borràs, F. E. , Breakefield, X. , Budnik, V. , Buzas, E. , Camussi, G. , Clayton, A. , Cocucci, E. , Falcon‐Perez, J. M. , Gabrielsson, S. , Gho, Y. S. , Gupta, D. , Harsha, H. C. , … Mathivanan, S. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. Plos Biology, 10, 8–12. 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar, S. , Chisanga, D. , Ariyaratne, D. , Al Saffar, H. , Anand, S. , Zhao, K. , Samuel, M. , Pathan, M. , Jois, M. , Chilamkurti, N. , Gangoda, L. , & Mathivanan, S. (2016). ExoCarta: A web‐based compendium of exosomal cargo. Journal of Molecular Biology, 428, 688–692. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.‐K. , Kang, B. , Kim, O. Y. , Choi, D. , Lee, J. , Kim, S. R. , Go, G. , Yoon, Y. J. , Kim, J. H. , Jang, S. C. , Park, K.‐S. , Choi, E.‐J. , Kim, K. P. , Desiderio, D. M. , Kim, Y.‐K. , Lötvall, J. , Hwang, D. , & Gho, Y. S. (2013). EVpedia: An integrated database of high‐throughput data for systemic analyses of extracellular vesicles. Journal of Extracellular Vesicles, 2, 20384. 10.3402/jev.v2i0.20384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.‐K. , Lee, J. , Kim, S. R. , Choi, D.‐S. , Yoon, Y. J. , Kim, J. H. , Go, G. , Nhung, D. , Hong, K. , Jang, S. C. , Kim, S.‐H. , Park, K.‐S. , Kim, O. Y. , Park, H. T. , Seo, J. H. , Aikawa, E. , Baj‐Krzyworzeka, M. , van Balkom, B. W. M. , Belting, M. , … Gho, Y. S. (2015). EVpedia: A community web portal for extracellular vesicles research. Bioinformatics, 31, 933–939. 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J. , & Raposo, G. (2014). The complex ultrastructure of the endolysosomal system. Cold Spring Harbor Perspectives in Biology, 6, e201808017 ‐ e201808039 . 10.1101/cshperspect.a016857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles, K. , Nunnari, J. , Korkut, C. , Barria, R. , Brewer, C. , Li, Y. , Leszyk, J. , Zhang, B. , & Budnik, V. (2012). Mechanism of evenness interrupted (Evi)‐exosome release at synaptic boutons. Journal of Biological Chemistry, 287, 16820–16834. 10.1074/jbc.M112.342667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski, C. C. , Davis, G. W. , & Goodman, C. S. (1996). A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science, 271, 1867–1870. 10.1126/science.271.5257.1867 [DOI] [PubMed] [Google Scholar]
- Koppen, T. , Weckmann, A. , Müller, S. , Staubach, S. , Bloch, W. , Dohmen, R. J. , & Schwientek, T. (2011). Proteomics analyses of microvesicles released by Drosophila Kc167 and S2 cells. Proteomics, 11, 4397–4410. 10.1002/pmic.201000774 [DOI] [PubMed] [Google Scholar]
- Korkut, C. , Ataman, B. , Ramachandran, P. , Ashley, J. , Barria, R. , Gherbesi, N. , & Budnik, V. (2009). Trans‐synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell, 139, 393–404. 10.1016/j.cell.2009.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut, C. , Li, Y. , Koles, K. , Brewer, C. , Ashley, J. , Yoshihara, M. , & Budnik, V. (2013). Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron, 77, 1039–1046. 10.1016/j.neuron.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal, J. , Arras, G. , Colombo, M. , Jouve, M. , Morath, J. P. , Primdal‐Bengtson, B. , Dingli, F. , Loew, D. , Tkach, M. , & Théry, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America, 113, E968–E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebel, P. W. , Majumdar, R. , Jenkins, L. M. , Senoo, H. , Wang, W. , Ammu, S. , Chen, S. , & Narayan, K. (2018). Extracellular vesicles direct migration by synthesizing and releasing chemotactic signals. Journal of Cell Biology, 217(8), 2891–2910. 10.1083/jcb.201710170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, F. A. , Benoit Bouvrette, L. P. , Perras, L. , Blanchet‐Cohen, A. , Garnier, D. , Rak, J. , & Lecuyer, E. (2016). Comparative transcriptomic analysis of human and Drosophila extracellular vesicles. Science Reports, 6, 27680–27694. 10.1038/srep27680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liégeois, S. , Benedetto, A. , Garnier, J. M. , Schwab, Y. , & Labouesse, M. (2006). The V0‐ATPase mediates apical secretion of exosomes containing Hedgehog‐related proteins in Caenorhabditis elegans . Journal of Cell Biology, 173, 949–961. 10.1083/jcb.200511072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford, A. , Yoshimura, S. I. , Bastos, R. N. , Langemeyer, L. , Gerondopoulos, A. , Rigden, D. J. , & Barr, F. A. (2012). Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Developmental Cell, 22, 952–966. 10.1016/j.devcel.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemannstöns, K. , Witte, L. , Karuna M, P. , Kittel, J. C. , Danieli, A. , Müller, D. , Nitsch, L. , Honemann‐Capito, M. , Grawe, F. , Wodarz, A. , & Gross, J. C. (2020). Ykt6‐dependent endosomal recycling is required for Wnt secretion in the Drosophila wing epithelium. Development (Cambridge, England), 147, dev185421–185436. 10.1242/dev.185421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz, P. , Mauvezin, C. , & Juhász, G. (2017). Exploring autophagy in Drosophila. Cells, 6, 22–36. 10.3390/cells6030022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C. I. J. , Yang, Y. , Kim, T. , Chen, C. H. , Polevoy, G. , Vissa, M. , Burgess, J. , & Brill, J. A. (2020). An early endosome‐derived retrograde trafficking pathway promotes secretory granule maturation. Journal of Cell Biology, 219, e201808017–e201808034. 10.1083/jcb.201808017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, P. , Fan, S.‐J. , Mendes, C. , Wainwright, S. M. , Harris, A. , Goberdhan, D. , & Wilson, C. (2020). Accessory ESCRT‐III proteins selectively regulate Rab11‐exosome biogenesis in Drosophila secondary cells. bioRxiv. 10.1101/2020.06.18.158725 [DOI]
- Mathieu, M. , Névo, N. , Jouve, M. , Valenzuela, J. I. , Maurin, M. , Verweij, F. J. , Palmulli, R. , Lankar, D. , Dingli, F. , Loew, D. , Rubinstein, E. , Boncompain, G. , Perez, F. , & Théry, C. (2021). Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nature Communication, 12, 4389–4407. 10.1038/s41467-021-24384-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan, S. , Fahner, C. J. , Reid, G. E. , & Simpson, R. J. (2012). ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Research, 40, D1241–D1244. 10.1093/nar/gkr828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusek, T. , Wendler, F. , Polès, S. , Pizette, S. , D'Angelo, G. , Fürthauer, M. , & Thérond, P. P. (2014). The ESCRT machinery regulates the secretion and long‐range activity of Hedgehog. Nature, 516, 99–103. 10.1038/nature13847 [DOI] [PubMed] [Google Scholar]
- Mauvezin, C. , Ayala, C. , Braden, C. R. , Kim, J. , & Neufeld, T. P. (2014). Assays to monitor autophagy in Drosophila. Methods, 68, 134–139. 10.1016/j.ymeth.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin, C. , Neisch, A. L. , Ayala, C. I. , Kim, J. , Beltrame, A. , Braden, C. R. , Gardner, M. K. , Hays, T. S. , & Neufeld, T. P. (2016). Coordination of autophagosome‐lysosome fusion and transport by a Klp98A‐Rab14 complex. [DOI] [PMC free article] [PubMed]
- McGough, I. J. , & Vincent, J.‐P. (2016). Exosomes in developmental signalling. Development (Cambridge, England), 143, 2482–2493. 10.1242/dev.126516 [DOI] [PubMed] [Google Scholar]
- Menck, K. , Scharf, C. , Bleckmann, A. , Dyck, L. , Rost, U. , Wenzel, D. , Dhople, V. M. , Siam, L. , Pukrop, T. , Binder, C. , & Klemm, F. (2015). Tumor‐derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. Journal of Molecular Cell Biology, 7, 143–153. 10.1093/jmcb/mju047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menck, K. , Sönmezer, C. , Worst, T. S. , Schulz, M. , Dihazi, G. H. , Streit, F. , Erdmann, G. , Kling, S. , Boutros, M. , Binder, C. , & Gross, J. C. (2017). Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. Journal of Extracellular Vesicles, 6, 1378056. 10.1080/20013078.2017.1378056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger, S. W. , Woo, S. S. , Sun, Z. , & Martin, T. F. J. (2018). A Ca2+‐stimulated exosome release pathway in cancer cells is regulated by Munc13‐4. Journal of Cell Biology, 217, 2877–2890. 10.1083/jcb.201710132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , Casagrande, J. T. , & Thomas, P. D. (2013). Large‐scale gene function analysis with the PANTHER classification system. Nature Protocols, 8, 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S. , Rani, S. , Mahanty, S. , Bissig, C. , Arora, P. , Azevedo, C. , Saiardi, A. , Van Der Sluijs, P. , Delevoye, C. , Van Niel, G. , Raposo, G. , & Setty, S. R. G. (2018). Rab4A organizes endosomal domains for sorting cargo to lysosome‐related organelles. Journal of Cell Science, 131, jcs216226. 10.1242/jcs.216226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelliot, A. , Bond, N. , & Hoshizaki, D. (2006). Fat‐body remodeling in Drosophila melanogaster . Genesis (New York, N.Y.: 2000), 44, 396–400. 10.1002/dvg [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Ostrowski, M. , Carmo, N. B. , Krumeich, S. , Fanget, I. , Raposo, G. , Savina, A. , Moita, C. F. , Schauer, K. , Hume, A. N. , Freitas, R. P. , Goud, B. , Benaroch, P. , Hacohen, N. , Fukuda, M. , Desnos, C. , Seabra, M. C. , Darchen, F. , Amigorena, S. , Moita, L. F. , & Thery, C. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology, 12, 19–30. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- Palmulli, R. , & van Niel, G. (2018). To be or not to be… secreted as exosomes, a balance finely tuned by the mechanisms of biogenesis. Essays in Biochemistry, 62(2), 177–191. [DOI] [PubMed] [Google Scholar]
- Panáková, D. , Sprong, H. , Marois, E. , Thiele, C. , & Eaton, S. (2005). Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature, 435, 58–65. 10.1038/nature03504 [DOI] [PubMed] [Google Scholar]
- Parchure, A. , Vyas, N. , Ferguson, C. , Parton, R. G. , & Mayor, S. (2015). Oligomerization and endocytosis of Hedgehog is necessary for its efficient exovesicular secretion. Molecular Biology of the Cell, 26, 4700–4717. 10.1091/mbc.E15-09-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan, M. , Fonseka, P. , Chitti, S. V. , Kang, T. , Sanwlani, R. , Van Deun, J. , Hendrix, A. , & Mathivanan, S. (2019). Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Research, 47, D516–D519. 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan, M. , Keerthikumar, S. , Ang, C.‐S. , Gangoda, L. , Quek, C. Y. J. , Williamson, N. A. , Mouradov, D. , Sieber, O. M. , Simpson, R. J. , Salim, A. , Bacic, A. , Hill, A. F. , Stroud, D. A. , Ryan, M. T. , Agbinya, J. I. , Mariadason, J. M. , Burgess, A. W. , & Mathivanan, S. (2015). FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics, 15, 2597–2601. 10.1002/pmic.201400515 [DOI] [PubMed] [Google Scholar]
- Pathan, M. , Keerthikumar, S. , Chisanga, D. , Alessandro, R. , Ang, C. , Askenase, P. , Batagov, A. O. , Benito‐Martin, A. , Camussi, G. , Clayton, A. , Collino, F. , Di Vizio, D. , Falcon‐Perez, J. M. , Fonseca, P. , Fonseka, P. , Fontana, S. , Gho, Y. S. , Hendrix, A. , Hoen, E. N. , … Mathivanan, S. (2017). A novel community driven software for functional enrichment analysis of extracellular vesicles data. Journal of Extracellular Vesicles, 6, 1321455–13214599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Riverol, Y. , Csordas, A. , Bai, J. , Bernal‐Llinares, M. , Hewapathirana, S. , Kundu, D. J. , Inuganti, A. , Griss, J. , Mayer, G. , Eisenacher, M. , Pérez, E. , Uszkoreit, J. , Pfeuffer, J. , Sachsenberg, T. , Yilmaz, Ş. , Tiwary, S. , Cox, J. , Audain, E. , Walzer, M. , … Vizcaíno, J. A. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research, 47, D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraki, S. , Alexander, B. , & Brückner, K. (2015). Assaying blood cell populations of the Drosophila melanogaster larva. Journal of Visualized Experiments: JoVE, 2015, 1–11. 10.3791/52733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil, S. , Akbar, M. A. , Ray, S. , Sevrioukov, E. A. , Haberman, A. S. , Rohrer, J. , & Krämer, H. (2005). Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. Journal of Cell Science, 118, 3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Raiborg, C. , & Stenmark, H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature, 458, 445–452. 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Redhai, S. , Hellberg, J. E. E. U. , Wainwright, M. , Perera, S. W. , Castellanos, F. , Kroeger, B. , Gandy, C. , Leiblich, A. , Corrigan, L. , Hilton, T. , Patel, B. , Fan, S. J. , Hamdy, F. , Goberdhan, D. C. I. , & Wilson, C. (2016). Regulation of dense‐core granule replenishment by autocrine BMP signalling in drosophila secondary cells. Plos Genetics, 12, 1–32. 10.1371/journal.pgen.1006366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis, S. , Sönnichsen, B. , & Zerial, M. (2002). Divalent Rab effectors regulate the sub‐compartmental organization and sorting of early endosomes. Nature Cell Biology, 4, 124–133. 10.1038/ncb744 [DOI] [PubMed] [Google Scholar]
- Rietveld, A. , Neutz, S. , Simons, K. , & Eaton, S. (1999). Association of sterol‐ and glycosylphosphatidylinositol‐linked proteins with Drosophila raft lipid microdomains. Journal of Biological Chemistry, 274, 12049–12054. 10.1074/jbc.274.17.12049 [DOI] [PubMed] [Google Scholar]
- Rink, J. , Ghigo, E. , Kalaidzidis, Y. , & Zerial, M. (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell, 122, 735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Rodrigues, M. L. , Nakayasu, E. S. , Oliveira, D. L. , Nimrichter, L. , Nosanchuk, J. D. , Almeida, I. C. , & Casadevall, A. (2008). Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic Cell, 7, 58–67. 10.1128/EC.00370-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden, C. T. , Schindelin, J. , Hiner, M. C. , DeZonia, B. E. , Walter, A. E. , Arena, E. T. , & Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics [Electronic Resource], 18, 1–26. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten, T. E. , Lindmo, K. , Juhász, G. , Sass, M. , Seglen, P. O. , Brech, A. , & Stenmark, H. (2004). Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Developmental Cell, 7, 179–192. 10.1016/j.devcel.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Saitou, N. , & Nei, M. (1987). The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4, 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Savina, A. , Fader, C. M. , Damiani, M. T. , & Colombo, M. I. (2005). Rab11 promotes docking and fusion of multivesicular bodies in a calcium‐dependent manner. Traffic (Copenhagen, Denmark), 6, 131–143. 10.1111/j.1600-0854.2004.00257.x [DOI] [PubMed] [Google Scholar]
- Savina, A. , Vidal, M. , & Colombo, M. I. (2002). The exosome pathway in K562 cells is regulated by Rab11. Journal of Cell Science, 115, 2505–2515. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J.‐Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , & Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH Image to ImageJ : 25 years of image analysis. Nature Methods, 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandala, T. , Woodcock, J. M. , Ng, Y. , Biggs, L. , Skoulakis, E. M. C. , a Brooks, D. , & Lopez, A. F. (2011). Drosophila 14‐3‐3ε has a crucial role in anti‐microbial peptide secretion and innate immunity. Journal of Cell Science, 124, 2165–2174. 10.1242/jcs.080598 [DOI] [PubMed] [Google Scholar]
- Silverman, J. M. , Clos, J. , De'Oliveira, C. C. , Shirvani, O. , Fang, Y. , Wang, C. , Foster, L. J. , & Reiner, N. E. (2010). An exosome‐based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. Journal of Cell Science, 123, 842–852. 10.1242/jcs.056465 [DOI] [PubMed] [Google Scholar]
- Sorvina, A. , Shandala, T. , & Brooks, D. A. (2016). Drosophila Pkaap regulates Rab4/Rab11‐dependent traffic and Rab11 exocytosis of innate immune cargo. Biology open, 5, 678–688. 10.1242/bio.016642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvina, A. , Shandala, T. , Wang, S. , Sharkey, D. J. , Parkinson‐Lawrence, E. , Selemidis, S. , & Brooks, D. A. (2020). CDKI‐73 is a novel pharmacological inhibitor of Rab11 cargo delivery and innate immune secretion. Cells, 9, 372–389. 10.3390/cells9020372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers, S. , Sem Wegner, C. , Stenmark, H. , & Brech, A. (2009). Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic (Copenhagen, Denmark), 10, 925–937. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- Takahashi, S. , Kubo, K. , Waguri, S. , Yabashi, A. , Shin, H. W. , Katoh, Y. , & Nakayama, K. (2012). Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. Journal of Cell Science, 125, 4049–4057. 10.1242/jcs.102913 [DOI] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants. Current Protocols in Cell Biology, 3.22, 1–29. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Regnault, A. , Garin, J. , Wolfers, J. , Zitvogel, L. , Ricciardi‐Castagnoli, P. , Raposo, G. , & Amigorena, S. (1999). Molecular characterization of dendritic cell‐derived exosomes: Selective accumulation of the heat shock protein hsc73. Journal of Cell Biology, 147, 599–610. 10.1083/jcb.147.3.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of extracellular vesicles, 7, 1535750–1535789. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todres, E. , Nardi, J. B. , & Robertson, H. M. (2000). The tetraspanin superfamily in insects. Insect Molecular Biology, 9, 581–590. 10.1046/j.1365-2583.2000.00222.x [DOI] [PubMed] [Google Scholar]
- Trajkovic, K. , Hsu, C. , Chiantia, S. , Rajendran, L. , Wenzel, D. , Wieland, F. , Schwille, P. , Brügger, B. , & Simons, M. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science, 319, 1244–1247. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- Ugrankar, R. , Bowerman, J. , Hariri, H. , Chandra, M. , Chen, K. , Bossanyi, M. F. , Datta, S. , Rogers, S. , Eckert, K. M. , Vale, G. , Victoria, A. , Fresquez, J. , McDonald, J. G. , Jean, S. , Collins, B. M. , & Henne, W. M. (2019). Drosophila snazarus regulates a lipid droplet population at plasma membrane‐droplet contacts in adipocytes. Developmental Cell, 50, 557–572.e5. 10.1016/j.devcel.2019.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari, T. , Rusten, T. E. , Menut, L. , Nezis, I. P. , Brech, A. , Stenmark, H. , & Bilder, D. (2009). Comparative analysis of ESCRT‐I, ESCRT‐II and ESCRT‐III function in Drosophila by efficient isolation of ESCRT mutants. Journal of Cell Science, 122, 2413–2423. 10.1242/jcs.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, F. J. , Revenu, C. , Arras, G. , Dingli, F. , Loew, D. , Pegtel, M. D. , Follain, G. , Allio, G. , Goetz, J. G. , Zimmermann, P. , Herbomel, P. , Del Bene, F. , Raposo, G. , & van Niel, G. (2019). Live tracking of inter‐organ communication by endogenous exosomes in vivo. Developmental Cell, 0, 1–17. 10.1016/j.devcel.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Witte, L. , Linnemannstöns, K. , Schmidt, K. , Honemann‐Capito, M. , Grawe, F. , Wodarz, A. , & Gross, J. C. (2020). The kinesin motor Klp98A mediates apical to basal Wg transport. Development, 147, dev186833–186848. 10.1242/dev.186833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H. , Koga, H. , Katoh, Y. , Takahashi, S. , Nakayama, K. , & Shin, H. W. (2010). Functional cross‐talk between Rab14 and Rab4 through a dual effector, RUFY1/Rabip4. Molecular Biology of the Cell, 21, 2746–2755. 10.1091/mbc.E10-01-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Zavec, A. B. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Silva, A. C. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles, 4, 1–60. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. M. , Ellis, S. , Sriratana, A. , Mitchell, C. A. , & Rowe, T. (2004). Sec15 is an effector for the Rab11 GTPase in mammalian cells. Journal of Biological Chemistry, 279, 43027–43034. 10.1074/jbc.M402264200 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Bilbao, A. , Bruderer, T. , Luban, J. , Strambio‐De‐Castillia, C. , Lisacek, F. , Hopfgartner, G. , & Varesio, E. (2015). The use of variable Q1 isolation windows improves selectivity in LC–SWATH–MS acquisition. Journal of Proteome Research, 14, 4359–4371. 10.1021/acs.jproteome.5b00543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Supporting information.
Supporting information.
Supporting information.


