Abstract
Aberrant self-assembly and toxicity of wild-type and mutant superoxide dismutase 1 (SOD1) has been widely examined in silico, in vitro and in transgenic animal models of amyotrophic lateral sclerosis. Detailed examination of the protein in disease-affected tissues from amyotrophic lateral sclerosis patients, however, remains scarce.
We used histological, biochemical and analytical techniques to profile alterations to SOD1 protein deposition, subcellular localization, maturation and post-translational modification in post-mortem spinal cord tissues from amyotrophic lateral sclerosis cases and controls. Tissues were dissected into ventral and dorsal spinal cord grey matter to assess the specificity of alterations within regions of motor neuron degeneration.
We provide evidence of the mislocalization and accumulation of structurally disordered, immature SOD1 protein conformers in spinal cord motor neurons of SOD1-linked and non-SOD1-linked familial amyotrophic lateral sclerosis cases, and sporadic amyotrophic lateral sclerosis cases, compared with control motor neurons. These changes were collectively associated with instability and mismetallation of enzymatically active SOD1 dimers, as well as alterations to SOD1 post-translational modifications and molecular chaperones governing SOD1 maturation. Atypical changes to SOD1 protein were largely restricted to regions of neurodegeneration in amyotrophic lateral sclerosis cases, and clearly differentiated all forms of amyotrophic lateral sclerosis from controls. Substantial heterogeneity in the presence of these changes was also observed between amyotrophic lateral sclerosis cases.
Our data demonstrate that varying forms of SOD1 proteinopathy are a common feature of all forms of amyotrophic lateral sclerosis, and support the presence of one or more convergent biochemical pathways leading to SOD1 proteinopathy in amyotrophic lateral sclerosis. Most of these alterations are specific to regions of neurodegeneration, and may therefore constitute valid targets for therapeutic development.
Keywords: superoxide dismutase-1, neurodegeneration, mislocalization, amyotrophic lateral sclerosis, post-translational modifications
Trist et al. profile the mislocalization, accumulation and deposition of immature SOD1 conformers in post-mortem spinal cord tissues of familial and sporadic ALS cases. They attribute these features to altered SOD1 maturation and post-translational modifications, also identified in these cases, which may represent valid therapeutic targets for ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal adult-onset neurodegenerative disease characterized by the progressive loss of motor neurons in the cortex, brain stem and ventral horns of the spinal cord. Ten percent of ALS cases are hereditary (familial; fALS), while the remaining ∼90% lack an overt familial history, referred to as sporadic (s)ALS.1 The gene encoding superoxide dismutase 1 (SOD1) was the first dominantly inherited genetic risk factor identified for ALS.2 and over 200 SOD1 mutations have since been documented in fALS patients (http://alsod.iop.kcl.ac.uk). These mutations disrupt key structural or functional motifs within SOD1 protein, resulting in structurally disordered mutant proteins that are strongly implicated in motor neuron death.3 A number of studies have also identified structurally disordered wild-type SOD1 proteinopathy in the spinal cord of non-SOD1-linked fALS and sALS patients.4–11 This finding, however, is not consistently reported,12–14 making it unclear whether wild-type SOD1 contributes to motor neuron death in ALS patients lacking SOD1 mutations.
Pathways hypothesized to underlie the self-assembly and toxicity of both wild-type and mutant SOD1 protein have been identified in silico and using in vitro systems, and in cellular and animal models of ALS.3 Evidence of these pathways in ALS patients in vivo or post-mortem, however, is scarce. These data are vital to ascertain whether SOD1 proteinopathy may indeed contribute to motor neuron death in familial and sporadic ALS patients through similar mechanisms to those identified in well-characterized disease models. Such knowledge may reveal productive treatment avenues, and improve the development and choice of preclinical models for therapeutic development, factors that are proposed to underlie recurrent failures to translate promising interventions into the clinic.15
Data from transgenic mouse models, cultured cells and in vitro proteomic assays demonstrate SOD1 mutations disrupt copper and zinc binding, intramolecular disulphide bond formation and protein dimerization (recently reviewed).3 These perturbations promote the accumulation of immature structurally disordered SOD1 (disSOD1) conformers, which elicit mitochondrial dysfunction,16 bioenergetic failure,17 reactive oxygen species accumulation18 and ultimately neurodegeneration in preclinical models. Non-genetic factors can also provoke structural and/or functional alterations to wild-type SOD1 protein analogous to those associated with SOD1 mutations. These include post-translational modifications (PTM) to key amino acid residue side chains, as well as disruptions to molecular chaperones involved in SOD1 protein maturation. These alterations confer toxic properties to wild-type SOD1 mirroring those of some mutant proteins in vitro,4 implying convergent molecular pathways resulting in SOD1 proteinopathy, irrelevant of SOD1 mutation status.19 Despite this abundance of data from preclinical models, it is unclear whether abnormal SOD1 proteinopathy is unique to SOD1-fALS patients, or constitutes a shared feature amongst all forms of ALS.
In this study, we characterized SOD1 proteinopathy in post-mortem spinal cord tissues from familial and sporadic ALS patients, and profiled soluble and aggregated SOD1 protein biochemistry within these same tissues for evidence of non-genetic alterations to SOD1 that may underlie any proteinopathy. We show that structurally disordered, immature SOD1 conformers becomes mislocalized from the ER–Golgi network and nucleus to the cytoplasm in SOD1-linked (SOD1-)fALS, non-SOD1-linked (non-SOD1-)fALS and sALS spinal cord motor neurons, where they accumulate and often form large inclusions. These findings were associated with the de-metallation and destabilization of enzymatically active mature SOD1 protein, as well as alterations to SOD1 PTMs and molecular chaperones governing SOD1 maturation. Heterogeneity in the presence of these alterations between ALS cases reinforces the likelihood that SOD1 proteinopathy arises from multiple biochemical pathways in ALS patients. Our data suggest some of these alterations may contribute to motor neuron death in ALS patients by promoting neurotoxic SOD1 proteinopathy, implying these pathways may constitute valid targets for therapeutic development.
Materials and methods
Human post-mortem tissues
Formalin-fixed and fresh frozen human post-mortem brain and spinal cord tissues from patients with SOD1-associated familial ALS (SOD1-fALS; n = 3), non-SOD1-associated fALS (non-SOD1-fALS; n = 4), sporadic ALS (sALS; n = 9) and age-matched controls (n = 10) were obtained from the MRC London Neurodegenerative Diseases Brain Bank (King’s College, London, UK) and the University of Maryland Brain and Tissue Bank, a biorepository of the NIH NeuroBioBank (Maryland, USA). Diagnoses of ALS were determined clinically using patient histories received from the donors’ physicians. Pathological identification of motor neuron loss in the anterior horn of the spinal cord by brain bank neuropathologists confirmed clinical findings. All ALS cases were free of other neurological or neuropathological conditions. Genotyping confirmed SOD1 and C9orf72 mutation status of fALS cases, as described next. Age-matched control cases were free of any clinically diagnosed neurological disorders and neuropathological abnormalities. Ethics approval was obtained from the University of Sydney Human Research Ethics Committee (approval number 2019/309). Demographic and clinical information for all cases is detailed in Supplementary Table 1. Diagnostic groups were matched for age and post-mortem interval (Kruskal–Wallis test, P > 0.05; Supplementary Table 2). A principal component analysis (PCA) of all data collected in this study revealed neither of these factors significantly affect variables of interest in the ventral spinal cord (Supplementary Fig. 1, further details in statistical analyses). This analysis also found no effect of sex on measured variables, despite differences in the ratio of males to females between diagnostic groups (Supplementary Fig. 2).
Fixed and fresh frozen tissues from the cervical and thoracic spinal cord represented regions of severe motor neuron degeneration in ALS, samples of occipital cortex represented a non-degenerating control region external to the spinal cord. Tissue availability varied between cases (Supplementary Table 1). Formalin-fixed tissues were embedded in paraffin and 7 µm sections prepared for immunohistochemical analyses. Fresh frozen tissues were trimmed to remove most of the white matter. Fresh frozen spinal cord tissues were bisected into dorsal and ventral regions representing degenerating and non-degenerating spinal cord tissues in ALS respectively. Paraffin-embedded formalin-fixed and fresh frozen tissue samples were randomly numbered by a secondary investigator (B.G.T. or S.G.) before experimentation to blind primary investigators (B.G.T., S.G., S.R., A.R. or J.A.F) to case diagnoses.
SOD1 and C9orf72 genotyping
SOD1 and C9orf72 genotyping was performed in ALS and control cases as previously described.20 DNA was extracted from fresh frozen human brain tissue from the occipital cortex using the DNeasy DNA extraction kit (catalogue no. 69506; Qiagen), according to manufacturer’s instructions. All five exons of SOD1, and at least 10 bp of flanking sequence were sequenced using polymerase chain reaction amplification and Sanger sequencing. The repeat primed polymerase chain reaction method was used to amplify the C9orf72 hexanucleotide repeat sequence and was analysed by fragment analysis.20 All results were independently analysed by two team members. Positive samples were repeated twice to confirm results.
Immunohistochemistry
Fixed tissue sections used for 3,3′-diaminobenzidine (DAB) staining and brightfield microscopy were deparaffinated and antigen retrieval performed in citrate buffer (pH 6; Fronine) at 95°C for 30 min. Sections were then washed with 50% ethanol, incubated with 3% H2O2 (Fronine) in 50% ethanol to quench non-specific peroxidase activity and blocked (0.5% casein, 1% bovine serum albumin in PBS) for 1.5 h at room temperature. Sections were then incubated overnight with appropriate primary antibodies diluted in blocking solution (Supplementary Table 3). Primary antibodies were detected using biotinylated IgG secondary antibodies (2 h, room temperature, 1:200 diluted in blocking solution) (Vector Laboratories) followed by Vector Elite Kit tertiary antibody complex (2 h, room temperature, 1:100 diluted in PBS) (Vector Laboratories), and visualized using a DAB (Sigma-Aldrich) solution containing cobalt and nickel ions, changing the chromogen colour from brown to black to enable distinction of DAB staining from pigmented neurons in the SNc. Sections were finally dehydrated and mounted with DPX, visualized and imaged using an Olympus VS 120 Slide Scanner at ×40 magnification (Olympus-Life Science), and images processed using OlyVIA (v.3.1; Olympus-Life Science) or Fiji [National Institute of Health (NIH)] software.
A lack of consistency between immunostaining protocols investigating mammalian SOD1 misfolding may underlie variability in misfolded SOD1 detection within ALS post-mortem tissues and transgenic SOD1 mice.9 Accordingly, our DAB immunostaining protocol for misfolded SOD1 protein adhered to guidelines proposed by Pare and colleagues,9 except for performing haematoxylin/eosin staining on adjacent serial tissue sections, which could not be performed due to limited tissue availability. A no primary negative control (processed as before in the absence of primary antibodies) was also included for each diagnostic group to identify protocol- and tissue-related artefacts independent of primary antibody immunoreactivity.
Multiplexed immunofluorescence
Fixed tissue sections used for fluorescent staining and confocal microscopy were deparaffinated, antigen retrieved, quenched, blocked and incubated with appropriate primary antibodies (Supplementary Table 3) as before. Multiplexed fluorescent immunolabelling was then performed using OPAL fluorophores (Akoya Biosciences) according to the manufacturer’s instructions. Antibody stripping between OPAL fluorophore incubations was performed at 95°C for 40 min using AR6 commercial buffer (Akoya Biosciences). Following incubation with the final fluorophore sections were washed with PBS and cover-slipped with 80% glycerol. Images were collected at ×60 magnification using a Nikon C2+ Confocal Microscope System and Nikon NIS-elements software (v.5.20.02; Nikon, Japan), and were viewed and analysed using Fiji software (NIH).
As before, a no primary negative control was included for each diagnostic group to identify non-specific fluorescent signals originating from autofluorescent tissue features (e.g. lipofuscin) or direct binding of secondary antibodies to tissues (Supplementary Fig. 3). During acquisition of double- or triple-immunolabelled fluorescent images a sequential protocol was used where each fluorophore was excited and the corresponding channel was individually acquired, before moving onto the next fluorophore and channel, to prevent any spectral cross-contamination between channels. Negligible spectral overlap was identified between OPAL650, OPAL620 and OPAL520 fluorophores using this protocol (Supplementary Fig. 3), indicating an absence of fluorophore cross-contamination across channels.
Neuronal quantification
Spinal cord motor neurons were quantified in DAB-immunostained serial 7 µm fixed tissue sections, whereby cells immunopositive for neuron-specific class III β-tubulin (TUJ-1) in ventral horn grey matter were counted in every fifth tissue section for a total of four sections. Quantification was completed by a single investigator (BT) blinded to sample diagnosis. Grey matter area (mm2) was recorded in Fiji software to enable conversion of neuron counts to neuronal densities (counts per mm2). Due to the absence of neuronal cell bodies in dorsal horn grey matter of the spinal cord, the average area (mm2) of grey matter in this region was used to approximate alterations to neuronal tract density. An index of neuronal loss was calculated for each investigated region of each case by normalizing neuronal density for each region/case to the mean neuronal density of the same investigated region for the healthy control group; Sample index = 1 − (sample neuronal density/healthy control mean neuronal density for sample region). Higher index values are associated with lower neuronal densities or greater neuron death. This index was used to explore correlations between neuron loss and quantified pathological features within investigated regions.
Quantification of pathological features of interest
Immunostaining for SOD1 and copper chaperone for SOD1 (CCS) was performed in ventral horn grey matter of the cervical and thoracic spinal cord as before, using appropriate primary antibodies (Supplementary Table 3). Pan and conformation-specific primary antibodies were used for SOD1 immunostaining. Pathologies of interest were SOD1 deposits, diffuse cytosolic SOD1, granular cytoplasmic SOD1, CCS deposits and diffuse cytosolic CCS. Counts of each pathology were expressed as the proportion of neurons exhibiting evidence of that pathology to account for differences in motor neuron number between cervical and thoracic spinal cord levels. Conformation-specific SOD1 antibodies yielded a 10-fold improvement in the identification of motor neuron inclusions compared with pan-SOD1 antibodies, supporting the strong involvement of disSOD1 conformers in SOD1 aggregation. Quantification of diffuse cytosolic disSOD1 staining was performed in sections stained with UβB conformation-specific SOD1 antibody and DAB. Using Fiji software, average DAB stain intensity was measured throughout the cross section of the neuron and was normalized to the average stain intensity within an area of similar size located immediately adjacent to the motor neuron of interest.
Fresh frozen tissue homogenization and fractionation
Fresh frozen tissue samples were homogenized in 5 volumes (µl) of homogenization buffer (20 mM Tris-base, pH 7.6, 1:100 EDTA-free protease inhibitor [Sigma-Aldrich), 1:10 EDTA-free phosphatase inhibitor (PhosSTOP; Roche)] per mg of tissue at 4°C using a hand-held tissue grinder with metal-free polypropylene probes (Kontes Pellet Pestle®, Sigma-Aldrich). Homogenates were aliquoted and stored at −80°C, or were centrifuged (16 000g, 30 min, 4°C) and the supernatant and pellet collected, aliquoted and stored at −80°C. Protein concentrations within homogenates and supernatants were quantified using a Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific). Supernatant and pellet tissue fractions will subsequently be referred to as soluble and insoluble tissue extracts, respectively. Measures were undertaken to prevent metal contamination of tissue extracts, including the use of plastic forceps for handling tissues, and the removal of trace metal impurities from buffers and glassware using Chelex resin (Bio-Rad) and a metal-chelating rinse solution (0.37% EDTA in 2.25 mM K3PO4; Sigma-Aldrich), respectively. These measures were also incorporated into native isoelectric focusing experiments and SOD1 specific activity assays.
SOD1 specific activity
SOD1 antioxidant activity was quantified in soluble tissue extracts using a commercial SOD Assay Kit (Cat. no. 19160, Sigma-Aldrich) and potassium cyanide (KCN), as previously described.21 KCN is a potent inhibitor of SOD1 antioxidant activity.22,23 Duplicate samples containing 2 µg protein were incubated with 15 mM KCN or 20 mM Tris buffer (pH 7.6; Sigma-Aldrich) for 60 min at room temperature, and SOD activity (U/ml) measured in both samples using the SOD Assay Kit, according to the manufacturer’s instructions. SOD1 activity (U/ml) was calculated by subtracting SOD activity measured in the KCN duplicate (non-SOD1 SOD activity) from the SOD activity measured in the Tris buffer duplicate (total SOD activity). As validation of complete SOD1 inhibition in KCN duplicates, incubation of commercial SOD1 (0–200 U/ml) with 15 mM KCN for 60 min at room temperature completely inhibited SOD1 activity up to ∼50 U/ml (Supplementary Fig. 4), whereas the average total SOD activity in age-matched control samples was only 3.91 U/ml in Tris buffer duplicates. The specific activity of SOD1 (U/ml/SOD1 protein) was determined by normalizing SOD1 activity to SOD1 protein levels measured by immunoblotting.
Immunoblotting
Here, 10 μg (SOD1) or 30 μg (CCS, GFAP) total protein from soluble tissue extracts was incubated in loading buffer [100 mM dithiothreitol (DTT), 3% SDS, 10% glycerol, 0.05% bromophenol blue, 62.5 mM Tris-base, pH 7.4; Sigma-Aldrich] at 95°C for 15 min. Extracts were then loaded into Any kD Mini-PROTEAN TGX Precast Protein Gels (15-well, 15 µl; Bio-Rad) and separated using the Bio-Rad Mini-PROTEAN Tetra Cell system (180 V, 40 min, 4°C). Separated proteins were transferred to Immun-blot PVDF membrane (0.2 µm pore size; Bio-Rad) overnight at 9 V and 4°C, before membranes were dried for 2 h at room temperature and proteins stained with Sypro Ruby Protein Blot Stain (ThermoFisher Scientific), according to the manufacturer’s instructions. Sypro Ruby-stained membranes were imaged using a Chemi-Doc XRS imaging system (630BP30nm filter, Bio-Rad), and total protein in each sample quantified by densitometry using ImageLab software (v.5.2, Bio-Rad). Membranes were then blocked for 2 h with 5% skim milk diluted in PBS-T (PBS containing 0.1% Tween 20; Sigma-Aldrich) and incubated overnight at 4°C with primary antibodies against SOD1, CCS and GFAP proteins (Supplementary Table 3) diluted in blocking solution. Primary antibodies were detected using appropriate horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) diluted in blocking solution (1:2000) for 2 h at room temperature. Protein signals were developed using ECL western blotting substrate (Bio-Rad), detected using a Chemi-Doc XRS imaging system (Bio-Rad) and quantified by densitometry using ImageLab software (v.5.2, Bio-Rad), all according to the manufacturer’s instructions. Protein levels of SOD1, CCS and GFAP were normalized to total protein levels within the same sample to correct for variations in protein loading, and an internal standard (pooled from all tissue samples) was included on every gel to correct for variability between gels and immunoblot runs. SOD1 protein amount was confirmed to exhibit a linear correlation with SOD1 immunoblot densitometry (Supplementary Fig. 5) from 0 to 1 µg using a recombinant human SOD1 protein standard. This encompasses the protein loading amounts used in our experiments, validating our SOD1 quantification method and accuracy. SOD1 and CCS reside within neurons and glia,21,24 and thus quantification of SOD1 and CCS protein levels must take into account neuron loss and astrogliosis within degenerating regions in ALS, reported herein and elsewhere.25 SOD1 and CCS protein levels were therefore normalized to a combined cellular index, which was generated by multiplying indices of neuronal loss (quantified by immunohistochemistry, Fig. 1H) and astrogliosis (quantified by GFAP immunoblot, Supplementary Fig. 6).
Figure 1.
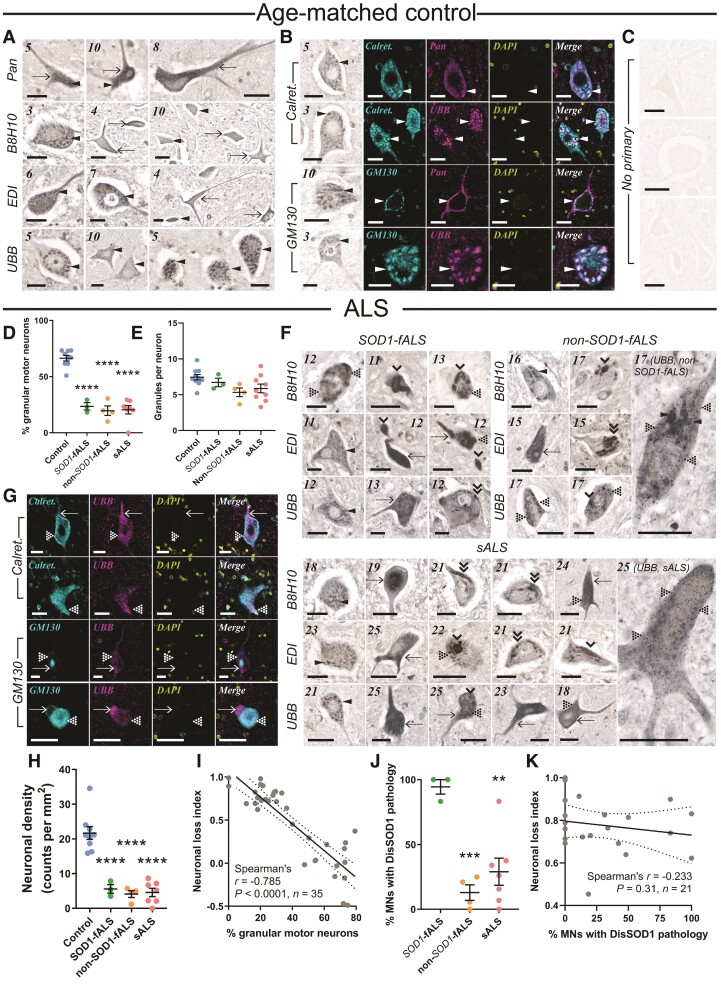
DisSOD1 accumulation, aggregation and mislocalization in the vulnerable ventral spinal cord of ALS cases. (A) Pan-SOD1 antibodies, as well as conformation-specific antibodies raised against disordered mutant SOD1 (B8H10), disSOD1 with an exposed dimer interface and disSOD1 with an unfolded β-barrel (UBB) were used to profile the distribution of mature SOD1 and disSOD1 conformers within control (n = 10) ventral spinal cord tissues. These SOD1 antibodies (Supplementary Table 3) detected granular cytoplasmic staining (arrowheads) and diffuse cytosolic staining (arrows) within control motor neurons. (B) Visualization of granular SOD1 immunostaining within control spinal cord motor neurons using the UBB conformation-specific SOD1 antibody and DAB chromogen. Granular SOD1 immunostaining strongly colocalized with the ER–Golgi markers Calreticulin and GM130 in control motor neurons. DAPI was also used to visualize cell nuclei. (C) No immunostaining was observed in spinal cord tissue sections processed as before in the absence of primary antibodies. (D) Motor neurons possessing granular SOD1 immunostaining were less abundant in all ALS subgroups compared with controls (one-way ANOVA: P < 0.0001, F = 55.82; Dunnett’s multiple comparisons post hoc tests: P < 0.0001 for all comparisons). (E) Granule numbers within surviving ALS motor neurons exhibiting granular SOD1 immunostaining were equivalent to those within control motor neurons (Kruskal–Wallis H-test, P = 0.073, H = 6.975). (F) In addition to granular and diffuse cytosolic immunostaining, conformation-specific antibodies labelled a variety of disSOD1 inclusions in ventral spinal cord motor neurons of SOD1-fALS (n = 3), non-SOD1-fALS (n = 4) and sALS (n = 9) cases; punctate inclusions (dotted arrowheads), globular inclusions (single downwards arrowheads) and fibrillar skein-like inclusions (double downwards arrowheads). Case numbers (Supplementary Table 1) are listed in the top left corner of each panel in A and F. (G) DisSOD1 conformers immunolabelled by the UBB SOD1 antibody were not colocalized with the ER–Golgi markers Calreticulin and GM130 in motor neurons of ALS cases. DAPI was also used to visualize cell nuclei. Scale bars = 25 µm in A–C, F and G. (H) Significant reductions in motor neuron density were observed in all ALS subgroups compared with controls (one-way ANOVA: P < 0.0001, F = 30.65; Dunnett’s multiple comparisons post hoc tests: P < 0.0001 for all comparisons). (I) The proportion of granular spinal cord motor neurons was correlated with great indices of motor neuronal loss in the ventral spinal cord. (J) The proportion of motor neurons (MNs) with punctate, globular and skein-like disSOD1 inclusions was higher in SOD1-fALS cases compared with non-SOD1-fALS and sALS cases (one-way ANOVA: P = 0.0011, F = 13.39; Dunnett’s multiple comparisons post hoc tests: non-SOD1-fALS: P = 0.0008; sALS: P = 0.002). Data in D, E, H and J represent mean ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001. (K) The proportion of spinal cord motor neurons with disSOD1 inclusion pathology was not correlated with neuronal loss in the ventral spinal cord. Spearman’s r coefficient, the P-value and the number of XY pairs analysed (n) are stated within I and K. A correlation is strong if Spearman’s r = 0.5 or higher.
Measurement of enzymatically active SOD1 metallation
The metal content of enzymatically active, dimeric SOD1 was quantified using size-exclusion chromatography (SEC) coupled with native isoelectric focusing (nIEF), synchrotron radiation X-ray fluorescence (SXRF) analyses and particle-induced X-ray emission spectroscopy (PIXE), according to our published method.26 Briefly, spinal cord and brain tissue homogenates were subjected to SEC to purify mature SOD1 dimers from post-mortem tissues, and SOD1 identified within collected fractions using nano-liquid chromatography–tandem mass spectrometry at a high sequence coverage of 92%. SOD1-containing fractions were then applied along immobilized pH-gradient (IPG) gels in duplicate and nIEF performed to further purify dimeric SOD1 according to its isoelectric point (pI). SEC-nIEF collectively yielded a 99-fold enrichment of SOD1 protein in post-mortem tissue extracts. After the pI of enzymatically active, dimeric SOD1 was identified within the first duplicate IPG gel using NBT staining and mass spectrometry, metal quantification was performed on the second duplicate IPG gel at the pI of active SOD1 using SXRF and PIXE. The SXRF analyses were performed in the microprobe hutch of the Hard X-ray Micro/Nano-Probe beamline P06 at the synchrotron PETRA III (DESY) in Hamburg (Germany). The analyses were carried out with an X-ray beam of 12 keV photon energy. The PIXE analyses were performed with 3 meV proton beam (focused down a spot size of 2 µm) produced at the AIFIRA facility of the CENBG (Gradignan, France) using a 3.5 MV singletron in-line particle accelerator (High Voltage Engineering Europe). For both techniques, triplicates were recorded for each sample and mean Cu/Zn ratios were calculated. Proteomic analyses using mass spectrometry identified minimal other metalloproteins within IPG gels at the pI of mature SOD1,26 and their extremely low abundance compared with SOD1 (≤0.57%) in protein extracts suggests negligible metal contamination from these proteins. Neither SEC nor nIEF altered the pI or the Cu:Zn ratio of a commercially available human SOD1 control,26 indicating preservation of endogenous metal-binding stoichiometry in purified SOD1 protein.
Native isoelectric focusing
Soluble tissue extract aliquots (40 μg total protein) were supplemented with DNase and RNase (Sigma-Aldrich) to a final concentration of 10 μg/ml and diluted to 140 μl using trace metal-free 20 mM Tris-base, pH 7.6. Duplicate IPG gels (pH 4.5–5.5, 7 cm; Zoom® IPG Strip, ThermoFisher Scientific) were rehydrated passively using 140 μl of diluted soluble extract in an IPG reswelling tray (Bio-Rad) for 2 h at 10°C. The backing of the IPG gels was covered in mineral oil (Bio-Rad) to prevent drying during rehydration. Gels were then focused in a Protean IEF Cell (Bio-Rad) for 12 kVh at 10°C (50 µA/gel; Supplementary Table 4). Immediately following nIEF, SOD1 activity was detected in IPG gels using nitroblue tetrazolium, as described previously,21 and the pI of SOD1 in each sample determined. One IPG strip rehydrated with commercially available human SOD1 (5 μg, ≥2500 units activity/mg protein; Sigma-Aldrich) was included per sample run to control for variability between runs.
Tissue preparation for SOD1 aggregate metal quantification
The metal content of SOD1 aggregates in the ventral spinal cord of ALS cases was quantified in fresh frozen tissue sections using XFM at the Australian Synchrotron (Clayton, Victoria, Australia), according to our published method.27 Briefly, frozen spinal cord tissue blocks from three SOD1-fALS, one non-SOD1-fALS and one sALS case underwent serial cryosectioning (20 µm) at −20°C. Immunohistochemical identification of SOD1 aggregates was performed on the first of two facing serial sections, using B8H10 conformation-specific SOD1 primary antibody-coupled with DAB staining on a Superfrost™ microscope slide (ThermoFisher Scientific), as described before. The adjacent section was reversed and mounted on a Si3N4 window (5 and 5 mm, 200 nm film thickness, 200 mm frame thickness; Australian National Fabrication Facility, QLD, Australia) with the matching tissue face exposed for synchrotron radiation analyses. We used an Olympus VS120 slide scanner (Olympus) to map the locations of SOD1 aggregates within immunolabelled sections using brightfield microscopy, and to map unlabelled facing sections mounted on Si3N4 windows using differential interference contrast imaging. Images of facing sections were overlaid using OlyVIA v.2.9 software (Olympus) and x,y pixel coordinates of SOD1 aggregates identified in unlabelled sections from their location in DAB-immunolabelled sections, often using distinct tissue features (blood vessels, dorsal or ventral horn boundaries) to triangulate aggregate locations in unlabelled sections. Pixel coordinates of aggregates were recorded and later transferred to motor stage positions at the beamline facility.
Australian Synchrotron XFM beamline operating parameters
The XFM beamline at the Australian Synchrotron28 is an in-vacuum undulator hard X-ray microprobe with a Maia 384 detector,29 which allows the rapid imaging of large samples. A Kirkpatrick–Baez mirror microprobe was used to focus monochromatic 15.8 keV X-rays to a 2 µm spot. Samples were mounted on Perspex frame and scanned in fly-scan mode with 1 µm step size and 5 ms dwell per pixel. Total XRF emission spectra were collected by the Maia detector. Pixel coordinates from OlyVIA v.2.9 were directly imported into the beamline using the integrated motor control system described previously.28
Australian Synchrotron XFM data analysis
Elemental maps were produced by fitting the XRF emission energies to the total XRF spectrum from each pixel. Model spectrum was produced with the assumption that dried tissues was 5 µm thick. This quantitative deconvolution of elemental fluorescence was performed using GeoPIXE v.7.2, which applies a first-order ‘matrix’ correction to estimate the reabsorption of the elemental fluorescence by the tissue.30 Images were exported as 32-bit tiff files for further analysis in Fiji software (National Institutes of Health, USA) as previously described.27 Images were analysed as described previously.27 In brief, limit of detection for each element was determined and each pixel corrected for background. A threshold of the upper 1% was also excluded from analysis. Aggregates were outlined by aligning brightfield microscopy photomicrographs with XFM images. Images were then normalized by pixel-wise division of Compton scatter emission to correct for variation in surface density and self-absorption effects, and aggregate measurements extracted for stacked XFM elemental maps. Extracted, corrected and normalized XFM data were imported into SPSS Statistics v.26 (International Business Machines, Illinois, USA) with matching subject demographic data. Outliers were defined by SPSS as ‘extreme values’ ≥3× the interquartile range (or 2 SD) and excluded from analysis. A two-sided t-test was used to compare elemental levels in SOD1 aggregates with background tissue and active SOD1. A significant difference in Cu:Zn ratio from the expected 1:1 was inferred from a 95% confidence interval that did not span 1 (Wilcoxon signed rank test).
Immunoprecipitation
SOD1 protein was immunoprecipitated from post-mortem spinal cord tissue homogenates using Dynabeads M-280 Tosylactivated (Invitrogen), according to the manufacturer’s instructions. Ten milligrams of Dynabeads M-280 Tosylactivated were conjugated to 100 µg polyclonal SOD1 antibody (Enzo Life Sciences; Supplementary Table 3) diluted to 40 mg beads/ml in coupling buffer (0.1 M boric acid, pH 9.5; 1.2 M ammonium sulphate) overnight at 37°C. Dynabeads were blocked with 0.5% BSA in PBS (pH 7.4), washed with 0.1% BSA in PBS (pH 7.4) and incubated with ventral spinal cord tissue homogenates (200 µg) diluted in PBS (pH 7.4) to 40 mg beads/ml overnight at 4°C. Following PBS washes, immunoprecipitated proteins were eluted from Dynabeads using successive 10 min incubations with 0.1 M glycine (pH 3), eluants neutralized using an equivalent volume of ammonium bicarbonate (pH 8) and extracts dried under pressure using a vacuum concentrator. We observed minimal reductions in the amount of SOD1 protein captured over successive rounds of immunoprecipitation on reuse of antibody-coupled Dynabeads (Supplementary Fig. 7), and hence beads were reused for three successive rounds of immunocapture. No measurable SOD1 protein was bound to antibody-coupled Dynabeads post elution (Supplementary Fig. 7), negating the possibility of cross-contamination between samples assigned to successive rounds of immunocapture. No alterations to the antioxidant activity or metal content of a commercially available human SOD1 standard were observed following immunoprecipitation (Supplementary Fig. 7),31 indicating SOD1 protein was probably isolated from post-mortem tissues having retained its endogenous metal occupancy and protein conformation. SOD1 protein was also not identified in immunoprecipitates prepared using Dynabeads that were not conjugated to our capture antibody (Supplementary Fig. 7).
Sample preparation for mass spectrometry
Dried SOD1 immunoprecipitates were resuspended in 50 mM ammonium bicarbonate (pH 8) containing 6 M urea, reduced with DTT (10 mM final) for 30 min at 56°C, alkylated with iodoacetamide (IAA; 20 mM final) for 30 min at room temperature in the dark and finally quenched with a further 10 mM DTT for 30 min at room temperature. Samples were diluted 5-fold using 50 mM ammonium bicarbonate (pH 8) to reduce the concentration of urea to 1.2 M and acetonitrile added (10% final), before in-solution digestion performed overnight at room temperature using 0.2 µg sequencing-grade modified trypsin (Promega). Samples were then acidified using trifluoroacetic acid, desalted using Pierce C18 Tips (ThermoFisher Scientific) according to the manufacturer’s instructions and dried under pressure using a vacuum concentrator. Samples were resuspended in loading buffer (0.1% formic acid, 3% ACN) and transferred to high-performance liquid chromatography vials immediately before mass spectrometry analyses.
Cysteine residues are highly susceptible to oxidation compared with other amino acids,32 therefore sample preparation for cysteine redox analysis utilized separate tissues prepared under a global reductive protein preparation strategy to limit the introduction of artefactual modifications during sample processing.33 Samples were homogenized as described before, and homogenates incubated with N-ethylmaleimide (NEM; 20 mM final) for 10 min at room temperature to alkylate free thiol groups. Excess NEM was quenched with DTT (25 mM final) for 15 min at room temperature, after which sample pH was adjusted above 7 using sodium hydroxide and samples reduced for 1 h at room temperature. Homogenates were then incubated with MMTS (60 mM final) for 1.5 h at 30°C to quench DTT and alkylate nascent thiols, and proteins precipitated using chloroform/methanol precipitation. Proteins were resuspended in 50 mM ammonium bicarbonate (pH 8) containing 6 M urea, diluted 5-fold using 50 mM ammonium bicarbonate (pH 8) and protein concentration quantified using a BCA assay according to manufacturer’s instructions. Acetonitrile was then added (10% final), and in-solution digestion performed overnight at room temperature using sequencing-grade modified trypsin (1:50 trypsin:protein; Promega). Samples were then acidified using trifluoroacetic acid, dried under pressure using a vacuum concentrator, resuspended in loading buffer (0.1% formic acid, 3% ACN) and transferred to high-performance liquid chromatography vials immediately before mass spectrometry analyses.
Mass spectrometry data acquisition
Label-free Fourier Transform Mass Spectrometry was used to analyse immunoprecipitated protein extracts at Sydney Mass Spectrometry (Sydney, New South Wales, Australia). Analyses were performed using an UltiMate 3000 RSLCnano system (ThermoFisher Scientific) coupled online via a Nanospray Ion Source (ThermoFisher Scientific) to an Orbitrap Fusion Tribrid Mass Spectrometer (ThermoFisher Scientific). Peptide digests were loaded onto an in-house packed ReproSil-Pur 120 C18-AQ analytical column (75 µm id × 40 cm, 1.9 µm particle size; Dr. Maisch GmbH) regulated to 60°C using a PRSO-V2 Sonation column oven. A binary gradient of solvent A (0.1% formic acid in MilliQ water) to solvent B (0.1% formic acid in 80% ACN diluted with MilliQ water) was used for peptide elution at a separation flow rate of 300–450 nl/min over 90 min (Supplementary Table 5). The mass spectrometer operated in positive ion mode at a 2.4 kV needle voltage. Data were acquired using Xcalibur software (v.4.4.16.14, ThermoFisher Scientific) in a data-dependent mode. MS scans (m/z 300–1650) were recorded at a resolution of R = 60 000 (m/z 200) and a standard automatic gain control target within 50 m. Dynamic exclusion was set to 9 s with a mass tolerance of ±10 ppm and top speed fragmentation in higher-energy collisional dissociation mode was performed for the top five most abundant precursor ions. MS/MS scans with a target value of 200% were collected in the ion trap with a maximum fill time of 28 ms. Additionally, only +2 to +5 charged ions were selected for higher-energy collisional dissociation fragmentation. Other settings were as follows: heated capillary temperature, 300°C; normalized higher-energy collisional dissociation collision energy of 30% and an isolation width of 1.6 m/z. Monoisotopic precursor selection was set to Peptide and an intensity threshold was set to 2 × 104.
For cysteine redox analysis, liquid chromatography–mass spectrometry was conducted on a Q-Exactive HFX mass spectrometer (ThermoFisher Scientific) coupled to an UltiMate 3000 RSLCnano UHPLC (ThermoFisher Scientific). Peptides were separated on a home-packed column (1.9 μm, 40 cm × 75 μm 120 Å pore size) of ReproSil-Pur C18 AQ beads (Dr. Maisch GmbH) regulated at 60°C with a PRSO-V2 Sonation column oven. A binary gradient of Buffer A (0.1% formic acid) to Buffer B (90% ACN 0.1% formic acid) from 5 to 98% Buffer B was used to elute peptide over 90 min at a rate of 300 nl/min. The mass spectrometer was coupled to a Nanospray Ion Source (ThermoFisher Scientific) and was operated in positive ion data dependant acquisition mode with a needle voltage of 2.4 kV and ion transfer capillary temperature of 300°C. Precursor ions were surveyed with an automatic gain control target of 1 × 106, maximum fill time of 50 ms and the orbitrap operating between 300–1650 m/z at a resolution of R = 60 000 at 200 m/z. The top 20 precursor ions with charges 2+–4+ were taken for higher collisional energy dissociation with an isolation window of 1.4 m/z, AGC target of 3 × 106, maximum ion fill time of 90 ms and normalized collision energy of 27 (N)CE. This occurred with a dynamic exclusion window of 15 s. Fragment ions were detected with a resolution of R = 15 000 at 200 Sm/z. For parallel reaction monitoring experiments, the mass spectrometer was operated in data independent acquisition mode consisting of a precursor ion scan followed by a loop of the scheduled inclusion list (Supplementary Table 6). Peptides were monitored in 16 min acquisition windows. The precursor ion scan occurred with a resolution of R = 30 000 at 200 m/z and fragment ion scans occurred with an isolation window of 0.7 Da and a maximum fill time of 65 ms, otherwise mass spectrometry parameters were as stated before (n = 3 injection replicates per sample).
Mass spectrometry data analysis
Raw data generated from immunoprecipitated protein extracts was processed using MaxQuant software (v.2.0.1.0, Max Planck Institute of Biochemistry, Planegg, Germany) under default settings.34 Spectra were screened against Uniprot entry P00441 (SODC-HUMAN), corresponding to human SOD1, with peptides higher than 4600 Da or lower than seven amino acids excluded from further analyses. Search parameters were as follows: mass accuracy of the monoisotopic precursor selection and peptide fragments was set to 10 ppm and 0.6 Da, respectively. Only b- and y-ions were considered for mass calculation. Oxidation of methionines (+16 Da) and protein N-terminal acetylation (+42 Da) were considered as variable modifications for protein quantification, whereas carbamidomethylation of cysteines (+57 Da) as well as carbamylation (+43 Da) of lysines and residue N termini were included as variable modifications for peptide identification and quantification. Given the presence of I113T and D101G mutations in SOD1-fALS samples, Xle->Thr and Asp->Gly were also included as variable modifications. PTMs of interest for comparison between ALS subgroups and controls were analysed in separate analysis batches, and included; 3-deoxyglucosone-derived hydroimidazolone (R; 144.0423), α-amino adipic acid (K; +14.9633), acetylation (K; +42.0106), allysine (K; −1.0316), carboxyethyllysine (K; +72.0211), carboxymethyllysine (K; 58.0055), deamidation (N/Q; +0.9840), glycation (K/R; +108.0211 with neutral loss of three water molecules),35 methylglyoxal-derived hydroimidazolone (R; +54.0106), nitration (W; +44.9976) oxidation (H/W; +15.9949), phosphorylation (S/T; +79.9663), succinylation (K; +100.0160) and ubiquitination (K; 114.0429). A maximum of five modifications per peptide was allowed. Two missed trypsin cleavages were allowed. Label-free quantification protein abundance was calculated using the MaxLFQ algorithm embedded in MaxQuant, requiring at least two ratio counts of unique peptides. Peptide validation was performed using the Andromeda algorithm and peptides were excluded if they possessed an Andromeda score lower than 80 or a localization score <0.8. The false discovery rates (FDR) for both peptides and proteins were adjusted to <1% using a target and decoy approach. The ‘match between runs’ option was enabled using a time window of 0.7 min. We achieved an average sequence coverage of 97.2% for SOD1 protein identified in sample immunoprecipitates. SOD1 protein was not identified in negative control immunoprecipitates prepared using Dynabeads that were not conjugated to our capture antibody, suggesting negligible false discovery of SOD1 protein in ALS and control protein extracts. Modified peptide intensities were normalized to SOD1 protein label-free quantification intensities to account for protein and/or sample concentration variance. No differences in the relative levels of PTMs of interest were identified between immunoprecipitated and non-immunoprecipitated commercial SOD1 protein (Supplementary Table 7), implying our immunoprecipitation protocol did not significantly alter PTMs of interest. Strong correlations between the abundance of SOD1 protein in technical replicates, as well as the abundance of modified peptides in these replicates, highlights the reproducibility of our experimental workflow (Supplementary Fig. 8).
Raw data generated from samples prepared for cysteine redox analysis was processed using Proteome Discoverer (ThermoFisher Scientific, v.2.4) and was searched against a Homo Sapiens Uniprot database (release 2020_05) with an in-house Mascot server (Matrix Science, London, UK) allowing for two missed trypsin cleavages and a minimum peptide length of six residues. Cys-N-ethylmaleimide (+125.05 Da) was set as a fixed modification and Cys-methylthio (+45.99 Da), Met oxidation (+16.00 Da) and N/Q deamidation (+0.98 Da) as variable modifications. A precursor mass tolerance of 20 ppm and fragment mass tolerance of 0.2 Da was used with a strict FDR cut-off of 1% and relaxed FDR cut-off of 5%. The results from the DDA experiment were then used to build a spectral library in Skyline (v.20.02). Chromatogram integration was performed in Skyline and summed transition peak areas for each peptide were taken for further processing. Each modified Cys peptide was normalized to two non-modified SOD peptides to account for protein and/or sample concentration variance. Rather than averaging these values, which would weight the normalization more towards one of the two non-modified peptides, normalized peak areas for both non-modified peptides were expressed as a ratio compared with the control diagnostic group, and were averaged to obtain final values.
Glutathione quantification
Glutathione (GSH) concentrations were quantified in soluble extracts using a published Tietze enzymatic recycling assay,36 modified to be compatible with a microtitre plate spectrophotometer. Protein was precipitated using 5% 5-sulphosalicylic acid, samples promptly centrifuged (10 000g, 10 min, 4°C), and supernatant collected and diluted 5-fold using 20 mM Tris buffer (pH 8) to reduce 5-sulphosalicylic acid to 1% before assay commencement. All assay steps were undertaken within an Atmosbag glove bag (catalogue no. Z530212, Sigma-Aldrich) under anaerobic conditions (<1% O2) using ultra-high purity nitrogen gas (99.999%, BOC Healthcare), except for spectrophotometry measurements where plates were sealed with airtight adhesive strips and transported to the microplate plate reader. Adhesive strips did not alter sample absorption (Supplementary Fig. 9). Anaerobic experimental conditions, together with storage of post-mortem tissues and extracts at −80°C before analysis, greatly reduces GSH degradation, ensuring measurements are as close to endogenous GSH levels as possible.37
Inductively coupled plasma–mass spectrometry
Metal levels in soluble and insoluble tissue extracts were quantified using inductively coupled plasma–mass spectrometry (ICP–MS), according to our group’s published methods.38 Soluble extracts were thawed on ice and diluted with 1% nitric acid (1:10 v/v; Suprapur grade, Merk Millipore) before analysis. Insoluble extracts were freeze-dried overnight, weighed and digested overnight using concentrated nitric acid (50 µl, 70%, Suprapur grade, Merk Millipore) at room temperature. Samples were then digested for a further 30 min at 70°C, and incubated with concentrated hydrogen peroxide (30%, VWH International) for 60 min at 70°C, and diluted with 1% nitric acid (1:10 v/v; Suprapur grade, Merk Millipore) before analysis. Total metal levels in each sample were measured in triplicate using an Agilent Technologies 7700 ICP–MS system with a Teflon MiraMist concentric nebulizer and Scott-type double-pass spray chamber (Glass Expansion). Buffer controls containing 1% nitric acid were incorporated every 20 samples. Helium (3 ml/min) was used as a collision gas for removal of polyatomic interferences. Measured mass-to-charge (m/z) ratios were 44 (Ca), 55 (Mn), 56 (Fe), 63 (Cu), 66 (Zn), 78 (Se) and 111 (Cd). External calibration was performed using multi-element standards (Sigma-Aldrich) diluted in 1% HNO3 and yttrium (Y; m/z = 89; Accustandard) was used as reference element via online introduction with a Teflon T-piece. Measurements were background corrected to metal levels in buffer controls, adjusted for dilution factors and were standardized against original wet tissue weights. Samples below the instrument’s limits of detection were excluded from analyses.
Statistical analyses
Statistical analyses were performed using IBM SPSS Statistics (v.27, IBM, Armonk, New York, USA) and GraphPad prism (v.7.02, Graphpad, San Diego, CA, USA). Parametric tests or descriptive statistics with parametric assumptions (standard two-way and one-way ANOVA, Pearson’s R and t-test) were used for variables meeting the associated assumptions, with data normality assessed using either the D’Agostino-Pearson (omnibus K2) normality test or the Shapiro–Wilk (Royston) normality test. Two- and one-way ANOVAs were paired with Sidak’s and Dunnett’s multiple comparisons post hoc tests, respectively, to assess pair-wise comparisons between select diagnostic groups for a given variable. Non-parametric tests or statistics (Kruskal–Wallis test, Spearman’s r and Mann–Whitney U-test) were used for variables where the observed data did not fit the assumptions of parametric tests, with Kruskal–Wallis tests paired with Dunn’s multiple comparisons post hoc tests to assess pair-wise comparisons between select diagnostic groups for a given variable. Where appropriate, non-parametric data were transformed using X = log(X) or X = SQRT(X) to meet parametric assumptions required for grouped analyses using two-way ANOVA. Outliers were identified using the combined robust regression and outlier removal method with a maximum FDR of 5%. Final values were reported as mean ± SEM. A P-value of <0.05 was accepted as the level of significance. Details of the statistical tests used (test statistics, sample sizes, P-values) for each variable of interest are included in the corresponding figure legend or results text section. An n of nine cases per diagnostic group was strongly powered to detect differences in SOD1 misfolding and conformational change (97.9–98.5% power, two-tailed t-test, α = 5%; SPSS software, IBM, Armonk, NY, USA) in a preliminary power analysis of pilot data describing the pI of wild-type SOD1 protein in the ventral spinal cord of a subset of sALS cases (n = 3) and age-matched controls (n = 3). Difficulties in sourcing and obtaining appropriate tissues from SOD1-fALS and non-SOD1-fALS cases limited the sample sizes of these diagnostic groups, however, retrospective power analyses reveal data from both groups were still strongly powered to detect differences in many measured variables (SOD1 pI, % motor neurons exhibiting granular disSOD1, SOD1 protein levels, SOD1 spec act.) compared with age-matched controls due to the large effect sizes observed for these variables (77.2–100% power).
Principal component analysis
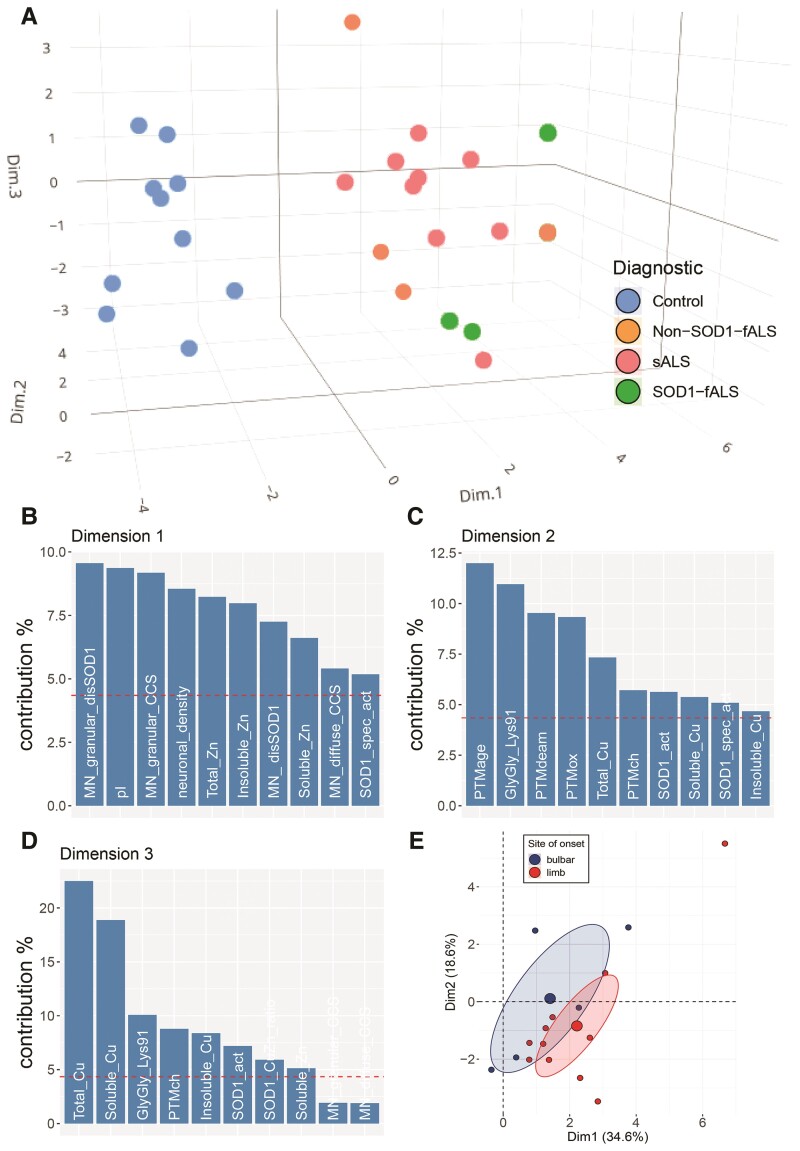
PCA was performed, and corresponding plots constructed, using RStudio v.1.4.1717, R 4.1.0 (RStudio Team, RStudio: Integrated Development Environment for R, Boston, MA, USA) and associated packages FactoMinerR v.2.4,39 missMDA v.1.18,40 factoextra v.1.0.741 and tidyverse v.1.3.1.42 The full reproducible code is available from the corresponding author on reasonable request. Active variables (n = 23) for the PCA are: percentage of motor neurons containing granular SOD1 (MN_granular_disSOD1), SOD1 inclusions (MN_disSOD1), granular CCS (MN_granular_CCS) and diffuse cytoplasmic CCS (MN_diffuse_CCS), motor neuron density (neuronal_density), total (SOD1_act) and specific SOD1 activity (SOD1_spec_act), SOD1 (SOD1_level) and CCS protein levels (CCS_level), Cu:Zn atomic ratio within enzymatically active soluble mature SOD1 (SOD1_CuZn_ratio), active SOD1 pI (pI), GSH levels and copper and zinc levels in whole tissues (Total_Cu, Total_Zn), as well as soluble and insoluble tissue extracts (Soluble_Cu, Soluble_Zn, Insoluble_Cu, Insoluble_Zn). SOD1 PTM data were also included, although these were grouped on the basis of similarities in expected functional consequences; PTMox—oxidation His, Trp, Cys, nitration Trp; PTMch—acetylation Lys, Phosphorylation Ser; PTMdeam—deamidation Asn; PTMage—CEL Lys; GlyGly_Lys91. Supplementary variables (case no., clinical diagnostic, age, sex, post-mortem interval, site of onset) do not influence on the PCA analysis and allow for easier interpretation and visualization of the results. The data were scaled before PCA and missing values were processed with the missMDA package. Our PCA revealed no significant differences in the relative levels of variables of interest between thoracic and cervical spinal cord levels (Supplementary Fig. 10). Thoracic and cervical spinal cord values were therefore averaged for cases possessing data for both spinal cord levels, yielding one data point per case for grouped statistical analyses.
Results
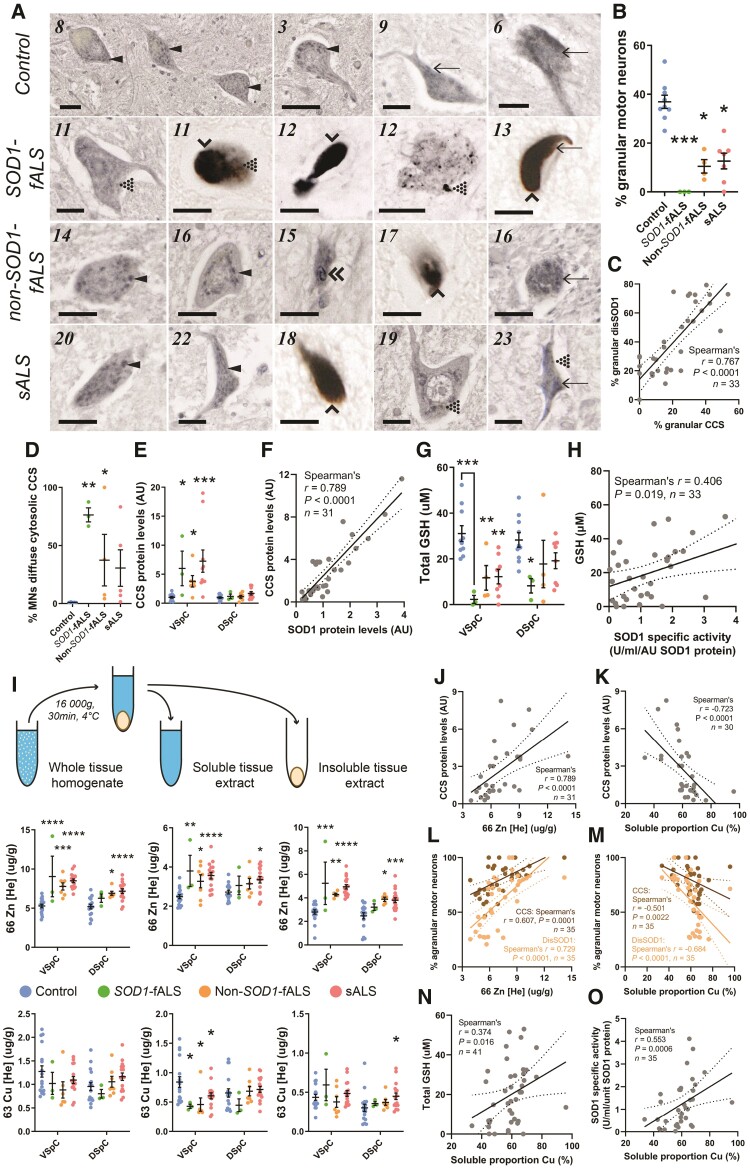
Disordered SOD1 conformers are mislocalized and accumulate in spinal cord motor neurons of ALS cases
Nascent, unfolded human SOD1 is structurally disordered and lacks antioxidant activity.43 Maturation of these immature species to enzymatically active, mature SOD1 dimers is multifaceted, and hence SOD1 protein exists within healthy neurons as a complex mixture of immature, intermediate and mature conformers.3,44 The accumulation of structurally disordered immature and intermediate conformers, hereon referred to as disordered (dis)SOD1, underlies the self-assembly and toxicity of mutant and wild-type SOD1 in vitro and in transgenic SOD1 mice.45,46
We used immunohistochemistry to profile the subcellular distributions of mature and disSOD1 conformers in 7 µm fixed post-mortem ventral spinal cord tissue sections collected from ALS patients and age-matched controls (Supplementary Table 1). Monoclonal and polyclonal pan-SOD1 antibodies recognizing full-length human SOD1 (Supplementary Table 3) identified diffuse cytosolic SOD1 immunoreactivity in 94% of spinal cord motor neurons in age-matched control cases (Fig. 1A), consistent with the primary subcellular localization of enzymatically active mature SOD1.47 Faint SOD1-immunopositive cytoplasmic granules (2.5–6 µm diameter) were observed within 10–20% of control spinal cord motor neurons (Fig. 1A), which colocalized with cis-Golgi (GM130; Fig. 1B) and endoplasmic reticulum (ER; calreticulin; Fig. 1B) markers, indicating partial localization of SOD1 to these compartments in control spinal cord motor neurons. Nuclear SOD1 immunostaining was also observed, consistent with a role for SOD1 as a nuclear transcription factor regulating antioxidant response genes.48 Due to the broad amino acid sequence coverage of their binding epitopes, pan-SOD1 antibodies immunolabel multiple mature and immature SOD1 conformers, thus it is unclear which specific conformers exist in each of these compartments.
Conformation-specific SOD1 antibodies can selectively detect immature disSOD1 conformers in human post-mortem tissues. We probed adjacent control spinal cord sections with exposed dimer interface, B8H10 and UβB conformation-specific antibodies (Supplementary Table 3), producing strong granular cytoplasmic SOD1 immunostaining within 66% of motor neurons, respectively, across all control cases (Fig. 1A). Colocalization of granular cytoplasmic SOD1 immunostaining with GM130 and calreticulin in sections probed with conformation-specific SOD1 antibodies (Fig. 1B) identified disSOD1 conformers within the ER–Golgi network in control motor neurons, constituting a physiological population of unfolded SOD1 in this compartment. A greater intensity of nuclear SOD1 immunostaining is these sections compared with those probed with pan SOD1 antibodies (Fig. 1A) suggests immature conformers may mediate the transcriptional activity of SOD1, rather than the mature, enzymatically active protein. In contrast, low-intensity diffuse cytosolic immunoreactivity was present within only 34% of control spinal cord motor neurons in these tissue sections (Fig. 1A). This suggests disSOD1 conformers exist either transiently, or in significantly lower abundance, within the cytosol of control motor neurons under physiological conditions. Overall, we show that mature SOD1 conformers probably constitute the dominant SOD1 species in the cytoplasm of healthy spinal cord motor neurons, while immature SOD1 conformers are present in relatively greater amounts in the nucleus and ER–Golgi network of these neurons.
Using control motor neuron immunostaining as a baseline, we applied pan and conformation-specific immunostaining to ventral spinal cord tissues from SOD1-fALS, non-SOD1-fALS and sALS cases to evaluate mature and immature SOD1 deposition and subcellular localization. The proportion of motor neurons possessing granular SOD1 immunostaining was reduced by 64–70% in all ALS subgroups compared with controls (Fig. 1D and E), and was replaced by high-intensity diffuse cytosolic disSOD1 and/or disSOD1 protein inclusions in the perikarya and neurites of these neurons in all ALS cases (Fig. 1F and Table 1). Nuclear disSOD1 was also absent in ALS spinal cord motor neurons at end-stage disease (Fig. 1F). Given the broad combined epitope coverage of all antibodies used, these data signify a subcellular redistribution of disSOD1 conformers within vulnerable motor neurons in ALS; indeed, disSOD1 conformers were primarily no longer localized to the ER–Golgi network in these patients (Fig. 1G). Reductions in spinal cord motor neuron density (Fig. 1H) were strongly correlated with lower proportions of granular disSOD1 motor neurons (Fig. 1I), but not higher proportions of motor neurons possessing disSOD1 inclusions (Fig. 1J and K). This is consistent with the involvement of smaller disSOD1 assemblages, rather than larger insoluble SOD1 deposits, in SOD1-mediated neuronal toxicity in ALS models.46 Collectively, our findings indicate that immature disSOD1 conformers are mislocalized to the cytoplasm from the nucleus and ER–Golgi network in ALS spinal cord motor neurons by end-stage disease, where they accumulate and often form large inclusions. These alterations are a reliable feature of all forms of ALS, which may be associated with motor neuron degeneration.
Table 1.
Characterization and quantification of SOD1 inclusions, as well as diffuse cytosolic and granular cytoplasmic disSOD1 staining, in ventral spinal cord tissue sections from all post-mortem tissue cases
| Case no. | Diagnostic group | Disordered SOD1 inclusions | Intense diffuse staining | Quantification (%MNs) | |||
|---|---|---|---|---|---|---|---|
| Globular | Skein-like | Punctate | Inclusions + diffuse staining | Granular | |||
| 1 | Age-matched control | − | − | − | ✓ | + | +++ |
| 2 | Age-matched control | NA | NA | NA | NA | NA | NA |
| 3 | Age-matched control | − | − | − | − | − | +++ |
| 4 | Age-matched control | − | − | − | ✓ | + | +++ |
| 5 | Age-matched control | − | − | − | ✓ | + | +++ |
| 6 | Age-matched control | − | − | − | − | − | +++ |
| 7 | Age-matched control | − | − | − | ✓ | ++ | +++ |
| 8 | Age-matched control | − | − | − | ✓ | ++ | +++ |
| 9 | Age-matched control | − | − | − | − | − | +++ |
| 10 | Age-matched control | − | − | − | ✓ | + | +++ |
| 11 | fALS (SOD1, I113T) | ✓ | ✓ | ✓ | ✓ | ++++ | ++ |
| 12 | fALS (SOD1, I113T) | ✓ | − | ✓ | ✓ | ++++ | + |
| 13 | fALS (SOD1, D101G) | ✓ | ✓ | ✓ | ✓ | ++++ | + |
| 14 | fALS (C9ORF72) | − | − | ✓ | ✓ | + | + |
| 15 | fALS (unknown) | ✓ | − | ✓ | ✓ | ++ | ++ |
| 16 | fALS (C9ORF72) | ✓ | − | − | ✓ | + | + |
| 17 | fALS (C9ORF72) | ✓ | − | ✓ | ✓ | + | + |
| 18 | sALS | − | − | ✓ | − | ++++ | + |
| 19 | sALS | − | − | − | ✓ | + | − |
| 20 | sALS | − | − | − | ✓ | ++ | + |
| 21 | sALS | ✓ | ✓ | − | ✓ | + | + |
| 22 | sALS | ✓ | − | ✓ | ✓ | + | + |
| 23 | sALS | − | − | − | ✓ | ++ | ++ |
| 24 | sALS | − | − | ✓ | ✓ | + | + |
| 25 | sALS | − | − | ✓ | ✓ | ++ | ++ |
| 26 | sALS | NA | NA | NA | NA | NA | NA |
Staining was performed using 7 µm paraffin-embedded formalin-fixed tissue sections and B8H10, exposed dimer interface and UBB conformation-specific SOD1 primary antibodies. Fixed tissues were not available (NA) for some cases. The four morphologies of disSOD1 pathology were noted as present (✓) or absent (−), and the proportion of motor neurons exhibiting any of the four morphologies quantified. The number of motor neurons exhibiting granular disSOD1 immunostaining was also quantified within each spinal cord ventral horn, expressed as a proportion of the total number of motor neurons in that ventral horn. Quantification classifications: − = 0%, + = >0–25%, ++ = 26–50%, +++ = 51–75%, ++++ = 76–100%. MN = motor neuron; NA = tissue not available.
Enzymatically dysfunctional SOD1 conformers are more abundant in the ventral spinal cord of ALS cases
Efficient superoxide detoxification by SOD1 is highly dependent on correct protein folding and maturation.3 To assess whether the cytoplasmic mislocalization and accumulation of immature disSOD1 conformers affects superoxide clearance by SOD1, we quantified SOD1 enzymatic activity in fresh frozen tissue from the ventral and dorsal spinal cord of our post-mortem cohort.21 Total SOD1 activity was unchanged in the ventral and dorsal spinal cord in all ALS subgroups compared with controls (Fig. 2A), indicating similar superoxide buffering capacities within these regions in ALS cases. These data are surprising considering oxidative stress is widely reported within the ALS spinal cord18 an environment that would be expected to result in an increased proportion of mature, enzymatically active SOD1 to increase superoxide clearance.
Figure 2.
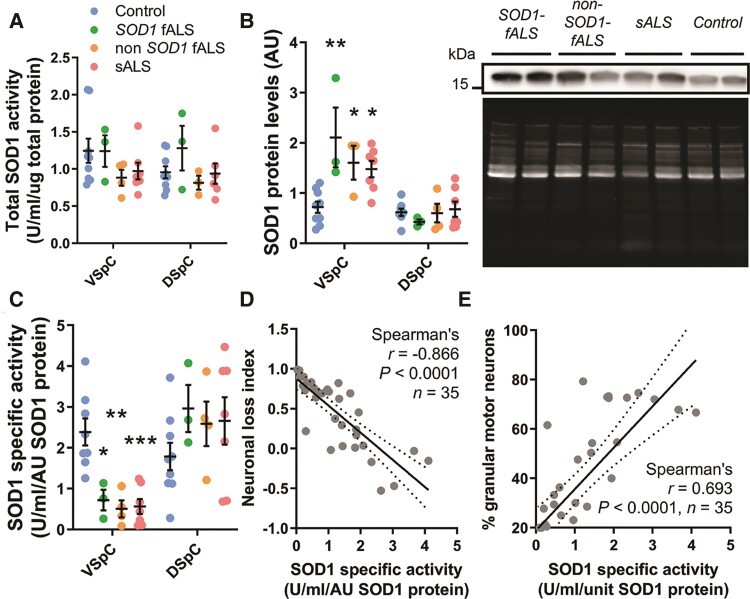
Alterations to SOD1 protein levels and SOD1 specific activity, but not total SOD1 activity, in the vulnerable ventral spinal cord of all ALS cases. (A) Total SOD1 antioxidant activity was unchanged in the ventral and dorsal spinal cord of ALS cases compared with controls [two-way ANOVA; F(3,36) = 1.888, P = 0.1491]. (B) SOD1 protein levels were significantly increased in the ventral spinal cord (VSpC) (two-way ANOVA with Sidak’s multiple comparisons post hoc test; SOD1-fALS: P = 0.0035; non-SOD1-fALS: P = 0.03; sALS: P = 0.0065), but not DSpC (SOD1-fALS: P = 0.677; non-SOD1-fALS: P = 0.974; sALS: P = 0.999), of all ALS subgroups compared with controls. Representative SOD1 immunoblots (SOD12 antibody, Supplementary Table 3) and Sypro Ruby blot staining for total protein in the VSpC are shown. (C) SOD1 specific activity (antioxidant activity per unit of SOD1 protein) was significantly decreased in the VSpC (two-way ANOVA with Sidak’s multiple comparisons post hoc test; SOD1-fALS: P = 0.0488; non-SOD1-fALS: P = 0.0011; sALS: P = 0.0002), but not DSpC (two-way ANOVA with Sidak’s multiple comparisons post hoc test; SOD1-fALS: P = 0.495; non-SOD1-fALS: P = 0.661; sALS: P = 0.724), of all ALS subgroups compared with controls. Data in A–C represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. (D and E) Reductions in SOD1 specific activity were strongly correlated with higher indices of neuron loss (D) and greater proportions of motor neurons lacking granular staining with UBB conformation-specific SOD1 antibody (E). Spearman’s r coefficient, P-values and the number of XY pairs analysed (n) are stated within D and E. A correlation is strong if Spearman’s r = 0.5 or higher.
Therefore, to investigate whether SOD1 protein production and/or maturation are dysregulated in ALS, we quantified SOD1 protein levels using immunoblotting and calculated SOD1 enzymatic activity per unit of protein (specific activity). Ventral spinal cord SOD1 protein levels were moderately increased in all ALS subgroups compared with controls (2–2.9-fold; Fig. 2B), however, SOD1 specific activity was reduced by 55–75% in this region of all ALS cases (Fig. 2C). These data are consistent with reductions in SOD1 activity measured in red blood cells, fibroblast and lymphoblast cell lines derived from SOD1-fALS patients.49 Levels of total SOD1 protein and specific activity were unchanged within the non-degenerating (Supplementary Fig. 11) dorsal spinal cord (Fig. 2B and C) of all ALS cases, indicating these alterations are restricted to regions of neuron death. Lower SOD1 specific activity was strongly correlated with decreased motor neuron density and greater disSOD1 mislocalization to the cytoplasm in the ventral spinal cord of ALS cases (Fig. 2E and E), consistent with a reduced viability of SOD1-deficient neurons under conditions of redox imbalance in Sod1+/− mice.18,49 Overall, these data demonstrate enzymatically dysfunctional SOD1 conformers selectively accumulate within vulnerable regions of the spinal cord in ALS (Fig. 2F).
Mature SOD1 destabilization may contribute to disordered SOD1 conformer accumulation in ALS spinal cord
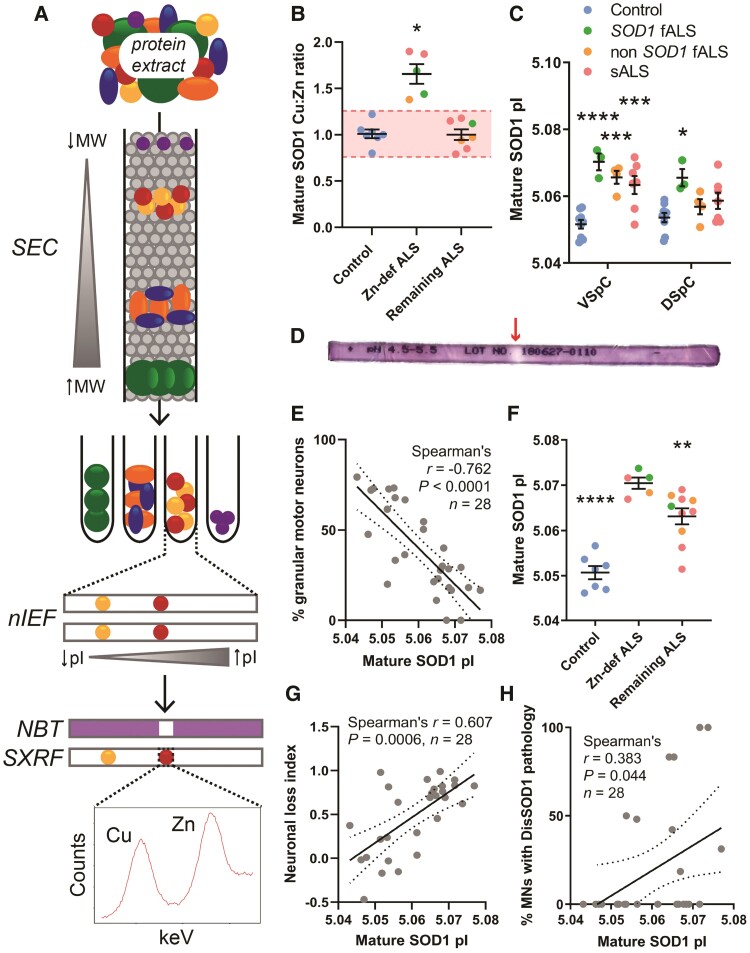
The accumulation of immature disSOD1 conformers in ALS motor neurons could result from disruptions to pathways governing SOD1 protein maturation, or alternatively from the destabilization of mature, enzymatically active SOD1 dimers, hereon referred to as mature SOD1. Mature SOD1 contains one zinc and copper ion per monomer, which together underlie its exceptional structural stability and enzymatic activity. Immature SOD1, on the other hand, exhibits a reduction in the binding of its metal cofactors, which underlies it is greater structural disorder and reduced enzymatic activity.3 We evaluated mature SOD1 stability and functionality by quantifying metals bound to enzymatically active SOD1 dimers (Fig. 3A).26 Mature SOD1 purified from the ventral and dorsal spinal cord of control cases contained copper and zinc in an exact 1:1 ratio (Fig. 3B),26 consistent with the theoretical Cu:Zn ratio bound to a mature SOD1 dimer. By contrast, the Cu:Zn ratio of mature SOD1 was significantly elevated in the ventral spinal cord of 42% of ALS cases (5-of-12; 1 Cu: 0.6 Zn) compared with controls (Fig. 3B), including two SOD1-fALS (I113T and D101G SOD1 mutants), one non-SOD1-fALS (C9ORF hexanucleotide repeat expansion mutant) and two sALS cases. Mature SOD1 metallation was unchanged in the non-degenerating occipital cortex of affected cases (Supplementary Fig. 12), suggesting perturbed SOD1 zinc binding results from biochemical factors unique to the ventral spinal cord. Our data provide the first evidence of alterations to the metallation of mature mutant and wild-type SOD1 in a proportion of familial and sporadic ALS patients, which may contribute to SOD1 pathology in these patients.
Figure 3.
Mature SOD1 metallation and surface charge are altered in the vulnerable ventral spinal cord of ALS cases. (A) Experimental workflow for measuring mature SOD1 metal content after purification of SOD1 from post-mortem tissue protein extracts.26 SEC and nIEF yielded over 99-fold enrichment of enzymatically active, SOD1 protein dimers by separating protein extracts according to molecular weight (MW) and pI, respectively. The absence of purple nitroblue-tetrazolium (NBT) staining identified enzymatically active SOD1 in the first of two duplicate IPG gels, which informed on the location of mature SOD1 in the second unstained IPG gel for SXRF analysis of metal content. (B) Mature SOD1 exhibited an increased Cu:Zn atomic ratio in 5 of 12 ALS cases compared with controls (Kruskal–Wallis H-test: P = 0.0014, H = 10.59; Dunn’s multiple comparisons post hoc tests: Zn-def ALS: P = 0.0118; remaining ALS: P = 0.9999). Control n = 7, remaining ALS n = 7. For ALS cases to be included in the Zn-deficient (Zn-def) group, their soluble mature SOD1 Cu:Zn ratio had to lie outside of 2 SD from the mean control Cu:Zn ratio. This range is commonly used to statistically delineate outliers and is represented by the red shaded band in this panel. (C) Mature SOD1 pI was significantly elevated in the ventral spinal cord (VSpC) of all ALS subgroups (two-way ANOVA with Sidak’s multiple comparisons post hoc test; SOD1-fALS: P < 0.0001; non-SOD1-fALS: P = 0.0002; sALS: P = 0.0002), as well as in the dorsal spinal cord (DSpC) of SOD1-fALS cases (two-way ANOVA with Sidak’s multiple comparisons post hoc test; SOD1-fALS: P = 0.0036; non-SOD1-fALS: P = 0.65; sALS: P = 0.163), compared with controls. (D) Representative nitroblue-tetrazolium-stained IEF gel, with enzymatically active SOD1 clearly visible by an achromatic gel band (red arrow). (E) The pI of mature SOD1 in cases exhibiting an increased Cu:Zn ratio of enzymatically active SOD1 dimers was significantly elevated compared with both controls, and ALS cases in which alterations to mature SOD1 metal content were absent (one-way ANOVA: P < 0.0001, F = 29.13; Holm–Sidak’s multiple comparisons post hoc test: control: P < 0.0001; remaining ALS: P = 0.009). Data in B, C and E represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (F) Increases in mature SOD1 pI were significantly correlated with lower proportions of spinal cord motor neuron possessing granular SOD1 staining. (G and H) Increases in mature SOD1 pI were significantly correlated with greater loss of spinal cord motor neurons (G) and more disSOD1 deposition (H) in these same post-mortem tissue cases. Spearman’s r coefficient, the P-value and the number of XY pairs analysed (n) is stated within F–H. A correlation is strong if Spearman’s r = 0.5 or higher.
Alterations to SOD1 protein pI reflect conformational changes50 that can alter both its aggregation propensity and the electrostatic guidance of anionic superoxide towards the active site. Mature SOD1 pI was significantly elevated in the ventral spinal cord in all ALS subgroups, and in the dorsal spinal cord of SOD1-fALS cases, compared with controls (Fig. 3C and D), suggestive of conformational changes to mature SOD1 in these regions. While elevated pIs in both spinal cord subregions of SOD1-fALS cases suggests SOD1 mutations may contribute to these alterations in SOD1-fALS, only one of the two mutant proteins examined in this study exhibits a reduced surface charge compared with wild-type SOD1,51 implicating alternative unknown factors. Likewise, the selectivity of this increase to the ventral spinal cord in other ALS cases indicates non-genetic factors unique to this region underlie such changes in these cases. Subcellular mislocalization of SOD1 to the cytoplasm within spinal cord motor neurons, for example, may contribute to the reduction in mature SOD1 surface charge in this region (Fig. 3E). Mature SOD1 pI was also increased in the ventral spinal cord of ALS cases exhibiting SOD1 zinc deficiency (n = 5), compared with control (n = 10) and remaining ALS cases (n = 11; Fig. 3F). While conformational change in mature SOD1 was not associated with reduced enzymatic activity (data not shown; Spearman’s r = −0.296, P = 0.13), it was correlated with greater motor neuron loss and deposition of SOD1 in the ventral spinal cord of ALS cases (Fig. 3G and H), consistent with data demonstrating reduced surface charge of SOD1 dimers promotes neurotoxic SOD1 aggregation from a native-like dimeric state in vitro.3 Overall, these results indicate that conformational change and mismetallation of mature SOD1 may contribute to the accumulation of dysfunctional, immature SOD1 conformers in the ventral spinal cord of ALS patients.
Zinc-deficient mature SOD1 is unlikely to undergo self-assembly from a native-like dimeric state in ALS cases
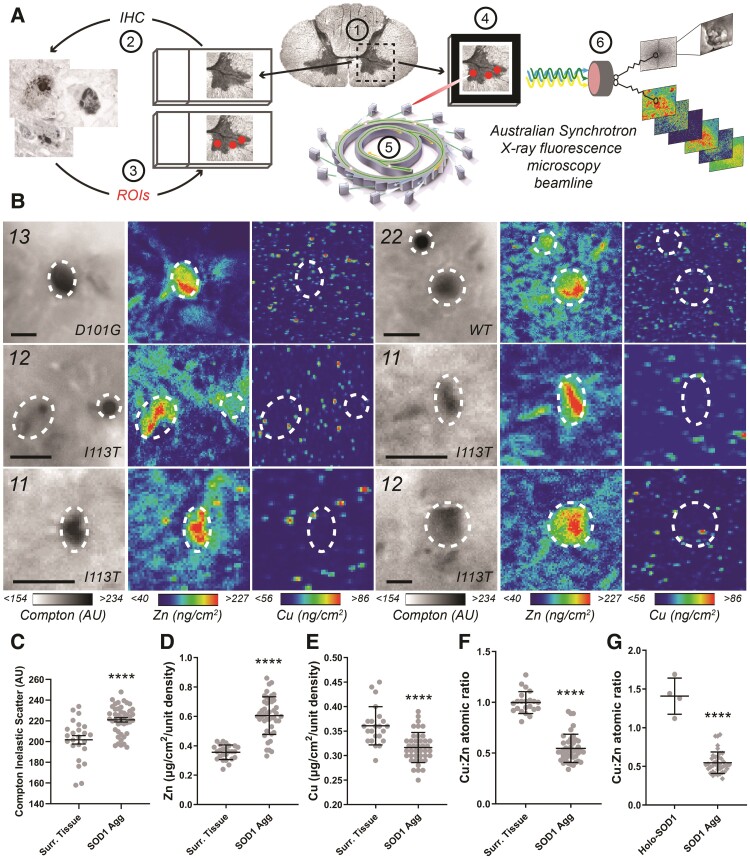
To ascertain whether zinc-deficient mature SOD1 may indeed aggregate directly from a disordered dimer-like state in vivo, we assessed whether the metal content of ventral spinal cord disSOD1 deposits reflects the elevated Cu:Zn ratio of soluble mature SOD1 in this region in select ALS patients. Copper and zinc levels were quantified within 45 individual SOD1 aggregates, ranging from 2.5 to 30 µm in diameter, and 25 areas of surrounding grey matter tissue of a similar size, in frozen unstained ventral spinal cord sections from three SOD1-fALS and one sALS case (Fig. 4A and B). Compton inelastic scattering was also measured simultaneously as an index of tissue matter density, revealing increased density within SOD1 aggregates compared with surrounding tissues (Fig. 4C). Normalization of metal levels to Compton scatter values ensured that varying tissue matter densities between aggregates, as well as between aggregates and surrounding tissues, did not confound interpretation of metal data. We identified a 70% increase in zinc (Fig. 4D), and a 12% reduction in copper (Fig. 4E), within SOD1 aggregates compared with surrounding grey matter tissues, constituting a significant decrease in the Cu:Zn ratio within SOD1 aggregates compared with surrounding tissues (Fig. 4F) and the Cu:Zn ratio of soluble mature SOD1 measured in these same cases (Fig. 4G). We speculate that these data indicate zinc-deficient, copper-containing mature SOD1 may not form aggregates from a native-like dimeric state in vivo. Instead, we propose that zinc deficiency promotes mature SOD1 dissolution into alternative metal-deficient disSOD1 conformers, which subsequently aggregate, as reported elsewhere.52 We recognize that this may also involve the recruitment of additional zinc-binding proteins, including CCS,21 which contribute to the reduced Cu:Zn ratio within SOD1 aggregates.
Figure 4.
Cu:Zn ratios within individual SOD1 aggregates are distinct from purified soluble mature SOD1. (A) Experimental workflow for identifying and analysing SOD1 aggregate metal content in post-mortem spinal cord tissues from ALS patients. Following serial sectioning of frozen post-mortem spinal cord tissues onto alternating Superfrost slides and Si3N4 synchrotron grids (1), SOD1 aggregates were immunostained on Superfrost slides using B8H10 conformation-specific SOD1 antibody (2) and their locations mapped as regions of interest (ROI) using an Olympus Slide Scanner (3). Tissue maps containing regions of interest were superimposed over differential interference contrast images of facing, serial, unstained frozen tissue sections mounted on Si3N4 grids (4). Synchrotron radiation was directed at regions of interest within unstained tissues mounted on Si3N4 grids using the XRF microscopy (XFM) beamline of the Australian Synchrotron (5) and XRF emission spectra and Compton scatter spectra collected by the Maia detector (6). (B and C) Compton scatter and XFM images of representative SOD1 aggregates in the ventral spinal cord (VSpC) of three SOD1-fALS and one sALS case (B). Scale bars = 10 µm, regions of interest are marked by white dashed circles. Case numbers and SOD1 mutations are listed in the top left corner of each panel. Higher Compton scatter values, represented by darker pixels, signify greater tissue matter densities in SOD1 aggregates compared with surrounding ventral horn grey matter tissue (Surr. Tissue; C; Unpaired t-test; P < 0.0001, t = 4.731, df = 66). (D–G) Zinc was increased by 70% (D; Unpaired t-test; P < 0.0001, t = 9.339, df = 67) and copper reduced by 12% (E; Unpaired t-test; P < 0.0001, t = 5.012, dfCu:Zn = 62), within SOD1 aggregates compared with surrounding grey matter tissues, resulting in a significant decrease in the atomic ratio of within SOD1 aggregates compared with surrounding tissues (F; Mann–Whitney U-test; P < 0.0001, U = 8.5) and the Cu:Zn ratio of soluble mature SOD1 measured in these cases (G; Mann–Whitney U-test; P < 0.0001, U = 0). Data in C–G represent mean ± SEM. ****P < 0.0001.
SOD1 protein PTMs are altered specifically in the vulnerable ventral spinal cord of ALS cases
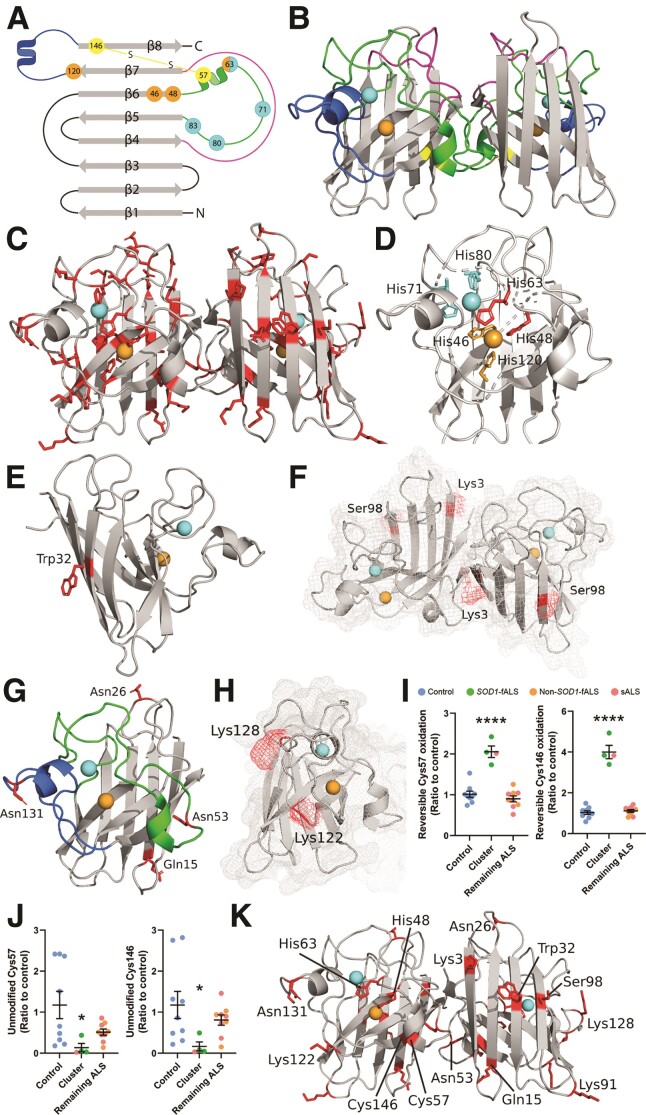
The PTM of key amino acid residue side chains within SOD1 protein regulates its structure (Fig. 5A and B), maturation, metallation, subcellular localization and enzymatic function.3,53 Recognizing that disSOD1 proteinopathy in ALS patients may result from altered SOD1 PTMs, we constructed post-translational fingerprints of SOD1 protein immunoprecipitated in its native state from post-mortem ventral and dorsal spinal cord of ALS patients and controls, identifying 42 individual PTMs to 29 residues (19%) of SOD1 protein across all diagnostic groups (Fig. 5C and Supplementary Tables 8 and 9).
Figure 5.
SOD1 protein structure and locations of PTMs which are significantly altered in the ventral spinal cord of fALS and sALS cases. (A and B) Mature SOD1 is dimeric, with each monomer comprising an eight-stranded β-barrel (grey) with one bound Cu (orange) and Zn (cyan) ion. The electrostatic loop (blue) contains charged and polar residues important for guiding anionic superoxide towards Cu in the active site. Three histidine residues and one aspartic acid residue (cyan) within the metal-binding loop (green) facilitate Zn coordination, while four histidine residues (orange) mediate Cu coordination. The disulphide loop (yellow) is a substructure within the metal-binding loop, containing one of two cysteine residues that form an intramolecular disulphide bond within SOD1 protein (yellow). The Greek key loop (pink) forms a plug at one pole of the β-barrel and contributes to dimer interface stability. (C) Distribution of all residues identified as sites of PTMs in SOD1 protein isolated from the ventral spinal cord (VSpC) of ALS cases and controls. Significant differences in the oxidation of His48 and His63 (D) oxidation and nitration of Trp32 (E) acetylation of Lys3 (F) phosphorylation of Ser98 (F) deamidation of Gln15, Asn26, Asn53 and Asn131 (G) and in the levels of carboxyethyllysine at Lys122 and Lys128 (H) were identified between a proportion of ALS cases and controls. Residues are labelled in wild-type SOD1 using three letter amino acid codes and the side chains (D–H) of altered residues are highlighted in red. Details of specific ALS cases exhibiting alterations to each PTM are presented in Supplementary Table 9. Reversible PTMs to Cys57 and Cys146 (I) were significantly increased in SOD1 protein isolated from three SOD1-fALS cases and one sALS case (cluster), compared with controls and remaining ALS cases (one-way ANOVA: Cys 57, P < 0.0001, F = 36.72; Cys146, P < 0.0001, F = 120.2; Dunnett’s multiple comparisons post hoc tests: P < 0.0001 when comparing both Cys57 and Cys146 to controls and remaining ALS cases). Corresponding significant reductions in unmodified Cys57 and Cys146 (J) residues were also identified in these cases compared with controls and remaining ALS cases (Kruskal–Wallis H-test with Dunn’s multiple comparisons post hoc tests for both residues; Cys 57—cluster versus control: P = 0.03; Cys146—cluster versus control: P = 0.04). Complete details of statistical analyses presented in I and J are presented in Supplementary Table 8. Data in I and J represent mean ± SEM. *P < 0.05, ****P < 0.0001. (K) Distribution of all residues whose post-translational modification is significantly altered in ALS cases.
Oxidation of SOD1 His48 and His63 was significantly increased (2-fold) in the ventral spinal cord of ALS cases possessing Zn-deficient soluble SOD1, compared with controls and remaining ALS cases (Supplementary Tables 8 and 9). This finding is consistent with a role for oxidation of metal-coordinating His residues in inducing SOD1 mismetallation in vitro54 (Fig. 5D). Significant increases in Trp32 oxidation (1.7-fold) and nitration (3.4-fold) were also identified within SOD1 protein isolated from the ventral spinal cord of three SOD1-fALS and five sALS cases, respectively, compared with controls (Supplementary Tables 8 and 9). Considering the role of this residue in redox-induced aggregation and propagation of disSOD1 in cellular and animal models of ALS55 (Fig. 5E), these modifications may contribute to the development of SOD1 proteinopathy in affected cases.
While Lys succinylation was unchanged between ALS cases and controls (Supplementary Table 8), Lys3 acetylation and Ser98 phosphorylation were significantly reduced in six and four sALS cases, respectively, compared with controls (Supplementary Tables 8 and 9). Although reduced Lys3 acetylation may increase the susceptibility of Lys3 to destabilizing PTMs, we did not detect any increase in oxidation of this residue between diagnostic groups (Supplementary Table 8). This reduction may, however, underlie the elevated active SOD1 pI in these cases (Fig. 3), which is proposed to promote SOD1 aggregation and intercellular propagation in vitro.56 Functionally, neither alteration is likely to underlie reduced SOD1 specific activity within the ventral spinal cord of sALS patients given their respective locations within SOD1’s dimer interface and β-barrel (Fig. 5F). Reduced Ser98 phosphorylation may, however, impede SOD1-mediated transcription of protective antioxidant genes by impairing SOD1 nuclear translocation.48
Deamidation of Gln15, Asn26, Asn53 and Asn131 was significantly increased in mutant SOD1 protein isolated from the ventral spinal cord of all SOD1-fALS cases compared with controls (Supplementary Tables 8 and 9). Comparatively mild (2-fold) increases in Asn26 deamidation may destabilize local hydrogen bonding networks in loop II, with similar increases in Gln15 deamidation capable of disrupting hydrogen bonding across the dimer interface to diminish dimer stability (Fig. 5G). Excessive (>10-fold) deamidation of Asn53 within the disulphide loop GDNT motif, and Asn131 within the electrostatic loop, on the other hand, can significantly affect coupling of SOD1 to CCS and greatly reduce the stability of mature SOD1 dimers3,57 (Fig. 5G). This may promote SOD1 aggregation in these patients.
While no sites of glycation were detected within SOD1 protein isolated from ALS cases and controls, levels of the advanced glycation end-product carboxyethyllysine were elevated at electrostatic loop residues Lys122 and Lys128 in mutant SOD1 protein isolated from ventral spinal cord of all SOD1-fALS cases compared with controls (Supplementary Tables 8 and 9). In vitro data suggest charge alterations resulting from these PTMs may disrupt superoxide diffusion towards the active site58 and alter stabilizing bonds between the electrostatic and disulphide loops, promoting structural disorder and aggregation59 (Fig. 5H).
Tryptic digestion of ubiquitin-conjugated proteins results in Gly-Gly dipeptide remnants on conjugated lysine residues, which serve as indicators of protein ubiquitination.60 Consistent with immunohistochemical analyses of SOD1 aggregates previously reported by our group,21 we identified a significant 2.5-fold increase in ubiquitination of mutant SOD1 at Lys91 in all SOD1-fALS cases compared with controls (Supplementary Tables 8 and 9), indicative of efforts by the ubiquitin-proteosome system to clear mutant disSOD1 from the ventral spinal cord of these cases. It is unclear why similar elevations in ubiquitination were not observed in non-SOD1-fALS or sALS cases exhibiting wild-type disSOD1 deposition, which suggests wild-type disSOD1 does not elicit the same proteostatic response from the UPS as mutant SOD1.
Analysis of free thiol groups in whole protein extracts prepared specially for cysteine redox analysis33 revealed 2- and 4-fold increases in reversible PTMs to Cys57 and Cys146, respectively, in mutant SOD1 protein isolated from the ventral spinal cord of all SOD1-fALS cases, and in wild-type SOD1 protein isolated from the ventral spinal cord of one sALS case, compared with controls and remaining ALS cases (Fig. 5I and Supplementary Tables 8 and 9). While the identity of these modifications remains unclear, increases in any candidate modification—sulphenic acid, S-nitrosylation, S-glutathionylation, S-palmitoylation, disulphide bonding—negatively affects the subcellular localization, structure and/or aggregation propensity of SOD1 protein in vitro.61–63 Indeed, all four cases exhibiting elevated reversible cysteine modifications possessed significantly higher SOD1 aggregate densities compared with other ALS cases examined (Supplementary Table 9), consistent with a role for redox-induced aggregation of SOD1 in these cases.64 It is unclear whether these modifications are upstream or downstream of redox imbalance documented in this region,18 however, we speculate that they probably form a cycle culminating in oxidative stress and a deteriorating antioxidant capacity within ALS spinal cord motor neurons. While we did not identify irreversible Cys PTMs in these cases (sulphinic/sulphonic acid), the greater magnitude of corresponding reductions in unmodified Cys57 (9.3-fold) and Cys146 (8.8-fold) residues (Fig. 5J and Supplementary Table 8) compared with reversible PTMs (2-to-4-fold) in these cases suggests they may be present but are undetectable in complex whole protein extracts.
In summary, we identified significant differences in levels of 15 PTMs to 14 SOD1 protein residues (9%) across 69% of ALS cases compared with controls (Fig. 5K and Supplementary Table 8 and 9). Importantly, only one of these alterations was also present in the dorsal spinal cord of affected cases (Supplementary Table 8), suggesting that the unique biochemical environment of the diseased ventral spinal cord may underlie alterations to SOD1 PTMs in these cases. Further examinations into these unknown predisposing factors for each altered PTM are warranted.
Disruptions to pathways governing SOD1 protein maturation may contribute to SOD1 pathology in ALS cases
In addition to direct alteration of SOD1 protein biochemistry, the impairment of key molecular chaperones facilitating SOD1 protein maturation promotes disSOD1 proteinopathy in vitro.65,66 To evaluate whether disrupted CCS may contribute to disSOD1 pathology in our ALS cases we profiled the subcellular distribution of CCS protein within spinal cord motor neurons and quantified CCS protein levels. Monoclonal and polyclonal antibodies recognizing full-length human CCS (Supplementary Table 3) identified CCS protein within cytoplasmic granules of a similar size and shape to those described for immature disSOD1 in ∼37% of control spinal cord motor neurons (Fig. 6A), consistent with preferential binding of CCS to immature SOD1 to promote metal insertion and maturation to mature SOD1.65,66 Low-intensity diffuse cytosolic CCS immunoreactivity was virtually absent within control spinal cord motor neurons (Fig. 6A and Supplementary Table 10).
Figure 6.
Alterations to the subcellular localization and levels of CCS protein, as well as in the levels of reduced GSH, copper and zinc, in the vulnerable ventral spinal cord of ALS cases. (A) Pan-CCS antibodies were used to profile the distribution of CCS within ventral spinal cord (VSpC) motor neurons from SOD1-fALS (n = 3), non-SOD1-fALS (n = 4) and sALS (n = 9) cases, and controls (n = 10). CCS antibodies (Supplementary Table 3) detected granular cytoplasmic staining (arrowheads) and diffuse cytosolic staining (arrows) within ALS and control motor neurons. In addition to granular and diffuse cytosolic immunostaining, CCS antibodies labelled motor neuron inclusions of punctate (dotted arrowheads), globular (single downwards arrowheads) and fibrillar skein-like morphologies (double downward arrowheads). Case numbers (Supplementary Table 1) are listed in the top left corner of each panel. Scale bars = 25 µm. (B) Motor neurons lacking granular CCS immunostaining were more abundant in all ALS cases compared with controls (Kruskal–Wallis H-test: P = 0.0005, H = 17.71; Dunn’s post hoc tests: SOD1-fALS: P = 0.0008; non-SOD1-fALS: P = 0.036; sALS: P = 0.011 compared with controls). (C) The proportion of spinal cord motor neurons lacking granular CCS and SOD1 immunostaining were strongly correlated. (D) The proportion of motor neurons possessing diffuse cytosolic CCS immunoreactivity was significantly higher in SOD1-fALS and non-SOD1-fALS cases compared with controls (Kruskal–Wallis H-test: P = 0.0014, H = 11.33; Dunn’s post hoc tests: SOD1-fALS: P = 0.006; non-SOD1-fALS: P = 0.05), with a strong trend for a similar increase in sALS cases compared with controls (sALS: P = 0.07). (E) CCS protein levels were significantly increased in the ventral, but not dorsal, spinal cord of all ALS subgroups [two-way ANOVA: F(3,58) = 11.89, P < 0.0001; Sidak’s multiple comparisons post hoc tests: VSpC—SOD1-fALS: P = 0.002; non-SOD1-fALS: P = 0.0011; sALS: P < 0.0001; DSpC—SOD1-fALS: P = 0.931; non-SOD1-fALS: P = 0.995; sALS: P = 0.163]. (F) CCS and SOD1 protein levels were significantly correlated within the VSpC. (G) GSH concentrations were significantly reduced in the ventral and DSpC of SOD1-fALS cases, and in the ventral spinal cord of non-SOD1-fALS and sALS cases (Two-way ANOVA: F (3, 42) = 10.52, P < 0.0001; Sidak’s multiple comparisons post hoc tests: VSpC—SOD1-fALS: P = 0.0007; non-SOD1-fALS: P = 0.0127; sALS: P = 0.002; DSpC—SOD1-fALS: P = 0.022; non-SOD1-fALS: P = 0.291; sALS: P = 0.231). (H) Reduced cellular GSH concentrations are correlated with reduced SOD1 specific activity in the VSpC. (I) Levels of zinc and copper were measured in whole tissue homogenates, as well as in Tris buffer-soluble and Tris buffer-insoluble tissue extracts, prepared from fresh frozen ventral and DSpC tissues of ALS cases and controls. Tissues were homogenized in Tris buffer and homogenates centrifuged at 16 000g for 30 min at 4°C to separate soluble and insoluble tissue fractions. Metal data are given as micrograms Cu or Zn per gram wet weight tissue. Complete details of statistical tests performed and test statistics are reported in Supplementary Table 11. Data in B, D, E, G and I represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Strong correlations were identified between biometal level changes and alterations to CCS protein levels (J and K) as well as increases in the proportions of motor neurons lacking granular disSOD1 or CCS immunostaining (L and M) in the VSpC. Reductions in the proportion of copper in the soluble tissue fraction were also associated with reduced GSH concentrations (N) and SOD1 specific activity (O) in the VSpC. Spearman’s r coefficient, the P-value and the number of XY pairs analysed (n) are stated within C, F, H and J–O. A correlation is strong if Spearman’s r = 0.5 or higher.
The proportion of motor neurons possessing granular CCS immunostaining was significantly reduced in all ALS subgroups compared with controls (Fig. 6A and B), indicating alterations to the subcellular localization of CCS within these neurons. The proportions of motor neurons lacking granular CCS and disSOD1 immunostaining were strongly correlated (Fig. 6C), suggesting CCS either contributes, or responds, to disSOD1 mislocalization to the cytoplasm. Diffuse CCS cytoplasmic staining was more intense and more prevalent in motor neurons of ALS cases compared with controls (Fig. 6A and D), and was associated with strong diffuse disSOD1 pathology (Supplementary Fig. 13). CCS protein levels were significantly increased in the ventral, but not dorsal, spinal cord of ALS cases compared with controls (Fig. 6E), and CCS and SOD1 protein levels were strongly correlated in the ventral spinal cord (Fig. 6F). CCS-immunopositive protein inclusions of varying morphologies were also observed in the perikarya and neuritic processes of 67–88% of SOD1-fALS motor neurons, as well as occasionally (<10%) in motor neurons of two non-SOD1-fALS and three sALS cases (Fig. 6A and Supplementary Table 10).
Glutathione in its reduced redox state (GSH) can also facilitate SOD1 copper loading and disulphide bond formation.67 We quantified a 2.5- to 14-fold reduction in GSH concentrations between all ALS subgroups compared with controls (Fig. 6G). Considering reductions in GSH were correlated with reductions in SOD1 specific activity (Fig. 6H), we speculate that alterations to CCS-independent SOD1 maturation pathways may contribute to immature disSOD1 accumulation in ALS patients by attenuating SOD1 copper loading.
Aside from disruptions to molecular chaperones governing SOD1 maturation, reduced copper or zinc bioavailability may contribute to SOD1 mismetallation in ALS cases (Figs 3 and 4). We identified a 47–71% increase in whole tissue zinc levels in the ventral and dorsal spinal cord of all ALS subgroups except the SOD1-fALS dorsal spinal cord (Fig. 6I), suggesting mature SOD1 zinc deficiency in select ALS cases is not likely to arise from a bulk reduction in zinc bioavailability. Copper levels remained unchanged in these tissues (Fig. 6H and Supplementary Table 11). To assess whether the compartmentalization of copper or zinc may also be altered in these patients, we partitioned whole tissue extracts into cytosolic and interstitial components (soluble fraction), and those encased within membranes, cellular organelles and other compartments (insoluble fraction).38 While zinc levels remained significantly elevated in both tissue fractions (Fig. 6I), copper was reduced 28–49% selectively in the soluble ventral spinal cord tissue fraction of all ALS subgroups compared with controls (Fig. 6I). Our data suggest zinc is elevated globally in all subcellular compartments of both spinal cord subregions, while copper is reduced selectively within the cytosol of ALS spinal cord motor neurons and glia, and the surrounding interstitium. Strong correlations between these biometal alterations and CCS protein levels reinforce a regulatory role for these metals in CCS expression68 (Fig. 6J and K), while additional correlations with higher proportions of motor neurons lacking granular disSOD1 or CCS immunostaining (Fig. 6L and M) suggests immature disSOD1 and CCS mislocalization to the cytoplasm are associated with biometal dysregulation in the ventral spinal cord. Copper redistribution within this region was also associated with reduced GSH (Fig. 6N), consistent with a role for GSH in cellular copper handling,69 which may affect SOD1 specific activity in ALS patients by reducing the availability of SOD1’s catalytic copper cofactor (Fig. 6O).
Collectively, we provide the first post-mortem tissue-derived evidence that alterations to cellular biometals and molecular chaperones facilitating SOD1 protein maturation may contribute to SOD1 mismetallation, aggregation, enzymatic dysfunction and subcellular redistribution in the ventral spinal cord of ALS patients.
Alterations to SOD1 biochemistry clearly demarcate ALS cases from controls
Alterations to SOD1 biochemistry, metallochaperones and tissue biometal levels, were identified in all ALS cases compared with controls, indicating such factors may constitute biomarkers for ALS. By applying a PCA to 23 parameters examined in this study we were able to accurately distinguish ALS cases from controls (Fig. 7A). The first three dimensions of this analysis accounted for most (64.6%) variation between cases, with the clear separation of ALS cases from controls largely a result of dimension 1. This suggests that the variables that contributed most to this dimension should be investigated further as potential biomarkers of ALS; disSOD1 and CCS subcellular localization, disSOD1 deposition, active SOD1 surface charge (pI) and tissue zinc levels (Fig. 7B).
Figure 7.
PCA of variables measured in this study. Active variables for this PCA were: percentage of motor neurons containing granular SOD1 (MN_granular_disSOD1), SOD1 inclusions (MN_disSOD1), granular CCS (MN_granular_CCS) and diffuse cytoplasmic CCS (MN_diffuse_CCS), motor neuron density (neuronal_density), total (SOD1_act) and specific SOD1 activity (SOD1_spec_act), SOD1 (SOD1 level) and CCS protein levels (CCS level), Cu:Zn atomic ratio within enzymatically active soluble mature SOD1 (SOD1_CuZn_ratio), active SOD1 pI (pI), GSH levels (GSH) and copper and zinc levels in whole tissues (Total_Cu, Total_Zn), as well as soluble (Soluble_Cu, Soluble_Zn) and insoluble tissue extracts (Insoluble_Cu, Insoluble_Zn). SOD1 PTM data were also included, although these were grouped on the basis of similarities in expected functional consequences; PTMox—oxidation His, Trp, Cys, nitration Trp; PTMch—acetylation Lys, Phosphorylation Ser; PTMdeam—deamidation Asn; PTMage—CEL Lys; GlyGly_Lys91. Each point represents a sample of ventral spinal cord (VSpC) sample in this study—that is, each dot expresses every measurement obtained for that particular sample as a single value in two-dimensional space. Large dots with coloured ellipses represent the centroid for that category or grouping and its 95% confidence interval. (A) PCA plot of SOD1-fALS (green), non-SOD1-fALS (orange), sALS (red) and control (blue) cases across dimension 1 (x-axis), dimension 2 (z-axis) and dimension 3 (y-axis). (B–D) The percentage contribution of the top 10 most influential variables comprising dimensions 1–3. The mean percentage contribution of all variables to each dimension is indicated by a red dotted line. (E) PCA plot of ALS cases grouped according to site of onset; bulbar (blue) or limb (red).
Importantly, we did not observe a clear separation of SOD1-fALS, non-SOD1-fALS and sALS cases across the three primary PCA dimensions (Fig. 7A). This is consistent with mounting data demonstrating similarities in the biochemistry of aberrant wild-type and mutant SOD1 proteinopathy in familial and sporadic forms of ALS.3 When considered as a single cohort however, we note substantial dispersion of ALS cases across dimensions 2 and 3 of our analysis. This is indicative of substantial heterogeneity in alterations to SOD1 biochemistry between individual ALS cases, in particular whole tissue copper levels, multiple SOD1 PTMs, SOD1 activity and soluble SOD1 metallation (Fig. 7C and D). It is possible that such dispersion of ALS cases may arise from differences in the site of disease onset (limb versus bulbar; Fig. 7E), or from differences between ALS patient subgroups (SOD1-fALS versus sALS).
Despite substantial heterogeneity in alterations to SOD1 biochemistry between ALS patients, our data suggest SOD1 biochemistry and other variables measured in this study may be able to distinguish ALS cases from controls, warranting further studies examining these factors in larger patient cohorts post-mortem and in the clinic.
Discussion
Despite several decades of intensive research into ALS-associated disease mechanisms in cellular and animal models of SOD1 proteinopathy, this is the first study to construct distinct profiles of the subcellular localization, relative abundance and deposition of mature and immature SOD1 conformers in spinal cord motor neurons of familial and sporadic ALS cases, and age-matched controls. We demonstrate that while large SOD1 protein inclusions are not reliable hallmarks of disease in spinal cord motor neurons of non-SOD1-linked fALS and sALS, the mislocalization of immature, disSOD1 from the nucleus and ER–Golgi network to the cytoplasm are characteristic of this vulnerable region in all forms of ALS. The presence of soluble or insoluble disSOD1 in non-SOD1-fALS and sALS cases is currently controversial.4–12,24 We and others9 posit this largely derives from methodological differences between studies. Accordingly, our disSOD1 immunostaining protocol adhered to logical guidelines recently proposed by Pare and colleagues for the use of conformation-specific SOD1 antibodies,9 which aim to minimize false-positives while maximizing tissue antigenicity to avoid false-negatives.
Accompanying our profiles of mature and immature SOD1 abundance, subcellular localization and deposition, we provide the first evidence that immature disSOD1 accumulation in the cytoplasm of ALS spinal cord motor neurons probably arises from both the destabilization of mature SOD1 and from abnormalities in SOD1 maturation in this region of ALS patients. Our novel SEC-nIEF workflow is the first to ensure metallation analyses focus selectively on enzymatically active mature SOD1,26 identifying reduced zinc binding and conformational change within mature SOD1 dimers; a finding that has yet to be reported in any transgenic ALS model. We are also the first to conduct such analyses on SOD1 protein isolated in it is native state from human tissues, a factor that we propose is likely to underlie discrepancies between our results and those previously reported from human SOD1-overexpressing transgenic animal and cellular models of ALS. While such over-expression is able to generate a robust ALS phenotype in these models, even wild-type human SOD1 is frequently isolated from these systems exhibiting reduced metallation,70,71 questioning whether changes identified accurately resemble those characterizing human ALS patients. Indeed, Lelie and colleagues70 found copper and zinc binding by endogenous mouse SOD1 was much closer to the expected 1:1 ratio compared with wild-type (Cu:Zn ∼1:3) and mutant (Cu:Zn ∼1:2.2) human SOD1 isolated from the spinal cord of transgenic mice of the same strain. These discrepancies are proposed to arise from the over-saturation of endogenous metallochaperones driving SOD1 metallation, or from dissimilarities between endogenous and transgenic human SOD1 proteins, warranting further investigation.
We also provide preliminary evidence of disruptions to SOD1’s molecular chaperones, CCS and GSH, as well as atypical PTM of structural motifs (e.g. GDNT) governing the maturation of SOD1 in ALS post-mortem tissues. While a previous study by Watanabe and colleagues72 only identified CCS-immunopositive inclusions in lumbar spinal cord motor neurons of one of 17 sALS cases, and neither of two SOD1-fALS cases, we highlight substantial differences in methodology between this and our study; spinal cord subregions examined, antibodies and dilutions used, and number of spinal cord sections analysed per patient. Perhaps most importantly for SOD1-fALS cases, we analysed tissues from patients carrying I113T and D101G SOD1 mutations, which probably affect SOD1-CCS coupling and metal transfer differently compared with A4V and unknown SOD1 mutants examined by Watanabe et al.72 This has already been demonstrated to some extent in vitro,73 whereby CCS is reported to possess a reduced efficacy in promoting zinc binding to mutant disSOD1 compared with wild-type SOD1.73 We speculate that this suggests structural disorder in immature mutant or wild-type proteins could subsequently result in under-metallation of mature SOD1 or SOD1 aggregate precursors, warranting further investigation. CCS also influences the subcellular compartmentalization of disSOD1 within organelles such as mitochondria by modulating SOD1 disulphide status.74 This suggests CCS mislocalization may precede and drive that of disSOD1 in ALS spinal cord motor neurons, although alterations to CCS biochemistry have not been thoroughly investigated in ALS post-mortem cases to date. Further profiling of chaperone-SOD1 interactions under physiological and pathological conditions is required to confirm the potential roles of CCS and GSH in the development of atypical SOD1 proteinopathy in ALS.
Many of the altered SOD1 PTMs in this study promote disSOD1 accumulation,48,75 self-assembly and mislocalization in vitro,3,53 and are thus also likely to contribute to the development of disSOD1 proteinopathy in the vulnerable ventral spinal cord of ALS patients. Indeed, immature disSOD1 protein is thought to exist on the precipice of aggregation, with even small disturbances in charge or hydrophobicity capable of surpassing small energy barriers between folding states to promote self-assembly.76 Additionally, the absence of all but one of these alterations in the dorsal spinal cord of our ALS cases suggests SOD1 proteinopathy is promoted selectively within the ventral spinal cord by endogenous biochemical factors within this region.
Importantly, while most biochemical changes to SOD1 identified in this study are specific to the ventral spinal cord, this does not necessarily equate to an aetiological contribution of all of these factors in motor neuron degeneration. Using our post-mortem data alone, it is difficult to establish whether alterations to SOD1 biochemistry precede, or follow, motor neuron death in ALS patients. It is indeed possible that some of the observed changes are attributable to cell death pathways activated by alternative aetiological factors, rather than any important SOD1-dependent mechanism. This is certainly the case for factors we report herein for the first time or which are seldom investigated elsewhere to date—disSOD1 and CCS subcellular mislocalization, and alterations to SOD1 phosphorylation, deamidation and acetylation. While reported structural and/or functional consequences of such changes for SOD1 may often imply associations with neuronal damage,48,57,74 direct examination of the cytotoxicity of these perturbations is required to examine any causal relationships. For other alterations, however, there are ample data from cellular and animal models of ALS to suggest an aetiological link with ALS; hence the inclusion of these variables for investigation in this study. Soluble disSOD1 conformers are widely linked to the protein’s toxicity in cellular and animal models of ALS,45,46,77 especially those that are metal-deficient52,78 or are atypically oxidized79 or glycated.80 Taken together, we posit that our data implicate mechanisms of SOD1-dependent toxicity in the death of spinal cord motor neurons in ALS patients.
Interestingly, substantial heterogeneity in the post-translational profile of SOD1 protein between ALS cases (Supplementary Table 9) suggests molecular mechanisms resulting in SOD1 proteinopathy and toxicity are variable between individual ALS patients, which may contribute to variability in disease progression in affected individuals. A similar concept was proposed by Wang and colleagues,81 who reported biochemical differences between mutant SOD1 proteins accounted for 69% of variability in SOD1-fALS patient survival times. Heterogeneity in additional disease-linked mechanisms between our ALS cases may combine with disSOD1 proteinopathy to influence rates of disease progression, including TDP-43 mislocalization and deposition, reported in a related study. The presence of multiple PTM alterations to SOD1 in 63% of cases examined (Supplementary Table 9) also indicates several pathways resulting in SOD1 self-assembly may be present in many patients. Mechanistically, we speculate that structural disorder induced by SOD1 mutations, or early-stage alterations to SOD1 PTMs, may promote subsequent alterations by increasing the solvent accessibility of new PTM sites. This may explain the greater number of alterations to mutant SOD1 in our SOD1-fALS cases compared with wild-type SOD1 in other ALS cases, although this may differ for other mutations not examined herein. Considering the similar disSOD1 conformations identified between cases using conformation-specific disSOD1 antibodies, we posit that these distinct pathways often culminate in common structural endpoints, reinforcing existing studies demonstrating similar disordered motifs within mutant and wild-type SOD1 proteins in ALS patients.4
Heterogeneity in ALS-linked pathologies has no doubt contributed to the limited success of past clinical trials in this disorder, suggesting multimodal or personalized treatment approaches may be required to meaningfully attenuate disease progression. For example, we reason that the cysteine-reactive compound Ebselen is unlikely to improve SOD1 folding and maturation in patients exhibiting redox modification of SOD1’s cysteine residues,82 and cases exhibiting increased oxidation of key proteins such as SOD1 are likely to respond better to antioxidant therapies. Similar categorization of SOD1-fALS patients into metal-binding-region and wild-type-like SOD1 mutants has already been proposed to identify those most likely to respond to copper supplementation using diacetylbis(4-methylthiosemicarbazonato)copperII [CuII(atsm)], given the negligible protection reported in cultured cells expressing metal-binding-region SOD1 mutants compared with wild-type-like mutants.83 A notable caveat of this proposal is the current lack of clinical diagnostic assays capable of differentiating these patient subgroups, which will be key to the successful implementation of such personalized approaches. While our study provides preliminary evidence that heterogenous SOD1 biochemistry between ALS patients may enable such stratification of these patients into subgroups that are more amenable to specific therapeutic approaches, difficulties in sourcing greater numbers of familial (SOD1, C9ORF) ALS cases prevents any concrete conclusions being drawn. Further studies examining these factors in larger patient cohorts are warranted.
Overall, we provide post-mortem evidence of pathways of SOD1 neurotoxicity in the spinal cord of ALS patients, which have previously only been characterized in cellular and animal models of ALS. Altered SOD1 biochemistry clearly differentiates all forms of ALS from controls, indicating SOD1 probably constitutes a valuable therapeutic target in all patient groups, irrelevant of SOD1 gene status.
Supplementary Material
Acknowledgements
Tissues were received from the University of Maryland Brain and Tissue Bank, a biorepository of the NIH NeuroBioBank, as well as from the Sydney Brain Bank, which is supported by Neuroscience Research Australia and the University of New South Wales, and from The London Neurodegenerative Diseases Brain Bank, which receives funding from the Medical Research Council and the Brains for Dementia Research programme, jointly funded by Alzheimer’s Research UK and Alzheimer’s Society. The authors thank Heather McCann of the Sydney Brain Bank for her help in processing fixed tissues. The authors acknowledge the facilities, as well as the scientific and technical assistance, of the Australian Microscopy and Microanalysis Research Facility (http://ammrf.org.au) node at the University of Sydney, as well as those of the Sydney Mass Spectrometry Facility at the University of Sydney. Research was also undertaken on the XFM beamline at the Australian Synchrotron, part of ANSTO. We acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities. Parts of this research were carried out at PETRA III beamline P06. Beamtime was allocated for proposal I-20180611. The research leading to this result has been supported by the project CALIPSOplus under the Grant Agreement 730872 from the EU Framework Programme for Research and Innovation HORIZON 2020. We thank the Biochemistry and Biophysics Platform (BioProt) of the Bordeaux Neurocampus facilities at the University of Bordeaux funded by the LABEX BRAIN (ANR-10-LABX-43) with the help of N. Sans and M. Sainlos. The AIFIRA facility is financially supported by CNRS/IN2P3, University of Bordeaux and the Région Nouvelle Aquitaine.
Abbreviations
- ALS =
amyotrophic lateral sclerosis
- CCS =
copper chaperone for SOD1
- disSOD1 =
structurally disordered SOD1
- ER =
endoplasmic reticulum
- f/sALS =
familial/sporadic ALS
- GSH =
glutathione
- IPG =
immobilized pH-gradient
- nIEF =
native isoelectric focusing
- PIXE =
particle-induced X-ray emission spectroscopy
- PTM =
post-translational modification
- SEC =
size-exclusion chromatography
- SXRF =
synchrotron radiation X-ray fluorescence
Contributor Information
Benjamin G Trist, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Sian Genoud, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Stéphane Roudeau, University Bordeaux, CNRS, CENBG, UMR 5797, F-33170 Gradignan, France.
Alexander Rookyard, School of Life and Environmental Sciences, Faculty of Science, The University of Sydney, Sydney 2006, Australia.
Amr Abdeen, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Veronica Cottam, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Dominic J Hare, School of Biosciences, The University of Melbourne, Parkville 3010, Australia; Atomic Medicine Initiative, University of Technology Sydney, Broadway 2007, Australia.
Melanie White, School of Life and Environmental Sciences, Faculty of Science, The University of Sydney, Sydney 2006, Australia.
Jens Altvater, Sydney Mass Spectrometry, The University of Sydney, Sydney 2006, Australia.
Jennifer A Fifita, Centre for Motor Neuron Disease Research, Macquarie Medical School, Faculty of Medicine, Health and Human Sciences, Macquarie University, Sydney, Australia.
Alison Hogan, Centre for Motor Neuron Disease Research, Macquarie Medical School, Faculty of Medicine, Health and Human Sciences, Macquarie University, Sydney, Australia.
Natalie Grima, Centre for Motor Neuron Disease Research, Macquarie Medical School, Faculty of Medicine, Health and Human Sciences, Macquarie University, Sydney, Australia.
Ian P Blair, Centre for Motor Neuron Disease Research, Macquarie Medical School, Faculty of Medicine, Health and Human Sciences, Macquarie University, Sydney, Australia.
Kai Kysenius, Department of Biochemistry and Pharmacology, The University of Melbourne, Parkville 3010, Australia.
Peter J Crouch, Department of Biochemistry and Pharmacology, The University of Melbourne, Parkville 3010, Australia.
Asuncion Carmona, University Bordeaux, CNRS, CENBG, UMR 5797, F-33170 Gradignan, France.
Yann Rufin, Plateforme Biochimie, University of Bordeaux, Bordeaux, France.
Stéphane Claverol, Plateforme Protéome, University of Bordeaux, Bordeaux, France.
Stijn Van Malderen, Deutsches Elektronen-Synchrotron DESY, Notkestr. 85, 22607 Hamburg, Germany.
Gerald Falkenberg, Deutsches Elektronen-Synchrotron DESY, Notkestr. 85, 22607 Hamburg, Germany.
David J Paterson, Australian Synchrotron, ANSTO, Clayton 3168, Australia.
Bradley Smith, Maurice Wohl Clinical Neuroscience Institute and the Institute of Psychiatry, Psychology and Neuroscience, King’s College London, SE5 9RT London, UK.
Claire Troakes, UK Dementia Research Institute at King’s College London, London SE5 9RT, UK.
Caroline Vance, Maurice Wohl Clinical Neuroscience Institute and the Institute of Psychiatry, Psychology and Neuroscience, King’s College London, SE5 9RT London, UK.
Christopher E Shaw, UK Dementia Research Institute at King’s College London, London SE5 9RT, UK.
Safa Al-Sarraj, London Neurodegenerative Diseases Brain Bank, Department of Basic and Clinical Neuroscience, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, SE5 8AF London, UK.
Stuart Cordwell, School of Life and Environmental Sciences, Faculty of Science, The University of Sydney, Sydney 2006, Australia.
Glenda Halliday, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Richard Ortega, University Bordeaux, CNRS, CENBG, UMR 5797, F-33170 Gradignan, France.
Kay L Double, Brain and Mind Centre and School of Medical Sciences (Neuroscience), Faculty of Medicine and Health, The University of Sydney, Sydney 2006, Australia.
Data Availability
The data and materials that support the findings of this study are available from the corresponding author on reasonable request.
Funding
This study was supported by ForeFront, a large collaborative research group dedicated to the study of neurodegenerative diseases and funded by the National Health and Medical Research Council of Australia Program Grant (1132524), Dementia Research Team Grant (1095127) and CogSleep Centre of Research Excellence (1152945). B.G.T. and K.L.D. are funded by the National Health and Medical Research Council of Australia (1181864), Parkinson’s NSW, The University of Sydney, MND Research Australia and The Michael J. Fox Foundation for Parkinson’s Research in partnership with the Shake It Up Australia Foundation. J.A.F. is funded by MND Research Australia (Bill Gole Fellowship). P.J.C. and K.K. were supported by funds from The Motor Neurone Disease Research Institute of Australia (Betty Laidlaw MND Research Project; Jenny Barr Smith MND Research Project), FightMND (Translational Research Grant) and the University of Melbourne. C.V. is funded by Motor Neurone Disease Association UK. G.H. is an NHMRC Senior Leadership Fellow (1176607).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Mejzini R, Flynn LL, Pitout IL, Fletcher S, Wilton SD, Akkari PA.. ALS genetics, mechanisms, and therapeutics: Where are we now? Front Neurosci. 2019;13:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide-dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature.1993;362:59–62. [DOI] [PubMed] [Google Scholar]
- 3. Trist BG, Hilton JB, Hare DJ, Crouch PJ, Double KL. Superoxide dismutase 1 in health and disease: How a frontline antioxidant becomes neurotoxic. Angew Chem Int Ed Engl. 2021;60:9215–9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guareschi S, Cova E, Cereda C, et al. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc Natl Acad Sci USA. 2012;109:5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tokuda E, Takei YI, Ohara S, Fujiwara N, Hozumi I, Furukawa Y. Wild-type Cu/Zn-superoxide dismutase is misfolded in cerebrospinal fluid of sporadic amyotrophic lateral sclerosis. Mol Neurodegener. 2019;14:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosco DA, Morfini G, Karabacak NM, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rotunno MS, Bosco DA. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front Cell Neurosci. 2013;7:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maier M, Welt T, Wirth F, et al. A human-derived antibody targets misfolded SOD1 and ameliorates motor symptoms in mouse models of amyotrophic lateral sclerosis. Sci Transl Med. 2018;10:eaah3924. [DOI] [PubMed] [Google Scholar]
- 9. Pare B, Lehmann M, Beaudin M, et al. Misfolded SOD1 pathology in sporadic amyotrophic lateral sclerosis. Sci Rep. 2018;8:14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forsberg K, Graffmo K, Pakkenberg B, et al. Misfolded SOD1 inclusions in patients with mutations in C9orf72 and other ALS/FTD-associated genes. J Neurol Neurosurg Psychiatry. 2019;90:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forsberg K, Jonsson PA, Andersen PM, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE. 2010;5:e11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Da Cruz S, Bui A, Saberi S, et al. Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathol. 2017;134:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerman A, Liu H-N, Croul S, et al. Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta Neuropathol. 2010;119:335–344. [DOI] [PubMed] [Google Scholar]
- 14. Liu HN, Sanelli T, Horne P, et al. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2009;66:75–80. [DOI] [PubMed] [Google Scholar]
- 15. van der Worp HB, Howells DW, Sena ES, et al. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7:e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pickles S, Destroismaisons L, Peyrard SL, et al. Mitochondrial damage revealed by immunoselection for ALS-linked misfolded SOD1. Hum Mol Genet. 2013;22:3947–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richardson K, Allen SP, Mortiboys H, et al. The effect of SOD1 mutation on cellular bioenergetic profile and viability in response to oxidative stress and influence of mutation-type. PLoS ONE. 2013;8:e68256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barber SC, Shaw PJ. Oxidative stress in ALS: Key role in motor neuron injury and therapeutic target. Free Radic Biol Med. 2010;48:629–641. [DOI] [PubMed] [Google Scholar]
- 19. Trist BG, Hare DJ, Double KL. A proposed mechanism for neurodegeneration in movement disorders characterized by metal dyshomeostasis and oxidative stress. Cell Chem Biol. 2018;25:807–816. [DOI] [PubMed] [Google Scholar]
- 20. McCann EP, Williams KL, Fifita JA, et al. The genotype-phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin Genet. 2017;92:259–266. [DOI] [PubMed] [Google Scholar]
- 21. Trist BG, Davies KM, Cottam V, et al. Amyotrophic lateral sclerosis-like superoxide dismutase 1 proteinopathy is associated with neuronal loss in Parkinson’s disease brain. Acta Neuropathol. 2017;134:113–127. [DOI] [PubMed] [Google Scholar]
- 22. Reddy CD, Venkaiah B. Purification and characterization of Cu, Zn-superoxide dismutases from mungbean (Vigna radiata) seedlings. J Biosci. 1984;6:115–123. [Google Scholar]
- 23. Iqbal J, Whitney P. Use of cyanide and diethyldithiocarbamate in the assay of superoxide dismutases. Free Radic Biol Med. 1991;10:69–77. [DOI] [PubMed] [Google Scholar]
- 24. Forsberg K, Andersen PM, Marklund SL, Brannstrom T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vargas MR, Johnson JA. Astrogliosis in amyotrophic lateral sclerosis: Role and therapeutic potential of astrocytes. Neurotherapeutics. 2010;7:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roudeau S, Trist BG, Carmona A, et al. Native separation and metallation analysis of SOD1 protein from the human central nervous system: A methodological workflow. Anal Chem. 2021;93:11108–11115. [DOI] [PubMed] [Google Scholar]
- 27. Genoud S, Jones MWM, Trist BG, et al. Simultaneous structural and elemental nano-imaging of human brain tissue. Chem Sci. 2020;11:8919–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howard DL, de Jonge MD, Afshar N, et al. The XFM beamline at the Australian Synchrotron. J Synchrotron Radiat. 2020;27:1447–1458. [DOI] [PubMed] [Google Scholar]
- 29. Ryan CG, Kirkham R, Hough RM, et al. Elemental X-ray imaging using the Maia detector array: The benefits and challenges of large solid-angle. Nucl Instrum Methods Phys Res A. 2010;619:37–43. [Google Scholar]
- 30. Ryan CG, Etschmann BE, Vogt S, et al. Nuclear microprobe-synchrotron synergy: Towards integrated quantitative real-time elemental imaging using PIXE and SXRF. Nucl Instrum Methods Phys Res B. 2005;231:183–188. [Google Scholar]
- 31. Kysenius K, Paul B, HIlton JB, Liddell JR, Hare DJ, Crouch PJ. A versatile quantitative microdroplet elemental imaging method optimised for integration in biochemical workflows for low-volume samples. Anal Bioanal Chem. 2019:411:603–616. [DOI] [PubMed] [Google Scholar]
- 32. Ahmad S, Khan H, Shahab U, et al. Protein oxidation: An overview of metabolism of sulphur containing amino acid, cysteine. Front Biosci (Schol Ed). 2017;9:71–87. [DOI] [PubMed] [Google Scholar]
- 33. Rookyard AW, Paulech J, Thyssen S, et al. A global profile of reversible and irreversible cysteine redox post-translational modifications during myocardial ischemia/reperfusion injury and antioxidant intervention. Antioxid Redox Signal. 2021;34:11–31. [DOI] [PubMed] [Google Scholar]
- 34. Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keilhauer EC, Geyer PE, Mann M. HCD fragmentation of glycated peptides. J Proteome Res. 2016;15:2881–2890. [DOI] [PubMed] [Google Scholar]
- 36. Ngamchuea K, Batchelor-McAuley C, Compton RG. Rapid method for the quantification of reduced and oxidized glutathione in human plasma and saliva. Anal Chem. 2017;89:2901–2908. [DOI] [PubMed] [Google Scholar]
- 37. Lin S, Tsai S-M, Huang J-C, et al. Effects of storage time and temperature on the stability of glutathione in deproteinized blood sample. J Food Drug Anal. 2006;14:141–146. [Google Scholar]
- 38. Genoud S, Roberts BR, Gunn AP, et al. Subcellular compartmentalisation of copper, iron, manganese, and zinc in the Parkinson’s disease brain. Metallomics. 2017;9:1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lê S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- 40. Josse J, Husson F. missMDA: A package for handling missing values in multivariate data analysis. J Stat Softw. 2016;70:1–31. [Google Scholar]
- 41. Kassambara A, Mundt F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Version R Package Version 1.0.7. 2020. https://CRAN.R-project.org/package=factoextra
- 42. Wickham H, Averick M, Bryan J, et al. . Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- 43. Banci L, Bertini I, Boca M, et al. Structural and dynamic aspects related to oligomerization of apo SOD1 and its mutants. Proc Natl Acad Sci USA. 2009;106:6980–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Culik RM, Sekhar A, Nagesh J, et al. Effects of maturation on the conformational free-energy landscape of SOD1. Proc Natl Acad Sci USA. 2018;115:E2546–E2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McAlary L, Aquilina JA, Yerbury JJ. Susceptibility of mutant SOD1 to form a destabilized monomer predicts cellular aggregation and toxicity but not in vitro aggregation propensity. Front Neurosci. 2016;10:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gill C, Phelan JP, Hatzipetros T, et al. SOD1-positive aggregate accumulation in the CNS predicts slower disease progression and increased longevity in a mutant SOD1 mouse model of ALS. Sci Rep. 2019;9:6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci USA. Feb. 1995;92:954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XF. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5:3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saccon RA, Bunton-Stasyshyn RK, Fisher EM, Fratta P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain. 2013;136:2342–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pergande MR, Cologna SM. Isoelectric point separations of peptides and proteins. Proteomes. 2017;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum Mol Genet. 2009;18:3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roberts BR, Lim NK, McAllum EJ, et al. Oral treatment with Cu(II)(atsm) increases mutant SOD1 in vivo but protects motor neurons and improves the phenotype of a transgenic mouse model of amyotrophic lateral sclerosis. J Neurosci. 2014;34:8021–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Banks CJ, Andersen JL. Mechanisms of SOD1 regulation by post-translational modifications. Redox Biol. 2019;26:101270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mulligan VK, Kerman A, Laister RC, Sharda PR, Arslan PE, Chakrabartty A. Early steps in oxidation-induced SOD1 misfolding: Implications for non-amyloid protein aggregation in familial ALS. J Mol Biol. 2012;421:631–652. [DOI] [PubMed] [Google Scholar]
- 55. Crown A, McAlary L, Fagerli E, et al. Tryptophan residue 32 in human Cu-Zn superoxide dismutase modulates prion-like propagation and strain selection. PLoS ONE. 2020;15:e0227655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abdolvahabi A, Shi Y, Rhodes NR, Cook NP, Marti AA, Shaw BF. Arresting amyloid with Coulomb’s law: Acetylation of ALS-linked SOD1 by aspirin impedes aggregation. Biophys J. 2015;108:1199–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shi Y, Rhodes NR, Abdolvahabi A, et al. Deamidation of asparagine to aspartate destabilizes Cu. Zn superoxide dismutase, accelerates fibrillization, and mirrors ALS-linked mutations. J Am Chem Soc. 2013;135:15897–15908. [DOI] [PubMed] [Google Scholar]
- 58. Fujii J, Myint T, Okado A, Kaneto H, Taniguchi N. Oxidative stress caused by glycation of Cu, Zn-superoxide dismutase and its effects on intracellular components. Nephrol Dial Transplant. 1996;11:34–40. [DOI] [PubMed] [Google Scholar]
- 59. Kato S, Takikawa M, Nakashima K, et al. New consensus research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 (SOD1) gene mutations: Inclusions containing SOD1 in neurons and astrocytes. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:163–184. [DOI] [PubMed] [Google Scholar]
- 60. Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. [DOI] [PubMed] [Google Scholar]
- 61. Valle C, Carri MT. Cysteine modifications in the pathogenesis of ALS. Front Mol Neurosci. 2017;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Redler RL, Wilcox KC, Proctor EA, Fee L, Caplow M, Dokholyan NV. Glutathionylation at Cys-111 induces dissociation of wild type and FALS mutant SOD1 dimers. Biochemistry. 2011;50:7057–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Toichi K, Yamanaka K, Furukawa Y. Disulfide scrambling describes the oligomer formation of superoxide dismutase (SOD1) proteins in the familial form of amyotrophic lateral sclerosis. J Biol Chem. 2013;288:4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rakhit R, Cunningham P, Furtos-Matei A, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem. 2002;277:47551–47556. [DOI] [PubMed] [Google Scholar]
- 65. Luchinat E, Barbieri L, Banci L. A molecular chaperone activity of CCS restores the maturation of SOD1 fALS mutants. Sci Rep. 2017;7:17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Banci L, Barbieri L, Bertini I, et al. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat Chem Biol. 2013;9:297–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carroll MC, Girouard JB, Ulloa JL, et al. Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc Natl Acad Sci USA. 2004;101:5964–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bertinato J, Iskandar M, L’Abbe MR. Copper deficiency induces the upregulation of the copper chaperone for Cu/Zn superoxide dismutase in weanling male rats. J Nutr. 2003;133:28–31. [DOI] [PubMed] [Google Scholar]
- 69. Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. [DOI] [PubMed] [Google Scholar]
- 70. Lelie HL, Liba A, Bourassa MW, et al. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J Biol Chem. 2011;286:2795–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hayward LJ, Rodriguez JA, Kim JW, et al. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–15931. [DOI] [PubMed] [Google Scholar]
- 72. Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8:933–941. [DOI] [PubMed] [Google Scholar]
- 73. Boyd SD, Ullrich MS, Calvo JS, Behnia F, Meloni G, Winkler DD. Mutations in superoxide dismutase 1 (SOD1) linked to familial amyotrophic lateral sclerosis can disrupt high-affinity zinc-binding promoted by the copper chaperone for SOD1 (Ccs). Molecules. 2020;25:1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kawamata H, Manfredi G.. Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid Redox Signal. 2010;13:1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tafuri F, Ronchi D, Magri F, Comi GP, Corti S. SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front Cell Neurosci. 2015;9:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sen Mojumdar S, N. Scholl Z, Dee DR, et al. Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat Commun. 2017;8:1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brotherton TE, Li Y, Glass JD. Cellular toxicity of mutant SOD1 protein is linked to an easily soluble, non-aggregated form in vitro. Neurobiol Dis. 2013;49:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hilton J, White A, Crouch P. Metal-deficient SOD1 in amyotrophic lateral sclerosis. J Mol Med (Berl). 2015;93:481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem. 2007;102:170–178. [DOI] [PubMed] [Google Scholar]
- 80. Sirangelo I, Vella FM, Irace G, Manco G, Iannuzzi C. Glycation in demetalated superoxide dismutase 1 prevents amyloid aggregation and produces cytotoxic ages adducts. Front Mol Biosci. 2016;3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Capper MJ, Wright GSA, Barbieri L, et al. The cysteine-reactive small molecule Ebselen facilitates effective SOD1 maturation. Nat Commun. 2018;9:1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Farrawell NE, Yerbury MR, Plotkin SS, McAlary L, Yerbury JJ. CuATSM protects against the in vitro cytotoxicity of wild-type-like copper-zinc superoxide dismutase mutants but not mutants that disrupt metal binding. ACS Chem Neurosci. 2019;10:1555–1564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials that support the findings of this study are available from the corresponding author on reasonable request.