Abstract
Nucleotide excision repair (NER) counteracts the onset of cancer and aging by removing helix-distorting DNA lesions via a ‘cut-and-patch’-type reaction. The regulatory mechanisms that drive NER through its successive damage recognition, verification, incision and gap restoration reaction steps remain elusive. Here we show that the RAD5-related translocase HLTF facilitates repair through active eviction of incised damaged DNA together with associated repair proteins. Our data shows a dual incision-dependent recruitment of HLTF to the NER incision complex, mediated by HLTF’s HIRAN domain that binds 3’-OH single-stranded DNA ends. HLTF’s translocase motor subsequently promotes dissociation of the stably damage-bound incision complex together with the incised oligonucleotide, allowing efficient PCNA loading and initiation of repair synthesis. Our findings uncover HLTF as an important NER factor that actively evicts DNA damage, thereby providing an additional quality control by coordinating the transition between the excision and DNA synthesis steps to safeguard genome integrity.
Keywords: HLTF, TFIIH, DNA damage, Nucleotide Excision Repair, genome stability, damage eviction, repair synthesis, post-replication repair, UV damage response
INTRODUCTION
Genomic integrity is continuously threatened by a diverse array of DNA lesions that interfere with vital cellular processes such as replication and transcription (Hoeijmakers, 2009; Jackson and Bartek, 2009). Inadequate removal of such DNA lesions may induce tumorigenesis or accelerated aging. To prevent these severe consequences, cells have evolved sophisticated networks of DNA repair pathways to remove DNA damage. One such DNA repair pathway that is particularly involved in the restoration of a wide variety of helix-distorting DNA lesions, such as those induced by chemotherapeutics and UV-light, is Nucleotide Excision Repair (NER). Defects in NER have been implicated in various genetic human diseases, including the cancer-prone disorder Xeroderma Pigmentosum and the premature aging-characterized Cockayne Syndrome, underscoring the importance of this repair pathway in human physiology (Marteijn et al., 2014).
NER relies on the concerted action of more than 30 proteins to remove DNA lesions from the genome and is divided into two sub-pathways by two distinct mechanisms of damage recognition (Marteijn et al., 2014; Schärer, 2013). Global genome NER (GG-NER) is initiated by damage recognition throughout the entire genome via the damage-sensing XPC and UV-DDB complexes (Puumalainen et al., 2016; Sugasawa, 2016), whereas damage-induced RNA polymerase II stalling initiates transcription coupled NER (TC-NER) (Lans et al., 2019). Damage recognition by either NER sub-pathway results in recruitment of TFIIH, a multisubunit transcription and repair complex (Compe and Egly, 2016) that, together with XPA, opens a local repair bubble and provides kinetic proofreading of NER substrates (Li et al., 2015; Sugasawa et al., 2009). This complex, further stabilized by the incorporation of RPA on the opposing undamaged strand, forms a scaffold for the sequential recruitment and correct positioning of the structure-specific endonucleases XPG and ERCC1-XPF (de Laat et al., 1998). Full assembly of the NER incision complex initiates coordinated dual incision of damage-containing DNA, triggered first by ERCC1-XPF 5’ of the lesion and subsequently by XPG 3’ of the lesion (Staresincic et al., 2009). After damage removal, the resulting 22–30 nucleotide single-stranded DNA (ssDNA) gap is restored through the combined action of PCNA, RFC and various DNA polymerases and is finally sealed by DNA ligation factors (Ogi et al., 2010; Sertic et al., 2018).
Intriguingly, although the incision complex is stably associated with the lesion and the surrounding chromatin, even covering an extensive stretch of DNA that extends beyond the 5’ edge of the repair bubble (Kokic et al., 2019), TFIIH and XPG dissociate from the chromatin in complex with the incised damage-containing oligonucleotide (Hu et al., 2013; Kemp et al., 2012). Therefore, the dissociation of damage-containing oligonucleotides after dual incision from the stable complex suggests active displacement rather than a spontaneous release. Such active eviction of the damaged DNA may also facilitate efficient gap filling synthesis and thereby represent a delicately controlled transition between the incision and DNA synthesis steps. In line with this hypothesis, upon damage incision, bacterial NER requires the specialized translocase UvrD to achieve lesion dissociation, thereby coordinating the initiation of the ensuing repair synthesis (Kisker et al., 2013). However, no such active damage eviction mechanism that uncouples the dual incision from the actual damage removal has been reported for mammalian NER thus far.
Here, we identify the RAD5-related SF2 translocase Helicase-Like Transcription Factor (HLTF) as an important NER factor. To date, HLTF has been predominantly known for its functions in post-replication repair (PRR), where it was shown to function as an E3 ubiquitin ligase for PCNA and stimulate replication fork reversal in response to replication stress (Neelsen and Lopes, 2015; Poole and Cortez, 2017). We show that HLTF plays an unexpected additional role in maintaining genome stability, namely by stimulating NER through catalyzing the release of damage-containing oligonucleotides incised by the NER machinery. HLTF is recruited to the lesion-bound NER incision complex via its HIRAN domain that binds 3’-OH ssDNA ends generated by NER-mediated incision, after which the ATP-dependent translocase motor of HLTF promotes active removal of damage-containing oligonucleotides together with the incision complex members TFIIH, XPG and ERCC1-XPF. This HLTF activity is crucial to allow the stable loading of downstream factors like PCNA and DNA polymerase δ to enable subsequent DNA synthesis to complete the NER reaction.
RESULTS
Identification of HLTF as an UV-induced TFIIH interaction partner
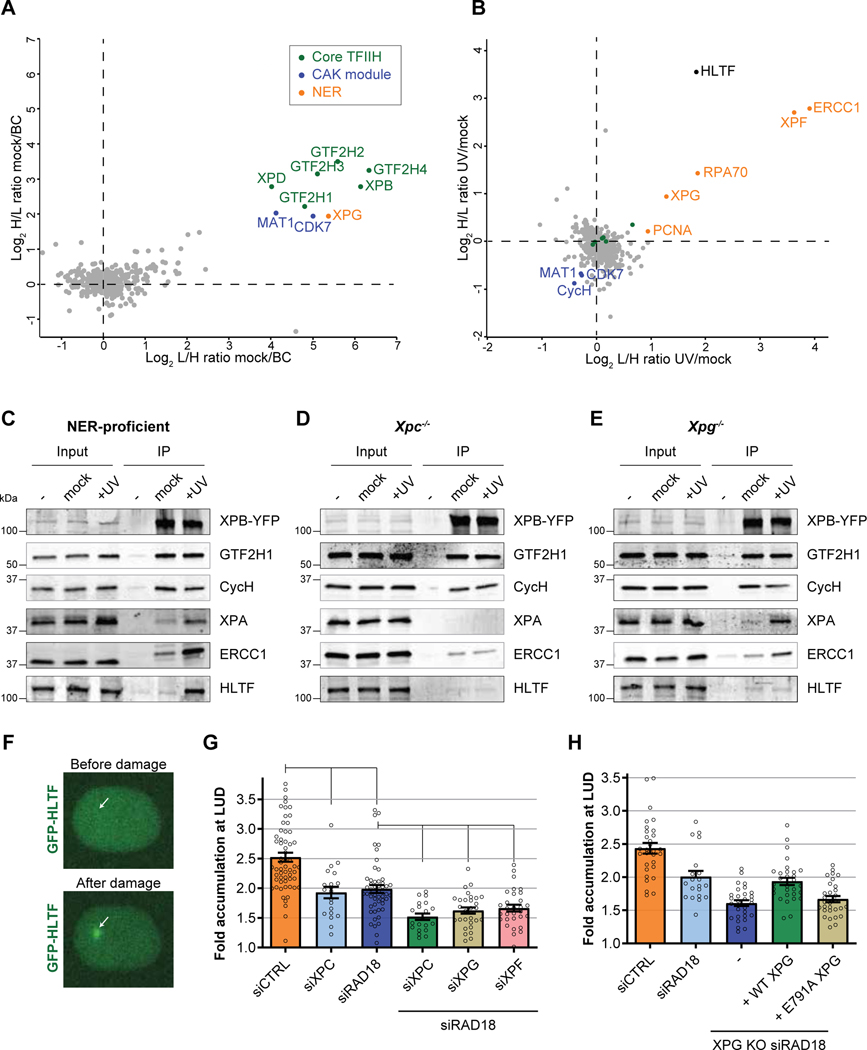
Since TFIIH is a central factor during NER and is both part of the incision complex and dissociates together with excised damage-containing oligonucleotides, we performed SILAC-based quantitative interaction proteomics to identify novel TFIIH interaction partners that could be involved in the active removal of incised oligonucleotides. TFIIH-interacting proteins were isolated from Xpb-YFP knock-in (KI) mouse dermal fibroblasts (MDFs) expressing a fully functional YFP-tagged version of this largest TFIIH subunit at endogenous levels (Giglia-Mari et al., 2009). We used a crosslinked immunoprecipitation approach as this was previously shown to successfully isolate chromatin bound NER complexes (Schwertman et al., 2012; Theil et al., 2011). Proof-of-principle for this approach was shown by the selective enrichment of multiple core TFIIH subunits (XPD, GTF2H1 through GTF2H4), as well as members of the ternary CAK subcomplex (Greber et al., 2017) and the XPG endonuclease (Ito et al., 2007) under unperturbed conditions (Fig 1A; Table S1). DNA damage-induced TFIIH interactors were identified by comparing the TFIIH interactome of unperturbed cells with that of cells exposed to UV-induced DNA damage. In line with previous studies, we observed a DNA damage-dependent increase in the association of TFIIH with the core NER factors ERCC1, XPF, XPG and RPA70 (de Laat et al., 1998; Mocquet et al., 2008; Volker et al., 2001), while the CAK module dissociated (Coin et al., 2008) (Fig 1B; Table S1). Interestingly, our approach also identified the SF2 family translocase HLTF (Mailand et al., 2013; Motegi et al., 2008; Neelsen and Lopes, 2015; Unk et al., 2008) as an UV-induced TFIIH interactor, with similar SILAC ratios as bona fide NER proteins. HLTF was also identified as a UV-induced TFIIH interactor using a non-crosslinked proteomics procedure (Fig S1A; Table S1). TFIIH immunoprecipitation experiments followed by immunoblotting confirmed that HLTF specifically interacts with TFIIH after UV-induced DNA damage (Fig 1C), like the core incision complex members ERCC1 and XPA.
Figure 1. Dual incision-dependent recruitment of HLTF to the NER incision complex.
(A-B) Scatter plots depicting log2 SILAC ratios of XPB interactors identified across 2 independent proteomics experiments including a label swap. XPB interacting proteins were isolated from Xpb-YFP knock-in mouse dermal fibroblasts (MDFs) and identified under either (A) unperturbed (mock-treated) conditions (BC; binding control), or (B) 1 hour after UV-induced DNA damage (16 J/m2).
(C-E) Crosslinked XPB-YFP immunoprecipitations followed by immunoblotting for the indicated proteins in (C) NER-proficient, or in NER-deficient (D) Xpc−/− or (E) Xpg−/− Xpb-YFP knock-in MDFs. (– lane: binding control, UV: 16 J/m2, 1 hour)
(F) Live cell images of HLTF accumulation at local UV-C laser (266 nm) induced DNA damage (arrow) in GFP-HLTF knock in U2OS cells.
(G-H) Relative GFP-HLTF accumulation at LUD 40–45 sec after damage induction in (G) GFP-HLTF knock-in U2OS cells transfected with indicated siRNAs, or (H) in siRNA-transfected XPG KO cells expressing wild-type or catalytically inactive (E791A) XPG where indicated. All live cell accumulation data was background corrected and normalized to pre-damage fluorescence, which was set at 1. n≥20 from at least two independent experiments, mean ± SEM are plotted.
Dual incision-dependent recruitment of HLTF to the NER incision complex
While HLTF, the closest mammalian orthologue to S. Cerevisiae RAD5, has been described to function in the UV-DNA damage response by promoting the template switching sub-pathway of post-replication repair (PRR) (Achar et al., 2015; Kile et al., 2015), it has not yet been functionally linked to NER. To test whether HLTF is indeed recruited to TFIIH as part of an active NER complex, we immunoprecipitated TFIIH from NER-deficient cells (Giglia-Mari et al., 2009). As expected, the UV-induced interaction between TFIIH and both XPA and ERCC1 was absent in MDFs deficient for the GG-NER damage sensor XPC, as GG-NER is not initiated (Fig 1D). Importantly, the UV-induced interaction between TFIIH and HLTF was also lost in these Xpc−/− MDFs, indicating that HLTF is specifically recruited to NER-engaged TFIIH (Fig 1D). Interestingly, HLTF was also not recruited to TFIIH in UV-irradiated Xpg−/− MDFs, where TFIIH is still able to successfully bind DNA lesions and initiate assembly of the incision complex by recruiting XPA and ERCC1 (Fig 1E) (Coin et al., 2008; Volker et al., 2001). Since physical presence of XPG is required for incision on either side of the damage (Staresincic et al., 2009), this suggests that HLTF is incorporated into a late-stage NER incision complex, likely after incision. However, the HLTF-TFIIH interaction is established prior to gap filling synthesis, as inhibition of this reaction step by hydroxyurea and cytarabine (Overmeer et al., 2011) (HU/AraC) did not prevent HLTF from associating with TFIIH (Fig S1B).
To precisely determine during which reaction step HLTF is recruited to the NER complex, we generated homozygous GFP-HLTF KI cells (Figs S1C–E) and quantified HLTF recruitment to sites of local UV damage (LUD) using live cell imaging (Dinant et al., 2007). In line with the UV-induced interaction of HLTF with TFIIH during NER (Figs 1B–C), we observed a clear HLTF accumulation at LUD (Figs 1F, S1F). Moreover, this HLTF accumulation was significantly reduced by siRNA-mediated depletion of XPC (Figs 1G, S1F–G), indicating that HLTF accumulates in a NER-dependent manner. Chemical inhibition of TFIIH by either degrading XPB (Alekseev et al., 2014) or inhibiting its ATPase activity (Titov et al., 2011) using spironolactone or triptolide, respectively, elicited a similar decreased HLTF accumulation at LUD (Figs S1H–I). Despite efficient inhibition of the NER reaction, residual HLTF accumulation remained detectable at LUD. As HLTF is also recruited to UV-stalled replication forks in conjunction with the RAD18-RAD6 complex to regulate PRR pathway choice (Masuda et al., 2018; Motegi et al., 2008; Unk et al., 2008), we postulated that the residual HLTF accumulation may be PRR-related. Indeed, RAD18 knockdown (Fig S1F–G) markedly decreased the accumulation of HLTF at LUD (Fig 1G), which acted additive to XPC depletion. These results indicate that HLTF is recruited to DNA damage via two distinct mechanisms, mediated via either the PRR or NER pathway. Therefore, to study NER-dependent HLTF accumulation we henceforth used RAD18-depleted cells and found that the additional depletion of either XPF or XPG impeded HLTF recruitment to the NER complex (Fig 1G), implying that HLTF recruitment is dependent on formation of the complete incision complex. To investigate whether dual or only a single DNA incision is required for HLTF recruitment, we knocked out XPG in GFP-HLTF KI cells, and subsequently ectopically expressed either wild-type (WT) or a binding-competent but catalytically dead (E791A) XPG mutant (Fig S1J). Both WT and E791A XPG were recruited to NER complexes (Fig S1K), and were therefore able to recruit ERCC1-XPF to the incision complex and facilitate 5’ incision (Staresincic et al., 2009). However, only WT XPG successfully complemented the UV sensitivity of the XPG KO cells (Figs S1L), as E791A XPG is unable to execute the 3’ incision required for damage removal. Interestingly, while WT XPG was able to fully restore the NER-associated HLTF accumulation that was lost in XPG KO cells, E791A XPG could not (Fig 1H), indicating that neither the presence of XPG nor a single 5’ incision by ERCC1-XPF is sufficient for HLTF recruitment. PRR-independent HLTF accumulation was further confirmed in non-replicating fibroblasts, which showed NER-dependent HLTF accumulation that was significantly reduced by either spironolactone treatment or in XP-A fibroblasts (Fig S1M). Taken together, these biochemical and live cell imaging data demonstrate a DNA damage-induced recruitment of HLTF to the NER incision complex following dual incision, but prior to gap filling synthesis.
HLTF stimulates dissociation of lesion-bound TFIIH
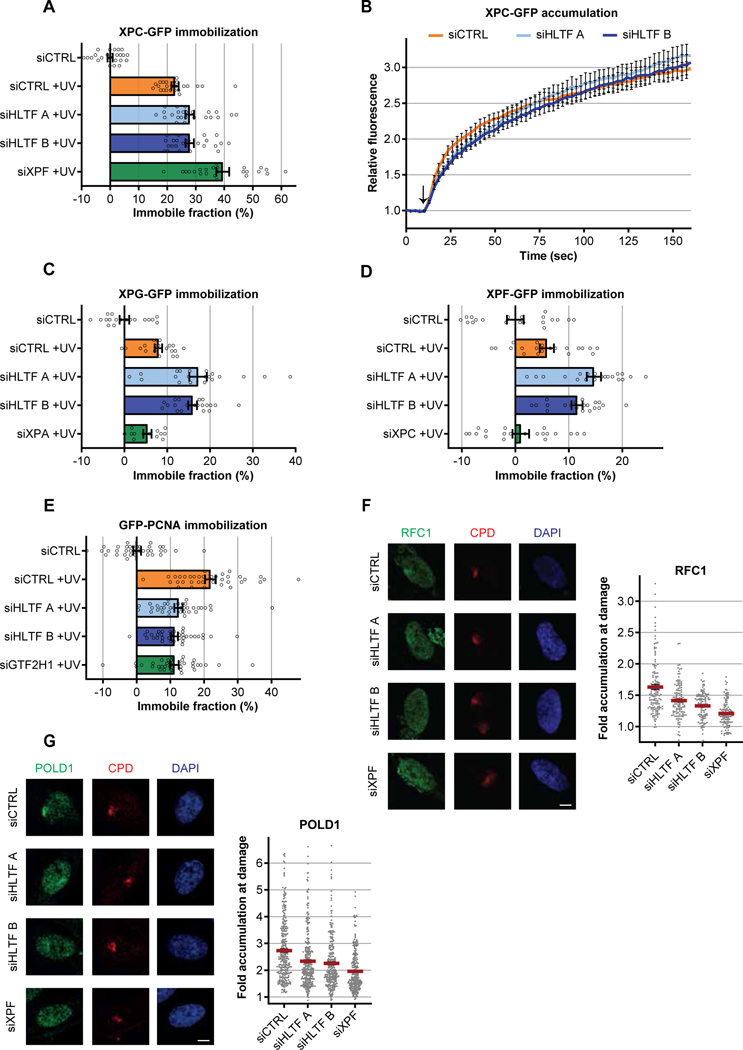
Next, we set out to study the putative function of HLTF during this late, post-incision NER reaction step. As we identified HLTF as a NER-dependent interactor of the TFIIH complex, we first studied how HLTF influenced TFIIH behavior at DNA damage. Interestingly, the recruitment of TFIIH to LUD, determined by immunofluorescence of endogenous XPB (Fig 2A) and GTF2H1 (Fig S2A), was significantly increased upon HLTF depletion, while TFIIH expression levels remained unaffected (Fig S2B). Loss of HLTF did not permanently sequester TFIIH at sites of NER, as TFIIH accumulation at LUD decreased over time, although at a reduced rate compared to control cells (Figs 2A, S2A). Similar results were observed using two independent HLTF KO clones, confirming that TFIIH release seems to be retarded, rather than fully blocked, in absence of HLTF (Figs S2C–D). The effect of HLTF on TFIIH accumulation were also independent of replication-associated DNA damage tolerance mechanisms, as a similar increase in TFIIH accumulation was found in non-replicating fibroblasts (Fig 2B). To more quantitatively study the effect of HLTF on TFIIH recruitment, we monitored XPB-GFP recruitment kinetics to UV-C laser (266 nm) induced DNA damage in live cell imaging experiments (Dinant et al., 2007). In this setup, XPB-GFP accumulates at LUD in a NER-dependent manner, as its recruitment was severely reduced upon XPC depletion. Upon HLTF depletion we observed an increased TFIIH recruitment, which was especially evident at later time points (>50 sec) (Figs 2C, S2E). To study whether this enhanced TFIIH recruitment was mainly due to an increased association (Kon) or reduced release (Koff), we performed inverse fluorescence recovery after photobleaching (iFRAP) to determine TFIIH dissociation rates at LUD (van Cuijk et al., 2015) (Fig S2F). iFRAP showed that TFIIH release from sites of DNA damage was nearly two-fold delayed in HLTF-depleted cells compared to control transfected cells, with an increase in t1/2 from 13 to 22 seconds (Fig 2D). To confirm this increased residence time of TFIIH, we determined the steady-state TFIIH chromatin binding following UV-induced DNA damage by XPB-GFP FRAP (Hoogstraten et al., 2002). UV-induced DNA damage results in a TFIIH immobilization, which is NER dependent as this immobilization was severely reduced upon XPC depletion. HLTF knockdown resulted in strong increase of the UV-induced TFIIH immobilization as shown by the FRAP curves (Fig 2E) and the subsequent calculated respective immobilized fractions (Fig 2F). Of note, TFIIH mobility was not affected by depletion of HLTF in unperturbed conditions. Similar to the increased accumulation at LUD, the increased TFIIH immobilization in the absence of HLTF had a transient nature, as TFIIH mobility recovered over time (Fig S2G). Depletion of RAD18, which in addition to PRR (Lin et al., 2011) has also been implicated in regulating the polymerase composition of post-incision NER complexes (Ogi et al., 2010; Sertic et al., 2018), did not influence TFIIH immobilization after UV damage (Figs 2G, S2H–I), further indicating that HLTF affects TFIIH in a PRR and RAD18-independent manner. Overall, our data shows an increase in damage-bound TFIIH in the absence of HLTF that could be explained by the accumulation of a NER intermediate due to an impaired reaction step.
Figure 2. HLTF stimulates incision to post-incision complex handover.
(A) Left: Representative immunofluorescence images showing co-localization of XPB with CPDs (UV-induced photolesions) at the indicated timepoints after local UV irradiation (60 J/m2) of U2OS cells. Scale bar: 5 μm. Right: XPB accumulation at LUD in U2OS cells transfected with the specified siRNAs, relative to the average nuclear fluorescence outside the damaged area which was set at 1. Mean ± SEM of n≥325 cells per condition, measured in three independent experiments.
(B) Left: Representative immunofluorescence images showing the recruitment of XPB to CPDs at LUD (60 J/m2, 30 min) in non-replicating C5RO fibroblasts. Scale bar: 5 μm. Right: Relative XPB levels at LUD were quantified following transfection with the indicated siRNAs. Mean ± SEM of n≥150 cells per condition, measured in two independent experiments.
(C) Relative XPB-GFP accumulation at sites of LUD using a 266nm UV-C laser following transfection of XPCS2BA cells with the indicated siRNAs. Accumulation values were background-corrected and normalized to pre-damage fluorescence, which was set at 1. Arrow marks the moment of UV-C induction. Plotted are mean ± SEM of n≥17 cells from at least two independent experiments.
(D) Inverse FRAP analysis (see Figure S2E) of XPB-GFP expressed in XPCS2BA cells at LUD (120 J/m2 UV). Relative loss of fluorescence at LUD during continuous bleaching outside the damage was monitored. The fluorescence level at LUD before bleaching was set as 1. Curves represent mean ± SEM from n≥20 cells measured in two independent experiments.
(E) FRAP analysis of XPB-GFP mobility in mock treated or globally UV irradiated (directly after 10 J/m2) XPCS2BA cells transfected with indicated siRNAs. XPB-GFP fluorescence was background-corrected and normalized to average pre-bleach values, which were set at 1.
(F-G) Calculated immobile fractions of XPB-GFP, as indicated by the dotted square in Figure 2D. XPB-GFP expressing XPCS2BA cells were transfected with control siRNA or siRNAs targeting either HLTF, XPC (F) or RAD18 (G). Plotted values represent mean ± SEM from n≥20 cells measured in two independent experiments.
HLTF facilitates incision to post-incision complex handover
To pinpoint at which step the NER reaction could be compromised, we subsequently determined the effects of HLTF depletion on the UV-induced immobilization of the NER factors XPC (damage recognition), XPG, XPF (dual incision) and PCNA (gap filling synthesis) using FRAP (Essers et al., 2005; Sabatella et al., 2018; van Cuijk et al., 2015). siRNA-mediated depletion of HLTF only marginally increased the UV-induced immobilization of XPC (Figs 3A, S3A–B) and did not significantly affect DNA damage recognition as determined by XPC recruitment to LUD (Fig 3B). This indicates that HLTF does not affect damage recognition and acts on TFIIH complexes no longer associated with XPC. Conversely, the UV-dependent immobilization of both endonucleases XPG and XPF was increased to similar extent as TFIIH after HLTF knockdown (Figs 3C–D, S3A and S3C–D), implicating that HLTF depletion results in the retention of the entire NER incision complex. The effects of HLTF depletion on the chromatin binding of the NER endonucleases was further confirmed by an enhanced accumulation of XPF at LUD in the absence of HLTF (Fig S3E). In striking contrast to factors of the incision complex, the UV-induced immobilization of the post-incision factor PCNA, whose UV-induced immobilization in non-replicating cells is fully NER-dependent (Fig S3F–G), was strongly reduced in the absence of HLTF (Fig 3E). In line with the reduced PCNA loading, recruitment of the upstream PCNA-loading RFC complex as well as the accumulation of the downstream DNA polymerases δ (polδ) and κ (polκ) (Ogi and Lehmann, 2006; Ogi et al., 2010) to NER sites was also reduced upon HLTF depletion in non-replicating cells (Fig 3F–G, S3I). The increase in chromatin binding of members of the incision complex combined with a reduced recruitment of factors involved in repair synthesis suggests that HLTF stimulates the handover from incision to post-incision complex by promoting the release of TFIIH, XPG and ERCC1-XPF.
Figure 3. HLTF stimulates incision to post-incision complex handover.
(A) Calculated immobilized fractions of XPC-GFP in either mock or UV-treated (10 J/m2) XP4PA cells after transfection with the indicated siRNAs. Plotted values represent mean ± SEM from n≥20 cells measured in at least two independent experiments.
(B) Relative XPC-GFP accumulation in XP4PA cells transfected with the indicated siRNAs was determined by live cell imaging after irradiation with a 266nm UV-C laser. The moment of UV-C damage induction is marked by the arrow. XPC accumulation was background-corrected and normalized to pre-damage fluorescence, which was set at 1. Plotted values are averages ± SEM of n≥20 cells from two independent experiments.
(C-E) Calculated immobilized fractions of (C) XPG-GFP, (D) XPF-GFP and (E) GFP-PCNA expressed in mock or UV-treated XPCS1RO, U2OS XPF KO and serum-deprived C5RO cells, respectively, as determined by FRAP analysis. Where indicated, cells were analyzed directly after 10 J/m2 UV-C irradiation, except for GFP-PCNA which was analyzed 30 min after irradiation. Plotted values represent mean ± SEM from n≥20 cells measured in at least two independent experiments.
(F-G) Left panels: Representative immunofluorescence images showing the co-localization of the endogenous RFC complex (F) or polδ (G), as determined by staining of, respectively, RFC1 or the catalytic POLD1 subunit, with CPDs at LUD (60 J/m2, 30 min after irradiation). Scale bar: 5 μm. Right panels: Relative RFC1 and polδ accumulation at LUD in non-cycling C3RO cells transfected with the indicated siRNAs. Mean ± SEM of n≥185 cells per condition, measured in at least two independent experiments.
Release of incised oligonucleotides is mediated by HLTF
Thus far, it was assumed that dual incision by the XPG and ERCC1-XPF endonucleases in itself was sufficient to directly release damage-containing oligonucleotides (Mu et al., 1996; Riedl et al., 2003). However, our data shows that depletion of HLTF, which is only recruited to NER complexes after dual incision (Figs 1C–G), results in an increased accumulation of XPF, TFIIH and XPG (Figs 2F, 3C–D). Since the latter two factors are released simultaneously and in complex with damage-containing oligonucleotides (Hu et al., 2013; Kemp et al., 2012), this suggests that the dually incised oligonucleotide may also be retained at NER sites and may be physically restrained within the incision complex in absence of HLTF. Based on these data, we speculated that HLTF, which contains a dsDNA translocase motor domain capable of displacing DNA-bound proteins (Achar et al., 2011), might be responsible for active eviction of these incised damage-containing oligonucleotides together with factors of the incision complex.
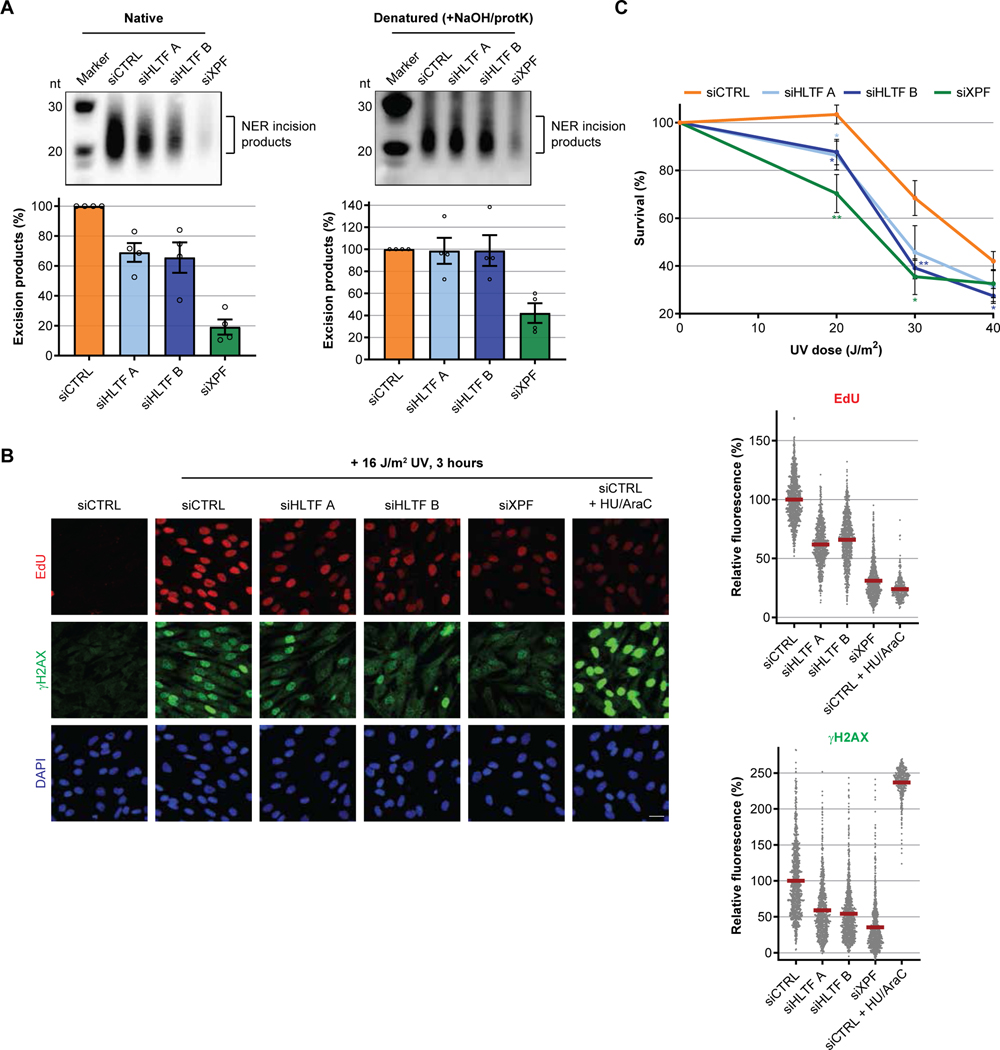
To test whether HLTF catalyzes the eviction of damage-containing incised oligonucleotides, we purified NER-excised oligonucleotides following UV irradiation using an UV lesion-specific (6–4PP) immunoprecipitation approach (Choi et al., 2013). Using this method, we could clearly observe the evicted ~20–30 nt long oligonucleotides, which were processed by NER as these were not observed without UV irradiation (Fig S4A) and almost completely absent upon XPF depletion (Fig 4A left panel). HLTF-depleted cells yielded significantly lower amounts of these evicted and thus soluble damage-containing oligonucleotides (Fig 4A left panel and S4B), confirming that HLTF promotes the eviction of incised oligonucleotides that likely otherwise would have remained stably embedded within the incision complex by protein-DNA interactions. To verify this, excised 6–4PP-containing oligonucleotides were extracted under denaturing conditions, which disrupt protein-DNA interactions. Indeed, under these conditions the number of incised oligonucleotides upon UV in HLTF depleted cells could be restored to levels observed in control transfected cells (Fig 4A, right panel, Fig S4A). Together, this comparative analysis confirms that HLTF is not required for dual incision per se, since this would have strongly reduced the detection of incised oligonucleotides under both conditions, as was observed upon XPF depletion. Instead HLTF stimulates the eviction of chromatin-bound NER complexes associated with incised damaged DNA into the nucleoplasm, of which the release could also be mimicked using denaturing conditions.
Figure 4. HLTF stimulates eviction of dually incised oligonucleotides from NER sites.
(A) Analysis of UV-induced (40 J/m2, 1 hour) 6–4PP-containing excision fragments isolated from MRC5 cells transfected with indicated siRNAs under either native (left) or denaturing (right) conditions. The resulting signals were quantified and compared to siCTRL-transfected cells, which were set at 100%. Values were calculated from four independent experiments ± SEM.
(B) Left: Representative images of EdU incorporation and γH2AX staining before and 3 hours after UV irradiation (16 J/m2) in non-replicating C5RO fibroblasts transfected with the indicated siRNAs or upon HU/AraC treatment (100 mM/10 μM, 30 min pre-treatment). Scale bar: 25 μm. Right: Relative UDS signal as determined by EdU incorporation and γH2AX signal were quantified and compared to siCTRL-transfected cells, which were set at 100%. n≥400 cells from at least two independent experiments.
(C) Survival of non-replicating C5RO fibroblasts transfected with indicated siRNAs following exposure to the indicated UV doses as determined by AlamarBlue staining. Undamaged samples were set at 100%. Plotted curves represent averages of five independent experiments ± SEM.
Retained oligonucleotides hamper repair synthesis, damage signaling and cellular viability
Next, we investigated the consequences associated with prolonged chromatin retention of these dually incised NER intermediates by testing how HLTF depletion affects gap filling synthesis and ssDNA repair patch-induced γH2AX signaling. Therefore, we simultaneously quantified the UV-induced unscheduled DNA synthesis (UDS) (Nakazawa et al., 2010) and γH2AX signaling in non-replicating fibroblasts (Hanasoge and Ljungman, 2007; Marteijn et al., 2009; Overmeer et al., 2011). As expected, absence of incision due to XPF depletion resulted in a loss of both damage-induced UDS and γH2AX staining, due to the reduced formation ssDNA repair intermediates (Fig 4B). Inhibition of gap-filling DNA polymerases by HU/AraC treatment resulted also in strong reduction in UDS, but now combined with a strong increase of γH2AX due to the accumulation of persisting ssDNA repair patches (Fig 4B). In line with the reduced loading of PCNA and polδ, HLTF knockdown strongly perturbed UDS (Fig 4B). Moreover, TC-NER-mediated UDS (Wienholz et al., 2017) was decreased to a similar extent (Fig S4C), indicating that HLTF is needed for efficient gap filling synthesis in both the GG- and TC-NER sub-branches. In addition to UDS, γH2AX signals were also markedly decreased upon HLTF depletion, confirming that absence of HLTF results in fewer ssDNA repair patches, which can be explained by a diminished eviction of incised damage-containing oligonucleotides. Next, we tested whether the timely displacement of damage-containing oligonucleotides is required to overcome the detrimental effects of UV-induced DNA damage. To exclude indirect effects of HLTF due to defects in the PRR pathway, we tested cell survival in response to UV damage in non-replicating cells. As expected, XPF depletion rendered non-cycling cells sensitive to UV irradiation. Interestingly, loss of HLTF gave rise to a similar effect, albeit to a lesser extent (Fig 4C). This increased UV-sensitivity is caused by HLTF’s role in NER, as no additional sensitivity was found upon HLTF depletion in non-replicating XP-A cells (Fig S4D). Together, these data indicate that HLTF-mediated damage eviction during NER is important to enable a proper cellular response to DNA damage and limit DNA damage-induced cytotoxicity.
HLTF employs a two-step mechanism to release incised oligonucleotides
HLTF exerts its function in PRR through combining its HIRAN-mediated 3’-OH ssDNA end binding, C3HC4-type RING finger ubiquitin ligase and dsDNA translocase (ATPase) activities during PRR (Achar et al., 2015; Hishiki et al., 2015; Kile et al., 2015). We set out to elucidate which of these activities are required for the eviction of damage-containing oligonucleotides during NER. To this end, we made use of the fact that TFIIH binding can be used as a read-out for the eviction of incised oligonucleotides, because these are released in tight complex with each other (Adar et al., 2016; Hu et al., 2013; Kemp et al., 2012). Therefore, we determined the effects of domain-specific HLTF mutants on TFIIH accumulation at LUD by re-expressing either WT HLTF, or HLTF carrying inactivating mutations in these different domains in HLTF KO cells (Fig S5A). The increased TFIIH accumulation at LUD in HLTF KO cells (Fig S2C), could be fully rescued by re-expression of WT HLTF (Fig 5A). RING-mutated HLTF also fully restored TFIIH accumulation levels, indicating that, unlike during template switching (Masuda et al., 2018; Motegi et al., 2008), HLTF’s function in NER does not rely on its E3 ligase activity. Interestingly, expression of either the HIRAN or translocase mutants failed to decrease TFIIH accumulation, revealing functions for these domains during NER. As dual incision precedes HLTF recruitment (Fig 1H) and generates two 3’-OH ssDNA ends, we speculated that the HIRAN domain might be required to position HLTF at 3’-OH termini embedded within the incision complex. This may subsequently enable the ATPase motor of HLTF, which acts in a 3’-to-5’ directional manner, to displace the incision complex proteins together with the incised oligonucleotide (Achar et al., 2011). Such a two-step ‘bind-and-remodel’ mechanism would be analogous to the action of HLTF during replication fork reversal (Chavez et al., 2018; Kile et al., 2015).
Figure 5. HIRAN- and translocase-dependent function of HLTF in NER.
(A) Left: Representative immunofluorescence images showing the co-localization of XPB with CPDs, 30 minutes after LUD induction (60 J/m2) in WT U2OS, U2OS HLTF KO and U2OS HLTF KO cells expressing the indicated HLTF mutants. Scale bar: 5 μm. Right: (top) Schematic representation of human HLTF showing the position of inactivating mutations in the indicated domains. (bottom): Quantification of relative XPB accumulation at LUD. Plotted values are averages ± SEM of n≥175 cells per condition, measured in three independent experiments.
(B) Top: Representative images showing the accumulation of the indicated GFP-tagged HLTF constructs at UV-C (266nm) laser-induced LUD. Bottom: Relative accumulation of GFP-HLTF mutants at LUD in siRAD18-transfected U2OS HLTF KO cells. HLTF accumulation was normalized to pre-damage fluorescence, which was set at 1. Arrow indicates the moment of damage induction. Plotted are averages ± SEM of n≥20 cells from two independent experiments. Arrow marks the moment of UV-C induction.
(C) Left: Representative images of PLA signal between endogenously expressed GFP-HLTF with either XPF or XPG. Where indicated, GFP-HLTF KI U2OS cells were treated with UV (20 J/m2, 30 min) and/or spironolactone (10 μM, 2 hours pre-treatment) before fixation. Right: Quantification of PLA foci per cell. n≥150 cells per condition from two independent experiments. Scale bar: 5 μm.
To test this hypothesis, we first quantified the recruitment of GFP-tagged HLTF mutants to LUD in a NER-dependent manner using live cell imaging (Figs 5B, S5B). Whereas RING mutant HLTF showed similar recruitment kinetics as WT protein, the accumulation of the HIRAN mutant was almost completely abrogated, implying that HLTF is indeed recruited to NER complexes via HIRAN-mediated binding to free 3’-OH ssDNA ends. In contrast, ATPase-dead HLTF showed an increased accumulation over time, suggesting that while this mutant can still efficiently bind to NER-generated 3’-OH ssDNA ends, it cannot dismantle the incision complex and release the incised oligonucleotide due to a lack of ATP hydrolysis capability. This will subsequently result in the accumulation of a HLTF-bound intermediate of the incision complex.
Having established that HIRAN is the principal substrate-recognition domain to recruit HLTF to NER sites, we next asked whether HIRAN targets the 3’-OH end generated by ERCC1-XPF (which is located 5’ of the lesion) or that generated by XPG (located 3′ of the lesion) after dual incision. To try to discriminate between these possibilities, we employed proximity ligation assay (PLA), which detects protein-protein interactions in close vicinity. We did not observe UV-induced PLA signal between the distally located XPF and XPG endonucleases, indicating that the two edges of the repair bubble are sterically hindered from generating PLA signal (Fig S5C). HLTF binding to the incision complex was exemplified by the UV-induced increase in PLA signal of HLTF with TFIIH (Fig S5D). Notably, XPF showed a UV-induced increase in PLA foci with HLTF (Fig 5C), which was NER-specific as it was fully lost upon degradation of XPB by spironolactone. However, no such signal was observed between XPG and HLTF, while compatibility of the XPG antibody for PLA is shown by its UV-induced signal with XPA (Fig S5E). Collectively, these data might suggest that HLTF is recruited to the 3’-OH end 5’ of the lesion in an HIRAN-dependent manner, where the ATPase activity subsequently mediates the release of the incision complex. This in turn results in the removal of incised damage-containing oligonucleotide, which is crucial for proper DNA damage-induced repair synthesis.
HLTF-independent incision complex dissociation is mediated by RFC
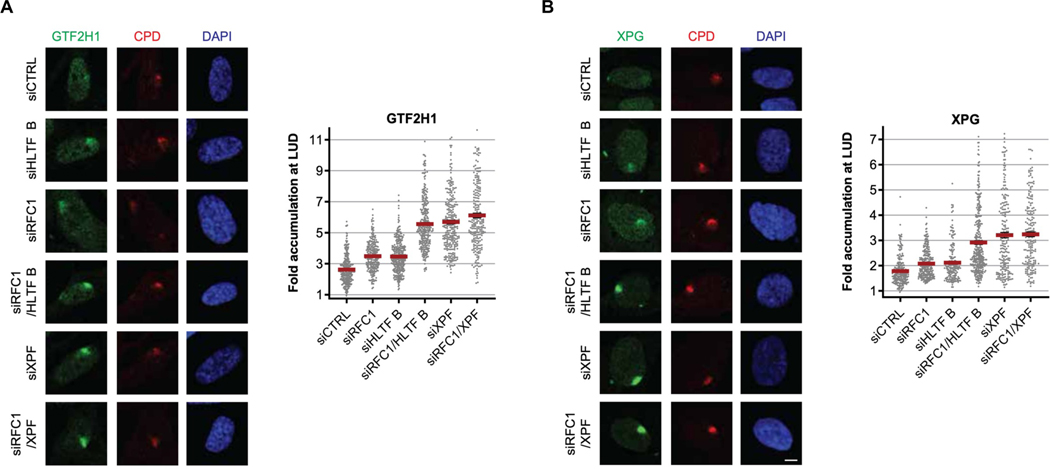
Absence of HLTF gives rise to a significant but partial NER defect that seems to slowly recover over time, since both TFIIH accumulation (Fig S2C) and gap filling synthesis (S6A) are most severely affected at early time points. This indicates that while inefficient, damage eviction can occur in an HLTF-independent manner, either spontaneously due to intrinsic instability of the incised complex, or by virtue of a redundant eviction mechanism. To find putative factors contributing to HLTF-independent damage eviction, we first tested whether the HLTF-related SF2 translocases SMARCAL1 and ZRANB3 affected UV-induced TFIIH and PCNA immobilization. However, neither SMARCAL1 nor ZRANB3 significantly affected the binding of TFIIH or PCNA in UV-irradiated cells, indicating that these proteins, which lack HIRAN domains, are not involved in the incision-to-repair synthesis transition (Fig S6B–D). Next, we considered downstream factors involved in the initiation of repair synthesis as alternative options for lesion eviction. The canonical RFC complex is the first core post-incision factor to be recruited, binding the 3’-OH located 5’ of the lesion in a manner similar as suggested for HLTF (Overmeer et al., 2010). Strikingly, knockdown of the catalytic RFC1 subunit in non-cycling fibroblasts gave rise to a similar increase in TFIIH accumulation at LUD as was observed in HLTF-depleted cells (Fig 6A). Co-depletion of both RFC1 and HLTF even further enhanced this effect to levels similar as observed in XPF-depleted cells (Fig 6A), and comparable effects were found for both XPA and XPG (Figs 6B and S6E). Taken together, these data indicate that both RFC1 and HLTF independently contribute to damage eviction and highlight the tightly regulated coordination of damage eviction and DNA repair synthesis during NER.
Figure 6. RFC1 can stimulate incision complex dissociation independent from HLTF.
(A-B) Left: Representative images of (A) GTF2H1 (rabbit antibody) or (B) XPG accumulation at CPDs 30 minutes after LUD induction (60 J/m2) in non-cycling VH10-hTERT cells following transfection with the indicated siRNAs. Scale bar: 5 μm. Right: Quantification of relative GTF2H1 accumulation at LUD. Plotted values are averages ± SEM of n≥250 cells per condition, measured in three independent experiments.
DISCUSSION
A role for HLTF in nucleotide excision repair
The function of HLTF in genome stability was originally described as an E3 ubiquitin ligase that protects against UV sensitivity by polyubiquitylating PCNA and stimulating error-free post-replication repair (Motegi et al., 2008; Unk et al., 2008). Later, HLTF was also found to possess fork reversal activity in the broader context of replication stress, such as in response to nucleotide depletion (Achar et al., 2015; Bai et al., 2020; Kile et al., 2015). In our current study, we identify HLTF as an important NER factor, adding yet another critical activity by which HLTF contributes to genome stability. Strikingly, the function of HLTF in NER appears to be completely independent of both replication and RAD18 (Figs 2B, 2G), another multifunctional E3 ligase that is involved in both NER and PRR (Lin et al., 2011; Ogi et al., 2010). Moreover, whereas RAD18 relies on its PCNA monoubiquitylating activity for both pathways, we show that in contrast to PRR, HLTF’s RING domain is dispensable for its function in NER and that HLTF thus differentially employs its activities to cope with UV lesions (Fig 5A). This dualistic function of HLTF in response to UV may explain the phenotypical characteristics associated with HLTF deficiency, which can arise from combined defects in both NER and PRR. HLTF expression has been found to be deregulated in multiple types of cancer, most notably in colon cancer, where HLTF expression is lost in ~40% of analyzed cases through promotor hypermethylation (Dhont et al., 2016; Moinova et al., 2002). It will therefore be interesting to investigate the contribution of HLTF’s functions in NER and PRR during tumorigenesis and to determine whether loss of HLTF sensitizes cancer cells to chemotherapeutics that are counteracted by NER.
From incision to excision: controlling incision to post-incision complex handover
NER is an intricate multiprotein repair system characterized by consecutively executed reaction steps that orchestrate the repair of a wide variety of helix-distorting lesions dispersed within a highly complex and dynamic genomic environment (Marteijn et al., 2014; Schärer, 2013). To allow efficient repair, it is crucial that the recruitment and release of proteins involved in the sequential damage detection, verification, incision and repair synthesis processes are carefully choreographed to ensure a smooth transition between these consecutive reaction steps. Indeed, several recent examples have highlighted the importance of a tightly organized interplay between the damage recognition and verification or incision complexes (Nakazawa et al., 2020; Rüthemann et al., 2017; van Cuijk et al., 2015). However, thus far, the involved mechanisms in the handover of the incision complex to the final gap restoration steps of the NER reaction have remained far less understood. We show that HLTF contributes to a delicately controlled transition between the incision and DNA synthesis steps of NER, thereby avoiding the occurrence of cytotoxic repair intermediates while supporting proper DNA damage signaling (Figs 4B–C). We propose that the tightly-bound incision complex that is retained in absence of HLTF poses a steric obstruction for efficient RFC-mediated PCNA loading after incision, a notion that is supported by overlapping binding regions and competitive binding of ERCC1-XPF and PCNA (Mocquet et al., 2008). HLTF does not seem to be required for the loading of a specific DNA polymerase during NER, as depletion of HLTF reduces accumulation of both polδ and polκ (Figs 3G, S3I). Interestingly, HLTF itself was recently also shown to dynamically compete with polδ for binding at primer/template junctions (Masuda et al., 2018). It is therefore likely that HLTF also needs to rapidly dissociate upon disassembly of the incision complex at NER sites for efficient polδ loading. In line with such an assumption, we observed that ATPase dead HLTF, which is binding-proficient but deficient in its translocase activity, shows increased accumulation at LUD over time, whereas WT HLTF readily reaches steady-state accumulation (Fig 5B). This may indicate that WT HLTF binds in a transient manner, and, in contrast to the ATPase mutant, rapidly reaches an association-dissociation equilibrium. Moreover, while HLTF depletion affects the dissociation of the entire incision complex, our data does not unequivocally rule out the possibility that HLTF mediates the dissociation of a specific NER factor. Release of such factor could then either destabilize the remaining incision complex or allow loading of PCNA and DNA polymerases to trigger displacement synthesis of the incised damage-containing oligonucleotide together with TFIIH and XPG.
A molecular sweeper facilitating DNA damage eviction
We found that HLTF is crucial to timely displace the NER incision complex and incised damage-containing oligonucleotide in vivo. We propose a model (Fig 7) in which HLTF is recruited via its HIRAN domain to the 3’-OH terminus generated by ERCC1-XPF 5’ of the lesion during NER. HIRAN-mediated binding is then followed up by displacement of the incised NER intermediate via the ATP-dependent dsDNA translocase activity of HLTF. As HLTF employs 3’-to-5’ translocase activity, this scenario would imply that the translocation movement diverges HLTF from the lesion itself. Surprisingly, such a directional activity suggests that HLTF does not disrupt DNA-protein interactions of incision complex factors with the damaged DNA, which is in line with observations that TFIIH and XPG remain bound to the damage-containing oligonucleotide upon its eviction (Hu et al., 2013; Kemp et al., 2012). Recently reported cryo-EM structures of TFIIH engaged in NER show DNA-interacting structural elements of both XPB and XPA extending 5’ of the repair bubble, likely associated with ERCC1-XPF (Kokic et al., 2019). Therefore, we propose that HLTF’s translocase activity is required to disrupt dsDNA-protein interactions of XPF, XPA and TFIIH 5’ of the repair bubble. These dsDNA-protein interactions may otherwise stabilize and physically restrain the damage-containing oligonucleotide within the incision complex, even after dual incision. Although the incised damaged DNA is restrained in the chromatin, it is not as stable as its non-incised counterpart, illustrated by its delayed, but not fully blocked, release in absence of HLTF (Figs S2C and S2G). This transient nature of the incised fragment could be explained by the fact that it is only stabilized by relatively short-lived DNA-protein interactions that may be disrupted by DNA transacting processes, or that an alternative mechanism is in place to stimulate damage eviction. In line with the latter, we found that RFC1 stimulates release of the incision complex in a manner additive to HLTF, suggesting that these proteins contribute to incision complex release independently from each other (Figs 6A–B). Which of these proteins is utilized to mediate eviction could be determined by relative expression levels of RFC and HLTF, or by the conformation of the repair site, which may be influenced by the surrounding chromatin environment, transcription status chromatin binding of other DNA transacting proteins. Open repair complex conformations may prefer RFC-mediated damage eviction followed by direct PCNA loading, while HLTF-mediated damage eviction might be required to allow the much larger RFC complex to access more occluded repairs sites and subsequently load PCNA. Additionally, the coordination between the ERCC1-XPF and XPG incisions might influence the involvement of HLTF, which may explain why HLTF is not recruited in absence of XPG incision, while UDS can be detected (Staresincic et al., 2009). A single incision by ERCC1-XPF most likely generates an open flap structure, as it strongly initiates DNA damage signaling (Overmeer et al., 2011; Staresincic et al., 2009). This might allow direct binding of RFC, resulting in HLTF-independent repair synthesis. Although the incision complex might thus eventually dissociate in absence of HLTF, deferred removal of this intermediate vastly diminishes repair and damage signaling, and consequently results in reduced cellular survival upon DNA damage (Figs 4A–C), illustrating the importance of this reaction step.
Figure 7. Model showing the function HLTF during NER.
In both GG- and TC-NER, lesion recognition is followed by recruitment of TFIIH and assembly of the incision complex, which also includes RPA, XPA, XPG and ERCC1-XPF (step 1). After dual incision, HLTF is recruited to 3’-OH ends in an HIRAN-dependent manner, most likely binding 5’ of the lesion (step 2). The activity of the ATPase domain subsequently drives the dissociation of ERCC1-XPF and the eviction of the incised oligo, which remains bound to TFIIH and XPG (step 3). This enables RFC-mediated loading of the PCNA sliding clamp at the primer/template junction and recruitment of DNA polymerase δ/κ to efficient fill and seal the generated ssDNA gap (step 4). We hypothesize that certain open repair conformations allow for HLTF-independent PCNA loading by RFC, which also results in dissociation of the incision complex.
Active damage eviction is an evolutionary conserved NER mechanism
The identification of HLTF reveals an interesting analogy between mammalian and bacterial NER, which operates using a simplified yet mechanistically comparable ‘cut-and-patch’-style reaction (Kisker et al., 2013). During bacterial GG-NER, lesions are recognized and verified by the respective actions of the UvrA and UvrB subunits of the UvrA2-UvrB2 tetramer, which fulfil similar roles as XPC and TFIIH. UvrB verifies the damage and UvrA dissociates from the complex, after which the UvrC endonuclease binds to UvrB to form the bacterial NER incision complex. Subsequent dual incision by the UvrB-UvrC complex results in a stable repair intermediate that obstructs the downstream repair reaction (Kisker et al., 2013). UvrD (also known as Helicase II) is essential to connect the incision and post-incision steps in bacterial NER by displacing this incision product from the NER site (Orren et al., 1992). Similar as suggested for HLTF, UvrD associates with 5’ incision sites and displaces the NER proteins and incision fragments in a 3’-to-5’ directional manner. Moreover, like TFIIH in mammals, bacterial NER products are released in complex with UvrB, and UvrD is required for efficient cycling of UvrB to newly identified lesions (Husain et al., 1985; Kemp et al., 2012; Van Houten et al., 1988). Additionally, while mechanistically poorly understood, S. cerevisiae RAD7 and RAD16 have also been suggested to stimulate eviction of damage-containing oligonucleotides (Reed et al., 1998; Yu et al., 2004), and RAD5 was shown to influence the excision of certain DNA adducts as well (Kiakos et al., 2002). Together this suggests that prokaryotic and eukaryotic cells have convergently evolved most NER reaction steps, including active damage removal.
In conclusion, we reveal a previously unknown role for active removal of DNA damage during eukaryotic NER, mediated by the translocase HLTF, which orchestrates a timely handover of dually incised repair intermediates to the gap filling machinery. Cells lacking such a quality control system jeopardize repair efficiency and may face increased accumulation of genotoxic repair intermediates. This uncoupling of damage incision and excision exemplifies the multifaceted and tight regulation of multistep repair mechanisms that are employed to preserve genome integrity.
Limitations of the Study
Our study identifies HLTF as a NER factor that stimulates the transition from incision to repair synthesis through displacement of incised damage-containing oligonucleotides and incision complex components. Based on our PLA data and the involvement of the 3’-OH binding HIRAN domain to recruit HLTF combined with the 3’-to-5’ directionality of its ATPase domain, we favor a model in which HLTF acts at the 3’OH ssDNA end located 5’ of the lesion to subsequently remove TFIIH and XPF from the incision complex together with DNA damage. However, while no PLA signal between XPF and XPG could be observed, PLA has an expected working range up to 30–40nm, which could theoretically cover the entire incision complex. Taking this limitation of the PLA assay in mind, we are not able to determine the exact binding site of HLTF at NER sites in an unequivocal manner. Determining the docking site of HLTF in more detail remains of importance to fully understand the mechanistic details of the transition from incision to repair synthesis, which minimizes the exposure of vulnerable repair intermediates.
Interestingly, no clear effects of HLTF-mediated eviction of incised damage-containing oligonucleotides in vitro using a reconstituted system combined with immobilized DNA substrates were observed (Fig S7). The absence of a HLTF function in vitro, while clearly detected in cellular assays, could be explained by the fact that HLTF functions in specific chromatin environments or genomic locations that are not accurately recapitulated in vitro. Alternatively, unknown factors or post-translation modifications for example of HLTF might strongly influence HLTF’s function in NER in vivo. Identifying such factors will be important to understand the mechanism of damage eviction in greater detail.
STAR METHODS
Resource Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jurgen Marteijn (J.Marteijn@erasmusmc.nl).
Materials Availability
All unique/stable reagents generated in this study are available from the lead contact upon request.
Data and Code Availability
The mass spectrometry data reported in this paper have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031243. Uncropped images of immunoblots and microscopy images are available from Mendeley Data 10.17632/h2nr6yp67j.1
This study did not generate code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Models and Subject Details
Cell lines, culture and treatments
U2OS, U2OS stably expressing CPD photolyase-mCherry (Steurer et al., 2019), U2OS XPF KO cells including those stably expressing XPF-GFP (Sabatella et al., 2018) (all female), SV40-immortalized XPCS2BA (male XP-B, stably expressing XPB-GFP) (Hoogstraten et al., 2002), XP4PA (male XP-C, stably expressing XPC-GFP) (Ribeiro-Silva et al., 2018), XPCS1RO (male XP-G, stably expressing XPG-GFP) (Zotter et al., 2006) and MRC5 (male) cells all were grown in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM; Gibco) and Ham’s F10 (Invitrogen), supplemented with 10% fetal calf serum (FCS; Biowest) and antibiotics. Xpb-YFP KI MDFs (NER-proficient, Xpc−/− and Xpg−/−) (Giglia-Mari et al., 2009), XP186LV (female XP-C) (Wienholz et al., 2017), hTERT-immortalized XP25RO (male XP-A) and WT C3RO, C5RO (female, including those expressing GFP-PCNA) (Essers et al., 2005) and VH10 (male) fibroblasts were maintained in Ham’s F10 containing 15% FCS and antibiotics. MDFs were grown at 3% O2, while all other cell lines were maintained at 20% O2. To reduce the number of S-phase cells, serum-deprivation (0.5% FCS) was initiated 48h before carrying out select experiments. All cell lines were cultured in humidified incubators at 37 °C and 5% CO2.
For SILAC labeling, cells were grown in DMEM without lysine and arginine (AthenaES) supplemented with 10% dialyzed FCS (Gibco), antibiotics and either 73 μg/mL light [12C6]-lysine and 42 μg/mL [12C6,14N4]-arginine (Sigma) or with heavy [13C6]-lysine and [13C6,15N4]-arginine (Cambridge Isotope Laboratories) for at least 5 passages before carrying out proteomics experiments.
GFP-HLTF knock-in cells were generated by co-transfecting U2OS cells with a Lenti-CRISPRv2 plasmid containing a sgRNA targeting the first exon of the HLTF gene and a homology-directed repair template containing 200 nucleotide long homology arms. Transfected cells were selected with puromycin (InvivoGen, 1 μg/mL) for 2 days, FACS sorted for GFP-positive cells and single cell seeded in 96-well plates. GFP-positive clones were selected and screened for correct insertion of the GFP cassette by PCR-based genotyping and expression of the full-length fusion protein by immunoblotting. HLTF and XPG KO cells were generated by transfecting cells with the appropriate sgRNAs followed by puromycin selection (1 μg/mL) for 2 days and single cells seeding. Frameshift mutations were confirmed using Sanger sequencing and absence of the protein was confirmed by immunoblotting.
Triptolide (1 μM, 1 hour), spironolactone (10 μM, 2 hours), hydroxyurea (100 mM, 30 min) and cytarabine (10 μM, 30 min) were purchased from Sigma and diluted to appropriate concentrations in culture medium.
Plasmids and RNA interference
Full-length Flag-HLTF expression construct was obtained from Dr. Lajos Haracska (Achar et al., 2015) (Hungarian Academy of Sciences, Hungary). Mutant expression constructs were generated by site directed mutagenesis, details are available upon request. Constructs were subsequently inserted into pLenti-CMV-Puro and transfected in HEK293T (female) cells for lentivirus production. The lentiviruses were subsequently utilized to infect U2OS HLTF KO cells. Full-length wild-type and catalytically dead (E791A) human XPG cloned in pWPXL-EF1α were kindly provided by Dr. Orlando D. Schärer (Staresincic et al., 2009) (UNIST, Republic of Korea) and expressed in XPG KO cells by lentiviral transduction. Full-length GFP-POLK construct was obtained from Dr. Tomoo Ogi (Nagoya University, Japan) (Ogi et al., 2010) and transiently expressed in the respective U2OS cell lines using X-tremeGENE 9 DNA transfection reagent (Roche) according to the manufacturer’s instructions.
siRNAs listed in the Key Resources Table were purchased from Horizon Discovery or ThermoScientific and transfected overnight using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-XPA | Santa Cruz | sc-853 |
| Rabbit anti-XPA | Genetex | GTX103168 |
| Rabbit anti-XPB | Santa Cruz | sc-293 |
| Rabbit anti-XPC | Bethyl | A301-112A |
| Mouse anti-XPD | Abcam | ab54676 |
| Mouse anti-XPF | Santa Cruz | sc-136153 |
| Mouse anti-XPG | Abcam | ab46 |
| Rabbit anti-XPG | Bethyl | A301-484A |
| Mouse anti-Cyclin H (2D4) | ThermoScientific | MA3-025 |
| Rabbit anti-RAD18 | Bethyl | A301-340A |
| Mouse anti-PCNA | Abcam | ab29 |
| Mouse anti-GTF2H1 | Sigma-Aldrich | 1F12-1B5 |
| Rabbit anti-GTF2H1 | Novus Biologicals | NBP-38556 |
| Rat anti-HA (3F10) | Roche | 12158167001 |
| Mouse anti-FLAG (M2) | Sigma-Aldrich | F1804 |
| Rabbit anti-HLTF | Santa Cruz | sc-366563 |
| Rabbit anti-HLTF | Abcam | ab155031 |
| Rabbit anti-HLTF | ThermoScientific | PA5-79371 |
| Mouse anti-6-4PP (64M2) | Cosmo Bio | CAC-NM-DND-002 |
| Mouse anti-CPD (TDM2) | Cosmo Bio | CAC-NM-DND-001 |
| Rabbit anti-ERCCI | Abcam | ab129267 |
| Mouse anti-XAB2 | Santa Cruz | sc-271037 |
| Mouse anti-Tubulin (B512) | Sigma-Aldrich | T5168 |
| Goat anti-Ku70 | Santa Cruz | sc1487 |
| Mouse anti-GAPDH | Abcam | ab8245 |
| Mouse anti-YH2AX | Millipore | JWB301 |
| Rabbit anti-GFP | Abcam | ab290 |
| Rabbit anti-POLDI | Abcam | ab126407 |
| Mouse anti-SMARCALI | Santa Cruz | sc-376377 |
| Rabbit anti-ZRANB3 | Proteintech Group | 23111-1-AP |
| Rabbit anti-RFC1 | Invitrogen | PA5-35965 |
| Goat anti-rabbit CF™ IRDye 770 | Sigma-Aldrich | sab4600215 |
| Goat anti-rabbit CF™ IRDye 680 | Sigma-Aldrich | sab4600200 |
| Goat anti-mouse CF™ IRDye 770 | Sigma-Aldrich | sab4600214 |
| Goat anti-mouse CF™ IRDye 680 | Sigma-Aldrich | sab4600199 |
| Donkey-anti-goat CF™ IRDye 770 | Sigma-Aldrich | sab4600375 |
| Goat anti-rat CF™ IRDye 770 | Sigma-Aldrich | sab4600479 |
| Goat anti-rabbit AlexaFluor® 488 | Invitrogen | A11034 |
| Goat anti-mouse AlexaFluor® 488 | Invitrogen | A11001 |
| Goat anti-rabbit AlexaFluor® 594 | Invitrogen | A11012 |
| Goat anti-mouse AlexaFluor® 594 | Invitrogen | A11032 |
| Goat anti-rat AlexaFluor® 555 | Invitrogen | A21434 |
| Donkey anti-rabbit AlexaFluor® 647 | Invitrogen | A31573 |
| Duolink® anti-Mouse MINUS PLA® Probe | Sigma-Aldrich | DU092004 |
| Duolink® anti-Rabbit PLUS PLA® Probe | Sigma-Aldrich | DU092005 |
| Bacterial and virus strains | ||
| One Shot™ Top1G Chemically competent cells | Invitrogen | C4040-06 |
| Chemicals, peptides, and recombinant proteins | ||
| Triptolide | Sigma-Aldrich | T3652 |
| Spironolactone | Sigma-Aldrich | S3378 |
| Hydroxyurea | Sigma-Aldrich | H8627 |
| Cytarabine | Sigma-Aldrich | C3350000 |
| Puromycin | InvivoGen | ant-pr-1 |
| 5-ethynyl-2'-deoxyuridine (EdU) | Invitrogen | A10044 |
| 5-fluoro-2’-deoxyuridine (FuDR) | Sigma-Aldrich | 856657 |
| Lipofectamine™ RNAiMAX | Invitrogen | 13778500 |
| DMEM without lysine and arginine | AthenaES | 0420 |
| Dialyzed fetal calf serum | Gibco | 26400044 |
| [12C6]-lysine | Silantes | 211004102 |
| [12C6,14N4]-arginine | Silantes | 201003902 |
| [13C6]-lysine | Cambridge Isotope Laboratories | CNLM-291-H-1 |
| [13C6,15N4]-arginine | Cambridge Isotope Laboratories | CNLM-539-H-1 |
| FuGENE® 6 Transfection Reagent | Promega | E2691 |
| X-tremeGENE® 9 DNA transfection reagent | Roche | 6365787001 |
| AlamarBlue® | Invitrogen | DAL1025 |
| 2x Laemmli sample buffer | Sigma-Aldrich | S3401 |
| Benzonase® Nuclease | Novagen/Millipore | 70664 |
| Atto 594 Azide | Atto Tec | AD594-105 |
| 4',6-diamidino-2-phenylindole (DAPI) | Sigma-Aldrich | D9542 |
| Aqua-Poly/Mount | Polysciences, Inc. | 18606-20 |
| RNase A | ThermoScientific | EN0531 |
| RNase T1 | ThermoScientific | EN0541 |
| Proteinase K | ThermoScientific | EO0491 |
| Critical commercial assays | ||
| Tyramide Signal Amplification kit with AlexaFluor™ 488 Tyramide Reagent | Invitrogen | B40932 |
| Duolink® In Situ Detection Reagents Red | Sigma-Aldrich | DUO92008 |
| Pierce™ BCA Protein Assay | ThermoScientific | 23225 |
| Deposited data | ||
| Experimental models: cell lines | ||
| U2OS | ATCC | |
| U2OS CPD photolyase-mCherry | (Steurer et al., 2019) | |
| U2OS XPF KO | (Sabatella et al., 2018) | |
| U2OS XPF KO + GFP-XPF | (Sabatella et al., 2018) | |
| XPCS2BA-sv40 + XPB-GFP | (Hoogstraten et al., 2002) | |
| XP4PA-sv40 + XPC-GFP | (Ribeiro-Silva et al., 2018) | |
| XPCS1RO-sv40 + XPG-GFP | (Zotter et al., 2006) | |
| MRC5-sv40 | ATCC | |
| Xpb-YFP KI MDFs | (Giglia-Mari et al., 2009) | |
| Xpb-YFP KI MDFs Xpc−/− | (Giglia-Mari et al., 2009) | |
| Xpb-YFP KI MDFs Xpg−/− | (Giglia-Mari et al., 2009) | |
| XP186LV | (Wienholz et al., 2017) | |
| C3RO-hTERT | ||
| C5RO-hTERT | ||
| VH10-hTERT | ||
| XP25RO-hTERT | ||
| U2OS GFP-HLTF KI | This paper | N/A |
| U2OS GFP-HLTF KI XPG KO | This paper | N/A |
| U2OS GFP-HLTF KI XPG KO + wt HA-XPG | This paper | N/A |
| U2OS GFP-HLTF KI XPG KO + EZ91A HA-XPG | This paper | N/A |
| U2OS HLTF KO A | This paper | N/A |
| U2OS HLTF KO B | This paper | N/A |
| HEK293T | ||
| Oligonucleotides | ||
| sgHLTF (5’-CCATGTCCTGGATGTTCAAG-3’) | Integrated DNA technologies | N/A |
| sgXPG (5’-GCTGGAGTGCTCCGGGCGGC-3’) | Integrated DNA technologies | N/A |
| siCTRL | Horizon Discovery | N/A |
| siHLTF A A (5’-GGUGCUUUGGCCUAUAUCAUU-3’) | Horizon Discovery | N/A |
| siHLTF B (5’-GGAAACAAUACCAGUGAAAUU-3’) | Horizon Discovery | N/A |
| siXPF (5’-AAGACGAGCUCACGAGUAUUU-3’) | Horizon Discovery | N/A |
| siXPC (5’-CUGGAGUUUGAGACAUAUCUU-3’) | Horizon Discovery | N/A |
| siXPG (SmartPool) | Horizon Discovery | M-006626-00 |
| siGTF2H1 (SmartPool) | Horizon Discovery | L-010924-00 |
| siRAD18 (5’-GCUCUCUGAUCGUGAUUUAUU-3’) | Horizon Discovery | N/A |
| siRFCI | ThermoScientific | M-009290-01 |
| siSMARCALI (5’-GCUCUCUGAUCGUGAUUUAUU-3’) | Horizon Discovery | N/A |
| siZRANB3 (5'-GAUCAGACAUCACACGAUUU-3') | Horizon Discovery | N/A |
| HIRAN mutant primer Forward (5’-CAACGAGAT CCT GCTGCCCCTTAT GATAAG-3') | Integrated DNA Technologies | N/A |
| HIRAN mutant primer Reverse (5’-CTTATCATAAGGGGCAGCAGGATCTCGTTG-3’) | Integrated DNA Technologies | N/A |
| RING mutant primer Forward (5’-GGTT CAGAT GAGGAAT CTGCAA1 1 1GCCTGGATTC-3’) | Integrated DNA Technologies | N/A |
| RING mutant primer Reverse (5’-GAATCCAGGCAAATTGCAGATTCCTCATCTGAACC-3’) | Integrated DNA Technologies | N/A |
| ATPase mutant primer Forward (: 5’-AGAGTGATCCTGGCTGCAGGACATGCCATACG-3’) | Integrated DNA Technologies | N/A |
| ATPase mutant primer Reverse (5’-CGTATGGCATGTCCTGCAGCCAGGATCACTCT-3’) | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pLenti-CMV-Puro | Addgene | 1Z452 |
| pLenti-CRISPRv2 | Addgene | 52961 |
| pLenti-CMV-Puro-GFP-HLTF | This paper | N/A |
| pLenti-CMV-Puro-Flag-HLTF | This paper | N/A |
| pWPXL-EF1 a-XPG (WT) | (Staresincic et al., 2009) | N/A |
| pWPXL-EF1 a-XPG (EZ91A) | (Staresincic et al., 2009) | N/A |
| pEGFP-POLK | (Ogi et al., 2010) | N/A |
| Software and algorithms | ||
| MaxQuant software suite (version 1.5.3.8) | (Cox et al., 2011) | N/A |
| Fiji ImageJ | N/A | |
| Prism GraphPad (version 9.0) | GraphPad software Inc. | N/A |
| Leica LAS | N/A | |
| LAS AF (version 27.4.10100) | Leica Microsystems | |
| Other | ||
| TUV lamp (UV-C) | Phillips | N/A |
| Isopore membrane filter 5 μm pores TMTP | Millipore | TMTP04Z00 |
| Isopore membrane filter 8 μm pores TETP | Millipore | TETP04Z00 |
| Glomax® Multimode reader | Promega | |
| Bioruptor Sonicator | Diagenode | |
| Soniprep 150 | MSE | |
| GFP-Trap-A® agarose bead slurry | ChromoTek | gta-100 |
| Agarose binding control beads | ChromoTek | bab-20 |
| Mini-PROTEAN TGX™ Precast Protein Gels | BioRad | 456-1084 |
| Orbitrap Fusion Tribid mass spectrometer | ThermoScientific | |
Methods Details
UV-C irradiation
For UV-C damage infliction, cells were rinsed with PBS and irradiated under a germicidal UV-C lamp with a 254 nm peak (TUV lamp, Phillips) at the indicated dose. Local UV irradiation was applied through an isopore membrane filter (Millipore) containing 5-μm pores, or 8-μm pores for Figure S1H.
Cell viability assay
40.000 siRNA-transfected C5RO hTERT cells were seeded at low (0.5%) serum in triplicates in 96-well plates and UV-irradiated at the indicated doses 30h later. At 72h after UV irradiation, AlamarBlue® (Invitrogen) was added and fluorescence was measured at 570 nm 2h later according to the manufacturer’s instructions using a Promega Glomax® Multimode reader. Data were background-corrected and normalized to mock-treated conditions.
Crosslinked immunoprecipitation
Crosslinked immunoprecipitation was performed as described previously (Wienholz et al., 2019), with minor adjustments. Briefly, cells were crosslinked with 1% formaldehyde in serum-free culture medium for 10 min at RT with constant shaking. Crosslinked cells were quenched by adding a final concentration of 0.125M glycine for 5 min, collected by scraping and chromatin was purified as described. Subsequently, the nuclear suspension was sonicated using either the Bioruptor Sonicator (Diagenode) with 15 cycles of 15 s on/60 s off using the High setting or Soniprep 150 (MSE) with 9 cycles of 20 sec on/60 sec off at amplitude 6. Equal amounts of crosslinked chromatin were pre-cleared for 30 minutes using agarose binding control beads (ChromoTek) and subsequently incubated with 30 μl GFP-Trap-A agarose bead slurry (ChromoTek) overnight at 4°C. Beads were collected by centrifugation, washed five times with ice cold IP buffer and bound proteins were eluted by incubation at 95 °C for 20 min in 2× Laemmli sample buffer (Sigma). Samples were loaded on 4–15% Mini-PROTEAN TGX™ Precast Protein Gels (BioRad) and either processed for immunoblotting or fixed and stained using using Roti®-Blue (Carl Roth) for mass spectrometry experiments.
Native immunoprecipitation
Cells were harvested by trypsinization, quenched with culture medium, washed with cold PBS and pelleted by 5 minutes of centrifugation at 1300 rpm and 4 °C. The cell pellets were resuspended in native IP buffer (30 mM HEPES pH 7.6, 130 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100 and EDTA-free Protease Inhibitor Cocktail (Roche)), and 1000 U of Benzonase® Nuclease (Novagen) was added to the lysates. After a 30 minutes incubation at 4 °C, 1000 U of additional Benzonase was added and another round of 30 minutes incubation at 4 °C was performed. The lysates were then cleared by centrifugation (15 minutes at 13200 rpm and 4 °C) and the supernatants were collected and the amount of cell lysates were equalized using Pierce BCA Protein Assay (ThermoScientific), according to the manufacturer’s protocol. The equalized lysates were subjected to immunoprecipation with GFP-Trap-A agarose beads overnight at 4°C. Beads were collected by centrifugation, washed five times with cold native IP buffer and bound proteins were eluted by incubation at 95°C for 5 min in 2× Laemmli SDS sample buffer. Samples were loaded on 4–15% Mini-PROTEAN TGX™ Precast Protein Gels, and fixed and stained by Colloidal Coomassie staining using Roti®-Blue, according to the manufacturer’s protocol for mass spectrometry experiments.
Mass spectrometry
SDS-PAGE gel lanes were cut into 2-mm slices and subjected to in-gel reduction with dithiothreitol, alkylation with iodoacetamide and digested with trypsin (sequencing grade; Promega), as described previously (Schwertman et al., 2012). Alternatively, proteins were directly digested with trypsin while still bound to Chromotek GFP-Trap-A beads, after several washes in 50 mM ammonium bicarbonate solution and subsequent reduction and alkylation with chloroacetamide).
Peptides were washed, eluted from C18 STAGE tips and analyzed by nanoflow liquid chromatography tandem mass spectrometry (nLC-MS/MS), performed on an EASY-nLC coupled to an Orbitrap Fusion Tribid mass spectrometer (ThermoScientific) operating in positive mode. Peptides were separated on a ReproSil-C18 reversed-phase column (Dr Maisch; 15 cm × 50 μm) using a linear gradient of 0–80% acetonitrile (in 0.1% formic acid) during 60 or 90 min at a rate of ~200 nL/min. The elution was directly sprayed into the electrospray ionization source of the mass spectrometer. Spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent mode by HCD.
Raw mass spectrometry data were analyzed with the MaxQuant software suite (Cox et al., 2011) (version 1.5.3.8) as described previously (Schwertman et al., 2012). A false discovery rate (FDR) of 0.01 for proteins and peptides and a minimum peptide length of 7 amino acids were set. The Andromeda search engine was used to search the MS/MS spectra against the UniProt database (taxonomy: Mus musculus, release June 2013) concatenated with the reversed versions of all sequences. A maximum of two missed cleavages was allowed. The peptide tolerance was set to 10 ppm and the fragment ion tolerance was set to 0.6 Da for HCD spectra. The enzyme specificity was set to trypsin and cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation, N-terminal protein acetylation, and di-glycine-lysine were chosen as variable modifications. Both the PSM and protein FDR were set to 0.01. In case the identified peptides of two proteins were the same or the identified peptides of one protein included all peptides of another protein, these proteins were combined by MaxQuant and reported as one protein group. Before further statistical analysis, known contaminants and reverse hits were removed.
Live cell confocal imaging
Live cell confocal laser-scanning microscopy was carried out as described before (van Cuijk et al., 2015), with minor adjustments. All live cell imaging experiments were performed at a controlled environment with 37 °C and 5% CO2. To study accumulation kinetics after local UV-C irradiation, a 2 mW pulsed (7.8 kHz) diode-pumped solid state laser emitting at 266 nm (Rapp Opto Electronic, Hamburg GmbH) coupled to a Leica SP5 laser-scanning confocal microscope was used (Dinant et al., 2007). Cells were grown on quartz coverslips and imaged and irradiated through a Ultrafluar quartz 100×/1.35 NA glycerol immersion lens (Carl Zeiss Micro Imaging Inc.). Accumulation curves were background-corrected and normalized to pre-damage values, set at 1.
To test the accumulation of exogenously expressed HLTF-mutants (Fig 4c), we selected cells exciting similar levels of GFP fluorescence as GFP-HLTF knock-in cells. In Figure 1F–G, we plotted the average GFP-HLTF accumulation at t=39 to 45 seconds, as exemplified in Figure S1F.
Fluorescence recovery after photobleaching (FRAP) was performed by bleaching a narrow strip (512×16 pixels) at high 488 nm-laser power and subsequently monitoring the recovery of the fluorescence signal in the strip at low 488 nm-laser power until a steady-state level was reached. All FRAP data was collected on Leica SP5 microscopes using 63×/1.4NA HCX PL APO CS and 40×/1.25 NA HCX PL APO CS oil immersion lenses. Fluorescent intensities were corrected for background values and normalized to pre-bleach values, which were set at 1. Immobile fractions (Fimm) were calculated using the individual and average (indicated by <brackets>) fluorescence intensities after bleaching (Ibleach) and fluorescence intensities after recovery from the bleaching (Irecovery):
For Inverse FRAP (iFRAP), XPB-GFP expressing XPCS2BA cells were siRNA-transfected and local damage was induced by 120 J/m2 UV-C through a 5 μm filter. After 15 min, the undamaged part of selected cells was continuously bleached at high 488-nm laser and the fluorescence decrease in the local damage was determined over time. The resulting dissociation curves were background-corrected and normalized to pre-bleach values, set at 1.
Immunofluorescence
Cells were grown on 24-mm glass coverslips and fixed for 15 min in PBS supplemented with 2% formaldehyde and 0.1% Triton X-100. Subsequently, cells were permeabilized with PBS containing 0.1% Triton X-100 and blocked with 2% bovine serum albumin (BSA) in PBS for 30 minutes at RT. For staining DNA damage with lesion-specific antibodies, DNA was denatured with 70 mM NaOH in PBS for 10 min. Thereafter, cells were incubated with the appropriate antibodies diluted in PBS overnight at 4 °C. Cells were washed with PBS containing 0.1% Triton X-100 and incubated with AlexaFluor™-conjugated secondary antibodies (Invitrogen) and DAPI for 1 hour at RT. After repeated washing with PBS including 0.1% Triton X-100, cells were mounted using Aqua-Poly/Mount and digital images were acquired using an LSM700 microscope equipped with a 40x Plan-apochromat 1.3 NA oil immersion lens (Carl Zeiss). Fluorescence intensities were determined using Fiji and an ImageJ macro. Fold accumulation of the respective proteins were calculated by dividing the average fluorescent intensity at LUD through the average fluorescent intensity inside the rest of the nucleus. Primary and secondary antibodies used for immunofluorescence are listed in the Key Resources Table, respectively.
Unscheduled DNA synthesis and TC-NER specific unscheduled DNA synthesis
UDS and TCR-UDS were performed as described elsewhere (van Cuijk et al., 2015; Wienholz et al., 2017). In short, for UDS C5RO fibroblasts were seeded on 24-mm coverslips, serum-deprived and transfected with the indicated siRNAs 48 h before global UV-C irradiation (16 J/m2). After irradiation, cells were incubated in medium containing 20 μM 5-ethynyl-2’-deoxyuridine (EdU, Invitrogen) and 1 μM 5-fluoro-2’-deoxyuridine (Sigma) for 3 h. Cells were subsequently rinsed with PBS, fixed with 3.6% formaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS. EdU incorporation was visualized using Click-iT chemistry with 60 μM Atto 594 Azide (Atto Tec.), 50 mM Tris-HCl pH 7.6, 4 mM CuSO4*5H2O (Sigma) and 10 mM ascorbic acid (Sigma) for 30 min at RT in the dark. Where indicated, cells were subsequently blocked with 2% BSA in PBS and γH2AX was stained as described in the immunofluorescence section. DNA was counterstained using 4′,6-diamidino-2-phenylindole (DAPI) and slides were mounted with using Aqua-Poly/Mount (Polysciences, Inc.).
For TC-NER specific unscheduled DNA synthesis, serum-starved XPC-deficient XP186LV cells were EdU pulse-labeled for 4 hours after irradiation with 16 J/m2 UV-C. EdU incorporation was visualized using Click-iT chemistry and amplified using Tyramide Signal Amplification kit with AlexaFluor™ 488 Tyramide Reagent (ThermoScientific) as described elsewhere (Wienholz et al., 2017).
All digital images were acquired using an LSM700 microscope equipped with a 40x Plan-apochromat 1.3 NA oil immersion lens (Carl Zeiss) and UDS and TC-UDS levels were quantified by determining the average nuclear fluorescence intensities with a Fiji macro. Background (no UV) fluorescent intensities were subtracted and data were normalized to control cells, which were set at 100%.
In situ proximity ligation assay
Cells were grown on 12 mm coverslips and fixed with 3.6% formaldehyde in PBS for 15 minutes. Cells were permeabilized with PBS containing 0.1% Triton X-100, blocked with 2% BSA in PBS for 30 minutes at RT, and incubated with the appropriate antibodies overnight at 4 °C. PLA was performed using the Duolink anti-Mouse MINUS and anti-Rabbit PLUS In Situ PLA probes (Sigma) and the Duolink In Situ Detection Reagents Red (Sigma), according to the to manufacturer’s instructions. Digital images were acquired using an LSM700 microscope equipped with a 40x Plan-apochromat 1.3 NA oil immersion lens (Carl Zeiss) and nuclear foci were quantified using an ImageJ macro.
In vivo excision assay
Cells that were transfected with the indicated siRNAs were washed with PBS, exposed to UV-C irradiated and harvested 1 h later. Cell pellets were resuspended in Triton X-100 lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100) and incubated for 15 min at 4°C. Soluble cell lysates were separated from insoluble pellets containing genomic DNA by centrifugation (20,000 x g) for 1 h at 4°C. Lysates were then incubated with RNase A and RNase T1, and treated with proteinase K (ThermoScientific).
For complete DNA denaturation during cell lysis, where indicated, the whole cell lysates containing genomic DNA were incubated with RNases, treated with 0.15 mg/ml of proteinase K for 30 min at 55°C, and further treated with 0.3 M NaOH for 30 min at 42°C to achieve DNA denaturation. Soluble cell lysates were then prepared by centrifugation.
After phenol-chloroform extraction and ethanol precipitation, the extracted DNA samples were subjected to immunoprecipitation with anti-(6–4)PP antibody, and processed for chemiluminescent detection as previously described (Choi et al., 2013; Choi et al., 2015; Hu et al., 2013)
Immunoblotting
Cells were lysed in RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, and protease inhibitors), cleared by centrifugation and diluted with equal amounts of 2× Laemmli sample buffer before samples were heated to 95 °C for 5 minutes. Lysates were separated by SDS-PAGE and transferred onto PVDF membranes (0.45 μm, Merck Millipore) overnight at 4 °C. Membranes were blocked with 5% skimmed milk powder (Sigma) in PBS and probed with the appropriate primary antibodies in the Key Resources Table. Membranes were subsequently washed extensively with PBS containing 0.05% Tween-20 and incubated with IRDye-conjugated secondary antibodies (LI-COR, Key Resources Table) to visualize proteins using an Odyssey CLx infrared scanner (LI-COR).
In vitro reconstituted excision assay
146-mer oligonucleotides containing an internal dG-Acetylaminofluorene (AAF) lesion and modified with a terminal biotin tag and complementary oligos were prepared by phosphorylation/ligation methods using T4 polynucleotide kinase/T4 DNA ligase as previously described (Hess et al., 1997). The biotinylated DNA was immobilized on Streptavidin beads (Dynabeads M-280 Streptavidin magnetic beads, Invitrogen). The following proteins were >95% pure and produced as previously described: XPC-RAD23B (Cheon et al., 2019); XPG (Hohl et al., 2003), ERCC1-XPF (Enzlin and Schärer, 2002). RPA was a gift from Walter Chazin (Vanderbilt University), TFIIH was a gift from Chi-Lin Tsai (MD Anderson Cancer Center). HLTF-encoding Baculovirus was prepared using pFL-HIS-HLTF-FLAG and the protein was subsequently expressed in insect cells and purified by sequential application of Ni-NTA, anti-FLAG affinity and SP sepharose chromatography.
The immobilized substrate was incubated with purified NER proteins and HLTF. For each reaction, 3.3 nM of XPC-RAD23B, 6.6 nM of TFIIH, 13.3 nM of XPA, 26.6 nM of RPA, 18 nM of XPG, 8.6 nM of XPF-ERCC1, and a variable amount of HLTF (0, 6.6, 13.3, 26.6 nM) was used. The reactions were carried out in repair buffer containing 45 mM HEPES-KOH pH 7.8, 5 mM MgCl2, 0.3 mM EDTA, 40 mM phosphocreatine (di-Tris salt, Sigma), 2 mM ATP, 1 mM DTT, 2.5 μg BSA, 0.5 μg creatine phosphokinase (Sigma), and NaCl (to a final concentration of 70 mM) in a volume of 15μL (Cheon et al., 2019). Mixtures were pre-warmed at 30 °C for 10 min, added to 12.5 fmol of immobilized substrate and the mixture incubated at 30 °C for 45 min. Following incubation, the beads were immobilized using a magnet, the supernatant removed and isolated and the beads washed with PBS (15μL). The excised DNA fragments of 24–32 oligonucleotides were 32P-labeled and detected in the supernatant,bead and total fractions as previously described (Yeo et al., 2012). The reaction products were visualized using a Phosphorimager (Amersham Typhoon RGB, GE Healthcare Bio-Sciences).
Quantification and Statistical Analysis
Statistical analysis was performed using Prism9 (GraphPad Software). Details on how data and error bars are presented can be found in the figure legends. Unpaired, two-tailed t-tests were used to calculate significance between samples. P values expressed as * P < 0.05; ** P < 0.01 and *** P < 0.001 were considered to be significant. ns, non-significant. For data presented in Figures 2D and 3B, a ROC curve analysis was performed to determine significance.
Supplementary Material
Table S1. Related to Figure. Results of quanftitative interaction proteomics after TFIIH pulldown from Xpb-YFP knock-in mouse dermal fibroblasts.
ACKNOWLEDGEMENTS
We thank the Optical Imaging Centre (OIC) of the Erasmus MC for support with microscopes. We thank Lajos Haracska (Hungarian Academy of Sciences, Hungary) for providing HLTF expression constructs and Tomoo Ogi (Nagoya University, Japan) for GFP-POLK expression construct. We would also like thank Minyong Kee and Jung-Eun Yeo (UNIST, Republic of Korea) for providing dG-AAF containing oligonucleotides, Chi-Lin Tsai (MD Anderson, United States) for providing purified TFIIH protein, Walter Chazin (Vanderbilt University, United States)) for purified RPA protein and Miaw-Sheue Tsai (LBNL, United States) for help with expressing XPC-RAD23B, ERCC1-XPF and XPG. This work is part of the Oncode Institute which is partly financed by the Dutch Cancer Society and was funded by a grant from the Dutch Cancer Society (KWF grant 10506). This work was funded by the Dutch organization for Scientific Research (NWO-ALW) VIDI (864.13.004) and VICI (VI.C.182.025) and by the NWO Graduate Program Erasmus MC – Medical Genetics Grant [022.004.002]. This work was also funded by the Korea Research Institute of Standards and Science (KRISS-2019-GP2019-0009), the Korean Institute for Basic Science (IBS-R022-A1 and IBS-R022-D1) and the NCI (US) (P01CA092584) and Global PhD Fellowship (NRF-2017H1A2A1044961).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Achar YJ, Balogh D, and Haracska L. (2011). Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proceedings of the National Academy of Sciences 108, 14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achar YJ, Balogh D, Neculai D, Juhasz S, Morocz M, Gali H, Dhe-Paganon S, Venclovas Č, and Haracska L. (2015). Human HLTF mediates postreplication repair by its HIRAN domain-dependent replication fork remodelling. Nucleic acids research 43, 10277–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adar S, Hu J, Lieb JD, and Sancar A. (2016). Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proceedings of the National Academy of Sciences 113, E2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseev S, Ayadi M, Brino L, Egly J-M, Larsen Annette K., and Coin F. (2014). A Small Molecule Screen Identifies an Inhibitor of DNA Repair Inducing the Degradation of TFIIH and the Chemosensitization of Tumor Cells to Platinum. Chemistry & Biology 21, 398–407. [DOI] [PubMed] [Google Scholar]
- Bai G, Kermi C, Stoy H, Schiltz CJ, Bacal J, Zaino AM, Hadden MK, Eichman BF, Lopes M, and Cimprich KA (2020). HLTF Promotes Fork Reversal, Limiting Replication Stress Resistance and Preventing Multiple Mechanisms of Unrestrained DNA Synthesis. Molecular Cell 78, 1237-1251.e1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez DA, Greer BH, and Eichman BF (2018). The HIRAN domain of helicase-like transcription factor positions the DNA translocase motor to drive efficient DNA fork regression. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon NY, Kim HS, Yeo JE, Scharer OD, and Lee JY (2019). Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res 47, 8337–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-H, Gaddameedhi S, Kim S-Y, Hu J, Kemp MG, and Sancar A. (2013). Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Research 42, e29–e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Kim SY, Kim SK, Kemp MG, and Sancar A. (2015). An Integrated Approach for Analysis of the DNA Damage Response in Mammalian Cells: NUCLEOTIDE EXCISION REPAIR, DNA DAMAGE CHECKPOINT, AND APOPTOSIS. The Journal of biological chemistry 290, 28812–28821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, and Egly JM (2008). Nucleotide Excision Repair Driven by the Dissociation of CAK from TFIIH. Molecular Cell 31, 9–20. [DOI] [PubMed] [Google Scholar]
- Compe E, and Egly J-M (2016). Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and Beyond. Annual Review of Biochemistry 85, 265–290. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M. (2011). Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. Journal of Proteome Research 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NGJ, and Hoeijmakers JHJ (1998). DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes & Development 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhont L, Mascaux C, and Belayew A. (2016). The helicase-like transcription factor (HLTF) in cancer: loss of function or oncomorphic conversion of a tumor suppressor? Cellular and Molecular Life Sciences 73, 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, de Jager M, Essers J, van Cappellen WA, Kanaar R, Houtsmuller AB, and Vermeulen W. (2007). Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. Journal of Cell Science 120, 2731. [DOI] [PubMed] [Google Scholar]
- Enzlin JH, and Schärer OD (2002). The active site of the DNA repair endonuclease XPF–ERCC1 forms a highly conserved nuclease motif. The EMBO Journal 21, 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, and Vermeulen W. (2005). Nuclear Dynamics of PCNA in DNA Replication and Repair. Molecular and Cellular Biology 25, 9350–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari P-O, Mourgues S, Nonnekens J, Andrieux LO, de Wit J, Miquel C, Wijgers N, Maas A, et al. (2009). Differentiation Driven Changes in the Dynamic Organization of Basal Transcription Initiation. PLOS Biology 7, e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Nguyen THD, Fang J, Afonine PV, Adams PD, and Nogales E. (2017). The cryo-electron microscopy structure of human transcription factor IIH. Nature 549, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanasoge S, and Ljungman M. (2007). H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis 28, 2298–2304. [DOI] [PubMed] [Google Scholar]
- Hess MT, Gunz D, Luneva N, Geacintov NE, and Naegeli H. (1997). Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Molecular and Cellular Biology 17, 7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A, Hara K, Ikegaya Y, Yokoyama H, Shimizu T, Sato M, and Hashimoto H. (2015). Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance. J Biol Chem 290, 13215–13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JHJ (2009). DNA Damage, Aging, and Cancer. New England Journal of Medicine 361, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, and Schärer OD (2003). Structural Determinants for Substrate Binding and Catalysis by the Structure-specific Endonuclease XPG. Journal of Biological Chemistry 278, 19500–19508. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Nigg AL, Heath H, Mullenders LHF, van Driel R, Hoeijmakers JHJ, Vermeulen W, and Houtsmuller AB (2002). Rapid Switching of TFIIH between RNA Polymerase I and II Transcription and DNA Repair In Vivo. Molecular Cell 10, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Hu J, Choi J-H, Gaddameedhi S, Kemp MG, Reardon JT, and Sancar A. (2013). Nucleotide Excision Repair in Human Cells: FATE OF THE EXCISED OLIGONUCLEOTIDE CARRYING DNA DAMAGE IN VIVO. Journal of Biological Chemistry 288, 20918–20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I, Van Houten B, Thomas DC, Abdel-Monem M, and Sancar A. (1985). Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proceedings of the National Academy of Sciences 82, 6774–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly J-M, and Tanaka K. (2007). XPG Stabilizes TFIIH, Allowing Transactivation of Nuclear Receptors: Implications for Cockayne Syndrome in XP-G/CS Patients. Molecular Cell 26, 231–243. [DOI] [PubMed] [Google Scholar]
- Jackson SP, and Bartek J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Reardon JT, Lindsey-Boltz LA, and Sancar A. (2012). Mechanism of Release and Fate of Excised Oligonucleotides during Nucleotide Excision Repair. Journal of Biological Chemistry 287, 22889–22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiakos K, Howard TT, Lee M, Hartley JA, and McHugh PJ (2002). Saccharomyces cerevisiae RAD5 Influences the Excision Repair of DNA Minor Groove Adducts*. Journal of Biological Chemistry 277, 44576–44581. [DOI] [PubMed] [Google Scholar]
- Kile Andrew C., Chavez Diana A., Bacal J, Eldirany S, Korzhnev Dmitry M., Bezsonova I, Eichman Brandt F., and Cimprich Karlene A. (2015). HLTF’s Ancient HIRAN Domain Binds 3′ DNA Ends to Drive Replication Fork Reversal. Molecular Cell 58, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C, Kuper J, and Van Houten B. (2013). Prokaryotic Nucleotide Excision Repair. Cold Spring Harbor Perspectives in Biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokic G, Chernev A, Tegunov D, Dienemann C, Urlaub H, and Cramer P. (2019). Structural basis of TFIIH activation for nucleotide excision repair. Nature Communications 10, 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Hoeijmakers JHJ, Vermeulen W, and Marteijn JA (2019). The DNA damage response to transcription stress. Nature Reviews Molecular Cell Biology 20, 766–784. [DOI] [PubMed] [Google Scholar]
- Li C-L, Golebiowski Filip M., Onishi Y, Samara Nadine L., Sugasawa K, and Yang W. (2015). Tripartite DNA Lesion Recognition and Verification by XPC, TFIIH, and XPA in Nucleotide Excision Repair. Molecular Cell 59, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-R, Lin J-R, Zeman, Michelle K, Chen J-Y, Yee M-C, and Cimprich, Karlene A. (2011). SHPRH and HLTF Act in a Damage-Specific Manner to Coordinate Different Forms of Postreplication Repair and Prevent Mutagenesis. Molecular Cell 42, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Gibbs-Seymour I, and Bekker-Jensen S. (2013). Regulation of PCNA–protein interactions for genome stability. Nature Reviews Molecular Cell Biology 14, 269–282. [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, and Vermeulen W. (2009). Nucleotide excision repair–induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. The Journal of Cell Biology 186, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, and Hoeijmakers JHJ (2014). Understanding nucleotide excision repair and its roles in cancer and ageing. Nature Reviews Molecular Cell Biology 15, 465. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Mitsuyuki S, Kanao R, Hishiki A, Hashimoto H, and Masutani C. (2018). Regulation of HLTF-mediated PCNA polyubiquitination by RFC and PCNA monoubiquitination levels determines choice of damage tolerance pathway. Nucleic Acids Research 46, 11340–11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocquet V, Lainé JP, Riedl T, Yajin Z, Lee MY, and Egly JM (2008). Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. The EMBO Journal 27, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moinova HR, Chen W-D, Shen L, Smiraglia D, Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R, et al. (2002). HLTF gene silencing in human colon cancer. Proc Natl Acad Sci U S A 99, 4562–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A, Liaw H-J, Lee K-Y, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JHJ, et al. (2008). Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proceedings of the National Academy of Sciences 105, 12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Hsu DS, and Sancar A. (1996). Reaction Mechanism of Human DNA Repair Excision Nuclease. Journal of Biological Chemistry 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hara Y, Oka Y, Komine O, van den Heuvel D, Guo C, Daigaku Y, Isono M, He Y, Shimada M, et al. (2020). Ubiquitination of DNA Damage-Stalled RNAPII Promotes Transcription-Coupled Repair. Cell 180, 1228-1244.e1224. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Yamashita S, Lehmann AR, and Ogi T. (2010). A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair 9, 506–516. [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, and Lopes M. (2015). Replication fork reversal in eukaryotes: from dead end to dynamic response. Nature Reviews Molecular Cell Biology 16, 207–220. [DOI] [PubMed] [Google Scholar]
- Ogi T, and Lehmann AR (2006). The Y-family DNA polymerase κ (pol κ) functions in mammalian nucleotide-excision repair. Nature Cell Biology 8, 640–642. [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. (2010). Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Molecular Cell 37, 714–727. [DOI] [PubMed] [Google Scholar]
- Orren DK, Selby CP, Hearst JE, and Sancar A. (1992). Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. Journal of Biological Chemistry 267, 780–788. [PubMed] [Google Scholar]
- Overmeer RM, Gourdin AM, Giglia-Mari A, Kool H, Houtsmuller AB, Siegal G, Fousteri MI, Mullenders LHF, and Vermeulen W. (2010). Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Molecular and cellular biology 30, 4828–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeer RM, Moser J, Volker M, Kool H, Tomkinson AE, van Zeeland AA, Mullenders LHF, and Fousteri M. (2011). Replication protein A safeguards genome integrity by controlling NER incision events. The Journal of Cell Biology 192, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LA, and Cortez D. (2017). Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit Rev Biochem Mol Biol 52, 696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puumalainen M-R, Rüthemann P, Min J-H, and Naegeli H. (2016). Xeroderma pigmentosum group C sensor: unprecedented recognition strategy and tight spatiotemporal regulation. Cell Mol Life Sci 73, 547–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SH, You Z, and Friedberg EC (1998). The Yeast RAD7 and RAD16 Genes Are Required for Postincision Events during Nucleotide Excision Repair: IN VITRO AND IN VIVO STUDIES WITHrad7 AND rad16 MUTANTS AND PURIFICATION OF A Rad7/Rad16-CONTAINING PROTEIN COMPLEX*. Journal of Biological Chemistry 273, 29481–29488. [DOI] [PubMed] [Google Scholar]
- Ribeiro-Silva C, Aydin ÖZ, Mesquita-Ribeiro R, Slyskova J, Helfricht A, Marteijn JA, Hoeijmakers JHJ, Lans H, and Vermeulen W. (2018). DNA damage sensitivity of SWI/SNF-deficient cells depends on TFIIH subunit p62/GTF2H1. Nature Communications 9, 4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T, Hanaoka F, and Egly J-M (2003). The comings and goings of nucleotide excision repair factors on damaged DNA. The EMBO journal 22, 5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthemann P, Balbo Pogliano C, Codilupi T, Garajovà Z, and Naegeli H. (2017). Chromatin remodeler CHD1 promotes XPC-to-TFIIH handover of nucleosomal UV lesions in nucleotide excision repair. The EMBO Journal 36, 3372–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatella M, Theil AF, Ribeiro-Silva C, Slyskova J, Thijssen K, Voskamp C, Lans H, and Vermeulen W. (2018). Repair protein persistence at DNA lesions characterizes XPF defect with Cockayne syndrome features. Nucleic Acids Research 46, 9563–9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer OD (2013). Nucleotide Excision Repair in Eukaryotes. Cold Spring Harbor Perspectives in Biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JHJ, Demmers JAA, Fousteri M, Vermeulen W, et al. (2012). UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nature Genetics 44, 598–602. [DOI] [PubMed] [Google Scholar]
- Sertic S, Mollica A, Campus I, Roma S, Tumini E, Aguilera A, and Muzi-Falconi M. (2018). Coordinated Activity of Y Family TLS Polymerases and EXO1 Protects Non-S Phase Cells from UV-Induced Cytotoxic Lesions. Molecular Cell 70, 34–47.e34. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, and Schärer OD (2009). Coordination of dual incision and repair synthesis in human nucleotide excision repair. The EMBO Journal 28, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer B, Turkyilmaz Y, van Toorn M, van Leeuwen W, P. Escudero-Ferruz, and Marteijn JA (2019). Fluorescently-labelled CPD and 6–4PP photolyases: new tools for live-cell DNA damage quantification and laser-assisted repair. Nucleic Acids Research 47, 3536–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K. (2016). Molecular mechanisms of DNA damage recognition for mammalian nucleotide excision repair. DNA Repair 44, 110–117. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Akagi J. i., Nishi R, Iwai S, and Hanaoka F. (2009). Two-Step Recognition of DNA Damage for Mammalian Nucleotide Excision Repair: Directional Binding of the XPC Complex and DNA Strand Scanning. Molecular Cell 36, 642–653. [DOI] [PubMed] [Google Scholar]
- Theil AF, Nonnekens J, Wijgers N, Vermeulen W, and Giglia-Mari G. (2011). Slowly Progressing Nucleotide Excision Repair in Trichothiodystrophy Group A Patient Fibroblasts. Molecular and Cellular Biology 31, 3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He Q-L, Bhat S, Low W-K, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. (2011). XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature Chemical Biology 7, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdú I, Fátyol K, Hurwitz J, Yoon J-H, Prakash L, Prakash S, and Haracska L. (2008). Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proceedings of the National Academy of Sciences 105, 3768–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cuijk L, van Belle GJ, Turkyilmaz Y, Poulsen SL, Janssens RC, Theil AF, Sabatella M, Lans H, Mailand N, Houtsmuller AB, et al. (2015). SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nature Communications 6, 7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B, Gamper H, Hearst JE, and Sancar A. (1988). Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. Journal of Biological Chemistry 263, 16553–16560. [PubMed] [Google Scholar]
- Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JHJ, van Driel R, van Zeeland AA, and Mullenders LHF (2001). Sequential Assembly of the Nucleotide Excision Repair Factors In Vivo. Molecular Cell 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wienholz F, Vermeulen W, and Marteijn JA (2017). Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Research 45, e68–e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholz F, Zhou D, Turkyilmaz Y, Schwertman P, Tresini M, Pines A, van Toorn M, Bezstarosti K, Demmers JAA, and Marteijn JA. (2019). FACT subunit Spt16 controls UVSSA recruitment to lesion-stalled RNA Pol II and stimulates TC-NER. Nucleic Acids Research 47, 4011–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J-E, Khoo A, Fagbemi AF, and Schärer OD (2012). The Efficiencies of Damage Recognition and Excision Correlate with Duplex Destabilization Induced by Acetylaminofluorene Adducts in Human Nucleotide Excision Repair. Chemical Research in Toxicology 25, 2462–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Owen-Hughes T, Friedberg EC, Waters R, and Reed SH (2004). The yeast Rad7/Rad16/Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair. DNA Repair 3, 277–287. [DOI] [PubMed] [Google Scholar]
- Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JHJ, van Driel R, Vermeulen W, and Houtsmuller AB (2006). Recruitment of the Nucleotide Excision Repair Endonuclease XPG to Sites of UV-Induced DNA Damage Depends on Functional TFIIH. Molecular and Cellular Biology 26, 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure. Results of quanftitative interaction proteomics after TFIIH pulldown from Xpb-YFP knock-in mouse dermal fibroblasts.
Data Availability Statement
The mass spectrometry data reported in this paper have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031243. Uncropped images of immunoblots and microscopy images are available from Mendeley Data 10.17632/h2nr6yp67j.1
This study did not generate code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.