Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) regulates cell growth and catabolism in response to nutrients through phosphorylation of key substrates. The tumor suppressor folliculin (FLCN) is a RagC/D guanosine triphosphatase (GTPase)–activating protein (GAP) that regulates mTORC1 phosphorylation of MiT-TFE transcription factors, controlling lysosome biogenesis and autophagy. We determined the cryo–electron microscopy structure of the active FLCN complex (AFC) containing FLCN, FNIP2, the N-terminal tail of SLC38A9, the RagAGDP:RagCGDP.BeFx- GTPase dimer, and the Ragulator scaffold. Relative to the inactive lysosomal FLCN complex structure, FLCN reorients by 90°, breaks contact with RagA, and makes previously unseen contacts with RagC that position its Arg164 finger for catalysis. Disruption of the AFC-specific interfaces of FLCN and FNIP2 with RagC eliminated GAP activity and led to nuclear retention of TFE3, with no effect on mTORC1 substrates S6K or 4E-BP1. The structure provides a basis for regulation of an mTORC1 substrate-specific pathway and a roadmap to discover MiT-TFE family selective mTORC1 antagonists.
The cryo-EM structure of the active FLCN RagC GAP complex opens the way to selectively activate MiT-TFE transcription factors.
INTRODUCTION
The mechanistic target of rapamycin complex 1 (mTORC1) plays a central role in response to fluctuations in nutrients, growth factors, and energy in cells by altering the balance between cell growth and catabolism. In nutrient-rich environments, mTORC1 activates anabolic processes including protein and lipid synthesis while inhibiting catabolic ones such as autophagy (1, 2). mTORC1 regulates its proanabolic programs through phosphorylation of key targets, some of which contain a conserved TOR signaling (TOS) motif (3, 4). The microphthalmia (MiT-TFE) family of transcription factors, including similarly regulated transcription factors EB and E3 (TFEB and TFE3), activate transcription of genes involved in autophagy (5) and lysosome biogenesis (6) and are regulated by mTORC1 (7–9). Among well-known mTORC1 substrates, MiT-TFE family members lack the TOS motif responsible for docking some other substrates onto mTORC1 via its Raptor subunit. Also, in contrast to other mTORC1 substrates, MiT-TFE phosphorylation and negative regulation are completely dependent on the activity of the RagC guanosine triphosphatase (GTPase)–activating protein (GAP), folliculin (FLCN) (10–12).
The Rag-Ragulator complex consists of two Rag GTPases (commonly referred to as Rags) that function as obligate heterodimers and are anchored to the lysosomal membrane by the pentameric scaffold Ragulator (13, 14). The Rags respond to lysosomal and cytoplasmic nutrient levels, which regulate the proportion of inactive (RagA or BGDP:RagC or DGTP) and active (RagA or BGTP:RagC or DGDP) Rag dimers (15). Conversion between active and inactive states is regulated by the RagA/B-specific GAP, GAP activity towards Rags 1 (GATOR1), and by the RagC/D-specific GAP, FLCN:FLCN-interacting protein 2 (FNIP2) (16, 17). In response to amino acid levels, as sensed by the transceptor SLC38A9 among other mechanisms (18–20), FLCN stimulates the conversion of Rags to their active form enabling mTORC1 recruitment and interaction with MiT-TFE members. Upon phosphorylation, MiT-TFE members bind to 14-3-3 proteins and are sequestered in the cytosol where they remain inactive. The regulation of TOS motif–containing mTORC1 substrates (3), including regulators of mRNA translation and cell proliferation, such as 4E-BP1 and S6K1, is unaffected (10, 11, 21). Together, FLCN directly and selectively controls mTORC1 regulation of MiT-TFE members but not other substrates (10, 11). These findings support the existence of separate mechanisms for control of mTORC1-dependent anabolic versus catabolic programs (22). This highlights a therapeutic potential for FLCN to be targeted as a means of mTOR inhibition for treating LSDs and neurodegenerative diseases, with fewer toxic effects on cell proliferation and translation.
The transient nature of the GAP interaction with Rag-Ragulator has proven challenging for visualizing details necessary for deducing the mechanism of GAP activity. Previous structural investigation into the basis for FLCN localization to lysosomes during amino acid starvation (23) revealed a stable inhibitory FLCN-Rag-Ragulator complex, known as the lysosomal FLCN complex (LFC) (11, 24). This begged the question as to how FLCN binds to RagC to promote nucleotide hydrolysis and what are the interfaces in FLCN essential for stimulating catalysis. We sought to understand how these residues interact with the RagC nucleotide-binding pocket and what makes FLCN specific for RagC. We previously found that the amino acid transporter, SLC38A9, promotes FLCN GAP activity by inserting its cytosolic domain in the cleft between the RagA and RagC G domains and thereby disrupting the LFC (19). Here, we trapped FLCN in its GAP-competent binding mode using a fusion construct of FNIP2 and the N terminus of SLC38A9 (SLC38A9NT), determined the cryo–electron microscopy (cryo-EM) structure, and deduced the mechanism of FLCN GAP activity essential for interrogating this interaction during drug development.
RESULTS
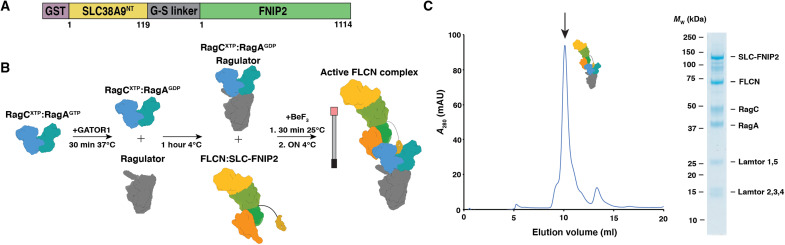
To provide a structural framework for understanding the mechanism of FLCN GAP activity, we stabilized the transient complex and determined the cryo-EM structure, hereafter referred to as the active FLCN complex (AFC). We began by generating a stable complex of FLCN:FNIP2, Rags, Ragulator, and SLC38A9NT, using a mutant FLCNF118D known to disrupt the LFC without affecting GAP activity (11). To tether FLCN to the Rag-Ragulator complex, we designed a fusion construct containing a 10-residue glycine-serine linker between the C terminus of SLC38A9NT and the N terminus of FNIP2 (Fig. 1A). The C-terminal tail of SLC38A9NT contains ~20 disordered residues, effectively increasing the linker length to ~30 residues allowing for up to 90 Å in length. We used a phosphate analog (BeF3) during complex formation to capture FLCNF118D bound to Rag-Ragulator (25). Rags were loaded into a state mimicking starvation conditions (RagAGDP:RagCGTP) and incubated with Ragulator to form a stable complex. Next, FLCNF118D and the SLC38A9NT-FNIP2 (SLC-FNIP2) fusion protein were added to RagAGDP:RagCGTP-Ragulator immediately followed by the addition of BeF3 (Fig. 1B and fig. S1). Incubating the complex overnight, we generated a nucleotide ground-state analog that trapped FLCNF118D in an active conformation and allowed for isolation of a stable active FLCN complex via size exclusion chromatography (Fig. 1, B and C).
Fig. 1. Active FLCN complex formation.
(A) Construct design for the SLC38A9NT-FNIP2 fusion construct. (B) AFC complex formation workflow. (C) Size exclusion profile for AFC. Fraction containing all components of complex is labeled with a black arrow and visualized on adjacent gel.
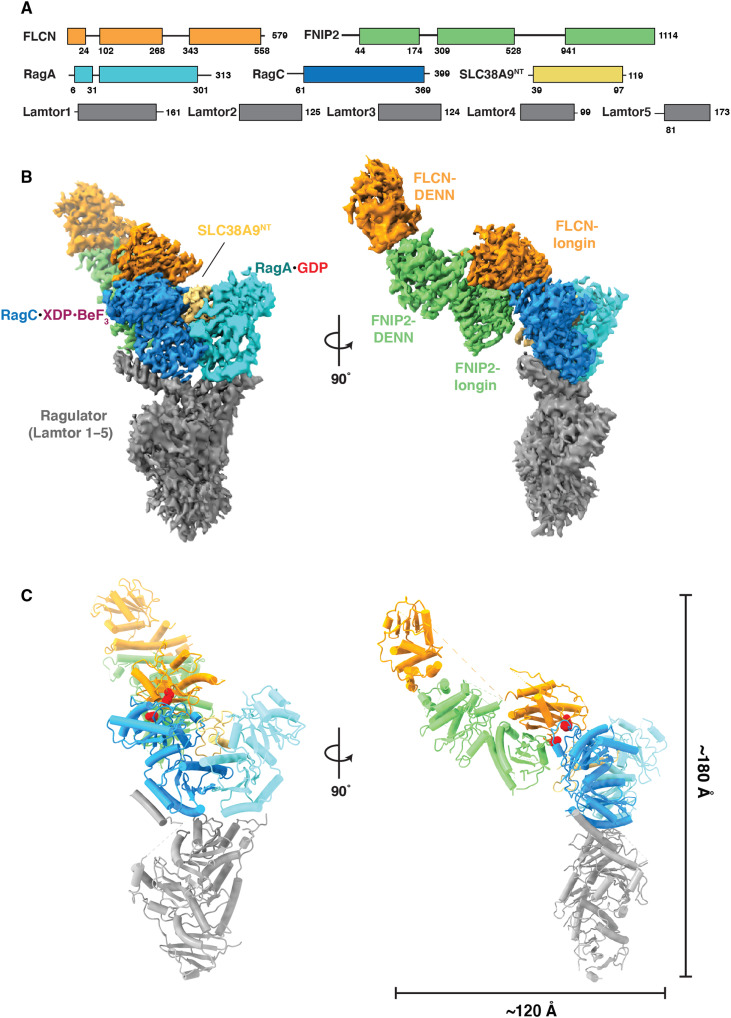
The cryo-EM structure of the AFC was determined to an overall resolution of 3.5 Å (fig. S2 and table S1). The cryo-EM density was of sufficient quality to model the ordered mass of the complex constituting the AFC (figs. S3 and S4). In the AFC, the longin domains of FLCNF118D and FNIP2 interact with the RagC G domain such that FLCNF118D:FNIP2 approaches Rag-Ragulator from the side (Fig. 2, B and C). SLC38A9NT sits in the cleft between the Rags and does not interact directly with FLCNF118D:FNIP2, suggesting that it does not play a role in positioning the GAP. The overall dimensions of the complex are ~120 Å by ~180 Å (Fig. 2C). To interact with RagC in a GAP-competent conformation, FLCNF118D:FNIP2 reorients 90° and transitions from its elongated position in the LFC to bind the side of RagC in the AFC (movie S1). In the LFC, FLCN:FNIP2 interacts with both RagC and RagA extensively, with 870- and 645-Å2 interfaces, respectively. In contrast, in the AFC conformation, FLCNF118D:FNIP2 interacts solely with RagC, burying 1141 Å2 of surface area with RagC, but none with RagA.
Fig. 2. Cryo-EM structure of the active FLCN complex.
(A) Domain organization for components of AFC. (B) Composite map for complex. Focused map of interface was combined with the overall map to produce a composite map containing the highest-resolution information for each component. (C) Reconstructed model for AFC with overall dimensions of the side view of the complex. Dotted regions in the model represent unstructured regions in FLCN connecting the two domains. Red spheres indicated locations of mutations in FLCN F10D and FNIP2 V146E.
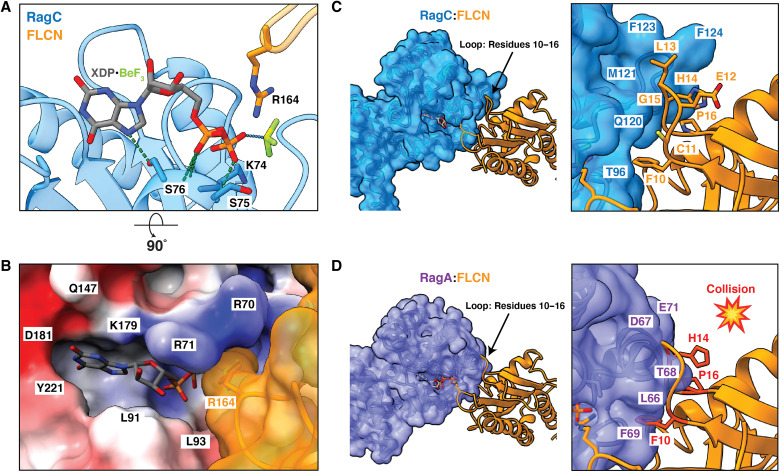
To visualize the AFC catalytic complex in as much detail as possible, we used local refinement at the FLCN-RagC interface to increase the resolution to 3.16 Å and allow for clear placement of the nucleotide analog and previously identified arginine finger (Arg164) in RagC nucleotide-binding domain (NBD) (figs. S2 and S3D) (11). As bound to GDP-BeF−3, RagC adopts an architecture analogous to GTP-bound RagC and Gtr2 (Fig. 3A) (26, 27). In this state, the switch I region is ordered and forms a lid at the top of the binding pocket. Additional density in the RagC NBD, adjacent to the BeF−3, aligned with the placement of the highly conserved Arg164. The position of FLCNF118D Arg164 resembled the geometry of previously identified arginine fingers in Rab1 GAP structures trapped with BeF−3 nucleotide analogs (fig. S5) (28, 29). The nucleotide-binding pocket is composed of primarily charged residues with a 534-Å2 surface area, while the overall volume of the larger surrounding pocket is 865 Å3 (Fig. 3B) (30).
Fig. 3. Specificity of FLCN-RagC GAP interaction in AFC.
(A) Nucleotide-binding pocket of RagC (blue) in AFC containing the XDP-BeF3 nucleotide analog. The location of FLCN (orange) arg finger is indicated. Hydrogen bonds between nucleotide and RagC residues (blue sticks) in the surrounding pocket are represented by dashed lines (green). (B) Surface representation of RagC binding site with FLCN arg finger (orange). RagC is colored on the basis of electrostatic potential. (C) RagC (blue) FLCN (orange) interface in AFC. Loop residues 10 to 16 in FLCN are indicated as sticks. (D) RagA (purple) FLCN interface modeled with GTP-bound RagA (Protein Data Bank: 6S6D). Residues in the FLCN loop that clash with RagA are shown in red.
To determine the basis for FLCN specificity for RagC over RagA, we modeled the GAP interaction with RagA-GTP. The FLCNF118D loop containing residues 10 to 16 was accommodated when interacting with RagC (Fig. 3C). In contrast, in the RagA model, FLCNF118D (Phe10, His14, and Pro16) clashed with RagA at the interface (Fig. 3D). We sought to further explore FLCN specificity for RagC by understanding why GATOR1 does not function as a GAP towards RagC. We modeled RagC with GATOR1 and observed residues in GATOR1 (Thr18 and Pro19) that clashed with RagC at the interface (fig. S6B). We also compared the RagC structure to that of Arf1, which is a substrate of another longin domain GAP, Smith-Magenis Syndrome Chromosomal Region Candidate 8 (SMCR8) (31, 32). In the Arf1 model, FLCNF118D (Leu13 and His14) clashes with Arf1 residues (fig. S6A), rationalizing specificity for RagC. To determine why other longin domain GAPs do not function to stimulate nucleotide hydrolysis of RagC, we modeled RagC with SMCR8, a longin GAP that acts on Arf GTPases. The binding between SMCR8 and RagC is incompatible due to clashes between SMCR8 (Glu62 and Phe63) and RagC (fig. S6C).
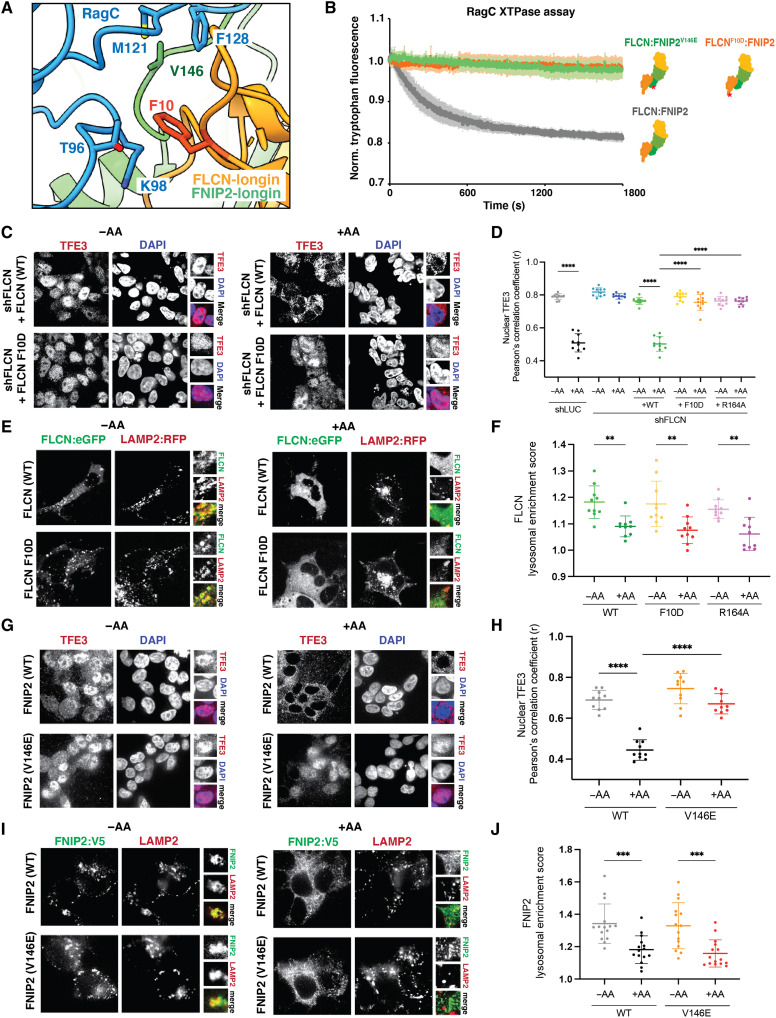
We analyzed the interface whereby FLCNF118D:FNIP2 interacts with RagC. Two hydrophobic residues FLCN Phe10 and FNIP2 Val146 form prominent contacts in the RagC binding interface interacting with Thr96, Lys98, Met121, and Phe128 (Fig. 4A). We hypothesized that these residues were critical for GAP activity. To validate the importance of these residues, we created two mutants FLCNF118D/F10D (F10D:Phe10→Asp) and FNIP2V146E (V146E:Val146→Glu). We used a tryptophan-based GAP activity assay to monitor the activity of FLCNF118D:FNIP2 and mutants FLCNF118D/F10D:FNIP2 and FLCNF118D:FNIP2V146E toward inactive Rags (RagAGDP:RagCGTP) (11, 19). The GAP activity of the FLCNF118D:SLC-FNIP2 fusion protein was identical to FLCNF118D:FNIP2 without the SLC38A9 N terminus fusion (fig. S7). Both FLCNF118D/F10D:FNIP2 and FLCNF118D:FNIP2V146E abolished GAP activity as compared to FLCNF118D:FNIP2, confirming their importance in facilitating GAP binding (Fig. 4).
Fig. 4. Hydrophobic residues in the FLCN:RagC interface necessary for GAP activity.
(A) RagC (blue) and the FLCN (orange):FNIP2 (green) interface in AFC. FLCN:FNIP2 residues mutated at the interface indicated (red). (B) Tryptophan fluorescence xanthosine triphosphatase (XTPase) assay with FLCN:FNIP2 mutants. FLCNWT:FNIP2 (gray), FLCNF10D:FNIP2 (orange), or FLCN:FNIP2V146E (green) was incubated with RagAGDP:RagCXTP. Plotted are means ± SEM. n = 3 replicates. (C) Immunofluorescence images of human embryonic kidney (HEK) 293T cells stably expressing short hairpin RNAs targeting the indicated genes and FLCN rescue constructs. Cells were starved for amino acids (−AA) or restimulated with complete Dulbecco’s modified essential medium (+AA). (D) Quantification of TFE3 nuclear localization for immunofluorescence images and the positive control FLCN164 identified previously (8) shown in fig. S8 (means ± SD, n = 10 fields). ****P < 0.0001. (E) Fluorescence images of HEK293A cells expressing the indicated FLCN:eGFP construct along with HA-FNIP2 and the lysosomal marker Lysosomal associated membrane protein-2 (LAMP2):RFP. Cells were starved for amino acids (−AA) or starved and then restimulated with amino acids for 15 min (+AA). (F) Quantification of the lysosomal localization of overexpressed FLCN for immunofluorescence images (means ± SD, n = 15 cells). **P < 0.01. (G) Immunofluorescence images of HEK293T cells stably overexpressing the FNIP2WT-eGFP-V5 or FNIP2V146E-eGFP-V5 constructs. Cells were starved for amino acids (−AA) or maintained in complete media (+AA). (H) Quantification of TFE3 nuclear localization for immunofluorescence images (means ± SD, n = 10 fields). ****P < 0.0001. (I) Immunofluorescence images of HEK293T cells stably overexpressing the indicated FNIP2-eGFP-V5 constructs. LAMP2 was used as a lysosomal marker. Cells were starved for amino acids (−AA) or maintained in complete media (+AA). (J) Quantification of the lysosomal localization of overexpressed FNIP2 for immunofluorescence images (means ± SD, n = 15 cells). ***P < 0.001. DAPI, 4′,6-diamidino-2-phenylindole.
We sought to investigate whether these GAP-inactive mutants, FLCNF10D and FNIP2V146E, affected the ability of Rag-Ragulator to activate mTORC1 with respect to its various substrates. We used cell-based assays to image the localization of the MiT-TFE family transcription factor TFE3. In amino acid–deprived cells reconstituted with either WT FLCN (FLCNWT) or WT FNIP2 (FNIP2WT), both FLCNWT and FNIP2WT were localized to the lysosome (Fig. 4, E and I) and TFE3 was present in the nucleus (Fig. 4, C and G). Cells expressing FLCNF10D and FNIP2V146E during starvation resulted in lysosomal localization and nuclear localization of the MiT-TFE family member TFE3 (Fig. 4, C, E, G, and I). When cells were restimulated with amino acids, those reconstituted with FLCNWT and FNIP2WT resulted in translocation of TFE3 to the cytoplasm. In contrast, in cells expressing FLCNF10D and FNIP2V146E, TFE3 remained nuclear, phenocopying the catalytic loss-of-function mutant FLCNR164 (fig. S8) (11), and was not sequestered to the cytoplasm despite high cellular nutrients (Fig. 4, C and G). These results further supported that the mutants FLCNF10D and FNIP2V146E eliminated GAP activity and confirmed that loss of GAP activity resulted in misregulation of mTORC1 and TFE3. Immunoblot analysis confirmed that the mutants did not have any effect on the phosphorylation of the TOS motif–containing substrates S6K and 4E-BP1 (fig. S9).
DISCUSSION
The cryo-EM structure presented in this work reveals that FLCN:FNIP2 undergoes a marked reorientation to convert from the inactive LFC to the active AFC. The AFC structure confirmed the prediction that FLCN activates nucleotide hydrolysis of RagC through the catalytic arginine finger Arg164 (11, 33). We identified features on the FLCN surface that are complementary to RagC and confer the high degree of specificity that is seen biologically. Our structure revealed that the loop containing residues 10 to 16 in FLCN makes it incompatible as a GAP for other small GTPases. In the AFC structure, SLC38A9NT sits in the cleft between the Rags, while FLCN:FNIP2 binds only RagC. This confirms the previously proposed model that SLC38A9NT displaces inactive FLCN:FNIP2 in the LFC to allow for binding in the GAP competent mode (19). SLC38A9NT does not, however, play a role in GAP activity through interaction with FLCN or either Rag nucleotide-binding pocket in the AFC, as further supported by the near-identical RagC-GAP activity of FLCNF118D:FNIP2 with or without the SLC38A9NT fusion.
RagA is regulated by a different GAP complex, GATOR1. A recent cryo-EM structure of the GAP-competent interaction between Rag-Ragulator and GATOR1 suggested that GATOR1 is stabilized by both Rag GTPases in its GAP mode (34). The overall geometry and the role of the Arg finger in the active GATOR1 complex resemble that of FLCNF118D:FNIP2 seen here, as well as that of another longin domain–based GAP, the C9orf72-SMCR8 complex with ARF1 (31). In comparison to GATOR1, the structure of the AFC revealed that FLCN solely interacts with RagC to stimulate GTP hydrolysis. This finding is consistent with observations that while GATOR1 RagA GAP activity depends on the RagC nucleotide state, the RagC GAP activity of the AFC is independent of the RagA nucleotide state (19).
The past few years have seen a paradigm shift in understanding how the MiT-TFE family of transcription factors is regulated downstream of mTORC1 in a uniquely FLCN- and RagCGDP-dependent manner (10, 11). Numerous studies corroborate the discovery of a “noncanonical” mTORC1 pathway that specifically regulates MiT-TFE transcription factors (12, 21, 35–38). The new knowledge of FLCN structural mechanism provided by the AFC structure can potentially be exploited for therapeutic benefit. The AFC presents itself as an attractive avenue for the long-standing goal of developing substrate selective mTORC1 inhibitors that can up-regulate autophagy and lysosomal capacity without toxic consequences for immune function, epithelial renewal, and other physiological processes that require mTORC1-dependent anabolic programs. Such inhibitors could be highly specific for MiT-TFE up-regulation, in turn promoting autophagy and increasing lysosomal capacity. The latter could be of potential benefit for treating LSDs, while both could potentially benefit neurodegenerative diseases involving intracellular protein aggregates. On the other hand, excessive up-regulation of MiT-TFE transcription factors in FLCN loss of function has been shown to cause kidney cystogenesis and tumorigenesis and induce melanoma (10, 35). Therapeutics that promote FLCN activity, for instance, through inhibition of the LFC, might have potential for treating these conditions. An important consideration in therapeutic use of FLCN GAP antagonists would be to avoid the complete shutdown of FLCN activity and so avoid promoting tumorigenesis. The structural insights into the AFC presented here fulfill a key step necessary to leverage FLCN as a drug target and develop selective mTORC1 inhibitors or activators for MiT-TFE–dependent transcriptional up-regulation.
MATERIALS AND METHODS
Cloning and protein purification
The pCAG-GST-SLC38A9NT-FNIP2 fusion construct was designed to contain a 10-residue glycine-serine linker. Codon-optimized DNA coding for SLC38A9NT and the C-terminal linker was amplified by polymerase chain reaction. DNA containing SLC38A9NT and the C-terminal linker was subcloned into the pCAG-GST vector containing codon-optimized DNA coding for FNIP2. The pCAG-GST-FNIP2 vector was linearized using Kpn I and Xho I restriction sites. DNA containing SLC38A9NT and the C-terminal linker was inserted at the N-terminus of FNIP2 by Gibson assembly. Human embryonic kidney (HEK) 293–GNTI cells were transfected with 1 mg of DNA at a 2:1 ratio (FLCN:SLC38A9NT-FNIP2) and 3 mg of polyethyleneimine (PEI) per 1 liter of cells at 2 × 106 cells/ml. Cells were pelleted at 2000g for 20 min at 4°C and purified as described previously (11).
Mutants FLCN F10D and FNIP2 V146E were generated by site-directed mutagenesis using KAPA HiFi HotStart ReadyMix (Roche) from pCAG-FLCN and pCAG-GST-FNIP2, respectively. HEK293-GNTI cells were transfected with 1 mg of DNA at a 2:1 ratio (FLCN:FNIP2) and 3 mg of PEI per 1 liter of cells at 2 × 106 cells/ml. Cells were pelleted at 2000g for 20 min at 4°C and purified as described previously (11).
RagA:RagCD181N and Ragulator were expressed and purified from Sf9 cells via baculovirus infection as previously described (11). GATOR1 was expressed and purified from HEK293-GNTI cells as previously described (11).
Nucleotide loading
Purified RagA:RagCD181N heterodimers were diluted 1:10 (v/v) in 1× phosphate-buffered saline (PBS) containing 5 mM EDTA and 0.5 mM TCEP [tris(2-carboxyethyl)phosphine] for 10 min at 25°C. Desired triphosphate nucleotides were added at a 10-fold molar excess over Rags and incubated for 30 min at 25°C. Next, MgCl2 was added to a final concentration of 20 mM and incubated for 10 min at 25°C. Excess nucleotides were removed using a PD-10 desalting column (Cytiva) and wash buffer containing 25 mM Hepes, 130 mM NaCl, 2.5 mM MgCl2, 2 mM EGTA (pH 7.4), and 0.5 mM TCEP. To obtain the inactive state for Rag heterodimers, RagAGTP:RagCXTP was incubated with GATOR1 at a 1:100 GAP:Rag molar ratio at 37°C for 30 min.
Active FLCN complex assembly
To assemble the active FLCN complex, RagA:RagCD181N heterodimers in the desired nucleotide state were incubated for 1 hour at 25°C with a 1.2× molar excess of Ragulator. FLCN:SLC38A9NT-FNIP2 was added to the Rag-Ragulator complex at a twofold molar excess followed immediately by the addition of 0.5 M BeF3. BeF3 was prepared by mixing BeSO4 and NaF to a final concentration of 0.5 and 5 mM, respectively, incubating for 20 min at 25°C before addition to complex. The complex containing Rags, Ragulator, FLCN:SLC38A9NT-FNIP2, and BeF3 was incubated at 25°C for 30 min, and then at 4°C for 16 hours. Following incubation, the complex was centrifuged at 17,000g for 10 min, and then loaded onto a Superose 6 (GE Healthcare) column equilibrated in wash buffer containing 25 mM Hepes, 130 mM NaCl, 2.5 mM MgCl2, 2 mM EGTA (pH 7.4), and 0.5 mM TCEP. Fractions were analyzed via SDS-PAGE and those containing assembled active FLCN complex were collected, concentrated to 1 mg/ml, and immediately frozen onto grids for imaging via cryo-EM.
Cryo-EM grid preparation and imaging
For data collection of the active FLCN complex, 3 μl of sample [protein (1 mg/ml) in 25 mM Hepes, 130 mM NaCl, 2.5 mM MgCl2, 2 mM EGTA, and 0.5 mM TCEP (pH 7.4)] was deposited onto freshly glow-discharged (PELCO easiGlow, 45 s in air at 20 mA and 0.4 mbar) holey carbon grids (C-flat: 2/1-3Cu-T). FEI Virobot Mark IV was used to blot grids for 2 s with a blot force of 20 (Whatman 597 filter paper) at 4°C and 90 to 100% humidity and subsequently plunged into liquid ethane. The Titan Krios G3i microscope equipped with a Gatan Quantum energy filter (slit width 20 eV) and a K3 summit camera at a defocus of −1.0 to −2.0 μm was used to record 4968 movies. Automated image acquisition was performed using SerialEM (39) recording four movies per 2-μm hole with image shift. Image parameters are summarized in table S1.
Cryo-EM data processing
The data processing workflow is summarized in fig. S1. In short, the Relion 3.1.1 wrapped MotionCor2 program was used to motion-correct gain-corrected movies (40, 41). Motion-corrected micrographs were imported into cryoSPARC2 v3.3.1 (42). Patch-based CTF estimation (multi) was used for CTF determination, and two rounds of cryoSPARC2 blob picker with a diameter range of 150 to 220 Å were used to generate 1,165,732 and 1,025,735 particles. Particles were extracted with a box size of 440 × 440 pixels in cryoSPARC2. A series of 2D classifications followed by an ab initio reconstruction was used to generate three reference maps. The resulting 3D maps were used for consecutive rounds of heterogeneous refinement from the initial particle sets following a round of 2D classification to remove obvious “junk.” The resulting particle sets from round 1 and round 2 of picks were merged and duplicates were removed. The combined particle set contained 177,018 particles. A final round of homogeneous refinement resulted in a 3.47-Å map at 0.143 Fourier Shell Correlation (FSC). A mask was generated surrounding the interface between RagC and FLCN:FNIP2 using UCSF Chimera and imported into cryoSPARC2 v3.3.1 where it was low-pass–filtered and dilated (43). The mask was used for subsequent local refinement and resulted in a focused map reaching a resolution of 3.16 Å.
Atomic model building and refinement
The coordinates from RagC were based on the GTP-bound crystal structure (3LLU), the coordinates for FLCN and FNIP2 were based on the LFC (6NZD), and all five Ragulator, RagA, and SLC38A9NT were based on the Rag-Ragulator-SLC38A9NT structure (6WJ2). All coordinates were rigid body–fitted separately into the density map using UCSF Chimera. The interface was refined using a map generated from local refinement separately by iterative rounds of Phenix real-space refinement and manual correction in Coot (44). Secondary structure restraints were enabled during real-space refinement. The interface was combined with the remaining components of the complex in Coot and refined using iterative rounds of Phenix real-space refinement and ISOLDE (45). The final model was validated using MolProbity (46). Coordinate models can be found in the Protein Data Bank with accession code 8DHB.
Tryptophan fluorescence RagC XTPase assay
Tryptophan fluorescence experiments were performed in triplicate according to Fromm et al. (19).
Cell culturing
HEK293T and HEK293A cells were grown in Dulbecco’s modified essential medium (DMEM; Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS; VWR), penicillin (100 U/ml), and streptomycin (100 U/ml) (Life Technologies) and were maintained at 37°C and 5% CO2.
Reagents and antibodies
The following antibodies were used in this study: phospho-T389 S6K1 (9234S), S6K1 (2708S), phospho-S65 4EBP (9451S), 4EBP (9644S and 9452S), FLCN (3697), TFE3 (14779), TFEB (37785), V5 (13202), FLAG (14793) (all from Cell Signaling Technology), and LAMP2 (sc-18822) (from Santa Cruz Biotechnology).
RNAi
The control pLKO.1-LUC short hairpin RNA (shRNA) vector and the pLKO.1-FLCN lentiviral shRNA vector were obtained respectively from Addgene (Plasmid, #30324) and the RNA Interference (RNAi) Consortium (TRCN0000237886). Target sequences are the following:
shLUC: TCCTAAGGTTAAGTCGCCCTCG
shFLCN: GATGGAGAAGCTCGCTGATTT
RNAi experiments were prepared as described below.
Lentiviral infection and stable cell line generation
Lentivirus was generated using the PEI method. Five micrograms of a lentiviral vector (desired construct in pLJM1-PURO) was combined with 3.75 μg of psPAX2 and 1.25 μg of pCMG.2 viral packaging plasmids in 500 μl of optiMEM (Thermo Fisher Scientific), mixed, and then combined with 60 μl of PEI (1 mg/ml). This solution was incubated at room temperature (RT) for 30 min, and then added to a 10-cm dish containing 2 million recently plated 293T cells.
Twenty-four to 48 hours after that, media containing virus were collected from cells, spun down at 1300 rpm for 5 min to remove cell debris, and sterile-filtered. Cells were plated at a density of 200,000 to 300,000 cells per well in a six-well plate in the presence of polybrene transfection reagent (Sigma-Aldrich) and infected with 10% of the total virus from one 10-cm plate of the indicated constructs. Twenty-four hours later, media were supplemented with 200 nM puromycin.
Live cell treatments
HEK293T cells were seeded and allowed to attach overnight in complete DMEM. Twenty-four hours after seeding, cells were rinsed with PBS and incubated in amino acid–free RPMI for 2 hours (−AA) and restimulated with amino acids for 15 min or overnight with complete media (+AA). Amino acid solutions were prepared from powders and the final concentration of amino acids in the media was the same as in commercial RPMI. Torin1 treatment was performed for 4 hours in complete media.
Western blotting
HEK293T cells were plated in a six-well plate at 1,000,000 cells per well in complete media. After the treatments (see above), the medium was removed and 150 μl of lysis buffer [40 mM Hepes (pH 7.4), 4 mM EDTA, 1% Triton X-100, 10 mM sodium β-glycerol phosphate, 10 mM sodium pyrophosphate, and one tablet of EDTA-free protease inhibitors per 50 ml] was added to each well. Total extracts were collected from each well after scraping, rotated over-end for 10 min, and then centrifuged at 13,000g for 10 min at 4°C. Samples were normalized to a total concentration of 1 mg/ml protein, combined with protein sample buffer, and then boiled for 5 min at 95°C.
Ten micrograms of protein was loaded into each lane of tris-glycine precast gels (Thermo Fisher Scientific). Polyvinylidene difluoride membranes were blocked in 5% skim milk dissolved in tris-buffered saline with Tween 20 (TBST) for 1 hour at RT. Membranes were incubated overnight at 4°C with the indicated primary antibodies, all diluted in 5% skim milk prepared in TBST. Secondary antibodies conjugated to horseradish peroxidase were used for protein detection. β-Actin was used as loading controls.
Immunofluorescence
HEK293T cells were plated on fibronectin-coated glass coverslips in 12-well plates, at 300,000 cells per well. The following day, cells were subjected to amino acid depletion/or complete media restimulation (see above) and then fixed in 4% paraformaldehyde (in PBS) for 15 min at RT. The coverslips were rinsed twice with PBS and cells were permeabilized with 0.1% (w/v) saponin in PBS for 10 min. After rinsing twice with PBS, the slides were incubated overnight at 4°C with the indicated primary antibody in 5% normal donkey serum, then rinsed with PBS, and incubated with fluorophore-conjugated secondary antibodies produced in goat or donkey (diluted 1:500 in 5% normal donkey serum; Life Technologies) for 45 min at RT in the dark. Coverslips were then washed three times in PBS and mounted on glass slides using VECTASHIELD Antifade Mounting Medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole stain. All images were collected on a Nikon Ti-E inverted microscope (Nikon Instruments, Melville, NY) equipped with a Plan Apo 60× oil objective. Images were acquired using a Zyla 5.5 scientific complementary metal-oxide semiconductor camera (Andor Technology), using the iQ3 acquisition software (Andor Technology).
Live imaging
HEK293A cells were plated onto glass-bottom 35-mm culture dishes and transfected with the indicated FLCN-FLAG-eGFP, HA-FNIP2, and LAMP2-RFP-FLAG constructs. The following day, cells were subjected to amino acid depletion/or amino acids restimulation (see above). Starved cells were transferred to imaging buffer [10 mM Hepes, 136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, and 1.2 mM MgCl2 (pH 7.4)], while restimulated cells were transferred to imaging buffer supplemented with amino acids, 5 mM glucose, and 1% dialyzed FBS (+AA) and imaged using the Nikon Ti-E inverted microscope (Nikon Instruments, Melville, NY) as explained above.
Quantitation of lysosomal enrichment score
For immunofluorescence and live imaging datasets, a home-built MATLAB (MathWorks) script was used to determine the lysosomal enrichment of both V5-FNIP2 and FLAG-FLCN stains as previously described (9). Briefly, a single cell was manually selected in the lamp channel and its nucleus was excluded from further analysis. Then, a mask was created in the LAMP2 channel to segment cellular pixels into LAMP2 (lysosomal) or non-LAMP2 (cytosolic) pixels. This mask was then applied to the non-LAMP2 channel. The lysosomal enrichment score was determined by dividing the average intensity of pixels in the lysosomal region by the average intensity of the pixels in the cytosolic region. For each condition, at least 20 cells were analyzed from different multichannel images.
Quantitation of cytoplasmic/nuclear TFE3 ratio
To measure TFE3 nuclear localization, Pearson’s coefficient of at least 10 different fields was measured using the ImageJ plugin JACoP (47).
Acknowledgments
We thank Z. Cui and A. Joiner for discussions, A. Ballabio for comments on the manuscript, R. Hooy for workstation support, and D. Toso and J. Remis for electron microscope support.
Funding: This work was supported by the NIH grants R01 GM111730 (to J.H.H.) and R01 GM130995 (to R.Z.), the National Science Foundation Graduate Research Fellowship (to R.M.J.), and an Italian Cancer Research Association (AIRC) postdoctoral fellowship (to R.P.).
Author contributions: R.M.J. and J.H.H. conceived and designed research. R.M.J. and R.P. carried out research. A.L.Y. and S.A.F. trained R.M.J. in cryo-EM. R.Z. and J.H.H. supervised the research. R.M.J. and J.H.H. wrote the first draft, and all authors edited the manuscript.
Competing interests: J.H.H. is a cofounder and a shareholder of Casma Therapeutics and receives research funding from Casma Therapeutics, Genentech, and Hoffmann-La Roche. R.Z. is a cofounder and a shareholder of Frontier Medicines and receives research funding from Genentech. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Coordinates and density are deposited in the RCSB (PDB ID 8DHB) and EMDB (EMD-27435), respectively. The fusion construct used to generate the purified AFC is deposited in Addgene.org (189869). The DNA constructs used for FNIP2 localization will be provided on request pending a completed material transfer agreement. Requests for the FNIP2 DNA constructs should be submitted to R.Z. at rzoncu@berkeley.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence R. E., Zoncu R., The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J., TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278, 15461–15464 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Settembre C., Malta C. D., Polito V. A., Arencibia M. G., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A., TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Malta C. D., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A., A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Martina J. A., Chen Y., Gucek M., Puertollano R., MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S. U., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., Ballabio A., A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C., Ferguson S. M., The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napolitano G., Di Malta C., Esposito A., de Araujo M. E. G., Pece S., Bertalot G., Matarese M., Benedetti V., Zampelli A., Stasyk T., Siciliano D., Venuta A., Cesana M., Vilardo C., Nusco E., Monfregola J., Calcagnì A., Di Fiore P. P., Huber L. A., Ballabio A., A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature 585, 597–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence R. E., Fromm S. A., Fu Y., Yokom A. L., Kim D. J., Thelen A. M., Young L. N., Lim C.-Y., Samelson A. J., Hurley J. H., Zoncu R., Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 366, 971–977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada S., Neinast M., Jang C., Ibrahim Y. H., Lee G., Babu A., Li J., Hoshino A., Rowe G. C., Rhee J., Martina J. A., Puertollano R., Blenis J., Morley M., Baur J. A., Seale P., Arany Z., The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 30, 2551–2564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekiguchi T., Hirose E., Nakashima N., Ii M., Nishimoto T., Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276, 7246–7257 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M., Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen K., Choe A., Sabatini D. M., Intersubunit crosstalk in the Rag GTPase heterodimer enables mTORC1 to respond rapidly to amino acid availability. Mol. Cell 68, 552–565.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsun Z.-Y., Bar-Peled L., Chantranupong L., Zoncu R., Wang T., Kim C., Spooner E., Sabatini D. M., The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar-Peled L., Chantranupong L., Cherniack A. D., Chen W. W., Ottina K. A., Grabiner B. C., Spear E. D., Carter S. L., Meyerson M., Sabatini D. M., A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Tsun Z.-Y., Wolfson R. L., Shen K., Wyant G. A., Plovanich M. E., Yuan E. D., Jones T. D., Chantranupong L., Comb W., Wang T., Bar-Peled L., Zoncu R., Straub C., Kim C., Park J., Sabatini B. L., Sabatini D. M., Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fromm S. A., Lawrence R. E., Hurley J. H., Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9. Nat. Struct. Mol. Biol. 27, 1017–1023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rebsamen M., Pochini L., Stasyk T., de Araújo M. E. G., Galluccio M., Kandasamy R. K., Snijder B., Fauster A., Rudashevskaya E. L., Bruckner M., Scorzoni S., Filipek P. A., Huber K. V. M., Bigenzahn J. W., Heinz L. X., Kraft C., Bennett K. L., Indiveri C., Huber L. A., Superti-Furga G., SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K., Wada S., Gosis B. S., Thorsheim C., Loose P., Arany Z., Folliculin promotes substrate-selective mTORC1 activity by activating RagC to recruit TFE3. PLoS Biol. 20, e3001594 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napolitano G., Di Malta C., Ballabio A., Non-canonical mTORC1 signaling at the lysosome. Trends Cell Biol. , S0962–S8924 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Petit C. S., Roczniak-Ferguson A., Ferguson S. M., Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202, 1107–1122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen K., Rogala K. B., Chou H.-T., Huang R. K., Yu Z., Sabatini D. M., Cryo-EM structure of the human FLCN-FNIP2-Rag-ragulator complex. Cell 179, 1319–1329.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher A. J., Smith C. A., Thoden J. B., Smith R., Sutoh K., Holden H. M., Rayment I., X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry 34, 8960–8972 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Jeong J.-H., Lee K.-H., Kim Y.-M., Kim D.-H., Oh B.-H., Kim Y.-G., Crystal structure of the Gtr1p(GTP)-Gtr2p(GDP) protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J. Biol. Chem. 287, 29648–29653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anandapadamanaban M., Masson G. R., Perisic O., Berndt A., Kaufman J., Johnson C. M., Santhanam B., Rogala K. B., Sabatini D. M., Williams R. L., Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazdag E. M., Streller A., Haneburger I., Hilbi H., Vetter I. R., Goody R. S., Itzen A., Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep. 14, 199–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavriljuk K., Gazdag E.-M., Itzen A., Kötting C., Goody R. S., Gerwert K., Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 109, 21348–21353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian W., Chen C., Lei X., Zhao J., Liang J., CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su M. Y., Fromm S. A., Remis J., Toso D. B., Hurley J. H., Structural basis for the ARF GAP activity and specificity of the C9orf72 complex. Nat. Commun. 12, 3786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su M. Y., Fromm S. A., Zoncu R., Hurley J. H., Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD. Nature 585, 251–255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen K., Valenstein M. L., Gu X., Sabatini D. M., Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J. Biol. Chem. 294, 2970–2975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egri S. B., Ouch C., Chou H.-T., Yu Z., Song K., Xu C., Shen K., Cryo-EM structures of the human GATOR1-Rag-ragulator complex reveal a spatial-constraint regulated GAP mechanism. Mol. Cell 82, 1836–1849.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alesi N., Akl E. W., Khabibullin D., Liu H.-J., Nidhiry A. S., Garner E. R., Filippakis H., Lam H. C., Shi W., Viswanathan S. R., Morroni M., Ferguson S. M., Henske E. P., TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat. Commun. 12, 4245 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin J. M., Walkup W. G. 4th, Hooper K., Li T., Kishi-Itakura C., Ng A., Lehmberg T., Jha A., Kommineni S., Fletcher K., Garcia-Fortanet J., Fan Y., Tang Q., Wei M., Agrawal A., Budhe S. R., Rouduri S. R., Baird D., Saunders J., Kiselar J., Chance M. R., Ballabio A., Appleton B. A., Brumell J. H., Florey O., Murphy L. O., GABARAP sequesters the FLCN-FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis. Sci. Adv. 7, eabj2485 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gosis B. S., Wada S., Thorsheim C., Li K., Jung S., Rhoades J. H., Yang Y., Brandimarto J., Li L., Uehara K., Jang C., Lanza M., Sanford N. B., Bornstein M. R., Jeong S., Titchenell P. M., Biddinger S. B., Arany Z., Inhibition of nonalcoholic fatty liver disease in mice by selective inhibition of mTORC1. Science 376, eabf8271 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paquette M., Yan M., Ramírez-Reyes J. M. J., El-Houjeiri L., Biondini M., Dufour C. R., Jeong H., Pacis A., Giguère V., Estall J. L., Siegel P. M., Audet-Walsh É., Pause A., Loss of hepatic Flcn protects against fibrosis and inflammation by activating autophagy pathways. Sci. Rep. 11, 21268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolte S., Cordelieres F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 and S2
Movie S1