Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces mild or asymptomatic COVID-19 in most cases, but some patients develop an excessive inflammatory process that can be fatal. As the NLRP3 inflammasome and additional inflammasomes are implicated in disease aggravation, drug repositioning to target inflammasomes emerges as a strategy to treat COVID-19. Here, we performed a high-throughput screening using a 2560 small-molecule compound library and identified FDA-approved drugs that function as pan-inflammasome inhibitors. Our best hit, niclosamide (NIC), effectively inhibits both inflammasome activation and SARS-CoV-2 replication. Mechanistically, induction of autophagy by NIC partially accounts for inhibition of NLRP3 and AIM2 inflammasomes, but NIC-mediated inhibition of NAIP/NLRC4 inflammasome are autophagy independent. NIC potently inhibited inflammasome activation in human monocytes infected in vitro, in PBMCs from patients with COVID-19, and in vivo in a mouse model of SARS-CoV-2 infection. This study provides relevant information regarding the immunomodulatory functions of this promising drug for COVID-19 treatment.
Niclosamide is an inhibitor of both inflammasome activation and SARS-CoV-2 replication.
INTRODUCTION
Human coronavirus infections are typically mild and rarely associated with a severe clinical outcome, but a few highly pathogenic human betacoronaviruses were previously identified: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 (1). In 2019, a novel human-infecting betacoronavirus named as SARS-CoV-2 was identified as responsible for the coronavirus disease 2019 (COVID-19) pandemic (2). SARS-CoV-2 infection may be asymptomatic or cause a broad spectrum of symptoms, varying from mild upper respiratory tract symptoms in most individuals to a life-threatening disease (3). Severe and critical COVID-19 cases are associated with pneumonia, acute respiratory distress syndrome, and respiratory failure (4). Poor clinical outcome correlates with the overactivation and dysfunctional immune response, along with cytokine storm (2, 3, 5).
The inflammasomes are multiprotein signaling platforms that assemble in the cytosol in response to microbial infections and cell stress. They are recognized by their important role not only for the defense against infections but also for their participation in the genesis of many human inflammatory disorders (6). Among the proinflammatory cytokines, active interleukin-1β (IL-1β) and IL-18 are hallmarks of the inflammatory response mediated by the inflammasomes (3). Recent reports suggest a critical role of the inflammasomes in the progression of COVID-19 to severe outcomes. It was initially shown that the inflammatory cytokine IL-6 is associated with COVID-19 severity (7). Notably, increased plasma levels of IL-6 are associated with elevated serum levels of lactate dehydrogenase, a cytosolic protein that is released from dying cells during inflammatory processes (8). Furthermore, an elegant longitudinal study performed in patients with COVID-19 revealed a correlation between the late-stage pathology in COVID-19 and cytokines linked to the inflammasome pathway, including IL-1β and IL-18 (9). Agreeing to these findings, we and others have demonstrated that SARS-CoV-2 infection triggers the activation of the NLRP3 inflammasome and that the magnitude of inflammasome activation in patients with COVID-19 correlates with the disease outcome (10–12), indicating that inflammasomes are activated by SARS-CoV2 and worsen the clinical outcome of patients with COVID-19.
Immunomodulatory agents that antagonize IL-6 have shown weak effects on the COVID-19 disease, possibly because of the dual functions of IL-6 during infection (13, 14). Although a clinical trial using an anti–IL-1β failed to meet primary end point for efficacy in patients with severe COVID-19 patients (https://clinicaltrials.gov/ct2/show/NCT04362813), studies using a recombinant IL-1 receptor antagonist commonly used to treat autoinflammatory disorders demonstrated clinical improvement in most patients with COVID-19 (15–18), indicating that the inflammasome is as a potential therapeutic target for COVID-19. Because repositioning of existing drugs for new indications is an effective strategy to quickly respond to emerging infectious diseases, we developed a quick and efficient high-content screening to identify approved drugs that can target multiple inflammasomes. We identified three compounds that effectively inhibit cell death, multiple inflammasome activation, and gasdermin D activation. Two of them were effective in inhibiting SARS-CoV-2 replication and impairing inflammasome activation in cells from patients with COVID-19. Our best hit, niclosamide (NIC), is under phase 1 clinical trials, and this study provides essential information regarding the immunomodulatory mechanisms of this highly promising drug.
RESULTS
A high-throughput screening identifies FDA-approved drugs that inhibit inflammasome activation
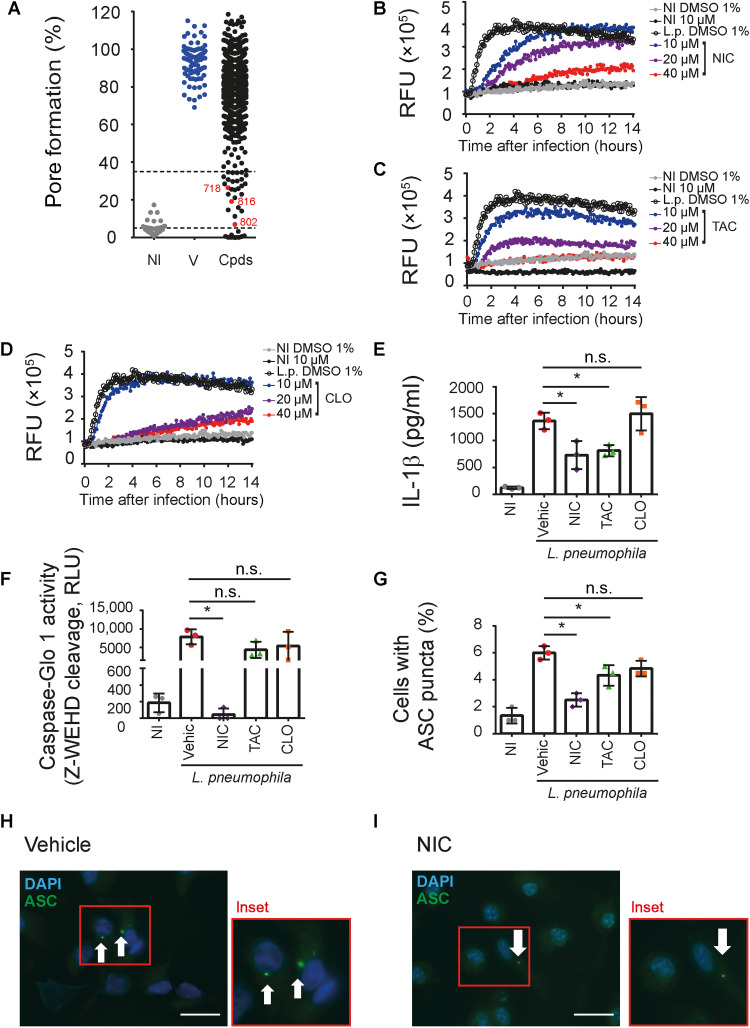
The Gram-negative bacteria Legionella pneumophila triggers multiple inflammasomes and is as a reliable model to investigate inflammasome activation in macrophages [reviewed in (19)]. Thus, we used membrane pore formation induced by L. pneumophila infection, a process dependent on inflammasome effector molecules such as caspase-1, caspase-11, and gasdermin D (19, 20), as a readout to screen for molecules that inhibit inflammasome activation in macrophages. Pore formation in macrophages infected with L. pneumophila occurs quickly after infection and can be easily assessed by probing the influx of propidium iodide (PI) to the cell nuclei (fig. S1A). Using this pore formation assay, we screened a compound library containing 2560 small molecules with known pharmacological properties. For the screening, murine bone marrow–derived macrophages (BMDMs) were pretreated with each library compound at 10 μM and infected with L. pneumophila, and pore formation was analyzed 3 hours after infection. We set 65 to 95% inhibition of pore formation as the cutoff criteria for compound selection, which allowed the selection of 26 compounds for further investigation (Fig. 1A). The screening Z′ factor of 0.85 is well above the generally accepted Z′ factor threshold (≥0.5) (21), indicating a robust and reliable assay (fig. S1B). From the selected 26 compounds, we excluded seven molecules not approved by the U.S. Food and Drug Administration (FDA). Next, we performed a revalidation assay with the 19 FDA-approved hits using three different concentrations for each compound (10, 20, and 40 μM) and by analyzing the inhibition of pore formation over a 14-hour infection period. Excluding antibiotics that may affect Legionella viability and on the basis of the dose-response reproducible validation, we selected three hit drugs: 718 (NIC; Fig. 1B), 802 [tannic acid (TAC); Fig. 1C], and 816 [closantel (CLO); Fig. 1D]. None of these three compounds reduced the nuclear count (fig. S1C), confirming that they are nontoxic for host cells. To further evaluate their inhibitory activity, we probed the effect of these drugs in other responses triggered by inflammasome activation, including caspase-1 activation, IL-1β production, and formation of ASC (apoptosis-associated speck-like protein containing a caspase activation and recruitment domain) puncta/specks 1 hour after infection, as previously described (22). We found that NIC, and to a lesser extent TAC, effectively inhibited the production of IL-1β (Fig. 1E), caspase-1 activation (Fig. 1F), and the formation of ASC puncta (Fig. 1, G to I). We also performed experiments in which NIC-treated macrophages were infected with Legionella for 1, 6, or 9 hours for the assessment of inflammasome activation and found that NIC treatment effectively inhibits IL-1β production, caspase-1 activation, and ASC puncta formation in all tested time points (fig. S2, A to C). We have previously described that, in the absence of caspase-1 activation, Legionella infection triggers a lytic cell death pathway that depends on caspase-8 and caspase-7 (22, 23). Thus, we believe that this pathway explains the detection of pore formation in the presence of NIC at longer time points (Fig. 1B) despite reduced activation of the inflammasome (Fig. 1, E to G, and fig. S2, A to C). Collectively, these data indicate that NIC is the most effective drug for inhibition of inflammasome activation in response to L. pneumophila, thus supporting additional investigation using this drug.
Fig. 1. A high-throughput screening identifies anti-inflammasome drugs.
(A) BMDMs were primed with lipopolysaccharide (LPS; 500 ng/ml) for 3 hours, treated with different library compound at 10 μM, and infected with L. pneumophila at a multiplicity of infection (MOI) of 10. Pore formation was quantified by PI uptake after 3 hours infection. Triton X-100 was used to determine 100% PI uptake. Each dot represents the activity of one compound. NI, noninfected controls; V, dimethyl sulfoxide (DMSO) controls; Cpds, tested compounds. The drugs 718 (NIC), 802 (TAC), and 816 (CLO) are highlighted in red. The dotted lines indicate the 65 to 95% range of pore formation inhibition used for the initial selection. (B to D) Kinetics of pore formation in BMDMs infected or NI with L. pneumophila for 14 hours. Pore formation was estimated by PI internalization [relative fluorescence units (RFU)]. (E and F) BMDMs were primed with LPS for 3 hours; treated with 10 μM NIC, TAC, or CLO or 1% DMSO (Vehic) for 1 hour and infected with L. pneumophila (MOI of 10). Cell supernatants (SNs) were assayed for IL-1β (E), caspase-1 activity (F), and percentage of cell containing ASC puncta (G). Representative images of ASC puncta (green) in L. pneumophila–infected BMDMs treated with vehicle (H) or NIC (I) are shown. Scale bars, 30 μm. *P < 0.05. n.s., nonsignificant as determined by Student’s t test. Data are presented from one representative experiment of three experiments performed with similar results.
NIC inhibits activation of multiple inflammasomes
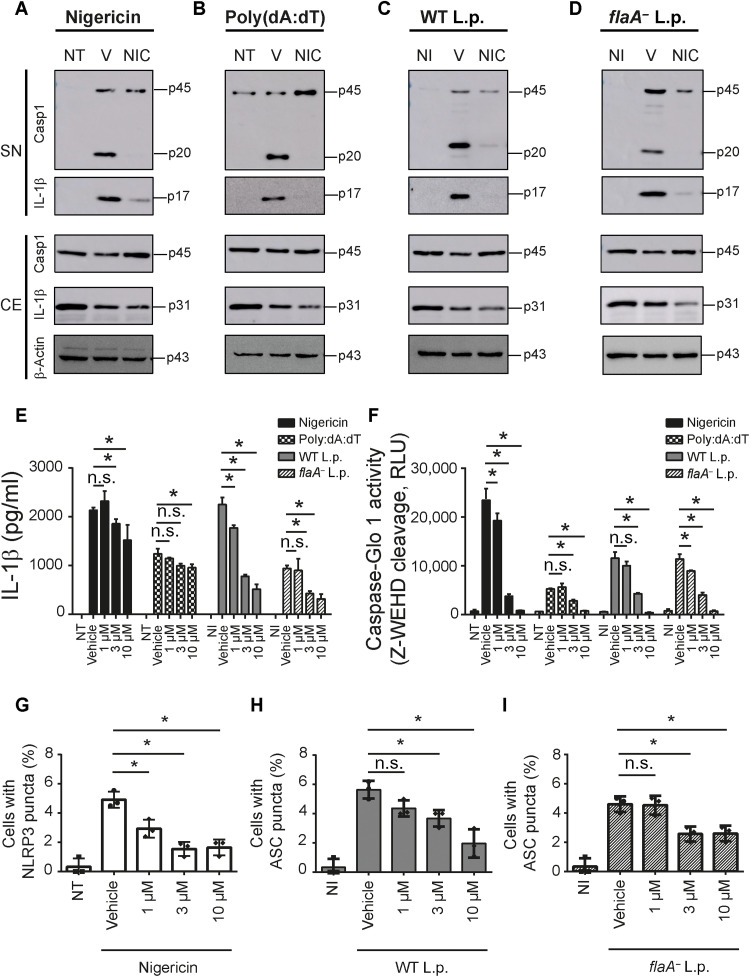
L. pneumophila infection activates multiple inflammasomes. Initially, the NAIP/NLRC4 inflammasome is strongly activated in response to bacterial flagellin (24). In the absence of flagellin (using flaA− bacteria), a robust activation of caspase-11 leads to noncanonical activation of the NLRP3 inflammasome (20). Thus, to test the inhibitory effect of NIC in different inflammasomes, we stimulated macrophages with nigericin or poly(dA:dT) and infected them with wild-type L. pneumophila (WT L.p.) or flaA− L. pneumophila (flaA− L.p.) to induce activation of NLRP3, AIM2, NAIP/NLRC4, and caspase-11 inflammasomes, respectively. We first tested the cleavage of caspase-1 and IL-1β by Western blot and found that NIC inhibits the inflammasome activation in response to canonical NLRP3, AIM2, and NAIP/NLRC4 and noncanonical NLRP3 activation mediated by caspase-11 (Fig. 2, A to D). By assessing IL-1β secretion by enzyme-linked immunosorbent assay (ELISA) and caspase-1 activation by Caspase-Glo 1 assay, we found that NIC promotes inhibition of inflammasome activation stimulated by nigericin, poly(dA:dT), WT L.p., and flaA− L.p. (Fig. 2, E and F). We detected a stronger inhibitory effect of NIC when we assessed inflammasome activation by Western blot and Caspase-Glo 1 assay compared to IL-1β quantification by ELISA. We speculate that the monoclonal antibody used to detect IL-1β in the ELISA kit is recognizing the pro–IL-1β in conditions where there is no processed IL-1β. This would explain the detection of IL-1β by ELISA in conditions where very low levels of IL-1β are present according to the Western blot assays.
Fig. 2. NIC inhibits multiple inflammasomes.
BMDMs were primed with LPS (500 ng/ml) for 3 hours; treated with vehicle (V) or NIC at 10 μM for 1 hour; and then left nontreated (NT) or stimulated with nigericin (20 μM) or poly(dA:dT) (5 ng/ml) or left noninfected (NI) or infected with WT L.p. (MOI of 10) or flaA− L.p. (MOI of 10). Western blot analysis showing caspase-1 p20 and IL-1β cleavage p17 in cell-free SNs and caspase-1 p45, IL-1β p31, and β-actin p43 in the cellular extract (CE) after stimulus with (A) nigericin (20 μM) or transfected with (B) poly(dA:dT) (5 ng/ml) or infected with (C) WT L.p. (MOI of 10) or (D) flaA− L.p. (MOI of 10). LPS-primed BMDMs were treated with 1, 3, or 10 μM NIC for 1 hour and then left NT or stimulated with nigericin (20 μM) or poly(dA:dT) (5 ng/ml) or left NI or infected with WT L.p. (MOI of 10) or flaA− L.p. (MOI of 10). SNs were assayed for (E) an IL-1β release or (F) a caspase-1 activity using the Caspase-1 Glo kit assay. The percentage of cell containing (G) NLRP3 puncta after nigericin treatment, (H) ASC puncta after WT L.p., or (I) ASC puncta after flaA− L.p. infection was estimated in cells. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Student’s t test. Data are presented from one representative experiment of three experiments performed with similar results.
Next, we tested the effect of NIC in NLRP3 and ASC puncta formation. We found inhibition of NLRP3 puncta induced by nigericin (Fig. 2G) and ASC puncta induced by WT L.p. (Fig. 2H) and flaA− L.p. (Fig. 2I). These results validate our initial screening findings and suggest that NIC inhibits the activation of multiple inflammasomes. Although our NIC treatment was always initiated after macrophage priming, we also performed experiments to evaluate the effect of NIC on the expression of pro–IL-1β induced during the macrophage priming. Thus, macrophages were treated with NIC for 1 hour and stimulated with lipopolysaccharide (LPS) or PAM3Cys for 1 and 3 hours. We found that NIC inhibited pro–IL-1β expression induced by priming (fig. S3A). In addition, we observed that NIC inhibited the secretion of cytokines that are rapidly produced after priming, including tumor necrosis factor–α (TNF-α), IL-10, and monocyte chemoattractant protein-1 (MCP-1) (fig. S3, B to D). This process occurs similarly in autophagy related 5 (ATG5)-deficient and ATG5-sufficient macrophages, indicating that autophagy is not required for the inhibitory activity of NIC in priming. Western blot using anti-ATG5 confirms ATG5 deletion in macrophages from ATG5flox/flox/LysMCre/+ mice (fig. S3E).
Autophagic machinery accounts for the inhibitory effects of NIC on NLRP3 inflammasome
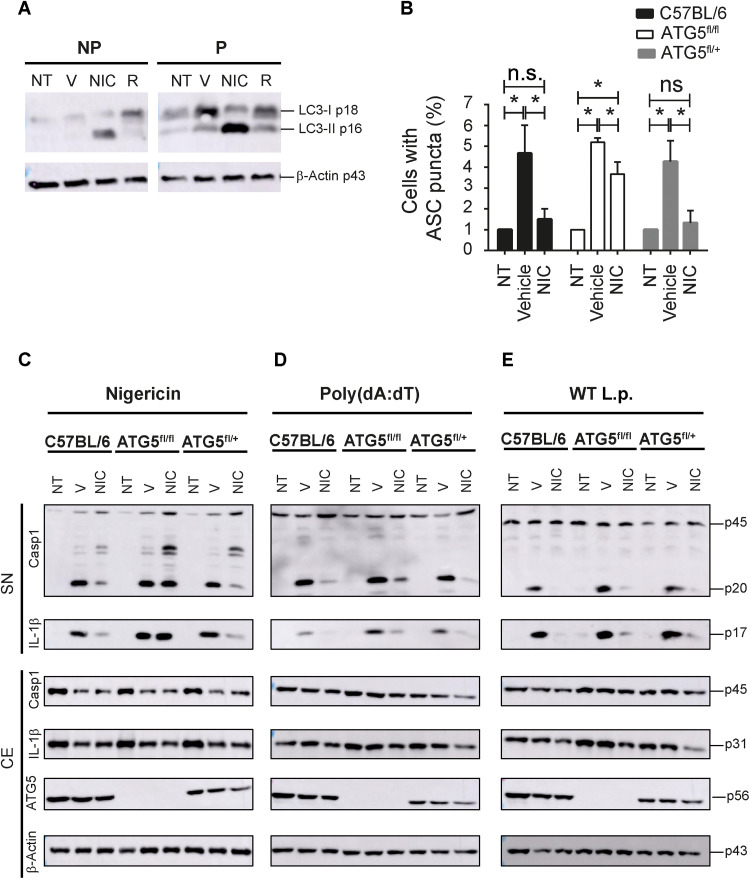
Next, we investigated the mechanisms by which NIC promotes inhibition of the inflammasome activation. NIC is able to trigger autophagy, a cell intrinsic mechanism that reduces the activation of the inflammasome (25–29). Thus, we initially tested whether NIC treatment is able to induce autophagy in macrophages by assessing the conversion of LC3-I to LC3-II, a biomarker of autophagy, by Western blot (Fig. 3A). Next, we tested whether the autophagic machinery is required for the inhibitory activity of NIC in inflammasomes. We performed experiments using macrophages from ATG5flox/flox/LysMCre/+ (and littermate controls) and found that ATG5 is required for the inhibition inflammasome activation as measured by the formation of ASC puncta induced by nigericin (Fig. 3B). In addition, NIC fails to inhibit cleavage of caspase-1 and IL-1β induced by nigericin in the absence of ATG5 (Fig. 3C). These data suggest that NIC impairs the activation of the NLRP3 inflammasome by inducing autophagy. We also investigated the importance of autophagy in the inhibitory activity of NIC in AIM2 and NAIP/NLRC4 inflammasomes. ATG5-sufficient and ATG5-deficient macrophages were treated with NIC, and the activation of AIM2 and NAIP/NLRC4 inflammasome was induced with poly(dA:dT) and L. pneumophila, respectively. We found that NIC inhibited the activation of inflammasome induced by poly(dA:dT) in ATG5-suficient cells, and this process were partially dependent on ATG5 (Fig. 3D). When we infected macrophages with L. pneumophila, we found that autophagy is dispensable for the inhibitory activity of NIC (Fig. 3E). In addition, the demonstration that NIC effectively inhibits the inflammasome activation induced by L. pneumophila, a bacterium that effectively inhibits autophagy (30), support the hypothesis that NIC inhibits the activation of NAIP5/NLRC4 inflammasome independently of autophagy. Collectively, our data indicated that autophagy is required for an NIC-mediated inhibition of the NLRP3 inflammasome, partially required for the inhibition of the AIM2 inflammasome and dispensable for inhibition of the NAIP5/NLRC4 inflammasome. These data indicate that multiple mechanisms may operate in the NIC-mediated inhibition of inflammasomes.
Fig. 3. NIC induces autophagy and autophagic machinery is required for inhibitory activity of NIC in inflammasomes.
BMDMs from C57BL/6 mice were not primed (NP) or primed (P) with LPS (500 ng/ml) for 3 hours and then left NT or treated with vehicle (V), NIC, or rapamycin (R) at 10 μM for 1 hour. (A) Western blot analysis showing LC3-I p18, LC3-II p16, and β-actin p43 in the CE after treatments. BMDMs of C57BL/6, ATG5flox/flox/LysMCRE/+ (ATG5fl/fl), and ATG5fl/+/LysMCRE/+ (ATG5fl/+) mice were primed with LPS (500 ng/ml) for 3 hours, treated with vehicle (V) or NIC at 10 μM for 1 hour, and then left NT or stimulated with nigericin (20 μM). (B) The percentage of cell containing ASC puncta after nigericin treatment was estimated in BMDMs of C57BL/6, ATG5fl/fl, and ATG5fl/+. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Student’s t test. (C to E) Western blot analysis showing caspase-1 and IL-1β in cell-free SNs and caspase-1 p45, IL-1β p31, ATG5 p56, and β-actin p43 in the CE after stimulation with nigericin (C) or poly(dA:dT) (D) or infection with WT L.p. (E). Data are presented from one representative experiment of three experiments performed with similar results.
NIC inhibits inflammasome activation in response to SARS-CoV-2 infection
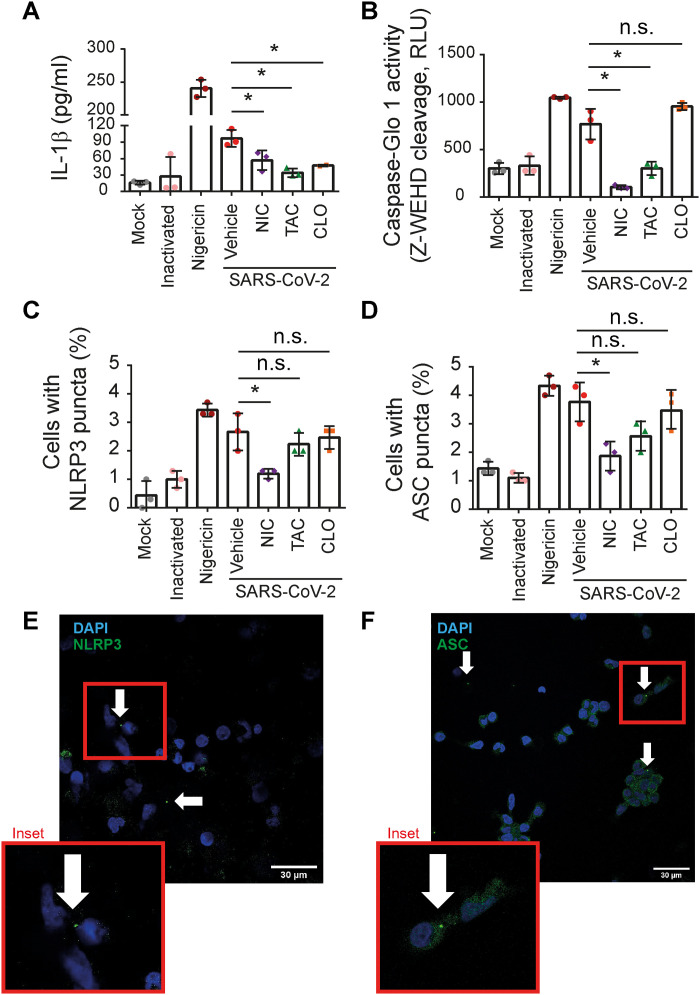
We previously demonstrated that primary human monocytes trigger the NLRP3 activation in response to SARS-CoV-2 infection and that inflammasome activation is associated with the poor clinical outcome of COVID-19 (10). Thus, we tested the effect of NIC, TAC, and CLO in the inhibition of inflammasome activation by SARS-CoV-2. Initially, we assessed inflammasome activation in primary human monocytes from healthy donors infected in vitro with SARS-CoV-2 and treated with 10 μM NIC, TAC, and CLO for 24 hours. For comparison, nigericin was used as a positive control. In agreement with our results from BMDMs infected with L. pneumophila, we found that NIC was the most effective drug to target inflammasome activation in CD14+ human monocytes infected with SARS-CoV-2. This was evident when we assessed inflammasome-mediated IL-1β secretion (Fig. 4A), caspase-1 activation (Fig. 4B), and NLRP3 puncta (Fig. 4C) and ASC (Fig. 4D) puncta formation. Images of NLRP3 and ASC puncta in SARS-CoV-2–infected monocytes are shown in Fig. 4 (E and F).
Fig. 4. NIC inhibits inflammasome activation triggered by SARS-CoV-2 in primary human monocytes.
Human CD14+ monocytes were primed with PAM3Cys (300 ng/ml) for 4 hours and infected with SARS-CoV-2 at an MOI of 10. After viral adsorption, cells were left untreated or treated with 10 μM NIC, TAC, and CLO or with 1% DMSO (vehicle) for 24 hours. Mock and ultraviolet (UV)–inactivated viruses were used as negative infection control, and nigericin was used as a positive control for NLRP3 inflammasome activation. SNs were assayed for (A) IL-1β release or (B) caspase-1 activity using the Caspase-1 Glo kit assay measured by relative luminescence units (RLU). The percentage of cell containing (C) NLRP3 puncta or (D) ASC puncta was estimated in cells. Representative images of (E) NLRP3 and (F) ASC puncta (green) in SARS-CoV-2–infected monocytes. Nuclei were stained in blue. Inset highlights an image clipping containing puncta. Scale bar, 30 μm. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Student’s t test. Data are presented from one representative experiment of three experiments performed with similar results.
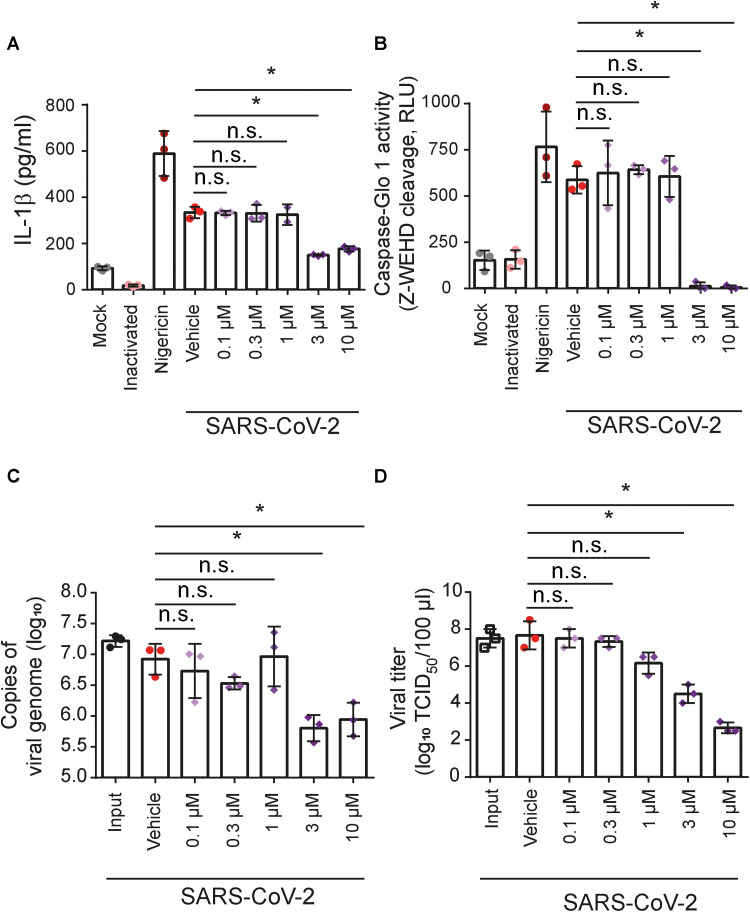
Our results presented so far indicate that NIC inhibits inflammasome activation in human monocytes infected with SARS-CoV-2. Thus, we investigated whether this drug could also show antiviral activity in such cells. To this end, primary human monocytes were infected in vitro with SARS-CoV-2 and treated with 0.1 to 10 μM NIC. Cell supernatants were collected, and inflammasome activation and viral loads were assessed 24 hours after treatment. We found that the IL-1β release (Fig. 5A) and caspase-1 activation (Fig. 5B) induced by SARS-CoV-2 infection were inhibited by treatment with 3 and 10 μM. NIC treatment also reduced the viral loads at 3 and 10 μM, as determined by real-time polymerase chain reaction (PCR; Fig. 5C) and median tissue culture infectious dose (TCID50) assay (Fig. 5D). In agreement with previous findings, similar viral loads were detected in the input- and vehicle-treated cells 24 hours after infection, corroborating that SARS-CoV-2 replicates poorly in primary monocytes (31).
Fig. 5. NIC inhibits SARS-CoV-2 replication and inflammasome activation in primary human monocytes.
Human CD14+ monocytes were primed with PAM3Cys (300 ng/ml) for 4 hours and infected with SARS-CoV-2 at an MOI of 10. After viral adsorption, cells were left untreated or treated with 0.1 to 10 μM NIC or with 1% DMSO (vehicle) for 24 hours. Mock and UV-inactivated viruses were used as negative infection control, and nigericin was used as a positive control for NLRP3 inflammasome activation. SNs were assayed for (A) IL-1β release or (B) caspase-1 activity using the Caspase-1 Glo kit assay measured by RLU. Supernatants were also assayed for (C) quantification of the viral genome by real-time PCR or (D) viral titer by TCID50. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Student’s t test. Data are presented from one representative experiment of three experiments performed with similar results.
NIC inhibits SARS-CoV-2 replication in Vero cells
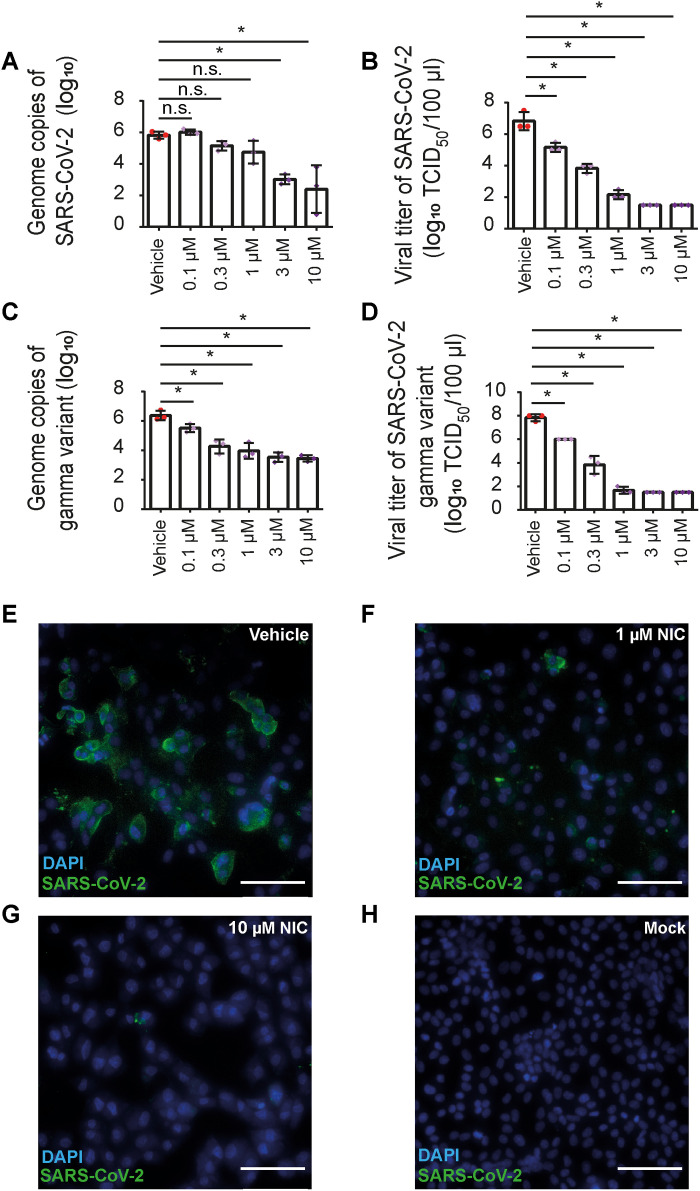
Next, we investigated the effect of NIC in the viral replication using immortalized cell lines that do not express components of the inflammasome machinery. To perform this assay, we first constructed a simian kidney epithelial Vero CCL81 cell line expressing human angiotensin-converting enzyme 2 (hACE2), a protein that facilitates viral entrance (32). To test an antiviral activity of NIC in hACE2-Vero cells, we infected cultures with SARS-CoV-2 and treated them with 0.1 to 10 μM NIC for 48 hours. Quantification of viral genomes by quantitative reverse transciption PCR (qRT-PCR) indicated that infected cells treated with vehicle contained higher viral loads than cells treated with NIC at 3 and 10 μM (Fig. 6A). We also measured viral titers by TCID50 and found a dose-dependent reduction of viral loads (Fig. 6B), suggesting that NIC effectively inhibits viral replication in Vero cells. NIC was also very effective in restricting the replication of gamma variant (P.1) of SARS-CoV-2, as measured by qRT-PCR (Fig. 6C) and by TCID50 (Fig. 6D).
Fig. 6. NIC inhibits viral replication in hACE2-Vero cells infected with ancestral/Wuhan and gamma variant of SARS-CoV-2.
Vero cells stably expressing hACE2 (hACE2-Vero) were infected with SARS-CoV-2 or gamma variant (P.1) at an MOI of 10. After viral adsorption, cells were washed and then left untreated or treated with 0.1 to 10 μM NIC or with 1% DMSO (vehicle) for 48 hours. Mock and UV-inactivated viruses were used as negative infection control. Cell supernatants were assayed for the quantification of (A) viral genome of SARS-CoV-2 by real-time PCR, (B) viral titer of SARS-CoV-2 by TCID50, (C) viral genome of gamma variant by real-time PCR, or (D) viral titer of gamma variant by TCID50. (E to H) hACE2-Vero cells were stained for SARS-CoV-2 using convalescent serum from a patient with COVID-19. A representative image of hACE2-Vero cells infected with SARS-CoV-2 (green) and treated with vehicle (E), 1 μM NIC (F), or 10 μM NIC (G) or mock treated (H). Nuclei are stained in blue. Scale bars, 85 μm. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Student’s t test. Data are presented for one representative experiment of three experiments performed with similar results.
We validated the antiviral effect of NIC by performing immunofluorescence analyses. We detected a robust viral replication in infected hACE2-Vero cells treated with the vehicle (Fig. 6E), and NIC treatment substantially reduced viral replication in cells (Fig. 6, F and G). No antibody cross-reaction was detected when we performed mock infection (Fig. 6H). Together, our data demonstrate that NIC inhibits inflammasome activation and viral replication in host cells.
NIC effectively inhibits inflammasome activation in PBMC from patients with COVID-19
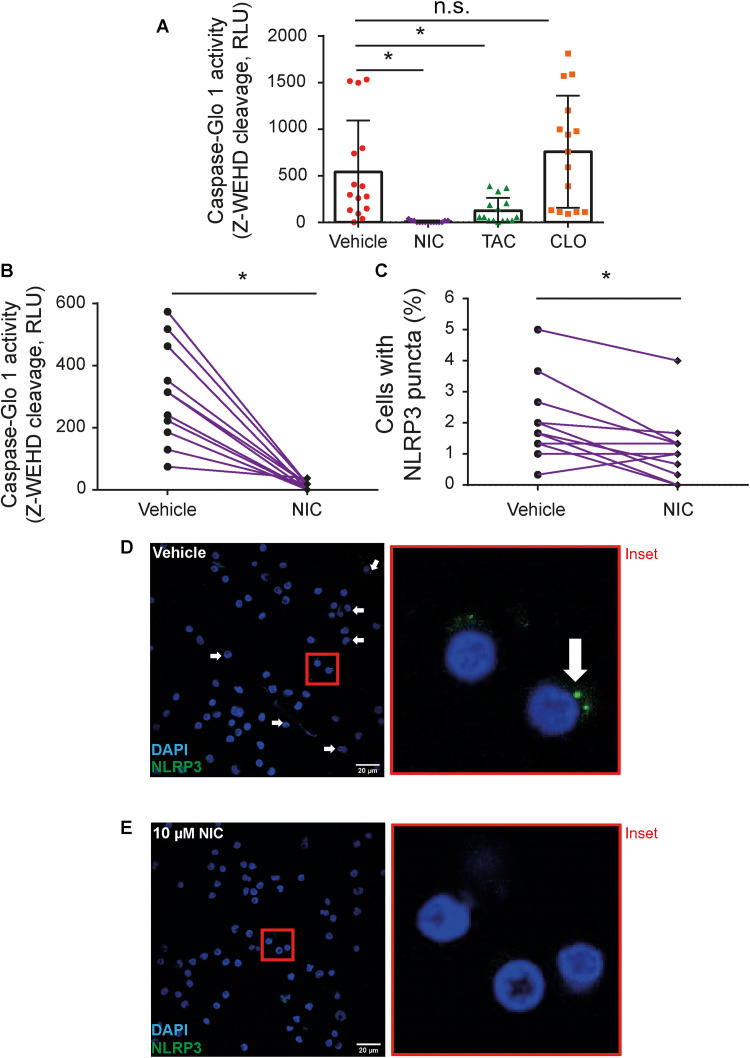
Our results demonstrate that NIC inhibits both inflammasome activation and SARS-CoV-2 replication, suggesting that it is a promising drug candidate for COVID-19 treatment. Thus, we assessed the effect of NIC in peripheral blood mononuclear cells (PBMCs) obtained from patients with COVID-19, which contains active inflammasomes (10). Initially, PBMCs were collected from 15 patients and treated with vehicle and 10 μM NIC, TAC, and CLO. Cells were treated for 24 hours, and active caspase-1 in the PBMC supernatants was measured by using a luminescent assay. Consistent with our in vitro observations, NIC and TAC strongly reduced active caspase-1 in patient PBMCs, while CLO did not interfere with caspase-1 activation (Fig. 7A). We also performed another assay where PBMCs from other 11 patients were collected and treated with 10 μM NIC or vehicle. We confirmed the highly efficient effect of NIC in the inhibition of caspase-1 activation in PBMCs from patients with COVID-19 (Fig. 7B). In this same experiment, we also quantified the formation of NLRP3 puncta in PBMCs. We found a significant reduction in NLRP3 puncta in NIC-treated cells as compared to vehicle (Fig. 7C). Representative images of NLRP3 puncta in PBMCs from patients with COVID-19 treated with vehicle (Fig. 7D) and NIC (Fig. 7E) are shown.
Fig. 7. NIC inhibits inflammasome activation in PBMCs of patients with COVID-19.
PBMCs were isolated from fresh blood of patients with COVID-19 and treated with 10 μM NIC, TAC, and CLO or with 1% DMSO (vehicle) for 24 hours. (A) SNs were assayed for caspase-1 activity using the Caspase-1 Glo kit assay measured by RLU after treatment with 10 μM NIC, TAC, and CLO or 1% DMSO (vehicle) (patients with COVID-19, n = 15; all tested positive by real-time PCR) or (B) after treatment with 10 μM NIC or 1% DMSO (vehicle) (patients with COVID-19, n = 11; all tested positive by real-time PCR). (C) PBMCs from patients with COVID-19 were stained for anti-NLRP3 for determination of active NLRP3 inflammasome (puncta). Representative images of infected PBMCs containing NLRP3 puncta (green) in cells treated (D) with 1% DMSO (vehicle) or (E) 10 μM NIC. The inset highlights an image clipping. Nuclei were stained in blue. Scale bars, 20 μm. *P < 0.05, compared to vehicle; n.s., nonsignificant as determined by Wilcoxon test. Each dot represents the value form a single individual.
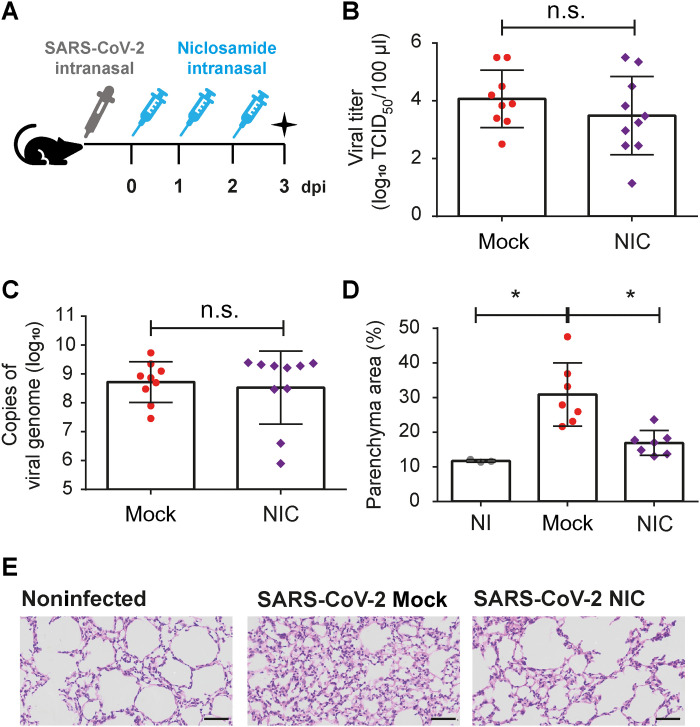
Next, we tested the effect of NIC in a mouse infection model. Initially, we infected restrictive C57BL/6 mice with L. pneumophila and treated them every 24 hours with NIC (0.3 mg/kg per day; or vehicle) starting after 4 hours of infection. We found that mice treated with NIC increased the replication of WT L.p. in the lungs 48 and 72 hours after infection (fig. S4A). We also performed infections with flaA− L.p., which bypass NAIP5/NLRC4-mediated growth restriction and replicate in mouse lungs. We found that NIC did not interfere with the replication of flaA− mutants in mouse lungs (fig. S4B). These data indicates that NIC can be used in vivo, as this compound was able to inhibit NAIP5/NLRC4 inflammasome–mediated restriction of L. pneumophila replication in mice lungs. Next, we used the K18-hACE2 transgenic mice that express hACE2 under regulation of cytokeratin-18 promoter and provide a reliable model of mice for infections with SARS-CoV-2 (33). We infected K18-hACE2 mice with 2 × 104 plaque-forming units (PFU) of SARS-CoV-2 and treated them with NIC (0.3 mg/kg per day) every 24 hours for up to 3 days after infection (Fig. 8A). After 3 days of infection, the lungs were collected for determination of viral load and quantification of pulmonary parenchyma area. We found that NIC treatment intranasally did not affect viral replication in the lungs, as measured by TCID50 assay and qRT-PCR (Fig. 8, B and C). However, NIC significantly ameliorated the inflammatory infiltrate in mouse lungs, as assessed by determination of the parenchyma area in lung histological sections of mice infected for 3 days (Fig. 8, D and E). We also quantified the production of cytokines in the lungs of infected mice and found that NIC did not significantly affect the production of interferon-γ (IFN-γ), TNF-α, MCP-1, IL-10, IL-6, or IL-12 (fig. S5). Together, these data confirm the beneficial role of intranasal treatment with NIC in vivo for reducing the inflammatory process, despite the fact that it does not interfere with cytokine production and viral replication.
Fig. 8. NIC reduces inflammation in lung parenchyma of ACE2-humanized mice infected with SARS-CoV-2.
(A) Transgenic K18-hACE2 mice were intranasally infected with 2 × 104 SARS-CoV-2 per mice and were treated with NIC (0.3 mg/kg per day) for up to 3 days. The lungs were collected to viral load and parenchyma area quantification. On day 3 of infection, lung homogenate was assayed for the quantification of viral genome by real-time PCR (B) or SARS-CoV-2 viral titer of by TCID50 (C). (D) The percentage of parenchyma area in NI-, mock-, or NIC-treated mice. (E) Representative images of hematoxylin and eosin staining of lung sections of NI or SARS-CoV-2–infected mock- or NIC-treated mice after 3 days of infection [days post infection (dpi)]. Scale bars, 50 μm. *P < 0.05, compared to untreated; n.s., nonsignificant as determined by Student’s t test. Data presented are a pool of two independent experiments performed with similar results.
DISCUSSION
Our high-throughput screening for compounds that target multiple inflammasomes revealed three FDA-approved drugs with inflammasome inhibitory activity. NIC, our best hit, was a potent inflammasome inhibitor that is also a highly effective inhibitor of SARS-CoV-2 replication in vitro. As an FDA-approved anthelminthic drug, NIC has been used for over 50 years to treat tapeworm infections, and it is currently listed on the World Health Organization’s list of essential medicines (34–36). Recent studies have suggested very broad clinical applications of NIC for the treatment of infectious diseases other than those caused by parasites [reviewed in (37)]. Accordingly, NIC was able to inhibit chikungunya virus entry and transmission (38), and years before the COVID-19 pandemic, NIC was identified as an inhibitor of SARS-CoV replication (39). In addition, NIC exerts anti-MERS activity, inhibiting S-phase kinase-associated protein 2 (SKP2) function and impairing MERS-CoV replication (40). In light of this information, the potential use of NIC against SARS-CoV-2 has been highly speculated on the basis of virtual screenings experimentally validated in vitro (36, 41–45). In agreement with these reports, our results using hACE2-Vero cells demonstrated that NIC effectively inhibits SARS-CoV-2 replication as measured by immunofluorescence, TCID50 assays, and qRT-PCR.
NIC exhibits a remarkably potent inhibitory activity against SARS-CoV-2 in Vero cells with activity at nanomolar concentrations (44, 46). Multiple mechanisms underlie the biological effects of NIC. The drug has been shown to exhibit antiviral activity due to its ability to perturb the pH-dependent membrane fusion required for virus entry (47). In addition, it dampens calcium variation and membrane conductance by suppressing the activity of TMEM16F (anoctamin 6), thus blocking spike-induced syncytia formation (48). Using a target-based screening, NIC was identified as a promising candidate to inhibit receptor binding domain uptake and spike pseudovirus infection (45). In addition, NIC has been shown to inhibit SKP2, which results in decreased ubiquitination and degradation of Beclin-1 and enhanced autophagic flux, augmenting autophagy and favoring restriction of viral replication (40). Thus, it is proposed that NIC-mediated autophagy induction is associated with the inhibition of SARS-CoV-2 infection (42). According to this observation, it is well reported in literature that NIC triggers autophagy (26–29). In this study, we confirmed that NIC induces autophagy, and this process is required for NIC-induced inhibition of NLRP3 inflammasome. However, different from the NLRP3, autophagy was only partially required for NIC-induced inhibition of AIM2 inflammasome and dispensable for inhibition of NAIP5/NLRC4 inflammasome. Thus, additional mechanisms besides autophagy operate in NIC-induced inhibition of inflammasome. In this work, we also demonstrated that NIC inhibit LPS- and PAM3Cys-induced expression of pro–IL-1β, TNF-α, IL-10, and MCP-1, revealing an additional mechanism operating in the anti-inflammatory properties of NIC. However, these mechanisms may not influence the NIC-mediated inhibition of inflammasome activation because, in our studies, NIC treatment initiated after macrophage priming. Furthermore, activation of the NAIP/NLRC4, which is effectively inhibited by NIC, does not require priming (20), supporting the notion that additional mechanisms, besides autophagy and inhibition of priming, are involved in the inhibitory activities of NIC. Collectively, our data indicate that the inhibitory effect of NIC on priming together with the autophagy-mediated inhibitory effect on inflammasome activation and additional unknown mechanisms operate for the anti-inflammatory activity of NIC and may benefit the treatment of patients with COVID-19.
The inhibitory activity of autophagy in inflammasome activation has been previously reported (25) and may participate in the inhibitory effects of NIC on NLRP3 inflammasome activation in macrophages. A previous report, not consistent with our data, suggested that NIC might function as an NLRP3 activator (49). The use of an immortalized macrophage-like cell THP-1, in addition to differences in the LPS used for priming, may explain these differences, as LPS is a potent activator of noncanonical NLRP3 inflammasome (50, 51). Note that ours and other studies demonstrated that inflammasome activation is associated with a poor outcome in patients with severe COVID-19 (9–12). Thus, despite an absence of a role for NIC in the restriction of viral replication under our in vivo experimental settings, it is possible that NIC targets both viral replication and inflammasome activation in patients with COVID-19, making NIC a very promising drug candidate for COVID-19. We do not envisage a prophylactic use of NIC, given that the drug can inhibit additional inflammasomes that may be protective against opportunistic fungi and bacterial infections, as is the case for L. pneumophila (fig. S4). Instead, individuals infected with SARS-CoV-2 undergoing moderate-to-severe inflammatory conditions may benefit from NIC treatment. Despite the fact that the drug is well tolerated and does not influence vital organ functions, one specific challenge for oral use of NIC is the low aqueous solubility and poor oral bioavailability (34, 36, 41). To minimize this problem, efforts have been made to develop inhalation and nasal spray systems to deliver the drug directly in the upper respiratory mucosa (52). Whether the delivery of NIC in nasal mucosa, rather than in the lower respiratory system, will be sufficient to inhibit inflammasome activation and ameliorate severe cases of COVID-19 is unknown. Nonetheless, prospective clinical trials including NIC already indicated the preliminary yet positive effects of NIC for treatment of hospitalized patients (53, 54). There are currently more than 20 ongoing clinical trials using NIC in different conditions, some of them registered for COVID-19 in the ClinicalTrials.gov clinical trials registry (55). Regardless of the possible approval of NIC for human treatment of COVID-19, our study provides key information regarding inhibitory activity of NIC in inflammasomes. This feature may be beneficial for drug effectiveness in patients with COVID-19, as the treatment may mitigate both SARS-CoV-2 replication and also the inflammatory processes initiated and amplified by the inflammasomes.
MATERIALS AND METHODS
Study approval
The procedures followed in the study were approved by the National Ethics Committee, Brazil CONEP (Comissão Nacional de Ética em Pesquisa), CAAE (Certificado de Apresentação de Apreciação Ética): (30248420.9.0000.5440). Written informed consent was obtained from the recruited patients.
Mice
C57BL/6 and ATG5-transgenic mice were bred and maintained at the Animal Facility of the Medical School of Ribeirão Preto/University of São Paulo (FMRP/USP). Atg5flox/flox and Atg5flox/+ mice were breed with LysMCre/+ mice (JAX) to generate LysMCre/+/Atg5flox/flox (here called as ATG5fl/fl) and the littermate controls LysMCre/+/Atg5Fl/+ (here called as ATG5fl/+). K18-hACE2 [B6.Cg-Tg(K18-ACE2)2Prlmn/J; the Jackson Laboratory strain no.: 034860], male or female mice ranging from 8 to 10 weeks old were infected with SARS-CoV-2 in a biosafety level 3 (BSL3) facility at the University of São Paulo, FMRP/USP. The mice protocols were previously approved by the Institutional Ethics Committee for Animal Care and Research (CETEA-FMRP/USP no. 085/2015).
Bacterial strains
L. pneumophila strains used were JR32 (WT L.p.) and isogenic clean deletion mutants for flagellin (flaA− L.p.) (24, 56). All bacteria were grown on buffered charcoal-yeast extract (BCYE) agar plates [1% yeast extract, 1% Mops, 3.3 mM l-cysteine, 0.33 mM Fe(NO3)3, 1.5% Bacto agar, and 0.2% activated charcoal (pH 6.9)] at 37°C (57).
Macrophages
BMDMs were harvested from femurs and tibias of C57BL/6, ATG5fl/fl, or ATG5fl/+ mice. Cells were differentiated with RPMI 1640 (Gibco) containing 20% fetal bovine serum (FBS) (Invitrogen) and 30% L-929 cell-conditioned medium (LCCM), 2 mM l-glutamine (Sigma-Aldrich), and penicillin-streptomycin (100 U/ml; Sigma-Aldrich) for 7 days, at 37°C with 5% CO2. Cells were detached with a cold phosphate-buffered saline (PBS) (Gibco), resuspended in RPMI 1640 supplemented with 10% FBS and 5% LCCM, seeded at the desired a concentration of cells per well according to each assay, and allowed to adhere for 24 hours before the infections.
Drug library
For the anti-inflammasome screening was used a commercial drug library containing 2560 drugs diluted at 10 mM in dimethyl sulfoxide (DMSO) (MicroSource Spectrum Collection, MicroSource Discovery Systems, Gaylordsville, CT). Those drugs were diluted at 1 mM DMSO and then diluted at the desired concentration for each assay in Dulbecco’s minimum essential medium (DMEM) without phenol red (Gibco FluoroBrite DMEM) or RPMI 1640 without phenol red (Gibco) getting at the assays a final concentration of 1% DMSO.
Anti-inflammasome screening
In vitro infections and pore formation inhibition
The anti-inflammasome screening was performed in BMDMs pretreated with the drugs (10 μM) from the library and infected with WT L.p. at a multiplicity of infection (MOI) of 10. Briefly, BMDMs were seeded at 5 × 104 cells per well in 96-well plates (Corning 96-well Flat Clear Bottom Black Polystyrene TC-Treated Microplates) and allowed to adhere for 24 hours. BMDMs were washed with PBS, and a solution of DMEM without phenol red containing PI (5 μg/ml) and drugs (10 μM) were added. One hour after treatment, 10 μl of a solution of DMEM without phenol red containing WT L.p. (MOI of 10), 1:1000 rabbit anti-Legionella antibody, and PI (5 μg/ml) was added to each well. Plates were centrifuged at 200g for 5 min and kept at 37°C with 5% CO2 for 3 hours. After that, cells were visualized and counted in MiniMax Imaging Cytometer SpectraMax i3x Multi-Mode Microplate (Molecular Devices), and four images per well were taken, where nuclei stained with PI were counted using SoftMax Pro Software. In each assay, noninfected BMDMs with/without 1% DMSO were used as negative control. L. pneumophila–infected BMDMs with/without 1% DMSO were used as positive control. The assay was performed in triplicate. Total pore formation was determined by lysing cells with 1.3% Triton X-100 for normalization and for the determination of pore formation inhibition in the conditions treated with drugs.
Kinetics of pore formation inhibition in NLRC4 inflammasome
The 19 compounds that caused at least 65% of pore formation inhibition were revalidated at three different concentrations (10, 20, and 40 μM) against WT L.p. Briefly, BMDMs were seeded at 5 × 104 cells per well in 96-well plates (Corning 96-well Flat Clear Bottom Black Polystyrene TC-Treated Microplates) and allowed to adhere for 24 hours. A solution of DMEM without phenol red containing PI (5 μg/ml) and drugs (10, 20, and 40 μM) were added. One hour after treatment, 10 μl of a solution of DMEM medium without phenol red containing WT L.p. (MOI of 10) was added to each well and plates were centrifuged at 200g for 5 min. Right after infection, cells were visualized and counted in MiniMax Imaging Cytometer, SpectraMax i3x Multi-Mode Microplate (Molecular Devices). PI was excited at 538 nm, and emission was measured at 617 nm every 5 min for a period of 14 hours; parameters of “relative fluorescence units (RFU)” were determined.
In vitro revalidation in BMDMs infected with WT L.p.
The revalidation assay was performed in BMDMs, which were seeded at 5 × 105 cells per well in 24-well plates (Corning 24-well Flat Clear Bottom Polystyrene TC-Treated Microplates) and allowed to adhere for 24 hours. BMDMs were primed with LPS (500 ng/ml) for 3 hours. Then, cells were pretreated with 10 μM NIC, TAC, and CLO or 1% DMSO for 1 hour. After treatment, 10 μl of a solution of RPMI 1640 without phenol red containing WT L.p. (MOI of 10) was added to each well. Plates were centrifuged at 200g for 5 min and kept at 37°C with 5% CO2 for 2 hours. After that, cells were processed for immunofluorescence assays, and the supernatant was collected to determine caspase-1 activity using the Caspase-Glo 1 assay and cytokine production. The assay was performed in triplicate.
Caspase-1, IL-1β, ATG5, and β-actin Western blot
To measure caspase-1 and IL-1β cleavage by Western blot, 8 × 105 cells per well of BMDMs were plated in 48-well plates (Corning 48-well Flat Clear Bottom Polystyrene TC-Treated Microplates) and allowed to adhere for 24 hours. Cells were primed with LPS (500 ng/ml) for 3 hours. Then, cells were washed with PBS and treated with 10 μM NIC. One hour after treatment, cells were treated with nigericin (20 μM), transfected with poly(dA:dT) (5 ng/ml), or infected with WT L.p. (MOI of 10), or flaA− L.p. (MOI of 10), and kept at 37°C with 5% CO2 for 2 hours. After inflammasome stimulus, supernatants were collected, and cell extracts were lysed with a radioimmunoprecipitation assay (RIPA) buffer [25 mM tris-HCl (pH 7.6) (Hexis Científica), 150 mM NaCl (J. T. Bacher), 1% NP-40 (Fluka Biochemika, USA), and 1% sodium deoxycholate (Sigma-Aldrich), 0.1% SDS] for 10 min on ice. Supernatants and lysates were resuspended in a sample buffer containing 50 mM tris (pH 6.8), 2% SDS (USB Corporation, USA), 0.1% bromophenol blue (Synth, USA), 10% glycerol (USB Corporation), and 2.5% β-mercaptoethanol.
After that, proteins were separated in a 12% SDS–polyacrylamide gel electrophoresis (PAGE) gel, and then proteins were transferred to a nitrocellulose membrane (GE Healthcare, USA) using Trans-Blot Turbo Transfer System (Bio-Rad) at 15 V for 30 min. After transfer, membranes were blocked for 1 hour at 25°C in a tris buffer [tris-buffered saline (TBS)–25 mM tris (pH 7.4) (Hexis Científica, Brazil), 0.3 mM KCl (J. T. Backer), and 140 mM NaCl (J. T. Bacher)] containing 0.1% Tween 20 (TBS-T) and 5% nonfat dry milk. After blocking, membranes were stained with rat monoclonal anti–caspase-1 antibody (1:500; Genentech, clone 4B4), goat monoclonal antibody against IL-1β (1:500; Sigma-Aldrich, I3767), rabbit monoclonal antibody anti-ATG5 (1:1000; Abcam), or rabbit monoclonal antibody against β-actin (1:1000; Sigma-Aldrich, A2066) diluted in 5% nonfat dry milk in TBS 1× with 0.01% Tween 20 overnight at 4°C. Membranes were washed with TBS-T and incubated for 1 hour at 25°C with appropriate peroxidase-conjugated secondary antibody (1:3000) (KPL, USA). For the detection of proteins linked to specific antibodies Amersham Imager 600 (GE Healthcare, Illinois) was used. The assay was performed in triplicate.
LC3-I and LC3-II Western blot
To measure the LC3-I and LC3-II cleavage by Western blot, 8 × 105 cells per well of BMDMs were plated in 48-well plates (Corning 48-well Flat Clear Bottom Polystyrene TC-Treated Microplates) and allowed to adhere for 24 hours. Cells were left nontreated or treated with 10 μM vehicle or NIC. One hour after, cells were primed with LPS (500 ng/ml) for 3 hours. Cell extracts were lysed with a RIPA buffer, and then lysates were resuspended in a sample buffer containing 50 mM tris (pH 6.8), 2% SDS (USB Corporation, USA), 0.1% bromophenol blue (Synth, USA), 10% glycerol (USB Corporation), and 2.5% β-mercaptoethanol.
After that, proteins were separated in a 15% SDS-PAGE gel, and then proteins were transferred to a nitrocellulose membrane (GE Healthcare, USA) using Trans-Blot Turbo Transfer System (Bio-Rad) at 15 V for 30 min. After transfer, membranes were blocked for 2 hours at 25°C in a tris buffer containing 5% of bovine serum albumin (BSA). After blocking, membranes were stained with rabbit monoclonal anti-LC3 antibody (1:1500; Sigma-Aldrich, L8918) and diluted in a tris buffer containing 1% of BSA for 2 hours at 25°C. Membranes were washed and incubated for 1 hour at 25°C with an appropriate peroxidase-conjugated secondary antibody (1:3000) (KPL, USA). For the detection of proteins linked to specific antibodies Amersham Imager 600 (GE Healthcare, Illinois) was used. The assay was performed in triplicate.
In vitro anti–SARS-CoV-2 assays
Vero cells expressing hACE2
The hACE2 gene was amplified by PCR using hACE2F primer 5′-AACGTACGATGTCAAGCTCTTCCTGG-3′ and hACE2R primer 5′-AAACCGGTATGTCAAGCTCTTCCTGG-3′, and the vector K18-hACE2 was used as template (58). hACE2 was cloned into a hemagglutinin-tagged vector (pRH-HA) that was modified from the original pCLXSN backbone from Imgenex. Human embryonic kidney (HEK) 293 cells were transfected with the lentiviral vector pRH-hACE2-HA and packing plasmids pspax2 and pMD2G, and, 60 hours after transfection, a lentivirus-containing supernatant from HEK293 cells was used to transduce Vero cells [American Type Culture Collection (ATCC), CCL-81]. Forty-eight hours after transduction, supernatant was replaced with fresh media containing hygromycin B (300 μg/ml) for selection. After 3 days, an hACE2 expression was confirmed by Western blot.
Viruses
The SARS-CoV-2 used was a Brazil/SPBR-02/2020 strain, isolated from the first Brazilian case of COVID-19. hACE2-Vero cells cultured in DMEM supplemented with 10% FBS were used for viral stock production. Virus stock, propagated under BSL3 conditions, were produced after infecting hACE2-Vero cells in the presence of trypsin-L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK) (1 μg/μl) in serum-free DMEM for 48 hours at 37°C in a 5% CO2 atmosphere as previously described (10) Virus stocks were titrated by standard limiting dilution to confirm the TCID50 (59) in Vero cells (ATCC CCL-81).
Purification of monocytes from healthy donors
Magnetic nanoparticles (BD) were used to purify monocytes (CD14+ cells). PBMCs were labeled with BD IMag Anti-human CD14 Magnetic Particles DM, and cells were separated through magnetic field. Labeled cells migrated toward the magnet (positive fraction), whereas unlabeled cells drew off (negative fraction). A separation was repeated twice to increase the purity of the positive fraction. The CD14+ monocytes resulting from this process were cultured in RPMI 1640 without phenol red (Gibco) containing 1% glutamine and 5% FBS.
In vitro infection of human monocytes
Purified human monocytes (2 × 105) were plated in 48-well plates and infected with SARS-CoV-2 at an MOI of 10. After 2 hours of viral adsorption, a new medium (RPMI 1640 without phenol red containing 1% glutamine and 2% FBS) (Gibco) containing 10 μM NIC, TAC, and CLO or vehicle (1% DMSO) was added. Cells were incubated for 24 hours at 37°C in the presence of 5% CO2 atmosphere. After incubation, cells were processed for immunofluorescence assays, and the supernatant was collected for the determination of active caspase-1, cytokine production. The assay was performed in triplicate. Another assay was performed in a similar way; however, a range of concentration (0.1 to 10 μM) of NIC or vehicle (1% DMSO) was added to the treatment of infected cells. The assay was performed in triplicate.
In vitro infection of hACE2-Vero cells
For in vitro infections, 2 × 104 cells per well were plated in 96-well plates (Corning 96-well Flat Clear Bottom Black Polystyrene TC-Treated Microplates) and infected with SARS-CoV-2 at an MOI of 10. After 2 hours of viral adsorption, cells were washed with PBS, and a new medium [RPMI 1640 without phenol red, 1% glutamine, and 2% FBS (pH 7.2)] (Gibco) containing a range of concentration (0.1 to 10 μM) of NIC or vehicle (1% DMSO) was added. Cells were incubated for 48 hours at 37°C in the presence of 5% CO2 atmosphere. After incubation, cells were processed for immunofluorescence assays, and the supernatant was collected for determination of viral loads by real-time PCR and TCID50. The assay was performed in triplicate.
COVID-19 patient assays
PBMCs isolation
A total of 26 patients with COVID-19 that were tested positive using real-time PCR have whole blood samples collected at Clinical Hospital of Ribeirão Preto–USP in tubes containing EDTA (BD Vacutainer CPT), as previously described (10). Briefly, the material was centrifuged at 400g for 10 min at room temperature. Then, the plasma was discarded, and the cell pellet was resuspended in PBS 1×. The cells were applied to the Ficoll-Paque PLUS gradient column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). A cell suspension was centrifuged at 640g for 30 min at room temperature to obtain the purified mononuclear fraction. The cells were washed, and the pellet was resuspended in RPMI 1640 for the following analysis as described (10).
Cultured PBMCs
Isolated PBMCs were plated (2 × 105 cells per well) in 48-well plates, and then cells were treated with 10 μM NIC, TAC, and CLO or vehicle (DMSO 1%). Cells were incubated for 24 hours at 37°C in the presence of 5% CO2 atmosphere. After incubation, cells were processed for immunofluorescence assays, and the supernatant was collected for the evaluation of active caspase-1.
In vivo assays
In vivo SARS-CoV-2 infection
C57BL/6 K18-hACE2 mice from 8 to 10 weeks old were used. The animals were anesthetized with ketamine (50 mg/kg) and xylazine (10 mg/kg) intraperitoneally and intranasally infected with SARS-CoV-2 (2 × 104 PFUs) contained in 20 μl of PBS. From day 0 to 2, animals were intranasally treated with NIC (0.3 mg/kg per day). After 3 days of infection, mice were euthanized, and lungs were collected for following analyses.
In vivo L. pneumophila infection
C57BL/6 mice from 8 to 10 weeks old were used. The animals were anesthetized with ketamine (50 mg/kg) and xylazine (10 mg/kg) intraperitoneally and intranasally infected with WT or flaA− L.p. (1 × 105 PFUs) contained in 20 μl of ultrapure water. From day 0 to 2, animals were intranasally treated with NIC (0.3 mg/kg per day). Mice were euthanized at 4, 48, and 72 hours after infection. The lungs were harvested and homogenized in 5 ml of sterile water for 30 s using a tissue homogenizer (Fisher Scientific). Lung homogenates were diluted in water, plated on CYE agar plates, and incubated for 4 days at 37°C for CFU determination.
Sample processing
Immunofluorescence staining of hACE2-Vero cells
For staining infected hACE2-Vero cells plated in 96-well plates, tissue culture supernatants were removed, and cells were fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. PFA was removed, and cells were washed with PBS and then blocked and permeabilized using PBS with goat serum and 0.05% saponin for 1 hour at room temperature. A convalescent serum from a patient with COVID-19 (1:300) was used as primary antibody, diluted in blocking solution, and added to each well. After 1 hour of incubation, the samples were washed with PBS, and secondary antibodies were added and incubated for 1 hour at room temperature. Secondary antibody used was rabbit anti-human immunoglobulin (whole molecule)–fluorescein isothiocyanate (FITC) (1:2000; Sigma-Aldrich). Cells were washed, and then, 1 mM 4′,6-diamidino-2-phenylindole (DAPI) was added in distilled water. Cells were visualized in the ImageXpress Micro XLS System (Molecular Devices LLC), and six images per well were taken in ×40 magnification. For image acquisition of the cell nuclei, the DAPI filter (λex, 377 nm and λem, 447 nm) was used, and for the SARS-CoV-2, the FITC filter (λex, 490 nm and λem, 525 nm) was used.
Immunofluorescence staining of isolated cells
For staining human isolated monocytes and PBMC from patients with COVID-19, cells were plated in Lab-Tek Chamber Slides (Nunc). Samples were fixed and processed as described for immunofluorescence. The primary antibodies used were a mix of rabbit monoclonal antibody anti-NLRP3 (1:1000; Cell Signaling Technology) and rabbit polyclonal antibody anti-ASC (1:2000; AdipoGen), which were diluted in blocking solution. Secondary antibodies used were goat anti-rabbit 488 (1:3000; Invitrogen). Slides were washed and mounted using DAPI (1 mM) and ProLong (Thermo Fisher Scientific).
Evaluation of active caspase-1 activity in cultured cells
To measure caspase-1 activity, the supernatants were collected and incubated with the Luciferin WEHD substrate provided by the Caspase-Glo 1 assay (Promega, WI, USA). After 1 hour of incubation at room temperature, luminescence was measured using SpectraMax i3x Multi-Mode Microplate (Molecular Devices).
Cytokine quantification
IL-1β from cell culture supernatants of BMDMs infected with L. pneumophila and from culture supernatants of human monocytes infected with SARS-CoV-2 was quantified by ELISA (R&D Systems) following the manufacturer’s instructions. TNF-α, IL-6, IL-12, IL-10, IFN-γ, and MCP-1 were quantified in mice lung tissue homogenates by CBA (cytometric bead array) (CBA Mouse Inflammation Kit, BD) following the manufacturer’s instructions.
qRT-PCR for SARS-CoV-2 genome quantification and TCID50
Detection of SARS-CoV-2 by qRT-PCR was performed with primer probe sets as previously described (10). Briefly, the genes N2 and E and ribonuclease P housekeeping gene were tested by one-step real-time PCR using total nucleic acids extracted with TRIzol (Invitrogen, CA) from 50 μl of cells supernatants to measure the genome viral load. Real-time PCR assays were done on a StepOnePlus real-time PCR thermocycler (Applied Biosystems, Foster City, CA, USA). For determination of TCID50, the supernatants of hACE2-Vero cells infected with SARS-CoV-2 were collected. Then, the samples were homogenized and diluted 1:10 in the presence of trypsin-TPCK (1 μg/μl) in serum-free DMEM (Gibco) and seeded onto a monolayer of Vero cells (ATCC CCL-81) in 96-well plates for 96 hours at 37°C in a 5% CO2 atmosphere. The virus-induced cytopathic effect was observed, and the standard limiting dilution to confirm the TCID50 was determined according to (59).
Histological evaluation
Mice lung tissue samples were fixed in 10% formalin, paraffin-embedded, and stained with standard hematoxylin and eosin in 3-μm sections. The tissue slides were scanned on a VS120 Olympus with high resolution to perform histological analysis of parenchyma area.
Statistics
Statistical significance was determined by either a two-tailed paired or unpaired Student’s t test for normal data. In the case of non-normal data, Wilcoxon test was applied. All analyses were performed in R (version 4.0.2) using RStudio (version 1.3.1056). Graph plots were performed with GraphPad Prism 6 software.
Acknowledgments
We thank M. Nakamura, D. Perucello, A. Zuin, and R. Sales for technical support.
Funding: This work was supported by Fundação de Amparo à Pesquisa do Estado de Sao Paulo (FAPESP) grants 2013/08216-2 (CRID-Center for Research in Inflammatory Diseases), 2019/11342-6, and 2020/04964-8; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant 303021/2020-9; and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) grant 88887.507253/2020-00.
Author contributions: Conceptualization: L.d.A., A.L.N.d.S., and D.S.Z. Methodology: L.d.A., A.L.N.d.S., W.A.A., and R.B.M. Investigation: L.d.A., A.L.N.d.S., T.S.R., S.O., A.Y.I., K.S.G.d.S., A.B., W.A.A., N.P., R.B.M., J.d.P.S., R.C., A.A.S., C.C.S.C., M.A.A., and S.S.B. Visualization: L.d.A. and A.L.N.d.S. Formal analysis: L.d.A., A.L.N.d.S., and A.Y.I. Patient recruitment: R.D.R.d.O. and P.L.-J. Data discussion: T.M.C., F.Q.C., F.G.F., L.D.C., R.D.R.d.O., P.L.-J., E.A., and A.T.F. Funding acquisition: D.S.Z. and F.Q.C. Project administration: D.S.Z. Supervision: D.S.Z. Writing (original draft): L.d.A. and D.S.Z. Writing (review and editing): W.A.A., L.D.C., K.S.G.d.S., T.S.R., and R.D.R.d.O.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Hui D. S., Azhar E. I., Madani T. A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T. D., Memish Z. A., Drosten C., Zumla A., Petersen E., The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health–The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 91, 264–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S., Channappanavar R., Kanneganti T. D., Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 41, 1083–1099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merad M., Martin J. C., Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay M. Z., Poh C. M., Renia L., MacAry P. A., Ng L. F. P., The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Giamarellos-Bourboulis E. J., Netea M. G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M. E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A., Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 27, 992–1000.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q., Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Wong P., Klein J., Castro T. B. R., Silva J., Sundaram M., Ellingson M. K., Mao T., Oh J. E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A. L., Vogels C. B. F., Earnest R., Lapidus S., Ott I. M., Moore A. J., Muenker M. C., Fournier J. B., Campbell M., Odio C. D., Casanovas-Massana A., Yale I. T., Herbst R., Shaw A. C., Medzhitov R., Schulz W. L., Grubaugh N. D., Dela Cruz C., Farhadian S., Ko A. I., Omer S. B., Iwasaki A., Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues T. S., de Sa K. S. G., Ishimoto A. Y., Becerra A., Oliveira S., Almeida L., Goncalves A. V., Perucello D. B., Andrade W. A., Castro R., Veras F. P., Toller-Kawahisa J. E., Nascimento D. C., de Lima M. H. F., Silva C. M. S., Caetite D. B., Martins R. B., Castro I. A., Pontelli M. C., de Barros F. C., do Amaral N. B., Giannini M. C., Bonjorno L. P., Lopes M. I. F., Santana R. C., Vilar F. C., Auxiliadora-Martins M., Luppino-Assad R., de Almeida S. C. L., de Oliveira F. R., Batah S. S., Siyuan L., Benatti M. N., Cunha T. M., Alves-Filho J. C., Cunha F. Q., Cunha L. D., Frantz F. G., Kohlsdorf T., Fabro A. T., Arruda E., de Oliveira R. D. R., Louzada-Junior P., Zamboni D. S., Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 218, e20201707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junqueira C., Crespo A., Ranjbar S., de Lacerda L. B., Lewandrowski M., Ingber J., Parry B., Ravid S., Clark S., Schrimpf M. R., Ho F., Beakes C., Margolin J., Russell N., Kays K., Boucau J., Das Adhikari U., Vora S. M., Leger V., Gehrke L., Henderson L. A., Janssen E., Kwon D., Sander C., Abraham J., Goldberg M. B., Wu H., Mehta G., Bell S., Goldfeld A. E., Filbin M. R., Lieberman J., FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 606, 576–584 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sefik E., Qu R., Junqueira C., Kaffe E., Mirza H., Zhao J., Brewer J. R., Han A., Steach H. R., Israelow B., Blackburn H. N., Velazquez S., Chen Y. G., Halene S., Iwasaki A., Meffre E., Nussenzweig M., Lieberman J., Wilen C. B., Kluger Y., Flavell R. A., Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 606, 585–593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosas I. O., Raby B., Tsai L., Interleukin-6 receptor antagonists in critically ill patients with covid-19. Reply. N. Engl. J. Med. 385, 1147–1149 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Salama C., Han J., Yau L., Reiss W. G., Kramer B., Neidhart J. D., Criner G. J., Kaplan-Lewis E., Baden R., Pandit L., Cameron M. L., Garcia-Diaz J., Chavez V., Mekebeb-Reuter M., Lima de Menezes F., Shah R., Gonzalez-Lara M. F., Assman B., Freedman J., Mohan S. V., Tocilizumab in patients hospitalized with covid-19 pneumonia. N. Engl. J. Med. 384, 20–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Din C. T., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L., Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: A retrospective cohort study. Lancet Rheumatol. 2, e325–e331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J. M., Bezie Y., Laplanche S., Le Berre A., Le Pavec J., Salmeron S., Emmerich J., Mourad J. J., Chatellier G., Hayem G., Anakinra for severe forms of COVID-19: A cohort study. Lancet Rheumatol. 2, e393–e400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyriazopoulou E., Panagopoulos P., Metallidis S., Dalekos G. N., Poulakou G., Gatselis N., Karakike E., Saridaki M., Loli G., Stefos A., Prasianaki D., Georgiadou S., Tsachouridou O., Petrakis V., Tsiakos K., Kosmidou M., Lygoura V., Dareioti M., Milionis H., Papanikolaou I. C., Akinosoglou K., Myrodia D. M., Gravvani A., Stamou A., Gkavogianni T., Katrini K., Marantos T., Trontzas I. P., Syrigos K., Chatzis L., Chatzis S., Vechlidis N., Avgoustou C., Chalvatzis S., Kyprianou M., van der Meer J. W., Eugen-Olsen J., Netea M. G., Giamarellos-Bourboulis E. J., An open label trial of anakinra to prevent respiratory failure in COVID-19. eLife 10, e66125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cauchois R., Koubi M., Delarbre D., Manet C., Carvelli J., Blasco V. B., Jean R., Fouche L., Bornet C., Pauly V., Mazodier K., Pestre V., Jarrot P. A., Dinarello C. A., Kaplanski G., Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc. Natl. Acad. Sci. U.S.A. 117, 18951–18953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascarenhas D. P., Zamboni D. S., Inflammasome biology taught by Legionella pneumophila. J. Leukoc. Biol. 101, 841–849 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Case C. L., Kohler L. J., Lima J. B., Strowig T., de Zoete M. R., Flavell R. A., Zamboni D. S., Roy C. R., Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc. Natl. Acad. Sci. U.S.A. 110, 1851–1856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Mascarenhas D. P. A., Cerqueira D. M., Pereira M. S. F., Castanheira F. V. S., Fernandes T. D., Manin G. Z., Cunha L. D., Zamboni D. S., Inhibition of caspase-1 or gasdermin-D enable caspase-8 activation in the Naip5/NLRC4/ASC inflammasome. PLOS Pathog. 13, e1006502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves A. V., Margolis S. R., Quirino G. F. S., Mascarenhas D. P. A., Rauch I., Nichols R. D., Ansaldo E., Fontana M. F., Vance R. E., Zamboni D. S., Gasdermin-D and caspase-7 are the key caspase-1/8 substrates downstream of the NAIP5/NLRC4 inflammasome required for restriction of Legionella pneumophila. PLOS Pathog. 15, e1007886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren T., Zamboni D. S., Roy C. R., Dietrich W. F., Vance R. E., Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLOS Pathog. 2, e18 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Carvalho R. V. H., Lima-Junior D. S., da Silva M. V. G., Dilucca M., Rodrigues T. S., Horta C. V., Silva A. L. N., da Silva P. F., Frantz F. G., Lorenzon L. B., Souza M. M., Almeida F., Cantanhede L. M., Ferreira R. G. M., Cruz A. K., Zamboni D. S., Leishmania RNA virus exacerbates Leishmaniasis by subverting innate immunity via TLR3-mediated NLRP3 inflammasome inhibition. Nat. Commun. 11, 203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M., Khambu B., Zhang H., Kang J. H., Chen X., Chen D., Vollmer L., Liu P. Q., Vogt A., Yin X. M., Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J. Biol. Chem. 288, 35769–35780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S. J., Shin J. H., Kang H., Hwang J. J., Cho D. H., Niclosamide induces mitochondria fragmentation and promotes both apoptotic and autophagic cell death. BMB Rep. 44, 517–522 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Ren X. R., Piao H., Zhao S., Osada T., Premont R. T., Mook R. A. Jr., Morse M. A., Lyerly H. K., Chen W., Niclosamide-induced Wnt signaling inhibition in colorectal cancer is mediated by autophagy. Biochem. J. 476, 535–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L. X., Zhao H. J., Sun D. L., Gao S. L., Zhang H. M., Ding X. G., Niclosamide attenuates inflammatory cytokines via the autophagy pathway leading to improved outcomes in renal ischemia/reperfusion injury. Mol. Med. Rep. 16, 1810–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choy A., Dancourt J., Mugo B., O’Connor T. J., Isberg R. R., Melia T. J., Roy C. R., The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338, 1072–1076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boumaza A., Gay L., Mezouar S., Bestion E., Diallo A. B., Michel M., Desnues B., Raoult D., La Scola B., Halfon P., Vitte J., Olive D., Mege J. L., Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: The clue for coronavirus disease 2019 immunoparalysis. J Infect Dis 224, 395–406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J., Wong L. R., Li K., Verma A. K., Ortiz M. E., Wohlford-Lenane C., Leidinger M. R., Knudson C. M., Meyerholz D. K., McCray P. B. Jr., Perlman S., COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrews P., Thyssen J., Lorke D., The biology and toxicology of molluscicides, bayluscide. Pharmacol. Ther. 19, 245–295 (1982). [DOI] [PubMed] [Google Scholar]
- 35.WHO, World Health Organization Model List of Essential Medicines, 21st List, 2019 (2019); https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf.
- 36.Xu J., Shi P. Y., Li H., Zhou J., Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 6, 909–915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W., Mook R. A. Jr., Premont R. T., Wang J., Niclosamide: Beyond an antihelminthic drug. Cell. Signal. 41, 89–96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y. M., Lu J. W., Lin C. C., Chin Y. F., Wu T. Y., Lin L. I., Lai Z. Z., Kuo S. C., Ho Y. J., Antiviral activities of niclosamide and nitazoxanide against chikungunya virus entry and transmission. Antiviral Res. 135, 81–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C. J., Jan J. T., Chen C. M., Hsieh H. P., Hwang D. R., Liu H. W., Liu C. Y., Huang H. W., Chen S. C., Hong C. F., Lin R. K., Chao Y. S., Hsu J. T., Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 48, 2693–2696 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gassen N. C., Niemeyer D., Muth D., Corman V. M., Martinelli S., Gassen A., Hafner K., Papies J., Mosbauer K., Zellner A., Zannas A. S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Muller-Myhsok B., Drosten C., Muller M. A., Rein T., SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat. Commun. 10, 5770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arshad U., Pertinez H., Box H., Tatham L., Rajoli R. K. R., Curley P., Neary M., Sharp J., Liptrott N. J., Valentijn A., David C., Rannard S. P., O’Neill P. M., Aljayyoussi G., Pennington S. H., Ward S. A., Hill A., Back D. J., Khoo S. H., Bray P. G., Biagini G. A., Owen A., Prioritization of anti-SARS-Cov-2 drug repurposing opportunities based on plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 108, 775–790 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gassen N. C., Papies J., Bajaj T., Emanuel J., Dethloff F., Chua R. L., Trimpert J., Heinemann N., Niemeyer C., Weege F., Honzke K., Aschman T., Heinz D. E., Weckmann K., Ebert T., Zellner A., Lennarz M., Wyler E., Schroeder S., Richter A., Niemeyer D., Hoffmann K., Meyer T. F., Heppner F. L., Corman V. M., Landthaler M., Hocke A. C., Morkel M., Osterrieder N., Conrad C., Eils R., Radbruch H., Giavalisco P., Drosten C., Muller M. A., SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat. Commun. 12, 3818 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ianevski A., Yao R., Biza S., Zusinaite E., Mannik A., Kivi G., Planken A., Kurg K., Tombak E. M., Ustav M. Jr., Shtaida N., Kulesskiy E., Jo E., Yang J., Lysvand H., Loseth K., Oksenych V., Aas P. A., Tenson T., Vitkauskiene A., Windisch M. P., Fenstad M. H., Nordbo S. A., Ustav M., Bjoras M., Kainov D. E., Identification and tracking of antiviral drug combinations. Viruses 12, 1178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon S., Ko M., Lee J., Choi I., Byun S. Y., Park S., Shum D., Kim S., Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 64, e00819-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prabhakara C., Godbole R., Sil P., Jahnavi S., Gulzar S. E., van Zanten T. S., Sheth D., Subhash N., Chandra A., Shivaraj A., Panikulam P., U I., Nuthakki V. K., Puthiyapurayil T. P., Ahmed R., Najar A. H., Lingamallu S. M., Das S., Mahajan B., Vemula P., Bharate S. B., Singh P. P., Vishwakarma R., Guha A., Sundaramurthy V., Mayor S., Strategies to target SARS-CoV-2 entry and infection using dual mechanisms of inhibition by acidification inhibitors. PLOS Pathog. 17, e1009706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mostafa A., Kandeil A., Elshaier Y. A. M. M., Kutkat O., Moatasim Y., Rashad A. A., Shehata M., Gomaa M. R., Mahrous N., Mahmoud S. H., GabAllah M., Abbas H., Taweel A. E., Kayed A. E., Kamel M. N., Sayes M. E., Mahmoud D. B., El-Shesheny R., Kayali G., Ali M. A., FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals (Basel) 13, 443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U. F., Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLOS Pathog. 8, e1002976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeno J. M., Ortega-Prieto A. M., Bussani R., Cannata A., Rizzari G., Collesi C., Schneider E., Arosio D., Shah A. M., Barclay W. S., Malim M. H., Burrone J., Giacca M., Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 594, 88–93 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thi Tran U., Kitami T., Niclosamide activates the NLRP3 inflammasome by intracellular acidification and mitochondrial inhibition. Commun. Biol. 2, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hagar J. A., Powell D. A., Aachoui Y., Ernst R. K., Miao E. A., Cytoplasmic LPS activates caspase-11: Implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kayagaki N., Wong M. T., Stowe I. B., Ramani S. R., Gonzalez L. C., Akashi-Takamura S., Miyake K., Zhang J., Lee W. P., Muszynski A., Forsberg L. S., Carlson R. W., Dixit V. M., Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Backer V., Sjobring U., Sonne J., Weiss A., Hostrup M., Johansen H. K., Becker V., Sonne D. P., Balchen T., Jellingso M., Sommer M. O. A., A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: A broad spectrum antiviral candidate for treatment of COVID-19. Lancet Reg. Health Eur. 4, 100084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdulamir A. S., Gorial F. I., Saadi S. J., Maulood M. F., Hashim H. A., Alnuaimi A. S., Abdulrrazaq M. K., A randomised controlled trial of effectiveness and safety of Niclosamide as add on therapy to the standard of care measures in COVID-19 management. Ann. Med. Surg. (Lond) 69, 102779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apaydin C. B., Cinar G., Cihan-Ustundag G., Small-molecule antiviral agents in ongoing clinical trials for COVID-19. Curr. Drug Targets 22, 1986–2005 (2021). [DOI] [PubMed] [Google Scholar]
- 55.ClinicalTrials.gov. (U.S. National Library of Medicine, 2022); https://clinicaltrials.gov. [DOI] [PubMed]
- 56.Pereira M. S., Morgantetti G. F., Massis L. M., Horta C. V., Hori J. I., Zamboni D. S., Activation of NLRC4 by flagellated bacteria triggers caspase-1-dependent and -independent responses to restrict Legionella pneumophila replication in macrophages and in vivo. J. Immunol. 187, 6447–6455 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Feeley J. C., Gibson R. J., Gorman G. W., Langford N. C., Rasheed J. K., Mackel D. C., Baine W. B., Charcoal-yeast extract agar: Primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10, 437–441 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCray P. B. Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H. P., Halabi C., Sigmund C. D., Meyerholz D. K., Kirby P., Look D. C., Perlman S., Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5