Abstract
IFNα is a key regulator of the dialogue between pancreatic β cells and the immune system in early type 1 diabetes (T1D). IFNα up-regulates HLA class I expression in human β cells, fostering autoantigen presentation to the immune system. We observed by bulk and single-cell RNA sequencing that exposure of human induced pluripotent-derived islet-like cells to IFNα induces expression of HLA class I and of other genes involved in antigen presentation, including the transcriptional activator NLRC5. We next evaluated the global role of NLRC5 in human insulin-producing EndoC-βH1 and human islet cells by RNA sequencing and targeted gene/protein determination. NLRC5 regulates expression of HLA class I, antigen presentation–related genes, and chemokines. NLRC5 also mediates the effects of IFNα on alternative splicing, a generator of β cell neoantigens, suggesting that it is a central player of the effects of IFNα on β cells that contribute to trigger and amplify autoimmunity in T1D.
NLRC5 is a key mediator of the effects of IFNα on human β cells and may contribute to trigger and amplify autoimmunity in T1D.
INTRODUCTION
Type 1 diabetes (T1D) is caused by an autoimmune attack against pancreatic β cells that provokes islet inflammation and progressive β cell loss. This takes place in the context of a dialogue between invading immune cells and the targeted β cells that culminate with CD8+ T cell–mediated β cell death (1–4). Recent evidence suggests that stress pathways triggered within β cells initiate and/or accelerate autoimmune β cell destruction (3, 5). Multiple layers of evidence implicate interferon α (IFNα) signaling as a key component of the early stages of T1D pathophysiology (6). Type I IFN is expressed in pancreatic islets from people with T1D, and islets obtained from living donors with recent-onset T1D show an IFN-stimulated gene signature (3, 7). IFNα induces endoplasmic reticulum (ER) stress, insulitis, and a massive human leukocyte antigen (HLA) class I overexpression in human β cells—three histological hallmarks of T1D (8, 9). This induction of HLA class I is long-lasting, persisting for several days after the removal of IFNα and, together with induction of ER stress and changes in mRNA splicing, may contribute to neoantigen presentation (10). However, the molecular mechanisms regulating HLA class I expression in β cells and neoantigen presentation—decisive steps for CD8-mediated β cell death—have never been clarified.
The functional maturation of pancreatic β cells and other nutrient-sensing cells is acquired during the first months of life, a period when the organism must adapt to both a changing environment and a changing source of nutrients (i.e., from the supportive intra utero environment to the mother’s breast milk and subsequently to solid foods) (11). These environmental and nutritional cues affect β cell maturation and function (12, 13). This is also the period when, in many cases, the autoimmunity process against the pancreatic β cells that will eventually lead to T1D starts. How the β cell’s maturation/adaptation to the new challenges described above cross-talks with the initial steps of autoimmunity [which probably starts with local triggering of an innate immune response (3)] is not known. Most modeling of the interplay between β cells and the immune cells is based on studies of adult human islets. To obviate this limitation and gain knowledge on the cross-talk between immune mediators and maturing β cells, we presently took advantage of β-like cells derived from induced pluripotent stem cells (iPSCs) (14–17). These cells already express Insulin and Pancreatic And Duodenal Homeobox 1 (PDX1) mRNAs, receptors to IFNα and other key proinflammatory cytokines (18) but are not yet fully mature (14–18). Thus, iPSCs provide an interesting model to study the impact of “early cytokines” such as IFNα on the developing β cells and to improve our understanding of T1D pathogenesis. Against this background, and to define the global pattern of IFNα-induced gene expression on these cells, we performed in parallel bulk and single-cell RNA sequencing (scRNA-seq) of iPSC-derived islet-like cells at the latest stage of in vitro development, i.e., stage 7 in the protocol used in our laboratory (19, 20). The data obtained (see Results) indicate relevant differences in the α and β cell responses to inflammation and a major impact of IFNα on the expression of HLA class I and other key genes involved in antigen processing and presentation by β cells. The bulk RNA-seq identified IFNα-induced expression of the transcription activator HLA class I transactivator (CITA), also known as “NLRC5” [NOD-like receptor (NLR) family, caspase recruitment (CARD) domain containing], a key regulator of HLA class I expression (21). NLRC5 expression is up-regulated in β cells from organ donors with T1D (22).
Nlrc5-deficient mice have impaired constitutive and inducible expression of both the classical murine major histocompatibility complex (MHC) class I genes and of critical genes involved in the antigen presentation pathway, including β2-microglobulin (B2m), immunoproteasome component 2 (Lmp2), and transporter associated with antigen processing 1 (Tap1), suggesting a critical role of Nlrc5 in regulating the expression of genes involved in the MHC class I antigen presentation machinery and in triggering MHC class I–dependent CD8+ T cell activation (23). NLRC5 is involved in nuclear factor κB (NF-κB) activation and type I IFN-regulated pathways during innate immune response. On the other hand, silencing NLRC5 suppresses the induction of IFNα after virus infection or polyinosinic:polycytidylic acid [poly (I:C)] treatment, while overexpression of NLRC5 protein results in up-regulation of IFNα (24–26). These diverse roles of NLRC5 on type I IFN signaling are cell- and context-dependent, and the available information is mostly based on murine data. Against this background, we further investigated the global role of NLRC5 expression in human insulin-producing EndoC-βH1 cells (a model that enables the study of β-like cells only without interference by other islet cells). We depleted NLRC5 gene expression using specific small interfering RNAs (siRNAs) and then performed deep RNA-seq (>200 million reads), followed by confirmation in EndoC-βH1 cells, iPSC-derived islet-like cells, and primary human islets. We observed that NLRC5 silencing not only down-regulates IFNα-induced expression of HLA class I and other genes involved in antigen presentation and recruitment of immune cells (e.g., chemokines) but also attenuates the alternative splicing changes triggered by IFNα. This double role of NLRC5 on HLA class I expression and regulation of alternative splicing suggests that this transcriptional activator may play a key role both in the generation of neoantigens and in their presentation to the immune system.
RESULTS
scRNA-seq of IFNα-treated iPSC-derived islet-like cells shows overexpression of genes related to antigen presentation in the context of HLA class I
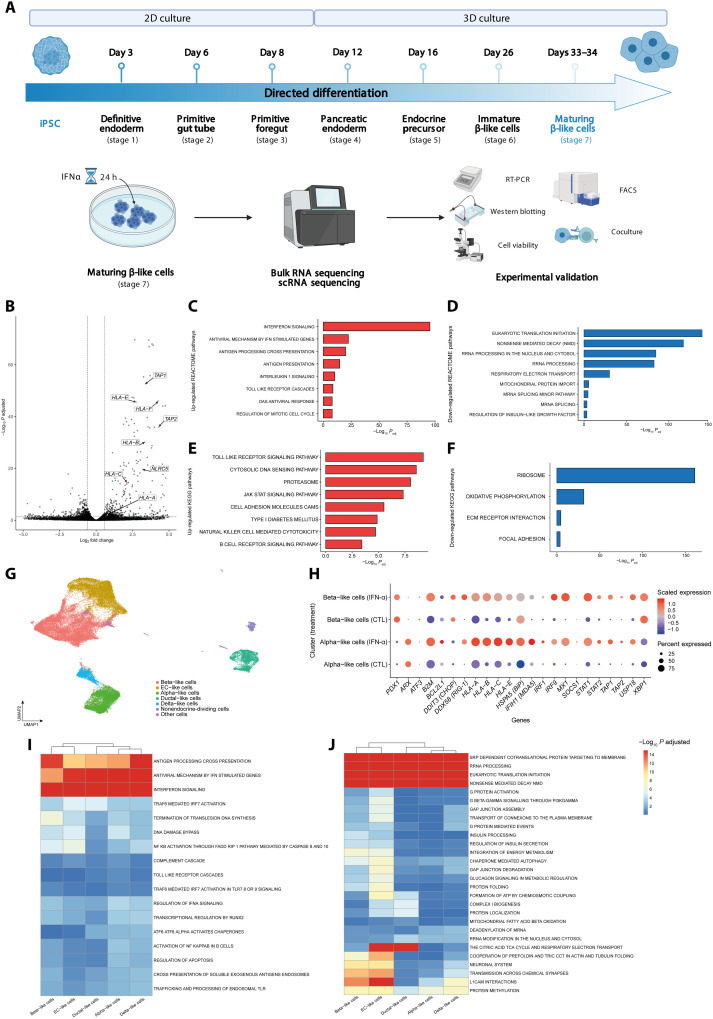
Human iPSC-derived islet-like cells at stage 7 of differentiation (Fig. 1A) had around 50% insulin positive and 10% glucagon or polyhormonal positive cells (fig. S1A). Similar to our previous observations (19, 20), these iPSC-derived β-like cells were not yet fully mature, as indicated by a 2-fold only increase in insulin release at 20 mM glucose but a >5-fold increase in insulin release following exposure to 20 mM + forskolin (fig. S1B). As previously described by our group (18), exposure of these cells to IFNα (2000 U/ml) for 24 hours induced a mild (~5%) increase in apoptosis (fig. S1C) and up-regulation of HLA-ABC, CXCL10, and PDL1 expression (fig. S1, D to F). The global gene expression of these iPSC-derived islet-like cells was characterized in parallel by bulk and scRNA-seq (Fig. 1A). There was a predominance of IFNα up-regulated genes (761 up-regulated versus 302 down-regulated) as indicated by the bulk RNA-seq data. Several key genes related to antigen presentation in the context of HLA class I are among the most overexpressed genes (Fig. 1B). Globally, gene set enrichment analysis (GSEA) using Kyoto Encyclopedia of Genes and Genomes (KEGG) and REACTOME (Fig. 1, C and D) databases showed an enrichment in antigen processing and presentation pathways. As expected, pathways related to IFN signaling, Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling, and antiviral responses were also identified (Fig. 1, E and F). These pathways, including “type 1 diabetes mellitus,” were in general similar to previous observations made in β cells obtained from patients affected by T1D, from adult human islets, or from EndoC-βH1 cells exposed to IFNα (9, 27, 28). Among the down-regulated pathways (Fig. 1, D and F), there were pathways related to mitochondrial function and oxidative phosphorylation and pathways related to mRNA splicing, indicating the impact of IFNα on this β cell function (9, 29).
Fig. 1. Single-cell and bulk RNA-seq of iPSC-derived islet-like cells.
(A) Experimental design of the iPSCs differentiation and subsequent exposure to IFNα. Stage 7 cells were exposed to IFNα during 24 hours; n = 5. FACS, fluorescence-activated cell sorting. (B) Volcano plot of the differentially expressed genes induced by IFNα after 24 hours of exposure. Paired experiments (n = 5) were analyzed using DESeq2 to determine differentially expressed genes. Each gene was plotted as (log2FC) (x axis) and −log10(adjusted P) (y axis). The horizontal dashed line indicates cutoff of −log10(adjusted P) > 1.3 (adjusted P < 0.05), and the horizontal dashed lines indicate |log2FC| > 0.58. (C to F) GSEA analysis following differential gene expression (DGE) analysis of β-like cells exposed to IFNα; REACTOME (C and D) and KEGG (E and F) show enriched and depleted gene sets. Selected pathways are shown as bar plots with −log10(P) on the x axis. (G) Uniform Manifold Approximation and Projection (UMAP) embedding of 73,834 integrated cells exposed, or not, to IFNα. (H) Expression of key IFNα-induced genes in β-like and α-like cells. The size of the dots indicates % of cells expressing the gene of interest, while the color scale indicates level of expression. (I and J) GSEA analysis in iPSC β-like cells following exposure to IFNα shows a heterogeneous response. After differential gene expression using MAST, results were input in fGSEA to determine enriched (I) or depleted (J) REACTOME pathways. −Log10(adjusted P) values were capped at 15. Hierarchical clustering based on those values was performed to highlight the (dis)similarity in response of identified cell types.
scRNA-seq of the iPSC-derived islet-like cells (Fig. 1, G to J; fig. S2, A and B; and data files S1 and S2) showed a predominancy of not only β- and α-like cells (Fig. 1G and figs. S2A and S3) but also enterochromaffin-like cells, all identified based on unsupervised clustering and previously described cell markers (Fig. 1H) (30, 31). In line with the bulk RNA-seq analysis (see above), GSEA analysis performed on the scRNA-seq data identified, among the top up-regulated pathways, antigen processing and presentation and antiviral mechanisms (Fig. 1I), while among the down-regulated pathways there were insulin processing and secretion, several pathways related to mitochondrial metabolism and protein folding, and pathways related to mRNA and ribosomal RNA (rRNA) processing (Fig. 1J). IFNα-induced expression of HLA class I A, B, C, E, and F and of the antiviral response gene MX Dynamin Like GTPase 1 (MX1) were present not only in the β-like cell cluster but also in nearly all other clusters (fig. S2B). This is in line with the previously described up-regulation of HLA class I in most islet cells, not only β cells, during insulitis (7). Notably, functional enrichment analysis indicated that β-like cells, as compared to α-like cells, had a more significant IFNα-induced antigen processing but a less significant expression of antiviral mechanisms (Fig. 1I). Because β cells, but not α cells, are destroyed by the autoimmune assault in T1D, we examined closely the expression of selected IFNα-induced genes in both β-like and α-like cells (Fig. 1H). As expected, PDX1 expression was higher in β-like cells, while Aristaless Related Homeobox (ARX), a transcription factor specifically expressed in glucagon-producing α cells, was much higher in α-like cells. β-Like cells had higher expression of the proapoptotic ER stress marker DDIT3 (also known as CHOP) [log2 fold change (log2FC) = 2.20, adjusted P << 0.0001] and also XBP1 (log2FC = 0.70, adjusted P << 0.0001) [in line with (32)], while α-like cells showed higher expression of the protective chaperone HSPA5 (also known as BiP) (log2FC = 0.13, adjusted P << 0.0001). Compared to β-like cells, α-like cells also had a higher expression of the antiapoptotic family member BCL2L1 (also known as Bcl-XL) (log2FC = 0.13, adjusted P << 0.0001), of the viral sensor IFIH1 (MDA5) (log2FC = 0.97, adjusted P << 0.0001), and of the inhibitory variant HLA-E (log2FC = 0.76, adjusted P << 0.0001). The higher HLA-E expression in α-like cells compared to β-like cells is in line with our previous observations in islets from patients affected by T1D (27). Unexpectedly, however, expression of HLA-A, HLA-B, and HLA-C was higher in α-like cells exposed to IFNα as compared to β-like cells (Fig. 1H), suggesting a similar impact of IFNα on the capacity of β and α cells to present antigens to CD8+ T cells.
NLRC5 KD decreases IFNα-induced HLA-ABC and related gene expression in human pancreatic β cells
The observations described above emphasized the key role for IFNα on the up-regulation of genes related to antigen processing and presentation by β cells in the context of HLA class I, including the induction of the potentially key regulatory transcriptional activator NLRC5. Hence, we further investigated the role of NLRC5 expression in human insulin-producing EndoC-βH1 cells, selected as a pure β-like cell model without interference by other islet cell types. Key findings were confirmed by experiments in primary human islets and iPSC-derived islet-like cells as described below.
We first performed a time-course and dose-response analysis of IFNα-treated EndoC-βH1 cells (fig. S4, A to O) to determine the best dose and time point for the follow-up experiments. The IFNα concentrations with the most intense and long-lasting mRNA expression were 200 and 2000 U/ml. The first genes to be induced were the transcription factors STAT1 and STAT2 (fig. S4, A and B) paralleled by the downstream transcription factors IRF1 and IRF9 (fig. S4, C and D). IRF9 was responsive to very low doses of IFNα, showing a significant increase at 0.2 U/mL of IFNα by 8 to 24 hours. Next, there was up-regulation of two genes previously shown to provide negative feedback on IFN signaling in β cells, namely, SOCS1 (33–35) and USP18 (36). SOCS1 was already increased by IFNα at 1 hour, with an expression peak at 2 to 4 hours (fig. S4E), while USP18 was increased by 2 hours and remained up-regulated up to 24 hours (fig. S4F). USP18 was up-regulated by relatively low concentrations of IFNα, namely, by 20 U/ml after 4 hours. The next “protective” gene to be induced is PDL1, which may inhibit T cell–induced toxicity to the β cells in T1D (3, 37), and it was up-regulated by 4 hours with an expression peak at 8 hours for IFNα both at 200 and 2000 U/ml (fig. S4G). Additional genes that were up-regulated early, i.e., by 4 hours, and at relatively low doses of IFNα (starting with 2 to 20 U/ml) are genes involved in the antiviral responses (38), namely, the viral double-stranded RNA receptor MDA5 (fig. S4H) and MX1 (fig. S4I). The transcriptional activator NLRC5 peaked at 8 hours and then returned to baseline by 24 hours for all IFNα concentrations except for 2000 U/ml (fig. S4J); NLRC5 induction preceded up-regulation of HLA class I ABC, which was only detected at 16 to 24 hours and at the highest concentrations of IFNα (fig. S4K). HLA-E (fig. S4L), which plays a protective role in β cells against T and natural killer (NK) cells attack (3), was already induced after 4 hours and remained up-regulated—at least at the two highest IFNα concentration—up to 16 to 24 hours (fig. S4L), similar to the other protective genes discussed above, namely, SOCS1, USP18, and PDL1. On the other hand, two genes that regulate either antigen presentation (HLA class I ABC; fig. S4K) or homing of immune cells to the islets (CXCL10; fig. S4M) (3) were only induced after 8 to 16 hours, suggesting that a short and/or low-intensity IFNα-mediated innate immune response will not necessarily translate into a subsequent adaptive immune response as early and sensitive regulatory/inhibitory mechanisms may prevail. As previously described, IFNα also induced expression of mRNAs related to the unfolded protein response, namely, ATF3 (fig. S4N) and, at late stage and for the highest doses of IFNα, the chaperone BiP (fig. S4O).
On the basis of this time-course/dose-response series, IFNα (2000 U/ml) at 8 and 24 hours was chosen for the follow-up experiments, focusing on the role for NLRC5. This was also the concentration of IFNα used in our previous experiments (Fig. 1 and figs. S1 to S3) (9), thus enabling data comparison.
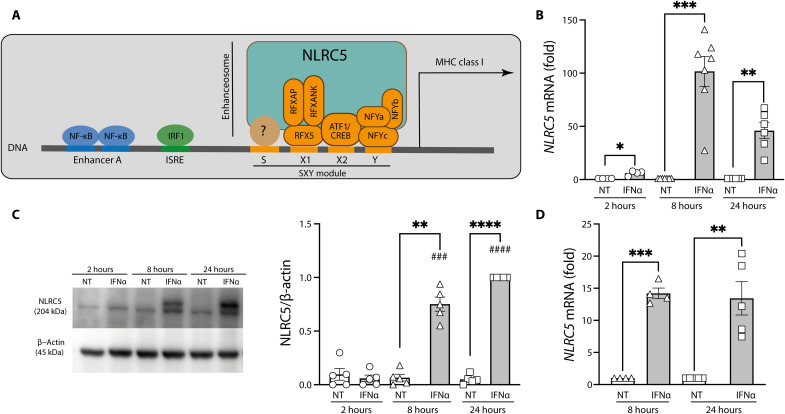
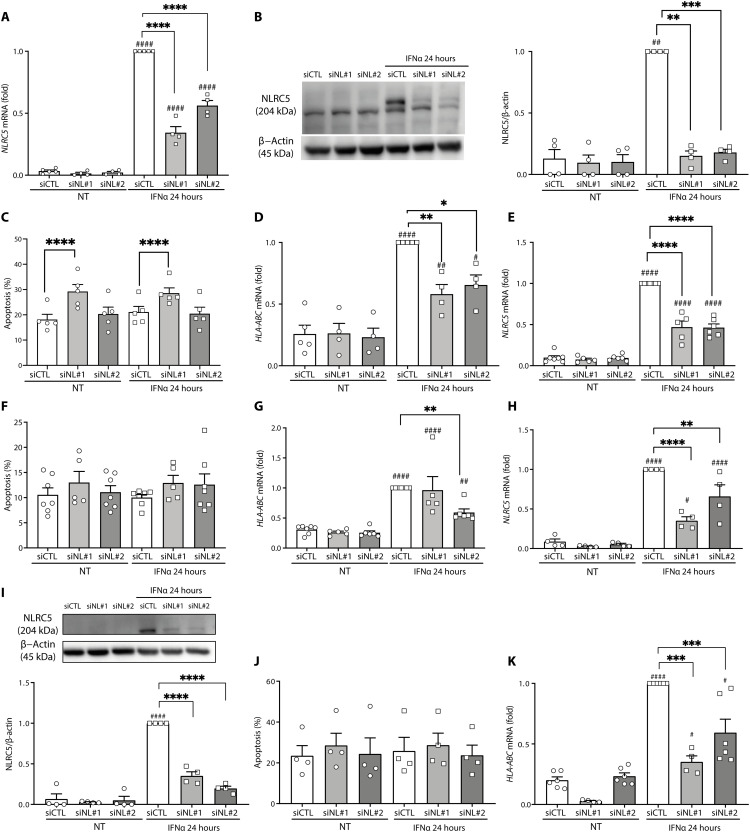
After its nuclear translocation, NLRC5 participates in the formation of the HLA class I enhanceosome together with RFX protein members and ATF1/CREB and NFY complexes. This complex binds to the SXY module to activate MHC class I transcription. In addition, IRF1 binds and activates ISRE elements present in the MHC class I promoter. NF-κB binding to enhancer A region also contributes to the activation of MHC class I promoter (Fig. 2A) (21, 39). IFNα, similarly to the observations made in fig. S4J, induced NLRC5 mRNA expression at 8 and 24 hours in both EndoC-βH1 cells (Fig. 2B; this was confirmed at the protein level Fig. 2C) and primary human islets (Fig. 2D). We achieved a 50 to 70% knockdown (KD) of IFNα-induced NLRC5 mRNA with the use of two separate siRNAs (Fig. 3A). The KD was even more marked at the protein level, with the two siRNAs abolishing >80% of IFNα-induced NLRC5 expression in EndoC-βH1 cells (Fig. 3B). As previously described (8, 9), IFNα alone did not induce apoptosis in EndoC-βH1 cells (Fig. 3C), but the first siRNA (siNL#1) induced an increase in cell death independently of the presence of IFNα; this deleterious effect was not present with the second siRNA (siNL#2). Both siRNAs induced a significant but partial inhibition of HLA-ABC expression (Fig. 3D). siRNAs #1 and #2 also inhibited NLRC5 mRNA expression in human islets (Fig. 3E), without significantly affecting viability (Fig. 3F) but leading to a decrease in HLA-ABC expression (Fig. 3G). These siRNAs induced a similar effect on iPSC-derived islet-like cells exposed to IFNα (Fig. 3, H to K). Notably, and as observed with the EndoC-βH1 cells (Fig. 3, A and B), the siRNAs induced a more marked effect on NLRC5 protein expression than on the mRNA expression (compare Fig. 3, H and I). On the basis of the facts that both siRNAs effectively inhibited NLRC5 expression but that siNL#2 did not affect cell viability in any of the models tested, all subsequent experiments in the study were performed with siNL#2.
Fig. 2. NLRC5 is a key MHC class I enhanceosome member, and it is induced by IFNα in human pancreatic β cells.
(A) Schematic representation of the MHC class I promoter region. (B to D) Cells were left untreated or exposed to IFNα (2000 U/ml) for 2, 8, and 24 hours (EndoC-βH1) (B and C) or for 8 and 24 hours (dispersed human islets) (D). mRNA (B) and protein (C) expression of NLRC5 were analyzed by RT-PCR and Western blotting in EndoC-βH1 cells, respectively. mRNA expression of NLRC5 was analyzed by RT-PCR in dispersed human islets (D). mRNA expression was analyzed by RT-PCR and normalized by the geometric mean of GAPDH and β-actin and then by the nontreated (NT) condition for each time point considered as 1. NLRC5 protein was quantified by densitometry and normalized by the housekeeping protein β-actin and then by the highest value of each experiment considered as 1. Results are means ± SEM of four to seven independent experiments. ###P < 0.001 and ####P < 0.0001 versus same condition at 2 hours; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 versus each untreated condition [analysis of variance (ANOVA) followed by Bonferroni correction for multiple comparisons].
Fig. 3. NLRC5 KD decreases IFNα-induced HLA-ABC expression in human pancreatic β cells.
(A to D) EndoC-βH1 cells, (E to G) dispersed human islets, and (H to K) iPSC-derived β-like cells were transfected with siRNA control (siCTL, white bars) or with two different siRNAs targeting NLRC5 (siNL#1 and siNL#2, gray bars). Cells were left untreated or treated with IFNα (2000 U/ml) for 24 hours. Expression of NLRC5 mRNA was measured by RT-PCR (A, E, and H), and NLRC5 protein was determined by Western blotting (B and I). The percentage of dead cells was counted by Hoechst and propidium iodine staining (C, F, and J). NLRC5 depletion decreased IFNα-induced expression of HLA-ABC (D, G, and K). The mRNA expression was analyzed by RT-PCR and normalized by the geometric mean of GAPDH and β-actin and then by siCTL treated with IFNα considered as 1. NLRC5 protein was quantified by densitometry and normalized by the housekeeping protein β-actin and then by the highest value of each experiment considered as 1. Results are means ± SEM of four to seven independent experiments. #P < 0.05, ##P < 0.01, and ####P < 0.0001 versus untreated and transfected with the same siRNA; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, as indicated by bars (ANOVA followed by Bonferroni correction for multiple comparisons).
The global impact of NLRC5 on IFNα-modulated genes
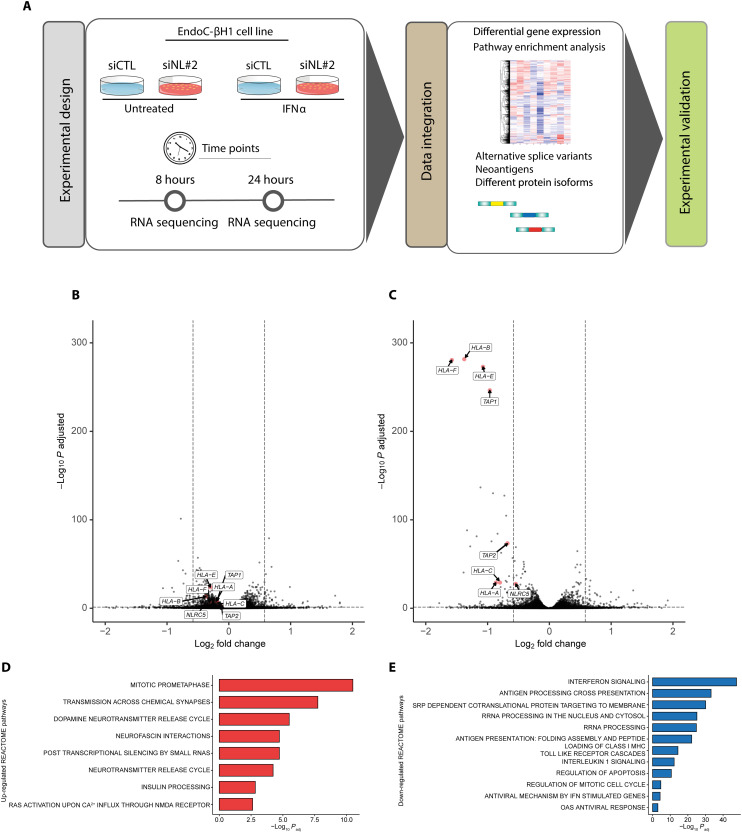
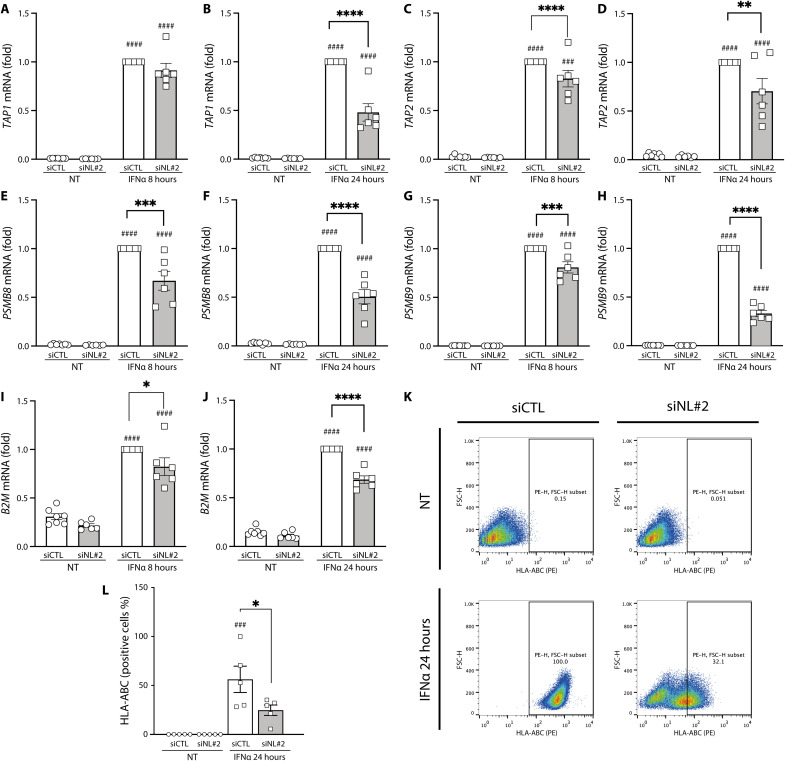
The effects of NLRC5 are cell/tissue specific and may go beyond the regulation of HLA class I and related genes (24–26). To address its role in a pure human β cell line, we next evaluated by bulk RNA-seq the mRNA expression of EndoC-βH1 cells exposed for 8 or 24 hours to IFNα (2000 U/ml) with or without KD of NLRC5 by siNL#2 (Fig. 4A). The inhibitory effects of the siRNA against NLRC5 were evaluated by Western blot ahead of the RNA-seq analysis, confirming near total inhibition of IFNα-induced NLRC5 expression at 24 hours (fig. S5). Analysis of the RNA-seq data indicated that the most marked effects of NLRC5 inhibition were observed after 24 hours of IFNα exposure (Fig. 4, B and C), with the differential gene expression (DGE) analysis showing that NLRC5 KD prevented the up-regulation of not only HLA class I A, B, C, and F but also TAP1 and TAP2, two key peptide transporters that help to load antigens on HLA class I molecules and to stabilize HLA at the cell surface (40). GSEA analysis based on REACTOME database (Fig. 4, D and E) indicated that decreasing NLRC5 expression prevented the inhibitory effects of IFNα on mitosis and insulin processing, among others (Fig. 4D). NLRC5 depletion ahead of IFNα exposure prevented IFN signaling, antigen processing and presentation, rRNA processing, and, of particular relevance, antigen presentation including folding assembly and peptide loading of MHC class I (Fig. 4E), which is in line with the observed decrease of TAP1 and TAP2 gene expression (Fig. 4C).
Fig. 4. RNA-seq of EndoC-βH1 cells after IFNα exposure preceded or not by NLRC5 depletion.
(A) Methodologic approach for the RNA-seq experiments. EndoC-βH1 were transfected with siRNA control (siCTL) or with an siRNA targeting NLRC5 (siNL#2). The cells were left untreated or treated with IFNα (2000 U/ml) for 8 or 24 hours. The RNA-seq was performed for all the conditions (n = 6), and DGE and alternative splicing analysis were performed as described in Materials and Methods. (B and C) Volcano plot of differentially expressed genes in EndoC-βH1 after 8 hours (B) and 24 hours (C) of exposure to IFNα with KD of NLRC5. Quantification of the RNA-seq data was performed using Salmon with GENCODE v36 as the genome reference. DGE was performed using DESeq2. (D and E) GSEA analysis. Results from the DGE analysis were input in fGSEA. Selected enriched pathways (D) showed a functional recovery of EndoC-βH1 cells, while selected depleted pathways (E) indicated that the previously IFNα-induced signature was reversed following KD of NLRC5. The output of the fGSEA analysis is provided in data file S3.
In β cells obtained from patients affected by T1D and normoglycemic individuals, NLRC5 expression was positively correlated with the expression HLA class I A, B, and C, TAP1, TAP2, B2M, and PSMB8, all genes that contribute to antigen presentation in the context of MHC class I (fig. S6, A to C). In agreement with these findings, NLRC5 expression was also correlated with the expression of the same genes in EndoC-βH1 cells exposed to IFNα with or without previous KD of NLRC5 (fig. S6B). To further evaluate whether there is an induction of NLRC5 in β cells during T1D, we downloaded scRNA-seq samples from T1D and normoglycemic donors (table S1) publicly available via the Human Pancreas Analysis Program (HPAP) portal (41). After reduction of the scRNA-seq data into “pseudo-bulk,” we detected the expression of NLRC5 in 16 of the 23 samples; NLRC5 expression was higher in β cells from patients affected by T1D when compared to normoglycemic donors (log2FC = 2.23; P < 0.0006) (fig. S6C).
Rank-rank hypergeometric overlap (RRHO) analysis of the data from Fig. 4 indicated that inhibition of NLRC5 ahead of IFNα exposure induced a highly significant anticorrelated signature with genes up-regulated by IFNα in EndoC-βH1 cells exposed to IFNα (fig. S6D), confirming a partial reversion of the IFNα-induced signature by NLRC5 inhibition. The analysis of the RNA-seq data confirmed the impact of NLRC5 KD on preventing IFNα-induced genes related to loading of antigens on HLA class I and their expression at the surface of pancreatic β cells, particularly after 24 hours of treatment with the cytokine (fig. S7, A to J). These effects were confirmed in independent experiments involving both EndoC-βH1 cells (Fig. 5, A to J) and, at least for some of the genes, primary human islets (fig. S7, K to O). Inhibition of NLRC5 expression decreased by >50% HLA-ABC at the pancreatic β cell surface, as evaluated by flow cytometry (Fig. 5, K and L), confirming that the observed changes in HLA-related genes had an impact on the subcellular localization of HLA class I.
Fig. 5. NLRC5 depletion prevents the expression on IFNα-induced genes related to the antigen presentation machinery in EndoC-βH1 cells.
EndoC-βH1 cells were transfected with an siRNA control (siCTL, white bars) or with an siRNA targeting NLRC5 (siNL#2, gray bars). EndoC-βH1 cells were then left untreated or treated with IFNα (2000 U/ml) for 8 or 24 hours. NLRC5 depletion decreased the expression of (A and B) TAP1 gene, (C and D) TAP2, (E and F) PSMB8, (G and H) PSMB9, and (I and J) B2M after exposure to IFNα for 8 and 24 hours. (K) The HLA-ABC expression at the β cell surface was evaluated by flow cytometry. (L) HLA-ABC quantification of five independent experiments. The mRNA expression was analyzed by RT-PCR and normalized by geometric mean of GAPDH and β-actin and then the condition of siCTL treated with IFNα considered as 1. Results are means ± SEM of five to six independent experiments. ###P < 0.001 and ####P < 0.0001 versus untreated and transfected with the same siRNA; *P < 0.05, **P < 0.01, *** P < 0.001, and ****P < 0.0001, as indicated by bars (ANOVA followed by Bonferroni correction for multiple comparisons).
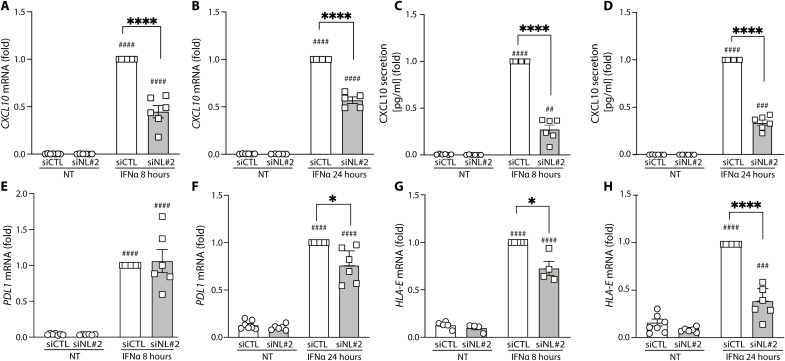
The role of NLRC5 is not, however, limited to the regulation of HLA-related genes. Inhibition of NLRC5 prevented IFNα-induced expression of the chemokine CXCL10 mRNA and its secretion to the medium at both 8 and 24 hours following addition of IFNα in EndoC-βH1 cells (Fig. 6, A to D), and led to a 55% inhibition on CXCL10 expression in dispersed human islets (n = 6, P < 0.001). NLRC5 KD did not affect PDL1 expression at 8 hours of IFNα exposure but induced a partial (24%) decrease on the expression of this mRNA at 24 hours (Fig. 6, E and F). PDL1 expression is up-regulated in β cells from patients affected by T1D (37) and plays a protective role against infiltrating CD8+ T cells (3), and it is of interest that its expression can be partially dissociated from the proinflammatory HLA class I or CXCL10 expression by NLRC5 inhibition. On the other hand, NLRC5 KD led to decreased expression of HLA-E, another potentially β cell protective gene (9), at both 8 and 24 hours after IFNα exposure in EndoC-βH1 (Fig. 6, G and H), and in human islets (45% decrease at 24 hours, n = 6, P < 0.001).
Fig. 6. Independent confirmation that NLRC5 depletion prevents induction of several key genes induced by IFNα in EndoC-βH1 cells.
(A to H) EndoC-βH1 cells were transfected with an siRNA control (siCTL, white bars) or with an siRNA targeting NLRC5 (siNL#2, gray bars). The cells were then left untreated or treated with IFNα (2000 U/ml) for 8 or 24 hours. NLRC5 depletion prevented IFNα-induced CXCL10 gene expression after 8 hours (A) and 24 hours (B) of IFNα exposure. CXCL10 secretion was evaluated by enzyme-linked immunosorbent assay (ELISA) (C and D). NLRC5 depletion did not affect PDL1 mRNA expression after 8 hours of IFNα exposure (E) but decreased PDL1 mRNA expression at 24 hours of IFNα exposure (F). NLRC5 depletion prevented IFNα-induced HLA-E gene expression after 8 hours (G) and 24 hours (H) of IFNα exposure. The mRNA expression levels were analyzed by RT-PCR and normalized by geometric mean of GAPDH and β-actin and then by siCTL treated with IFNα considered as 1. Results are means ± SEM of four to seven independent experiments. ##P < 0.01, ###P < 0.001, and ####P < 0.0001 versus untreated and transfected with the same siRNA; *P < 0.05 and ****P < 0.0001, as indicated by bars (ANOVA followed by Bonferroni correction for multiple comparisons).
NLRC5 inhibition on β-like cells decreases T cell activation in a coculture experiment
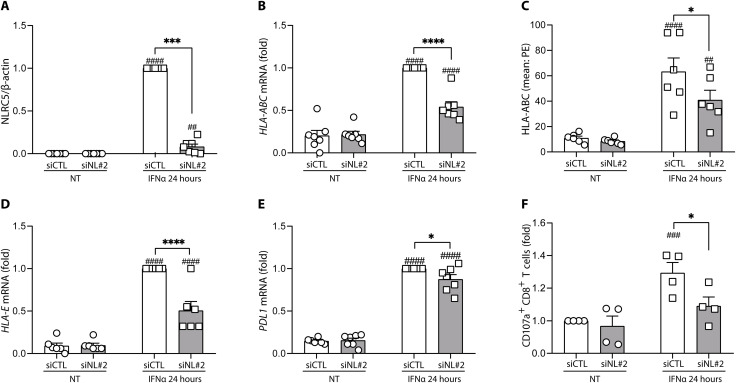
Using preproinsulin (PPI)–directed CD8+ T cells (42), we have assessed the effect of NLRC5 KD on the immunogenicity of HLA-A2 H1-derived islet-like cells. As observed in 1023A-derived islet-like cells, silencing of NLRC5 in H1-derived islet-like cells led to >90% decrease of NLRC5 protein expression (Fig. 7A) and decreased expression of both the HLA-ABC gene expression and its protein cell surface expression (Fig. 7, B and C). In addition, expression of the protective molecules HLA-E and PDL1 were reduced after NLRC5 silencing (Fig. 7, D and E). After coculture with PPI-directed T cells, we observed an increased T cell activation following IFNα treatment (determined by the percentage of CD107a+ CD8+ T cells) despite an overrepresentation of PPI-derived peptides in basal condition (Fig. 7F). In line with our observations on the role of NLRC5 in regulating HLA expression in IFNα conditions, NLRC5 KD decreased the IFNα effect on T cell activation (Fig. 7F). The fact that the decrease in CD8+ T cell activation was not complete may be at least, in part, due to the parallel inhibition of PDL1 in the NLRC5 KD islet-like cells (Fig. 7E). The presently observed protective effects of NLRC5 inhibition are in line with a very recent publication, indicating that NLRC5 KD partially protects stem cells (SC)–derived islet-like cells (previously cultured at high glucose and in the presence of a cytokine mix and a chemical ER stressor) against apoptosis induced by allogenic T1D peripheral blood mononuclear cells (PBMCs) (43).
Fig. 7. NLRC5 KD in H1-derived islet-like cells decreases the T cell activation in coculture experiments.
H1-derived islet-like cells were transfected with an siRNA control (siCTL, white bars) or with an siRNA targeting NLRC5 (siNL#2, gray bars). Cells were then left untreated or treated with IFNα (2000 U/ml) for 24 hours. (A) NLRC5 protein expression was determined by Western blotting. NLRC5 protein was quantified by densitometry and normalized by the housekeeping protein β-actin and then by the highest value of each experiment considered as 1. NLRC5 depletion decreased the mRNA expression of HLA-ABC (B) and its cell surface expression, evaluated by flow cytometry (C). NLRC5 depletion decreased the mRNA expression of (D) HLA-E and (E) PDL1. mRNA expression was analyzed by RT-PCR and normalized by the geometric mean of GAPDH and β-actin and then by siCTL-treated cells exposed to IFNα and considered as 1. (F) T cell activation determined by the percentage of CD107a+ CD8+ T cells. Results are means ± SEM of four to seven independent experiments. ##P < 0.01, ###P < 0.001, and ####P < 0.0001 versus untreated and transfected with the same siRNA; *P < 0.05, ***P < 0.001, and ****P < 0.0001, as indicated by bars (ANOVA followed by Bonferroni correction for multiple comparisons).
NLRC5 inhibition prevents IFNα-induced alternative splicing
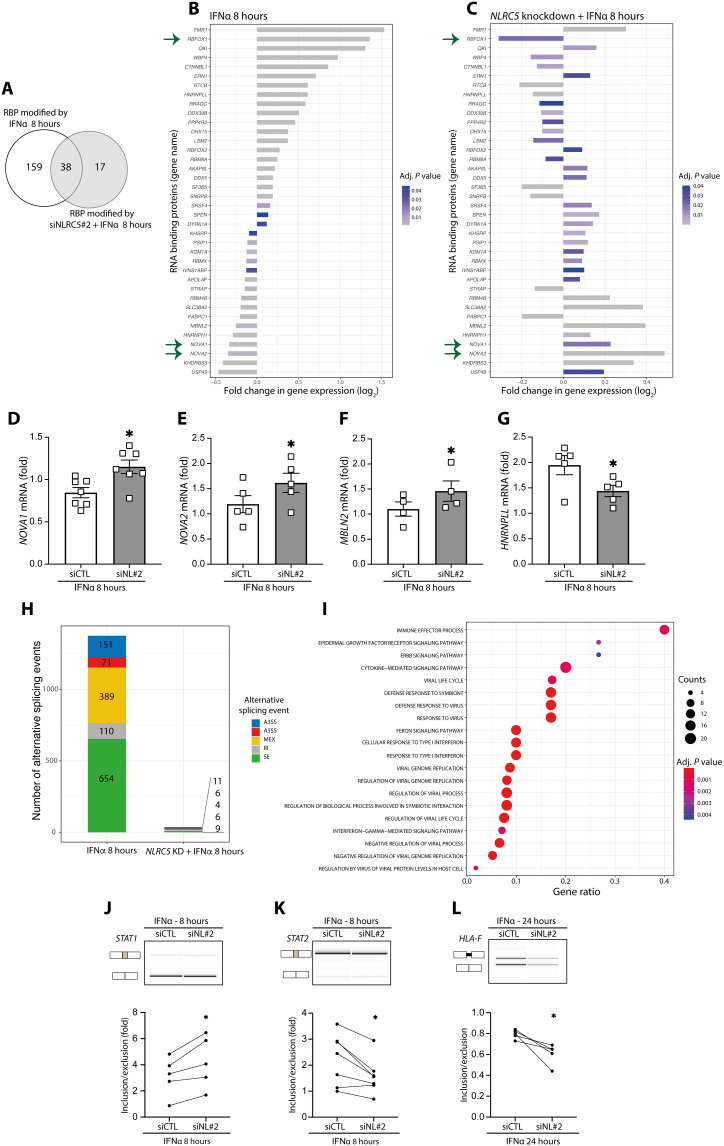
Exposure of EndoC-βH1 cells to IFNα induced a major change on the expression of RNA binding proteins (RBPs), particularly after 8 hours as compared to 24 hours (Fig. 8, A and B, and fig. S8), suggesting that early changes in splicing regulation, as we previously observed for chromatin opening (9), may be important in the innate immunity and antiviral effects of type I IFNs. Of the nearly 200 RBPs whose expression was modified by IFNα at 8 hours, 70% of them were restored to baseline or even expressed in the opposite way (i.e., changed from up- to down-regulated, or the other way around) when NLRC5 was KD ahead of IFNα exposure (Fig. 8, A to G). Of particular relevance were NOVA 1 and 2, which were down-regulated by IFNα alone (Fig. 8B) but become up-regulated when exposure to IFNα was preceded by NLRC5 KD (Fig. 8, C to E). NOVA1 and NOVA2 are key regulators of splicing in β cells, and their inhibition contribute to β cell apoptosis (44). In the context of IFNα exposure, the depletion of NOVA1 increased EndoC-βH1 apoptosis (fig. S9, A and D), while the parallel depletion of NLRC5 and NOVA1 reverted NOVA1 KD-induced apoptosis, suggesting that NLRC5 depletion attenuates NOVA1 KD-induced toxicity (fig. S9D). RBFOX1 followed an inverse pathway, i.e., it was up-regulated by IFNα alone (Fig. 8B) and then down-regulated in the case of NLRC5 KD plus IFNα (Fig. 8C). Inhibition of the splicing regulator RBFOX1 increases insulin synthesis and secretion (45), and this up-regulation by IFNα—an effect prevented by NLRC5 KD—may contribute to progressive β cell dysfunction in the early stages of T1D. We additionally confirmed the impact of NLRC5 KD on two other RBPs modified by IFNα, namely, MBNL2 (Fig. 8F) and HNRNPLL (Fig. 8G).
Fig. 8. NLRC5 depletion attenuates the impact of IFNα-induced (8 hours) alternative splicing modifications in EndoC-βH1 cells.
(A) Venn diagram of the overlap between RNA binding proteins (RBPs) modified by exposure to IFNα with or without NLRC5 silencing. (B and C) mRNA expression of RBPs in cells exposed to IFNα alone (B) or with NLRC5 silencing (C). Bar charts depict log2-transformed FC (adjusted P < 0.05). Arrows indicate relevant RBPs modified by IFNα (B) but prevented by NLRC5 KD (C). (D and E) mRNA expression analyzed by quantitative RT-PCR of selected RBPs regulated by IFNα and NLRC5 depletion: (D) NOVA1, (E) NOVA2, (F) MBNL2, and (G) HNRNPLL; mRNA expression was normalized by the geometric mean of GAPDH and β-actin; means ± SEM of four to seven independent experiments. *P < 0.05 (paired t test). (H) Cells were exposed to IFNα or to IFNα following NLRC5 silencing (NLRC5 KD). A3ss, alternative 3′ splice site; A5ss, alternative 5′ splice site; MEX, mutually exclusive exons; RI, intron retention; SE, skipped exons. (I) Gene Ontology (GO) enrichment of the genes with modified alternative splicing events in cells treated with IFNα for 8 hours. Gene ratio indicates % of total genes with AS modifications in the given GO term (adjusted P < 0.05). (J to L) Confirmation by semiquantitative RT-PCR of IFNα and NLRC5 KD-regulated splicing in selected genes (STAT1, STAT2, and HLA-F). Representative digital gel images (top) and quantifications with paired individual data points for five to seven independent experiments (bottom) are shown for each splicing event. *P < 0.05 (paired t test).
The IFNα-induced changes in RBPs were translated into major changes in mRNA splicing, with a total of 1375 alternative splicing events modified at 8 hours (Fig. 8H) and 844 events modified at 24 hours (fig. S8, A to D) following IFNα exposure. The most common alternative splicing event was skipped exons (SEs) followed by mutually exclusive exons (MXEs) (Fig. 8H and fig. S8D). Gene Ontology enrichment analysis of the genes with modified splicing at 8 hours showed, among others, pathways related to immune effector processes, defense responses against viruses, responses to IFNs, etc. (Fig. 8I). Many of these pathways, including defense responses against viruses, were not maintained at 24 hours (fig. S8E), reinforcing the concept that alternative splicing changes may be part of the arsenal of early innate immunity/antiviral responses induced by type I IFNs. The KD of NLRC5 had a major impact on the number of IFNα-modified events, decreasing them by 97% at 8 hours (Fig. 8H) and by 80% at 24 hours (fig. S8D), which is in agreement with the observation that inhibition of NLRC5 decreases the impact of IFNα on the expression of RBPs (Fig. 8, A to G, and fig. S8A).
Some of the observed splicing modifications affected STAT1, STAT2, and HLA-F coding transcripts (Fig. 8, J to L). The splicing modification involving the STAT1 transcript is predicted to generate an alternative first exon, and the inclusion of this exon was potentiated by NLRC5 KD after 8 hours of exposure to IFNα (Fig. 8J). The splicing modification affecting STAT2 involved the exclusion of exon 16. After 8 hours of exposure to IFNα, the depletion of NLRC5 induced the skipping of this exon (Fig. 8K). Using the software Exon Ontology (46), exon 16 is predicted to affect the coding of STAT2 protein binding domain. Regarding HLA-F splicing modifications, NLRC5 depletion followed by the exposure to IFNα during 24 hours led to a significant decrease in the retention of the first intron from the HLA-F pre-mRNA (Fig. 8L). As a “nonclassical” HLA class I molecule, HLA-F harbors some structural differences from the other HLA class I members, namely, the antigen recognition site (47). In addition, and contrary to the classical HLA class I members, HLA-F depends on its cytoplasmic tail to exit the ER, pointing to a potential functional impact of this splicing modification (48). Detailed functional studies are needed to clarify the functional impact of the splicing modifications described in Fig. 8 (J to L).
DISCUSSION
IFNα plays a key role on the early stages of T1D. This cytokine, together with its downstream signaling molecules, are now being tested in preclinical models as therapeutic targets to prevent and/or revert the disease (6). A key effect of IFNα is to up-regulate HLA class I expression in human β cells (4, 49), which augments the number and breadth of autoantigens presented to the immune system (50). We presently observed by bulk and scRNA-seq that exposure of iPSC-derived islet-like cells to IFNα induces expression of HLA class I and of several other genes involved in antigen presentation and attraction of immune cells to the islets.
IFNα induced a slightly higher up-regulation of HLA-A and HLA-B expression in α-like cells as compared to β-like cells. There were also important differences between these two cell types regarding expression of the following: (i) Bcl-XL, a key antiapoptotic gene that has a higher expression in α-like cells than in β-like cells; this is in agreement with our previous findings in rat islet cells, where high Bcl-xl expression renders α cells resistant to metabolic ER stress–induced apoptosis (51); (ii) ER stress–related genes, with the proapoptotic CHOP showing higher expression in β-like cells, while the protective chaperone BIP is higher in α-like cells; this may contribute to the α cell endurance to ER stress (51); (iii) viral recognition and innate immune response genes, such as MDA5, have higher expression in α-like cells; this is again in line with previous findings in rat islets showing that α cells are much more resistant than β cells to coxsackievirus B group 5 (CVB5) infection (52). It thus seems that the capacity for antigen presentation, as evaluated by HLA expression, is similar between β and α cells, but either α cells are somehow “less antigenic” than β cells [and it is of interest that CD8+ T cells invading the islets of patients affected by T1D are reactive to PPI but not to glucagon (53)] for reasons that remain to be clarified and/or α cells’ enhanced capacity to endure viral infections and ER stress allows them to better survive early stresses that may cause cell death and consequently amplify antigen presentation to the immune system.
The above-described increase in HLA class I expression in islet-like cells was accompanied by the increased expression of the transcriptional activator NLRC5, a regulator of HLA class I expression. We next evaluated the global role of NLRC5 in human insulin-producing EndoC-βH1 cells by RNA-seq, followed by confirmation experiments in both EndoC-βH1 cells and human islets. We observed that NLRC5 regulates not only HLA class I expression but also expression of genes involved on peptide loading and stabilization of HLA class I at the cell surface, as well as chemokines that may attract immune cells. The down-regulation of HLA class I mediated by NLRC5 KD led to a reduction in the percentage of activated CD8+ T cells in coculture experiments, suggesting that NLRC5 inhibition may protect β cells against CD8+ T cell attack. In line with this possibility, a very recent study indicates that inhibition of NLRC5 partially protects stressed SC-derived islet-like cells against apoptosis induced by allogenic PBMCs (43).
NLRC5 works similarly to the MHC class II transactivator protein CIITA (54): CIITA does not bind directly to DNA, but it is instead recruited to the MHC class II promoter region through protein-protein interaction with a promoter binding complex, the MHC class II enhanceosome (55). Similarly, NLRC5 induces gene expression in an MHC class I enhanceosome–dependent manner—its binding is indirect and dependent on the highly cooperative protein complex, the MHC class I enhanceosome (54).
Unexpectedly, we found that NLRC5 is a major mediator of the effects of IFNα on alternative splicing, a potential generator of β cell neoantigens (3, 50). These observations raise the intriguing possibility that NLRC5 is a central mediator of many of the key effects of IFNα on β cells that contribute to trigger and amplify autoimmunity in T1D. This effect on mRNA splicing might be explained by the direct or indirect impact of NLRC5 on the expression of different RBPs. We presently observed that NLRC5 depletion modifies IFNα-induced splicing changes in STAT1-, STAT2-, and HLA-F–encoding transcripts, supporting the RNA-seq data.
We confirmed the relevance for NLRC5 on the regulation of HLA class I and related gene expression in our three experimental models at different degrees of human β cell differentiation, namely, iPSC-derived β-like cells, insulin-producing EndoC-βH1 cells, and adult human islets. This NLRC5 up-regulation was also observed in pseudo-bulk data from scRNA-seq of β cells from T1D donors compared to normoglycemic donors (fig. S6C), although this has been difficult to confirm by histology (7, 56) perhaps due to issues with the available antibodies. As a whole, these observations suggest an early role of NLRC5 in β cell antigen presentation, starting during the first months of life, a period when the autoimmunity process against these cells is probably triggered in many of the individuals that will later develop T1D (57). NLRC5 is already induced by relatively low concentrations of IFNα, e.g., 20 U/ml, a concentration that also induces the protective protein PDL1 but not HLA class I or chemokines (present data). This, and the fact that a nearly complete KD of NLRC5 at the protein level only decreases HLA class I expression by around 50%, indicates that the regulation of HLA class I in β cells involves additional mediators besides NLRC5. This observation raises the hypothesis that deleterious signals (i.e., HLA class I and chemokine expression) can be potentially dissociated from the beneficial (i.e., PDL1) effects of IFNα exposure (see below). In line with this possibility, KD of the upstream transcription factor STAT2 inhibits IFNα-induced HLA class I expression (8) while inducing a paradoxical increase in PDL1 (37).
Potential limitations of the study include first its in vitro nature, i.e., all findings are based on three models of human β cells or human β-like cells under culture conditions. Models of systemic Nlrc5 KO mice have been generated (23–26, 58), but major limitations of using these models include the species-specific nature of mRNA splicing [i.e., alternative splicing findings observed in human islets are often not reproduced in rodent islets (59)], the absence of a β cell–specific NLRC5 KO model, and the difficulties of translating results obtained in mouse models of T1D to the human disease (60), which emphasizes the need for research on human β cells and, when available, fully humanized mouse models. Second, the RNA-seq following NLRC5 inhibition was performed in a human β cell line, namely, EndoC-βH1 cells. Although cytokine-induced whole gene expression is well conserved between EndoC-βH1 cells and primary human islets (9, 61), future validation of our global findings in primary human islets is important particularly in the context of specific alternative splicing. Some of the specific findings made in EndoC-βH1 cells were presently confirmed in human islets, but all donors were older than 60 years, and we cannot exclude that donor’s age affected the data obtained. Third, it would be crucial to identify the specific splice variants regulated by NLRC5 in human β cells that are recognized by CD8+ T cells obtained from T1D patients. This would serve as a “proof of concept” for the role for NLRC5-regulated splicing in human T1D. This will require the combination of deep RNA-seq with peptidomics (50) in human β cells KD, or not, to NLRC5 followed by exposure to IFNα, a highly time- and resource-demanding approach that goes beyond the objectives of the present study. Last, the dual role of NLRC5, which makes data interpretation difficult, must be considered. On one hand, NLRC5 depletion decreases IFNα-induced HLA class I expression and alternative splicing, which may be β cell protective by potentially decreasing neoantigen presentation to the immune system. On the other hand, we observed that NLRC5 KD decreases IFNα-induced expression of protective molecules such as HLA-E and PDL1, which may hamper the ability of β cells to evade—at least for a time—the immune assault. Future work should aim to determine how to therapeutically dissociate the potentially beneficial from deleterious effects of NLRC5 inhibition.
Perhaps the most intriguing finding of the present study is the discovery that NLRC5 is a key mediator of the effects of IFNα on alternative splicing. KD of NLRC5 prevented by more than 80% IFNα-induced changes in the expression of RBPs and, most importantly, downstream splicing events. Among the identified RBPs that are regulated by IFNα via NLRC5 are NOVA1, NOVA2, and RBFOX1, previously shown by us to modulate β cell viability and function (44, 45). Splicing alterations might contribute to pancreatic β cell demise and its recognition by the immune system in the context of T1D (29, 50, 62) and other autoimmune diseases (6, 63) and may also have an impact on the role for candidate genes in T1D and other diseases (29, 64, 65). Alternative splicing plays also a regulatory role for β cell identity, development, function, and survival (29, 44, 64, 66–69). It will be thus of high interest to further characterize the potential role for NLRC5 in β cell neoantigen generation and presentation in the early stages of T1D.
MATERIALS AND METHODS
Experimental model and subject details
Human islets
Human islets were isolated from nondiabetic (ND) organ donors by collagenase digestion and density gradient purification with the approval of the local ethical committee in Pisa, Italy. The islets were characterized as previously reported (70) and cultured in M199 culture medium (5.5 mM glucose) ahead of transport to the Brussels’ laboratory for additional experiments. On arrival, the islets were dispersed and cultured in Ham’s F-10 medium containing 6.1 mM glucose (Gibco, Thermo Fisher Scientific), 10% fetal bovine serum (Gibco, Thermo Fisher Scientific), 2 mM GlutaMAX (Gibco, Thermo Fisher Scientific), 50 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich), 1% fatty acid–free bovine serum albumin (BSA) fraction V (Roche), and penicillin (50 U/ml) and streptomycin (50 mg/ml) (Lonza, Verviers, Belgium) (71). All the experimental replicates are from different donors. The characteristics of the different donors are listed in table S2.
EndoC-βH1 human β cell line
The human pancreatic β cell line EndoC-βH1 was provided by R. Scharfmann (Institut Cochin, Université Paris, Paris, France) (72). The EndoC-βH1 cells were cultured in Dulbecco’s modified Eagle medium with 5.6 mM glucose (Gibco, Thermo Fisher Scientific), 2% fatty acid–free BSA fraction V, 50 μM 2-mercaptoethanol (Gibco, Thermo Fisher Scientific), 10 mM nicotinamide (Sigma-Aldrich), transferrin (5.5 μg/ml; Sigma-Aldrich), selenite (6.7 ng/ml; Sigma-Aldrich), and penicillin (100 U/ml) + streptomycin (100 μg/ml) in Matrigel-fibronectin–coated plates (71). All the experiments performed with EndoC-βH1 cells refer to independent experiments, i.e., EndoC-βH1 cells from different passages.
SC lines
The iPSC lines HEL115.6 and 1023A were derived from human healthy fibroblasts and bone marrow, respectively. The human embryonic SC line H1 was obtained from WiCell, Wisconsin Materials [provider scientist: M. Sander, University of California; stock WA01 (H1)] and was used for coculture experiments because it expresses the HLA-A2. The SCs had normal karyotype (both XY), classical SC colony morphology, and expressed pluripotency markers, as shown in (19, 20, 73). SCs were maintained in E8 medium (Life Technologies) in matrigel-coated plates. SCs were differentiated into pancreatic β-like cells using a seven-step protocol as previously described (18, 20, 73, 74). After differentiation, stage 7 aggregates were dispersed. Briefly, aggregates were incubated in phosphate-buffered saline (PBS; Lonza) solution with 0.5 mM EDTA at room temperature for 6 min, exposed to Accumax (Sigma-Aldrich) for 8 min, and then dispersed by gentle pipetting. Knockout serum (Gibco, Thermo Fisher Scientific) was added to quench the dissociation process, and cells were seeded at 5 × 104 cells per 6.4-mm well in stage 7 medium. All the results shown for SC-derived β-like cells refer to independent biological samples, i.e., independent differentiations.
Immunofluorescence
The percentage of β cells in primary human islets and in SC-derived islet-like cells was assessed by immunofluorescence. The cells were fixed in 4% formaldehyde for 20 min, permeabilized with 0.5% Triton X-100 for 10 min, blocked with UltraV block (Thermo Fisher Scientific) for 10 min, and incubated with primary antibodies (table S3) diluted in 0.1% Tween in PBS overnight at 4°C. Following incubation with secondary antibodies for 1 hour at room temperature, samples were mounted with Vectashield with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and covered with glass coverslips.
Insulin secretion and content
Forty stage 7 aggregates were washed with glucose-free Krebs buffer (Univercell Biosolutions) in low-adhesion plates and preincubated in Krebs buffer for 30 min. Aggregates were exposed to 0 mM glucose, 20 mM glucose, or 20 mM glucose plus 10 μM forskolin for 1 hour. Aggregates were lysed by sonication and acid ethanol for determination of insulin content. Supernatants were stored at −80°C, and human insulin levels were measured by ELISA (Mercodia) according to the manufacturer’s instructions.
Cell preparation and sequencing for the scRNA-seq
HEL115.6-derived islet-like aggregates were washed once with Ham’s F-10 medium containing 3.75 g of free fatty acid–free BSA, 2.5 ml of GlutaMAX, and penicillin-streptomycin (100 U/ml) and transferred into a new 10-cm low-adherence dish containing 20 ml of the medium. Dishes were kept under constant agitation on an OrbiShaker CO2 (Benchmark Scientific). After 1 hour of rest, IFNα (2000 U/ml; PeproTech) was added or not (negative control). After 24 hours, aggregates were dispersed as described above. After that, cells were filtered using a 30-μm cell strainer (Corning) and resuspended in 200 μl of Dead Cell Removal MicroBeads (Dead Cell Removal kit, Miltenyi Biotec). The dead cells were removed according to the manufacturer’s instructions, using LS columns and MidiMACS separator (Miltenyi Biotec). The final cell suspension was centrifuged at 300 relative centrifugal force (RCF) for 3 min and resuspended into 1 ml of PBS + 5% BSA. The cell count and the cell viability were determined using a Countess II Automated Cell Counter and trypan blue solution (Thermo Fisher Scientific). The cells were used for scRNA-seq only if the following conditions were met: cell viability equal or superior to 80% and cell density between 700 and 1200 cells/μl. The single-cell gene expression analysis was conducted using a 10X Chromium single-cell system (10X Genomics Inc.) and a NovaSeq 6000 sequencer (Illumina Inc.) at the Center for Genomics and Bioinformatics, Indiana University, School of Medicine. More than 10,000 targeted cell recovery per sample were applied to a single-cell master mix with lysis buffer and reverse transcription reagents, following the Chromium NextGEM Single-Cell 3′ Reagent Kits User Guide, CG000204 Rev D (10X Genomics Inc.). Along with the single-cell gel beads and partitioning oil, the single-cell master mixture containing the single-cell suspension was dispensed onto Single-Cell Chip G in separate wells, and the chip was loaded to the Chromium Controller for GEM generation and barcoding, followed by cDNA synthesis and library preparation. At each step, the quality of cDNA and library was examined by Bioanalyzer (Agilent Technologies) and Qubit (Thermo Fisher Scientific). The resulting library was sequenced in a custom program for 28b plus 91b paired-end sequencing on Illumina NovaSeq 6000. This resulted in the sequencing of a range of 21,000 to 49,000 reads per cell over 8000 to 16,000 cells, depending on the sample.
Bulk RNA-seq from iPSC-derived islet-like cells
Total RNA was extracted from at least 200,000 cells collected at the end of dead cell removal step, using an RNeasy Plus mini kit according to the manufacturer’s instructions (Germantown). Total RNA was eluted in 30 μl of RNA-free water. Genomic DNA was removed by using deoxyribonuclease I (Thermo Fisher Scientific), and the purified RNA was recovered using the SurePrep RNA Cleanup and Concentration Kit, according to the manufacturer’s instructions (Fisher Scientific). The genomic DNA removal and cleanup step was repeated a second time. The purity of the finally obtained total RNA was evaluated using Bioanalyzer. Sequencing was performed on NovaSeq 6000 at the Center for Genomics and Bioinformatics, Indiana University, School of Medicine.
scRNA-seq processing and analysis
Raw scRNA-seq data (fastq) were aligned with Cell Ranger v6.0 (75) on the human genome GRCh38 in accordance with the instructions provided by 10X Genomics. In total, we obtained on average 390 million reads per sample (reads mapped confidently to genome > 94%). For each sample, we performed an initial clustering using the Seurat (76) pipeline, followed by a decontamination of the ambient mRNA [false discovery rate (FDR) > 0.01] with SoupX (77) using genes representing the major cell types preidentified in the initial clustering and other potential contaminant (INS, GCG, SST, TTR, IAPP, PYY, KRT19, TPH1, FEV, HLA-A, HLA-B, HLA-C, HLA-E, and B2M). Doublets were removed from the filtered counts with scDblFinder (78).
The decontaminated data were imported with the following thresholds: number of genes expressed > 1000, fraction of mitochondrial content < 10%, and total counts per cell > 3000. The new single-cell gene expression profiles were normalized with scran (79), and we scaled the data with Seurat. For each biological replicate, the top 1000 variable genes in each condition were identified using the “vst” method implemented in Seurat. In total, 2032 combined unique variable genes were used to perform a principal components analysis (PCA) analysis on all samples. The top 50 PCA components were selected to integrate the data with Harmony (80) with sample ID and stimulation as confounding factors (max.iter.harmony = 20, max.iter.cluster = 40). Integrated components were then used to build a Uniform Manifold Approximation and Projection (UMAP) (min.dist = 0.05) embedding of the 73,834 integrated cells. Unsupervised clustering of the integrated data was performed with the Louvain algorithm to determine the major cell types (resolution = 0.2, k = 10). Hallmark genes described in (30, 31) were used to validate the unsupervised clustering (see Fig. 1, G and H) along the “FindAllMarkers” function implemented in Seurat as an unbiased validation.
To evaluate the IFNα-induced changes on the cells, a DGE analysis was done on each identified cluster with MAST (Model-based Analysis of Single-Cell Transcriptomics) (81) (via the wrapper function available in Seurat) over genes expressed in at least 10% of the cell in either condition (control or IFNα-treated). Output results from the MAST analysis are available in data file S1.
Small RNA interference and treatment
NLRC5 gene expression was silenced using two independent siRNAs [Thermo Fisher Scientific; siNL#1 (siRNA HSS130675): 5′-GGACACCUGGCAGUCUUUCAUUCAU-3′; siNL#2 (siRNA HSS130676): 5′-GCAGUUGGCAGAGUCUCUCGUUCUU-3′]. AllStars Negative Control siRNA (siCTL) (Qiagen) was used as a negative control; the siRNA control does not interfere with β cell gene expression, function, or viability (82). NOVA1 gene expression was silenced using an siRNA [Thermo Fisher Scientific; siNO1 (siRNA HSS143142): 5′-UUUGCAACUGAACAAUUGUCUGUCC-3′] (44). Cells were transfected using the Lipofectamine RNAiMAX lipid reagent (Invitrogen, Life Technologies) in Opti-MEM (Gibco, Thermo Fisher Scientific) reduced serum medium, according to the manufacturer’s instructions. EndoC-βH1 was transfected using 30 nM of each siRNA, overnight. Dispersed human islets and SC-derived β-like cells were transfected with 60 nM of each siRNA, also during an overnight incubation period. After transfection, EndoC-βH1, dispersed human islets, and SC-derived β-like cells were kept in culture for a 48-hour recovery period and subsequently exposed, or not, to human IFNα (2000 U/ml; PeproTech) for 8 or 24 hours. These conditions are based not only on previously published time dose-response experiments (8) but also on time course and dose response experiments presented in this article (fig. S4).
Cell viability assessment
Cell viability was determined by fluorescence microscopy using the nuclear dye propidium iodide (10 μg/ml; Sigma-Aldrich) and Hoechst 33342 (10 μg/ml; Sigma-Aldrich), as previously described (82, 83). A minimum of 500 cells was counted per condition. Viability was evaluated by two independent observers, one of them being unaware of sample identity. The agreement between the two observers was >90%. The results are expressed as percentage of apoptosis, calculated as the number of apoptotic cells/total number of cells.
Quantitative real-time PCR and validation of splicing events
Poly(A)+ mRNA was isolated using the Dynabeads mRNA DIRECT Kit (Invitrogen) and reverse-transcribed using the Reverse Transcriptase Core Kit (Eurogentec), following the manufacturer’s protocol. The quantitative RT-PCR amplification reactions were performed with IQ SYBR Green Supermix (Bio-Rad Laboratories) and ran in the CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories). The product quantification was performed using the standard curve method (84). The target gene concentration was expressed as copies per microliter. The cycling conditions used were 95°C from 3 min, followed by 40 cycles of 95°C for 15 s, and 58°C for 20 s, followed by a final step of 95°C for 1 min, 70°C for 5 s, and 95°C for 50 s. For each gene, the melting curve was analyzed to confirm amplification of a single PCR product. Gene expression values were normalized by the geometric mean of the housekeeping genes β-actin and GAPDH. The geometric mean of β-actin and GAPDH was not modified by the IFNα treatment except for a mild decrease in human islets after 24 hours, which may be due to cell loss (EndoC-βH1: 5.47 × 104 ± 4.68 × 103 copies/μl in nontreated condition versus 5.71 × 104 ± 4.75 × 103 copies/μl in IFNα condition; human islets: 1.71 × 105 ± 3.37 × 104 copies/μl in nontreated condition versus 1.18 × 105 ± 2.22 × 104 copies/μl in IFNα condition (P < 0.01); iPSC-derived β-like cells: 4.77 × 104 ± 8.73 × 103 copies/μl in nontreated condition versus 4.93 × 104 ± 7.95 × 103 copies/μl in IFNα condition). Selected alternative splicing events were validated by semiquantitative RT-PCR using specifically designed primers encompassing the putative splicing change as described (67, 68). The primers were manually designed to target the constitutive flanking exons of the region of interest, allowing the simultaneous amplification of the different splice variants that were further distinguished based on amplicon size. The semiquantitative-PCR was performed using the RedTaq DNA polymerase (Bioline) following the manufacturer’s recommendations. The resulting PCR products were separated using the LabChip electrophoretic Agilent 2100 Bioanalyzer system using the DNA 100 LabChip Kit (Agilent Technologies). The sequence of primers used for quantitative RT-PCR and semiquantitative-PCR is shown in table S4.
Protein extraction and Western blotting analysis
Cells were first washed with PBS buffer and then lysed using 1× Laemmli sample buffer [60 mM tris[hydroxymethyl]aminomethane (pH 6.8), 10% glycerol, 1.5% dithiothreitol, 1.5% 2-mercaptoethanol, 2% SDS, and 0.005% bromophenol blue] with protease and phosphatase inhibitor cocktails (Roche). Total protein was resolved on 10% SDS–polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes. The membranes were probed using specific antibodies against NLRC5 (anti-NLRC5 antibody, clone 3H8, Sigma-Aldrich), diluted 1:500, and anti–β-actin (Cell Signaling Technology) as a housekeeping protein, diluted 1:5000. The antibodies were diluted in 1× tris-buffered saline and 0.1% Tween 20 with 5% BSA. The primary antibodies were incubated overnight at 4°C, and then the membranes were probed with peroxidase-conjugated secondary antibodies (listed in table S3) for 1 hour at room temperature. The immunoreactive bands were detected using a chemiluminescent substrate (SuperSignal West Femto, Thermo Fisher Scientific) in a Bio-Rad ChemiDoc XRS+ system (Bio-Rad Laboratories). The ImageLab software (Bio-Rad Laboratories) was used for densitometric quantification of the bands. The values obtained were normalized by the housekeeping protein β-actin after background subtraction and then by the highest value of each experiment considered as 1.
Flow cytometry
The expression of HLA-ABC at the β cell surface was quantified by flow cytometry after transfection with siCTL or siNL#2, under nontreated condition and after IFNα (2000 U/ml) exposure for 24 hours. Briefly, the cells were blocked with Hanks’ balanced salt solution (HBSS; Gibco, Thermo Fisher Scientific) with 2% BSA for 30 min on ice. After blocking, the cells were incubated with conjugated primary antibody anti–HLA-ABC [phycoerythrin (PE) mouse anti-human HLA ABC; BD Biosciences] diluted 1:50 in HBSS + 2% BSA. The primary antibody was incubated for 30 min at 4°C to prevent HLA-ABC internalization. The cells were then washed in cold HBSS + 2% BSA and fixed with 4% paraformaldehyde (PFA). The cells were detached using Accutase (Capricorn) for 5 min at 37°C. Before the analysis, the cells were resuspended in PBS with 5 mM EDTA. Cells were analyzed using a BD LSRFortessa X-20 (BD Biosciences) flow cytometer. Data analysis and graphical representation were performed using FlowJo software version v10 (Tree Star, Ashland). In summary, the cells were identified on the basis of forward scatter (FSC) and side scatter (SSC) parameters, which allow the exclusion of cell debris and nonviable cells, due to the relative differences in, respectively, size and granularity of the cells. Next, the relation between FSC width and FSC height was used to eliminate cell doublets. Last, the number of HLA-ABC–positive cells was assessed in the dot blots. Positive and negative thresholds were set using an isotype immunoglobulin (Ig) control (PE mouse IgG1κ isotype control; BD Biosciences) with the same fluorophore (fig. S10A).
Analysis of CXCL10 secretion by ELISA
ELISA (Human CXCL10/IP-10 Immunoassay, Quantikine ELISA kit, R&D Systems) was used to quantify CXCL10 secretion to the culture medium (by 45,000 EndoC-βH1 cells/200 μl of culture medium and by 30,000 human islet cells/200 μl; culture time as indicated in the legends for the figures). The cell culture supernatants’ were collected and immediately stored at −80°C until samples’ processing. EndoC-βH1 and dispersed human islet samples under nontreated conditions, after transfection with siCTL or siNL#2, at 8- and 24-hour time points, were diluted twofold. EndoC-βH1, transfected with siCTL and siNL#2 after exposure to IFNα for 8 and 24 hours, was diluted 20-fold. The dispersed human islets transfected with siCTL and siNL#2 after exposure to IFNα for 8 and 24 hours were diluted 50-fold. The assay procedure and calculation of the results were conducted following the manufacturer’s recommendation.
RNA-seq from EndoC-βH1 cell line
Total RNA was isolated from EndoC-βH1 samples using the RNeasy Plus Micro Kit (Qiagen) and quantified using NanoDrop ND-1000 (Thermo Fisher Scientific). A minimum of 500 ng of high-quality RNA with RNA integrity number > 9 (as determined by Agilent Bioanalyzer) per sample and replicate was used for library preparation. RNA-seq was performed on NovaSeq 6000 (Eurofins).
Bulk RNA-seq processing and analysis
Raw RNA-seq data [2 × 150-bp paired-end with a depth of >200 million reads for EndoC-βH1 cells (Eurofins Genomics Europe Sequencing GmbH, Konstanz, Germany) and 2 × 100-bp paired-end with a depth of >50 million reads for iPSC-derived islet-like cells (Center for Genomics and Bioinformatics, Indiana University, School of Medicine)] were quality-checked with fastp (85) to remove reads with poor quality and trim remaining adapters. The quality control (QC) of the passing reads was assessed with FastQC (Babraham Bioinformatics). One paired experiment for the IFNα treatment at 24 hours did not pass our QC (insufficient read quality) and was removed from the subsequent analyses.
Gene expression was quantified in TPM (transcripts per million) using Salmon v1.4 (86) with additional parameters “--seqBias --gcBias –validateMappings.” We used GENCODE v35 (87) as the reference genome and indexed it with default k-mer values. DGE analysis was performed with DESeq2 v1.30.1 (88). Briefly, for each gene included in the DESeq2 model, a log2FC value was obtained on the basis of the comparison performed (e.g., EndoC-βH1 cells exposed to IFNα during 24 hours with or without siNL#2). Each of those values is associated to a Wald test statistic, a P value, and an adjusted P value (Benjamini-Hochberg correction). The thresholds to consider a gene as differentially expressed were |log2FC| > 0.58 and adjusted P < 0.05. As all samples were paired by biological replicates, we used the experiment number as a confounding factor in the general linear model of DESeq2.
The identification of genes associated with splicing regulation was based on the association with the Gene Ontology term GO: 0008380 “RNA splicing.” No |FC| cutoff was applied for the identification of differentially expressed splicing regulators.
Gene set enrichment analysis
GSEA was done using the previously generated DGE results in bulk and scRNA-seq data. The enrichment or depletions in metabolic pathways were assessed with fGSEA (89) using the KEGG (90) and REACTOME (91) databases as references. For the bulk RNA-seq data analysis, genes were preranked according to the value of their Wald test statistics obtained through the DESeq2 analysis (data file S2). For scRNA-seq, we preranked the data using the average log2FC obtained with MAST (81) (data file S3). In both analyses, an up- or down-regulated pathway (depending on the direction of their enrichment score) was considered significant when adjusted P < 0.05.
For the genes affected by alternative splicing modifications after IFNα exposure alone or NLRC5 depletion followed by IFNα exposure at different time points, the functional enrichment analysis was performed with clusterProfiler (92) for Gene Ontology. Genes identified by RNA-seq in control or after NLRC5 KD and IFNα with normalized read counts >1 were considered as expressed and hence used as background. The standard parameters were applied, and adjusted P < 0.05 was considered to be statistically significant.
RRHO analysis
The inversion of the IFNα-induced signature in EndoC-βH1 cells following NLRC5 KD was computed with the RRHO algorithm (93). RRHO is a method to highlight concordance and discordance between two ranked lists. In short, we ranked genes in each analysis from the most up-regulated to the most down-regulated. An intersection of both lists was then performed to ensure that the same elements were present and the list of similar length. Last, the analysis of the order of the ranked genes was performed with a hypergeometric test. The RRHO map interpretation guidelines are illustrated in fig. S6D.
Pseudo-bulk analysis
Comparison between scRNA-seq of β cells from ND and T1D donors was performed by reprocessing the raw data available on the HPAP portal (41). Briefly, we downloaded raw fastq files from 15 ND donors and 8 T1D donors and processed the data with the scRNA-seq pipeline described above, except for the mRNA decontamination that was performed on another set of genes (INS, TTR, GCG, IAPP, SST, and KRT19). β Cells were identified after Louvain clustering (resolution = 0.3), and data were merged into pseudo-bulk (i.e., aggregation of the counts of the same cell type at the sample level). DGE was then performed on the preidentified β cell cluster as described for bulk RNA-seq.
Coculture between SC-derived islet-like cells and CD8+ T cells
H1-derived islet-like cells were transfected with siCTL or siNL#2 and were then left untreated or treated with IFNα (2000 U/ml) for 24 hours. The cells were detached using Accutase for 5 min at 37°C, and then the HLA-ABC expression at the cell surface was evaluated by flow cytometry and the NLRC5 KD efficiency by Western blotting. For the coculture, 100,000 H1-derived islet-like cells were cultured with 50,000 HLA-A2 PPI-specific CD8+ T cells (42) in human islet medium supplemented with IL-2 (25 U/ml; R&D Systems), IL-15 (5 ng/ml; PeproTech), and anti-human CD107a–fluorescein isothiocyanate (Thermo Fisher Scientific) in ultralow attachment U-bottom plates (Corning). The cocultured cells were then incubated at 37°C for 4 hours. Cells were washed with flow cytometry buffer (2% BSA in PBS with 5 mM EDTA) and then incubated with mouse anti-human CD8 allophycocyanin (APC) (BD Biosciences) at 4°C for 30 min. Last, the cells were fixed with 4% PFA at 4°C for 20 min and analyzed using a BD LSRFortessa X-20 flow cytometer (San Jose, CA, USA).
The percentage of CD107a+ of the CD8+ T cells was calculated using FlowJo software version v10. Briefly, the single cells were identified as described above, and then CD8+ T cells were first selected to allow the calculation of the percentage of CD107a+ (fig. S10B).
Alternative splicing analysis
Samples were aligned with STAR v2.7.6a (94) to GENCODE v35 reference genome (87), in agreement with the guidelines provided by rMATS-Turbo (v4.1.1) (95). rMATS-Turbo was run over the BAM files generated by STAR as paired experiments with the recommended defaults parameters. We specified read type, read length, and strand specificity with “--variable-read-length --readLength 150 –paired-stats --libType fr-firststrand.”
The alternative splicing events classified by the rMATS software were identified on the basis of junction-spanning reads. They included SE, alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), MXEs, and retained intron (RI). The alternative splicing event significance level was FDR < 0.05. Potential candidates were further filtered by the absolute change in their “percent spliced-in” (PSI) value (|ΔPSI| > 0.05).
Statistical analysis
Data are shown as means ± SEM. Significant differences between experimental conditions were determined by paired t test or by analysis of variance (ANOVA) followed by Bonferroni correction as indicated. P < 0.05 was considered statistically significant. The statistical tools used for the analysis of the RNA-seq experiments are described above.
Acknowledgments
We are grateful to I. Millard, A. Musuaya, N. Pachera, C. Ying, and M. Depessemier (ULB Center for Diabetes Research) for providing excellent technical support; O. Ballew (Indiana Biosciences Research Institute, Indianapolis) for helpful discussions; Y. Liu, X. Xuei, M. Luthra, P. McGuire, and J. Li (Center for Genomics and Bioinformatics, Indiana University, School of Medicine) for help and advice in the scRNA-seq of iPSC-derived islet-like cells; and E. Bosi (Department of Experimental and Clinical Medicine, Pancreatic Islets Laboratory, University of Pisa, Pisa) for helpful advice for the scRNA-seq analysis. Figure 1A was made with BioRender.
Funding: D.L.E. acknowledges the support of grants from the Welbio-FNRS (Fonds National de la Recherche Scientifique) (WELBIO-CR-2019C-04), Belgium; the Dutch Diabetes Research Foundation (Innovate2CureType1), Holland; the JDRF (3-SRA-2022-1201-S-B); the National Institutes of Health Human Islet Research Network Consortium on Beta Cell Death and Survival from Pancreatic β-Cell Gene Networks to Therapy (HIRN-CBDS) (grant U01 DK127786); and the Innovative Medicines Initiative 2 Joint Undertaking under grant agreements 115797 (INNODIA) and 945268 (INNODIA HARVEST), supported by the European Union’s Horizon 2020 research and innovation programme. These joint undertakings receive support from the European Union’s Horizon 2020 research and innovation programme and European Federation of Pharmaceutical Industries and Associations (EFPIA), JDRF, and the Leona M. and Harry B. Helmsley Charitable Trust. F.S. was supported by a Research Fellow (Aspirant) fellowship from the Fonds National de la Recherche Scientifique (FNRS, Belgium) (grant FC 38603). P.M. was supported by the Italian Ministry of University and Research, PRIN 2017 (2017KAM2R5_005).
Author contributions: D.L.E., Â.C., F.S. S.M.-C., and M.I.A. conceived the study and planned the experiments. D.L.E. supervised the whole project. S.D. prepared the iPSC-derived islet-like cells for bulk RNA-seq and scRNA-seq. Â.C. and M.I.A. prepared the EndoC-βH1 samples for bulk RNA-seq. F.S., M.I.A., and M.L.C. performed bioinformatics analysis. Â.C., S.M.-C., M.I.A., A.O.d.B., and S.D. performed confirmation experiments for the RNA-seq. S.T. and A.Z. provided the immune cells for the coculture and supervised the experiments and analysis. L.M. and P.M. provided human islets and performed experiments with these cells. D.L.E. wrote the first draft of the manuscript, which was revised and approved by all authors.
Competing interests: D.L.E. received grant support from Eli Lilly and Co., Indianapolis, IN, for research on new approaches to protect pancreatic β cells in T1D (not directly related to the present study). The other authors declare no competing interests.
Data and materials availability: All raw and processed scRNA-seq and bulk RNA-seq data that were generated during this study have been deposited in NCBI Gene Expression Omnibus (GEO) under the superseries GSE203385 (subseries are as follows: GSE202852 for RNA-seq of EndoC-βH1, GSE203430 for RNA-seq of iPSC-derived islet-like cells, and GSE203384 for scRNA-seq of iPSC-derived islet-like cells). scRNA-seq data from T1D and ND donors are publicly available via the HPAP portal (https://hpap.pmacs.upenn.edu/), and sample identifiers used for the present study are available in table S1. All the software used for the present analysis are free and open source. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.Coppieters K. T., Dotta F., Amirian N., Campbell P. D., Kay T. W. H., Atkinson M. A., Roep B. O., von Herrath M. G., Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 209, 51–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pugliese A., Autoreactive T cells in type 1 diabetes. J. Clin. Invest. 127, 2881–2891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eizirik D. L., Pasquali L., Cnop M., Pancreatic β-cells in type 1 and type 2 diabetes mellitus: Different pathways to failure. Nat. Rev. Endocrinol. 16, 349–362 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Carre A., Richardson S. J., Larger E., Mallone R., Presumption of guilt for T cells in type 1 diabetes: Lead culprits or partners in crime depending on age of onset? Diabetologia 64, 15–25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallone R., Eizirik D. L., Presumption of innocence for beta cells: Why are they vulnerable autoimmune targets in type 1 diabetes? Diabetologia 63, 1999–2006 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Eizirik D. L., Szymczak F., Alvelos M. I., Martin F., From pancreatic β-cell gene networks to novel therapies for type 1 diabetes. Diabetes 70, 1915–1925 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson S. J., Rodriguez-Calvo T., Gerling I. C., Mathews C. E., Kaddis J. S., Russell M. A., Zeissler M., Leete P., Krogvold L., Dahl-Jørgensen K., von Herrath M., Pugliese A., Atkinson M. A., Morgan N. G., Islet cell hyperexpression of HLA class I antigens: A defining feature in type 1 diabetes. Diabetologia 59, 2448–2458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marroqui L., dos Santos R. S., op de beeck A., Coomans de Brachène A., Marselli L., Marchetti P., Eizirik D. L., Interferon-α mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia 60, 656–667 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Colli M. L., Ramos-Rodríguez M., Nakayasu E. S., Alvelos M. I., Lopes M., Hill J. L. E., Turatsinze J. V., Coomans de Brachène A., Russell M. A., Raurell-Vila H., Castela A., Juan-Mateu J., Webb-Robertson B. J. M., Krogvold L., Dahl-Jorgensen K., Marselli L., Marchetti P., Richardson S. J., Morgan N. G., Metz T. O., Pasquali L., Eizirik D. L., An integrated multi-omics approach identifies the landscape of interferon-α-mediated responses of human pancreatic beta cells. Nat. Commun. 11, 2584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coomans de Brachene A., Castela A., de Beeck A. O., Mirmira R. G., Marselli L., Marchetti P., Masse C., Miao W., Leit S., Evans-Molina C., Eiziri D. L., Preclinical evaluation of tyrosine kinase 2 inhibitors for human beta-cell protection in type 1 diabetes. Diabetes Obes. Metab. 22, 1827–1836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande S., Ward Platt M., The investigation and management of neonatal hypoglycaemia. Semin. Fetal Neonatal Med. 10, 351–361 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Jacovetti C., Matkovich S. J., Rodriguez-Trejo A., Guay C., Regazzi R., Postnatal β-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat. Commun. 6, 8084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elghazi L., Blandino-Rosano M., Alejandro E., Cras-Meneur C., Bernal-Mizrachi E., Role of nutrients and mTOR signaling in the regulation of pancreatic progenitors development. Mol. Metab. 6, 560–573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rezania A., Bruin J. E., Arora P., Rubin A., Batushansky I., Asadi A., O’Dwyer S., Quiskamp N., Mojibian M., Albrecht T., Yang Y. H. C., Johnson J. D., Kieffer T. J., Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Pagliuca F. W., Millman J. R., Gürtler M., Segel M., van Dervort A., Ryu J. H., Peterson Q. P., Greiner D., Melton D. A., Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balboa D., Saarimaki-Vire J., Otonkoski T., Concise review: Human pluripotent stem cells for the modeling of pancreatic β-cell pathology. Stem Cells 37, 33–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brusko T. M., Russ H. A., Stabler C. L., Strategies for durable β cell replacement in type 1 diabetes. Science 373, 516–522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demine S., Schiavo A. A., Marín-Cañas S., Marchetti P., Cnop M., Eizirik D. L., Pro-inflammatory cytokines induce cell death, inflammatory responses, and endoplasmic reticulum stress in human iPSC-derived beta cells. Stem Cell Res. Ther. 11, 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cosentino C., Toivonen S., Diaz Villamil E., Atta M., Ravanat J. L., Demine S., Schiavo A. A., Pachera N., Deglasse J. P., Jonas J. C., Balboa D., Otonkoski T., Pearson E. R., Marchetti P., Eizirik D. L., Cnop M., Igoillo-Esteve M., Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 46, 10302–10318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Franco E., Lytrivi M., Ibrahim H., Montaser H., Wakeling M. N., Fantuzzi F., Patel K., Demarez C., Cai Y., Igoillo-Esteve M., Cosentino C., Lithovius V., Vihinen H., Jokitalo E., Laver T. W., Johnson M. B., Sawatani T., Shakeri H., Pachera N., Haliloglu B., Ozbek M. N., Unal E., Yıldırım R., Godbole T., Yildiz M., Aydin B., Bilheu A., Suzuki I., Flanagan S. E., Vanderhaeghen P., Senée V., Julier C., Marchetti P., Eizirik D. L., Ellard S., Saarimäki-Vire J., Otonkoski T., Cnop M., Hattersley A. T., YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J. Clin. Invest. 130, 6338–6353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meissner T. B., Li A., Kobayashi K. S., NLRC5: A newly discovered MHC class I transactivator (CITA). Microbes Infect. 14, 477–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell M. A., Redick S. D., Blodgett D. M., Richardson S. J., Leete P., Krogvold L., Dahl-Jørgensen K., Bottino R., Brissova M., Spaeth J. M., Babon J. A. B., Haliyur R., Powers A. C., Yang C., Kent S. C., Derr A. G., Kucukural A., Garber M. G., Morgan N. G., Harlan D. M., HLA class II antigen processing and presentation pathway components demonstrated by transcriptome and protein analyses of islet β-cells from donors with type 1 diabetes. Diabetes 68, 988–1001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staehli F., Ludigs K., Heinz L. X., Seguín-Estévez Q., Ferrero I., Braun M., Schroder K., Rebsamen M., Tardivel A., Mattmann C., MacDonald H. R., Romero P., Reith W., Guarda G., Tschopp J., NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J. Immunol. 188, 3820–3828 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Yao Y., Wang Y., Chen F., Huang Y., Zhu S., Leng Q., Wang H., Shi Y., Qian Y., NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 22, 836–847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuenzel S., Till A., Winkler M., Häsler R., Lipinski S., Jung S., Grötzinger J., Fickenscher H., Schreiber S., Rosenstiel P., The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J. Immunol. 184, 1990–2000 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Shi T., Li J., NLRC5: A paradigm for NLRs in immunological and inflammatory reaction. Cancer Lett. 451, 92–99 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Colli M. L., Szymczak F., Eizirik D. L., Molecular footprints of the immune assault on pancreatic beta cells in type 1 diabetes. Front. Endocrinol. (Lausanne) 11, 568446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szymczak F., Colli M. L., Mamula M. J., Evans-Molina C., Eizirik D. L., Gene expression signatures of target tissues in type 1 diabetes, lupus erythematosus, multiple sclerosis, and rheumatoid arthritis. Sci. Adv. 7, eabd7600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvelos M. I., Juan-Mateu J., Colli M. L., Turatsinze J. V., Eizirik D. L., When one becomes many—Alternative splicing in β-cell function and failure. Diabetes Obes. Metab. 20 (Suppl. 2), 77–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veres A., Faust A. L., Bushnell H. L., Engquist E. N., Kenty J. H. R., Harb G., Poh Y. C., Sintov E., Gürtler M., Pagliuca F. W., Peterson Q. P., Melton D. A., Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balboa D., Barsby T., Lithovius V., Saarimäki-Vire J., Omar-Hmeadi M., Dyachok O., Montaser H., Lund P.-E., Yang M., Ibrahim H., Näätänen A., Chandra V., Vihinen H., Jokitalo E., Kvist J., Ustinov J., Nieminen A. I., Kuuluvainen E., Hietakangas V., Katajisto P., Lau J., Carlsson P.-O., Barg S., Tengholm A., Otonkoski T., Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cell. Nat. Biotechnol. 40, 1042–1055 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dettmer R., Niwolik I., Cirksena K., Yoshimoto T., Tang Y., Mehmeti I., Gurgul-Convey E., Naujok O., Proinflammatory cytokines induce rapid, NO-independent apoptosis, expression of chemotactic mediators and interleukin-32 secretion in human pluripotent stem cell-derived beta cells. Diabetologia 65, 829–843 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cottet S., Dupraz P., Hamburger F., Dolci W., Jaquet M., Thorens B., SOCS-1 protein prevents Janus Kinase/STAT-dependent inhibition of beta cell insulin gene transcription and secretion in response to interferon-gamma. J. Biol. Chem. 276, 25862–25870 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Flodstrom M., Maday A., Balakrishna D., Cleary M. M., Yoshimura A., Sarvetnick N., Target cell defense prevents the development of diabetes after viral infection. Nat. Immunol. 3, 373–382 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Flodström-Tullberg M., Yadav D., Hägerkvist R., Tsai D., Secrest P., Stotland A., Sarvetnick N., Target cell expression of suppressor of cytokine signaling-1 prevents diabetes in the NOD mouse. Diabetes 52, 2696–2700 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Santin I., Moore F., Grieco F. A., Marchetti P., Brancolini C., Eizirik D. L., USP18 is a key regulator of the interferon-driven gene network modulating pancreatic beta cell inflammation and apoptosis. Cell Death Dis. 3, e419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colli M. L., Hill J. L. E., Marroquí L., Chaffey J., dos Santos R. S., Leete P., Coomans de Brachène A., Paula F. M. M., op de Beeck A., Castela A., Marselli L., Krogvold L., Dahl-Jorgensen K., Marchetti P., Morgan N. G., Richardson S. J., Eizirik D. L., PDL1 is expressed in the islets of people with type 1 diabetes and is up-regulated by interferons-α and-γ via IRF1 induction. EBioMedicine 36, 367–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Op de Beeck A., Eizirik D. L., Viral infections in type 1 diabetes mellitus—Why the β cells? Nat. Rev. Endocrinol. 12, 263–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neerincx A., Castro W., Guarda G., Kufer T. A., NLRC5, at the heart of antigen presentation. Front. Immunol. 4, 397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggensperger S., Tampe R., The transporter associated with antigen processing: A key player in adaptive immunity. Biol. Chem. 396, 1059–1072 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Kaestner K. H., Powers A. C., Naji A.; HPAP Consortium, Atkinson M. A., NIH initiative to improve understanding of the pancreas, islet, and autoimmunity in type 1 diabetes: The Human Pancreas Analysis Program (HPAP). Diabetes 68, 1394–1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skowera A., Ellis R. J., Varela-Calviño R., Arif S., Huang G. C., van-Krinks C., Zaremba A., Rackham C., Allen J. S., Tree T. I., Zhao M., Dayan C. M., Sewell A. K., Unger W. W., Drijfhout J. W., Ossendorp F., Roep B. O., Peakman M., CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 118, 3390–3402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leite N. C., Pelayo G. C., Melton D. A., Genetic manipulation of stress pathways can protect stem-cell-derived islets from apoptosis in vitro. Stem Cell Rep. 17, 766–774 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villate O., Turatsinze J. V., Mascali L. G., Grieco F. A., Nogueira T. C., Cunha D. A., Nardelli T. R., Sammeth M., Salunkhe V. A., Esguerra J. L. S., Eliasson L., Marselli L., Marchetti P., Eizirik D. L., Nova1 is a master regulator of alternative splicing in pancreatic beta cells. Nucleic Acids Res. 42, 11818–11830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juan-Mateu J., Rech T. H., Villate O., Lizarraga-Mollinedo E., Wendt A., Turatsinze J. V., Brondani L. A., Nardelli T. R., Nogueira T. C., Esguerra J. L. S., Alvelos M. I., Marchetti P., Eliasson L., Eizirik D. L., Neuron-enriched RNA-binding proteins regulate pancreatic beta cell function and survival. J. Biol. Chem. 292, 3466–3480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tranchevent L. C., Aubé F., Dulaurier L., Benoit-Pilven C., Rey A., Poret A., Chautard E., Mortada H., Desmet F. O., Chakrama F. Z., Moreno-Garcia M. A., Goillot E., Janczarski S., Mortreux F., Bourgeois C. F., Auboeuf D., Identification of protein features encoded by alternative exons using Exon Ontology. Genome Res. 27, 1087–1097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geraghty D. E., Wei X. H., Orr H. T., Koller B. H., Human leukocyte antigen F (HLA-F). An expressed HLA gene composed of a class I coding sequence linked to a novel transcribed repetitive element. J. Exp. Med. 171, 1–18 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle L. H., Gillingham A. K., Munro S., Trowsdale J., Selective export of HLA-F by its cytoplasmic tail. J. Immunol. 176, 6464–6472 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Morgan N. G., Richardson S. J., Fifty years of pancreatic islet pathology in human type 1 diabetes: Insights gained and progress made. Diabetologia 61, 2499–2506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Duque S., Azoury M. E., Colli M. L., Afonso G., Turatsinze J.-V., Nigi L., Lalanne A. I., Sebastiani G., Carré A., Pinto S., Culina S., Corcos N., Bugliani M., Marchetti P., Armanet M., Diedisheim M., Kyewski B., Steinmetz L. M., Buus S., You S., Dubois-Laforgue D., Larger E., Beressi J.-P., Bruno G., Dotta F., Scharfmann R., Eizirik D. L., Verdier Y., Vinh J., Mallone R., Conventional and neo-antigenic peptides presented by β cells are targeted by circulating naive CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab. 28, 946–960.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Marroqui L., Masini M., Merino B., Grieco F. A., Millard I., Dubois C., Quesada I., Marchetti P., Cnop M., Eizirik D. L., Pancreatic α cells are resistant to metabolic stress-induced apoptosis in type 2 diabetes. EBioMedicine 2, 378–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marroqui L., Lopes M., dos Santos R. S., Grieco F. A., Roivainen M., Richardson S. J., Morgan N. G., Op de beeck A., Eizirik D. L., Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α and β cells. eLife 4, e06990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson A. M., Landry L. G., Alkanani A. A., Pyle L., Powers A. C., Atkinson M. A., Mathews C. E., Roep B. O., Michels A. W., Nakayama M., Human islet T cells are highly reactive to preproinsulin in type 1 diabetes. Proc. Natl. Acad. Sci. U.S.A. 118, e2107208118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neerincx A., Rodriguez G. M., Steimle V., Kufer T. A., NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J. Immunol. 188, 4940–4950 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Choi N. M., Majumder P., Boss J. M., Regulation of major histocompatibility complex class II genes. Curr. Opin. Immunol. 23, 81–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skog O., Korsgren S., Wiberg A., Danielsson A., Edwin B., Buanes T., Krogvold L., Korsgren O., Dahl-Jørgensen K., Expression of human leukocyte antigen class I in endocrine and exocrine pancreatic tissue at onset of type 1 diabetes. Am. J. Pathol. 185, 129–138 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Knip M., Luopajarvi K., Harkonen T., Early life origin of type 1 diabetes. Semin. Immunopathol. 39, 653–667 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Kumar H., Pandey S., Zou J., Kumagai Y., Takahashi K., Akira S., Kawai T., NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J. Immunol. 186, 994–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Flamez D., Roland I., Berton A., Kutlu B., Dufrane D., Beckers M. C., de Waele E., Rooman I., Bouwens L., Clark A., Lonneux M., Jamar J. F., Goldman S., Maréchal D., Goodman N., Gianello P., van Huffel C., Salmon I., Eizirik D. L., A genomic-based approach identifies FXYD domain containing ion transport regulator 2 (FXYD2)gammaa as a pancreatic beta cell-specific biomarker. Diabetologia 53, 1372–1383 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Roep B. O., Atkinson M., von Herrath M., Satisfaction (not) guaranteed: Re-evaluating the use of animal models of type 1 diabetes. Nat. Rev. Immunol. 4, 989–997 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Ramos-Rodriguez M., Raurell-Vila H., Colli M. L., Alvelos M. I., Subirana-Granés M., Juan-Mateu J., Norris R., Turatsinze J.-V., Nakayasu E. S., Webb-Robertson B.-J. M., Inshaw J. R. J., Marchetti P., Piemonti L., Esteller M., Todd J. A., Metz T. O., Eizirik D. L., Pasquali L., The impact of proinflammatory cytokines on the β-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nat. Genet. 51, 1588–1595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eizirik D. L., Sammeth M., Bouckenooghe T., Bottu G., Sisino G., Igoillo-Esteve M., Ortis F., Santin I., Colli M. L., Barthson J., Bouwens L., Hughes L., Gregory L., Lunter G., Marselli L., Marchetti P., McCarthy M. I., Cnop M., The human pancreatic islet transcriptome: Expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLOS Genet. 8, e1002552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren P., Lu L., Cai S., Chen J., Lin W., Han F., Alternative splicing: A new cause and potential therapeutic target in autoimmune disease. Front. Immunol. 12, 713540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nogueira T. C., Paula F. M., Villate O., Colli M. L., Moura R. F., Cunha D. A., Marselli L., Marchetti P., Cnop M., Julier C., Eizirik D. L., GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLOS Genet. 9, e1003532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki A., Terao C., Yamamoto K., Linking of genetic risk variants to disease-specific gene expression via multi-omics studies in rheumatoid arthritis. Semin. Arthritis Rheum. 49, S49–S53 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Dlamini Z., Mokoena F., Hull R., Abnormalities in alternative splicing in diabetes: Therapeutic targets. J. Mol. Endocrinol. 59, R93–R107 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Juan-Mateu J., Alvelos M. I., Turatsinze J. V., Villate O., Lizarraga-Mollinedo E., Grieco F. A., Marroquí L., Bugliani M., Marchetti P., Eizirik D. L., SRp55 regulates a splicing network that controls human pancreatic β-cell function and survival. Diabetes 67, 423–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alvelos M. I., Brüggemann M., Sutandy F. X. R., Juan-Mateu J., Colli M. L., Busch A., Lopes M., Castela Â., Aartsma-Rus A., König J., Zarnack K., Eizirik D. L., The RNA-binding profile of the splicing factor SRSF6 in immortalized human pancreatic β-cells. Life Sci. Alliance 4, e202000825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moss N. D., Sussel L., mRNA processing: An emerging frontier in the regulation of pancreatic β cell function. Front. Genet. 11, 983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marselli L., Piron A., Suleiman M., Colli M. L., Yi X., Khamis A., Carrat G. R., Rutter G. A., Bugliani M., Giusti L., Ronci M., Ibberson M., Turatsinze J. V., Boggi U., de Simone P., de Tata V., Lopes M., Nasteska D., de Luca C., Tesi M., Bosi E., Singh P., Campani D., Schulte A. M., Solimena M., Hecht P., Rady B., Bakaj I., Pocai A., Norquay L., Thorens B., Canouil M., Froguel P., Eizirik D. L., Cnop M., Marchetti P., Persistent or transient human β cell dysfunction induced by metabolic stress: Specific signatures and shared gene expression with type 2 diabetes. Cell Rep. 33, 108466 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Brozzi F., Nardelli T. R., Lopes M., Millard I., Barthson J., Igoillo-Esteve M., Grieco F. A., Villate O., Oliveira J. M., Casimir M., Bugliani M., Engin F., Hotamisligil G. S., Marchetti P., Eizirik D. L., Cytokines induce endoplasmic reticulum stress in human, rat and mouse beta cells via different mechanisms. Diabetologia 58, 2307–2316 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P., Scharfmann R., A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 121, 3589–3597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lytrivi M., Senée V., Salpea P., Fantuzzi F., Philippi A., Abdulkarim B., Sawatani T., Marín-Cañas S., Pachera N., Degavre A., Singh P., Derbois C., Lechner D., Ladrière L., Igoillo-Esteve M., Cosentino C., Marselli L., Deleuze J. F., Marchetti P., Eizirik D. L., Nicolino M., Chaussenot A., Julier C., Cnop M., DNAJC3 deficiency induces β-cell mitochondrial apoptosis and causes syndromic young-onset diabetes. Eur. J. Endocrinol. 184, 455–468 (2021). [DOI] [PubMed] [Google Scholar]
- 74.Igoillo-Esteve M., Oliveira A. F., Cosentino C., Fantuzzi F., Demarez C., Toivonen S., Hu A., Chintawar S., Lopes M., Pachera N., Cai Y., Abdulkarim B., Rai M., Marselli L., Marchetti P., Tariq M., Jonas J. C., Boscolo M., Pandolfo M., Eizirik D. L., Cnop M., Exenatide induces frataxin expression and improves mitochondrial function in Friedreich ataxia. JCI Insight 5, e134221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng G. X., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., Farland C. M., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young M. D., Behjati S., SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Germain P., Lun A., Macnair W., Robinson M., Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lun A. T., McCarthy D. J., Marioni J. C., A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P. R., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M., Linsley P. S., Gottardo R., MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moore F., Cunha D. A., Mulder H., Eizirik D. L., Use of RNA interference to investigate cytokine signal transduction in pancreatic beta cells. Methods Mol. Biol. 820, 179–194 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Moore F., Colli M. L., Cnop M., Esteve M. I., Cardozo A. K., Cunha D. A., Bugliani M., Marchetti P., Eizirik D. L., PTPN2, a candidate gene for type 1 diabetes, modulates interferon-gamma-induced pancreatic beta-cell apoptosis. Diabetes 58, 1283–1291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Overbergh L., Valckx D., Waer M., Mathieu C., Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 11, 305–312 (1999). [DOI] [PubMed] [Google Scholar]
- 85.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frankish A., Diekhans M., Ferreira A. M., Johnson R., Jungreis I., Loveland J., Mudge J. M., Sisu C., Wright J., Armstrong J., Barnes I., Berry A., Bignell A., Carbonell Sala S., Chrast J., Cunningham F., di Domenico T., Donaldson S., Fiddes I. T., García Girón C., Gonzalez J. M., Grego T., Hardy M., Hourlier T., Hunt T., Izuogu O. G., Lagarde J., Martin F. J., Martínez L., Mohanan S., Muir P., Navarro F. C. P., Parker A., Pei B., Pozo F., Ruffier M., Schmitt B. M., Stapleton E., Suner M. M., Sycheva I., Uszczynska-Ratajczak B., Xu J., Yates A., Zerbino D., Zhang Y., Aken B., Choudhary J. S., Gerstein M., Guigó R., Hubbard T. J. P., Kellis M., Paten B., Reymond A., Tress M. L., Flicek P., GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.G. Korotkevich, V. Sukhov, N. Budin, B. Shpak, M. N. Artyomov, A. Sergushichev, Fast gene set enrichment analysis. bioRxiv 060012 [Preprint]. 1 February 2021. 10.1101/060012. [DOI]
- 90.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C. D., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Viteri G., Weiser J., Wu G., Stein L., Hermjakob H., D’Eustachio P., The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plaisier S. B., Taschereau R., Wong J. A., Graeber T. G., Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shen S., Park J. W., Lu Z. X., Lin L., Henry M. D., Wu Y. N., Zhou Q., Xing Y., rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S4
Data S1 to S3