Abstract
The processing of DNA double-strand breaks is a critical event in nucleic acid metabolism. This is evidenced by the severity of phenotypes associated with deficiencies in this process in multiple organisms. The core component involved in double-strand break repair in eukaryotic cells is the Mre11-Rad50 protein complex, which includes a third protein, p95, in humans and Xrs2 in yeasts. Homologues of Mre11 and Rad50 have been identified in all kingdoms of life, while the Nbs1 protein family is found only in eukaryotes. In eukaryotes the Mre11-Rad50 complex has nuclease activity that is modulated by the addition of ATP. We have isolated the Mre11 and Rad50 homologues from the thermophilic archaeon Pyrococcus furiosus and demonstrate that the two proteins exist in a large, heat-stable complex that possesses single-strand endonuclease activity and ATP-dependent double-strand-specific exonuclease activity. These findings verify the identification of the P. furiosus Rad50 and Mre11 homologues and demonstrate that functional homologues with similar biochemical properties exist in all kingdoms of life.
The ability to properly repair DNA double-strand breaks (DSBs) is of paramount importance for cellular survival and maintenance of genomic stability. These breaks can be caused by ionizing radiation or genotoxic chemicals, or they can occur spontaneously during DNA synthesis or as part of a number of programmed cellular event such as mating type switching and meiotic crossing over in Saccharomyces cerevisiae and V(D)J recombination in the mammalian immune system (8, 12, 24). The importance of the repair pathways involved in processing DSBs has been demonstrated by the severe phenotypes observed in a number of organisms associated with the inability to repair this type of DNA damage (7, 17). The genes of the S. cerevisiae RAD52 epistasis group (RAD50 to 59, MRE11, XRS2, and RPA) have been determined to be the main effectors of DSB repair in this organism (7). The understanding of the functions of the gene products of this epistasis group has led to a greater understanding of DSB repair in multiple organisms, given that most of the members of the RAD52 epistasis group are conserved in a number of species including mammals (17).
Among the members of the RAD52 epistasis group, MRE11, RAD50, and XRS2 have been shown to function as a stable multiprotein complex that possesses nuclease activity (Mre11) and ATP-dependent DNA binding activity (Rad50) (6, 13, 18). In humans, the homologues of the MRE11 and RAD50 gene products have been previously isolated and found to be members of a multiprotein complex that possesses nuclease activity, DNA binding activity, and DNA unwinding activity (15, 16, 22). A third member of the human complex, p95, has been isolated and found to be the protein that is defective in the chromosomal instability disorder Nijmegen breakage syndrome (NBS) (3, 23). Furthermore, mutations in the hMRE11 gene lead to a disease similar to ataxia telangiectasia termed ataxia telangiectasia-like disorder (ATLD) (21). Patients with ATLD are similar to those with NBS in that they display an abnormal response to the induction of DNA DSBs. Genetic studies in S. cerevisiae have implicated the complex in the repair of DSBs by both the homologous recombination repair pathway and the nonhomologous end-joining pathway (9). Examination of the homologues of Mre11, Rad50, and p95 by sequence database survey has shown that Rad50 and Mre11 are conserved in all kingdoms of life, while functional homologues of Nbs1 exist only in eukaryotes (1). Furthermore, purification of the Escherichia coli Mre11 and Rad50 homologues SbcD and SbcC shows that there do not appear to be any additional factors associated with these proteins (4). Thus, it appears that Mre11 and Rad50 comprise the core enzymatic members of this conserved multiprotein machine. To date there has been no characterization of this evolutionarily conserved complex from an archaeon. While Mre11 homologues are readily observed in archaea, the identity of an archaeal Rad50 is unclear because the protein is a member of the structural maintenance of chromosomes (SMC) family of proteins, of which there are several homologues in a given organism.
We report here the isolation and initial characterization of the homologues of MRE11 and RAD50 from the thermophilic archaeon Pyrococcus furiosus. Our results demonstrate that the P. furiosus Mre11 (pfMre11) and Rad50 (pfRad50) proteins show conservation within critical core domains and form a large stable complex (pfMR50) that possesses both single-strand endonuclease activity and double-strand exonuclease activity with a 3′-to-5′ polarity. Despite the lack of overall sequence similarity, the archaeal Mre11-Rad50 complex is functionally similar to those from bacteria and eukaryotes.
MATERIALS AND METHODS
Identification and cloning of P. furiosus Mre11 and Rad50.
The sequence of human Mre11 (hMre11; AAC78721) was used in a BLAST search (tblastn at http://www.ncbi.nlm.nih.gov/blast/unfinishedgenome.html) to identify a homologue of hMre11 in the genome of P. furiosus. A single hit with an E (expect) value of 3e-07 was found, and this reading frame (pfMre11) was amplified by PCR from P. furiosus genomic DNA using the oligonucleotide primers 5′-AAAAAAAAAAACATATGAAGTTTGCTCACTTAGCCGATATTC-3′ and 5′-AAAAAAGGATCCCTATTATCTCGCACCACCAAGCCAGCTATC-3′. The resulting PCR product was digested with BamHI and NdeI and cloned into pET15b (Novagen) to create pET15b-pfMre11.
To identify a probable homologue to human Rad50 (hRad50) in archaea, we used the sequence of hRad50 (AAB07119) to perform a BLAST search (see above) against available archaeal genomes. In general, we found two hits per genome with E values in the range of 1e-26 and 1e-15 and which spanned the entire sequence of 1,300 residues of hRad50. One hit was generally annotated as “chromosome segregation protein,” whereas the other was annotated as “putative purine NTPase” (nucleoside triphosphatase). In particular, we obtained two isolated hits with E values of 6e-17 and 4e-17, respectively, in the genome of P. furiosus. Comparison with annotated homologues in other archaeal genomes showed that the former hit has the highest homology with chromosome segregation proteins whereas latter has the highest homology with a putative purine NTPase. The gene for the latter (encoding pfRad50) was found to be adjacent to the gene for pfMre11 in the P. furiosus genome. This arrangement is similar to that observed for the Mre11 and Rad50 homologues from E. coli, SbcC and SbcD (14). Based on this, we hypothesized that this putative purine NTPase from P. furiosus was the hRad50 homologue. The pfRad50 open reading frame was amplified by PCR from P. furiosus genomic DNA using the oligonucleotide primers 5′-AAAAAAAAAAAACATATGAAATTGGAGAGAGTGACTGTGAAAAAC-3′ and 5′-AAAAAAAAACGGCCGCTATTAAGAGACCACCTCCACCTTGGAG-3′. The resulting PCR product was digested with NdeI and EagI and cloned into pET27b (Novagen) to create pET27b-pfRad50.
Plasmid construction.
A segment of DNA carrying the ribosome binding site (RBS), the start codon, and the coding sequence of pfMre11 was amplified by PCR using pET15b-pfMre11 as the template and oligonucleotide primers 5′-AAAAAACGGCCGGATCCCGGGTACCGCGGCCGCGTCGAAATAATTTTGTTTAACTTTAAGAAGGAGAT-3′ and 5′-GCTAGTTATTGCTCAGCGGTGGCAGCC-3′. The resulting PCR product was digested with EagI and Bpu1102I and ligated into pET27b-pfRad50 to create pET27b-pfRad50-pfMre11. This bicistronic expression vector carries the T7 promoter, an RBS for pfRad50, the open reading frame of pfRad50, a short linker, the RBS for pfMre11, the open reading frame of pfMre11, and the T7 terminator. In this construct, only pfMre11 contains a six-histidine tag, present on the N terminus of the protein.
Expression and purification of the pfMR50.
Competent E. coli BL12(DE3) cells were transformed with pET27B-pfRad50-pfMre11; a single colony was used to inoculate 500 ml of Luria broth containing kanamycin, (50 μg/ml) and this culture was grown overnight. Then 120 ml of this overnight culture was used to seed 6 liters of Luria broth containing kanamycin sulfate (50 μg/ml), and cells were grown at 37°C with shaking to an optical density at 550 nm of 0.8. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.8 mM, and the culture was further incubated at 37°C. After 5 h, the cells were harvested by centrifugation and shock frozen in liquid nitrogen. To isolate the protein, the cells (typically 20 g) were resuspended in 40 ml of a mixture containing 50 mM sodium phosphate (pH 8.0), 500 mM NaCl, 1 mM imidazole, and 1 tablet of Complete EDTA-free protease inhibitor mix (Roche Molecular Biochemicals). Cells were disrupted by sonication, and insoluble matter was pelleted by centrifugation at 30,000 × g. The supernatant was saved and heat shocked at 75°C for 10 min. Precipitated protein was removed by centrifugation at 30,000 × g. The supernatant from this step was loaded on 5 ml of Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen, Carlsbad, Calif.), and the column was washed with 50 ml of 50 mM phosphate (pH 8.0)–500 mM NaCl–1 mM imidazole followed by 50 ml of 20 mM phosphate (pH 8.0)–200 mM NaCl–1 mM imidazole–5% glycerol. Bound protein was eluted with a linear gradient of 1 to 200 mM imidazole in 20 mM phosphate (pH 8.0)–200 mM NaCl–5% glycerol. Fractions from this column were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining; pfMre11 and pfRad50 were found to coelute from this column. The pfMre11- and pfRad50-containing fractions were pooled, titrated to pH 7.5, diluted by 2 with water, and loaded onto 10 ml of phosphocellulose column (Whatman) equilibrated with 20 mM Tris (pH 7.5)–100 mM NaCl–1 mM EDTA–5% glycerol. The column was washed with 50 ml of equilibration buffer, and the protein was eluted with a linear gradient from 10 to 250 mM phosphate in 20 mM Tris (pH 7.5)–100 mM NaCl–1 mM EDTA–5% glycerol. pfMre11 and pfRad50 eluted at ∼50 mM phosphate, and the pfMre11- and pfRad50-containing fractions were pooled. The pool was dialyzed against 20 mM Tris–500 mM NaCl–1 mM EDTA–5% glycerol. The fractions from the phosphocellulose column contained significantly more pfMre11 than pfRad50; thus, an aliquot of this pool was subjected to further purification using a Superose 6 HR 10/30 column and a fast protein liquid chromatography system. Proteins were injected on to a Superose 6 column and separated in 50 mM Tris (pH 8.0)–300 mM NaCl–1 mM dithiothreitol (DTT)–10% glycerol at a flow rate of 18 ml/h.
Nuclease assays.
One microgram of purified pfMre11 or pfMR50 was incubated with 1 μg of circular single-stranded φX174 DNA in 25 mM HEPES (pH 7.0)–25 mM NaCl–5 mM MnCl2 or 5 mM MgCl2–1 mM DTT at 50°C; aliquots were removed at the indicated times, and reactions were stopped by the addition of EDTA to 10 mM. One-tenth volume of 10× loading dye containing 3% SDS was added to each aliquot, and the reaction products were separated on 1.0% agarose–1× Tris-borate-EDTA gels containing 500 ng of ethidium bromide per ml.
Exonuclease activity was measured by monitoring the release of trichloroacetic acid-soluble mononucleotides from restriction enzyme-linearized 3H-pUC19 DNA. Briefly, 3H-pUC19 was digested with HincII and used as substrate in 300-μl reactions containing 25 mM HEPES-KOH (pH 7.0), 50 mM NaCl, 5 mM MnCl2, 5 mM MgCl2, 1 mM DTT, 1 nmol (total nucleotide) of 3H-pUC19, 1 mM ATP or adenylyl-imidodiphosphate (AMP-PNP), and 100 nM pfMR50. Reactions were performed at 50°C; 50-μl aliquots were removed, reactions were stopped by the addition of EDTA to 10 mM, and the aliquots were placed on ice. Following a 1-h incubation, 10 μl of salmon sperm DNA (2 mg/ml) and 100 μl of 10% trichloroacetic acid was added to each aliquot. Nucleic acid was precipitated for 10 min on ice, the samples were centrifuged for 10 min at 14,000 × g, and the supernatants were assayed for the released 3H-nucleoside monophosphate residues by liquid scintillation counting. Polarity was determined by labeling a 39-mer oligonucleotide at the 5′ end with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) as described by the manufacturer. The same oligonucleotide was 3′-end labeled by incubation with terminal deoxynucleotidyltransferase (Roche Biochemicals, Indianapolis, Ind.) in the presence of [α-32P]ddATP (Amersham, Chicago, Ill.) as described by the manufacturer. The labeled oligonucleotide was annealed to its complement and used in exonuclease assays. Exonuclease assays contained 50 mM HEPES (pH 7.0), 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 2 mM ATP, 1 mM DTT, 6 pmol of substrate oligonucleotide, and 6 pmol of pfMR50. Reactions were incubated at 50°C, and aliquots were removed at various times, mixed with loading dye containing 50% formamide, heat denatured, and resolved on 10% polyacrylamide–7 M urea gels. Reaction products were visualized by autoradiography.
RESULTS
Isolation of the P. furiosus MRE11 and RAD50 genes.
The P. furiosus homologue of Mre11 was found by BLAST search, and the sequence coding for this protein was similar to sequences of other Mre11 family members. pfMre11 is a 426-amino-acid protein with a predicted molecular mass of 49 kDa. pfMre11 is 29% identical and 42% similar to hMre11 in the conserved N-terminal domains of the two proteins. The four domains proposed to be critical for nuclease activity are conserved in pfMre11, suggesting that it possesses nuclease activity (Fig. 1A).
FIG. 1.
Conservation of pfMre11 and pfRad50. (A) Alignment of pfMre1 with homologues from humans (hMre11), S. cerevisiae (scMre11), and E. coli (EcSbcD), using the program CLUSTALW. The four conserved nuclease domains that have been described for the Mre11 family (I, II, III, and IV) are shown with key residues in bold. (B) Similar alignment of pfRad50, carried out using Rad50 homologues from humans (hRad50), S. cerevisiae (scRad50), and E. coli (EcSbcC). The two conserved ATP binding domains are shown with the key residues in bold (23a). Numbers indicate amino acid positions of the motifs; numbers in parentheses are the number of residues between conserved domains.
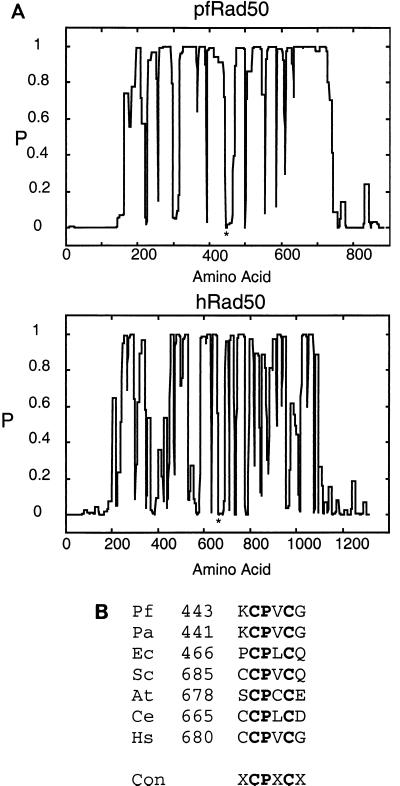
The gene encoding pfRad50 was found to be adjacent to the pfMre11-coding sequence within the P. furiosus genome, similar to that observed with the E. coli homologues SbcC and SbcD (14). This Rad50 gene homologue codes for an 882-amino-acid protein with a predicted molecular mass of 104 kDa that shows weak (19%) homology to the human protein. The key residues of the Walker A and B ATP boxes are conserved between the putative pfRad50 and other Rad50 homologues from a variety of species (Fig. 1B) (20). Analysis of pfRad50 and hRad50 with the program COILS (www.ch.embnet.org/software/COILS_form.html) shows that the central portion of each protein has a high probability to form an alpha-helical coiled-coil structure (Fig. 2A). pfRad50 contains only two cysteine residues that surprisingly occur in a conserved motif (Fig. 2B) at the center of the coiled-coil domain. Despite overall low sequence homology in this region, this motif is conserved in all Rad50 family members, and the predicted probability of forming a coiled-coil structure at these positions is zero. This suggests that this motif may be a hinge region in the center of the coiled-coil domain.
FIG. 2.
Structural conservation of pfRad50 and hRad50. (A) The human and P. furiosus Rad50 homologues were examined for the potential to form alpha-helical coiled-coil structures using the program COILS (see Materials and Methods). The plot shows the probability (P) of a given amino acid residue being present in an alpha-helical coiled-coil domain and indicates that the overall structure of the two proteins is conserved. The asterisk indicates the position of a conserved CPXC motif in each protein. (B) Sequence comparison of a number of Rad50 homologues shows the presence of a conserved (Con) CPXC motif in the central portion of each molecule. This region of the molecule is in a position that has a low probability of forming a coiled-coil structure (A), implicating it as a potential hinge region in the coiled-coil domain. Organisms listed are P. furiosus (Pf), Pyrococcus abyssi (Pa), E. coli (Ec), S. cerevisiae (Sc), Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), and Homo sapiens (Hs).
Expression and purification of pfMR50.
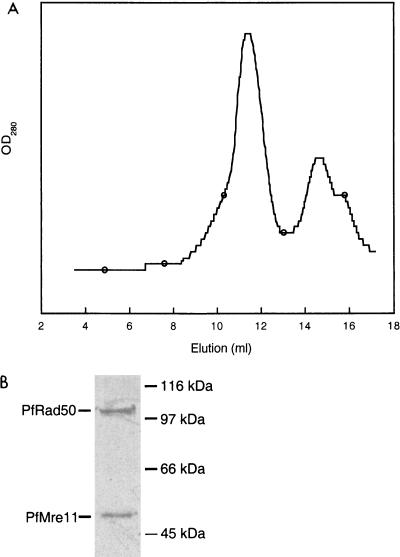
To test for the physical interaction of the two proteins, pfMre11 containing an N-terminal histidine tag and pfRad50 were coexpressed in E. coli using a bicistronic vector that produces a single mRNA encoding both proteins. After induction of expression, a soluble extract was made and heated to 75°C to denature endogenous E. coli proteins. Following centrifugation to remove denatured proteins, this heat-soluble extract was loaded on to an Ni2+-NTA-Sepharose column and bound proteins were eluted with an imidazole gradient. Proteins in the eluted fractions were separated by SDS-PAGE, and a band of the predicted molecular weight of pfMre11 was observed by Coomassie blue gel staining along with a coeluting band of the predicted molecular weight of pfRad50 (data not shown). In this fraction, pfMre11 was present at a greater concentration than pfRad50, suggesting that pfMre11 was expressed to a greater level than pfRad50. To separate the pfMR50 complex from excess pfMre11, fractions from the Ni2+ column were pooled, and an aliquot was subjected to gel filtration chromatography. Upon separation on a Superose 6 column, the Ni2+ fraction gave rise to two peaks, one containing the pfMR50 complex and the other containing only pfMre11. This confirmed that pfMre11 is produced in excess in this system (Fig. 3A). An aliquot of pfMR50 from the Superose 6 column was subjected to SDS-PAGE and Coomassie blue staining. Quantitation of pfMre11 and pfRad50 indicates that the two proteins are present in equimolar quantities (Fig. 3B). Thus, pfMre11 and pfRad50 form a heat-stable protein complex that appears to contain multiple copies of each subunit in equimolar amounts.
FIG. 3.
(A) Gel filtration analysis of pfMR50. pfMre11 and pfRad50 were coexpressed in E. coli and purified by Ni2+-affinity chromatography. This purified fraction was separated on a Superose 6 gel filtration column. The first peak that eluted from the gel filtration column is the coelution of the complex of pfMre11 and pfRad50, while the second peak corresponds to the elution of excess pfMre11. Comparison with native molecular weight markers indicates a native molecular mass of 106 Da for pfMR50. Purification through a heating step (see Materials and Methods), Ni2+-NTA-Sepharose chromatography, and coelution from a gel filtration column indicate that the two proteins exist in a stable complex. (B) SDS-PAGE of pfMR50 and Coomassie blue staining followed by quantitation of the bands by densitometry indicates that pfMre11 and pfRad50 are present in equimolar amounts.
Characterization of pfMR50.
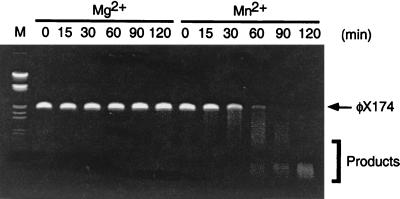
As other members of the Mre11 family have been reported to possess single-stranded DNA-specific endonuclease activity, we examined pfMre11 for this type of activity (4–6, 13, 15, 22). Purified protein were incubated at 50°C with single-stranded φX174 DNA, and the amount of degradation was assessed by gel electrophoresis and ethidium bromide staining. Using pfMre11 alone, we found that the protein was able to efficiently digest single-stranded DNA over the course of a standard 2-h incubation in the presence of Mn2+ (Fig. 4). No digestion was observed in the presence of Mg2+. We also examined the heterodimeric pfMR50 complex and found similar single-strand-specific endonuclease activity (data not shown).
FIG. 4.
Single-strand endonuclease activity of pfMre11. Based on the presence of the conserved nuclease domains in the N terminus of pfMre11, we examined the pfMre11 complex for nuclease activity. Purified protein was incubated with φX174 single-stranded DNA, and the extent of single-strand endonuclease activity was assessed by gel electrophoresis. The reactions were carried out in the presence of either 5 mM MgCl2 (Mg2+) or 5 mM MnCl2 (Mn2+). The results indicate that pfMre11 is a Mn2+-dependent single-strand endonuclease. Identical results were obtained with pfMR50 (data not shown).
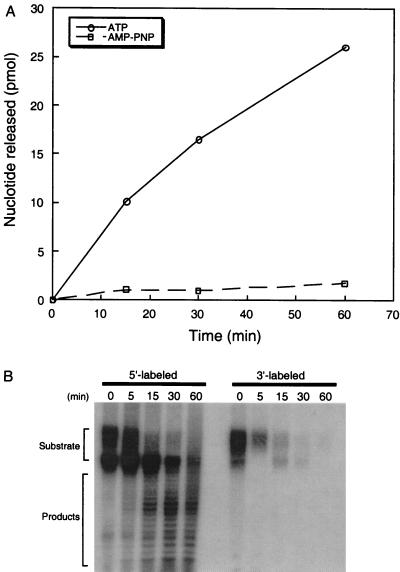
Studies of the E. coli Rad50 and Mre11 homologues SbcC and SbcD, respectively, have shown that the digestion of linear DNA by this complex requires the presence of ATP (4). Further work with the human Mre11-Rad50-Nbs1 complex has shown that ATP is required for the full spectrum of activities, including the ability to nick DNA hairpins and the ability to unwind a short DNA duplex (16). However, in both instances the hydrolysis of ATP could not be detected by direct means. We also examined the digestion of linear plasmid DNA by pfMR50. pfMR50 was able to digest linear plasmid DNA, and this nuclease activity absolutely required the presence of a hydrolyzable form of ATP (Fig. 5A). Reactions carried out in the presence of the nonhydrolyzable ATP analogue AMP-PNP failed to show any appreciable release of mononucleotides (Fig. 5). Furthermore, assays carried out in the absence of ATP did not show any release of acid-soluble mononucleotides above background levels (data not shown). Thus, the exonuclease activity of pfMR50 requires ATP hydrolysis, and the small amount of digestion observed in the presence of AMP-PNP but not in the absence of ATP suggests that ATP hydrolysis is necessary for release following binding and digestion.
FIG. 5.
Double-strand exonuclease activity of pfMR50. (A) Purified pfMR50 was incubated with linear 3H-pUC19 plasmid and 2 mM ATP (solid line) or 2 mM AMP-PNP (dashed line) for 1 h, and aliquots were removed at the times indicated. The release of acid-soluble mononucleotide at each time point was measured, and the results indicate that the exonuclease digestion requires the presence of ATP. Identical reactions done in the absence of ATP failed to show any release of mononucleotide above background levels (data not shown). (B) Purified pfMR50 was incubated with a double-strand oligonucleotide substrate labeled at the 5′ and 3′ ends (as indicated), and reaction aliquots were taken at the times indicated. Analysis of the products by denaturing PAGE and autoradiography indicates the presence of a ladder of reaction products with the 5′-labeled substrate, but no similar DNA laddering is observed with a 3′-labeled substrate. Thus, pfMR50 functions as a 3′-to-5′ exonuclease.
To examine the polarity of the exonuclease activity, an oligonucleotide substrate was differentially labeled on the 5′ and 3′ ends and annealed to its complement. The time course of digestion of this substrate revealed the presence of exonuclease breakdown products with the 5′-labeled substrate but not with the 3′-labeled substrate, indicating that the exonuclease proceeds in a 3′-to-5′ manner (Fig. 5B).
DISCUSSION
The activities described in this paper for pfMR50 are similar to those of the E. coli SbcCD complex and the human Mre11-Rad50-Nbs1 complex (4, 5, 15, 22). Additionally, the active site sequences of both pfRad50 and pfMre11 have been conserved as well as the predicted coiled-coil domain of pfRad50 (20). Thus, despite a lack of overall sequence conservation, it appears that the structure and function of the Mre11-Rad50 complex have been conserved throughout evolution. This is notable since there are very few DNA repair proteins that are conserved across all three kingdoms (1).
Biochemical analysis of the Mre11-Rad50 complexes from all three kingdom has demonstrated that the complex possesses exonuclease activity with a 3′-to-5′ polarity. In the case of the bacterial SbcCD and pfMR50, the exonuclease activity requires the hydrolysis of ATP (4). While this same experiment has not been described for the human Mre11-Rad50-p95 complex, there are activities that are stimulated by the addition of ATP, in particular, the ability of the human complex to nick a fully complementary hairpin and to unwind a short stretch of duplex DNA. We have observed a similar stimulation of DNA binding by ATP for the catalytic domain of pfRad50 along with a substantial conformational change (10). The binding of ATP appears to generate a DNA binding channel in the structure of the pfRad50 protein; given the inability of the human Mre11-Rad50-p95 complex to unwind DNA of longer than 34 bp, this suggests that DNA binding by Rad50 in this channel leads to a local melting of small stretches of duplex DNA. As the yeast Rad50 has been shown to have ATP-dependent DNA binding activity, it appears that this mode of DNA binding may be a universal characteristic of Rad50 from all species. The molecular nature of the ATP-dependent DNA binding that leads to the DNA melting is currently under investigation.
The conservation of the nuclease activity of Mre11 suggests a critical conserved function. Although this nuclease activity was initially thought to not be necessary for any mitotic function (13), work with E. coli (5, 8a), and recent results for yeast implicate this activity in the metabolism of hairpin-containing DNA (19). Additionally, recent data have shown that a mutation near the active site of hMre11 leads to ATLD, a disease similar to ataxia telangiectasia (21). Although this mutation, which changes asparagine 117 to serine, has not been examined biochemically, its position adjacent to conserved active-site sequences suggests that it affects nuclease activity. Furthermore, the addition of a third component, Xrs2 in yeast and Nbs1 in humans, likely reflects the expansion of functionalities of the Mre11-Rad50 complex within eukaryotes. Unlike archaea and bacteria, eukaryotic systems have the additional burden of sexual reproduction, cell cycle control, and persistent DNA ends in the form of telomeres. Nbs1 and Xrs2 may have evolved to carry out these eukaryote-specific functions.
The details of the in vivo functions of the Mre11-Rad50 complex are not clearly understood. pfMR50 has been observed to be a dumbbell-like molecule similar to other SMC family members (10, 11). This suggests that the Mre11-Rad50 complex plays a structural role in its DNA repair and recombination functions. A general premise of DNA DSB repair is that the two free DNA ends must be maintained in reasonable proximity to one another for accurate repair to take place. This premise is probably true regardless of whether a particular break is being repaired by homologous recombination or nonhomologous end joining. Additionally, in homologous recombination the template for repair is typically the sister chromatid, and recent genetic results have demonstrated a function for Mre11 and Rad50 in mediating sister chromatid interactions (2, 25). Therefore, the dumbbell-like molecules of Mre11-Rad50 could be crucial for the interaction of sister chromatids or DNA ends in order to allow proper repair to take place.
ACKNOWLEDGMENTS
We thank the members of the Tainer and Carney laboratories for helpful discussions. We also thank R. DiGate and A. Cseresnyes for the generous gift of the 3H-pUC19 DNA.
This work was supported by Laboratory Directed Research and Development funds of LBNL (J.A.T. and J.P.C.) and American Cancer Society research project grant RPG-00-059-01-CCE (to J.P.C.). K.P. was supported by a BASF fellowship through the Studienstiftung des Deutschen Volkes, and J.P.C. was supported in part by NIH grant GM53918 (to William F. Morgan).
REFERENCES
- 1.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bressan D A, Baxter B K, Petrini J H. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates III J R, Hays L, Morgan W F, Petrini J H. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 4.Connelly J C, de Leau E S, Okely E A, Leach D R. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 5.Connelly J C, Leach D R F. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 6.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Game J C. DNA double strand breaks and the RAD50-RAD57 genes in Saccharomyces. Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 8.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 8a.Gibson F P, Leach D R F, Lloyd R G. Identification of sbcD mutations as cosuppressors of recBC that allow propogation of DNA palindromes in Escherichia coli K-12. J Bacteriol. 1992;174:1222–1228. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 10.Hopfner K P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 11.Melby T E, Ciampaglio C N, Briscoe G, Erickson H P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreau S, Ferguson J R, Symington L. The nuclease activity of Mre11 is required for meiosis but not mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naom I S, Morton S J, Leach D R, Lloyd R G. Molecular organization of sbcC, a gene that affects genetic recombination and the viability of DNA palindromes in Escherichia coli K-12. Nucleic Acids Res. 1989;17:8033–8045. doi: 10.1093/nar/17.20.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 16.Paull T T, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrini J H, Bressan D A, Yao M S. The RAD52 epistasis group in mammalian double strand break repair. Semin Immunol. 1997;9:181–188. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- 18.Raymond W E, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard G F, Goellner G M, McMurray C T, Haber J E. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharples G J, Leach D R. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 21.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G J, Raams A, Byrd P J, Petrini J H J, Taylor A M R. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 22.Trujillo K M, Yuan S S, Lee E Y, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 23.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, Stumm M, Weemaes C M, Gatti R A, Wilson R K, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 23a.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward J F. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and repairability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi-Iwai Y, Sonoda E, Sasaki M S, Morrison C, Haraguchi T, Hiraoka Y, Yamashita Y M, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]