Abstract
The discovery of non-chromosomal circular DNA offers new directions in linking genome structure with function in plant biology. Glyphosate resistance through EPSPS gene copy amplification in Palmer amaranth was due to an autonomously replicating extra-chromosomal circular DNA mechanism (eccDNA). CIDER-Seq analysis of geographically distant glyphosate sensitive (GS) and resistant (GR) Palmer Amaranth (Amaranthus palmeri) revealed the presence of numerous small extra-chromosomal circular DNAs varying in size and with degrees of repetitive content, coding sequence, and motifs associated with autonomous replication. In GS biotypes, only a small portion of these aligned to the 399 kb eccDNA replicon, the vehicle underlying gene amplification and genetic resistance to the herbicide glyphosate. The aligned eccDNAs from GS were separated from one another by large gaps in sequence. In GR biotypes, the eccDNAs were present in both abundance and diversity to assemble into a nearly complete eccDNA replicon. Mean sizes of eccDNAs were similar in both biotypes and were around 5kb with larger eccDNAs near 25kb. Gene content for eccDNAs ranged from 0 to 3 with functions that include ribosomal proteins, transport, metabolism, and general stress response genetic elements. Repeat content among smaller eccDNAs indicate a potential for recombination into larger structures. Genomic hotspots were also identified in the Palmer amaranth genome with a disposition for gene focal amplifications as eccDNA. The presence of eccDNA may serve as a reservoir of genetic heterogeneity in this species and may be functionally important for survival.
Introduction
Extra-chromosomal circular DNA (eccDNA) are nucleus limited ring-like DNA entities derived from the genome and have been found in a wide range of eukaryotic organisms including yeast, Drosophila, Xenopus, mice, and humans [1–4]. In yeast, eccDNAs with functional genes and sizes of up to 38 kb that cover 23% of the genome have been reported [5]. EccDNAs have been reported in normal healthy cells in humans [6, 7] with functions associated with aging and the formation of telomeric circles [8, 9], cancer progression, and therapeutic resistance [10–12]. EccDNAs have been implicated in approximately half of all human cancers contributing to genetic heterogeneity that enables aggressive tumors with a selective advantage; hence the higher prevalence in malignant tumors [13–15]. Sizes of cancer related eccDNA have been reported to range from several hundred base pairs to 600 kb encoded with functional oncogenes and their various regulatory elements [16, 17]. In plants, eccDNAs have been reported in Arabidopsis [18, 19], Oryza, Pisum, Secale, Triticum, and Vicia [20, 21] with sizes that range from 1 kb to 50kb. These eccDNAs contain coding sequences commonly found within the nucleus such as ribosomal genes, tRNAs, and transposons [19, 22, 23]. EccDNAs are thought to arise from linear chromosomes through repeat-mediated intrachromosomal homologous recombination that results in the ‘looping-out’ of circular structures. These focal amplifications are mediated by multimers corresponding to 5S ribosomal DNA, non-coding chromosomal high-copy tandem repeats, and telomeric DNA [1, 21, 22]. In Arabidopsis, eccDNA genesis is the result of recombination among inverted repeats upstream and downstream of the various tRNAs and transposons [19]. Several follow up studies using Arabidopsis and rice, have shown that defective RNA polymerase II (Pol II) activity and simultaneous inhibition of DNA methylation leads to the activation of retrotransposons which can induce eccDNA formation upon stress [11]. These studies suggest a possible relationship among epigenetic status, regulation of transposon bursts, and genomic focal amplifications.
The presence of eccDNA with functional genes in a cell can be a signature of a stress and/or function as a reservoir of genetic variation in which a cell may activate as a rapid response to stress. For example, oncogene amplification and expression via eccDNA in human cancers provides a unique mechanism for massive gene expression [16] and ultimately a reservoir of genetic heterogeneity by which cancer cells have a selective advantage for aggressive behavior and persistence [13].
Recently, the genetic entity conferring resistance to the herbicide glyphosate in Palmer amaranth (Amaranthus palmeri), now termed the eccDNA replicon, was revealed to be a massive, 399 kb extrachromosomal circular DNA (eccDNA) [24, 25]. Glyphosate resistance in Palmer amaranth is achieved through replicon amplification with simultaneous gene copy amplification and expression of the 5-enoylpyruvylshikimate-3-phosphate synthase (EPSPS) gene and its product, EPSP synthase [26], which is the herbicide target of glyphosate [27]. Glyphosate resistance may occur with as few as 5 copies of EPSPS. The increase in EPSPS functions to ameliorate the unbalanced or unregulated metabolic changes, such as shikimate accumulation, loss of aromatic amino acids, phenolic acids for lignin synthesis, and structural intermediates for plant growth regulators associated with glyphosate activity in sensitive plants [26, 28]. Isolation and single-molecule sequencing of the replicon resulted in a single copy of the EPSPS gene along with 58 other predicted genes whose broad functions traverse detoxification, replication, recombination, DNA binding, and transport [25, 29]. Gene expression profiling of the replicon under glyphosate treatment showed transcription of 41 of the 59 genes in GR biotypes, with high expression of EPSPS, aminotransferase, zinc-finger, and several uncharacterized proteins [25, 29].
Repeat sequences and mobile genetic elements have been associated with eccDNA formation [4, 6, 18, 20, 29, 30] in higher eukaryotes. The repeat landscape of the replicon is described as a complex arrangement of repeat sequences and mobile genetic elements interspersed among arrays of clustered palindromes which may function in stability, DNA duplication and/or a means of nuclear integration [25]. In a follow up study, sequence analysis identified a region in the replicon with elevated A+T content and an exact match to a conserved eukaryotic extended autonomous consensus sequence (EACS) [31]. Surrounding this sequence were multiple DNA unwinding elements (DUE), which together are often associated with DNA bending and origins of replication and typically found near EACS [32, 33]. Regions flanking these elements in the replicon were cloned into an ARS-less yeast plasmid which resulted in colony formation, suggesting autonomous replication as the mechanism for the replicon increases in copy number [34].
Initial low-resolution FISH analysis of GR A. palmeri showed the amplified EPSPS gene was randomly distributed in the genome, suggesting a possible transposon-based mechanism of mobility [26]. A follow up study using much longer bacterial artificial chromosome (BAC) probes coupled with high resolution fiber extension microscopy verified the eccDNA replicon and identified various structural polymorphisms including intact, circular, dimerized circular, and linear forms [24]. Additionally, this study resolved a critical question regarding the maintenance mechanism that explains uneven segregation of glyphosate resistance among progenies–genomic tethering. Analysis of fiber-FISH images with replicon probes and meiotic pachytene chromosomes revealed very clear, single signals [24]. If the replicon were integrated into the genome, then double signals would be evident, suggesting a tethering mechanism as a means of genomic persistence to daughter cells during cell division [24]. Other genetic entities that maintain genomic persistence through tethering include DNA viruses such as Epstein-Barr, Rhadinovirus, Papillomavirus, and others [35].
Glyphosate resistance in Palmer amaranth has been observed in individuals with EPSPS copy numbers that range from 5–150 copies [26, 36, 37]. Amplification of the EPSPS gene correlated with amplification of flanking genes and sequence [25, 29], which suggests a large amplification unit and genome size enlargement in cells with many replicon copies [29]. Flow cytometry verified significant genome expansion in plants with high copy numbers (eg. 11% increase in genome size with ~100 extra copies of the replicon), seemingly without fitness penalty [29].
Glyphosate resistance in Palmer amaranth was originally reported in Georgia in the early 2,000’s [38], and a recent analysis using whole genome shotgun sequencing verified that the replicon was present and intact in GR Palmer amaranth populations across the USA [39, 40]. This study also reported a lack of replicon SNP variation among GR eccDNAs from geographically distant states when aligned to the Mississippi replicon reference [25]. The replicon was not present in GS individuals, which supports a single origin hypothesis and spread of the replicon across the USA through mechanical means such as spread of GR pollen in contaminated plant products, on farm equipment, and cattle movement, or via pollen.
The genomic mechanisms, origins and how the replicon assembled and gave rise to eccDNA in Palmer amaranth remains elusive, but the above studies lead to a couple of hypotheses: 1) the eccDNA replicon formed through intramolecular recombination among distal parts of the nuclear genome in short evolutionary time, or 2) there may exist a reservoir of smaller eccDNAs that are basal in the cell that may have the ability to recombine to assemble larger units as part of a dynamic response to stress. In this study, we report the presence and sequence characterization of an abundant reservoir of eccDNAs in both GS and GR biotypes using single molecule sequencing and the CIDER-Seq approach [18]. We examine the similarities and differences among samples representing distant geographic locations reported in [40], quantitate their abundance and diversity and assess whether recombination may be possible to form larger multimeric units.
Results
EccDNA content and coding structure in geographically distributed A. palmeri
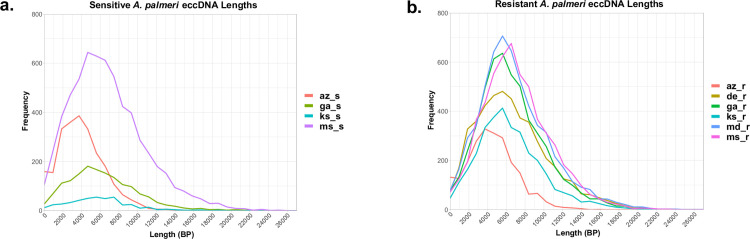
Following the general methods and recommended computational pipelines outlined in the CIDER-Seq single-molecule approach [18], we identified an extensive amount of variable-sized eccDNA in all samples of (GS) and GR) biotypes that were sequenced [Table 1]. The number of unique eccDNAs detected in GS samples ranged from 443 (ks_s) to 6,227 (ms_s) with a mean of 2,661 [Table 1]. Unique eccDNAs were in much higher abundance in GR samples and ranged from 2,200 (az_r) to 5,650 (ms_r), with a mean of 4,448, nearly double that of GS [Table 1]. Length distributions of eccDNA were similar among both GS and GR biotypes and ranged from 27bp to nearly 27kb, with mean lengths of around 6kb, [Table 1 and Fig 1]. Gene prediction resulted in eccDNAs both with and without complete open reading frames. In GS samples, the number of eccDNAs with predicted genes ranged from 76–505 with a mean of 272 eccDNAs with genes per sample. GR eccDNAs with predicted genes was nearly 4 times greater with a range of 263–1,179 and a mean of 718 eccDNA with genes per sample, suggesting that glyphosate stress influenced unique gene focal amplifications [Table 1]. Of the eccDNA with predicted genes, the number of predicted genes per eccDNA ranged from 1 to 10, with an average of 2 genes per eccDNA in both GS and GR [S1 and S2 Tables]. Transfer RNAs (tRNA) were predicted exclusively on eccDNA without CDS sequences and ranged widely from 46–715 (average of 350 per sample) in GS and samples and 130–528 (average of 364 per sample).
Table 1. EccDNA characterization of GS and GR biotypes.
| Samplebiotype | # eccDNA | # eccDNA with Genes | # eccDNA with tRNA | % eccDNA with CDS | Length Distribution | Mean length |
|---|---|---|---|---|---|---|
| ks_s | 443 | 76 | 46 | 17 | 60–22,237 | 5,906 |
| ga_s | 1564 | 257 | 183 | 16 | 57–22,477 | 6,273 |
| az_r | 2200 | 263 | 130 | 12 | 42–16,029 | 4461 |
| az_s | 2410 | 251 | 148 | 10 | 66–19,859 | 3999 |
| ks_r | 3234 | 578 | 267 | 18 | 35–23,960 | 6,400 |
| de_r | 4653 | 967 | 467 | 21 | 28–26,413 | 6668 |
| ga_r | 5138 | 978 | 528 | 19 | 30–23,670 | 6618 |
| ms_r | 5650 | 347 | 458 | 6 | 27–23,400 | 7,088 |
| md_r | 5817 | 1179 | 553 | 20 | 30–27,000 | 6,872 |
| ms_s | 6227 | 505 | 715 | 8 | 30–24,870 | 6,744 |
Fig 1. Frequency polygon graph for Lengths (bp) of A. palmeri eccDNAs.
A) Glyphosate sensitive samples were sourced from Arizona (az_s), Georgia (ga_s), Kansas (ks_s), and Mississippi (ms_s) A. palmeri plants. B) Glyphosate resistant samples were sourced from Arizona (az_r), Delaware (de_r), Georgia (ga_r), Kansas (ks_r), Maryland (md_r), and Mississippi (ms_r) plants.
Coding content of eccDNAs in glyphosate sensitive and resistant A. palmeri
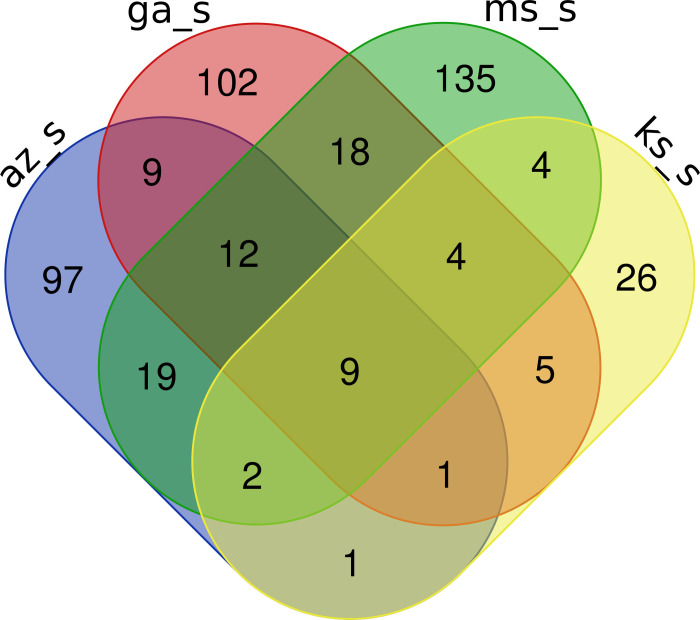
Gene content from both GS and GR biotypes was compared to identify unique and common functional protein coding domains among the geographically distant samples. In GS biotypes, 9 functional protein coding domains were discovered that are common among the each of the states [Fig 2]. These functional domains are annotated as ATP synthase, cytochrome P450, protein kinase, ribosomal protein, NADH dehydrogenase, Clp protease, and oxidoreductase [Table 2]. Various pairwise combinations of GS A. palmeri biotypes shared a range of 1 to 12 elements [Fig 2 and S3 Table]. Genes that regulate cell division, such as the Ras protein family and those involved in DNA replication (helicase) were common among Arizona, Georgia, and Mississippi GS eccDNA samples [S3 Table].
Fig 2. Venn diagram of PFAM elements shared by GS eccDNA samples.
Arizona (az_s), Georgia (ga_s), Kansas (ks_s), and Mississippi (ms_s) sensitive A. palmeri eccDNA shared 9 total PFAM elements.
Table 2. Gene elements shared by all sensitive eccDNA samples.
| Total | PFAM Accession | Annotation |
|---|---|---|
| 9 | PF00006 | ATP synthase alpha/beta family, nucleotide-binding domain |
| PF00067 | Cytochrome P450 | |
| PF00069 | Protein kinase domain | |
| PF00164 | Ribosomal protein S12/S23 | |
| PF00181 | Ribosomal Proteins L2, RNA binding domain | |
| PF00346 | Respiratory-chain NADH dehydrogenase, 49 Kd subunit | |
| PF00411 | Ribosomal protein S11 | |
| PF00574 | Clp protease | |
| PF01058 | NADH ubiquinone oxidoreductase, 20 Kd subunit |
Several abiotic/biotic resilience-related functional protein domains were found in Arizona and Mississippi GS samples that includes an oxysterol-binding protein, pectinesterase, NmrA-like family, and WRKY DNA-binding domain elements [S3 Table]. Also discovered were shared functional domains involved in DNA methylation and histone maintenance (H2A/H2B/H3/H4) [S3 Table]. Common between Georgia and Mississippi GS biotypes were ABC transporter and Cytochrome C oxidase subunit II (periplasmic domain) protein domains [S3 Table]. Unique to Arizona were response regulators such as trehalose-phosphatase, chalcone-flavanone isomerase, O-methyltransferase, Myb-like DNA-binding domain [S3 Table]. Hundreds of other unique functional domains in different GS biotypes were recorded in S3 Table. It is notable that the EPSPS gene was not found in any of the GS eccDNAs.
In GR biotypes, we identified a total of 20 functional protein domains that are shared among all 6 resistant samples [Table 3]. The shared GR domains had various cellular maintenance functions in addition to stress response domains that include ABC transporter, HSP70 protein, Ribosomal protein, WD domain, and Leucine rich repeats [Table 3]. A range of 1 to 9 protein family domains were shared by at least 5 of the GR biotypes [S4 Table]. No apical meristem (NAM) protein, peroxidase, TCP-1/cpn60 chaperonin family are among the stress response elements. Arizona, Delaware, Kansas, and Maryland GR biotypes all contained EPSP synthase (3-phosphoshikimate 1-carboxyvinyltransferase) and Arabidopsis phospholipase-like protein (PEARLI 4) functional domains, with 21 and 24 copies distributed across various eccDNA within these four samples respectively.
Table 3. Gene elements shared by all GR eccDNA samples.
| Total | PFAM Accession | Annotation |
|---|---|---|
| 20 | PF00004 | ATPase family associated with various cellular activities (AAA) |
| PF00005 | ABC transporter | |
| PF00006 | ATP synthase alpha/beta family, nucleotide-binding domain | |
| PF00012 | HSP70 protein | |
| PF00067 | Cytochrome P450 | |
| PF00069 | Protein kinase domain | |
| PF00076 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) | |
| PF00164 | Ribosomal protein S12/S23 | |
| PF00181 | Ribosomal Proteins L2, RNA binding domain | |
| PF00306 | ATP synthase alpha/beta chain, C terminal domain | |
| PF00346 | Respiratory-chain NADH dehydrogenase, 49 Kd subunit | |
| PF00400 | WD domain, G-beta repeat | |
| PF00411 | Ribosomal protein S11 | |
| PF00481 | Protein phosphatase 2C | |
| PF00574 | Clp protease | |
| PF01058 | NADH ubiquinone oxidoreductase, 20 Kd subunit | |
| PF02874 | ATP synthase alpha/beta family, beta-barrel domain | |
| PF03947 | Ribosomal Proteins L2, C-terminal domain | |
| PF07714 | Protein tyrosine and serine/threonine kinase | |
| PF13855 | Leucine rich repeat |
Gene ontology enrichment of A. palmeri eccDNA
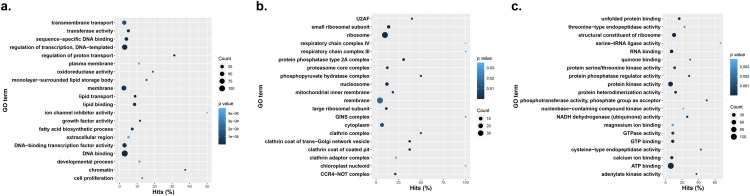
Gene ontology enrichment analysis of predicted coding elements on eccDNA of GS eccDNA revealed a variety of enriched biological processes, cellular components, and molecular functions encoded on eccDNA [Fig 3]. Enriched biological processes include regulation of transcription, membrane and lipid transport, DNA binding, fatty acid biosynthesis, protein phosphorylation, oxidation-reduction, chromatin maintenance, and protein translation [Fig 3A and S5 Table]. Cellular component and molecular function categories of interest include membrane and ribosome components [Fig 3B], cytoplasm, protein kinase activity, and ATP binding [Fig 3C].
Fig 3. Gene ontology enrichment terms and their prevalence among GS A. palmeri eccDNA samples.
A. Biological processes, B. cellular components, C. molecular functions.
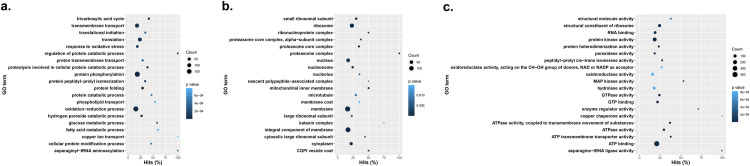
Glyphosate resistant eccDNAs showed similar, but slightly different enriched biological processes such as transmembrane transport, translation, protein phosphorylation, and oxidation-reduction process [Fig 4A]. Ribosome, nucleus, membrane, and integral component of membrane were also enriched in the cellular component category [Fig 4B]. Representative molecular functions for GR eccDNA were mainly in the ribosome and membrane categories, but ATP binding, protein kinase activity, and catalytic activity were enriched [Fig 4C and S6 Table].
Fig 4. Gene ontology enrichment terms and their prevalence among GR A. palmeri eccDNA samples.
A. Biological processes, B. cellular components, C. molecular functions.
Repeat structure of A. palmeri eccDNA
Repeat characterization revealed a high proportion of repetitive sequences among both GS and GR eccDNAs [S7 Table]. The most common repeat classes were simple repeats, long terminal repeats (LTR) from the Copia superfamily, low complexity regions, and LTR from the Gypsy superfamily [S7 Table]. Interestingly, simple repeat content varied drastically among the GR and GS states. For example, Arizona and Mississippi GS and GR pairs were closely balanced in terms of content, but Mississippi has nearly 6 times as many with ~17.5k compared to ~4k simple repeats [S7 Table]. The Long Terminal Repeats/Copia class was second in abundance among eccDNAs, followed by low complexity repeats and then Gypsy elements. DNA elements such as Stowaway, LINES, Cassandra, hAT-Tip100, MULE-MuDR, and helitrons were also identified in both GS and GR biotypes [S7 Table].
Similarity to the eccDNA replicon and replication origins on eccDNAs in A. palmeri
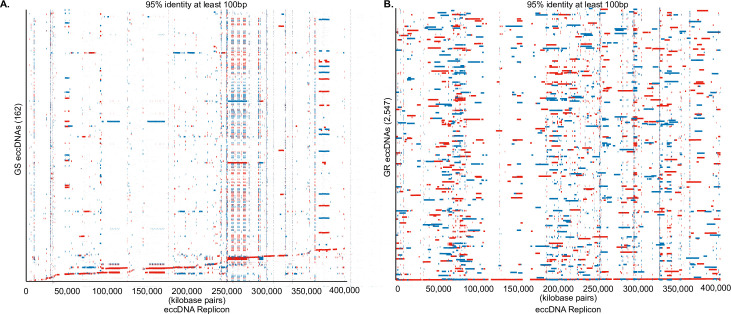
Alignment and comparative analysis for coding content and conserved sequence structure between GS and GR eccDNAs and the eccDNA replicon [25] identified a total of 162 GS eccDNA and 2,547 GR eccDNA with at matches at least 100 bp in length with a percent identify of at least 95% [Fig 5]. A total of 7 and 11 eccDNA replicon genes were predicted in GS and GR eccDNA, respectively [S8 Table]. Predicted eccDNA replicon genes in GS eccDNA include PEARLI4, Heat shock (HSP70), no apical meristem (NAM), replication factor-A, retrotransposon, zinc finger, and suppressor of gene silencing [S8 Table]. GR predicted replicon genes include: EPSPS, PEARLI4, Domain of unknown function (DUF), ethylene response factor, HSP70, NAM, replication factor A, and retrotransposon [S8 Table]. Interestingly, several GR eccDNA contained multiple copies of the EPSPS gene from Arizona, Delaware, Kansas, and Maryland, while the EPSPS gene was not present on any eccDNA in GS [Fig 6A].
Fig 5. Alignment of eccDNA to the replicon in GS and GR biotypes.
A. Alignment of 162 GS eccDNA to the eccDNA replicon. B. Alignment of 2,547 GR eccDNA to the eccDNA replicon. Red colors indicate indirect orientation and blue are direct. Alignments are filtered for matches of at least 95% identity and matches of at least 100 bp.
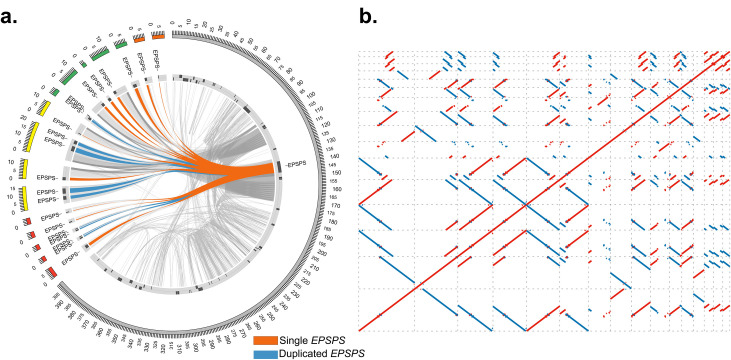
Fig 6. EPSPS gene copies in GR eccDNA.
A. Sequence similarity of GR eccDNA aligned to the eccDNA replicon. Blue and orange links indicate single or duplicated EPSPS genes. Grey links show broader sequence similarities. B. Self-alignment of the GR eccDNA containing multiple EPSPS copies. Blue dots indicate inverted repeat sequences and red dots indicate repetitive sequence in the forward direction.
In GR eccDNA we identified 5 eccDNA with 2 copies of the EPSPS gene and 11 eccDNA with a single EPSPS copy [Fig 6A]. A self-alignment of the GR EPSPS eccDNA shows many conserved direct and indirect repeats [Fig 6B] with very high sequence identity (>95% with at least 100bp). Palindromic repeats that flank the EPSPS gene, previously described as possible genome tethering sites [25], were also evident among various eccDNA (Grey links in A and on the top right corner of B) indicating the potential for recombination among these smaller eccDNA, relative to the replicon.
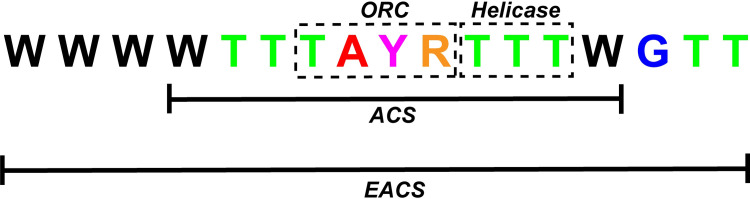
Previous work has implicated a 17bp extended autonomous consensus sequence (EACS) with a motif of WWWWTTTAYRTTTWGTT that contains a core 11bp autonomous consensus sequence (ACS) reported in yeast [41] as a sequence where replication machinery initiates autonomous replication in plants [32]; which was functionally verified in the eccDNA replicon [34] [Fig 7]. Analysis of the GS and GR eccDNA for autonomous consensus (ACS) sequences (ACS) [41] identified a total of 430 unique eccDNA with 16 of the 17 bp present in the EACS with the common missing base being the first ‘W’ (A or T), several of which had multiple EACS sequences [Fig 7 and S9 Table]. A total of 36,237 core ACS sites (11bp) were predicted within 18,679 unique eccDNA out of the total 37,336 predicted eccDNAs implicating this sequence as a possible common origin of replication sequence among smaller eccDNA in Amaranthus palmeri. Of the eccDNA that contained ARS sequences, 2,785 were predicted to contain coding sequences, whereas 16,048 eccDNA did not contain an ARS sequence.
Fig 7. Extended autonomous consensus sequence (EACS) presented in [33].
The core autonomous consensus sequence is highlighted with the TAYR motif highlighted as the origin of replication complex binding site (ORC) and the TTT motif highlighted as a helicase binding site. ‘W’ denotes A or T, ‘Y’ denotes C or T, ‘R’ denotes G or A.
Genomic origins of eccDNAs in A. palmeri
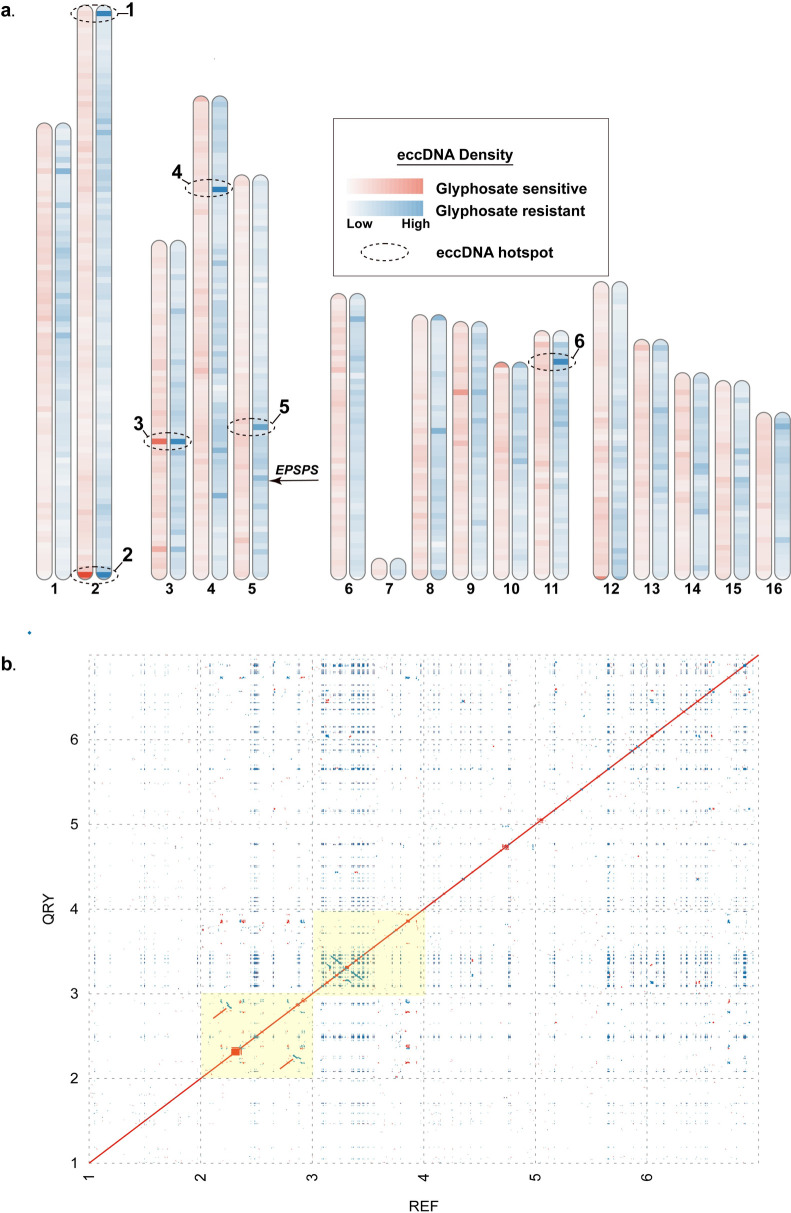
To determine the genomic origins of eccDNA and the possibility of genomic regions with a disposition for eccDNA formation, GS and GR eccDNA were mapped to the chromosome scaffolded Amaranthus palmeri assembly [42] and counted using non-overlapping genomic windows of 500kb [Fig 8]. We identified several regions of the genome with a very high disposition for focal amplifications that are conserved between GS and GR. These regions include the distal end of chromosome 2 and near the center of chromosome 3, and several other regions distributed throughout the genome [Fig 8A]. The 500kb window localized at the distal end of chromosome 2 contained 285 eccDNA from GS and 469 from GR [Fig 8A and S10 Table]. The center of chromosome 3 contained 225 GS and 449 GR eccDNA. The genomic region of eccDNA origin among GR samples with the most eccDNA was on chromosome 4 with 487 eccDNA and only 51 from GS, suggesting a possible signal of glyphosate stress. Extraction and self-alignment of the 6 genomic windows from the Palmer amaranth chromosome scale assembly from [42] revealed intricate arrays of repetitive sequence [Fig 8B]. Short, inverted repeats were the most common among all 6 regions [Fig 8B]. Clustered palindromes of various sizes were discovered in segments 2, 3, 4, and 5, as indicated by box-like structures. Regions 2 and 3 (highlighted in Fig 8B) contained more complex repetitive structure with larger direct repeats (region 2) and indirect repeats (region 3) [Fig 8B].
Fig 8.
A. Alignment and quantification of unique eccDNA of GS (red) and GR (blue) to the chromosome-scale Palmer amaranthus reference assembly in 500kb non-overlapping windows. Darker colors represent a larger abundance of mapped eccDNA. Regions with the dotted ellipses indicate a high abundance of mapped eccDNA. B. Self-alignment of the 6 highlighted regions from A. Red dots indicated direct repeats, while blue are indirect. Regions highlighted in yellow are derived from the 2 genomic regions (2 & 3 - 500kb each) with the highest abundance of mapped eccDNA.
PCR validation of eccDNA
To validate circular structure of eccDNA in A. palmeri, primers were designed in various configurations to exclude the possibility of linear DNA, [S1 Fig]. Random templates were selected, Ga_r_ecc_311 and Az_r_ecc_1037, and PCR conducted that exhibit amplification products of the expected size verifying a circular molecule [S1 Fig]. Because of the highly repetitive nature of eccDNA and the low complexity of the sequence, we did observe non-specific binding when using the Ga template, likely due to the repetitive nature of the eccDNA, but do observe amplification products close in size to the predicted sizes.
Discussion
Gene copy number variation is a predominant mechanism by which organisms respond to selective pressures in nature. Focal amplifications of transcriptionally active chromatin as eccDNAs have been found in both abundance and diversity across higher and lower order eukaryotic species underpinning their importance as a vehicle for gene copy amplification. Advancements of single molecule sequencing and approaches to purify and directly sequence circular DNA have led to evidence that eccDNA may have a fundamental role in the cell and function and also function as a source of genetic heterogeneity in response to environmental pressures [1–4, 18, 22, 25, 30]. Previous work in Palmer amaranth demonstrated that several genes in addition to EPSPS were co-amplified on a large eccDNA (~400kb) with sophisticated repetitive content and origins from distal segmental genomic regions [25]. This large eccDNA served as the vehicle for EPSPS gene copy amplification, but whether construction of this large eccDNA was the result of intramolecular recombination or recombination among a population of smaller eccDNA is unclear.
Using single molecule sequencing and the CIDER-Seq analytical pipeline [18], we identified diverse and abundant eccDNA species in both GS and GR biotypes collected from distal geographic regions that were previously reported [40]. The sizes of these eccDNA ranged from a few hundred base pairs to nearly 30kb in both biotypes and between 6 and 20% were predicted to contain genes which indicates that eccDNAs are present in Amaranthus palmeri without glyphosate exposure.
Gene enrichment analysis of both GS and GR eccDNA provided insight on biological processes and molecular functions enriched for activities related to a generalized stress response or important for rapid adaptation such as transcription regulation, development, chromatin, protein phosphorylation, oxidation-reduction, ribosomal and membrane components, protein kinase activity, and ATP binding. This indicates that eccDNAs may have a role in preserving important protein synthesis genes. Notably, transfer RNAs (tRNA) were predicted to reside on eccDNA in both GS and GR samples, but only on eccDNA that do not contain coding sequences. This was also shown by Wang et al., 2021 in Arabidopsis [19] and suggests that protein synthesis is a key attribute or component of the early response to stress and or the adaptive response. This finding also suggests that regulation of protein synthesis is perhaps as driven by eccDNA is an independent component of selection and directed gene focal amplifications as eccDNA. Furthermore, plants likely require additional copies of these protein synthesis genes for stress responses to produce significant immunity or defense products, as is the case for GR A. palmeri [1]. For example, transmembrane transport has been shown to plays an important role in adaptation of Arabidopsis to metalliferous soils [43], resource allocation and sensing under plant abiotic stress [44–46] and were enriched on GS Palmer amaranth eccDNA. Fatty acid biosynthesis is another category of enriched genes on GS eccDNA which has been implicated in signaling and plant defense to pathogens [47, 48].
At the gene level, there were a core set of 9 functional protein coding domains in common among the GS samples. Ribosomal proteins (circular rDNA), which are commonly reported as functional genes among eccDNA, were found among all 9 GS samples suggesting a common role for rDNAs as circular structures in plants [6, 30, 49, 50]. Interestingly, Cytochrome p450 and Clp protease domains were also present in each of the GS samples. Cytochrome p450s are a superfamily of genes that perform a suite of functions in plant development and protection from various stresses via multiple biosynthetic and detoxification pathways. Cytochrome p450 activity plays a central role detoxification of xenobiotics in various weed species [51–54], biosynthesis of hormones, fatty acids, sterols, cell wall components, biopolymers, and various defense compounds [55]. Clp proteases are proteolytic enzymes whose increased expression also play a protective role for the plant in both abiotic and biotic stress [56–58]. Clp proteases help maintain protein homeostasis in chloroplasts and remove nonfunctional proteins, which is essential during stress episodes when proteins tend to be more vulnerable to damage [20–22]. These core genes encoded on GS eccDNA may contribute Palmer amaranth’s innate ability to rapidly adapt.
GS biotypes shared the same 9 core functional domains as GS biotypes including Cytochrome p450 and Clp protease, in addition to 11 other domains indicating that eccDNAs are dynamic and their presence and coding structure may be the result of selective pressures. Notably, the additional functional domains in GR biotypes include additional ribosomal motifs, ABC transporters, HSP70 proteins, and leucine rich repeat (LRR) domains. ABC transporters are important for detoxification, environmental stresses and pathogen resistance [23] and may play a complementary role in glyphosate detoxification in addition to EPSP synthase over accumulation. The most abundant functional domain and conserved among all the samples is the HSP70 domain, which functions in protein maintenance and a wide variety of stress response mechanisms such as response to high temperatures [59], and was also a predicted gene on the eccDNA replicon [25]. Hsp70 have been reported to function by holding together protein substrates to help in movement, regulation, and prevent aggregation under physical and or chemical pressure in plants [59, 60] and have served as functional target in improving abiotic stress resilience in Arabidopsis [61] and other species. It is notable that the HSP70 is present in both GS and GR biotypes but is a core gene shared among all GR biotypes. The presence of Hsp70 on eccDNA suggests a possible role in glyphosate resistance, or perhaps, a genomic mechanism for rapid mitigation of heat and other abiotic stresses. Leucine rich repeat (LRR) domains are associated with protein-protein interactions, often as part of plant innate immune receptors [62]. Various transcription factors such as WRKY, bZIP, helicases, GATA (zinc finger), E2F, helix-loop-helix, TCP, and others were also predicted on A. palmeri eccDNA. Since transcription factor access to heterochromatin is limited by its compact structure, eccDNAs may provide a faster and more effective avenue for protein synthesis. Cancer cells with oncogenes encoded on eccDNAs appear to produce significantly more transcript copies compared to the same oncogenes encoded on linear DNA structures [14].
A primary question underlying the origins and structural dynamics of the large eccDNA replicon (~400kb) [24, 25] is the mechanism by which it is assembled. The most likely scenarios are long-range genomic interactions and a compounded building event over short evolutionary time; or intramolecular recombination between smaller eccDNA with newly selected genomic focal amplifications resulting from glyphosate stress to form the larger structure, again over short evolutionary time scales. Here we show a moderate degree of eccDNA replicon coverage with GS eccDNA [Fig 6A], however there are large, disconnected gaps in coverage. It is notable that the EPSPS gene was not found on any GS biotype eccDNA in this study, while several other replicon genes were. One of the primary drawbacks to the CIDER-Seq methodology is the limitation of eccDNA size to the read length of the Pacific Biosciences Sequel II instrument [18] which means eccDNAs larger than an average read length will not be sequenced intact, such as the eccDNA replicon [25]. This limitation prevented the complete assembly of the EPSPS replicon, however the EPSPS gene and most other predicted eccDNA replicon genes were found in GR biotype eccDNA and coverage of the replicon was practically complete, with only a few small gaps. Furthermore, the EPSPS gene was found on smaller eccDNA in GR biotypes in multiple copies, which corroborates the work of Koo et al., that observed the extra-chromosomal EPSPS gene vehicle as multi-meric forms. [24]. Together, these results suggest that eccDNA are present as a basal source of genetic heterogeneity or rapid response mechanism, are selectively amplified, and the large eccDNA structure reported to confer glyphosate resistance is likely built by recombination among smaller eccDNA over rapid evolutionary timescales.
Another important observation and similarity with the eccDNA replicon are the high abundance and seemingly random distribution of the core 11bp autonomous consensus sequence and a longer more conserved 16bp extended autonomous consensus sequence [41] among approximately half of the GS and GR eccDNA. The greater abundance seems to be on eccDNA without coding sequences. These sequences were previously verified to function in autonomous replication and may be regulated mechanism, perhaps epigenetic or other, to maintain gene copy numbers in A. palmeri. In the eccDNA replicon, there is a single copy of the 17bp consensus sequence and 46 copies of the 11bp sequence, seemingly randomly distributed among the replicon [25, 34]. This observation further supports the possibility that the eccDNA replicon is the result of recombination among smaller eccDNA. It is also possible that there are alternate mechanisms or origins of replication on eccDNA in A. palmeri that are used to maintain and amplify copy number. Previous work showed that the coding components of the eccDNA replicon seem to be derived from distal regions of the genome [25], and evidence presented here show that eccDNA in both GS and GR seem to originate from all over the genome, Fig 8A. Here, we also demonstrate that there are segments of the genome, or perhaps a genomic context, with a disposition for focal amplifications. These genomic ‘hotspots’ are comprised of various repeat structures that may have facilitate eccDNA formation. There are also regions of the genome that seem to be activated as ‘hotspots’ in response to glyphosate stress that suggests eccDNA formation may also be a directed event, rather than random. It is still unclear if genes need to be in the ‘right’ genomic context for a focal amplification to occur, or if other regulatory/initiation mechanisms exist. Validation of circular structure with overlapping PCR amplicons provided single PCR bands in most cases, but non-specific binding was also observed–likely due to the repetitive nature of eccDNA. This work provides evidence that eccDNA are a basal component of the cell and likely function as a reservoir of genetic heterogeneity in A. palmeri as part of the rapid adaptation program.
Materials and methods
Plant material and genomic DNA extraction
Seeds were collected from individual GR plants that had survived glyphosate application as previously described [29, 40]. Plants were grown in 9 × 9 × 9 cm plastic pots that contained a commercial potting mix (Metro-Mix 360; Sun Gro Horticulture, Bellevue, WA, USA). Seeds were sown on the potting mix surface and lightly covered with 2 mm of potting mix. Pots were sub-irrigated and maintained in a greenhouse set at a temperature regime of 30/25 ∘C (day/night) and a 15-h photoperiod under natural sunlight conditions supplemented with high-pressure sodium lights providing 400 μmol m−2 s−1. Sampling for whole genome sequencing was performed using a leaf from the third node of two representative plants from each population. Total DNA was extracted using a modified CTAB-based protocol with chloroform, isopropanol, and RNase A buffer [39]. Briefly, leaf material from each sample (approximately 20–100 mg) was ground into a fine powder using a mortar and pestle with liquid nitrogen, extracted with CTAB buffer, chloroform extracted, and ethanol precipitated. Total genomic DNA was resuspended in 50 μl of TE (10 mM Tris, 0.1 m MEDTA, pH 8.0) buffer containing RNaseA. The tube was incubated at 37°C for 30 minutes and stored at -20°C.
EccDNA enrichment and sequencing (CIDER-seq)
Circular DNA enrichment sequencing (CIDER-Seq) was used to enrich, sequence, and analyze eccDNAs from the leaf tissue DNA extraction samples according to the protocol by Mehta et al., [18]. Because we wanted to survey the landscape of eccDNA, we did not perform a size exclusion step prior to enrichment. Otherwise, the circular DNA amplification, debranching reaction, and DNA branch release and repair stages closely followed the methods of Mehta et al., [18]. Enriched eccDNA for each sample [10] was individually barcoded following the manufacturer’s recommended protocol (Pacific Biosciences), pooled in equimolar amounts, and sequenced on a Sequel II single molecule sequencer (Pacific Biosciences).
EccDNA sequence processing and analysis
Raw sequence reads were demultiplexed and circular consensus sequences analyzed with the SMRT link software (Pacific Biosciences). Parameters for CCS analysis were stringent and include: 1) predicted quality = 0.999; and 2) minimum read length = 1,000 bp. Processed reads were stored as .fastq files. Processed fastq files were analyzed with the packaged CIDER-seq software using the suggested approach to identify circular DNA. Predicted eccDNA were matched to the A. palmeri reference genome by Montgomery et al., [42]. After processing of predicted eccDNA, shorter duplicate eccDNAs were collapsed into the longest reference eccDNA with the CDhit software [63] with an identity threshold of 90%. Reference eccDNA were annotated for genuine open reading frames using the MAKER annotation pipeline [64] and evidence for genes derived from the A. palmeri published annotation [42]. Alignments to the reference genome were performed with the Minimap2 software [65] and comparative genome alignments performed with Mummer 4.0 [66]. Transfer RNAs were determined with the tRNAscan-SE software with default settings [67]. The A. palmeri reference assembly from [42] was divided into non-overlapping windows of 500kb and mapped eccDNA counted with BedTools [68].
PCR validation of circular DNA
Primer pairs of forward and reverse primers were selected to yield PCR products covering the entire circular DNA structure of several selected eccDNA sequences. The primers were designed with Geneious software and produced by Integrated DNA Technologies. Primers were resuspended in water at 100 uM concentrations. Aliquots of mixed primer pairs were prepared with 20 ul of forward primer, 20 ul of reverse primer, and 160 ul of water to yield 10 uM concentration for each primer pair. The PCR reactions contained 10 ul of 2x buffer, 1 ul of primer pair solution, 1 ul of genomic DNA matching the respective eccDNA origin, and 8 ul water. The thermal cycler settings were 98°C for 4 minutes, 98°C for 12 seconds, 52–58°C for 30 seconds, 72°C for 1 minute and 30 seconds, cycle to step two for 34 more times, 72°C for 2 minutes, and incubate at 10°C forever. After PCR, gel electrophoresis was performed to determine fragment size of the products.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
A./B. candidate eccDNA from Georgia resistant and Arizona sensitive biotypes with locations of primers. C. Agarose gel with amplicons D. Table of primers used, predicted and observed PCR amplicon sizes.
(TIF)
Data Availability
Sequence data supporting the paper are available at the Sequence Read Archive at NCBI (BioProject ID: PRJNA780915, and BioSample accessions: SAMN23211506 - SAMN23211515).
Funding Statement
Clemson University Research Foundation (CURF). Award number: 2013629; Recipient: Christopher A. Saski South Carolina Research Authority (SCRA). Award number: 2014083; Recipient: Christopher A. Saski Clemson University Experiment Station. Award number: SC-1700581; Recipient: Christopher A. Saski The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gaubatz JW. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res. 1990;237(5–6):271–92. Epub 1990/09/01. doi: 10.1016/0921-8734(90)90009-g . [DOI] [PubMed] [Google Scholar]
- 2.Cohen S, Regev A, Lavi S. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14(8):977–85. Epub 1997/02/27. doi: 10.1038/sj.onc.1200917 . [DOI] [PubMed] [Google Scholar]
- 3.Cohen S, Menut S, Mechali M. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol. 1999;19(10):6682–9. Epub 1999/09/22. doi: 10.1128/MCB.19.10.6682 ; PubMed Central PMCID: PMC84653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003;13(6A):1133–45. Epub 2003/06/12. doi: 10.1101/gr.907603 ; PubMed Central PMCID: PMC403641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moller HD, Parsons L, Jorgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. P Natl Acad Sci USA. 2015;112(24):E3114–E22. doi: 10.1073/pnas.1508825112 PubMed PMID: WOS:000356251800007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9(1):1069. Epub 2018/03/16. doi: 10.1038/s41467-018-03369-8 ; PubMed Central PMCID: PMC5852086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, et al. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015;11(11):1749–59. doi: 10.1016/j.celrep.2015.05.020 PubMed PMID: WOS:000356863600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16(10):1010–5. Epub 2009/10/08. doi: 10.1038/nsmb.1660 ; PubMed Central PMCID: PMC4041010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazzucco G, Huda A, Galli M, Piccini D, Giannattasio M, Pessina F, et al. Telomere damage induces internal loops that generate telomeric circles. Nature Communications. 2020;11(1). doi: ARTN 5297. doi: 10.1038/s41467-020-19139-4 PubMed PMID: WOS:000585935900004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hull RM, Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet. 2020;66(5):889–94. Epub 2020/04/17. doi: 10.1007/s00294-020-01069-9 ; PubMed Central PMCID: PMC7497353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Guo G, Huang J, Gao M, Zhu Q, Zeng S, et al. Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance. J Hematol Oncol. 2020;13(1):124. Epub 2020/09/16. doi: 10.1186/s13045-020-00960-9 ; PubMed Central PMCID: PMC7491193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Zhang H, Zhou Y, Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021;19(1):257. Epub 2021/06/12. doi: 10.1186/s12967-021-02927-x ; PubMed Central PMCID: PMC8194206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543(7643):122–5. Epub 2017/02/09. doi: 10.1038/nature21356 ; PubMed Central PMCID: PMC5334176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tandon I, Pal R, Pal JK, Sharma NK. Extrachromosomal circular DNAs: an extra piece of evidence to depict tumor heterogeneity. Future Sci OA. 2019;5(6):FSO390. Epub 2019/07/10. doi: 10.2144/fsoa-2019-0024 ; PubMed Central PMCID: PMC6609892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Chen X, Yu F, Ding H, Zhang Y, Wang K. Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer. Int J Biol Sci. 2021;17(4):1010–25. Epub 2021/04/20. doi: 10.7150/ijbs.54614 ; PubMed Central PMCID: PMC8040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575(7784):699–703. Epub 2019/11/22. doi: 10.1038/s41586-019-1763-5 ; PubMed Central PMCID: PMC7094777. WOS:000500036800066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. 2020;52(1):29–34. Epub 2019/12/18. doi: 10.1038/s41588-019-0547-z ; PubMed Central PMCID: PMC7008131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta D, Cornet L, Hirsch-Hoffmann M, Zaidi SS, Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat Protoc. 2020;15(5):1673–89. Epub 2020/04/05. doi: 10.1038/s41596-020-0301-0 . WOS:000523115900001. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Tian H, Wang L, Wang L, Tan Y, Zhang Z, et al. Deciphering extrachromosomal circular DNA in Arabidopsis. Comput Struct Biotechnol J. 2021;19:1176–83. Epub 2021/03/09. doi: 10.1016/j.csbj.2021.01.043 ; PubMed Central PMCID: PMC7899950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navratilova A, Koblizkova A, Macas J. Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol. 2008;8:90. Epub 2008/08/30. doi: 10.1186/1471-2229-8-90 ; PubMed Central PMCID: PMC2543021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita Y, Ohnishi N, Yamada Y, Kunisada T, Yamagishi H. Extrachromosomal Circular DNA from Nuclear Fraction of Higher-Plants. Plant Cell Physiol. 1985;26(7):1401–9. PubMed PMID: WOS:A1985ASZ0400021. [Google Scholar]
- 22.Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 2008;53(6):1027–34. Epub 2007/12/20. doi: 10.1111/j.1365-313X.2007.03394.x . [DOI] [PubMed] [Google Scholar]
- 23.Thieme M, Lanciano S, Balzergue S, Daccord N, Mirouze M, Bucher E. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 2017;18(1):134. Epub 2017/07/09. doi: 10.1186/s13059-017-1265-4 ; PubMed Central PMCID: PMC5501947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo DH, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. P Natl Acad Sci USA. 2018;115(13):3332–7. doi: 10.1073/pnas.1719354115 PubMed PMID: WOS:000428382400050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molin WT, Yaguchi A, Blenner M, Saski CA. The EccDNA Replicon: A Heritable, Extranuclear Vehicle That Enables Gene Amplification and Glyphosate Resistance in Amaranthus palmeri. Plant Cell. 2020;32(7):2132–40. doi: 10.1105/tpc.20.00099 PubMed PMID: WOS:000545974100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci U S A. 2010;107(3):1029–34. Epub 2009/12/19. doi: 10.1073/pnas.0906649107 ; PubMed Central PMCID: PMC2824275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Funke T, Han H, Healy-Fried ML, Fischer M, Schonbrunn E. Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci U S A. 2006;103(35):13010–5. Epub 2006/08/19. doi: 10.1073/pnas.0603638103 ; PubMed Central PMCID: PMC1559744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sammons RD, Gaines TA. Glyphosate resistance: state of knowledge. Pest Manag Sci. 2014;70(9):1367–77. Epub 2014/09/03. doi: 10.1002/ps.3743 ; PubMed Central PMCID: PMC4260172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molin WT, Wright AA, Lawton-Rauh A, Saski CA. The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: a repetitive path to resistance. BMC Genomics. 2017;18(1):91. Epub 2017/01/18. doi: 10.1186/s12864-016-3336-4 ; PubMed Central PMCID: PMC5240378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen S, Agmon N, Sobol O, Segal D. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mob DNA. 2010;1(1):11. Epub 2010/03/17. doi: 10.1186/1759-8753-1-11 ; PubMed Central PMCID: PMC3225859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282(5734):39–43. Epub 1979/11/01. doi: 10.1038/282039a0 . [DOI] [PubMed] [Google Scholar]
- 32.Eckdahl TT, Bennetzen JL, Anderson JN. DNA structures associated with autonomously replicating sequences from plants. Plant Mol Biol. 1989;12(5):507–16. Epub 1989/05/01. doi: 10.1007/BF00036965 . [DOI] [PubMed] [Google Scholar]
- 33.Kowalski D, Eddy MJ. The DNA Unwinding Element—a Novel, Cis-Acting Component That Facilitates Opening of the Escherichia-Coli Replication Origin. Embo J. 1989;8(13):4335–44. doi: 10.1002/j.1460-2075.1989.tb08620.x PubMed PMID: WOS:A1989CE21700044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molin WT, Yaguchi A, Blenner M, Saski CA. Autonomous replication sequences from the Amaranthus palmeri eccDNA replicon enable replication in yeast. BMC Res Notes. 2020;13(1):330. Epub 2020/07/12. doi: 10.1186/s13104-020-05169-0 ; PubMed Central PMCID: PMC7350638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feeney KM, Parish JL. Targeting mitotic chromosomes: a conserved mechanism to ensure viral genome persistence. Proc Biol Sci. 2009;276(1662):1535–44. Epub 2009/02/11. doi: 10.1098/rspb.2008.1642 ; PubMed Central PMCID: PMC2660980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaines TA, Shaner DL, Ward SM, Leach JE, Preston C, Westra P. Mechanism of resistance of evolved glyphosate-resistant Palmer amaranth (Amaranthus palmeri). J Agric Food Chem. 2011;59(11):5886–9. Epub 2011/02/19. doi: 10.1021/jf104719k . [DOI] [PubMed] [Google Scholar]
- 37.Kupper A, Borgato EA, Patterson EL, Netto AG, Nicolai M, de Carvalho SJP, et al. Multiple Resistance to Glyphosate and Acetolactate Synthase Inhibitors in Palmer Amaranth (Amaranthus palmeri) Identified in Brazil. Weed Sci. 2017;65(3):317–26. doi: 10.1017/wsc.2017.1 PubMed PMID: WOS:000405094400001. [DOI] [Google Scholar]
- 38.Culpepper AS, Grey TL, Vencill WK, Kichler JM, Webster TM, Brown SM, et al. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006;54(4):620–6. doi: 10.1614/Ws-06-001r.1 PubMed PMID: WOS:000239469200003. [DOI] [Google Scholar]
- 39.Molin WT, Wright AA, VanGessel MJ, McCloskey WB, Jugulam M, Hoagland RE. Survey of the genomic landscape surrounding the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the USA. Pest Manag Sci. 2018;74(5):1109–17. Epub 2017/07/08. doi: 10.1002/ps.4659 . WOS:000428524600013. [DOI] [PubMed] [Google Scholar]
- 40.Molin WT, Patterson EL, Saski CA. Homogeneity among glyphosate-resistant Amaranthus palmeri in geographically distant locations. PLoS One. 2020;15(9):e0233813. Epub 2020/09/10. doi: 10.1371/journal.pone.0233813 ; PubMed Central PMCID: PMC7480871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang F, May CD, Hoggard T, Miller J, Fox CA, Weinreich M. High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucleic Acids Res. 2011;39(15):6523–35. Epub 2011/05/12. doi: 10.1093/nar/gkr301 ; PubMed Central PMCID: PMC3159467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, et al. Draft Genomes of Amaranthus tuberculatus, Amaranthus hybridus, and Amaranthus palmeri. Genome Biol Evol. 2020;12(11):1988–93. Epub 2020/08/25. doi: 10.1093/gbe/evaa177 ; PubMed Central PMCID: PMC7643611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sailer C, Babst-Kostecka A, Fischer MC, Zoller S, Widmer A, Vollenweider P, et al. Transmembrane transport and stress response genes play an important role in adaptation of Arabidopsis halleri to metalliferous soils. Sci Rep. 2018;8(1):16085. Epub 2018/11/02. doi: 10.1038/s41598-018-33938-2 ; PubMed Central PMCID: PMC6208402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller I, Rodrigues CM, Neuhaus HE, Pommerrenig B. Improved resource allocation and stabilization of yield under abiotic stress. J Plant Physiol. 2021;257:153336. Epub 2020/12/29. doi: 10.1016/j.jplph.2020.153336 . [DOI] [PubMed] [Google Scholar]
- 45.Shabala S, Bose J, Fuglsang AT, Pottosin I. On a quest for stress tolerance genes: membrane transporters in sensing and adapting to hostile soils. J Exp Bot. 2016;67(4):1015–31. Epub 2015/10/29. doi: 10.1093/jxb/erv465 . [DOI] [PubMed] [Google Scholar]
- 46.Zhu JK. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167(2):313–24. Epub 2016/10/08. doi: 10.1016/j.cell.2016.08.029 ; PubMed Central PMCID: PMC5104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raffaele S, Leger A, Roby D. Very long chain fatty acid and lipid signaling in the response of plants to pathogens. Plant Signal Behav. 2009;4(2):94–9. doi: 10.4161/psb.4.2.7580 PubMed PMID: WOS:000213940700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim GH, Singhal R, Kachroo A, Kachroo P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu Rev Phytopathol. 2017;55:505–36. Epub 2017/08/05. doi: 10.1146/annurev-phyto-080516-035406 . [DOI] [PubMed] [Google Scholar]
- 49.Cao X, Wang S, Ge L, Zhang W, Huang J, Sun W. Extrachromosomal Circular DNA: Category, Biogenesis, Recognition, and Functions. Front Vet Sci. 2021;8:693641. Epub 2021/09/28. doi: 10.3389/fvets.2021.693641 ; PubMed Central PMCID: PMC8458813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91(7):1033–42. Epub 1998/01/15. doi: 10.1016/s0092-8674(00)80493-6 . [DOI] [PubMed] [Google Scholar]
- 51.Yanniccari M, Gigon R, Larsen A. Cytochrome P450 Herbicide Metabolism as the Main Mechanism of Cross-Resistance to ACCase- and ALS-Inhibitors in Lolium spp. Populations From Argentina: A Molecular Approach in Characterization and Detection. Front Plant Sci. 2020;11. doi: ARTN 600301. doi: 10.3389/fpls.2020.600301 PubMed PMID: WOS:000593945200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busi R, Vila-Aiub MM, Powles SB. Genetic control of a cytochrome P450 metabolism-based herbicide resistance mechanism in Lolium rigidum. Heredity (Edinb). 2011;106(5):817–24. Epub 2010/09/30. doi: 10.1038/hdy.2010.124 ; PubMed Central PMCID: PMC3186236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Fang Y, Li X, Zhang H, Liu M, Yang H, et al. Mechanism of the plant cytochrome P450 for herbicide resistance: a modelling study. J Enzyme Inhib Med Chem. 2013;28(6):1182–91. Epub 2012/10/13. doi: 10.3109/14756366.2012.719505 . [DOI] [PubMed] [Google Scholar]
- 54.Dimaano NG, Iwakami S. Cytochrome P450-mediated herbicide metabolism in plants: current understanding and prospects. Pest Manag Sci. 2021;77(1):22–32. Epub 2020/08/11. doi: 10.1002/ps.6040 . [DOI] [PubMed] [Google Scholar]
- 55.Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PVV, Jugulam M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants (Basel). 2020;9(5). Epub 2020/05/30. doi: 10.3390/antiox9050454 ; PubMed Central PMCID: PMC7278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ali MS, Baek KH. Protective Roles of Cytosolic and Plastidal Proteasomes on Abiotic Stress and Pathogen Invasion. Plants (Basel). 2020;9(7). Epub 2020/07/08. doi: 10.3390/plants9070832 ; PubMed Central PMCID: PMC7412383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8(4):397–403. Epub 2005/06/09. doi: 10.1016/j.pbi.2005.05.014 . [DOI] [PubMed] [Google Scholar]
- 58.Baek KH, Choi D. Roles of Plant Proteases in Pathogen Defense. Plant Pathology J. 2008;24(4):367–74. doi: 10.5423/Ppj.2008.24.4.367 PubMed PMID: WOS:000261334900001. [DOI] [Google Scholar]
- 59.Usman MG, Rafii MY, Martini MY, Yusuff OA, Ismail MR, Miah G. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol Genet Eng Rev. 2017;33(1):26–39. Epub 2017/06/27. doi: 10.1080/02648725.2017.1340546 . [DOI] [PubMed] [Google Scholar]
- 60.Alderson TR, Kim JH, Markley JL. Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes. Structure. 2016;24(7):1014–30. Epub 2016/06/28. doi: 10.1016/j.str.2016.05.011 ; PubMed Central PMCID: PMC4938735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masand S, Yadav SK. Overexpression of MuHSP70 gene from Macrotyloma uniflorum confers multiple abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol Biol Rep. 2016;43(2):53–64. Epub 2015/12/24. doi: 10.1007/s11033-015-3938-y . [DOI] [PubMed] [Google Scholar]
- 62.Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol. 2009;11(2):191–8. Epub 2008/11/20. doi: 10.1111/j.1462-5822.2008.01260.x ; PubMed Central PMCID: PMC2762402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9. Epub 2006/05/30. doi: 10.1093/bioinformatics/btl158 . [DOI] [PubMed] [Google Scholar]
- 64.Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–96. Epub 2007/11/21. doi: 10.1101/gr.6743907 ; PubMed Central PMCID: PMC2134774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. Epub 2018/05/12. doi: 10.1093/bioinformatics/bty191 ; PubMed Central PMCID: PMC6137996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marcais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. 2018;14(1):e1005944. Epub 2018/01/27. doi: 10.1371/journal.pcbi.1005944 ; PubMed Central PMCID: PMC5802927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan PP, Lowe TM. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol Biol. 2019;1962:1–14. Epub 2019/04/26. doi: 10.1007/978-1-4939-9173-0_1 ; PubMed Central PMCID: PMC6768409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 2 1–34. Epub 2014/09/10. doi: 10.1002/0471250953.bi1112s47 ; PubMed Central PMCID: PMC4213956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
A./B. candidate eccDNA from Georgia resistant and Arizona sensitive biotypes with locations of primers. C. Agarose gel with amplicons D. Table of primers used, predicted and observed PCR amplicon sizes.
(TIF)
Data Availability Statement
Sequence data supporting the paper are available at the Sequence Read Archive at NCBI (BioProject ID: PRJNA780915, and BioSample accessions: SAMN23211506 - SAMN23211515).