Abstract
Present work aimed to identify blood feeding sources and attempt to detect Leishmania DNA in Nyssomyia antunesi, suspected vector of Leishmania sp., from a park in the urban center of Belém, the capital of Pará State, in the Brazilian Amazon. Entire bodies and gut contents of Ny. antunesi engorged females, previously captured in the urban park with Centers for Disease Control (CDC) light traps and aspiration on tree bases, were subjected to Leishmania and vertebrate DNA detection through amplification of the Leishmania mini-exon and vertebrate cytochrome b (cyt b) gene regions, respectively. The quality of DNA extraction from entire bodies was ensured through amplification of the dipteran cyt b region. The vertebrate cyt b amplicons were sequenced and compared with those available on GenBank. A maximum likelihood phylogenetic tree was constructed to assess the clustering patterns of these sequences. Leishmania DNA was not detected. The sequences of 13 vertebrate cyt b amplicons were considered informative, exhibiting similarity and clustering with the following six vertebrate species: Dasyprocta leporina (1), Cuniculus paca (1), Tamandua tetradactyla (4), Choloepus didactylus (4), Pteroglossus aracari aracari (2), Homo sapiens (1). The samples of D. leporina and C. paca were obtained from the CDC canopy, whereas the others were by aspiration from tree bases. The present results revealed the eclectic and opportunist blood-feeding behavior of Ny. antunesi, with birds and mammals, these last ones acting as potential reservoirs for Leishmania species, distributed throughout the vertical forest strata.
Keywords: Phlebotominae, host, mammal, hematophagous insect, transmission
Phlebotomines (Diptera: Psychodidae) are insects worthy of medical importance mainly because some species are the proven vectors of Leishmania protozoans (Kinetoplastida: Trypanosomatidae), the etiological agents of leishmaniases (Ready 2013).
The life cycle of Leishmania involves close ecological interactions between vector and reservoir systems and niches which are often associated with silvatic ecotopes (Lainson and Shaw 2010). However, progressive changes in transmission patterns may occur either naturally or anthropogenically. In Brazil, several cities have experienced the occurrence and expansion of both visceral leishmaniasis (VL) and cutaneous leishmaniasis (CL) (Miguel et al. 2019). In the Belém Metropolitan Region (BMR), Pará State, CL is occasionally endemic, where the foci usually comprise forest fragments inhabited by anthropophilic phlebotomine populations (Ferreira et al. 2014), an essential condition for the establishment of Leishmania enzootics (Lainson and Shaw 2010). The etiology is attributed to the following species: Leishmania (L.) amazonensis, L. (Viannia) lainsoni, and L. (V.) lindenbergi, the latter accounting for approximately 40% of cases (Gonçalves et al. 2020).
Nyssomyia antunesi is an anthropophilic phlebotomine species widely distributed in South America, mainly in the Amazon Basin, and has been recorded in eight countries, including Brazil, where it occurs in ten Federated States (Aguiar and Vieira 2018). Ny. antunesi started gaining medical attention in the Pará State from the 1980s onward, when the first natural Leishmania-like suprapylaric infection was registered in this species in a VL endemic area of Marajó Island (Ryan et al. 1984). In the BMR, Ny. antunesi was also known to be infected with an unknown trypanosomatid (Silveira et al. 1991). Later, in the same region, it was regarded as a suspected vector of L. (V.) lindenbergi owing to being, by far, the most abundant anthropophilic species in the type locality of this parasite (Silveira et al. 2002). Since then, and chronologically congruent with the popularization of molecular detection techniques in vector ecology, Leishmania spp. DNA has been habitually found in Ny. antunesi (Vásquez-Trujillo et al. 2008, Trujillo et al. 2013, Thies et al. 2013, Ogawa et al. 2016, Leão et al. 2020, Araújo-Pereira et al. 2020, Da Silva Costa et al. 2021).
The arthropod-borne and zoonotic nature of Leishmania life cycles places the investigation of phlebotomines as vectors and mammals as their potential reservoir hosts on the priority list of epidemiological surveillance strategies. Currently, vector and parasite associations are inferred by tracking Leishmania DNA in phlebotomines. Among the eligible genomic regions, the mini-exon gene is considered to be unique and tandemly repeated. Moreover, the nontranscribed spacer region is distinct in length and sequence among Leishmania species (Fernandes et al. 1994). Likewise, identifying phlebotomine blood meal sources can be an alternative way to unravel potential Leishmania reservoirs (Roque and Jansen 2014, Kocher et al. 2017). Mitochondrial DNA genes, such as cytochrome b (cyt b), have been widely used as molecular markers, showing sufficient interspecific variation to distinguish between vertebrate host samples while exhibiting minimal intraspecific variation (Boakye et al. 1999, Steuber et al. 2005).
The present work aimed to provide knowledge of the blood feeding sources of Ny. antunesi from an urban park in the Brazilian Amazon region. Leishmania DNA detection in the blood was also attempted.
Materials and Methods
Samples of the present study were derived from an earlier survey conducted in an urban park in the Belém Metropolitan Region, Bosque Rodrigues Alves, Jardim Botânico da Amazônia (BRAJBA), in which the ecology of phlebotomines has been previously studied (Sánchez-Uzcátegui et al. 2020). The BRAJBA has a total area of 15 hectares, being 80% covered by primary forest with about 5,000 trees belonging from 300 species; vertebrate fauna account about 435 specimens, being 29 species living in captivity, and another 29 in freedom/semifreedom conditions (PMB 2015). Entomological captures were performed in the forest environment during four monthly occasions, from January to December 2018, with CDC light traps (n = 4) set from 6:00 p.m. to 6:00 a.m. about 1.5 m above ground (n = 2) and about 20 m above ground (n = 2) in the canopy strata, and aspiration of tree bases from 6:00 a.m. to 8:00 a.m.
Engorged females of Ny. antunesi captured were stored for further analysis. They were identified based on the external characteristics and morphology of female spermathecae (Galati 2018), dissected under fresh conditions, and/or slide-mounted in Berlese fluid. The Ny. antunesi samples destined for processing and analysis consisted of the entire bodies of female specimens that were not dissected in the field, and gut contents from dissected specimens previously examined for Leishmania-like flagellates. The samples were stored in 70% ethanol at −20°C until DNA extraction.
DNA from Ny antunesi females was extracted following a protocol adapted from Solano et al. (1997). Samples stored in ethanol were washed briefly in sterile distilled water. Then they were individually macerated within polypropylene tubes and homogenized with 100 µL of 5% Chelex 100 resin beads (Sigma-Aldrich). Each tube was heated at 95°C for 15 min and centrifuged at 13,000 rpm for 10 min. The supernatants were used for PCR.
Quality control of DNA extraction from each entire body sample was performed via amplification of a 540 bp fragment of the dipteran cytochrome b gene from the mitochondrial DNA (mtDNA) using the primers CB3-PDR (5ʹ-CA(T/C)ATTCAACC(A/T)GAATGATA-3ʹ), N1N/PDR (5ʹ-GGTA(C/T)(A/T)TTGCCTCGA(T/A)TTCG(T/A)TATGA-3ʹ), under PCR conditions described by Ready et al. (1997).
Leishmania DNA was searched in the samples of gut contents and dipteran cyt b-positive entire bodies, through the amplification of a fragment of the mini-exon gene repeat with the primers S1629 (5ʹ-GGGAATTCAATATAGTACAGAAACTG-3ʹ), S1630 (5ʹ- GGGAAGCTTCTGTACTTTATTGGTA-3ʹ), under PCR conditions described by Fernandes et al. (1994), which could distinguish the Leishmania ‘complexes’ based on the differences in their amplicon lengths, as follows: New World dermotropic species subgenus L. (Leishmania) (330 bp); New World dermotropic species subgenus L. (Viannia) (250 bp); Old/New World viscerotropic species (450 bp). The positive control consisted of DNA from L. (L.) amazonensis (IFLA/BR/1967/PH8), L. (L.) infantum chagasi (MHOM/BR/1974/PP75(M2682)), and L. (V.) braziliensis (MHOM/BR/1975/M2903).
DNA from gut contents and dipteran cyt b-positive samples were subjected to PCR to amplify a 360 bp fragment from the conserved region of the cyt b gene in vertebrate mitochondrial DNA, using the primers cytb1 (5ʹ-CCA TCCAACATCTCAGCATGATGAAA-3ʹ), cytb2 (5ʹ-GCCCCTCAGAATGATATT TGTCCTCA-3ʹ), under PCR conditions described by Steuber et al. (2005).
The amplified products were visualized by horizontal electrophoresis on a 1% agarose gel with ethidium bromide (0.5 mg/ml) staining. The products were recorded using an L-PIX STi (Loccus) electrophoresis gel photodocumenter.
The vertebrate cyt b PCR products were purified using a commercial kit (Wizard SV Gel and PCR Clean-up System) and quantified using a Quantus Fluorometer (Promega). Samples with more than 10 ng of DNA were sent for sequencing, in duplicate, by the Sanger method, using the ACTGene Análises Moleculares LTDA UFRGS/RS sequencing service. The alignment of the sequences, as well as the electropherograms, was analyzed using BioEdit 7.2.5 software (Hall 1999). The electropherograms were manually checked to remove primer residuals and trim noninformative segments. Consensus sequences were assembled, submitted to GenBank, and assigned the following accession numbers ON316828-ON316840.
Sequences generated were compared with those available in the National Institutes of Health (NIH) genetic sequence database GenBank (Benson et al. 2013), using the nucleotide Basic Local Alignment Search Tool (BLASTn) (Altschul et al. 1990). Sequences with query cover and percent identity values above 97% were considered reliable for association with the top hit ID organism. However, when the occurrence of a given vertebrate was well documented in the study area, identity values above 90% were considered acceptable.
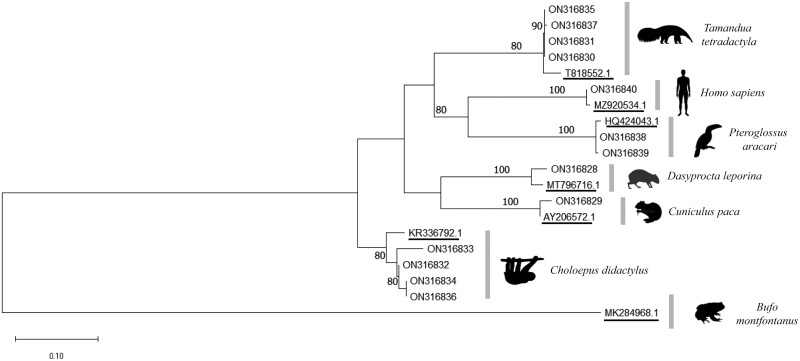
Maximum likelihood (ML) phylogenetic inference was conducted to improve the taxonomic determination of the target sequences. Sequence alignments were performed using the ClustalW algorithm (Thompson et al. 1994) hosted in MEGA X 10.1.6 software (Kumar et al. 2018). To assess the clustering pattern, an ML phylogenetic tree was constructed using the alignments of the generated sequences, together with those of the top hit from the BLAST searches and that from the hot creek toad Bufo montfontanus (Anura: Bufonidae) (MK284968.1), to the root as an outgroup. The following configurations were applied: a general time reversible substitution model with gamma distribution rates and the nearest neighbor interchange heuristic inference method, using a consensus bootstrap of 1,000 replications. Clades with bootstraps >90 were considered consistent (Dhar and Minin 2016). Clades were illustrated with VectorStock silhouettes.
Results
Amplification of dipteran cyt b was positive in 42/59 entire body samples, of which 16 were vertebrate cyt b positive and four samples had sufficient DNA for sequencing. The gut contents of 19/22 females were vertebrate cyt b-positive, and 12 samples had sufficient DNA for sequencing. Therefore, 16 samples were sequenced, 13 of which were informative, and exhibited similarities with the following six vertebrate species: red-rumped agouti, Dasyprocta leporina (Rodentia: Dasyproctidae) (1); lowland paca, Cuniculus paca (Rodentia: Cuniculiade) (1); anteater, Tamandua tetradactyla (Pilosa: Myrmecophagidae) (4); sloth, Choloepus didactylus (Pilosa: Megalonychidae) (4); toucan, Pteroglossus aracari (2); and man, Homo sapiens (Primates: Hominidae) (1). The samples of D. leporina and C. paca were obtained from the CDC canopy, whereas the others were obtained from aspiration on tree bases (Table 1). Phylogenetic ML reconstruction generated well-supported clades, which were in agreement with the BLAST-based identification of our generated sequences of blood sources of Ny. antunesi (Fig. 1). The 42 dipteran cyt b-positive samples and 22 gut contents were submitted for Leishmania DNA detection through amplification of the mini-exon gene region. No samples were found positive.
Table 1.
Vertebrate bloodmeal sources of engorged field-collected Nyssomyia antunesi females from the Bosque Rodrigues Alves - Jardim Botânico da Amazônia, Belém, Pará State, Brazil (2018)
| N | Capture method | Sample | Bloodmeal source | Accession number (generated sequences) | Query coverage (%) | E-value | Identity (%) | Accession number (top hit) |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC canopy | Entire body | Dasyprocta leporina | ON316828 | 98 | 1e-129 | 96.7 | MT796716.1 |
| 2 | CDC canopy | Entire body | Cuniculus paca | ON316829 | 99 | 4e-78 | 92.42 | AY206572.1 |
| 3 | Tree bases | Gut content | Tamandua tetradactyla | ON316830 | 98 | 8e-100 | 99.5 | KT818552.1 |
| 4 | Tree bases | Gut content | Tamandua tetradactyla | ON316831 | 98 | 8e-100 | 99.5 | KT626616.1 |
| 5 | Tree bases | Gut content | Choloepus didactylus | ON316832 | 99 | 3e-94 | 98 | AF232012.1 |
| 6 | Tree bases | Gut content | Choloepus didactylus | ON316833 | 100 | 2e-85 | 96.02 | AF232012.1 |
| 7 | Tree bases | Gut content | Choloepus didactylus | ON316834 | 100 | 1e-103 | 95.7 | AF232012.1 |
| 8 | Tree bases | Gut content | Tamandua tetradactyla | ON316835 | 99 | 8e-100 | 99.5 | KT818552.1 |
| 9 | Tree bases | Gut content | Choloepus didactylus | ON316836 | 100 | 1e-103 | 96.6 | AF232012.1 |
| 10 | Tree bases | Gut content | Tamandua tetradactyla | ON316837 | 98 | 8e-100 | 99.5 | MW752306.1 |
| 11 | Tree bases | Gut content | Pteroglossus aracari | ON316838 | 100 | 4e-98 | 100 | HQ424043.1 |
| 12 | Tree bases | Entire body | Pteroglossus aracari | ON316839 | 100 | 4e-98 | 100 | HQ424043.1 |
| 13 | Tree bases | Entire body | Homo sapiens | ON316840 | 99 | 2e-141 | 97.9 | LC088152.1 |
Fig. 1.
Maximum likelihood phylogenetic tree of partial vertebrate cytochrome b sequences of the phlebotomine Nyssomyia antunesi blood feeding sources from the Bosque Rodrigues Alves - Jardim Botânico da Amazônia (BRAJBA), Belém, Brazil, together with their respective identity top-hit from the BLAST searches (GenBank), rooted with Bufo montfontanus. Reference accession numbers are underlined. Values at the nodes represent bootstrap support of the ML tree (only nodes with >70% support are presented).
Discussion
In the present study, we identified blood feeding sources and attempted to detect Leishmania DNA in Ny. antunesi samples captured in a preserved environment, with diverse and already described fauna and flora, surrounded by urban areas (PMB 2015); this phlebotomine species is recognized as the dominant species in both canopy and ground strata (Sánchez-Uzcátegui et al. 2020). Preserved forest environments, such as the sampled area, generally offer a high variety of blood meal sources for phlebotomines, guaranteeing the maintenance of the gonotrophic cycle (Lainson et al. 1981). Blood feeding sources on a given phlebotomine species collected from numerous widely dispersed areas over a long period must eventually provide some indication of the most likely reservoir(s) of Leishmania, especially if they are related to the simultaneous detection of the parasite in dissected phlebotomines from the same area at the same time (Lainson and Shaw 1979).
The identification of bloodmeals from phlebotomines using partial sequences from PCR products of the mtDNA cyt b gene is suitable and widely used for this purpose (Jiménez et al. 2013, Carvalho et al. 2017, Pereira Junior et al. 2019, Doe et al. 2020, Leão et al. 2020, Remadi et al. 2020, Lozano-Sardaneta et al. 2021, Rodrigues et al. 2021, Torchitte et al. 2020). Six vertebrate species were found to be blood feeding sources of Ny. antunesi, all of which have already been documented in the BRAJBA (PMB 2015). The eclectic blood feeding repertoire observed for this phlebotomine species is consistent with recent literature. In the rural environment of the western Amazon, it has been found that females from the Ny. antunesi obtain bloodmeals from the following vertebrates: Bos taurus (Artiodactyla: Bovidae), Pecari tajacu (Artiodactyla: Tayassuidae), Plecturocebus bernhardi (Primates: Pitheciidae), Philander canus (Didelphimorphia: Didelphidae), Sus. scrofa (Artiodactyla: Suidae), and T. tetradactyla (Da Silva Costa et al. 2021), with the former also being a blood source for Ny. antunesi in a closely related region (Pereira Junior et al. 2019).
The most common blood feeding sources were two mammals of the order Pilosa: T. tetradactyla and C. didactylus. Both have recognized roles as Leishmania spp. reservoirs (Roque and Jansen 2014, Muñoz-García et al. 2019), with particular ecological associations with other phlebotomines of the genus Nyssomyia. Anteaters spend much of their time climbing up and down trees in search of termite nests and therefore, come into intimate contact with tree trunks inhabiting phlebotomines, such as Nyssomyia umbratilis and Nyssomyia whitmani (Lainson et al. 1981). In the same way, sloths have been proven as the primary blood feeding sources of Ny. umbratilis and Nyssomyia anduzei (Christensen et al. 1982).
Two vertebrate cyt b sequences were obtained from toucan. The role of birds in the life cycle of phlebotomines has been discussed, as their presence may influence fly population dynamics with increasing population density, and consequently contribute to Leishmania transmission dynamics (Teodoro et al. 1993, De Ávila et al. 2018). In the studied area, Ny. antunesi is a canopy-dominant species (Sánchez-Uzcátegui et al. 2020), spatially congruent with birds ecotopes. Similar results were observed with the also canopy dwelling Ny. umbratilis in the Guianan Amazon biome, where bird blood were detected as food sources (Vasconcelos dos Santos et al. 2018).
Two vertebrate cyt b sequences were from C. paca and D. leporina. These ground-dwelling rodents are common blood sources for phlebotomines (Kocher et al. 2017). In addition, C. paca is the only known potential reservoir of L. (V.) lainsoni (Roque and Jansen, 2014), a parasite species found in the enzootics of the studied area, but associated with other phlebotomine species, Trichophoromyia brachipyga (Sánchez-Uzcátegui et al. 2020, Vasconcelos dos Santos and Silveira 2020).
One vertebrate cyt b sequence belonged to the human genome, strengthening the well-documented anthropophilic behavior of Ny. antunesi (Silveira et al. 2002, Pereira Junior et al. 2019, Sánchez-Uzcátegui et al. 2020).
Arias et al. (1982) stated that the vertical habitat of a given phlebotomine species tends to be spatially congruent with that of its associated natural hosts. However, given the small sample size of the present study and considering that some blood sources, such as anteaters, sloths, and birds, frequently move between the canopy and ground level, it was not possible to determine whether feeding occurred arboreally (Leão et al. 2020). Conversely, DNA from terrestrial rodents has been found in Ny. antunesi females collected in the canopy, making it impossible to determine whether this phlebotomine species favors bloodmeals from a specific stratum. Based on these findings, it can be assumed that Ny. antunesi can feed on the ground and climb into the canopy.
Successful identification of blood feeding sources may have been prejudiced because of the small volume of blood and the time elapsed after feeding. Owing to the quick blood digestion process inherent to dipterans, only recently engorged phlebotomines are suitable for DNA analysis (Sant’Anna et al. 2008). Minuscule amounts of blood are ingested by phlebotomines (Rogers et al. 2002) and, especially in partially fed specimens, there may be insufficient DNA for detection. Moreover, the sizes of phlebotomines vary between species, and those from Nyssomyia are usually very small when compared, for instance, with Lutzomyia and Trichophoromyia. Despite these facts, most of the dipteran cyt b-positive samples were obtained from gut contents, instead of entire bodies, all of which were stored since 2018 to be processed in 2021–2022. The easy handling and long-term stability of DNA in blood samples make this methodology applicable in field conditions, where biological material is often collected far away and processed after a long time (Sant’Anna et al. 2008).
Leishmania DNA was not detected in either the entire body samples (dipteran cyt b-positive samples) or the gut contents, which is in accordance with previous reports, where no Ny. antunesi from BRAJBA were found to be naturally infected, despite other phlebotomine species known to harbor Leishmania at that site, with a general infection rate of 0.013 (Sánchez-Uzcátegui et al. 2020). All gut contents submitted to PCR were previously examined in the field and found negative for promastigotes. However, given the high sensitivity of PCR, the samples were re-examined to detect DNA tracks of dead and disintegrated promastigotes, not viewable through microscopic observation.
True vector importance of Ny. antunesi remains uncertain in BMR. However, the present results provide an advance in the biological knowledge of this phlebotomine species, revealing an eclectic and opportunist blood feeding behavior, including attraction to birds and various orders of mammals, these last ones acting as potential reservoirs of Leishmania species, distributed throughout the vertical forest strata.
Acknowledgments
We would like to acknowledge Edna de Freitas Leão, Fábio Márcio Medeiros da Silva Freire, Iorlando da Rocha Barata, Luciene Aranha da Silva Santos and Maria Sueli Barros Pinheiro for their technical support in the sampling; Lindomar Silva and the staff of Bosque Rodrigues Alves - Jardim Botânico da Amazônia, for their logistical facilities provided to access the park; and Hellen Thais Fuzii for her facilities with the DNA purification and quantification steps. This research was financially supported by the Universidade Federal do Pará and Instituto Evandro Chagas/ Ministry of Health, Brazil. ACP and YVSU received, respectively, Master and PhD scholarships from the ‘Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brasil’ (CAPES) - Financial code 001.
Contributor Information
Amanda Costa Pimentel, Programa de Pós Graduação em Doenças Tropicais, Núcleo de Medicina Tropical, Universidade Federal do Pará, Belém, Pará State, Brazil.
Yetsenia del Valle Sánchez Uzcátegui, Programa de Pós Graduação em Biologia de Agentes Infecciosos e Parasitários, Instituto de Ciências Biológicas, Universidade Federal do Pará, Belém, Pará State, Brazil; Seção de Parasitologia, Instituto Evandro Chagas, Ananindeua, Pará State, Brazil; Departamento de Biología, Facultad de Ciencias, Universidad de Los Andes, Mérida, Venezuela.
Ana Carolina Stocco de Lima, Seção de Parasitologia, Instituto Evandro Chagas, Ananindeua, Pará State, Brazil.
Fernando Tobias Silveira, Programa de Pós Graduação em Doenças Tropicais, Núcleo de Medicina Tropical, Universidade Federal do Pará, Belém, Pará State, Brazil; Seção de Parasitologia, Instituto Evandro Chagas, Ananindeua, Pará State, Brazil.
Thiago Vasconcelos dos Santos, Programa de Pós Graduação em Biologia de Agentes Infecciosos e Parasitários, Instituto de Ciências Biológicas, Universidade Federal do Pará, Belém, Pará State, Brazil; Seção de Parasitologia, Instituto Evandro Chagas, Ananindeua, Pará State, Brazil.
Edna Aoba Yassui Ishikawa, Programa de Pós Graduação em Doenças Tropicais, Núcleo de Medicina Tropical, Universidade Federal do Pará, Belém, Pará State, Brazil.
References Cited
- Aguiar, G. M. D., and Vieira V. R.. 2018. Regional distribution and habitats of Brazilian phlebotomine species, pp. 251–298. InRangel E. F., and Shaw J. J. (eds.), Brazilian sand flies: biology, taxonomy, medical importance and control. Brazilian Ministry of Health, Oswaldo Cruz Foundation, Rio de Janeiro. Springer, Cham. [Google Scholar]
- Altschul, S. F., Gish W., Miller W., Myers E. W., and Lipman D. J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Araujo-Pereira, T. D., Pita-Pereira D. D., Baia-Gomes S. M., Boité M., Silva F., Pinto I. D. S., De Souza R. L. T., Fuzari A., De Souza C., Brazil R., et al. 2020. An overview of the sandfly fauna (Diptera: Psychodidae) followed by the detection of Leishmania DNA and blood meal identification in the state of Acre, Amazonian Brazil. Memórias do Instituto Oswaldo Cruz. 115: e200157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias, J. R., and Freitas R. A. D.. 1982. On the vectors of cutaneous leishmaniasis in the Central Amazon of Brasil. 3. Phlebotomine sand fly stratification in a terra firme forest. Acta Amazonica. 12: 599–608. [Google Scholar]
- Benson, D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., and Sayers E. W.. 2013. GenBank. Nucleic Acids Res. 41: D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boakye, D. A., Tang J., Truc P., Merriweather A., and Unnasch T. R.. 1999. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Med. Vet. Entomol. 13: 282–287. [DOI] [PubMed] [Google Scholar]
- Carvalho, G. M. D. L., Rêgo F. D., Tanure A., Silva A. C. P., Dias T. A., Paz G. F., and Andrade Filho J. D.. 2017. Bloodmeal identification in field-collected sand flies from Casa Branca, Brazil, using the cytochrome b PCR method. J. Med. Entomol. 54: 1049–1054. [DOI] [PubMed] [Google Scholar]
- Christensen, H. A., Arias J. R., de Vasquez A. M., and de Freitas R. A.. 1982. Hosts of sandfly vectors of Leishmania braziliensis guyanensis in the central Amazon of Brazil. Am. J. Trop. Med. Hyg. 31: 239–242. [DOI] [PubMed] [Google Scholar]
- Da Silva Costa, G., Júnior A. M. P., Castro T. S., de Paulo P. F. M., Ferreira G. E. M., and Medeiros J. F.. 2021. Sand fly fauna and molecular detection of Leishmania species and blood meal sources in different rural environments in western Amazon. Acta Trop. 224: 106150. [DOI] [PubMed] [Google Scholar]
- De Ávila, M. M., Brilhante A. F., de Souza C. F., Bevilacqua P. D., Galati E. A. B., and Brazil R. P.. 2018. Ecology, feeding and natural infection by Leishmania spp. of phlebotomine sand flies in an area of high incidence of American tegumentary leishmaniasis in the municipality of Rio Branco, Acre, Brazil. Parasit. Vectors. 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, A., and Minin V. N.. 2016. Maximum likelihood phylogenetic inference, pp. 499–506. InEncyclopedia of evolutionary biology. Elsevier. [Google Scholar]
- Doe, E. D., Kwakye-Nuako G., and Egyir-Yawson A.. 2020. Identification of blood meal of sand flies in a cutaneous leishmaniasis endemic area, Volta Region-Ghana. Am. J. Biomed. Sci. Life Sci. 8: 69–75. [Google Scholar]
- Fernandes, O., Murthy V. K., Kurath U., Degrave W. M., and Campbell D. A.. 1994. Mini-exon gene variation in human pathogenic Leishmania species. Mol. Biochem. Parasitol. 66: 261–271. [DOI] [PubMed] [Google Scholar]
- Ferreira, J. V. S., Vasconcelos dos Santos T., Santos E. M. D., and Gorayeb I. D. S.. 2014. Phlebotomine sand flies (Diptera: Psychodidae) in forest fragments of Belém metropolitan area, Pará State, Brazil, with considerations on vectors of American cutaneous leishmaniasis agents. Revista Pan-Amazônica de Saúde. 5: 29–35. [Google Scholar]
- Galati, E. A. 2018. Phlebotominae (Diptera, Psychodidae): classification, morphology and terminology of adults and identification of American taxa, pp. 9–212. InRangel E. F. and Shaw J. J. (eds.), Brazilian sand flies: biology, taxonomy, medical importance and control. Brazilian Ministry of Health, Oswaldo Cruz Foundation, Rio de Janeiro. Springer, Cham. [Google Scholar]
- Gonçalves, L. P., Vasconcelos dos Santos T., Campos M. B., Lima L. V. D. R., Ishikawa E. A. Y., Silveira F. T., and Ramos P. K. S.. 2020. Further insights into the eco-epidemiology of American cutaneous leishmaniasis in the Belém metropolitan region, Pará State, Brazil. Rev. Soc. Bras. Med. Trop. 53: e20200255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, pp. 95–98. InNucleic Acids Symp. Ser., vol. 41. Elsevier. [Google Scholar]
- Jiménez, M., González E., Iriso A., Marco E., Alegret A., Fúster F., and Molina R.. 2013. Detection of Leishmania infantum and identification of blood meals in Phlebotomus perniciosus from a focus of human leishmaniasis in Madrid, Spain. Parasitol. Res. 112: 2453–2459. [DOI] [PubMed] [Google Scholar]
- Kocher, A., de Thoisy B., Catzeflis F., Valière S., Bañuls A. L., and Murienne J.. 2017. iDNA screening: disease vectors as vertebrate samplers. Mol. Ecol. 26: 6478–6486. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Stecher G., Li M., Knyaz C., and Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainson, R., and Shaw J. J.. 1979. The role of animals in the epidemiology of South American leishmaniasis, pp. 1–116. InLumsden W. H. R., and Evans D. A. (eds.), Biology of the Kinetoplastida, vol. 2. Academic Press, London, New York and San Francisco. [Google Scholar]
- Lainson, R., and Shaw J. J.. 2010. New world leishmaniasis. InTopley & Wilson’s microbiology and microbial infections. John Wiley & Sons. https://onlinelibrary.wiley.com/doi/book/10.1002/9780470688618. [Google Scholar]
- Lainson, R., Shaw J. J., and Povoa M.. 1981. The importance of edentates (sloth and anteaters) as primary reservoirs of Leishmania braziliensis guyanensis, causative agent of “pian-bois” in north Brazil. Trans. R Soc. Trop. Med. Hyg. 75: 611–612. [DOI] [PubMed] [Google Scholar]
- Leão, P. D. O., Pereira Júnior A. M., de Paulo P. F. M., Carvalho L. P. C., Souza A. B. N., da Silva M. S., Castro T. S., Souza Freitas M. T. D., Rodrigues M. M. D. S., Ferreira G. E. M., et al. 2020. Vertical stratification of sand fly diversity in relation to natural infections of Leishmania sp. and blood-meal sources in Jamari National Forest, Rondônia State, Brazil. Parasit. Vectors. 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Sardaneta, Y. N., Jiménez-Girón E. I., Rodríguez-Rojas J. J., Sánchez-Montes S., Álvarez-Castillo L., Sánchez-Cordero V., and Becker I.. 2021. Species diversity and blood meal sources of phlebotomine sand flies (Diptera: Psychodidae) from Los Tuxtlas, Veracruz, Mexico. Acta Trop. 216: 105831. [DOI] [PubMed] [Google Scholar]
- Miguel, C. D., and Guarnier C. D.. 2019. Canine and human leishmaniasis: disease progression to Brazilian urbanized areas. Int. J. Trop. Dis. 2: 23. [Google Scholar]
- Muñoz-García, C. I., Sánchez-Montes S., Villanueva-García C., Romero-Callejas E., Díaz-López H. M., Gordillo-Chávez E. J., Martínez-Carrasco C., Berriatua E., and Rendón-Franco E.. 2019. The role of sloths and anteaters as Leishmania spp. reservoirs: a review and a newly described natural infection of Leishmania mexicana in the northern anteater. Parasitol. Res. 118: 1095–1101. [DOI] [PubMed] [Google Scholar]
- Ogawa, G. M., Pereira Júnior A. M., Resadore F., Ferreira R. D. G. M., Medeiros J. F., and Camargo L. M. A.. 2016. Sandfly fauna (Diptera: Psychodidae) from caves in the state of Rondônia, Brazil. Revista Brasileira de Parasitologia Veterinária. 25: 61–68. [DOI] [PubMed] [Google Scholar]
- Pereira Júnior, A. M., Souza A. B. N., Castro T. S., da Silva M. S., de Paulo P. F. M., Ferreira G. E. M., and de Medeiros J. F.. 2019. Diversity, natural infection and blood meal sources of phlebotomine sandflies (Diptera, Psychodidae) in the western Brazilian Amazon. Memórias do Instituto Oswaldo Cruz. 114: e190170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PMB - Prefeitura Municipal de Belém/ SEMMA - Secretaria Municipal de Meio Ambiente - Bosque Rodrigues Alves. (2015). Conheça o bosque: Fauna. Retrieved from https://semma.belem.pa.gov.br/bosque/fauna/; accessed on 23/03/2022
- Ready, P. D. 2013. Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 58: 227–250. [DOI] [PubMed] [Google Scholar]
- Ready, P. D., Day J. C., De Souza A. A., Rangel E. F., and Davies C. R.. 1997. Mitochondrial DNA characterization of populations of Lutzomyia whitmani (Diptera: Psychodidae) incriminated in the peri-domestic and silvatic transmission of Leishmania species in Brazil. Bull. Entomol. Res. 87: 187–195. [Google Scholar]
- Remadi, L., Chargui N., Jimenez M., Molina R., Haouas N., González E., Chaabane-Banaouas R., Salah E. B., Haddaji M., Chaabouni Y., et al. 2020. Molecular detection and identification of Leishmania DNA and blood meal analysis in Phlebotomus (Larroussius) species. PLoS Negl. Trop. Dis. 14: e0008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, B. L., Costa G. D. S., and Shimabukuro P. H. F.. 2021. Identification of bloodmeals from sand flies (Diptera: Psychodidae) collected in the Parque Nacional do Viruá, State of Roraima, Brazil. J. Med. Entomol. 58: 2488–2494. [DOI] [PubMed] [Google Scholar]
- Rogers, M. E., Chance M. L., and Bates P. A.. 2002. The role of promastigote secretory gel in the origin and transmission of the infective stage of Leishmania mexicana by the sandfly Lutzomyia longipalpis. Parasitology. 124: 495–507. [DOI] [PubMed] [Google Scholar]
- Roque, A. L. R., and Jansen A. M.. 2014. Wild and synanthropic reservoirs of Leishmania species in the Americas. Int. J. Parasitol. Parasit. Wildlife. 3: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, L., Silveira F. T., Lainson R., and Shaw J. J.. 1984. Leishmanial infections in Lutzomyia longipalpis and Lu. antunesi (Diptera: Psychodidae) on the island of Marajó, Pará State, Brazil. Trans. R Soc. Trop. Med. Hyg. 78: 547–548. [DOI] [PubMed] [Google Scholar]
- Sánchez Uzcátegui, Y. D. V., Vasconcelos Dos Santos T., Silveira F. T., Ramos P. K., Dos Santos E. J. M., and Póvoa M. M.. 2020. Phlebotomines (Diptera: Psychodidae) from a urban park of Belém, Pará State, northern Brazil and potential implications in the transmission of American cutaneous leishmaniasis. J. Med. Entomol. 57: 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna, M. R., Jones N. G., Hindley J. A., Mendes-Sousa A. F., Dillon R. J., Cavalcante R. R., Alexander B., and Bates P. A.. 2008. Blood meal identification and parasite detection in laboratory-fed and field-captured Lutzomyia longipalpis by PCR using FTA databasing paper. Acta Trop. 107: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira, F. T., Souza A. A., Lainson R., Shaw J. J., Braga R. R., and Ishikawa E. E.. 1991. Cutaneous leishmaniasis in the Amazon region: natural infection of the sandfly Lutzomyia ubiquitalis (Psychodidae: Phlebotominae) by Leishmania (Viannia) lainsoni in Pará State, Brazil. Memórias do Instituto Oswaldo Cruz. 86: 127–130. [DOI] [PubMed] [Google Scholar]
- Silveira, F. T., Ishikawa E. A. Y., De Souza A. A. A., and Lainson R.. 2002. An outbreak of cutaneous leishmaniasis among soldiers in Belém, Pará State, Brazil, caused by Leishmania (Viannia) lindenbergi n. sp.-a new leishmanial parasite of man in the Amazon region. Parasite. 9: 43–50. [DOI] [PubMed] [Google Scholar]
- Solano, P., Duvallet G., Dumas V., Cuisance D., and Cuny G.. 1997. Microsatellite markers for genetic population studies in Glossina palpalis (Diptera: Glossinidae). Acta Trop. 65: 175–180. [DOI] [PubMed] [Google Scholar]
- Steuber, S., Abdel-Rady A., and Clausen P. H.. 2005. PCR-RFLP analysis: a promising technique for host species identification of blood meals from tsetse flies (Diptera: Glossinidae). Parasitol. Res. 97: 247–254. [DOI] [PubMed] [Google Scholar]
- Teodoro, U., La Salvia Filho V., Lima E. M. D., Spinosa R. P., Barbosa O. C., Ferreira M. E. M. C., and Lonardoni M. V. C.. 1993. Observações sobre o comportamento de flebotomíneos em ecótopos florestais e extraflorestais, em área endêmica de leishmaniose tegumentar americana, no norte do Estado do Paraná, sul do Brasil. Revista de Saúde Pública. 27: 242–249. [DOI] [PubMed] [Google Scholar]
- Thies, S. F., Ribeiro A. L. M., Michalsky E. M., Miyazaki R. D., Fortes-Dias C. L., Fontes C. J. F., and Dias E. S.. 2013. Phlebotomine sandfly fauna and natural Leishmania infection rates in a rural area of Cerrado (tropical savannah) in Nova Mutum, State of Mato Grosso in Brazil. Rev. Soc. Bras. Med. Trop. 46: 293–298. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins D. G., and Gibson T. J.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchitte, A. P. A., Pereira Júnior A. M., Paulo P. F. M., Costa G. S., Castro T. S., Ferreira R. G. M., and Medeiros J. F.. 2020. Identification of sand flies (Diptera: Psychodidae) and blood meal sources in periurban areas of Ji-Paraná municipality, Western Brazilian Amazon. Braz. J. Biol. 81: 225–227. [DOI] [PubMed] [Google Scholar]
- Trujillo, A. V., Reina A. E. G., Orjuela A. G., Suárez E. P., Palomares J. E., and Alvarez L. S. B.. 2013. Seasonal variation and natural infection of Lutzomyia antunesi (Diptera: Psychodidae: Phlebotominae), an endemic species in the Orinoquia region of Colombia. Memórias do Instituto Oswaldo Cruz. 108: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos dos Santos, T., and Silveira F. T.. 2020. Increasing putative vector importance of Trichophoromyia phlebotomines (Diptera: Psychodidae). Memórias do Instituto Oswaldo Cruz. 115: e190284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos dos Santos, T., Prévot G., Ginouvès M., Duarte R., Silveira F. T., Póvoa M. M., and Rangel E. F.. 2018. Ecological aspects of Phlebotomines (Diptera: Psychodidae) and the transmission of American cutaneous leishmaniasis agents in an Amazonian/Guianan bordering area. Parasit. Vectors. 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Trujillo, A., Santamaría-Herreño E., González-Reina A. E., Buitrago-Álvarez L. S., Góngora-Orjuela A., and Cabrera-Quintero O. L.. 2008. Lutzomyia antunesi as suspected vector of cutaneous leishmaniasis in the Orinoquian region of Colombia. Revista de Salud Pública. 10: 625–632. [DOI] [PubMed] [Google Scholar]