Abstract
Expression of the alcohol dehydrogenase gene, adhE, in Escherichia coli is anaerobically regulated at both the transcriptional and the translational levels. To study the AdhE protein, the adhE+ structural gene was cloned into expression vectors under the control of the lacZ and trpc promoters. Wild-type AdhE protein produced under aerobic conditions from these constructs was inactive. Constitutive mutants (adhC) that produced high levels of AdhE under both aerobic and anaerobic conditions were previously isolated. When only the adhE structural gene from one of the adhC mutants was cloned into expression vectors, highly functional AdhE protein was isolated under both aerobic and anaerobic conditions. Sequence analysis revealed that the adhE gene from the adhC mutant contained two mutations resulting in two amino acid substitutions, Ala267Thr and Glu568Lys. Thus, adhC strains contain a promoter mutation and two mutations in the structural gene. The mutant structural gene from adhC strains was designated adhE*. Fragment exchange experiments revealed that the substitution responsible for aerobic expression in the adhE* clones is Glu568Lys. Genetic selection and site-directed mutagenesis experiments showed that virtually any amino acid substitution for Glu568 produced AdhE that was active under both aerobic and anaerobic conditions. These findings suggest that adhE expression is also regulated posttranslationally and that strict regulation of alcohol dehydrogenase activity in E. coli is physiologically significant.
Escherichia coli and other facultative organisms respire using oxygen or alternative electron acceptors but can also grow in the absence of external electron acceptors by coupling reduction of metabolic intermediates to NADH oxidation, a process known as fermentation. This remarkable metabolic flexibility is tightly regulated in response to factors such as pH, redox potential, carbon source, and the availability of oxygen or other electron acceptors (4, 25, 33). In E. coli and other mixed acid fermenters, pyruvate is reduced to a mixture of fermentation products including lactate, succinate, acetate, and ethanol. Alcohol dehydrogenase (ADH; AdhE) converts acetyl coenzyme A (CoA) to acetaldehyde and then to ethanol in a two-step reduction that is coupled to oxidation of two NADH molecules. In addition, AdhE regulates pyruvate formate lyase (PFL) activity. Thus, this enzyme acts as an ADH (5, 31), a CoA-dependent acetaldehyde dehydrogenase (28, 29), and a PFL deactivase (16). Transcription of adhE is induced only under anaerobic conditions, largely in response to elevated levels of reduced NADH (23, 24), and adhE mutants cannot grow under fermentative conditions (4). Constitutive adhC mutants possess high levels of AdhE both aerobically and anaerobically (6). These adhC mutations map within the promoter region and affect transcription of adhE (21, 23, 24). In addition, the adhE message must be processed by RNase III before it can be translated (1). Thus, adhE expression is regulated at the transcriptional and translational levels. Here we show that adhC mutants contain mutations in the adhE structural gene (now designated adhE*) in addition to their promoter mutations. One of these mutations, Glu568Lys, is essential for AdhE* activity under aerobic conditions. These findings suggest that ADH expression in E. coli is also regulated posttranslationally.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains are derivatives of E. coli K-12. Aerobic cultures were grown at 37°C, with shaking, in Luria-Bertani (LB) medium (27) supplemented with ampicillin (100 μg/ml) or chloramphenicol (50 μg/ml) as indicated. Anaerobic cultures were grown in 250-ml bottles filled to the top with anaerobic glucose medium (30) and incubated at 37°C with gentle magnetic stirring. Transcription of cloned genes was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an A600 of 0.1, and cultures were then grown to an A600 of 1.5. Alcohol indicator plates (AIP) were used to detect ADH activity of bacterial colonies. On AIP, ADH activity is indicated by reduction of the indicator 2,3,5-triphenyl tetrazolium chloride, producing red colonies (3).
Amplification and cloning of chromosomal DNA.
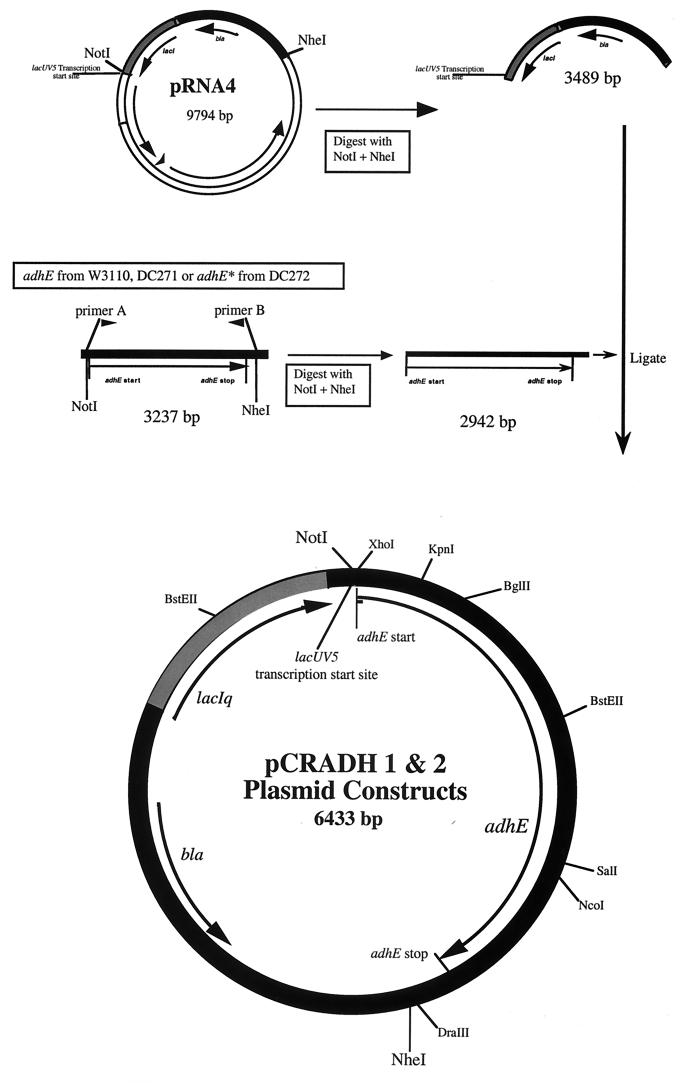
PCR employing two flanking primers was used to amplify adhE alleles. The 5′ primer (5′-ATGTGTGGAAGCGGCCGCTTTCAGGAGGCTCGAGAAATGGCTGTTACTAATGTCGCTGAA-3′) anneals to nucleotides −36 to +24 of the 5′ region of the adhE coding sequence and produces a NotI (underlined) restriction site in the resulting PCR product. The start codon is also underlined. The 3′ primer (5′-CTCGAGCGGGCTAGCAGGTGCGTCAGGCAGTGTTGTATC-3′) anneals 256 bp downstream of the adhE stop codon and produces an NheI restriction site (underlined) in the PCR product (Fig. 1). Template for the PCRs was prepared from 1 μl of overnight cultures by using the GeneReleaser protocol (BioVentures Inc., Murfreesboro, Tenn.). The PCR protocol consisted of an initial denaturation at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 3.5 min, followed by a final extension at 72°C for 10 min. PCR products containing the 3,237-bp amplified region were gel purified, digested with NotI and NheI, and ligated into a pACYC177-derived expression vector, pRNA4 (20), under the control of a lacUV5 promoter to give plasmids pW3110 (adhE3110), pCRADH1 (adhE271), and pCRADH2 (adhE272) (Fig. 1). The adhE genes from pCRADH1 and pCRADH2 were cloned into the XhoI and DraIII restriction sites of a pBR322 derivative, placing them under the control of the trpc constitutive promoter (20), resulting in the plasmids pCRADH7 (adhE271) and pCRADH8 (adhE272), thus alleviating the need for induction with IPTG. Ligation products were electroporated into strain DH5 or LEO42 (22). Transformants were grown in SOC (13) for 1 h and screened on AIP.
FIG. 1.
Construction of pCRADH1 and pCRADH2. The adhE and adhE* structural genes from E. coli were amplified using PCR and cloned into the plasmid pRNA4 as described in the text.
Fragment exchange experiments were performed by replacing sections of the wild-type gene with the corresponding fragments of the mutant gene. Three different fragment exchanges were performed. The first used BstEII (double cutter) and exchanged the 5′ region of the adhE gene. The second used the BglII and SalI cut sites and exchanged the middle of adhE, while the third used BglII and DraIII, which allowed exchange of the 3′ region of the gene. A construct containing only the mutation at position 267 of adhE (pMUT1) was made by transferring the BstEII fragment of pCRADH2 into pCRADH1. A construct with only the mutation at position 568 (pMUT2) was made by transferring the BstEII fragment from pCRADH1 (Fig. 1) into pCRADH2. Plasmids pMUT3 and pMUT4 were made by transferring the KpnI-NcoI fragments from pMUT1 and pMUT2, respectively, into pHIL145, placing the altered adhE genes under trpc control. All constructs were confirmed by sequencing.
Site-directed mutagenesis.
PCR was used to construct site-specific mutations of adhE (10, 14). For each set of mutations, two “outside” and two “inside” primers were used. For mutagenesis of Ala267 to Thr, two outside primers, ADH1 (5′-AAAGATGCCACCAACAAAGCG-3′) and ADH5 (5′-CGGACCAACGTTTTCAGAGAT-3′), were designed to anneal outside of the BglII and BstEII restriction sites. The inside primers were the mutagenic primer ADH*Thr (5′-GCCGTGGGTTGTAAAACGTTCA-3′) and ADH3 (5′-TTATGACGCTGTACGTGAACG-3′). The PCR product was gel purified, digested with BglII and BstEII, and ligated into pCRADH7. Transformants were screened for AdhE activity on AIP. For mutagenesis of Glu568, two outside primers, ADH13 (5′-ACGCGCTACCGGATTTTTAGACCC-3′) and ADH7 (5′-CAACCGGCTCGCGTTTCTTAC-3′), were designed to anneal to either side of the BstEII and NcoI sites of pCRADH7 and pCRADH8. To mutate Glu568, the primer ADH12 (5′-AGTGACTTCAGAACCTGTACCAGAAGTG-3′) was used together with one of four mutagenic primers: ADH*1 (5′-CACTTCGAASATCTGGCGCTGCGCTTTA-3′), which changes Glu to Asp or His; ADH*2 (5′-CACTTCGAACGTCTGGCGCTGCGCTTTA-3′), which changes Glu to Arg; ADH*3 (5′-CACTTCGAANNNCTGGCGCTGCGCTTTA-3′), which changes Glu to any amino acid; and ADH*4 (5′-CACTTCGAAKCGCTGGCGCTGCGCTTTA-3′), which changes Glu to Ala or Ser. The mutated PCR product was gel purified, digested with the restriction enzymes BstEII and NcoI, and ligated into either pCRADH7 or pCRADH8, which had been digested with the same restriction enzymes. Transformants were screened on AIP for the desired phenotype.
Chemical mutagenesis.
For mutagenesis by ethyl methanesulfonate (EMS), log-phase cells grown in LB medium plus 100 μg of ampicillin per ml were collected by centrifugation, washed twice, and resuspended in M9 glucose containing EMS (15 μg/ml) (Sigma Chemical Co.) at 37°C for 30 min. The mutagenized cells were washed, resuspended in M9 glucose, and grown aerobically overnight. The cells were then centrifuged, washed, resuspended in M9 ethanol, and incubated aerobically at 30°C for up to 1 week. Plasmids pEMS1 to pEMS5 were isolated from the resulting cultures.
RESULTS AND DISCUSSION
AdhE+ is active only anaerobically.
E. coli strains deficient in AdhE cannot grow by fermentation (4, 9). Our initial plasmid-borne clones of the wild-type adhE gene complemented adhE mutants anaerobically but did not show significant levels of ADH activity when grown aerobically (8). Since the adhE gene is expressed only anaerobically (21), this was not surprising. However, the constitutive adhC mutants of Clark and Cronan (6) produced large amounts of active ADH under both aerobic and anaerobic conditions, and the adhC mutant, DC272, was shown elsewhere to contain a mutation in the promoter region of adhE allowing constitutive transcription (35). Nonetheless, when a cloned adhE structural gene was placed under the control of the inducible lacUV5 promoter, the resulting strain failed to produce significant levels of ADH activity under aerobic conditions even when induced (data not shown). This suggested that AdhE protein was inactive under aerobic conditions. Presumably, the adhC mutants contained additional mutations affecting the activity of AdhE. To test this and identify any alterations in the coding sequence for AdhE, we amplified and cloned the adhE genes from DC272 (adhC mutant), DC271 (wild-type parent of DC272), and W3110 (source of the original cloned adhE+ gene) and put them under the control of the inducible lacUV5 promoter (Fig. 1). PCR was performed on genomic DNA with primers designed to amplify only the adhE structural gene plus 217 bp downstream of the stop codon. PCR products were gel purified, digested with NotI and NheI, and ligated behind the lacUV5 promoter, to give plasmids pW3110 (adhE3110), pCRADH1 (adhE271), and pCRADH2 (adhE272) (Fig. 1). These plasmids were transformed into PRC5 (adhE recA) and selected on LB medium plus ampicillin, and the plasmids were confirmed by restriction analysis. Transformants were then screened for ADH activity on AIP in the presence of IPTG. None of the clones carrying adhE from DC271 or W3110 showed ADH activity aerobically when induced with IPTG; however, 25 of 29 clones from DC272 produced functional AdhE aerobically when induced. The four DC272 clones that did not produce active ADH aerobically presumably contained PCR-induced mutations. Representative strains carrying cloned adhE genes were assayed for ADH activity (Table 1). Constructs carrying adhE from wild-type strains (e.g., CAS56) produced active ADH only anaerobically, but those with adhE from adhC mutants (e.g., CAS57) produced functional ADH under both aerobic and anaerobic conditions. The adhE+ strain CAS50, containing only the vector, had an ADH specific activity of <0.1 U aerobically and 1.9 U anaerobically. Much higher ADH levels were seen in CAS56, which has the chromosomal adhE gene deleted and carries a plasmid-borne wild-type adhE gene in multicopy. The vector-containing adhC strain, CAS51, had an ADH specific activity of 22 nmol/min/mg of protein aerobically and 20 nmol/min/mg of protein anaerobically. This is about twofold higher than that of CAS57, which has a chromosomal adhE deletion and a multicopy plasmid-borne adhE gene derived from an adhC mutant. The higher activity of the adhC mutant, with only a single copy of the adhE structural gene, is probably due to the very active adhC promoter, which is absent in the pCRADH2 construct (Fig. 1).
TABLE 1.
Specific activity of ADH clones
| Strain | Genotypea | Plasmidb | ADH activityc
|
|
|---|---|---|---|---|
| Aerobic | Anaerobic | |||
| CAS50 | DC271 (adhE+) | pACYC177 | <0.1 | 1.9 ± 0.53 |
| CAS51 | DC272 (adhC) | pACYC177 | 21.9 ± 0.1 | 20.1 ± 4.06 |
| CAS62 | Leo42 (ΔadhCE) | pACYC177 | <0.1 | ND |
| CAS56 | Leo42 (ΔadhCE) | pCRADH1 | 1.9 ± 0.5 | 7.8 ± 2.08 |
| CAS57 | Leo42 (ΔadhCE) | pCRADH2 | 9.3 ± 1.95 | 8.1 ± 1.84 |
Genotypes: DC271 is fadR mel tyrT (6), DC272 is fadR mel tyrT adhC (6), and Leo42 is fadR mel tyrT srl::Tn10 recA (ΔadhCE of DC272) (23).
Plasmids are described in the text and in Fig. 1.
ADH was assayed spectrophotometrically by monitoring the reduction of NAD+ to NADH at 340 nm (6). Activity is reported as nanomoles of NADH produced per minute per milligram of protein. ND, Leo42 is unable to grow anaerobically in the medium used to assay AdhE.
Aerobic activity of AdhE* from adhC mutants is due to a single mutation in the adhE structural gene.
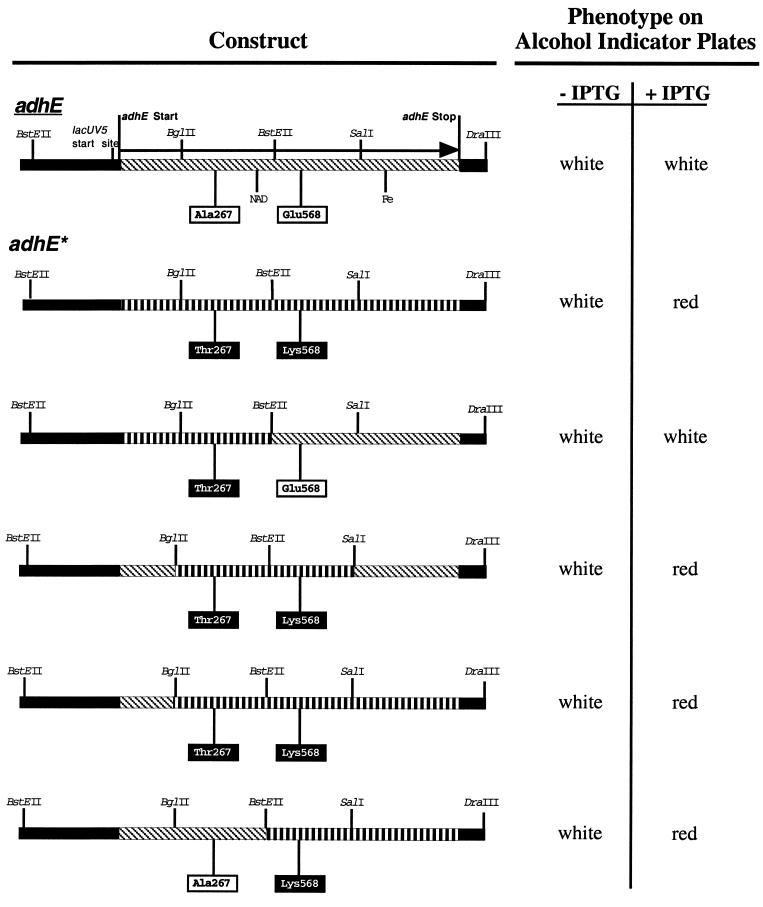
The differential aerobic expression of the adhE structural genes from adhC versus wild-type strains suggested that, in addition to alterations in the promoter region, adhC mutants have an altered adhE structural gene, which we will refer to as adhE*. To identify this mutation, the adhE+ gene on pCRADH1 and the adhE* gene on pCRADH2 were sequenced. Two mutations were discovered in the adhE* sequence, Ala267Thr and Glu568Lys. To determine if both mutations were required for aerobic activity of AdhE, the two mutations were separated by exchanging BstEII fragments between pCRADH1 and pCRADH2 (Fig. 1), creating the plasmids pMUT1 and pMUT2. Examination for AdhE activity on AIP (Fig. 2) showed that Glu568Lys alone is sufficient for aerobic activity of AdhE. To determine if other mutations in the AdhE protein allow aerobic activity, cultures containing pCRADH1 (adhE+) were mutagenized in vivo with EMS and colonies expressing active AdhE were selected on AIP under aerobic conditions. Five independent mutants were chosen and sequenced. All five contained the Glu568Lys mutation (Table 2). Two of the mutants contained additional silent mutations in the adhE gene. These data indicate that only the amino acid at position 568 affects the aerobic activity of AdhE.
FIG. 2.
Phenotypes of constructs containing hybrids of adhE+ and adhE* on alcohol indicator medium. Red colonies indicate complete oxidation of ethanol to acetyl-CoA. Slanted hash marks represent fragments from wild-type adhE+; straight hash marks represent fragments from the adhE* mutant. Restriction sites used in the fragment exchange experiments are indicated. The adhE* mutations discussed in the text are boxed. Putative NAD and iron binding sites (Fe) are shown.
TABLE 2.
Phenotypes of EMS and site-directed AdhE mutantsa
| Mutant type and AdhE clone | Position 267
|
Position 568
|
Other mutation
|
No. of occurrences | Phenotype | |||
|---|---|---|---|---|---|---|---|---|
| Amino acid | Codon | Amino acid | Codon | Amino acid | Mutation | |||
| EMS | ||||||||
| pCRADH1 (wt) | Ala | GCA | Glu | GAG | White | |||
| pCRADH2 (adhE*) | Thr | ACA | Lys | AAG | Red | |||
| pMUT1 | Thr | ACA | Glu | GAG | White | |||
| pMUT2 | Ala | GCA | Lys | AAG | Red | |||
| pEMS1 | Ala | GCA | Lys | AAG | Red | |||
| pEMS2 | Ala | GCA | Lys | AAG | Red | |||
| pEMS3 | Ala | GCA | Lys | AAG | Red | |||
| pEMS4 | Ala | GCA | Lys | AAG | Ala470Ala | GCG→GCA | Red | |
| pEMS5 | Ala | GCA | Lys | AAG | Arg463Arg | CGC→CGG | Red | |
| Site directed | ||||||||
| Hydrophobic | Ala | GCA | Ala | GCG | 1 | Red | ||
| Ala | GCA | Leu | CTG | 1 | Red | |||
| Polar | Ala | GCA | Asn | AAC | 1 | Red | ||
| Ala | GCA | Gly | GGC | 4 | Red | |||
| Ala | GCA | Gly | GGG | 12 | Red | |||
| Ala | GCA | Gly | GGT | 1 | Red | |||
| Ala | GCA | Gly | GGA | 5 | Red | |||
| Ala | GCA | Ser | TCT | 1 | Red | |||
| Basic | Ala | GCA | Arg | AGG | 7 | Red | ||
| Ala | GCA | Arg | CGG | 2 | Red | |||
| Ala | GCA | Arg | AGA | 1 | Red | |||
| Ala | GCA | His | CAT | 1b | Red | |||
| Ala | GCA | Lys | AAG | 1 | Red | |||
| Acidic | Ala | GCA | Asp | GAC | 2 | White | ||
| Ala | GCA | Asp | GAT | 1 | White | |||
| Ala | GCA | Glu | GAG | 16 | White | |||
| Ala | GCA | Glu | GAA | 5 | White | |||
| Nonsense | Ala | GCA | Stop | TGA | 11 | White | ||
AdhE phenotype and genotype of EMS mutants and site-directed mutants selected for ADH activity under aerobic conditions on alcohol indicator medium as described in the text. Red colonies on this medium indicate oxidation of ethanol by ADH and the coupled reduction of 2,3,5-triphenyl tetrazolium chloride. Mutated nucleotides for the EMS-mutagenized strains are underlined. All mutants derived from site-directed mutagenesis except the one containing His were isolated by random PCR mutagenesis of all three of the Glu568 codon nucleotides. Occurrence is the number of random mutants with the indicated codon. wt, wild type.
The His mutant was constructed by site-directed mutagenesis.
An acidic residue at position 568 causes aerobic inactivation of AdhE.
Using PCR, the codon specifying amino acid 568 was randomly mutated and cloned into either pCRADH1 (adhE+) or pCRADH2 (adhE*). Colonies with both active (red) and inactive (white) AdhE were selected on AIP under aerobic conditions, and their adhE genes were sequenced (Table 2). Several different amino acid substitutions were found in clones that produced AdhE that was active both aerobically and anaerobically. However, all of the mutants that produced active AdhE only under anaerobic conditions contained Asp or Glu at position 568. This suggests that an acidic residue at position 568 is necessary and sufficient for aerobic inactivation of AdhE.
Our results suggest the possibility of regulation of adhE at the posttranslational level. Only mutations resulting in substitution of a nonacidic residue at position 568 (such as Glu568Lys) of AdhE produce functional enzyme when the cultures are grown under aerobic conditions. Both mutants and wild type grew well under fermentative conditions, but only Glu568Lys-containing strains could grow on ethanol as sole carbon source in air, whereas Glu568 strains could not (data not shown). Since both ADH and acetaldehyde coenzyme A dehydrogenase activities are required for ethanol oxidation in vivo, these findings suggest that amino acid 568 affects both enzymatic activities in the E. coli AdhE protein. These data imply that the amino acid at position 568 is critical for the inactivation of AdhE in the presence of oxygen. Physiologically, such inactivation makes good sense: AdhE that had been synthesized anaerobically would be inactivated upon a shift to aerobic conditions, thereby avoiding the waste of substrate as ethanol when respiration is possible.
PFL converts pyruvate to acetyl-CoA plus formate (18), is induced anaerobically, and is posttranslationally activated by insertion of an organic free radical at Gly734 (34). Under aerobic conditions, PFL deactivase (the third function of AdhE) quenches the PFL radical, thereby inactivating PFL. Though the mechanism is unclear, it requires Fe2+, NAD+, and CoA (15). Whether aerobic inactivation of the ADH activity of AdhE requires any of these cofactors or affects its PFL deactivase activity is unknown. Propanediol oxidoreductase (FucO protein), which is homologous to the ADH domain of AdhE, is also oxidation sensitive (26). Mutations rendering FucO oxidation resistant (Ile7Leu and Leu8Val) also lowered its specific activity and appeared to act via changes in protein structure. In contrast, the only substitutions yielding aerobically active AdhE were those replacing Glu568. Moreover, these mutants showed increased specific enzyme activity. Thus, the mechanism of aerobic inactivation is probably different between FucO and AdhE. In anaerobically grown E. coli, AdhE exists as a complex that disassembles upon exposure of the cells to oxygen, resulting in loss of enzyme activity (15). Charge repulsion between adjacent acidic residues (e.g., Glu568) at the interface between two monomers of AdhE provides a possible mechanism for disassembly. It is uncertain how exposure to oxygen might alter the protein structure to bring such residues together or how Glu568→Lys would permit continued aerobic function. Iron-sulfur centers (32) are the oxidation-sensitive sites in several proteins, including the anaerobic regulator Fnr (11, 12). Though AdhE contains iron, it is not in an Fe-S cluster (7), and so this mechanism seems unlikely. It is thought that dissociation of Fnr dimers in the presence of oxygen is due to repulsion between the Asp154 residues of each polypeptide. Neutralization of charge repulsion by substitution with Ala154 allows the subunits to remain together in the presence of oxygen (2, 17, 19). It is not clear why the Asp154 residues come together only aerobically or how oxidation of the Fe-S center affects dimerization.
Our data show that virtually any amino acid substitution for Glu568 prevents loss of AdhE enzyme activity when cells are grown under aerobic conditions. The presence of Glu568 in wild-type AdhE and the previously reported regulation of the adhE gene at the transcriptional and translational levels suggest that strict regulation of ADH activity in E. coli has been selected as an important component of facultative metabolism.
ACKNOWLEDGMENTS
We thank Allen Nicholson and Laurie Boore for critical review of the manuscript.
This work was supported by National Institutes of Health grants GM55745 and GM52896.
REFERENCES
- 1.Aristarkhov A, Mikulskis A, Belasco J G, Lin E C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J Bacteriol. 1996;178:4327–4332. doi: 10.1128/jb.178.14.4327-4332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates D M, Lazazzera B A, Kiley P J. Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J Bacteriol. 1995;177:3972–3978. doi: 10.1128/jb.177.14.3972-3978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochner B R, Savageau M A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977;33:434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark D P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 5.Clark D P, Cronan J E., Jr Acetaldehyde coenzyme A dehydrogenase of Escherichia coli. J Bacteriol. 1980;144:179–184. doi: 10.1128/jb.144.1.179-184.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark D P, Cronan J E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980;141:177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins L A, Egan S M, Stewart V. Mutational analysis reveals functional similarity between NARX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J Bacteriol. 1992;174:3667–3675. doi: 10.1128/jb.174.11.3667-3675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham P. Ph.D. dissertation. Carbondale: Southern Illinois University; 1987. [Google Scholar]
- 9.Cunningham P, Clark D P. The use of suicide substrates to select mutants of Escherichia coli lacking enzymes of alcohol fermentation. Mol Gen Genet. 1986;205:487–493. doi: 10.1007/BF00338087. [DOI] [PubMed] [Google Scholar]
- 10.Erlich H A. PCR technology: principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. [Google Scholar]
- 11.Green J, Guest J R. Activation of FNR-dependent transcription by iron: an in vitro switch for FNR. FEMS Microbiol Lett. 1993;113:219–222. doi: 10.1111/j.1574-6968.1993.tb06517.x. [DOI] [PubMed] [Google Scholar]
- 12.Green J, Guest J R. A role for iron in transcriptional activation by FNR. FEBS Lett. 1993;329:55–58. doi: 10.1016/0014-5793(93)80192-w. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi R, Krummel B, Saiki R. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessler D, Herth W, Knappe J. Ultrastructure and pyruvate formate-lyase radical quenching property of the multienzymatic AdhE protein of Escherichia coli. J Biol Chem. 1992;267:18073–18079. [PubMed] [Google Scholar]
- 16.Kessler D, Leibrecht I, Knappe J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 1991;281:59–63. doi: 10.1016/0014-5793(91)80358-a. [DOI] [PubMed] [Google Scholar]
- 17.Khoroshilova N, Beinert H, Kiley P J. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci USA. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knappe J, Neugebauer F A, Blaschkowski H P, Ganzler M. Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc Natl Acad Sci USA. 1984;81:1332–1335. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 20.Lee K, Holland-Staley C A, Cunningham P R. Genetic analysis of the Shine-Dalgarno interaction: selection of alternative functional mRNA-rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- 21.Leonardo M R. Ph.D. dissertation. Carbondale: Southern Illinois University; 1993. [Google Scholar]
- 22.Leonardo M R, Clark D P. Locations of genes in the nar-adhE region of the Escherichia coli K-12 chromosome. J Bacteriol. 1991;173:1574–1575. doi: 10.1128/jb.173.5.1574-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonardo M R, Cunningham P R, Clark D P. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J Bacteriol. 1993;175:870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin E C C, Kuritzkes D R. Pathways for anaerobic electron transport. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 201–221. [Google Scholar]
- 26.Lu Z, Cabiscol E, Obradors N, Tamarit J, Ros J, Aguilar J, Lin E C C. Evolution of an Escherichia coli protein with increased resistance to oxidative stress. J Biol Chem. 1998;273:8308–8316. doi: 10.1074/jbc.273.14.8308. [DOI] [PubMed] [Google Scholar]
- 27.Luria S E, Burrous J W. Hybridization between Escherichia coli and Shigella. J Bacteriol. 1957;74:461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph F, Purich D, Fromm H. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. I. Partial purification, properties and kinetic studies of the enzyme. J Biol Chem. 1968;243:5536–5545. [PubMed] [Google Scholar]
- 29.Shone C, Fromm H. Steady state and presteady state kinetics of coenzyme A linked aldehyde dehydrogenase from Escherichia coli. Biochemistry. 1981;20:7494–7501. doi: 10.1021/bi00529a026. [DOI] [PubMed] [Google Scholar]
- 30.Smith M W, Neidhardt F C. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol. 1983;154:336–343. doi: 10.1128/jb.154.1.336-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Still J. Alcohol enzyme of Bacterium coli. Biochem J. 1940;34:1177–1182. doi: 10.1042/bj0341177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unden G, Becker S, Bongaerts J, Schirawski J, Six S. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Leeuwenhoek. 1994;66:3–23. doi: 10.1007/BF00871629. [DOI] [PubMed] [Google Scholar]
- 33.Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 34.Wagner A F V, Frey M, Neugebauer F A, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong P, Barrett E L. Aerobic and anaerobic alcohol dehydrogenases in Escherichia coli. FEMS Microbiol Lett. 1983;22:143–148. [Google Scholar]