Abstract
Background and study aims Artificial intelligence is currently able to accurately predict the histology of colorectal polyps. However, systems developed to date use complex optical technologies and have not been tested in vivo. The objective of this study was to evaluate the efficacy of a new deep learning-based optical diagnosis system, ATENEA, in a real clinical setting using only high-definition white light endoscopy (WLE) and to compare its performance with endoscopists.
Methods ATENEA was prospectively tested in real life on consecutive polyps detected in colorectal cancer screening colonoscopies at Hospital Clínic. No images were discarded, and only WLE was used. The in vivo ATENEA’s prediction (adenoma vs non-adenoma) was compared with the prediction of four staff endoscopists without specific training in optical diagnosis for the study purposes. Endoscopists were blind to the ATENEA output. Histology was the gold standard.
Results Ninety polyps (median size: 5 mm, range: 2–25) from 31 patients were included of which 69 (76.7 %) were adenomas. ATENEA correctly predicted the histology in 63 of 69 (91.3 %, 95 % CI: 82 %–97 %) adenomas and 12 of 21 (57.1 %, 95 % CI: 34 %–78 %) non-adenomas while endoscopists made correct predictions in 52 of 69 (75.4 %, 95 % CI: 60 %–85 %) and 20 of 21 (95.2 %, 95 % CI: 76 %–100 %), respectively. The global accuracy was 83.3 % (95 % CI: 74%–90 %) and 80 % (95 % CI: 70 %–88 %) for ATENEA and endoscopists, respectively.
Conclusion ATENEA can accurately be used for in vivo characterization of colorectal polyps, enabling the endoscopist to make direct decisions. ATENEA showed a global accuracy similar to that of endoscopists despite an unsatisfactory performance for non-adenomatous lesions.
Introduction
Colorectal cancer (CRC) is the third most common cancer in both sexes and the second leading cause of death in the world 1 . Screening for CRC and the removal of neoplastic polyps by colonoscopy has led to a substantial improvement in survival 2 .
Optical diagnosis aims to predict the histology of a polyp based on its endoscopic features. This practice could avoid pathological analysis in several cases and reduce the derived costs. The American Society for Gastrointestinal Endoscopy’s PIVI working group established diagnostic thresholds for real-time implementation of optical diagnosis for diminutive polyps (≤ 5 mm) 3 . However, PIVI criteria have not yet been met in community-based practices or in non-expert hands 4 5 . In this regard, the European Society of Gastrointestinal Endoscopy (ESGE) emphasizes the importance of being able to ensure and maintain competence in optical diagnosis as well as considering only the proportion of high-confidence diagnosis as a benchmark 6 .
During the past few decades, considerable technological advances have been made in the application of artificial intelligence (AI) to medicine. Computer-aided diagnosis (CADx) is a promising solution to overcome human variation in characterization of polyps by providing decision support. In this specific field, CADx approaches based on deep learning (DL) represent an advantage over previous machine learning by combining both the automatic extraction and classification of image characteristics using a multilayered system called convolutional neural networks (CNNs) 7 .
The quality and design of published CADx systems has varied over time. Initial studies were carried out retrospectively and were tested ex vivo using selected stored images 8 9 10 11 12 . More recent studies, most of them DL approaches, have been prospectively conducted reporting higher accuracy with faster processing times, which allows diagnosis in real time 13 14 15 16 . However, to date, only CADx systems using complex optical technologies as endocytoscopy have been tested in vivo 13 , that is, while the colonoscopy is being performed, which still hinders the adoption of this technology in daily practice. Furthermore, the newest CADx systems are currently using advanced imaging modalities, which clearly limits their implementation worldwide.
The aim of this study was to evaluate the efficacy of a new CADx system based on DL called ATENEA for in vivo optical diagnosis in consecutive patients using only white light endoscopy (WLE) and compare its performance with endoscopists.
Methods
Development of ATENEA
As a first step, images of any polyp from routine colonoscopies performed at Hospital Clínic of Barcelona from January 2016 to December 2020 were prospectively collected. The images were captured with high-definition colonoscopes (CF-HQ180, CF-HQ-185, CF-HQ-190 and EVIS EXERA III videoprocessor, Olympus Europe, Hamburg, Germany) using an external computer with a frame grabber to ensure image acquisition with the highest quality. Only white light images without magnification or chromoendoscopy were used. Data from morphology, location, and size of the polyp were collected and periodically transferred by an assistant into the database along with the histological category of the lesion obtained after pathological analysis.
The development of ATENEA consisted of two stages: feature extraction and image classification. Feature extraction was made by using a faster region-based CNN with ResNet50 as backbone. For system training purposes, a region of interest (ROI) delineating the polyp was manually defined by clinicians using a program called GTCreator ( Fig. 1 ) 17 . The extracted features, ROI, and actual histologic class of the polyp were used to train the system. ATENEA learned to classify images into adenoma or non-adenoma categories using an 80 % (high confidence) threshold value (predictions with a confidence value < 80 % were not considered valid to represent the actual performance of the system as they could not guarantee a robust performance).
Fig. 1.
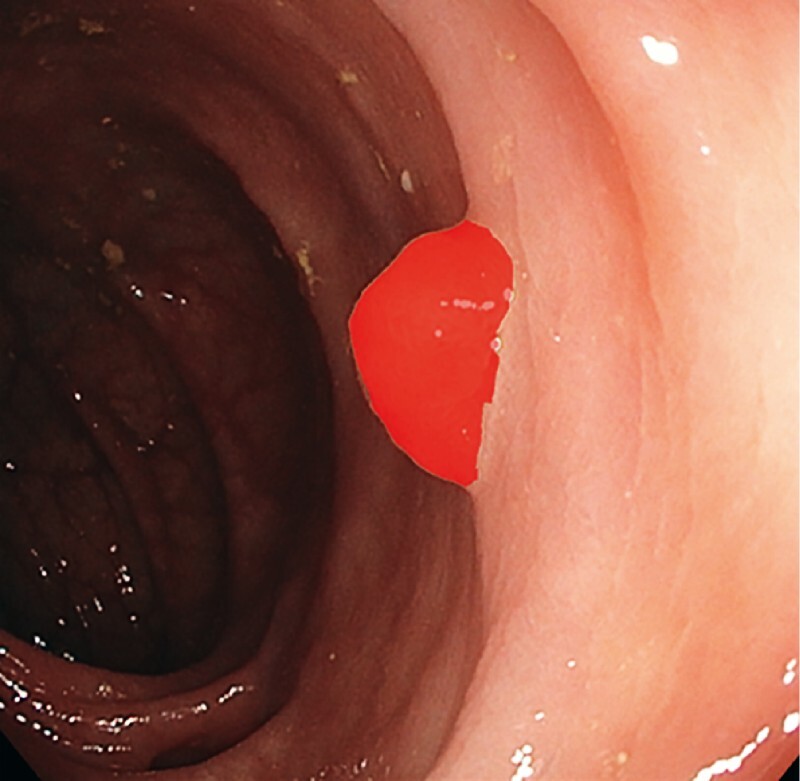
Example of a manually defined region of interest (ROI) delineating the polyp using GTCreator in the training phase.
ATENEA was trained and validated with a total of 1049 high-definition white light images of 483 polyps from 354 patients, with a maximum of three images of the same polyp (but with a different view or perspective). Images had variable quality but all had a visible mucosal pattern and only blurred images and polyps covered by mucus were excluded. Images containing patient data or without histological analysis were also excluded. About two-thirds were adenomas and one-third non-adenomas, following a similar proportion to what is found in real life, with a median size of 4.5 mm (range 2 mm-20 mm). These images were randomly split into training (n = 837) and validation sets (n = 212), with the condition that all the images of the same polyp were in the same set.
In vivo experiment
This observational prospective cohort study was performed in Hospital Clinic of Barcelona between January and March 2021 and included individuals from the fecal immunochemical test-based (cut-off of ≥ 20 μg of hemoglobin/g of feces) organized population CRC screening program in which all individuals aged 50 to 69 years are invited to participate. Colonoscopies were performed by four staff endoscopists with more than 1000 colonoscopies and adenoma detection rates of 47 %, 48%, 51 %, and 55 %, respectively (average in this program and time period was 49 %). No specific training in optical diagnosis was performed for the study purpose. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Hospital Clínic of Barcelona (HCB2017/0506, 7/18/2017) and informed consent was obtained from all patients involved in the study. It was also registered in ClinicalTrials.gov (NCT03775811).
All polyps detected and resected with a final pathological report were prospectively included regardless of image quality (not centered polyps, blurred or covered by mucus). Only the image that was considered adequate for the prediction (either by the orientation of the polyp within the image or by the proximity for its correct assessment) was selected by the endoscopist during the exploration and was analyzed. The following variables were collected: estimated size (in mm), location (rectum-sigma, descending, transverse, ascending) and morphology according to Paris classification 18 , which were properly noted in the colonoscopy report.
In real time, ATENEA classified each polyp as an adenoma or non-adenoma and provided the confidence value for each prediction. To avoid losing any prediction, we were more flexible than in train/validation phases and the minimum threshold was reduced to 50 %. Values between 50 % and 80 % were considered low-confidence predictions. In this phase, the system was fully automatic (without delineation of ROI), providing for each lesion a bounding box and its corresponding histological class. Time needed to acquire and process each image to obtain the automatic prediction was of 40 milliseconds (real time).
For the optical diagnosis process, the endoscopists were asked to categorize each lesion into two categories (adenoma and non-adenoma) based on its surface pattern and to provide its diagnostic confidence (high vs low) without any time limitation. Endoscopists were blinded to the ATENEA output. The decision was intuitive and based on their previous experience but without using chromoendoscopy or any of the existing classifications. Serrated sessile lesions (SSLs) were included in the non-adenoma category. Fig. 2 shows the set-up in the exploration room.
Fig. 2.
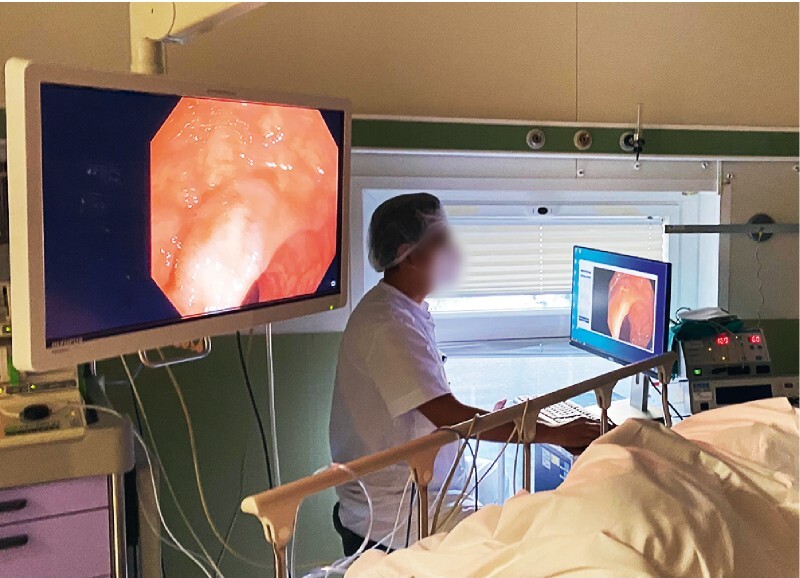
Setting in the endoscopy room showing the position of the assistant sitting in front the computer: the endoscopist is blind to the image displayed in the computer and ATENEA’s output.
Histopathology
All polyps were removed using the usual techniques and sent separately for further evaluation to the Pathology Department, which was the gold standard. The diagnosis of dysplasia in neoplastic polyps was made based on the Modified Vienna Classification 19 . The pathologist was blinded to the predictions of both endoscopist and ATENEA.
Statistical analysis
All the polyps were globally analyzed, independently of the endoscopist who performed the exploration. The numbers of polyps that were true positive (adenomatous polyps predicted to be adenomatous), true negative (non-adenomatous polyps predicted to be non-adenomatous), false positive (non-adenomatous polyps predicted to be adenomatous) or false negative (adenomatous polyps predicted to be non-adenomatous) were calculated. Sensitivity, specificity, positive predictive value, negative predictive value and accuracy with 95 % confidence intervals were calculated. Comparisons of these metrics between ATENEA and endoscopists were performed with Chi squared test.
Receiver operator characteristics (ROC) curves for use of different prediction confidence values to determine ATENEA in vivo performance for diagnosis of adenoma were constructed with Matlab. The area under the curve (AUC) and optimal operating point with its sensitivity and specificity and 95 % confidence intervals were calculated.
Two-sided P < 0.05 was considered statistically significant. All calculations were performed with R version 4.1.2 and Matlab for Windows version 2020.
Results
During the study period, 90 polyps from 31 consecutive patients were included. Sixty-nine (76.7 %) were adenomas with a median size of 5.0 mm (range: 2–25 mm). Characteristics of the polyps are described in Table 1 .
Table 1. Characteristics of the polyps included in the study.
| In vivo test (n = 90) | |
| Histological type | |
|
69 (76.7 %) |
|
21 (23.3 %) |
|
16 |
|
5 |
| Size (in mm) | |
|
52 (57.8 %) |
|
21 (23.3 %) |
|
17 (18.8 %) |
| Location | |
|
22 (24.4 %) |
|
68 (75.6 %) |
| Morphology | |
|
5 (5.5 %) |
|
30 (33.3 %) |
|
55 (61.1 %) |
ATENEA provided an output for all 90 polyps and was able to correctly predict the histology in 63 of 69 adenomas (sensitivity: 91.3 %, 95 % CI: 82 %–97 %) and 12 of 21 non-adenomas (specificity: 57.1 %, 95 % CI: 34 %–78 %). Endoscopists were able to provide a correct optical diagnosis in 52 of 69 adenomas (sensitivity: 75.4 %, 95 % CI: 64 %–85 %) and 20 of 21 non-adenomas (specificity: 95.2 %, 95 % CI: 76 %–100 %), with an accuracy of 83.3 % (95 % CI: 74 %–90 %) and 80 % (95 % CI: 70 %–88 %) for CADx and endoscopists, respectively ( Table 2 ).
Table 2. Performance characteristics of ATENEA and the endoscopists for diagnosis of adenoma.
| TP | FP | TN | FN | PPV | SENS | NPV | Spec | Accuracy | |
| ATENEA, n = 90 | 63 | 9 | 12 | 6 | 87.5 % (95 % CI: 78 %–94%) | 91.3 % (95 % CI: 82 %–97%) | 66.7 % (95 % CI: 41 %–87%) | 57.1 % (95 % CI: 34 %–78%) | 83.3 % (95 % CI: 74 %–90%) |
| Endoscopists, n = 90 | 52 | 1 | 20 | 17 | 98.1 % (95 % CI: 90 %–100%) | 75.4 % (95 % CI: 64 %–85%) | 54.0 % (95 % CI: 37 %–71%) | 95.2 % (95 % CI: 76 %–100%) | 80.0 % (95 % CI: 70 %–88%) |
| P value | 0.07 | 0.02 | 0.55 | 0.01 | 0.7 | ||||
| ATENEA diminutive polyps ≤ 5 mm, n = 52 | 30 | 7 | 11 | 4 | 81.1 % (95 % CI: 65 %–92%) | 88.2 % (95 % CI: 73 %–97%) | 73.3 % (95 % CI: 45 %–92%) | 61.1 % (95 % CI: 36 %–83%) | 78.8 % (95 % CI: 65 %–89%) |
| Endoscopists diminutive polyps ≤ 5 mm, n = 52 | 20 | 1 | 17 | 14 | 95.2 % (95 % CI: 76 %–100%) | 58.8 % (95 % CI: 41 %–75%) | 54.8 % (95 % CI: 36 %–73%) | 94.4 % (95 % CI: 73 %–100%) | 71.1 % (95 % CI: 57 %–83%) |
| P value | 0.27 | 0.01 | 0.38 | 0.04 | 0.5 | ||||
| ATENEA small polyps < 10 mm, n = 73 | 47 | 9 | 12 | 5 | 83.9 % (95 % CI: 72 %–92%) | 90.4 % (95 % CI: 79 %–97%) | 70.6 % (95 % CI: 44 %–90%) | 57.2 % (95 % CI: 34 %–78%) | 80.8 % (95 % CI: 70 %–89%) |
| Endoscopists small polyps < 10 mm, n = 73 | 36 | 1 | 20 | 16 | 97.3 % (95 % CI: 86 %–100%) | 69.2 % (95 % CI: 55 %–81%) | 55.5 % (95 % CI: 38 %–72%) | 95.2 % (95 % CI: 76 %–100%) | 76.7 % (95 % CI: 65 %–86%) |
| P value | 0.09 | 0.02 | 0.46 | 0.01 | 0.69 | ||||
TP, true positive; FP, false positive; TN, true negative; FN, false negative; PPV, positive predictive value; SENS, sensitivity; NPV, negative predictive value; SPEC, specificity.
With respect to diminutive polyps (≤ 5 mm), ATENEA was able to correctly predict the histology in 31 of 35 adenomas (sensitivity: 88.2 %, 95 % CI: 73 %–97 %) and 11 of 18 non-adenomas (specificity: 61.1 %, 95 % CI: 36 %–83 %) and endoscopists in 20 of 35 adenomas (sensitivity: 58.8 %, 95 % CI: 41%–75 %) and 17 of 18 non-adenomas (specificity: 94.4 %, 95 % CI: 73 %–100 %), respectively, with an accuracy of 78.8 % (95 % CI: 65 %–89 %) and 71.1 % (95 % CI: 57 %–83 %) for ATENEA and endoscopists, respectively ( Table 2 ). Results of prediction for small polyps (< 10 mm) are also shown in Table 2 .
ATENEA and the endoscopists disagreed in their prediction in 31 of 90 cases (34.5 %). Endoscopists made their prediction with high confidence in 79 cases (87 %) and with low confidence in 11. In all these 11 cases, ATENEA made a good prediction ( Fig. 3 ).
Fig. 3.
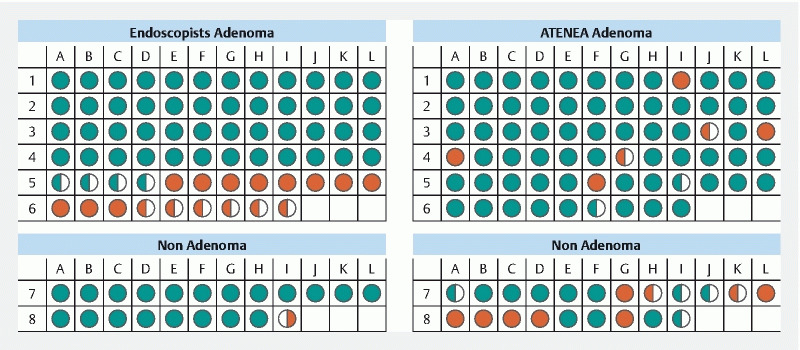
ATENEA and endoscopists’ predictions for all polyps. Each circle represents a polyp and colors correspond to a correct prediction (green) and incorrect prediction (red) with high confidence (full circle) or low confidence (half circle).
The ROC curve for ATENEA in vivo performance ( Fig. 4 ) showed an AUC of 0.782. The optimal operating point was achieved by using 74.2 % as the threshold value with a sensitivity of 86.9 % (95 % CI: 79.9 %–93.9 %) and a specificity of 66.7 % (95 % CI: 56.9 %–76.4 %).
Fig. 4.
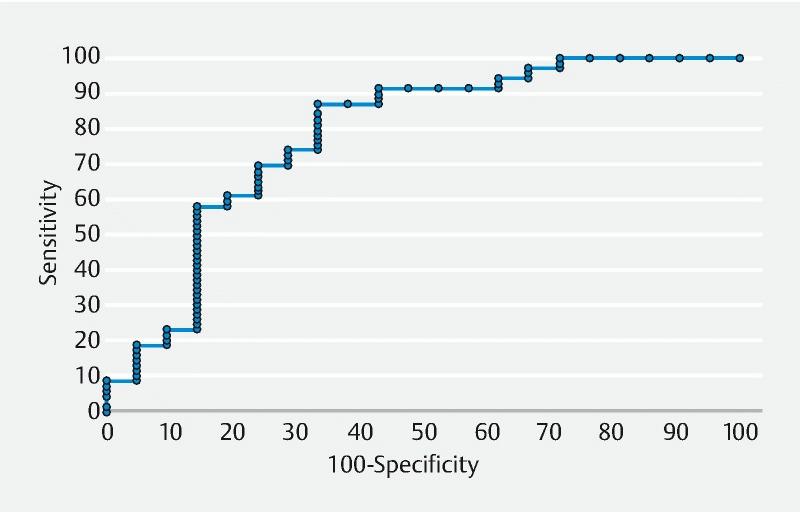
Receiver operating characteristic (ROC) curve for different prediction confidence values for ATENEA. The optimal operating point (defined as the point in the curve with better balance of specificity and sensitivity) was achieved by using 74.2 % as the threshold value.
Discussion
This study demonstrates that ATENEA, a fully automatic optical diagnostic system, can accurately predict polyp histology in an in vivo setting using only white light, which answers one of the 10 key research questions identified by international experts related to AI implementation in colonoscopy 20 . ATENEA is particularly useful for identifying adenomatous lesions, showing its readiness to be used in a clinical environment.
Until now, the majority of CAD systems developed for characterizing colorectal polyps that use AI have required advanced optical diagnostic equipment, such as narrow band imaging (NBI) 9 10 11 12 21 and endocytoscopy 8 22 23 . However, WLE. which is the most common endoscopic modality, has not been extensively studied in this context yet. For this reason, we used only white light images to allow for widespread use of ATENEA regardless of the manufacturer and model of endoscope used.
Our group previously developed a hand-crafted predictive model based on extraction of surface patterns (textons) over white light images. This system was validated in a small dataset containing only 225 high-quality images with a diagnostic accuracy of over 90 % 24 . Unfortunately, the system was not fully automatic and could not perform in real time and, consequently, it could not be used in an in vivo experiment. In contrast, ATENEA can perform in vivo automatic polyp classification due to its capability of calculating the result in less than 40 ms (real time). It is important to make a difference between what real-time and in vivo means: the first refers to the time the system takes to process an image, whereas the second is related to where and when the system is applied (in vivo: in the exploration room versus ex vivo: off-line experiments). These concepts are commonly confused in the literature and, in most of the cases, real-time is used instead of in vivo without mentioning the actual processing time 13 14 .
The results obtained in the present experiment showed that ATENEA had better global accuracy than endoscopists, particularly for adenomatous lesions. Conversely, clinicians performed better for the non-adenoma category. It has to be pointed out that they had more information than the system because they knew the location and size of the lesion (variables for which ATENEA was blind), so this could lead to a major pretest probability of better diagnosis of non-adenomatous lesions when faced with a small or diminutive polyp in the rectosigmoid colon. In contrast, ATENEA performance depended solely on the number and variability of examples from each of the classes in the training set.
The lower performance of ATENEA for non-adenomatous lesions could limit implementation of a “leave in situ” policy. However, it must be stated that the training of the AI system was greatly affected by the reduced number of examples of this class in the dataset. There are two reasons for this: first, the prevalence of non-adenomatous lesions is generally lower and second, they are not systematically removed when located in the rectosigmoid colon, making their collection difficult. Hence, it is necessary to enlarge the dataset both in quantity and percentage of examples of the minority class.
CADx systems are not intended to replace endoscopists, but rather, to help them in their tasks. As stated by the ESGE review on advanced endoscopic imaging, the most likely scenario is that the intelligent systems will be used as a “second reader” to support the endoscopist’s final diagnosis 25 . In this sense, an important result from our study was that if endoscopists with low confidence had followed the output of the system, all their predictions would have been correct. This shows the potential of ATENEA to assist clinicians with less expertise.
Similar to endoscopists, AI systems do also provide a confidence value in their predictions using a percentage instead of a binary assessment (high versus low). By varying the confidence value, we represented different performances of ATENEA using a ROC curve. Our results show that if the confidence value is low, the sensitivity of the system is lower as it provides more outputs, some of them erroneous. Conversely, if we increase the allowed confidence value, less but more confident outputs will be provided, with the risk of missing some of the lesions without a prediction. The so-called Simple Optical Diagnosis Accuracy or SODA criteria, which were recently published by the ESGE Curricula Working Group 26 , are more flexible than the PIVI criteria and emphasize the importance of not leaving any diminutive lesion with advanced neoplasia in situ. In accordance with SODA criteria, we considered sensitivity to be the most important outcome of the intelligent system. The AUC value obtained in our study is similar to the 0.84 reported in a recent meta-analysis using only WLE 27 (which is usually lower than the AUC in studies using chromoendoscopy or magnification techniques).
Unlike other studies in which low-quality images or the known “difficult for AI cases” were not included, the main strength of this study is that it was performed under real clinical practice conditions and polyps were included regardless of image quality (not centered polyps, blurred or covered by mucus). If we had excluded these polyps from the analysis, the ATENEA performance would have been better. Nevertheless, the lack of publicly available annotated datasets does not allow for a fair comparison between ATENEA and other CADx systems. In this sense, comparison of metrics in meta-analysis are difficult to understand as the number, quality, and class of the images are different in each study and affect training and testing stages of the validation of computational systems.
The study has the following limitations. First, the training dataset was small in terms of number of different polyps. To mitigate this, we used pre-trained weights from ImageNet 28 , which is a more general dataset and we applied data augmentation operations (color transformation, rotation, horizontal and vertical flip) to enlarge its size. The collaboration with other centers and the public availability of other datasets could also be of great use to both enlarge the dataset and perform multicenter validations, increasing the robustness of the results. Second, our study did not consider a separate class for SSL due to the low number of examples in the dataset. The problem of SSLs has not adequately been addressed in other previous studies. It is well known that SSLs are neoplastic lesions and there is not an ideal optical method for their characterization 29 . Due to the clinical relevance of SSLs, some studies propose a classification between neoplastic and non-neoplastic lesions instead of using adenoma vs. non-adenoma. Following this logic, if we had included all SSL in the same category as adenomas, four of five SSLs would have been correctly classified.
Conclusions
In conclusion, ATENEA, a CADx system based only on WLE data, is ready to be used for in vivo and real-time characterization of colorectal polyps, enabling the endoscopist to make direct decisions. ATENEA achieved a global accuracy similar to endoscopists, despite having lower performance for non-adenomatous lesions.
Acknowledgments
This work was supported by the Instituto de Salud Carlos III (PI17/00894 and PI19/01050), co-funded by the European Union; the Spanish Ministry for Science and Innovation (MCIN) (ALETHEIA project, PID2020-120311RB-I00 funded by MCIN/AEI/10.13039/501100011033 ); Fundació Marató de TV3 (201932–30); Secretaria d’Universitats i Recerca de la Generalitat de Catalunya (2014-SGR-1470, 2014-SGR-135, SGR-2017–1669 and SGR-2017–653), and by CERCA Programme/Generalitat de Catalunya. Ana García-Rodríguez received a personal grant from the Societat Catalana de Digestologia. The authors thank Dr. Andrés Cárdenas for his English medical writing assistance and Roger Borràs for performing the statistical analysis.
Footnotes
Competing interests Glòria Fernández-Esparrach was employed on an advisory board of Medtronic and is shareholder of MiWEndo Solutions S.L. Míriam Cuatrecases is shareholder of MiWEndo Solutions S.L. Maria Pellisé has served on clinical advisory boards for Fujifilm Europe; has served on the clinical advisory board and owns share options in MiWendo S.L.; reports speaker fees from Casen Recordati, Fujifilm, Medtronic and Olympus Europe; and has received research funding from Fujifilm Europe, Casen Recordati and Ziuz.
References
- 1.Bray F, Ferlay J, Soerjomataram I et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Levin B, Lieberman D A, McFarland B et al. Screening and Surveillance for the Early Detection of Colorectal Cancer and Adenomatous Polyps, 2008: A Joint Guideline From the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Abu Dayyeh BK, Thosani N, Konda V et al. ASGE technology committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:502–516. doi: 10.1016/j.gie.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Vu H T, Sayuk G S, Hollander T G et al. Resect and discard approach to colon polyps: real-world applicability among academic and community gastroenterologists. Dig Dis Sci. 2015;60:502–508. doi: 10.1007/s10620-014-3376-z. [DOI] [PubMed] [Google Scholar]
- 5.Rees C J, Rajasekhar P T, Wilson A et al. Narrow band imaging optical diagnosis of small colorectal polyps in routine clinical practice: The Detect Inspect Characterise Resect and Discard 2 (DISCARD 2) study. Gut. 2016;66:887–895. doi: 10.1136/gutjnl-2015-310584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker E, Houwen B BSL, Puig I et al. Curriculum for optical diagnosis training in Europe: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy. 2020;52:C10. doi: 10.1055/a-1264-2634. [DOI] [PubMed] [Google Scholar]
- 7.Lecun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 8.Misawa M, Kudo S E, Mori Y et al. Characterization of colorectal lesions using a computer-aided diagnostic system for narrow-band imaging endocytoscopy. Gastroenterology. 2016;150:1531–1.532E6. doi: 10.1053/j.gastro.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Byrne M F, Chapados N, Soudan F et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross S, Trautwein C, Behrens A et al. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354–1359. doi: 10.1016/j.gie.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Kominami Y, Yoshida S, Tanaka S et al. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643–649. doi: 10.1016/j.gie.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Chen P J, Lin M C, Lai M J et al. Accurate Classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Mori Y, Kudo S E, Misawa M et al. Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy a prospective study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Diaz E, Baffy G, Lo W K et al. Real-time artificial intelligence-based histologic classification of colorectal polyps with augmented visualization. Gastrointest Endosc. 2021;93:662–670. doi: 10.1016/j.gie.2020.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi H, Tamai N, Kamba S et al. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand J Gastroenterol. 2019;54:800–805. doi: 10.1080/00365521.2019.1627407. [DOI] [PubMed] [Google Scholar]
- 16.Zachariah R, Samarasena J, Luba D et al. Prediction of polyp pathology using convolutional neural networks achieves “resect and discard” thresholds. Am J Gastroenterol. 2020;115:138–144. doi: 10.14309/ajg.0000000000000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernal J, Histace A, Masana M et al. GTCreator: a flexible annotation tool for image-based datasets. Int J Comput Assist Radiol Surg. 2019;14:191–201. doi: 10.1007/s11548-018-1864-x. [DOI] [PubMed] [Google Scholar]
- 18.Lambert R, Lightdale C J. The Paris endoscopic classification of superficial neoplastic lesions: Esophagus, stomach, and colon – Paris, France November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3–S43. doi: 10.1016/s0016-5107(03)02159-x. [DOI] [PubMed] [Google Scholar]
- 19.Schlemper R J, Riddell R H, Kato Y et al. The vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad O F, Mori Y, Misawa M et al. Establishing key research questions for the implementation of artificial intelligence in colonoscopy: A modified Delphi method. Endoscopy. 2021;53:893–891. doi: 10.1055/a-1306-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takemura Y, Yoshida S, Tanaka S et al. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video) Gastrointest Endosc. 2012;75:179–185. doi: 10.1016/j.gie.2011.08.051. [DOI] [PubMed] [Google Scholar]
- 22.Mori Y, Kudo S E, Chiu P WY. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110–1118. doi: 10.1055/s-0042-113609. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Kudo S E, Mori K. Potential of artificial intelligence-assisted colonoscopy using an endocytoscope (with video) Dig Endosc. 2018;30:52–53. doi: 10.1111/den.13005. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez-Montes C, Sánchez F J, Bernal J et al. Computer-aided prediction of polyp histology on white light colonoscopy using surface pattern analysis. Endoscopy. 2019;51:261–265. doi: 10.1055/a-0732-5250. [DOI] [PubMed] [Google Scholar]
- 25.Bisschops R, East J E, Hassan C et al. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline – Update 2019. Endoscopy. 2019;51:1155–1179. doi: 10.1055/a-1031-7657. [DOI] [PubMed] [Google Scholar]
- 26.Houwen B BSL, Hassan C, Coupé V MH et al. Definition of competence standards for optical diagnosis of diminutive colorectal polyps: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy. 2022;54:88–99. doi: 10.1055/a-1689-5130. [DOI] [PubMed] [Google Scholar]
- 27.Lui T KL, Guo C G, Leung W K. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:11–2.2E7. doi: 10.1016/j.gie.2020.02.033. [DOI] [PubMed] [Google Scholar]
- 28.Deng J, Dong W, Socher R.ImageNet: a Large-Scale Hierarchical Image Database in 2009 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPR Workshops) Miami, FL: 2009pp. 248–255. 10.1109/CVPR.2009.5206848Corpus ID: 57246310 [DOI] [Google Scholar]
- 29.Bustamante-Balén M, Satorres C, Ramos-Soler D et al. Evaluation of the optical criteria for sessile serrated lesions of the colon: A prospective study on a colorectal cancer screening population. Endosc Int Open. 2021;9:E14–E21. doi: 10.1055/a-1293-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]


