Abstract
Bacteriophage λ uses a holin-endolysin system for host cell lysis. R, the endolysin, has muralytic activity. S, the holin, is a small membrane protein that permeabilizes the inner membrane at a precisely scheduled time after infection and allows the endolysin access to its substrate, resulting in host cell lysis. λ S has a single cysteine at position 51 that can be replaced by a serine without loss of the holin function. A collection of 27 single-cysteine products of alleles created from λ SC51S were tested for holin function. Most of the single-cysteine variants retained the ability to support lysis. Mutations with the most defective phenotype clustered in the first two hydrophobic transmembrane domains. Several lines of evidence indicate that S forms an oligomeric structure in the inner membrane. Here we show that oligomerization does not depend on disulfide bridge formation, since the cysteineless SC51S (i) is functional as a holin and (ii) shows the same oligomerization pattern as the parental S protein. In contrast, the lysis-defective SA52V mutant dimerizes but does not form cross-linkable oligomers. Again, dimerization does not depend on the natural cysteine, since the cysteineless lysis-defective SA52V/C51S is found in dimers after treatment of the membrane with a cross-linking agent. Furthermore, under oxidative conditions, dimerization via the natural cysteine is very efficient for SA52V. Both SA52V (dominant negative) and SA48V (antidominant) interact with the parental S protein, as judged by oxidative disulfide bridge formation. Thus, productive and unproductive heterodimer formation between the parental protein and the mutants SA52V and SA48V, respectively, may account for the dominant and antidominant lysis phenotypes. Examination of oxidative dimer formation between S variants with single cysteines in the hydrophobic core of the second membrane-spanning domain revealed that positions 48 and 51 are on a dimer interface. These results are discussed in terms of a three-step model leading to S-dependent hole formation in the inner membrane.
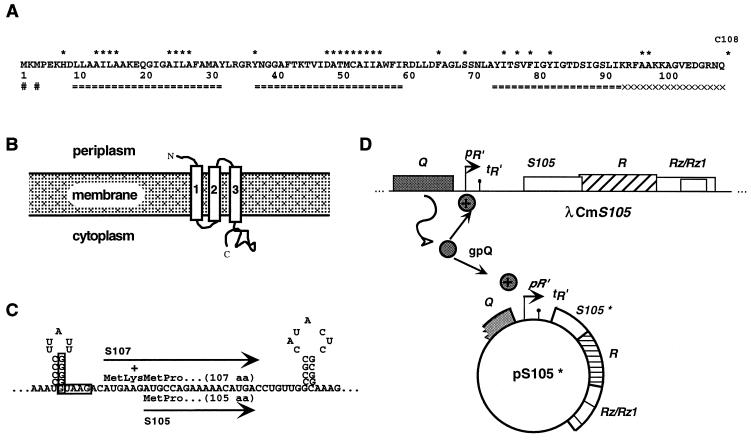
With the exception of filamentous phages, all bacteriophages terminate their infective cycles by causing lysis of the host cell (29). Double-stranded phages, like bacteriophage λ, use a holin-endolysin system for liberation of their progeny virions. Phage λ has four lysis genes, S, R, Rz, and Rz1, clustered in the λ lysis cassette and transcribed from the single late gene promoter, pR′ (7, 17, 20, 21, 30). Under standard laboratory conditions, only S and R are required for host cell lysis (11, 28). R, the endolysin gene, encodes a transglycosylase, which accumulates fully folded in the cytoplasm (3, 10). Lacking a signal sequence, the endolysin requires the function of S to get access to its substrate, the peptidoglycan (28). S, the holin, is a small inner membrane protein which causes the formation of a lethal membrane lesion at a precisely scheduled time after phage infection (1, 2, 12, 28). The membrane lesion terminates respiration and allows the escape of the muralytic enzyme to the periplasm. Genetic and biochemical data have shown that S has three transmembrane (TM) domains, with its N terminus located in the periplasm and its C terminus located in the cytoplasm (5, 13, 14) (Fig. 1A and B). A remarkable feature of λ S is that it encodes in its 107-codon sequence two proteins with opposing functions: the holin, S105, and the holin inhibitor, S107, synthesized as a result of independent translation initiation events at Met codons 3 and 1, respectively (Fig. 1A and C) (4, 6). Consequently, part of the timing mechanism of host cell lysis depends on the proportion of the two proteins, which is 2:1 in favor of the holin effector, S105 (9). Artificial alteration of this ratio in favor of the holin inhibitor or holin effector retards or accelerates the onset of lysis, respectively (6).
FIG. 1.
Primary structure, membrane topology, translational control region, and transactivation of λ S. (A) Primary structures of λ S. ===, transmembrane domains as predicted by the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM-1.0/) (25); XXX, the highly charged, dispensable C-terminal region (5); #, the two start codons of S. The positions of single-cysteine substitutions are indicated by asterisks above the sequence. (B) Membrane topology of λ S. The three α-helical transmembrane domains are numbered from 1 to 3. (C) Dual-start motif of λ S. The boxed sequences indicate the Shine-Dalgarno sequences for the dual translational start sites of S. The lengths of both protein products are given in amino acid (aa) residues. (D) The lambda lysis genes lie in an overlapping cluster in the lysis cassette downstream of the single late gene promoter pR′. Expression of the lysis genes from a prophage and/or transactivation plasmid is dependent on the protein Q that is required for antitermination of the terminator tR′. For dominance and antidominance tests, the holin S105 was expressed from a prophage and a second S105 variant (S105*) was expressed from a medium-copy-number transactivation plasmid (pS105*) (Table 2) (23, 24).
The small size of the holin gene and its lethality make the S gene ideal for genetic selection. Most S mutations which were selected for loss of lethality mapped to TM1, TM2, and the connecting cytoplasmic loop (19). TM2 appeared to be especially crucial for λ S function. For example, an Ala-to-Val change at position 48 or 52 generates a nonfunctional holin protein. Furthermore, replacement of the Ala at position 55 with a Thr confers a temperature-sensitive phenotype (16, 19, 20). Among this large collection of S− mutants, a number of dominant S alleles were found. These dominant mutants had a negative effect on lysis caused by the wild-type S gene, seen as a retardation of cell lysis. Some alleles exhibited a different phenotype, termed “antidominance” (previously called “early dominance” [19]). Antidominant S alleles are lysis defective when expressed alone; however, in the presence of wild-type S, these alleles accelerate lysis at least as well as a second wild-type allele (19) (Fig. 1D). It was originally suggested that this phenotype reflects differential effects of the lysis-defective S protein on the parental S105 holin and S107 holin inhibitor proteins. In this model, the mutant S protein was thought not to contribute to lysis directly but to titrate out the parental S107 inhibitor and thus indirectly accelerate the onset of lysis.
Phenotypic analysis of lysis-defective S alleles suggests that the S gene product must oligomerize to achieve its lethal membrane effect. Cross-linking experiments with membranes from cells where the wild-type gene, encoding S105 and S107, was expressed revealed that an S oligomeric ladder up to a 6-mer could be detected by Western blotting (31). Furthermore, in an assay with purified holin protein, the S protein allowed the release of a fluorescent dye from liposomes without the requirement for any additional factors, indicating that S forms a homo-oligomeric structure (23).
In this study, the molecular basis for the lysis defect of various S alleles was investigated by biochemical analysis. Furthermore, a site-directed cross-linking approach was used to localize intermolecular interactions. Helix proximity was assessed by using site-directed disulfide bridge formation of coexpressed S molecules, each with a single cysteine residue at a defined position in the second membrane-spanning domain. A detailed phenotypical and biochemical analysis of dominant and antidominant S alleles is described and discussed in the form of a revised model for dominance and antidominance.
MATERIALS AND METHODS
Strains, bacteriophages, plasmids, and growth media.
The Escherichia coli strains MC4100 and XL1-Blue, the lysis-defective thermoinducible prophages λCmΔSR and λKnΔSR, and the lysis-proficient thermoinducible prophages λRG1, λCmS105, and λCmS105τ94 have been described previously (19, 22, 23). λRG1 carries the wild-type S gene and thus generates S105 and S107, while the S alleles in λCmS105 and λCmS105τ94 generate S105 and the oligohistidine-tagged S105τ94, respectively. Media, growth conditions, and thermal induction of the λ lysis genes from prophages and plasmids have been described previously (9, 23).
Standard DNA manipulation, PCR, site-directed mutagenesis, and DNA sequencing.
Standard DNA manipulation, PCR, site-directed mutagenesis, and DNA sequencing have been described previously (14, 22, 23). The plasmids used in this study are listed in Table 1 and Table 2. Newly constructed plasmids were created by site-directed mutagenesis using the QuikChange kit from Stratagene (La Jolla, Calif.). All primer pairs used for site-directed mutagenesis contained 15 to 30 nucleotides of homology to either side of the altered nucleotides. In all constructs, TGC was used as the cysteine codon and base changes were verified by automated fluorescence sequencing as described previously (22).
TABLE 1.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pKB1 | Derivative of pOR19; λ lysis gene region (Sam7) cloned as HindIII/ClaI fragment (λ nt 4141–46440) | 18 |
| pS105 | S with M1L mutation (ATG→CTG); bears S105 only | 23 |
| pS105A52V | S105 with A52V mutation (GCC→GTC) | 24 |
| pS105A48V | S105 with A48V mutation (GCA→GTA) | This study |
| pS105C51S | S105 with C51S mutation (TGC→AGC) | 14 |
| pS105C51S/A52V | S105 with C51S and A52V mutations (TGC→AGC and GCC→GTC) | This study |
| pS105C51S/C108 | S105 with C51S mutation (TGC→AGC) and C-terminal addition of cysteine codon (TGC) | This study |
| pS105τ94 | Insertion of (G2H6G2) after F94 codon in S105 | 23 |
| pS105τ94C51S/I53C | S105τ94 with C51S/I53C mutations (TGC→AGC and ATT→TGC) | This study |
TABLE 2.
Plasmids, phages, and lysis times
| Plasmid | Position of single cysteine | Plasmid lysis time (min) | Phage | Phage lysis time (min) |
|---|---|---|---|---|
| pS105C51S/H7C | H7C | 55 | λCmS105C51S/H7C | 60 |
| pS105C51S/A12C | A12C | 35 | NDc | |
| pS105C51S/I13C | I13C | 30 | ND | |
| pS105C51S/L14C | L14C | 35 | ND | |
| pS105C51S/A15C | A15C | 20 | ND | |
| pS105C51S/A23C | A23C | 75 | λCmS105C51S/A23C | No lysis |
| pS105C51S/I24C | I24C | 35 | λCmS105C51S/I24C | 40 |
| pS105C51S/L25C | L25C | No lysis | λCmS105C51S/L25C | No lysis |
| pS105C51S/A26C | A26C | No lysis | λCmS105C51S/A26C | No lysis |
| pS105C51S/Y36C | Y36C | >80 (slow) | λCmS105C51S/Y36C | No lysis |
| pS105C51S/D47Ca | D47C | No lysis | λCmS105C51S/D47C | No lysis |
| pS105C51S/A48Ca | A48C | 60 | ND | |
| pS105C51S/T49Ca | T49C | 55 | λCmS105C51S/T49C | 75 (slow) |
| pS105C51S/M50Ca | M50C | No lysis | ND | |
| pS105a | C51 (wild type) | 35 | λCmS105 | 40 |
| pS105C51Sa | C51S | 30 | λCmS105C51S | 35–40 |
| pS105C51S/A52Ca | A52C | No lysis | ND | |
| pS105C51S/I53Cab | I53C | 45 | λCmS105C51S/I53C | 55 (slow) |
| pS105C51S/I54Ca | I54C | 65 | λCmS105C51S/I54Ca | No lysis |
| pS105C51S/A55Ca | A55C | 45 | ND | |
| pS105C51S/F64C | F64C | 40 | λCmS105C51S/F64C | 55 |
| pS105C51S/S68C | S68C | 55 | λCmS105C51S/S68C | 90 (slow) |
| pS105C51S/I74C | I74C | 55 | λCmS105C51S/I74C | 90 (slow) |
| pS105C51S/S76C | S76C | 15–20 | λCmS105C51S/S76C | 25 |
| pS105C51S/F78C | F78C | 40 | λCmS105C51S/F78C | 65 |
| pS105C51S/Y81C | Y81C | >75 (slow) | λCmS105C51S/Y81C | No lysis |
| pS105C51S/F94C | F94C | 35 | λCmS105C51S/F94C | 45 |
| pS105C51S/A95C | A95C | 30 | ND | |
| pS105C51S/C108 | C108 | 30 | ND |
S alleles for which lysis curves are shown in Fig. 2A.
The histidine-tagged version of this S allele was constructed (pS105τ94C51S/I53C) and recombined into the phage.
ND, not determined.
Placing S alleles from the plasmid in the phage context.
Recombinant phages were isolated from lysates of thermally induced MC4100(λCmΔSR) lysogens carrying pS105-derived plasmids. These lysates (100 μl) were added to 100 μl of an MC4100 overnight culture and incubated for 30 min at room temperature. After the addition of 3 ml of Luria-Bertani (LB) broth, the mixture was aerated for 2 h at 30°C, for 20 min at 42°C, and for another 60 min at 37°C. The enriched lysate was sterilized by the addition of chloroform at a final concentration of 2%. Cell debris was removed by centrifugation at 14,000 × g for 15 min at 4°C. This enrichment procedure was repeated a second time. One hundred microliters of the doubly enriched lysate was adsorbed for 30 min at room temperature to 100 μl of a freshly saturated culture of MC4100. After the addition of 1 ml of LB broth and further incubation for 30 min at room temperature, 200 μl of this mixture was spread on selective medium (LB-Cm). The plates were incubated overnight at the permissive temperature (30°C). The recombinant lysogens were screened on plates for Apr and temperature sensitivity. Testing for a functional R gene was done by thermal induction in liquid culture as described previously (9, 23). In the case of nonlytic S alleles, R function was tested by the addition of 1% chloroform to permeabilize the membrane.
Membrane protein preparation, oxidative disulfide bridge formation, SDS-polyacrylamide gel electrophoresis, Western blotting, and immunodetection.
Detergent-soluble preparations of inner membrane proteins were prepared as described previously (9, 22). Oxidative disulfide bridge formation was performed as described previously (15). Briefly, cultures expressing one S allele from a prophage and a second S allele from a plasmid (Fig. 1D) were induced as described above except that the A550 at induction was 0.3. After lysis was completed or 100 min after induction, 5 ml of culture was further disrupted by one passage through a large SLM-Aminco French pressure cell (Spectronic Instruments, Rochester, N.Y.) at 16,000 lb/in2. Oxidation of the French pressate was performed for 60 min at room temperature with 20 mM CuSO4 and 60 mM 1-10 phenanthroline. The reactions were stopped with N-ethylmaleimide at a final concentration of 0.1 M, followed by incubation at room temperature for 60 min. Membrane fractions were collected by ultracentrifugation at 100,000 × g for 60 min at 18°C. The membrane pellet was solubilized in 50 μl of membrane extraction buffer (1% Triton X-100, 10% glycerol, 0.5 M NaCl, 35 mM MgCl2, 20 mM Tris-HCl, pH 8.0) supplemented with 0.1 M N-ethylmaleimide for 12 to 14 h at 37°C. After solubilization of the membrane pellet, insoluble material was removed by ultracentrifugation at 100,000 × g for 45 min at 18°C. As a control to ensure that disulfide bond formation did not occur during sample preparation, two different plasmid-borne S alleles were induced in separate cultures and mixed shortly before disruption in the French pressure cell. These combined lysates were then subjected to centrifugation and detergent extraction as described above. For sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, the detergent-soluble fractions were diluted 1:1 with 2× protein sample buffer (110 mM Tris-HCl, 10% glycerol, 4.3% SDS, 0.002% bromphenol blue, pH 6.8) devoid of reducing agents. Protein samples were placed for 10 min at 37°C and then centrifuged at 14,000 × g for 5 min in a microcentrifuge at room temperature. The proteins were separated on a precast 16% Tris-Tricine minigel (Xcell II Minicell; Novex, San Diego, Calif.) following the manufacturer's instructions. Western blotting and immunodetection with S-specific antibodies were performed as described previously (14).
DSP cross-linking.
S expression from the plasmids pKB1, pS105, pS105C51S, pS105A52V, and pS105C51S/A52V was induced in the strain MC4100(λKnΔSR) at an A550 of 0.2. The λRG1-derived prophages bearing different S mutants are described by Raab et al. (19) and were induced in MC4100. After cell lysis was completed or 60 min after induction, 50 ml of the induced cultures was passed once through a large SLM-Aminco French pressure cell at 16,000 lb/in2. The membranes were collected by ultracentrifugation at 100,000 × g for 60 min at 18°C and resuspended in 300 μl of 0.1 M MOPS (morpholinepropanesulfonic acid)–20 mM NaCl, pH 7.6. The total protein concentrations of these suspensions were determined using the Bio-Rad (Hercules, Calif.) protein assay following the manufacturer's instructions. For the cross-linking reaction, a final protein concentration of 2.5 mg/ml and a final dithiobis(succinimidyl propionate) (DSP) concentration of 16 mM were used. A 100 mM stock solution of DSP in dimethylsulfoxide was prepared just prior to use. As a negative control, dimethyl sulfoxide without a cross-linker was added to the membrane samples. Typically, the final volume of a reaction mixture was 250 μl. The reaction mixtures were incubated for 30 min at room temperature with gentle shaking. The reactions were stopped by the addition of glycine at a final concentration of 100 mM and additional incubation for 5 min at room temperature. The membranes were collected by ultracentrifugation as described above, and the membrane proteins were solubilized in membrane extraction buffer (1% Triton X-100, 10% glycerol, 0.5 M NaCl, 35 mM MgCl2, 20 mM Tris-HCl, pH 8.0) for 12 to 14 h at 37°C in one-fifth (typically 50 μl) of the original reaction volume. The insoluble material was removed by ultracentrifugation at 100,000 × g for 45 min at 18°C, and the soluble fraction was mixed 1:1 with 2× protein sample buffer devoid of reducing agents. The proteins were separated on a 16% Tris-Tricine gel and analyzed by Western blotting as described previously (14).
RESULTS
Single-amino-acid changes in S have a strong effect on lysis timing.
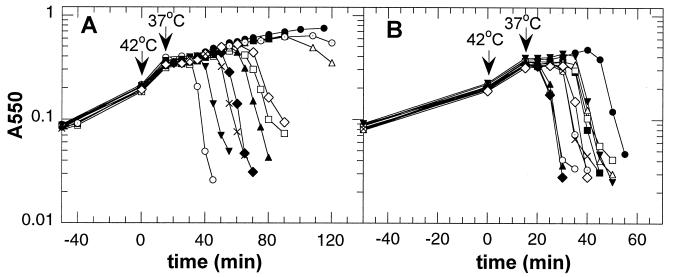
In a previous study, a collection of single-cysteine-containing S mutants was constructed to determine the membrane topology of λ S (14). Starting from the cysteineless S105C51S variant, single cysteines were introduced at 26 positions throughout the S protein, and one cysteine was added at the C terminus. The resulting mutants were expressed from a transactivation plasmid in the strain MC4100(λCmΔSR) and tested for their lysis phenotypes. Synthesis of the cysteineless S protein, S105C51S, where the native cysteine at position 51 was replaced by a serine, resulted in lysis 5 min earlier than with the parental S105 protein (Fig. 2 and Table 2) (15). Lysis curves of products of S alleles with single cysteines in positions 47 to 55 are shown in Fig. 2A. These lysis curves illustrate the general finding that, although most of the single-cysteine mutants retained their functions as holins, they differed greatly in the timing of cell lysis (Table 2). Many of the mutant S alleles were recombined back onto the phage and the products were tested for lysis function. A number of single-cysteine mutants exhibiting a late-lysis phenotype in the plasmid context were completely lysis defective in the phage context (Table 2). These results suggested that S expression from the medium-copy-number plasmid is somewhat higher than that from the prophage.
FIG. 2.
Lysis phenotypes of single-cysteine-containing S mutants. (A) MC4100(λCmΔSR) cells carrying the plasmids pKB1 (Sam7; ●), pS105 (wild type; C51; ▾), pS105C51S (○), pS105C51S/D47C (■), pS105C51S/A48C (□), pS105C51S/T49C (▴), pS105C51S/M50C (▵), pS105C51S/A52C (⊙), pS105C51S/I53C (⧫), pS105C51S/I54C (◊), and pS105C51S/A55C (X) were induced and monitored for turbidity until cell lysis was completed or for 110 to 120 min after induction. (B) Expression of single-cysteine-containing S mutants in trans to S105. MC4100(λCmS105) cells carrying the same plasmids as in panel A were induced and monitored for turbidity. All single-cysteine S mutants showed an antidominant lysis phenotype with lysis occurring as fast as or faster than with two copies of the parental S105 holin (▾).
Antidominance with S105 alleles.
Antidominant alleles are lysis defective when expressed by themselves, but in the presence of a wild-type S allele, expression of these alleles causes lysis as fast as or even faster than expression of two wild-type S alleles (19). As shown in Fig. 2B, S105 mutants with single cysteines from positions 47 to 55 displayed the antidominant lysis phenotype. Their expression in trans to S105 (Fig. 1D) resulted in cell lysis at least as early as expression of two S105 alleles (Fig. 2B). For some of the mutants, such as S105T49C or S105I53C, this antidominant lysis phenotype was particularly evident. All other single-cysteine S alleles listed in Table 2 showed the same antidominant lysis phenotype (data not shown).
Lysis deficiency of S105A52V is due to an oligomerization but not a dimerization defect.
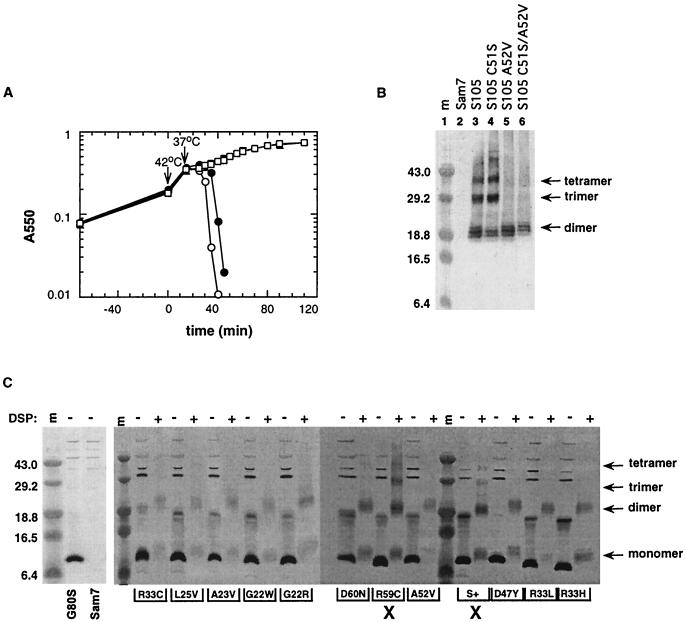
Treatment of membrane vesicles containing S protein (both S105 and S107) with the membrane-permeable cross-linker DSP results in the formation of S-specific oligomers (31). We have recently shown that S forms SDS-resistant disulfide-bridged dimers during membrane extraction with Triton X-100 (15). To distinguish between dimer formation via disulfide bridge formation during membrane extraction and DSP-dependent cross-linking in the cytoplasmic membrane, we performed a cross-linking experiment with the functional but cysteineless S105C51S. Both S105 and S105C51S holin proteins formed higher oligomers in the presence of DSP (Fig. 3A and B, lanes 3 and 4). S-specific bands up to tetramers could be detected by Western blotting. In contrast, S105A52V and S105C51S/A52V, which showed severe lysis defects when expressed from the transactivation plasmid (Fig. 3A), formed only dimers but not oligomers under the same conditions (Fig. 3B, lanes 5 and 6). Since the cysteineless lysis-defective S105C51S/A52V protein also formed dimers, dimerization cannot be due to artifactual disulfide bridge formation during membrane extraction with Triton X-100.
FIG. 3.
Lysis phenotype and DSP cross-linking pattern of different S variants. (A) MC4100(λKnΔSR) cells carrying the plasmids pS105 (●), pS105C51S (○), pS105A52V (■), and pS105C51S/A52V (□) were induced and monitored for turbidity. (B) 60 min after induction, membrane samples of MC4100(λKnΔSR) plus pKB1 (Sam7) and the same strains as in panel A were prepared and treated with 16 mM DSP cross-linker. The cross-linking experiment was performed as described in Materials and Methods and analyzed by Western blotting. Lane 1, molecular mass (m) marker; lane 2, pKB1(Sam7); lane 3, pS105; lane 4, pS105C51S; lane 5, pS105A52V; lane 6, pS105C51S/A52V. The masses of prestained molecular standards are given in kDa on the left. S-specific bands are marked by arrows. (C) Left gel, Triton X-100-soluble inner membrane samples of MC4100 cells carrying λRG1-derived prophages were prepared and analyzed by Western blotting as described in Materials and Methods with the alteration that the samples were mixed with 2× sample buffer containing 2.8 M β-mercaptoethanol. Lane 1, molecular mass standards (m); lane 2, SG80S; lane 3, Sam7. Right gel, membrane samples of induced MC4100 cells carrying λRG1- or λRG1-derived prophages bearing defective S alleles were prepared. DSP cross-linking and Western blot analysis were performed as described in Materials and Methods. The lanes labeled with + and − indicate the presence and absence, respectively, of the cross-linking agent during sample preparation. The amino acid change for each S variant is given below the panel. An X below the panel indicates S proteins which form higher oligomers upon DSP treatment. The masses of prestained molecular standards are given in kDa on the left. S monomer and oligomer bands are marked by arrows.
Most S alleles with a severe lysis defect show an oligomerization but not a dimerization defect.
Ten other absolute lysis-defective S proteins were tested for the ability to form higher oligomers in the bacterial membrane. All mutant proteins tested accumulated essentially normally and were able to form dimers (Fig. 3C). Only one variant, where the Arg at position 59 was replaced by a Cys, showed formation of higher oligomers comparable to the parental S protein (Fig. 3C). This indicates that SR59C is blocked in a later step than the other S− alleles. As shown in Fig. 3C (left gel), the high-molecular-weight bands are nonspecific immuno-cross-reactive species, as indicated by their presence in an S− background. Upon addition of DSP, these cross-reactive species are not observed (Fig. 3B), likely because treatment of bacterial membranes with the cross-linker prevents the subsequent extraction of these high-molecular-weight species with Triton X-100.
Dominant and antidominant lysis-defective alleles form dimers with the parental S105 allele in the bacterial membrane under oxidative conditions.
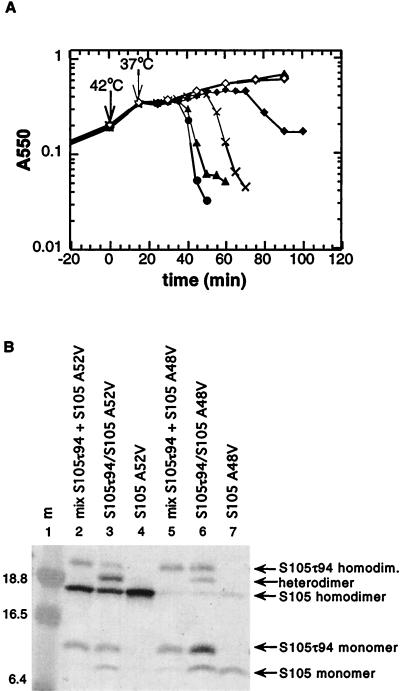
Expression of S105A52V in trans to S105 delayed the onset of lysis (Fig. 4A), indicating that the mutant and parental proteins were interacting in the membrane. Indeed, under oxidative conditions, S105A52V formed not only homodimers but also heterodimers with the histidine-tagged S105τ94 (Fig. 4B, lanes 3 and 4). The oxidizing method and disulfide bond formation via the single cysteine at position 51 have been used previously to determine a specific interaction between holin and holin inhibitor (15). The use of the histidine-tagged holin protein S105τ94 allowed us to discriminate between homodimer and heterodimer formation. We also tested the antidominant lysis-defective S105A48V protein for its ability to form heterodimers with S105τ94 (Fig. 4). S105A48V formed homodimers in the membrane, although not very efficiently (Fig. 4B, lane 7). In contrast, heterodimer formation between S105A48V and S105τ94 was very efficient (Fig. 4B, lane 6).
FIG. 4.
Expression of S105A48V and S105A52V in trans to S105 and dimerization with S105τ94. (A) MC4100(λCmΔSR) cells carrying the plasmid pS105A48V (▵) or pS105A52V (◊) and MC4100(λCmS105) cells carrying the plasmid pKB1 (Sam7; X), pS105 (●), pS105A48V (▴), or pS105A42V (⧫) were induced and monitored for turbidity. (B) Oxidation and sample preparation for Western blot analysis were performed as described in Materials and Methods. Lane 1, molecular mass (m) marker; lane 2, mixed cultures MC4100(λCmΔSR) plus pS105τ94 and MC4100(λCmΔSR) plus pS105A52V; lane 3, MC4100(λCmS105τ94) plus pS105A52V; lane 4, MC4100(λCmΔSR) plus pS105A52V; lane 5, mixed cultures MC4100(λCmΔSR) plus pS105τ94 and MC4100(λCmΔSR) plus pS105A48V; lane 6, MC4100(λCmS105τ94) plus pS105A48V; lane 7, MC4100(λCmΔSR) plus pS105A48V. Labeling of the panel is as in Fig. 3.
Strong dimer interaction on one side of the second membrane-spanning α-helix.
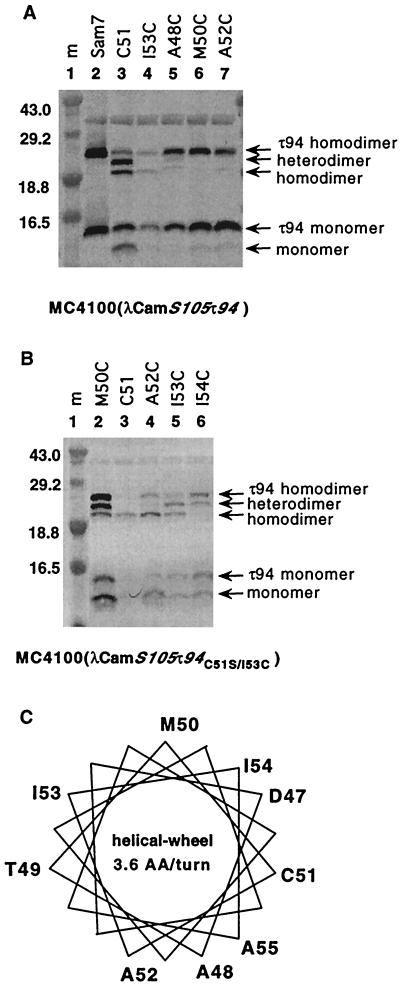
Under oxidative conditions, a disulfide bridge can be formed very efficiently between two S molecules with the natural single cysteine at position 51 (15) (Fig. 5A, lane 3). To map further interaction points in the second membrane-spanning domain, we tested S mutants with single cysteines from positions 47 to 55 for the ability to form heterodimers with S105τ94 (cysteine at position 51). In these experiments, the S105τ94 allele was expressed from the prophage and the single-cysteine S alleles were expressed from a transactivation plasmid. Heterodimer formation was seen only between positions 51 and 48 or 51 and 51 (Table 3 and Fig. 5A), indicating that residues at positions 48 and 51 lie on a dimer interface (Fig. 5C). In a similar fashion, we examined heterodimer formation between the histidine-tagged S105τ94C51S/I53C (single cysteine at position 53) and S proteins with single cysteines from positions 47 to 55. Heterodimer formation with the cysteine at position 53 was not as specific as with the cysteine at position 51 (Table 3 and Fig. 5B). Heterodimer formation between the cysteine at position 53 and cysteines at positions 50, 53, and 54 was stronger than with the other positions, 48, 51, and 52 (Table 3). Again, these positions (50, 53, and 54) cluster on one face of an α-helix (Fig. 5C).
FIG. 5.
Dimer formation between S molecules with single cysteines in TM2. S proteins were tested for heterodimer formation under oxidative conditions and analyzed by Western blotting as described in Materials and Methods. Samples were prepared from MC4100(λCmS104τ94) (A) and from MC4100(λCmS104τ94C51S/I53C) (B) cells harboring a transactivation plasmid. The lanes are labeled with the S allele on the transactivation plasmid. S-specific monomer and dimer bands, as well as the masses of prestained molecular mass (m) standards in kDa, are indicated. These Western blots show the heterodimer formation between different pairs of S molecules; the results are summarized in Table 3. (C) α-Helical wheel projection of the second transmembrane domain of λ S, with 3.6 amino acids per turn. The positions and original amino acids individually replaced by cysteine are indicated in single-letter code in this scheme.
TABLE 3.
Heterodimer formation
| Position of cysteine | Heterodimer formation with C51a | Heterodimer formation with C53a |
|---|---|---|
| 47 | − | ND |
| 48 | + | + |
| 49 | − | ND |
| 50 | − | +++ |
| 51 | +++ | − |
| 52 | − | + |
| 53 | − | ++ |
| 54 | − | +++ |
| 55 | − | ND |
Western blots were scored for heterodimer formation as follows: −, no heterodimer formation; +, weak heterodimer formation; ++, heterodimer formation; +++, strong heterodimer formation; ND, not determined (discrimination between homodimer and heterodimer formation was not possible, or Western blot signal was too weak for evaluation).
DISCUSSION
Intrinsic timing determinants and antidominance.
The λ S holin functions to allow the R endolysin access to its substrate, the cell wall, by somehow permeabilizing the cytoplasmic membrane. This system has been nearly universally adopted by double-stranded DNA phages instead of the simpler strategy of providing a signal sequence to the endolysin, and it presumably reflects the critical nature of the lysis timing system. For every host growing at an environmentally defined rate and for a given kinetics of virion assembly within the host, there is an ideal lysis time which ensures optimal spread of the phage within the prey population (26, 27). Thus, the critical features of the holin are that it triggers hole formation at a precisely defined time (optimized by evolution) and, once triggered, generates lesions which efficiently permeabilize the membrane to ensure that lysis is rapid and complete (27, 29). The “dual-start” motif that results in the production of both the holin effector, S105, and its cognate inhibitor, S107, is a significant component of the timing mechanism. Small changes in the proportions of holin and inhibitor lead to dramatic alterations in the timing of lysis (4, 8, 9). However, although this remarkable regulatory adaptation is widespread among holins (27, 29), it must be noted that it is a fine-tuning strategy. The fundamental timing mechanism is intrinsic to the primary structure of the holin, S105, as can be seen from the fact that even in the absence of S107, the S105 allele supports sharp and efficient lysis, albeit somewhat earlier than the parental S allele (6). Moreover, mutational analysis in this and previous studies have shown that single missense changes within the S reading frame can have a profound effect on both the process of host lysis and its timing (19).
Here we have shown that most of the 27 single-cysteine products of S105 alleles retain holin function. Induction of these mutants resulted in lysis at a precisely scheduled time point with a sharp decline in A550. However, most of these variants also exhibited significantly altered lysis times (Fig. 2A and Table 2). The parental S105 protein has been shown to be stable in a pulse-chase labeling experiment (4). The simplest rationale for the observation of delayed lysis would be reduced stability for the mutant S105 proteins. According to this view, each missense change would reduce the stability of the S105 product to a certain degree; the reduced stability would result in a decreased rate of accumulation and thus indirectly affect the timing by lengthening the period required to achieve a triggering concentration. However, this is not the case for all mutants. Although detailed pulse-chase stability assessments have not yet been done on the entire collection of mutants, Western blot analysis has shown that S accumulates to much higher than normal levels in several lysis delay alleles (e.g., A23C and A26C) but is reduced in another, L25C (A. Gründling and R. Young, unpublished data). Moreover, most alleles with absolute lysis defects do not show altered accumulation (Fig. 3C). We conclude that the wide variety of lysis timing phenotypes comes about, for many of the single-cysteine alleles, by alteration of the intrinsic clock mechanism of the holin.
It must be noted that phenotypic analysis of the single-cysteine mutant collection has falsified a model for another remarkable feature of S: the existence of early dominant, or antidominant, alleles. This phenomenon depends on the fact that S gene dosage affects lysis timing. Thus, a lysogen with two inducible S+ prophages lyses significantly earlier than a lysogen with a single prophage. Antidominant alleles are lysis defective on their own, but in the presence of the wild-type allele, they accelerate lysis at least as much as the parental allele (19). Dominance recessiveness analysis on the set of single-cysteine mutants reveals that some of the lysis delay alleles (e.g., T49C and I53C) display a very strong antidominant phenotype. That is, these alleles support delayed lysis in trans to a null allele (Fig. 2A) but support an acceleration of the lysis time better than the parental S105 allele (Fig. 2B). The original observation of this unique antidominance characteristic was made with S alleles which had the wild-type translational start and thus produced S105 and S107 in their normal proportions. This led to a model in which the antidominant lysis-defective alleles produced holins which were nonfunctional but, in trans to S+, could titrate out the S107 inhibitor and thus allow free wild-type S105 to accumulate and lead to early hole formation. However, in this study, the single-cysteine mutants were constructed in the context of the S105 allele, and thus, the antidominance phenotype must derive from interactions which affect the timing function intrinsic to the S105 sequence (see below).
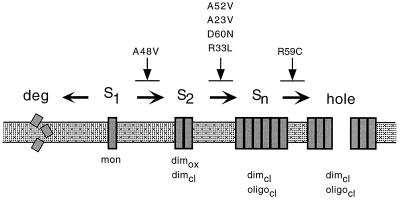
Some lysis-defective S proteins are blocked in a step beyond dimerization.
For functional analysis of a holin protein, it is important to use an expression system which closely mimics the expression levels of the vegetative phase. Unphysiologically high expression of these toxic membrane proteins might result in nonspecific host cell lysis. Even expression of the lysis genes under their cognate promoter, pR′, but cloned on a medium-copy-number plasmid results in a measurable difference in lysis timing compared to expression from the induced prophage, despite the fact that extensive DNA replication normally precedes the bulk of late gene expression (Table 2). However, expression of the lysis-defective S105A52V allele does not result in host cell lysis even in the context of a transactivation plasmid (Fig. 3A). At the molecular level, this lysis defect is associated with an oligomerization defect. In cross-linking experiments with the bifunctional agent DSP, we have shown that the parental S105 forms dimers and higher-order oligomers (Fig. 3C), as was previously demonstrated for the mix of S105 and S107 produced from the S+ allele (31). In contrast, the lysis-defective S105A52V can form dimers but not higher oligomers (Fig. 3B). An identical phenotype is observed for the cysteineless S105C51S/A52V, confirming that the dimerization observed with DSP cross-linking is not due to disulfide bonds formed during membrane extraction. Consequently, one can reasonably conclude that the ability of S molecules to dimerize is not sufficient for the lytic step in holin function. All other lysis-defective S mutants which were tested by DSP cross-linking also showed efficient dimer formation, and except for SR59C, all were deficient in oligomerization (Fig. 3C). SR59C shows an oligomerization pattern which is indistinguishable from that of the parental S (Fig. 3C). The simplest model to account for these phenotypes is that there must be at least three steps in holin function: first, dimerization; second, oligomerization; and third, a concerted conformational change which is equivalent to triggering of hole formation (Fig. 6). Moreover, we note that the total amount of S protein accumulating after induction of the S105A48V allele is much less than with S105 or S105A52V (Fig. 4B). Although definitive pulse-chase analysis has not been done, it is very unlikely that this reduced accumulation derives from a defect in synthesis, especially since the presence of the wild-type S105 protein relieves the defect in accumulation of S105A48V (Fig. 4B). More likely, this indicates that the S105A48V is proteolytically unstable. Not only is there much less S105A48V accumulated, a lower percentage of the total protein is in the disulfide dimer form (Fig. 4B), suggesting that a reduced efficiency in dimer formation may lead to proteolysis. This rationale leads to the prediction that the monomer species in the forced-oxidation experiments may not only derive from a membrane pool of S monomers but also reflect S molecules which are stabilized in an oligomeric form where the Cys51 disulfide bond formation is sterically unfavored. The model in Fig. 6 incorporates these findings into a working hypothesis for hole formation.
FIG. 6.
Model for hole formation. At least three steps, described in the text, are required for hole formation: first, dimerization of λ S; second, oligomerization; and third, a concerted conformational change which is equivalent to the triggering of hole formation. The S alleles which are blocked in different steps during the process of hole formation are indicated above the individual steps. S1, monomer; S2 dimer; Sn oligomer; deg, degradation of S molecules; dimcι and oligocι dimers and oligomers detected by DSP cross-linking; dimox, dimers detected by cysteine-specific disulfide bond formation.
The molecular basis for dominance and antidominance.
The dimerization ability of S105A52V can explain the dominant phenotype of this mutant protein. Under oxidative conditions, not only is homodimer formation by Cys51 disulfide linkages very efficient in this mutant but the mutant also forms disulfide heterodimers with the parental S105 (Fig. 4B), demonstrating that these proteins interact in the bacterial membrane. A reasonable model consistent with these results is that these heterodimers cannot oligomerize and participate in the process of hole formation. As a consequence, host cell lysis is retarded because the number of functional S105 molecules is reduced (Fig. 6). The same model of heterodimer formation between a mutant and the parental S protein can explain an antidominant lysis phenotype, if one assumes that heterodimers formed between the wild-type and the mutant S proteins can now functionally contribute to the pool of lysis-competent dimers (Fig. 6). Indeed, the antidominant S105A48V protein forms Cys51 disulfide heterodimers with S105 in the bacterial membrane (Fig. 4B and 6).
Rationale for an oligomerization but not a dimerization defect.
Chemical modification studies on the collection of single-cysteine mutants have provided evidence that both S105 and the lysis-defective, dimerization-proficient, oligomerization-deficient S105A52V assume an N-out, C-in topology with three transmembrane domains (Fig. 1C) (14). Moreover, the circular dichroism spectrum of the two proteins solubilized in detergent are identical, with, as predicted from the topology, more than 60 residues in α-helical conformation (J. Deaton and R. Young, unpublished data). In the simplest model to explain the oligomerization-defective, dimerization-proficient character of SA52V, the alanine at position 52 is located on a part of the S molecule which is important for oligomerization but not for dimerization. According to this view, the increase in side chain bulk associated with the A52V missense mutation would disrupt the oligomerization interface. Alternatively, this substitution so near the dimerization interface might result in an altered dimer structure and render it incapable of oligomerizing into higher-order structures. Either model is consistent with the fact that under oxidative conditions, disulfide dimers via Cys51 are more efficient with the mutant S105A52V than with the S105 protein (Fig. 4, lane 2). In our model, this reflects the accumulation of the mutant S proteins in the dimer intermediate (Fig. 6).
Dimer interaction along one face of the TM2 helix.
Using a cysteine-scanning approach, it has been shown that in λ S the natural cysteine at position 51 is in the core of the second membrane-spanning domain (14). Oxidative disulfide bond formation between S molecules with a cysteine at this position is very efficient (15). Here we have shown that heterodimer formation with the natural cysteine at position 51 is only seen with positions 48 and 51, which cluster on one face of the TM2 helix (Table 3 and Fig. 5C). Heterodimer formation with the cysteine at position 53 is not as specific as with the cysteine at position 51 (Table 3 and Fig. 5B). The strongest heterodimer formation with the cysteine at position 53 is seen with cysteines at positions 50, 53, and 54, which cluster on the opposite face of the α-helix (Table 3 and Fig. 5C). These data are consistent with the idea that a stable dimer is formed, mediated by the interaction along the face of the TM2 helix containing position 51. This type of analysis, coupled with the availability of mutants blocked in each step of hole formation, will be a powerful tool for understanding holin function at the molecular level.
ACKNOWLEDGMENTS
Support for this work was provided by PHS grant GM27099 and funds from the Robert A. Welch Foundation and Texas Agricultural Experiment Station.
We thank all the members of the Young laboratory for their support and Sharyll Pressley for her always-reliable secretarial assistance.
REFERENCES
- 1.Altman E, Altman R K, Garrett J M, Grimaila R J, Young R. S gene product: identification and membrane localization of a lysis control protein. J Bacteriol. 1983;155:1130–1137. doi: 10.1128/jb.155.3.1130-1137.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman E, Young K, Garrett J, Altman R, Young R. Subcellular localization of lethal lysis proteins of bacteriophages λ and φX174. J Virol. 1985;53:1008–1011. doi: 10.1128/jvi.53.3.1008-1011.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage λ is the murein transglycosylase. Mol Gen Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- 4.Bläsi U, Chang C-Y, Zagotta M T, Nam K, Young R. The lethal λ S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bläsi U, Fraisl P, Chang C-Y, Zhang N, Young R. The C-terminal sequence of the lambda holin constitutes a cytoplasmic regulatory domain. J Bacteriol. 1999;181:2922–2929. doi: 10.1128/jb.181.9.2922-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bläsi U, Nam K, Hartz D, Gold L, Young R. Dual translational initiation sites control function of the lambda S gene. EMBO J. 1989;8:3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell A, Campillo-Campbell A D. Mutant of bacteriophage lambda producing a thermolabile endolysin. J Bacteriol. 1963;85:1202–1207. doi: 10.1128/jb.85.6.1202-1207.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C-Y, Nam K, Bläsi U, Young R. Synthesis of two bacteriophage lambda S proteins in an in vivo system. Gene. 1993;133:9–16. doi: 10.1016/0378-1119(93)90218-r. [DOI] [PubMed] [Google Scholar]
- 9.Chang C-Y, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evrard C, Fastrez J, Declercq J P. Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes. J Mol Biol. 1998;276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- 11.Garrett J, Fusselman R, Hise J, Chiou L, Smith-Grillo D, Schulz R, Young R. Cell lysis by induction of cloned lambda lysis genes. Mol Gen Genet. 1981;182:326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- 12.Garrett J, Young R. Lethal action of bacteriophage lambda S gene. J Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graschopf A, Bläsi U. Molecular function of the dual-start motif in the λ S holin. Mol Microbiol. 1999;33:569–582. doi: 10.1046/j.1365-2958.1999.01501.x. [DOI] [PubMed] [Google Scholar]
- 14.Gründling A, Bläsi U, Young R. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J Biol Chem. 2000;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- 15.Gründling A, Smith D L, Bläsi U, Young R. Dimerization between the holin and holin inhibitor of phage λ. J Bacteriol. 2000;182:6075–6081. doi: 10.1128/jb.182.21.6075-6081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris A W, Mount D W A, Fuerst C R, Siminovitch L. Mutations in bacteriophage lambda affecting host cell lysis. Virology. 1967;32:553–569. doi: 10.1016/0042-6822(67)90032-3. [DOI] [PubMed] [Google Scholar]
- 17.Kedzierska S, Wawrzynow A, Taylor A. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene. 1996;168:1–8. doi: 10.1016/0378-1119(95)00712-1. [DOI] [PubMed] [Google Scholar]
- 18.Nam K. Translational regulation of the S gene of bacteriophage lambda. Ph.D. thesis. College Station: Texas A&M University; 1991. [Google Scholar]
- 19.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 20.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology. 1971;43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- 21.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology. 1971;43:623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- 22.Smith D L, Chang C-Y, Young R. The λ holin accumulates beyond the lethal triggering concentration under hyper-expression conditions. Gene Expr. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D L, Struck D K, Scholtz J M, Young R. Purification and biochemical characterization of the lambda holin. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith D L, Young R. Oligohistidine tag mutagenesis of the lambda holin gene. J Bacteriol. 1998;180:4199–4211. doi: 10.1128/jb.180.16.4199-4211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnhammer E L, von Heijne G, Krogh A. ISMB 6:175–182. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. [PubMed] [Google Scholar]
- 26.Wang I-N, Dykhuizen D E, Slobodkin L B. The evolution of phage lysis timing. Evol Ecol. 1996;10:545–558. [Google Scholar]
- 27.Wang I-N, Smith D L, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:798–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 28.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young R, Wang I-N, Roof W D. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 30.Young R, Way S, Yin J, Syvanen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132:307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 31.Zagotta M T, Wilson D B. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J Bacteriol. 1990;172:912–921. doi: 10.1128/jb.172.2.912-921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]