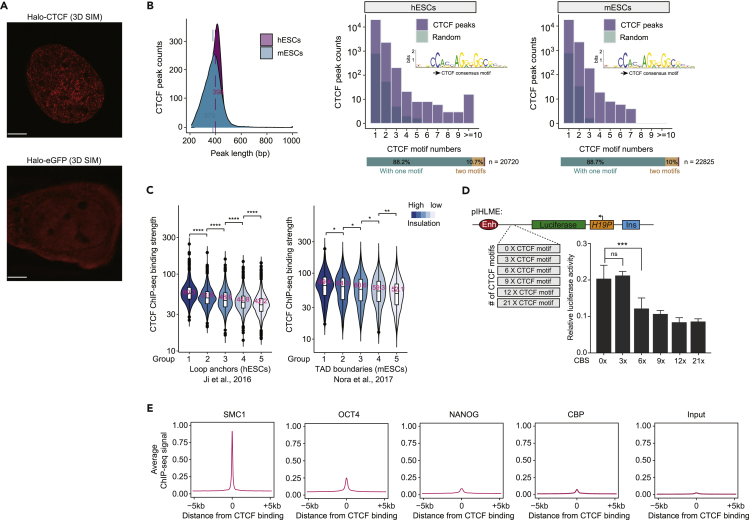
Figure 3.
Full-length CTCF forms small protein clusters and interacts with CTCF motif arrays occupied by low densities of transcriptional activators
(A) 3D SIM imaging of halo-tagged CTCF (top) or halo-eGFP (bottom) in U2OS cells. Scale bars, 5 μm.
(B) Left: CTCF peak length distributions of ChIP-seq data generated from human embryonic stem cells (hESCs) and ChIP-exo data derived from mouse embryonic stem cells (mESCs). Right: histogram displaying CTCF motif occurrence in CTCF ChIP-seq peaks identified in hESCs and mESCs, with the bottom bar indicating the percentage of occurrence frequency of this motif. The light purple and cyan colors indicate CTCF peaks and randomly selected regions in the corresponding human and mouse genomes, respectively. The position weight matrix for the canonical JASPAR CTCF motif finding is shown at the middle right.
(C) Left: violin plot of CTCF ChIP-Seq signals corresponding to different insulation strengths at loop anchors from cohesin ChIA-PET data in hESCs (1–5: insulation strength from high to low). The analyses of the left anchor are shown, and the right anchor behaves in the same way. Right: same as the left but for TAD anchors from Hi-C data in mESCs.
(D) Schematic illustrations of the pIHLME reporter used in the luciferase assay (top). Luciferase activities of the pIHLME construct in the presence of the indicated number of CTCF motif insertions in HEK293T cells (bottom right). p-values were calculated using two-tailed Student’s t-tests (∗∗∗<0.001, ∗<0.05). Data are represented as the mean ± SD. The CTCF motif sequences are shown in Table S5.
(E) Averaged ChIP-Seq signals of Cohesin (SMC1), OCT4, NANOG, CBP, and input control at CTCF-binding sites.