Abstract
As a key regulator of innate immunity, mitochondrial function is essential to maintain antiviral activities. Common mitochondrial DNA variants (haplogroups) have been associated with different physiological capacities and the nrisk of developing several diseases. Haplogroup H was associated with increased survival among sepsis patients, and lower risk of progression toward AIDS in HIV infected and lower manifestation of severe manifestation of herpex virus disease. We studied 316 Spanish with critical COVID-19, and found that the 7028C (haplogroup H) was protective among patients with early-onset disease (≤65 vs > 65 years, p = 0.01), while the ancestral 16223T was a risk factor for early-onset critical COVID-19 (OR = 3.36, 95 %CI = 1.49–7.54). Our work suggested that common mitochondrial variants may serve as predictors of COVID-19 severity. Additional studies to confirm this effect from other populations are of special interest.
Keywords: COVID-19, Mitochondria, Innate immunity, Mt haplogroups, Genetic association
1. Introduction
In addition to their primary function as energy producers and regulators of cellular processes such as apoptosis and ageing, mitochondria play an important role in the vertebrates innate immunity (Koshiba, 2013). The presence of virus inside the cell is detected by a group of cytosolic proteins that bind to and activate the mitochondrial antiviral-signaling protein (MAVS) located in the inner membrane of the mitochondria (Moore and Ting, 2008). Activated MAVS trigger the secretion of immunomodulators such as type I interferons and proinflammatory cytokine sthat would clear the viruses and limit the infection damage (Belgnaoui et al., 2011). The mitochondrion membrane potential is also essential to activate these immunological pathways (Schneider et al., 2019, Hu et al., 2019 Jun). Many viruses can interfere with mitochondrial function to impair the antiviral activity. For instance, the PB1-F2 influenza A protein targets the mitochondria and induces apoptosis and impaired cellular innate immunity (Yoshizumi et al., 2014, Zamarin et al., 2005).
Cytomegalovirus impairs MAVS through the viral apoptosis proteins that localizes in the mitochondria and reduces the pro-inflammatory response (Choi et al., 2018). Several SARS-CoV-1 proteins such as ORF3b and ORF-9b localizes into host mitochondria and suppresses innate immunity by manipulating the MAVS, while other mitochondrial localized viral proteins enhance infection by promoting viral replication (Shi et al., 2014, Chen et al., 2007).
Cells infected by SARS-CoV-2 exhibit a mitochondrial dysfunction with mitochondrial membrane depolarization, mitochondrial permeability transition pore opening and increased release of reactive oxygen species (ROS) (Shang et al., 2022). Interestingly, the SARS-CoV-2 membrane (M) protein would induce lung epithelial cells apoptosis by promoting the translocation of pro-apoptotic proteins into mitochondria (Yang et al., 2022). This would exacerbate the lung and other organs damage that characterizes the severe manifestation of COVID-19 (Mo et al., 2022, Costa et al., 2022).
The clinical features of COVID-19 caused by SARS-CoV-2 range from an asymptomatic state to severe pneumonia and multiorgan dysfunction with hospital admission (Zeng et al., 2021). The most severe cases would require respiratory support in the ICU and are at high risk of death. This heterogeneous symptoms might be partly explained by the individual’s hereditary susceptibility, and several nuclear-encoded genes have been associated with COVID-19 severity and death (COVID-19 Host Genetics Initiative, 2021, Wang et al., 2020). The mitochondria innate antioxidant and immune capacities could also play a role in the susceptibility, and in this regard the mitochondrial DNA variants might serve as susceptibilty markers for severe COVID-19. Common mtDNA variants define the mtDNA haplogroups and have been related with different mitochondrial-mediated capacities, such as ROS and ATP production (Kenney et al., 2014, Kenney et al., 2014, Krzywanski et al., 2016). These mtDNA variants have been associated with the risk of developing common diseases and might also contribute to define the lifespan (Yonova-Doing et al., 2021).
The effect of mtDNA variants on viral-mediated disease has been previously addressed. Among others, manifestations of AIDS, herpex, and COVID-19 could be regulated by the mtDNA variation (Hendrickson et al., 2008, Medrano et al., 2018, Hart et al., 2013, Levinson et al., 2016, Wu et al., 2021, Dirican et al., 2022).
Here, we determined the association of the common European haplogroups with severe COVID-19 and death among SARS-CoV-2 infected.
2. Patients and methods
We obtained the demographic and clinical data of 316 patients who required admission in the intensive care unit (ICU) due to COVID-19 (mean age 64, range 24–95). These patients were hospitalised between March-2020 and April-2021, period in which three pandemic waves took place in our community. They were followed till disease remission with hospital discharge or death. Following previously reported criteria we considered early-onset COVID-19 as an age < 65 years (Gentilotti et al., 2021).
All the participants were of European ancestry and from the region of Asturias (Northern Spain, total population 1 million). The study was approved by the Ethics Committee of Principado de Asturias (Oviedo, Spain). All the patients (or their next of kin) and controls gave their consent to participate in the study. The controls were recruited from the general population with the only purpose of defining the mtDNA variant frequencies, and no data about common traits or clinical manifestations were considered. Although we did not determine the existence of SARS-CoV-2 infection, none of the controls required hospitalization due to COVID-19. In order to avoid the posibility of age-bias we compared patients and controls within the same age-range.
2.1. Haplogroups classification
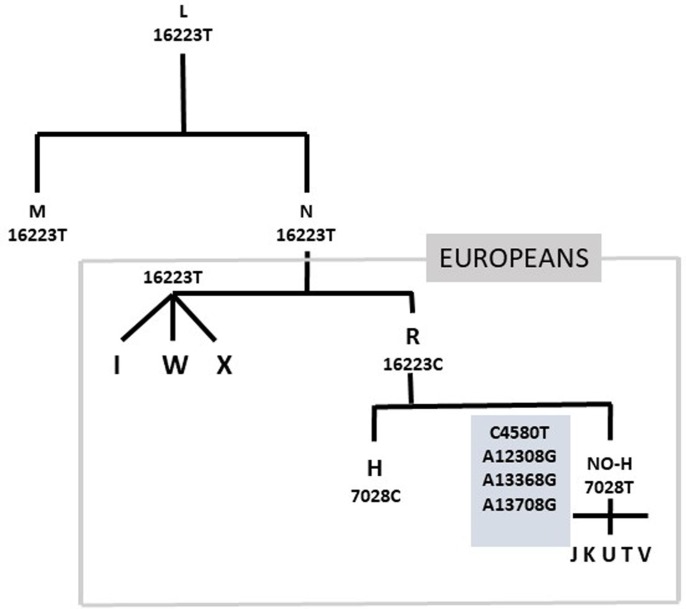
Five mtDNA single nucleotide polymorphisms (SNPs G4580A, C7028T, A12308G, G13368A, and G13708A) were used to determine the most common European mitochondrial haplogroups (see supplementary file methods and the mitomap database for haplogroups definition; www.mitomap.org). Individuals who were 7028C were considered as haplogroup H, and 7028T were further classified as J, KU, T, V, IWX, or other based on the variants combinations (supplementary table). These were also genotyped for the T16223C that differentiates the macro-haplogroup R (16223C) from the ancestral macro-haplogroup N (16223T) (Fig. 1 ).
Fig. 1.
Evolution of the common European haplogroups from the ancestral African L. N originated about 65,000 years ago and is the ancestral for the out-of-Africa haplogroups. The R lineage originated about 60,000 years ago in the Middle East and is characterised by several nucleotide changes, such as 16223C. Most of the common European haplogroups derived from R, icluding the H lineage (7028C) that surged about 20,000 years ago in Southwest Asia and is currently present in 40–50 % of Europeans.
2.2. Statistical analysis
The statistical analysis was performed with the R free software (www.r-project.org). The logistic regression (linear generalized model, LGM) was used to compare mean values and frequencies between the groups.
3. Results
The main characteristics of the patients are summarised in Table 1 . All the participants were first genotyped fro the C7028T that defines the most common European haplogroup H (7028C) (Table 2 ). 7028T patients had a significantly lower mean age than 7028C (63 vs 67 years; p = 0.002). We found a non-significantly higher frequency of the 7028T among the early-onset patients (≤65 years) compared to matched controls (p = 0.11), and a significantly higher frequency compared to the elderly patients p = 0.01). This difference was not attributable to an age-effect in the general population because we did not find differences between controls younger or older than 65 years (47 % and 45 %, respectively). The mtDNA 7028C (haplogroup H) was thus protective for developing severe COVID-19 at an age ≤ 65.
Table 1.
Main values in the 316 COVID-19 patients. PO2/FiO2 = ratio of arterial oxygen partial pressure (PaO2, mmHg) to fractional inspired oxygen (FiO2). Moderate/severe hypoxemia, <300.
|
≤65 years N = 148 |
>65 years N = 168 |
p-value | |
|---|---|---|---|
|
Male Female |
104 (70 %) (30 %) 44 |
125 (74 %) (26 %) 43 |
0.41 |
|
Age Median years (range) |
57 (25–65) |
73 (66–91) |
|
|
BMI median (range) |
29 (19–55) | 31(19–53) | 0.01 |
| BMI > 30 | 81 (41 %) | 68 (55 %) | 0.012 |
| Diabetes | 20 (13 %) | 44 (26 %) | 0.005 |
| Hypercholesterolemia | 51 (34 %) | 91 (54 %) | 0.0004 |
| Hypertension | 57 (39 %) | 115 (69 %) | <0.0001 |
| Death | 13 (9 %) | 54 (32 %) | <0.0001 |
| PO2/FiO2 < 300 | 26 (17 %) | 34 (20 %) | 0.43 |
Table 2.
Frequency of the mtDNA variants in the ICU patients and population controls. All them were genotyped for the 7028 variant (C = haplogroup H). The 7028T were genotyped for the G4580A, A12308G, G13368A, G13708A, and C16223T variants to determine the common non-H haplogroups (see suppl. file methods).
|
≤65 years |
>65 years |
|||
|---|---|---|---|---|
| Covid-19 N = 148 |
Controls N = 182 |
Covid-19 N = 168 |
Controls N = 181 |
|
| 7028C | 57 (39 %) | 86 (47 %) | 88 (52 %) | 82 (45 %) |
| 7028T | 91 (61 %) | 96 (53 %) | 80 (48 %) | 99 (55 %) |
| p-value (T)OR (95 %CI) |
p = 0.11 OR = 1.43 (0.92–2.22) |
p = 0.19 OR = 0.75 (0.49–1-15) |
||
| 16223T | 22 (15 %) | 9 (5 %) | 12 (8 %) | 9 (5 %) |
| 16,223C | 126 (85 %) | 173 (95 %) | 156 (92 %) | 172 (95 %) |
| p-value (T) OR (95 %CI) | p = 0.002 OR = 3.36 (1.49–7.54) | p = 0.39 OR = 1.47 (0.60–3.59) | ||
| J | 18 (12 %) | 28(15 %) | 12 (7 %) | 17 (9 %) |
| K + U | 32 (22 %) | 47(26 %) | 30 (18 %) | 46 (25 %) |
| T | 6 (4 %) | 7 (4 %) | 17 (10 %) | 19 (10 %) |
| V | 3 (2 %) | 2 (1 %) | 4(2 %) | 6 (3 %) |
| IWX | 22 (15 %) | 9(5 %) | 12 (9 %) | 9 (6 %) |
| OTHER | 10 (6 %) | 3(2 %) | 5 (2 %) | 2 (2 %) |
patients ≤ 65 vs. > 65 years: C7028T, p = 0.01; C16223T, p = 0.03.
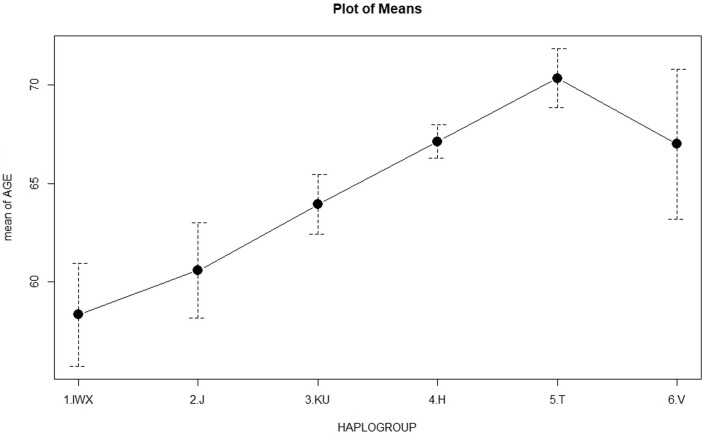
The 7028T were further genotyped for the mtDNA variants to determine the non-H haplogroups (J, KU, T, V, and WXI). We found a higher frequency of the IWX haplogroups in the patients compared to their age matched controls (p < 0.002, OR = 3.36, 95 %CI = 1.49–7.54). There was a significantly higher IWX frequency in the younger vs elderly patients (15 % vs 9 %, p = 0.03). Haplogroups frequencies were non significantly different between elderly patients and controls (Table 2). The IWX haplogroups are characterised by 16223T, that is the ancestral allele compared to the 16223C that characterises the macro-haplogroup R that rooted the main European haplogroups (Fig. 1). In our population, 16223T was a marker for early-onset severe COVID-19 with frequencies 15 % and 9 % in patients younger and older than 65 years, and mean ages of 57 years (16223T) and 66 years (16223C) (p = 0.006) (Fig. 2 ).
Fig. 2.
Mean ages for the common haplogroups in the critical COVID-19 patients. Haplogroups IWX (16223T) had.s a significantly lower mean age compared to the other haplogroups (16223C): 58 vs 66 years, p = 0.006. The frequency of 16223T was significantly higher among erly-onset (≤65 years) patients, p = 0.03. The frequency of 7028C (haplogroup H) was significantly lower among early-onset patients, p = 0.002.
We determined the association between the main cardiovascular traits and antropometric values and the mtDNA haplogroups. The multiple logistic regression including age as a covariate showed that none of the variables was associated with the mtDA variants in the whole cohort (supplementary table). The mtDNA variants were not associated with an increased risk of death, although the number of deceased patients in the younger group (n = 13) was too low to conclude statistical associations.
4. Discussion
The main finding of our study was the significant protective effect of the 7028C variant (haplogroup H) for early onset critical COVID-19. The risk effect of non-H haplogroups was further sttributed to an increased frequency of 16223T (haplogroups IWX) among the younger patients. The age-dependent association of nuclear gene variants with severe COVID-19 has been reported, including the main genetic risk factor in chromosome 3 (Nakanishi et al., 2021).
The 16223T differentiates the ancestral R and non-R haplogroups, represented by the WXI that is rare among the Europeans. Interestingly these mtDNA variants were associated with risk of herpex virus disease (Levinson et al., 20162016). In a large cohort (n = 9,691) of Caucasian Herpex patients haplogroup H was protective (OR = 0.82;95 % CI = 0.71–0.94) whereas the IWX clade was a risk factor for herpes zoster status (OR = 1.38; 95 % CI = 1.07–1.77).In HIV infected patients haplogroup H was associated with alower chance of developing AIDS and higher odds of having a better CD4 + recovery than patients without this haplogroup (Medrano et al., 2018, Guzmán-Fulgencio et al., 2013).Haplogroup R (16223C) and H (7028C) were associated with a significant protection against severe sepsis among Han Chinese an Europeans (Baudouin et al., 2005, Yang et al., 2008, Jiménez-Sousa et al., 2015). This sepsis-protective might confer a survival advantage for H-carriers, that could explain in part why H is the most common European haplogroup despite being the most recent.
In reference to COVID-19, to our knowledge only one study afforded the association with common mtDNA variants (Wu et al., 2021). In this Chinese based case–control study the authors concluded that mtDNA variants defining common haplogroups might contribute to individual’s risk of developing severe COVID-19. This finding was in agreement with our results, but the data are difficult to compare because the different haplogroup profile between Chinese and European populations. Additional studies to characterise the whole mtDNA variation in individuals with the risk haplogroups should be necessary to determine whether these populations share particular functional variants that might explain the association with severe COVID-19.
5. Study limitations
Our study was based on a limited number of patients and from a single population, and requires validation in larger cohorts and from different regions. Also, the variants that defined the haplogroups were not functional and an explanation of the putative mechanism linking the mtDNA variants with severe COVID-19 is not provided, beyond the reported difeerence in mitochondrial function between the different haplogroups (Kenney et al., 2014, Kenney et al., 2014, Krzywanski et al., 2016). This would require a complete sequencing of the mtDNA in patients, as well as functional studies to determine the putative effect on the control of viral infection.
6. Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. An Excel file with the raw data would be available for meta-analysis research.
7. Ethics and consent
This study was approved by the clinical research ethics committee of Hospital Universitario Central Asturias (HUCA). All the participants or they next of kin gave written or verbal consent. Data were handled in observance of Spanish legislation on data protection. The study complies with the principles of the Declaration of Helsinki (“Recommendations guiding doctors in biomedical research involving human subjects”).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a grant from the Spanish Plan Nacional de I + D + I Ministerio de Economía y Competitividad and the European FEDER, grants ISCIII-PI21/00971 (E.C.) and RICOR2040- RD21/0005/0011 (E.C.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mito.2022.09.001.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Baudouin S.V., Saunders D., Tiangyou W., et al. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005 Dec 17;366(9503):2118–2121. doi: 10.1016/S0140-6736(05)67890-7. PMID: 16360789. [DOI] [PubMed] [Google Scholar]
- Belgnaoui S.M., Paz S., Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 2011 Oct;23(5):564–572. doi: 10.1016/j.coi.2011.08.001. Epub 2011 Aug 22PMID: 21865020. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Ping, Y.H., Lee, H.C., et al., 2007. Open reading frame 8a of thehuman severe acute respiratory syndrome coronavirus not only promotesviral replication but also induces apoptosis. J. Infect. Dis.,196, 405–415. [DOI] [PMC free article] [PubMed]
- Choi H.J., Park A., Kang S., Lee E., Lee T.A., Ra E.A., Lee J., Lee S., Park B. Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat. Commun. 2018 Jan 9;9(1):125. doi: 10.1038/s41467-017-02624-8. PMID: 29317664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T.J., Potje S.R., Fraga-Silva T.F.C., da Silva-Neto J.A., Barros P.R., Rodrigues D., Machado M.R., Martins R.B., Santos-Eichler R.A., Benatti M.N., de Sá K.S.G., Almado C.E.L., Castro Í.A., Pontelli M.C., Serra L.L., Carneiro F.S., Becari C., Louzada-Junior P., Oliveira R.D.R., Zamboni D.S., Arruda E., Auxiliadora-Martins M., Giachini F.R.C., Bonato V.L.D., Zachara N.E., Bomfim G.F., Tostes R.C. Mitochondrial DNA and TLR9 activation contribute to SARS-CoV-2-induced endothelial cell damage. Vasc.Pharmacol. 2022;142:106946. doi: 10.1016/j.vph.2021.106946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature volume 600, pages472–477 (2021). [DOI] [PMC free article] [PubMed]
- Dirican, E., Savrun, Ş.T., Aydın, İ.E., Gülbay, G., Karaman, Ü., 2022. Analysis of mitochondrial DNA cytochrome-b (CYB) and ATPase-6 gene mutations in COVID-19 patients. J. Med. Virol., 2022 Mar 8. 10.1002/jmv.27704. Online ahead of print.PMID: 35258110. [DOI] [PMC free article] [PubMed]
- Gentilotti E., Savoldi A., Compri M., Górska A., De Nardo P., Visentin A., Be G., Razzaboni E., Soriolo N., Meneghin D., Girelli D., Micheletto C., Mehrabi S., Righi E., Tacconelli E. Assessment of COVID-19 progression on day 5 from symptoms onset. BMC Infect. Dis. 2021;21(1) doi: 10.1186/s12879-021-06596-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán-Fulgencio M., Jiménez J.L., García-Álvarez M., Bellón J.M., Fernández-Rodriguez A., Campos Y., Rodríguez C., González-Garcia J., Riera M., Viciana P., Muñoz-Fernández MÁngeles, Resino S. Mitochondrial haplogroups are associated with clinical pattern of AIDS progression in HIV-infected patients. J. Acquir. Immune Defic. Syndr. 2013;63(2):178–183. doi: 10.1097/QAI.0b013e3182893f74. [DOI] [PubMed] [Google Scholar]
- Hart A.B., Samuels D.C., Hulgan T. The other genome: a systematic review of studies of mitochondrial DNA haplogroups and outcomes of HIV infection and antiretroviral therapy. AIDS Rev. 2013;15(4):213–220. PMID: 24322381. [PMC free article] [PubMed] [Google Scholar]
- Hendrickson S.L., Hutcheson H.B., Ruiz-Pesini E., Poole J.C., Lautenberger J., Sezgin E., Kingsley L., Goedert J.J., Vlahov D., Donfield S., Wallace D.C., O'Brien S.J. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22(18):2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MengJie, Schulze K.E., Ghildyal R., Henstridge D.C., Kolanowski J.L., New E.J., Hong Y., Hsu A.C., Hansbro P.M., Wark P.AB., Bogoyevitch M.A., Jans D.A. Respiratory syncytial virus co-opts host mitochondrial function to favour infectious virus production. Elife. 2019 Jun;8:e42448. doi: 10.7554/eLife.42448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Sousa M.A., Tamayo E., Guzmán-Fulgencio M., Heredia M., Fernández-Rodríguez A., Gómez E., Almansa R., Gómez-Herreras J.I., García-Álvarez M., Gutiérrez-Junco S., Bermejo-Martin J.F., Resino S. Mitochondrial DNA haplogroups are associated with severe sepsis and mortality in patients who underwent major surgery. J. Infect. 2015;70(1):20–29. doi: 10.1016/j.jinf.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Kenney M.C., Chwa M., Atilano S.R., Falatoonzadeh P., Ramirez C., Malik D., Tarek M., Caceres-del-Carpio J., Nesburn A.B., Boyer D.S., Kuppermann B.D., Vawter M., Jazwinski S.M., Miceli M., Wallace D.C., Udar N. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum. Mol. Genet. 2014;23(13):3537–3551. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney M.C., Chwa M., Atilano S.R., Falatoonzadeh P., Ramirez C., Malik D., Tarek M., del Carpio J.C., Nesburn A.B., Boyer D.S., Kuppermann B.D., Vawter M.P., Jazwinski S.M., Miceli M.V., Wallace D.C., Udar N. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. BBA. 2014;1842(2):208–219. doi: 10.1016/j.bbadis.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T. Mitochondrial-mediated antiviral immunity. BBA. 2013 Jan;1833(1):225–232. doi: 10.1016/j.bbamcr.2012.03.005. Epub 2012 Mar 13PMID: 22440325. [DOI] [PubMed] [Google Scholar]
- Krzywanski D.M., Moellering D.R., Westbrook D.G., Dunham-Snary K.J., Brown J., Bray A.W., Feeley K.P., Sammy M.J., Smith M.R., Schurr T.G., Vita J.A., Ambalavanan N., Calhoun D., Dell’Italia L., Ballinger S.W. Endothelial Cell Bioenergetics and Mitochondrial DNA Damage Differ in Humans Having African or West Eurasian Maternal Ancestry. Circ. Cardiovasc. Genet. 2016;9(1):26–36. doi: 10.1161/CIRCGENETICS.115.001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson, R.T., Hulgan, T., Kalams, S.A., Fessel, J.P., 2016. Samuels DC. Mitochondrial Haplogroups as a Risk Factor for Herpes Zoster. Open Forum Infect. Dis., 3(4), ofw184. 10.1093/ofid/ofw184. eCollection 2016 Oct.PMID: 27807590. [DOI] [PMC free article] [PubMed]
- Medrano L.M., Gutiérrez-Rivas M., Blanco J., García M., Jiménez-Sousa M.A., Pacheco Y.M., Montero M., Iribarren J.A., Bernal E., Martínez O.J., Benito J.M., Rallón N., Resino S. Mitochondrial haplogroup H is related to CD4+ T cell recovery in HIV infected patients starting combination antiretroviral therapy. J. Transl. Med. 2018;16(1) doi: 10.1186/s12967-018-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, Y., To, K.K., Zhou, R., et al., 2022. Mitochondrial Dysfunction Associates With Acute T Lymphocytopenia and Impaired Functionality in COVID-19 Patients. Front Immunol., 12, 799896. 10.3389/fimmu.2021.799896. eCollection 2021.PMID: 35095881. [DOI] [PMC free article] [PubMed]
- Moore C.B., Ting J.P. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008 Jun;28(6):735–739. doi: 10.1016/j.immuni.2008.05.005. PMID: 18549796. [DOI] [PubMed] [Google Scholar]
- Nakanishi, T., Pigazzini, S., Degenhardt, F., et al., 2021. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. J. Clin. Invest., 131(23), e152386. 10.1172/JCI152386.PMID: 34597274. [DOI] [PMC free article] [PubMed]
- Schneider A., Kurz S., Manske K., Janas M., Heikenwälder M., Misgeld T., Aichler M., Weissmann S.F., Zischka H., Knolle P., Wohlleber D. Single organelle analysis to characterize mitochondrial function and crosstalk during viral infection. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-44922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, C., Liu, Z., Zhu, Y., et al., 2022. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol.,12, 780768. 10.3389/fmicb.2021.780768. eCollection 2021.PMID: 35069483. [DOI] [PMC free article] [PubMed]
- Shi C.-S., Qi H.-Y., Boularan C., Huang N.-N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innateimmunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 sig-nalosome. J. Immunol. 2014;193(6):3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., Xian W., Qian X., Li Z., Huang Y., Tang Q., Liu P., Chen R., Liu R., Li X., Tong X., Zhou X., Bai Y., Duan G., Zhang T., Xu X., Wang J., Yang H., Liu S., He Q., Jin X., Liu L. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.i., Wang X.-H., Li X.-H., Song L.-Y., Yu S.-L., Fang Z.-C., Liu Y.-Q., Yuan L.-Y., Peng C.-Y., Zhang S.-Y., Cheng W., Ma H.-C., Wang L.-F., Tang J.-M., Wang Y.-F., Ji F.-Y. Common mtDNA variations at C5178a and A249d/T6392C/G10310A decrease the risk of severe COVID-19 in a Han Chinese population from Central China. Mil. Med. Res. 2021;8(1) doi: 10.1186/s40779-021-00351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.i., Shou Z., Zhang P., He Q., Xiao H., Xu Y., Li C., Chen J. Mitochondrial DNA haplogroup R predicts survival advantage in severe sepsis in the Han population. Genet. Med. 2008;10(3):187–192. doi: 10.1097/GIM.0b013e318163c343. [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu Y., Meng X., Wang Z., Younis M., Liu Y.e., Wang P., Huang X.i. SARS-CoV-2 membrane protein causes the mitochondrial apoptosis and pulmonary edema via targeting BOK. Cell Death Differ. 2022;29(7):1395–1408. doi: 10.1038/s41418-022-00928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonova-Doing E., Calabrese C., Gomez-Duran A., Schon K., Wei W., Karthikeyan S., Chinnery P.F., Howson J.M.M. An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank. Nat. Genet. 2021;53(7):982–993. doi: 10.1038/s41588-021-00868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi T., Ichinohe T., Sasaki O., Otera H., Kawabata S.-I., Mihara K., Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014;5(1) doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- Zamarin, D., García-Sastre, A., Xiao, X., Wang, R., Palese, P., 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. Sep;1(1):e4. 10.1371/journal.ppat.0010004. Epub 2005 Sep 30.PMID: 16201016. [DOI] [PMC free article] [PubMed]
- Zeng, H., Ma, Y., Zhou, Z., et al., 2021. Spectrum and Clinical Characteristics of Symptomatic and Asymptomatic Coronavirus Disease 2019 (COVID-19) With and Without Pneumonia. Front. Med. (Lausanne), 8:645651. 10.3389/fmed.2021.645651. eCollection 2021.PMID: 33869253. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. An Excel file with the raw data would be available for meta-analysis research.