Abstract
Aggregation of amyloidogenic proteins is a characteristic of multiple neurodegenerative diseases. Atomic resolution of small molecule binding to such pathological protein aggregates is of interest for the development of therapeutics and diagnostics. Here we investigate the interaction between α-synuclein fibrils and anle138b, a clinical drug candidate for disease modifying therapy in neurodegeneration and a promising scaffold for positron emission tomography tracer design. We used nuclear magnetic resonance spectroscopy and the cryogenic electron microscopy structure of α-synuclein fibrils grown in the presence of lipids to locate anle138b within a cavity formed between two β-strands. We explored and quantified multiple binding modes of the compound in detail using molecular dynamics simulations. Our results reveal stable polar interactions between anle138b and backbone moieties inside the tubular cavity of the fibrils. Such cavities are common in other fibril structures as well.
Subject terms: Solid-state NMR, Molecular modelling, Molecular neuroscience, Protein aggregation
Understanding how small molecules bind to pathological aggregates is of importance for therapeutic and diagnostic development in diseases such as Parkinson’s Disease. Here, the authors reveal a binding site of anle138b to lipid-induced α-synuclein fibrils.
Introduction
The presence of α-synuclein fibrils in inclusion bodies is a hallmark of neurodegenerative diseases such as Parkinson’s Disease1, Multiple System Atrophy2,3 and Dementia with Lewy bodies4,5. α-Synuclein aggregation is thought to be involved in cellular pathology and spreading between cells6. To date, no disease-modifying treatment is available for any of those diseases7. Hence, atomic resolution structures of small molecule binding to such pathological protein aggregates are of interest for the development of therapeutics and diagnostics8,9. To address this issue, we studied the binding of the small molecule anle138b to α-synuclein fibrils.
Anle138b [3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)-1H-pyrazole] shows efficacy in animal models for several aggregation related diseases8,10–14. It was tested successfully in healthy volunteers15 and is currently undergoing phase Ib testing in Parkinson’s disease (NCT04685265). On a molecular level, anle138b was shown to interfere with α-synuclein oligomerization in vitro, inhibiting pore formation in membranes as well as cytochrome c leakage from mitochondria and depopulating α-synuclein aggregates in vivo8,14. Due to its highly lipophilic nature, it readily crosses the blood-brain-barrier11. Its tight binding (Kd = 190 ± 120 nM) to α-synuclein fibrils16 also makes it an attractive starting point for the development of diagnostic positron emission tomography (PET) tracer molecules9.
α-Synuclein is a 140 amino-acid intrinsically disordered protein, which has been suggested to participate in synaptic vesicle exocytosis, brain lipid metabolism, and neuronal survival because of its affinity to lipid membranes.17 In Parkinson’s disease-related Lewy bodies, α-synuclein fibrils are found to colocalize with lipids, indicating the importance of lipids in the α-synuclein aggregation cascade18. We recently studied the aggregation of α-synuclein grown in the presence of negatively charged liposomes consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and observed a segmental folding process19. Concordantly, we resolved the resulting protofilament fold L2 of lipid-bound α-synuclein fibrils using cryogenic electron microscopy (cryo-EM)20.
Results
Anle138b binds to fibril interior
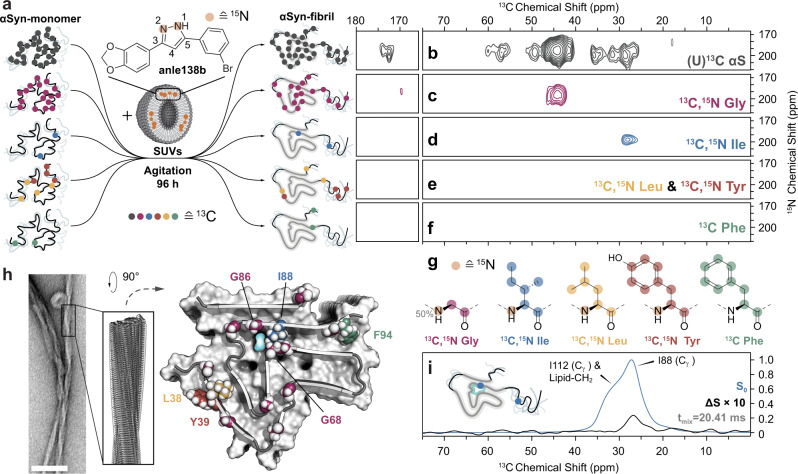
We located the binding site by isotope labeling anle138b with 15N, and directly detecting its interaction with fibrils grown from 13C-labeled full-length α-synuclein (Fig. 1a). Such interactions were recorded in two-dimensional (2D) NHHC nuclear magnetic resonance (NMR) spectra with signal enhancement via dynamic nuclear polarization (DNP) and magic-angle spinning (MAS) at 100 K21. During fibrillization, we used anle138b doped liposomes consisting of POPA and POPC (1:1). Proton–proton mixing in these experiments results in cross-peaks between nearby nuclei up to a distance of about 6 Å22 (Fig. 1b and Supplementary Fig. 1). The most prominent cross-peak at 44.8 ppm is consistent with the pyrazole NH of anle138b binding near Cβ of Phe, Leu, Tyr, or Ile or Cα of Gly. We prepared α-synuclein fibrils with selective 13C-labeling on these residues and used 2D 13C,13C RFDR for the assignment of resonances (Supplementary Fig. 2a, c–f). 2D NHHC spectra of these fibrils in the presence of 15N-labeled anle138b (Fig. 1a, b–g, Supplementary Fig. 2b, and Supplementary Table 1) establish Gly as responsible for the peak at 44.8 ppm and Ile as another part of the binding site (28 ppm peak). Interaction with other lipophilic residues, Leu, Tyr, and Phe, or the amine of lysine (Fig. 1e, f and Supplementary Fig. 1b), could not be detected. The interaction of the compound with Cγ of an Ile (Fig. 1d) establishes the binding site at I88, since the only other Ile at position 112 can be excluded based on its distinct chemical shift and broad line shape (Supplementary Fig. 2d). The sidechain of I88 is facing towards the protofilament interior and is part of a glycine-rich sequence that encloses a tubular hydrophobic cavity along α-synuclein protofilament L220 (Fig. 1h, PDB ID 8A4L). Rotational echo double resonance (REDOR) experiments confirm close proximity of anle138b to I88 (Fig. 1i). The observed interactions at I88 and Gly, as well as spectroscopic features of G68 and G86 (vide infra), therefore indicate binding of anle138b inside this cavity.
Fig. 1. Identification of anle138b binding inside α-synuclein fibrils by DNP-enhanced MAS NMR.
a Isotope labeling strategy used for the preparation of fibril samples for studies by DNP-enhanced MAS NMR. 13C-labeling of amino acids indicated by colored circles: unifom- (gray), Gly- (magenta), Ile- (blue), Leu- (yellow), Tyr- (red), and Phe-labeling (green). 15N-labeling on anle138b indicated by orange circles. b–f Slices of 2D NHHC spectra of α-synuclein fibrils prepared from protein with b uniform 13C-labeling and c–f amino-acid-specific isotope labeling on c Gly, d Ile, e Leu and Tyr, and f Phe in the presence of 1,2-15N-anle138b. The spectral region shown contains only cross-peaks between pyrazole NH of anle138b (195 ppm) and protein carbon atoms. Proton–proton mixing was 200 μs. g Chemical structures of amino acids used for specific isotope labeling with 13C- labeled nuclei indicated by colored circles (same as in a) and 15N-labeling indicated by orange circles. h Negative stain EM micrograph of fibrils grown in the presence of lipids (scale bar 100 nm) alongside cryo-EM structure of α-synuclein protofilament L220 with color coding of specific isotope-labeled residues. i Frequency selective REDOR spectrum (carrier frequency at 26 ppm, Supplementary Table 2) of Ile-labeled α-synuclein fibrils in the presence of 1,2-15N-anle138b. The difference spectrum (ΔS) is scaled up 10-fold with respect to the reference spectrum (S0) for clarity. Fibrils were prepared at a lipid to protein to anle138b molar ratio of 10:1:0.5. All spectra were recorded at 600 MHz at 100 K (see Supplementary Table 1) in the presence of TEMTriPol-1.
Bound anle138b changes the local structure
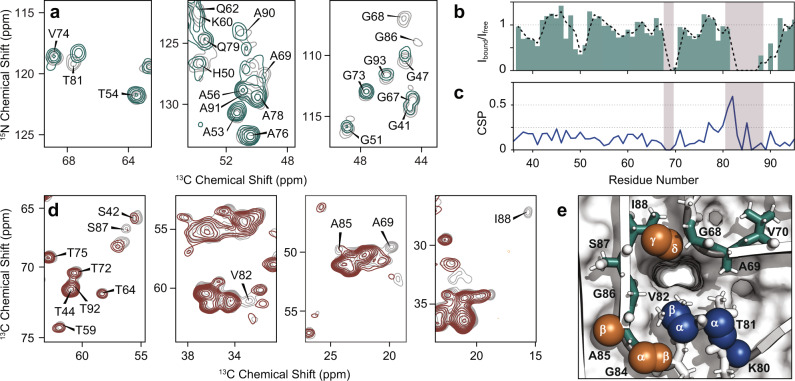
To probe potential changes in the α-synuclein protofilament structure induced by anle138b, we recorded room temperature MAS NMR spectra. Specifically, we acquired 2D 13C,13C correlation spectra with dipolar assisted rotational recoupling (DARR) to obtain correlations between carbon atoms, and 3D (H)CANH spectra to obtain correlations between protein backbone Cα, NH and HN. The 2D DARR spectra with long mixing (Supplementary Fig. 3) show similar protein long-range contacts in the presence and absence of anle138b, confirming an overall conserved α-synuclein protofilament fold. The presence of anle138b results in chemical shift perturbation (CSP) for 80KTV82, A85, and I88 (Fig. 2c and Supplementary Fig. 4a–c) as well as attenuation for residues 67GGAV70 and 82VEGAGSI88 as seen from 3D (H)CANH (Fig. 2a, b and Supplementary Fig. 4d) and 2D DARR spectra (Fig. 2d). This observation indicates increased dynamics at the affected residues compared to the absence of anle138b. Peak-broadening of Cα of G68 as well as G86 (Fig. 2a) and particularly Cγ of I88 (Fig. 2d) corroborate the contacts observed under DNP conditions. As these residues belong to the two stretches of the α-synuclein sequence which establish the aforementioned cavity in protofilament L2 (Fig. 2e), we conclude that anle138b is embedded within α-synuclein fibrils, while causing only local structural changes.
Fig. 2. Chemical shift perturbations comparing α-synuclein fibrils grown in the presence and absence of anle138b.
a Excerpts of carbon-nitrogen projections of 3D (H)CANH spectra of lipidic α-synuclein fibrils. Signal attenuation (Ibound/Ifree) and chemical shift perturbation (CSP) are observed comparing spectra of fibrils with anle138b (green, 800 MHz with 55 kHz MAS) and without anle138b (gray, 950 MHz with 100 kHz MAS). b Relative peak intensities from 3D (H)CANH spectra and c, average CSP from DARR and (H)CANH spectra induced by anle138b inside α-synuclein fibrils plotted per residue. Black dotted line represents a 2-residue moving average; highlighted areas indicate the cavity in protofilament L2. d Excerpts of 2D 13C,13C correlation spectra (20 ms DARR mixing) of α-synuclein fibrils with (red) and without (gray) anle138b acquired at 850 MHz with 17 kHz MAS. e Residues enclosing the cavity in α-synuclein protofilament L2 color-coded by nuclei experiencing significant CSP (blue), signal attenuation (green), or both (copper). Source data are provided as a Source Data file.
Polar contacts govern internal binding
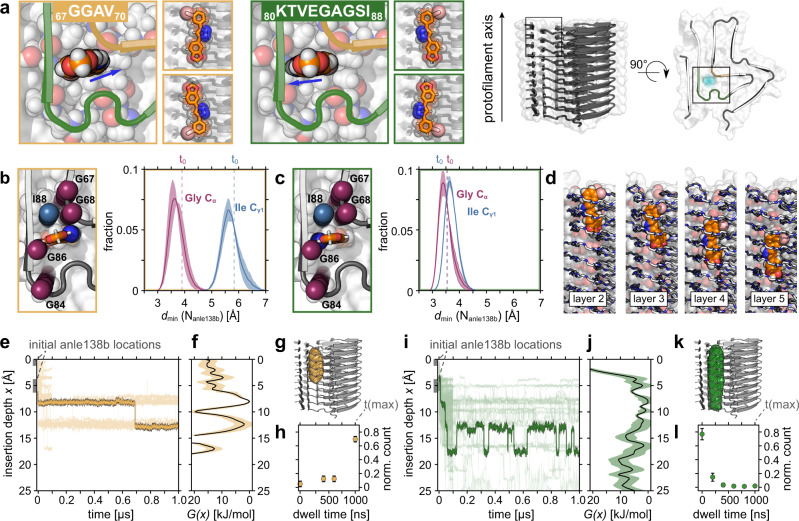
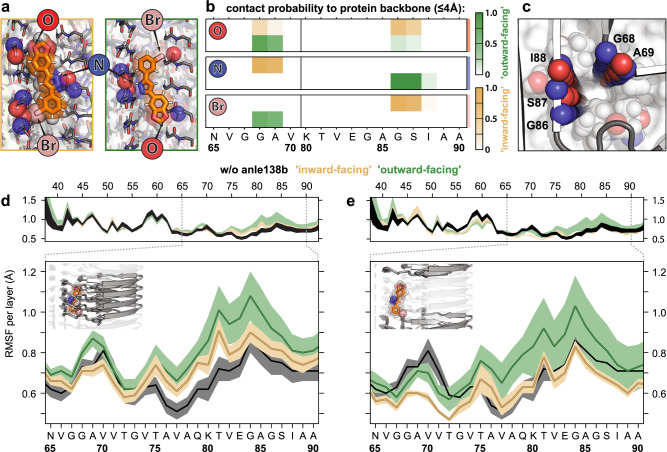
The rather narrow cavity of the α-synuclein protofilament L2 permits four principal internal binding poses for the planar anle138b molecule. Inside the protofilament cavity, the pyrazole nitrogen atoms of anle138b can face either “inward” or “outward” and the bromophenyl ring pointing “up” or “down” with respect to the direction of the long axis of the protofilament (Fig. 3a). Here, the protofilament axis was defined by the direction of the NH vector of every 2nd amino acid in the folded, N-terminal part of the protofilament, starting with the residue G36. All-atom molecular dynamics (MD) simulations (see “Methods” and Supplementary Table 3) were employed to probe if these anle138b-binding poses are indeed consistent with the observations made by NMR. To do so, both tautomers of the anle138b molecule were considered and simulated in the dominant conformer as determined by NMR, respectively (Supplementary Fig. 5). During 40 independent, 1-µs long MD simulations, no unbinding events from the cavity of the α-synuclein protofilament L2 structure were observed independent of the initial binding pose and tautomeric form of anle138b (Supplementary Fig. 6a). Intentionally placing the compound at the edge of the protofilament leads to spontaneous insertion into the internal binding site, demonstrating favorable binding (Supplementary Fig. 7a, b). The steric hindrance imposed by nonpolar side-chain groups (A69, T81, V82, and I88) surrounding the cavity did not allow for rotations of the molecule (Supplementary Fig. 6b). The Cγ of I88 showed close and persistent contacts to the anle138b pose with outward-facing pyrazole nitrogen atoms, while the Cα of either G67, G68, G84 or G86 did so for both anle138b poses in the simulations (Fig. 3b, c). This is most consistent with NHHC and REDOR spectra for outward-facing anle138b (Fig. 1c, d, i). Since short distances dominate REDOR data, the presence of the inward-facing pose, with larger separation of anle138b nitrogen atoms and I88 Cγ is compatible as well. We thus surmise that the outward-facing pose is present in the fibrils, as well as an undetermined amount of inward-facing anle138b. From the MD simulations, we observed a stepwise displacement of anle138b within the internal cavity and along the stacked β-strand layers of the protofilament (Fig. 3d and Supplementary Fig. 6d) on the tens to hundreds of ns timescale (Fig. 3e, h, i, l and Supplementary Movie S1), consistent with the disappearance of some NMR signals at ambient temperatures and the use of cryogenic DNP conditions to localize the compound. Relative free-energy binding profiles (Fig. 3f, g, j, k and Supplementary Fig. 7c) show multiple local energy minima as a function of anle138b insertion depth within the protofilament core for all tested binding poses. However, for inward-facing anle138b binding poses significantly higher energy barriers are found (~20 kJ/mol, Fig. 3f). In concordance with simulations on model systems23, anle138b showed characteristic polar interactions with exposed backbone epitopes (Fig. 4a). Polar ladders of backbone atoms lining the cavity (Fig. 4a, c and Supplementary Figs. S6e, S8) interact with anle138b either through hydrogen bonds via the pyrazole ring (outward-facing) or halogen bonds involving the bromine atom (inward-facing) (see “Methods”/Supplementary Information). The common pattern of close contacts between nitrogen atoms of anle138b and protein backbone atoms of G68, A69, or G86 and S87 (Fig. 4b and Supplementary Fig. 6e) agrees with 2D NHHC spectra (Fig. 1c and Supplementary Fig. 1b).
Fig. 3. Anle138b binds dynamically inside the α-synuclein protofilament.
a Starting structures for MD simulations for the four principal poses of anle138b: Pyrazole nitrogen atoms of anle138b (indicated by blue arrows) are oriented either to residues 67GGAV70 (inward-facing, yellow) or to residues 80KTVEGAGSI88 (outward-facing, green) inside the cavity (cyan) of α-synuclein protofilament L2. The bromophenyl moiety can point up or down the protofilament (small panels). A color code denotes the individual simulation sets for each binding pose in all following panels (inward-facing—yellow, outward-facing—green). b, c Orientation (left) and distributions of minimal distances (right) between nitrogen atoms of anle138b and Cγ1 of I88 (light-blue), and Cα of G67, G68, G84, G86 (magenta) as described in a. Dashed lines indicate the initial positions of anle138b at the start of the simulation. d Snapshots and e, i trajectories (representative trajectories as bold lines; others as thin lines for clarity) illustrate discrete translational motion along the protofilament axis and alignment of anle138b with individual β-strand layers. Relative free-energy binding profiles G(x) as a function of anle138b insertion depth x for combined simulation sets of f inward-facing and j outward-facing pose. Data are presented as mean values ± SEM (depicted by shading). g, k Isosurfaces of corresponding anle138b occupancy. h, l Cumulative histogram of dwell times calculated from anle138b displacements inside the tubular cavity for h inward-facing (n = 24) and l outward-facing (n = 218) pose. Data are presented as mean values ± SEM (indicated by error bars). Water and lipid molecules omitted for clarity in all renderings.
Fig. 4. Anle138b forms polar interactions with the protein backbone that alter local structural fluctuations in the protofilament.
a Polar contacts between anle138b atoms (indicated by arrows) and protein backbone atoms (blue circles—amide nitrogen; red circles—carbonyl oxygen) observed in MD simulations for inward- (yellow) and outward-facing (green) internal binding poses. b Contact probabilities for polar interactions with backbone atoms of individual residues for polar interactions defined in a. Scale bars (right) indicate contact probabilities. c Tubular cavity viewed up close down the long axis of the α-synuclein protofilament (shown without anle138b for clarity). Prominent contact sites in the backbone scaffold are represented by circles (colors as in a). d, e Average root mean squared fluctuations (RMSF) for d the four consecutive β-strands in contact with anle138b and e β-strands without close polar interactions. Average RMSF without anle138b present shown for comparison (black lines). β-strands of the fibril ends were not considered. Data are presented as mean values ± SEM (depicted by shading). Source data are provided as a Source Data file.
Moreover, when bound inside the protofilament, anle138b molecules markedly alter the structural fluctuations of residues around the cavity during MD simulations. All β-strand layers in proximity to the compound, especially when facing outward, (Fig. 4d) exhibit a higher averaged RMSF for residues 67GGA69, and 80KTVEGAGSI88 consistent with the attenuation and CSP observed by NMR for these perturbed regions (Fig. 2). A similar effect is also seen for layers not in contact with the compound (Fig. 4e).
Simulations with anle138b molecules starting outside the α-synuclein protofilament cavity demonstrate the affinity of anle138b for the lipid environment and pronounced colocalization of the compound around lipid binding sites on the protofilament exterior (Supplementary Figs. S9 and S10). In contrast to the stable internal binding, such binding events are transient, explaining the absence of exterior contacts in NMR spectra (Fig. 1).
Discussion
Long tubular cavities, as observed in the α-synuclein protofilament L2 and other α-synuclein folds such as PDB IDs 6SSX/6RT024, 7NCI, and 7NCH25, (Supplementary Fig. 11) as well as many other amyloid fibrils26,27 are a unique feature of the repetitive fibril architecture. This level of commonality across different fibrils could have implications for positron emission tomography radiotracer development. So far, tracer targets are usually identified in exterior binding sites similar to those reported for fibrils of HET-s28, amyloid-β (Aβ)29, and tau30,31. While theoretical studies have predicted the existence of binding sites in the core of fibrillar aggregates32, experimental data did not allow for a high-resolution structure33. End-to-end filling of tubular cavities in fibrils by hydrophobic molecules has been proposed before34. We find selective binding inside and translational motion of anle138b along the α-synuclein fibril cavity, consistent with this model. Our results emphasize that expanding radiotracer target identification to fibril cavities as well as structure-based refinement of cavity binding molecules, such as anle138b, could prove to be beneficial. Fibrils amplified from Parkinson’s Disease patient's brain exhibit a similar fold to the lipidic α-synuclein L2 fibrils and support the relevance of the protofilaments in the present study35,36. Further studies are needed to determine whether the internal binding mode of anle138b in lipidic fibrils, is also a prominent feature of anle138b interactions with early aggregates23.
We presented a high-resolution structural model for anle138b binding in a lipidic α-synuclein fibril. We explored how the linear arrangement of polar groups inside the continuous tubular cavity formed along the cross-β spine of the protofilament facilitates small molecule binding. It will be of significant interest whether the particular features of glycine enriched tight and hydrophobic cavities as reported here, can also be exploited for selective compound binding to fibrillar aggregates of other amyloidogenic proteins such as Aβ, tau, and prion protein which also bear these cavities.
Methods
Preparation of isotope-labeled protein
α-Synuclein was recombinantly produced in E. coli strain BL21(DE3) as previously described37. Uniformly 15N- and 13C,15N-labeled samples were expressed in a minimal medium supplemented with 15NH4Cl and 13C6-d-glucose (Cambridge Isotope Laboratories and Sigma Aldrich). For the production of amino-acid-specific forward-labeled protein (13C,15N-Leu/Tyr, 13C-Phe, 13C,15N-Ile), the labeled amino acids were added to the minimal medium 1 h before induction of protein expression. For forward labeling with 13C- and 13C,15N-Gly, protein expression was performed with the glycine auxotrophic E. coli strain DL39 (DE3) GlyA following a published protocol38. The protein was finally dialyzed against buffer (50 mM HEPES, 100 mM NaCl, pH 7.4) to obtain a 0.3 mM solution, and the resulting solution was stored at −80 °C until use.
Preparation of isotope-labeled anle138b
The general information for the synthesis of compounds was described previously8.
Preparation of [1,2-15N2]-3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)-1H-pyrazole
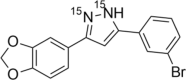
A mixture of [15N2] hydrazine sulfate (2.57 g, 19.5 mmol), EtOH (20 mL), water (1 mL), and 40% KOH aqueous solution (5.6 g, 40 mmol) was stirred in a sealed pressure flask at 80 °C for 1 h. The mixture was cooled, 3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)propane-1,3-dione8 (2.43 g, 7 mmol) was added, the reaction vessel was sealed again, and the heterogeneous mixture was stirred at 80 °C for 15 h. The cooled mixture was diluted with water (100 mL), the resulting precipitate was filtered off, washed with water, and air-dried. The crude product was crystallized from n-BuOH (10 mL) and vacuum dried to give the title compound as a white solid (1.95 g, 5.65 mmol, 80%). 1H NMR (400 MHz, DMSO-d6 + 0.5% TFA) d = 8.05 (s, 1H), 7.84 (d, J = 7.8 Hz, 1H), 7.52 (d, J = 8.1 Hz, 1H), 7.45-7.40 (m, 2H), 7.40-7.34 (m, 1H), 7.25 (d, J = 3.1 Hz, 1H), 7.01 (d, J = 8.1 Hz, 1H), 6.06 (s, 2H). 13C NMR (100.6 MHz, DMSO-d6 + 0.5% TFA) d = 147.9, 147.4, 146.5, 146.3, 133.9, 131.1, 130.6, 127.7, 124.4, 124.2, 122.3, 119.3, 108.8, 105.8, 101.3, 100.1. LC MS (RP18-100Å, gradient 50% CH3CN/50% H2O to 100% CH3CN in 30 min), RT 14.2 min and mass 345.1 (100%), 347.2 (98%), [M + H]+. TLC (SiO2, n-hexane/EtOAc = 2/1) Rf 0.39, m.p. 195–197 °C.
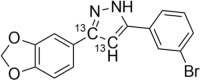
Preparation of [3,4-13C2]-3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)-1H-pyrazole
[1,2-13C2]-1-(1,3-benzodioxol-5-yl)ethanone
Starting material [1,2-13C2]-1-(1,3-benzodioxol-5-yl)ethanone was synthesized by Friedel-Crafts reaction of 1,3-benzodioxole and [1,2-13C2]-acetic acid (99% 13C) according to procedure described in39. In an atmosphere of nitrogen, triflic anhydride (5.0 g, 17.7 mmol) was added to [1,2-13C2]-acetic acid (99% 13C) (1 g, 16.1 mmol) with cooling (10 °C bath temperature). After stirring at room temperature for 5 min, 1,3-benzodioxole (1.97 g, 16.1 mmol) was added, and the stirring at room temperature was continued for 30 min. The mixture was quenched with saturated NaHSO4 solution (30 mL) and extracted with EtOAc (30 mL). The separated organic layer was washed with water, brine, dried over Na2SO4, and concentrated under reduced pressure. The residue was purified by flash chromatography on silica gel (n-hexane/EtOAc = 3/1) to afford the title compound (1.63 g, 9.8 mmol, 61%) as a white solid. 1H NMR (400 MHz, CDCl3) d = 7.55 (ddd, J = 8.1, 4.1 1.7 Hz, 1H), 7.44 (dd, J = 3.6, 1.7 Hz, 1H), 6.85 (d, J = 8.1 Hz, 1H), 6.04 (s, 2H), 2.54 (dd, J = 127.5, 5.9 Hz, 3H). 13C NMR (100.6 MHz, CDCl3) d = 196.2 (d, J = 43.1 Hz, 13C-enriched), 151.7, 148.2, 132.1 (dd, J = 53.9, 14.1 Hz, 13C-enriched), 124.7, 108.0, 107.8, 101.8, 26.4 (d, J = 43.1 Hz, 13C-enriched). LC MS (RP18-100Å, gradient 50% CH3CN/50% H2O to 100% CH3CN in 30 min), RT 6.2 min and mass 166.9 [M + H]+. TLC (SiO2, n-hexane/EtOAc = 3/1) Rf 0.5, m.p. 87–89 °C.
[3,4-13C2]-3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)-1H-pyrazole
The compound was synthesized according to the procedure described in8. To a solution of [1,2-13C2]-1-(1,3-benzodioxol-5-yl)ethanone (830 mg, 5.0 mmol) and methyl 3-bromobenzoate (1.35 g, 6.3 mmol) in DMSO (12 mL) and THF (3 mL) was added sodium hydride (60% in oil, 6.3 mmol, 252 mg). The reaction mixture was stirred at 20 °C for 24 h, then poured into 60 mL of ice water containing 85% H3PO4 (0.6 mL) and stirred for 1 h. The resulting precipitate was filtered off, washed with water (3.10 mL), MeOH (5 mL), hexane (10 mL), and air-dried to give the crude intermediate [1,2-13C2]-3-(1,3-benzodioxol-5-yl)-5-(3-bromophenyl)propane-1,3-dione (1.57 g) as a yellow solid which was used for the next step without purification. To a suspension of this in EtOH (20 mL) hydrazine hydrate (500 mg, 10 mmol) was added. The reaction mixture was stirred at 78 °C for 15 h, cooled down, and concentrated in vacuum. The crude product was crystallized from n-BuOH (10 mL) and vacuum dried to give 1.12 g of the title compound as a white solid (3.2 mmol, 65% over two steps). 1H NMR (400 MHz, DMSO-d6 + 0.5% TFA) d = 8.05 (s, 1H), 7.85 (d, J = 7.7 Hz, 1H), 7.52 (d, J = 8.1 Hz, 1H), 7.35–7.41 (m, 3H), 7.26 (dd, J = 176.3, 6.2 Hz, 1H, 13C-enriched), 7.01 (d, J = 8.1 Hz, 1H), 6.07 (s, 2H). 13C NMR (100.6 MHz, DMSO-d6 + 0.5% TFA) d = 147.9 (d, J = 5.8 Hz, 13C-enriched), 147.4, 146.6 (d, J = 61.0 Hz, 13C-enriched), 146.3 (d, J = 62.7 Hz, 13C-enriched), 133.8, 131.1, 130.7, 127.7, 124.4 (d, J = 62.5 Hz, 13C-enriched), 124.2, 122.3, 119.3, 108.9 (d, J = 3.9 Hz, 13C-enriched), 105.8, 101.3, 100.1 (d, J = 62.7 Hz, 13C-enriched). LC MS (RP18-100Å, gradient 50% CH3CN /50% H2O to 100% CH3CN in 30 min), RT 14.2 min and mass 345.1 (100%), 347.1 (98%), [M + H]+. TLC (SiO2, n-hexane/EtOAc = 2/1) Rf 0.39, m.p. 195–197 °C.
Preparation of α-synuclein fibrils
Samples of α-synuclein fibrils were prepared as previously reported19. In brief, vesicles were prepared by mixing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA), sodium salt and anle138b each dissolved in chloroform respectively and evaporating the solvent under a N2-stream and lyophilized overnight. SUVs were obtained by repeated sonication of a solution of 1.5 mM POPC, 1.5 mM POPA, and 150 μM anle138b. Vesicles were incubated with 70 μM α-synuclein in buffer (50 mM HEPES, 100 mM NaCl, pH 7.4) at a lipid to protein ratio of 10:1 and a compound to protein ratio of 1:2 and subjected to repeated cycles of 30-s sonication (20 kHz) at 37 °C followed by an incubation period of 30 min. After 96 h, fibrils were harvested by centrifuging at 152,460 × g (TLA-100.3 rotor in an Optima™ MAX-TL) for 1 h at 4 °C. The supernatant was removed, and the resulting pellet was washed with fresh buffer and centrifuged again 212,940 × g for 10 min at 18 °C. Excess moisture was removed, and the pellet was immediately packed into rotors for ssNMR.
ssNMR spectroscopy
For ssNMR studies, fibrils were grown from (U)-13C15N-α-synuclein in the presence of vesicles of POPA and POPC (1:1) containing 3,4-13C-anle138b at a L/P ratio of 5:1. For room temperature experiments, both samples with and without the compound were prepared at the same time and a single sample each was used for all 3D (H)CANH and 2D 13C13C-DARR experiments. Fibrils were packed into ssNMR rotors by cutting off the bottom of the tube and centrifuging the pellet directly into the rotor of choice through a custom-made filling device made from a truncated pipette tip. Last, the sample was centrifuged into the rotor in an ultracentrifuge packing device for 30 min at 98,381×g in a SW 32 Ti rotor in an Optima™ L-80 XP Ultracentrifuge (both Beckman Coulter)40.
3D (H)CANH experiments41 on fibrils in the presence of anle138b were recorded on an 800 MHz Bruker Avance III HD spectrometer at a magnetic field of 18.8 T equipped with a 1.3-mm magic-angle spinning (MAS) HCN probe and MAS at 55 kHz. The temperature of the cooling gas was set to 250 K, resulting in an estimated sample temperature of 20 °C.
3D (H)CANH spectra on fibrils in the absence of anle138b were recorded on a 950 MHz Bruker Avance III HD spectrometer at a magnetic field of 22.3 T equipped with a 0.7 mm HCDN probe and MAS at 100 kHz. The temperature of the cooling gas was set to 250 K, resulting in an estimated sample temperature of 20 °C.
2D 13C,13C-DARR spectra were acquired on an 850 MHz Avance III spectrometer with a 3.2-mm MAS HCN probe with mixing times of 200 ms at a magnetic field of 20.0 T and MAS at 17 kHz. The temperature of the cooling gas was set to 265 K, resulting in an estimated sample temperature of 20 °C.
3D (H)CANH spectra were acquired in three blocks of 32 h in the presence and four blocks of 19 h in the absence of anle138b. All 2D 13C,13C DARR spectra were acquired in 34 blocks of 7.2 h in the presence and 25 equivalent blocks in the presence of anle138b. All related blocks were corrected for linear drift of the static magnetic field using an in-house program executed from the command line in Bruker Topspin 4.0.842. The drift-corrected blocks were then averaged and processed as one spectrum. Spectra were analyzed using CcpNmr Analysis and NMRFAM-Sparky43. Peak intensities were determined by integrating cross-peaks using the Lorentzian fit algorithm in Sparky, not allowing peak center motion. Assignments on α-synuclein fibrils in the absence of anle138b had previously been reported19. Chemical shift perturbations were calculated as the average of HN, NH, Cα, and Cβ chemical shifts.
DNP-enhanced ssNMR
For DNP-enhanced ssNMR, fibrils were grown as for ssNMR measurements, but with isotopic labeling (see the section “Preparation of isotope-labeled protein”) as indicated in the text.
TEMTriPol-144 was synthesized based on published protocols45. A solution of 13C-depleted d8-Glycerol (70%-vol in water), containing 4–5 mM TEMTriPol was added to the resulting aggregate-lipid mixture and mixed thoroughly. Samples were packed into 3.2 mm zirconia ssNMR rotors via a custom-made filling device made from a truncated pipette tip. Finally, the sample was centrifuged into the rotor in an ultracentrifuge packing device for 30 min at 98,381 × g in a SW 32 Ti rotor in an Optima™ L-80 XP Ultracentrifuge.
All DNP spectra were recorded on a 600 MHz Bruker Avance III HD spectrometer at a magnetic field of 14.1 T equipped with a 3.2 mm low temperature (LT) HCN MAS probe. MAS frequencies are indicated in Supplementary Table 1. For DNP radical-proton transfers, 395 GHz microwave irradiation from a gyrotron oscillator was delivered to the sample through a corrugated waveguide. Samples were cooled with a second-generation BRUKER liquid nitrogen cold cabinet, operating at approx. 100 K.
The DNP ssNMR experiments were performed with a recycle delay of 2–4 s (1.2 × T1) for each sample and radio-frequency power levels of 100, 72, and 42 kHz for 1H, 13C, and 15N, respectively. During acquisition, during REDOR, as well as during radiofrequency driven recoupling (RFDR), SPINAL6446 decoupling was used with 100 kHz power on 1H.
2D 13C,13C spectrum, 1024 increments were acquired using 1.3 ms RFDR mixing using an xy-8 phase cycling and 83 kHz pi-pulses. 1H,13C cross-polarization (CP) was applied for 1.5 ms with a 90–100% ramp on the proton channel.
For NHHC spectra, 32 increments were acquired with 200 μs of proton–proton mixing (Supplementary Table 1). The 1H,13C CP was set to 300 μs to limit transfers to the direct proton environment, and 1H,15N CP was applied for 800 μs with an 80–100% ramp on the proton channel.
Frequency selective 13C,15N REDOR experiments47 were performed with 180° equivalent Q3 shape pulses on both 13C and 15N concurrently (Supplementary Table 2). The total time of the experiment was 4.67 h with a recycle delay of 2 s (4096 Scans).
Solution NMR spectroscopy
For determination of the conformation of anle138b we prepared a 20 mM solution of vesicles consisting of POPC and POPA (1:1) containing 1 mM anle138b in buffer (50 mM HEPES, 100 mM NaCl, pH 7.4) with 10% D2O and 100 μM 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (IUPAC: 3-(trimethylsilyl)propane-1-sulfonate, sodium salt). NOESY spectra were acquired with 72 scans using 256 increments in the indirect dimension and a relaxation delay of 2.3 s. The mixing time was set to 3 ms. Proton 90° flip pulses were 14.4 μs. Water suppression was achieved by presaturation at a power level of 100 Hz. Experiments were recorded on a Bruker 900-MHz spectrometer. The temperature during measurements was kept at 298 K. 2D datasets were processed in NMRPipe48 and analyzed in NMRFAM-Sparky43. Spectral traces were fitted to a mixed Gaussian and Lorentzian function using the line shapes tool in Bruker Topspin 4.0.8.
Molecular dynamics (MD) simulations
The GROMACS 2018.3 simulation software package49,50 was used to set up and carry out the MD simulations. For all simulations, the AMBER99SB*-ILDN51–53 force field and the ion parameters in ref. 54 for use with the TIP3P water model55 were employed. A modified version56 of the all-atom Slipids force field57–59 was used for the description of POPC lipids. The parameterization scheme for the small molecule compound anle138b is detailed elsewhere23. In order to speed up the simulations, aliphatic hydrogen atoms were converted to virtual sites60 and bonds between all atoms in protein, lipid, and ligand molecules were constrained using the P-LINCS61 algorithm. Water molecules were constrained using SETTLE62. The integration timestep was set to 4 fs. Neighbor lists were updated with the Verlet list scheme50,63. The long-range electrostatic interactions were treated using the Particle Mesh Ewald (PME) method64,65, with a 1.0 nm real-space cutoff. The van der Waals interactions were cut off at 1.0 nm. A dispersion correction for energy and pressure was applied. For production runs, the temperature was kept at 300 K by applying the velocity-rescaling66 algorithm. The pressure was held constant at 1 bar by using the Parrinello–Rahman barostat67. All simulations were carried out using periodic boundary conditions for the simulation box.
Setup of α-synuclein protofilament structure models
Simulations of α-synuclein β-sheet constructs were carried out based on structure coordinates from the α-synuclein fibril polymorph L2A (PDB ID 8A4L; EMDB ID EMD-15148)20 constituted by the residues 33-96, and the resolved parts of the N-terminus (residues 14-24). For simplicity, we refer to the protofilament structure of polymorph L2A as L2, since it is structurally identical to the co-occurring polymorph L2B. The simulation model (see Supplementary Table 2) consisted of one protofilament structure with ten β-strand layers stacked on top of each other, respectively. A rhombic dodecahedron box was used for all simulations with the distance between the solute and the box edge of at least 2.5 nm. The resulting box size dimensions converged to ~12.8 × 12.8 × 12.8 nm3 after initial equilibration prior to starting the production runs. For each simulation system, the titratable groups of the protein structure were protonated according to their standard protonation states at pH 7, and the N- and C-termini of the peptides were capped with acetyl and N-methyl groups, respectively. Counterions (Na+, Cl−) were added to yield an ionic strength of 150 mM and to neutralize the net system charge. In all simulations, 100 POPC molecules were present and initially placed randomly in the solvent.
In total, the simulation systems contained ~155,000 atoms, including ~41,000 water molecules and 316 ions.
The MD production runs
All production runs were preceded by a multi-step energy minimization and equilibration of the simulation systems. Initial velocities were taken according to the Maxwell-Boltzmann distribution at 300 K. Simulations were carried out in the presence and absence of anle138b. Two principal conditions were chosen for simulations with α-synuclein protofilament models and anle138b:
-
A.
One anle138b molecule was modeled inside the internal cavity such that its long axis aligned with the protofilament axis, and the solvent-facing atoms of the compound aligned with the surface of the protofilament tip. The compound was inserted without interatomic overlaps to the side-chain atoms of the cavity enclosing residues, in a total of four different poses: with the pyrazole nitrogens oriented toward residues 67GGAV70 (inward-facing) or residues 81TVEGAGSI88 (outward-facing), and with an up or down positioning of the bromophenyl moiety of anle138b with respect to direction of the closest edge of the protofilament. Each initial pose was energy minimized and subsequently equilibrated for 1 ns with position restraints acting on the protein-heavy atoms.
-
B.
Ten anle138b molecules were placed randomly in the solvent and at least 1 nm away from any protein-heavy atom.
The total simulation time for this study adds up to 100 μs, resulting in 50 μs sampling of the internally bound state of anle138b. From the individual simulation trajectories, snapshots were collected every 100 ps. Only the last 75% of the trajectory data were used for subsequent analyses as a measure to discard the initial equilibration phase and structural relaxation of the systems.
Analysis and visualization
The MD simulation trajectories were preprocessed and analyzed by tools provided by the GROMACS 2018.3 simulation software package49,50, as well as custom-made bash, awk, and python scripts. Renderings of atomic coordinates were carried out with the PyMOL molecular visualization system68. Graphs and figures were created using gnuplot 5.2 and inkscape 1.1.
Relative free-energy binding profile
For each simulation frame, the center of mass distance between the pyrazole ring of the anle138b molecule and the protofilament tip (edge β-strand layer) was determined (see e.g., Fig. 3e). The distances from each simulation trajectory set were combined and sorted into 1-d bins. The logarithm of the probability p = xi,j/xtotal was calculated for each bin i, j, where xi,j is the number of frames in the bin and xtotal is the total number of frames. Multiplying a constant factor (-RT, with T = 300 K) to p(x) results in G(x) in units of kJ/mol.
Contact analysis
The frequency of short-range interatomic interactions between the anle138b small molecules, POPC lipids, and the peptide aggregates were quantified with the g_contacts program69. An anle138b or POPC contact was considered as formed, if any heavy atom of the respective molecules was within a cutoff of 0.4 nm from any heavy atom of a protein residue. In addition, and according to the isotope labeling scheme in the NMR experiments, if the 1,2-N atoms of the anle138b molecule were found within a cutoff of 0.5 nm from any backbone carbon or nitrogen atom of a protein residue, a contact was considered formed. Contacts were averaged over time and over the individual trajectories of each simulation set. If not explicitly stated, only contacts to the core β-strand layers (neglecting the top β-strands on both protofilament edges) were considered for analysis. Hydrogen bond (HB) energies were calculated according to the empirical formula provided in ref. 70. For the definition of halogen bonds (XB) the following criteria were adopted from ref. 71: (a) distance between halogen atom and carbonyl oxygens ≤ 0.36 nm; (b) angle between XB and halogen–carbon bond ≥ 150°.
Anle138b-binding mode classification
Based on the previously described heuristic classification of anle138b interaction patterns to oligomeric forms of peptide aggregates in terms of atomic contact numbers and the presence of hydrogen bonds23, the following binding modes to fibrillar aggregates were defined: (I) “unbound”, no atomic contacts; (II) “partially bound”, fewer than half of the anle138b atoms in contact with peptide heavy atoms, mostly describing transient binding events; (III) “no HB or XB”, half or more of the anle138b atoms in contact with peptide heavy atoms, no HB or XB interactions to the backbone or side-chain atoms; (IV) “polar interactions”, at least one HB or XB between anle138b and the protofilament structure; (IVa) “pyrazole HB”, at least one HB between anle138b (via the pyrazole ring) and the protofilament structure; (IVb) “benzodioxole HB”, at least one HB between anle138b (via the benzodioxole ring) and the protofilament structure; (IVc) “bromophenyl XB”, at least one XB between anle138b (via the bromophenyl ring) and the protofilament structure.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary files
Acknowledgements
We thank M. Wegstroth and K. Giller for excellent technical help in protein expression and purification. We also thank G. Heim for the acquisition of negative stain EM images. We thank J. Schimpfhauser, J. Bienert, and V.N. Belov from the facility for Synthetic Chemistry at the Max Planck Institute for Multidisciplinary Sciences, Göttingen for synthesizing the TEMTriPol-1 radical. This work was supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy-EXC 2067/1-390729940 as well as the Emmy Noether program (grant AN1316/1-1 to L.B.A.).
Source data
Author contributions
L.A., D.M., B.L.dG, C.G., and L.B.A. conceptualized the project. S.B. oversaw protein expression and purification. A.L. synthesized isotope-labeled anle138b. L.A. and V.S. prepared fibril samples. L.A., R.D., E.N., and L.B.A. performed NMR experiments. B.F., C.D., and G.F.S. provided αSyn fibril atomic coordinates and discussion thereof. D.M. carried out MD simulations. L.A. and D.M. prepared figures and wrote the initial draft. S.R. and A.G. contributed to the interpretation of data in the medical context. B.L.dG., C.G., and L.B.A. supervised the project. All authors contributed to the writing of the manuscript.
Peer review
Peer review information
Nature Communications thanks Ashok Dasmahapatra, Alexey Potapov and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
NMR spectra raw data generated in this study, Negative stain EM micrographs for αSyn fibrils in the presence of anle138b, and mass spectrometry data for the synthesis of selectively labeled α-synuclein and 13C- and 15N-labeled anle138b, respectively, have been deposited in the open research data repository Edmond at [10.17617/3.9C6TEW]. The molecular dynamics data generated in this study have been deposited in the Edmond data repository at [10.17617/3.R4FJQG]. Assigned chemical shift data (HN, Cα, Cβ, and C′) for α-synuclein fibrils were deposited in the BMRB as updates under the accession number 50585. Atomic coordinates for L2A-fibrils of αSyn used for MD simulations and visualization in this manuscript are available in the Protein Data Bank under the accession code 8A4L. Previously published structures of αSyn fibrils used for a comparison of tubular fibril cavities are available in the Protein Data Bank under the accession codes 6SSX, 7NCI, and 7NCH. All other data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper.
Competing interests
A.G., A.L., and S.R. are employed by MODAG GmbH, which licensed WO/2010/000372 in which anle138b is described. A.G. and C.G. are shareholders of MODAG GmbH. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Leif Antonschmidt, Dirk Matthes, Rıza Dervişoğlu.
Contributor Information
Bert L. de Groot, Email: bgroot@gwdg.de
Christian Griesinger, Email: cigr@mpinat.mpg.de.
Loren B. Andreas, Email: land@mpinat.mpg.de
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-32797-w.
References
- 1.Spillantini MG, et al. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Graham JG, Oppenheimer DR. Orthostatic hypotension and nicotine sensitivity in a case of multiple system atrophy. J. Neurol. Neurosurg. Psychiatry. 1969;32:28. doi: 10.1136/jnnp.32.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanciulli A, Wenning GK. Multiple-system atrophy. N. Engl. J. Med. 2015;372:249–263. doi: 10.1056/NEJMra1311488. [DOI] [PubMed] [Google Scholar]
- 4.Gibb WRG, Esiri MM, Lees AJ. Clinical and pathological features of diffuse cortical Lewy body disease (Lewy body dementia) Brain. 1987;110:1131–1153. doi: 10.1093/brain/110.5.1131. [DOI] [PubMed] [Google Scholar]
- 5.Gómez-Tortosa E, Newell K, Irizarry MC, Sanders JL, Hyman BT. alpha-Synuclein immunoreactivity in dementia with Lewy bodies: morphological staging and comparison with ubiquitin immunostaining. Acta Neuropathol. 2000;99:352–357. doi: 10.1007/s004010051135. [DOI] [PubMed] [Google Scholar]
- 6.Spillantini MG, Goedert M. Synucleinopathies: past, present and future. Neuropathol. Appl Neurobiol. 2016;42:3–5. doi: 10.1111/nan.12311. [DOI] [PubMed] [Google Scholar]
- 7.Cole TA, et al. α-Synuclein antisense oligonucleotides as a disease-modifying therapy for Parkinson’s disease. JCI Insight. 2021;6:e135633. doi: 10.1172/jci.insight.135633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner J, et al. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013;125:795–813. doi: 10.1007/s00401-013-1114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuebler L, et al. [11C]MODAG-001—towards a PET tracer targeting α-synuclein aggregates. Eur. J. Nucl. Med Mol. Imag. 2021;48:1759–1772. doi: 10.1007/s00259-020-05133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin J, et al. The oligomer modulator anle138b inhibits disease progression in a Parkinson mouse model even with treatment started after disease onset. Acta Neuropathol. 2014;127:779–780. doi: 10.1007/s00401-014-1265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner J, et al. Reducing tau aggregates with anle138b delays disease progression in a mouse model of tauopathies. Acta Neuropathol. 2015;130:619–631. doi: 10.1007/s00401-015-1483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez Hernandez A, et al. The diphenylpyrazole compound anle138b blocks Aβ channels and rescues disease phenotypes in a mouse model for amyloid pathology. EMBO Mol. Med. 2018;10:32–47. doi: 10.15252/emmm.201707825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heras-Garvin A, et al. Anle138b modulates α-synuclein oligomerization and prevents motor decline and neurodegeneration in a mouse model of multiple system atrophy. Mov. Disord. 2019;34:255–263. doi: 10.1002/mds.27562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegrzynowicz M, et al. Depopulation of dense α-synuclein aggregates is associated with rescue of dopamine neuron dysfunction and death in a new Parkinson’s disease model. Acta Neuropathol. 2019;138:575–595. doi: 10.1007/s00401-019-02023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin J, et al. Safety, tolerability and pharmacokinetics of the oligomer modulator anle138b with exposure levels sufficient for therapeutic efficacy in a murine Parkinson model: a randomised, double-blind, placebo-controlled phase 1a trial. eBioMedicine. 2022;80:104021. doi: 10.1016/j.ebiom.2022.104021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deeg AA, et al. Anle138b and related compounds are aggregation specific fluorescence markers and reveal high affinity binding to α-synuclein aggregates. Biochim Biophys. Acta. 2015;1850:1884–1890. doi: 10.1016/j.bbagen.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Sulzer D, Edwards RH. The physiological role of alpha-synuclein and its relationship to Parkinson’s disease. J. Neurochem. 2019;150:475–486. doi: 10.1111/jnc.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahmoradian SH, et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 2019;22:1099–1109. doi: 10.1038/s41593-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 19.Antonschmidt L, et al. Insights into the molecular mechanism of amyloid filament formation: Segmental folding of α-synuclein on lipid membranes. Sci. Adv. 2021;7:eabg2174. doi: 10.1126/sciadv.abg2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frieg, B. et al. The 3D structure of lipidic fibrils of α-synuclein. Preprint at https://www.biorxiv.org/content/10.1101/2022.03.02.481946v1 (2022). [DOI] [PMC free article] [PubMed]
- 21.Lange A, Luca S, Baldus M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J. Am. Chem. Soc. 2002;124:9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 22.Loquet A, Gardiennet C, Böckmann A. Protein 3D structure determination by high-resolution solid-state NMR. C. R. Chim. 2010;13:423–430. doi: 10.1016/j.crci.2010.03.007. [DOI] [Google Scholar]
- 23.Matthes D, Gapsys V, Griesinger C, de Groot BL. Resolving the atomistic modes of Anle138b inhibitory action on peptide oligomer formation. ACS ChemNeurosci. 2017;8:2791–2808. doi: 10.1021/acschemneuro.7b00325. [DOI] [PubMed] [Google Scholar]
- 24.Guerrero-Ferreira R, et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife. 2019;8:e48907. doi: 10.7554/eLife.48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lövestam S, et al. Seeded assembly in vitro does not replicate the structures of α-synuclein filaments from multiple system atrophy. FEBS Open Bio. 2021;11:999–1013. doi: 10.1002/2211-5463.13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Jo H, DeGrado WF, Hong M. Water distribution, dynamics, and interactions with Alzheimer’s β-amyloid fibrils investigated by solid-state NMR. J. Am. Chem. Soc. 2017;139:6242–6252. doi: 10.1021/jacs.7b02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyer DR, et al. Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 2019;26:1044–1052. doi: 10.1038/s41594-019-0322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schütz AK, et al. The amyloid–Congo Red interface at atomic resolution. Angew. Chem. Int Ed. 2011;50:5956–5960. doi: 10.1002/anie.201008276. [DOI] [PubMed] [Google Scholar]
- 29.Niu Z, et al. Mapping the binding interface of PET tracer molecules and Alzheimer disease Aβ fibrils by using MAS solid-state NMR spectroscopy. ChemBioChem. 2020;21:2495–2502. doi: 10.1002/cbic.202000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease with PET ligand APN−1607. Acta Neuropathol. 2021;141:697–708. doi: 10.1007/s00401-021-02294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan P, et al. Binding sites of a positron emission tomography imaging agent in Alzheimer’s β-amyloid fibrils studied using 19F solid-state NMR. J. Am. Chem. Soc. 2022;144:1416–1430. doi: 10.1021/jacs.1c12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuang G, Murugan NA, Ågren H. Mechanistic insight into the binding profile of DCVJ and α-synuclein fibril revealed by multiscale simulations. ACS ChemNeurosci. 2019;10:610–617. doi: 10.1021/acschemneuro.8b00465. [DOI] [PubMed] [Google Scholar]
- 33.Pujols J, et al. Small molecule inhibits α-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. Proc. Natl Acad. Sci. USA. 2018;115:10481–10486. doi: 10.1073/pnas.1804198115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prade E, et al. Structural mechanism of the interaction of Alzheimer disease Aβ fibrils with the non-steroidal anti-inflammatory drug (NSAID) sulindac sulfide. J. Biol. Chem. 2015;290:28737–28745. doi: 10.1074/jbc.M115.675215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frieg, B. et al. Quaternary structure of patient-homogenate amplified α-synuclein fibrils modulates seeding of endogenous α-synuclein. Commun. Biol. (2022). In press. [DOI] [PMC free article] [PubMed]
- 36.Fan, Y. et al. Different structures and pathologies of α-synuclein fibrils derived from preclinical and postmortem patients of Parkinson’s disease. Preprint at https://www.biorxiv.org/content/10.1101/2021.11.02.467019v1 (2021).
- 37.Hoyer W, et al. Dependence of α-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 2002;322:383–393. doi: 10.1016/S0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 38.Lin B, et al. 17O NMR studies of yeast ubiquitin in aqueous solution and in the solid state. ChemBioChem. 2021;22:826–829. doi: 10.1002/cbic.202000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khodaei MM, Alizadeh A, Nazari E. Tf2O as a rapid and efficient promoter for the dehydrative Friedel–Crafts acylation of aromatic compounds with carboxylic acids. Tetrahedron Lett. 2007;48:4199–4202. doi: 10.1016/j.tetlet.2007.04.066. [DOI] [Google Scholar]
- 40.Böckmann A, et al. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 2009;45:319. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 41.Barbet-Massin E, et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Najbauer EE, Andreas LB. Correcting for magnetic field drift in magic-angle spinning NMR datasets. J. Magn. Reson. 2019;305:1–4. doi: 10.1016/j.jmr.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2014;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathies G, et al. Efficient dynamic nuclear polarization at 800 MHz/527 GHz with trityl-nitroxide biradicals. Angew. Chem. 2015;54:11770–11774. doi: 10.1002/anie.201504292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Villamena FA, Rockenbauer A, Song Y, Zweier JL. Structural factors controlling the spin–spin exchange coupling: EPR spectroscopic studies of highly asymmetric trityl–nitroxide biradicals. J. Am. Chem. Soc. 2013;135:2350–2356. doi: 10.1021/ja311571v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 47.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate 13C−15N distance measurements in uniformly 13C,15N-labeled peptides. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 51.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinforma. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix−coil transition of polypeptides. J. Phys. Chem. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Bioinforma. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith DE, Dang LX. Computer simulations of NaCl association in polarizable water. J. Chem. Phys. 1994;100:3757–3766. doi: 10.1063/1.466363. [DOI] [Google Scholar]
- 55.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 56.Melcr J, Bonhenry D, Timr Š, Jungwirth P. Transmembrane potential modeling: comparison between methods of constant electric field and ion imbalance. J. Chem. Theory Comput. 2016;12:2418–2425. doi: 10.1021/acs.jctc.5b01202. [DOI] [PubMed] [Google Scholar]
- 57.Jämbeck JPM, Lyubartsev AP. An extension and further validation of an all-atomistic force field for biological membranes. J. Chem. Theory Comput. 2012;8:2938–2948. doi: 10.1021/ct300342n. [DOI] [PubMed] [Google Scholar]
- 58.Jämbeck JPM, Lyubartsev AP. Derivation and systematic validation of a refined all-atom force field for phosphatidylcholine lipids. J. Phys. Chem. B. 2012;116:3164–3179. doi: 10.1021/jp212503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jämbeck JPM, Lyubartsev AP. Another piece of the membrane puzzle: extending slipids further. J. Chem. Theory Comput. 2013;9:774–784. doi: 10.1021/ct300777p. [DOI] [PubMed] [Google Scholar]
- 60.Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 61.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 62.Miyamoto S, Kollman PA. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 63.Verlet L. Computer “Experiments” on classical fluids. I. Thermodynamical properties of Lennard-Jones molecules. Phys. Rev. 1967;159:98–103. doi: 10.1103/PhysRev.159.98. [DOI] [Google Scholar]
- 64.Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 65.Essmann U, et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 66.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 67.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 68.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 2.0. (2015).
- 69.Blau C, Grubmuller H. g_contacts: fast contact search in bio-molecular ensemble data. Comput. Phys. Commun. 2013;184:2856–2859. doi: 10.1016/j.cpc.2013.07.018. [DOI] [Google Scholar]
- 70.Espinosa E, Molins E, Lecomte C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998;285:170–173. doi: 10.1016/S0009-2614(98)00036-0. [DOI] [Google Scholar]
- 71.Kolář MH, Hobza P. Computer modeling of halogen bonds and other σ-hole interactions. Chem. Rev. 2016;116:5155–5187. doi: 10.1021/acs.chemrev.5b00560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
NMR spectra raw data generated in this study, Negative stain EM micrographs for αSyn fibrils in the presence of anle138b, and mass spectrometry data for the synthesis of selectively labeled α-synuclein and 13C- and 15N-labeled anle138b, respectively, have been deposited in the open research data repository Edmond at [10.17617/3.9C6TEW]. The molecular dynamics data generated in this study have been deposited in the Edmond data repository at [10.17617/3.R4FJQG]. Assigned chemical shift data (HN, Cα, Cβ, and C′) for α-synuclein fibrils were deposited in the BMRB as updates under the accession number 50585. Atomic coordinates for L2A-fibrils of αSyn used for MD simulations and visualization in this manuscript are available in the Protein Data Bank under the accession code 8A4L. Previously published structures of αSyn fibrils used for a comparison of tubular fibril cavities are available in the Protein Data Bank under the accession codes 6SSX, 7NCI, and 7NCH. All other data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper.