SUMMARY
Healthy skin maintains a diverse microbiome and a potent immune system to fight off infections. Here, we discovered that the epithelial cell-derived antimicrobial peptides defensins activated orphan G protein-coupled receptors (GPCRs) Mrgpra2a/b on neutrophils. This signaling axis was required for effective neutrophil-mediated skin immunity and microbiome homeostasis. We generated mutant mouse lines lacking the entire Defensin (Def) gene cluster in keratinocytes or Mrgpra2a/b. Def and Mrgpra2 mutant animals both exhibited skin dysbiosis, with reduced microbial diversity and expansion of Staphylococcus species. Defensins and Mrgpra2 were critical for combatting S. aureus infections and the formation of neutrophil abscesses, a hallmark of antibacterial immunity. Activation of Mrgpra2 by defensin triggered neutrophil release of IL-1β and Cxcl2 which are vital for proper amplification and propagation of the antibacterial immune response. This study demonstrates the importance of epithelial-neutrophil signaling via the defensin-Mrgpra2 axis in maintaining healthy skin ecology and promoting antibacterial host defense.
Keywords: Antimicrobial peptide, neutrophil, innate immunity, skin microbiome, skin dysbiosis, GPCR, Mrgpr, IL-1β, Cxcl2
Graphical Abstract
In brief
Antimicrobial peptides (AMPs) are vital for host defense. Dong et al. discover that keratinocyte-derived defensins activate G protein-coupled receptors Mrgpra2a/b on neutrophils. This epithelial-neutrophil signaling axis prevents skin dysbiosis and is required for immunity against bacterial infections.
INTRODUCTION
The skin is the largest physical and immune barrier to the external environment. Under steady-state conditions, epithelial cells and skin-resident immune cells interact with commensal micro-organisms including bacteria, fungi and viruses and form a harmonious ecosystem (Chen et al., 2018; Grice and Segre, 2011). However, pathological disturbances can lead to an outgrowth of pathogenic bacteria such as Staphylococcus aureus at the cost of other diverse commensal species, a process termed dysbiosis (Byrd et al., 2018; Kobayashi et al., 2015; Nakatsuji et al., 2017; Williams and Gallo, 2017; Williams et al., 2017b). Despite the importance of microbiota in skin health, host mechanisms that prevent dysbiosis and disease remain poorly understood.
The host skin possesses an intricate immune system poised to respond to any barrier breaches and infections (Kabashima et al., 2019; Krishna and Miller, 2012; Miller and Cho, 2011; Ridder et al., 2022). Epithelial cells in the infected area are the first cells to encounter pathogens. Pathogen- and damage-associated molecular patterns (PAMPs and DAMPs) activate pattern recognition receptors (PRRs) on keratinocytes and trigger the release of pro-inflammatory signals (Nestle et al., 2009; Pasparakis et al., 2014; Yang et al., 2017). Tissue-resident immune cells rapidly amplify the danger signal and trigger a cytokine cascade that leads to rapid recruitment of a large number of immune cells from circulation (Kim and Luster, 2015; Ley et al., 2007). Neutrophils account for the vast majority of infiltrating immune cells and are indispensable for host defense (Cho et al., 2012; Miller and Cho, 2011; Sadik et al., 2011). In response to bacterial invasion, neutrophils form an abscess around the site of infection, which effectively restricts bacterial spread and facilitates bacterial killing (Borregaard et al., 2007; Cho et al., 2012; Corbin et al., 2008; Kobayashi et al., 2018; Miller and Cho, 2011). This highly efficient defense response requires coordinated signals between the epithelial and immune cells.
Here, we report a ligand-receptor signaling axis between the skin epithelium and neutrophils that is vital for maintaining microbiome homeostasis and antibacterial immunity. The ligands are a group of epithelial cell-derived antimicrobial peptides (AMPs) named defensins. AMPs are small peptides with antibacterial, antiviral and antifungal properties (Chessa et al., 2020; Ganz, 2003; Gerdol et al., 2020; Nakatsuji and Gallo, 2012). Defensins are the largest family of AMPs in mammalian genomes and are released by epithelial cells upon infection (Ali et al., 2001; Hinrichsen et al., 2008; Lee et al., 2005; Midorikawa et al., 2003; Miller and Cho, 2011; Pasparakis et al., 2014; Sumikawa et al., 2006). It has been shown that in addition to directly killing microbes, defensins can perform immune-modulatory functions and rapidly mobilize immune cells (Rohrl et al., 2010; Yang et al., 1999; Yang et al., 2017). However, due to the massive size of the gene family and highly redundant functions between members, the specific cellular and molecular mechanisms employed by defensins to orchestrate antibacterial immunity could not be evaluated in a genetic loss-of-function study (Morrison et al., 2002; Zhou et al., 2013). We overcame this problem by conditionally deleting the entire Defensin (Def) gene cluster from keratinocytes. Additionally, we identified two orphan G protein-coupled receptors (GPCRs), Mrgpra2a and Mrgpra2b, as defensin receptors specifically expressed in neutrophils. Ablation of defensins and their neutrophil receptors both resulted in decreased skin commensal diversity and severe immune defects against S. aureus infections. Further, we found that defensin-Mrgpra2 signaling mediated the release of key pro-inflammatory cytokine IL-1β and neutrophil autocrine chemokine Cxcl2. These signals are important for neutrophil recruitment, abscess formation, and antibacterial immunity. Collectively, these findings highlighted how the defensin-Mrgpra2 axis influenced skin ecology and critically promoted antibacterial host defense.
RESULTS
Mrgpra2a and Mrgpra2b were defensin receptors on neutrophils
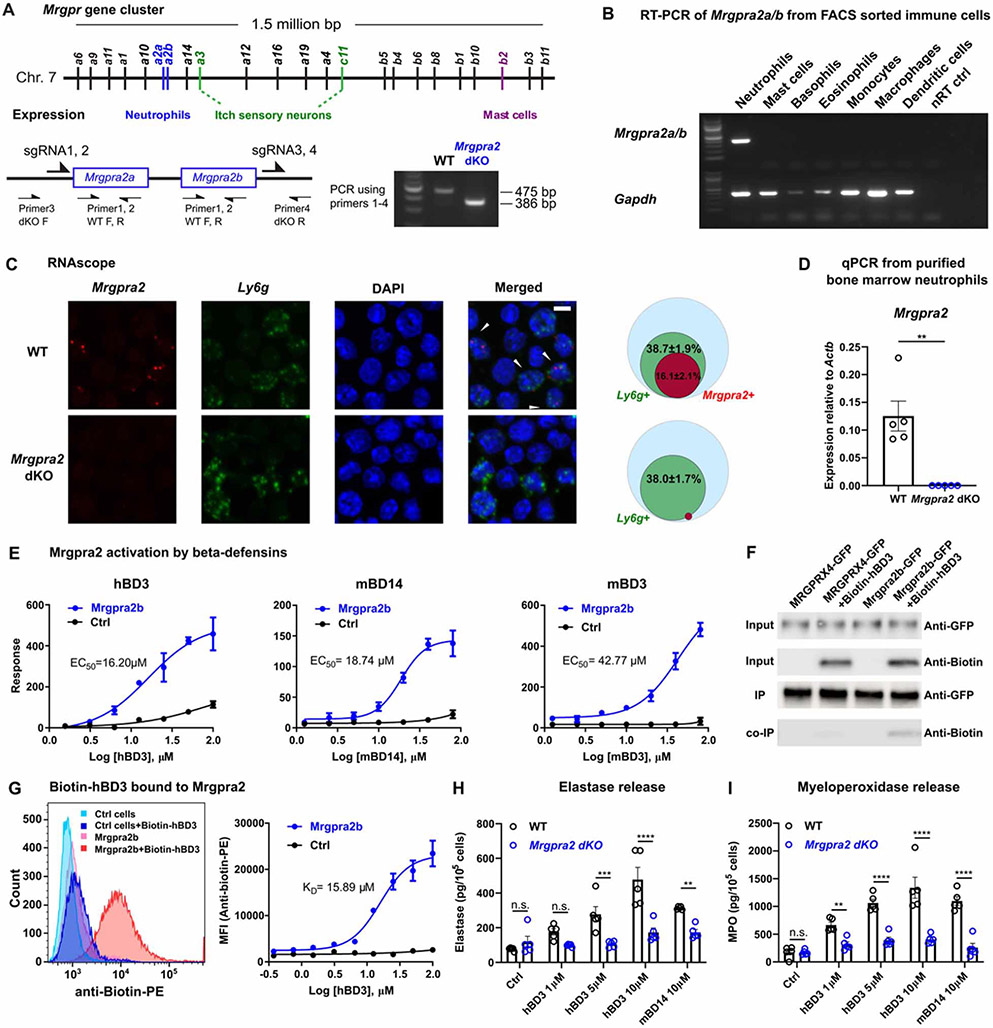
Previously, we and others have identified members of the Mas-related G protein-coupled receptors (Mrgprs) family (Figure 1A) as critical regulators of innate immunity (Arifuzzaman et al., 2019; Pundir et al., 2019; Subramanian et al., 2013a). Expression profiling by the Immunological Genome Project Consortium revealed that mouse neutrophils express a pair of previously uncharacterized Mrgprs, Mrgpra2a and Mrgpra2b (Dwyer et al., 2016; Ericson et al., 2014). The two genes, which reside next to each other within the Mrgpr gene cluster, encode proteins that differ only by two amino acids and likely arose by duplication (Figure 1A, Figure S1A) (Sievers et al., 2011). We will refer to them together as Mrgpra2. Reverse transcription-PCR (RT-PCR) of RNA purified from FACS-sorted innate immune cells confirmed that the Mrgpra2 genes are specifically expressed by neutrophils but not by other granulocytes (mast cells, basophils, eosinophils), monocytes, macrophages, or dendritic cells (Figure 1B). We generated Mrgpra2a/b double knockout (Mrgpra2a−/−Mrgpra2b−/−, referred to hereafter as Mrgpra2 dKO) mice using CRISPR/Cas9 and confirmed deletion of Mrgpra2 transcripts from neutrophils (Figure 1A, C, D). In situ hybridization with RNAscope revealed that roughly 40% of Ly6g+ neutrophils in wild-type (WT) bone marrow were Mrgpra2+ (16% of total bone marrow cells, Figure 1C) and that the Mrgpra2 transcripts were undetectable in Mrgpra2 dKO bone marrow cells (Figure 1D). Expression of other genes in bone marrow neutrophils was not affected in the Mrgpra2 dKO mice and neither were the percentages of neutrophils in the bone marrow, blood and spleen in baseline conditions (Figure S1B, C).
Figure 1. Mrgpra2a/b were neutrophil-specific receptors for defensi.
(A) Schematic illustration of the Mrgpr gene cluster on mouse Chromosome 7 and cellular expression specificities of a few well-studied members of the gene family. Mrgpra2a and a2b were deleted by CRISPR/cas9 using four gRNAs flanking the two genes, removing the entire genomic region. PCR genotyping using primers 1-4 as indicated in the diagram detects a WT band of 475 base pairs and an Mrgpra2 dKO mutant band of 386 base pairs.
(B) RT-PCR gel showing expression of Mrgpra2 and housekeeping gene Gapdh in FACS-sorted neutrophils, mast cells, basophils, eosinophils, monocytes, macrophages, dendritic cells, and the no reverse transcriptase control (nRT Ctrl) of the neutrophil sample.
(C) Representative RNAscope in situ hybridization images of bone marrow cells labeled using probes for Mrgpra2 (red), Ly6g (green), and DAPI (blue). Arrowheads point to Mrgpra2+; Ly6g+ neutrophils. Scale bar=5 μm. Venn diagrams depict percentages of Ly6g+ (green) and Mrgpra2+ (red) cells in all bone marrow cells (blue). n=3
(D) qPCR of mRNA from purified bone marrow neutrophils showed that Mrgpra2 were expressed highly in WT neutrophils but deleted from Mrgpra2 dKO neutrophils. n=5
(E) HEK cells expressing Mrgpra2b activation by hBD3 (EC50=16.2μM), mBD14 (EC50=18.74μM) and mBD3 (EC50=42.77μM) as determined by FLIPR intracellular calcium mobilization assay. n=4-9
(F) Western blot showing biotin-hBD3 being co-immunoprecipitated (co-IP) by Mrgpra2b-GFP. MRGPRX4-GFP (negative control) and Mrgpra2b-GFP were pulled down using anti-GFP beads, biotin-hBD3 was co-precipitated only with Mrgpra2b-GFP but not MRGPRX4-GFP.
(G) Dissociation constant (KD=15.89μM) between biotin-hBD3 and Mrgpra2b determined by flow cytometry. Left, representative flow cytometry histogram showing control HEK cells (blue) and Mrgpra2b-GFP-expressing cells (red) incubated with (darker shades) and without (lighter shades) biotin-hBD3 (100μM). Right, quantification of mean fluorescent intensity (MFI) of PE-anti-biotin showing the amount of biotin-hBD3 bound to the cells at each concentration. n=6
(H-I) hBD3 and mBD14 triggered WT neutrophils to release elastase (H) and myeloperoxidase (I) (black). Neutrophils purified from Mrgpra2 dKO animals did not respond to hBD3 stimuli (blue). n=5
Results are presented as mean ± SEM from at least three independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. not significant by two-tailed unpaired Student’s t test (D) or two-way ANOVA (H-I).
See also Figure S1.
Using a FLIPR intracellular Ca2+ mobilization assay, we found that several beta-defensins activate Mrgpra2 (Figure 1E). Human beta-defensin 3 (hBD3) and its mouse homologue mouse beta-defensin 14 (mBD14, 67% identity) evoked robust receptor activation with EC50s of 16.20μM and 18.74μM, respectively (Figure 1E) (Hinrichsen et al., 2008; Röhrl et al., 2008), while mouse beta-defensin 3 (mBD3) activated Mrgpra2 with a higher EC50 (42.77μM) (Figure 1E). Human peptides hBD1, 2 and 4, and mouse peptides mBD1, 2, 4 did not trigger receptor activation (Figure S1E). Raw FLIPR Ca2+ traces are shown in Figure S1D. Of the four human MRGPRX receptors, MRGPRX3 was shown to be expressed in human neutrophils (Human Protein Atlas, Figure S1F) (Uhlen et al., 2019) and was activated by hBD3 at a similar EC50 (10.76μM) (Figure S1G) but not by other human defensins tested (Figure S1H). To examine direct interaction between hBD3 and Mrgpra2, we synthesized N-terminal biotinylated hBD3 (biotin-hBD3), which allowed us to probe for this peptide using anti-biotin antibodies. We first used co-immunoprecipitation (co-IP) to examine direct interaction between biotin-hBD3 and Mrgpra2. Mrgpra2b-GFP was pulled down using anti-GFP beads, and we found that biotin-hBD3 could be co-precipitated (Figure 1F). MRGPRX4-GFP, which was not activated by hBD3 (Figure S1G), was used as negative control. We next used a flow cytometry-based method to determine the dissociation constant (KD) since biotin-hBD3 bound to Mrgpra2b-GFP-expressing HEK cells could be detected and quantified using a PE-conjugated anti-biotin antibody (Figure 1G). We observed that hBD3 associated with Mrgpra2b with a KD of 15.89μM, consistent with the EC50 determined using the FLIPR assay (Figure 1E, G). Similarly, hBD3 bound to human MRGPRX3 with a KD of 11.06μM, but did not bind to MRGPRX4 (Figure S1I).
Next, we evaluated this ligand-receptor interaction by stimulating neutrophils purified from WT and Mrgpra2 dKO mice. Neutrophils perform a variety of functions including the formation of neutrophil extracellular traps (NETs), production of reactive oxygen species (ROS), and release of granular contents which contain large amounts of bactericidal molecules (Mayadas et al., 2014). When applied to WT neutrophils, hBD3 did not cause chemotaxis, the formation of NETs, or ROS production (Figure S1J-L). However, enzyme-linked immunosorbent assay (ELISA) analysis revealed that hBD3 and its mouse homologue mBD14 triggered the neutrophils to release elastase and myeloperoxidase (MPO), two major components of primary granules (Borregaard et al., 2007; Cassatella et al., 2019) (Figure 1H, I). Neutrophils purified from Mrgpra2 dKO mice did not respond to hBD3 or mBD14 stimuli, suggesting that defensins activated neutrophils through Mrgpra2 (Figure 1H, I). These functional assays (Figure 1E-I), together with the expression pattern of Mrgpra2 (Figure 1B-D), supported the role of Mrgpra2 as neutrophil-specific defensin receptors.
Generation of Defensin conditional knockout mice
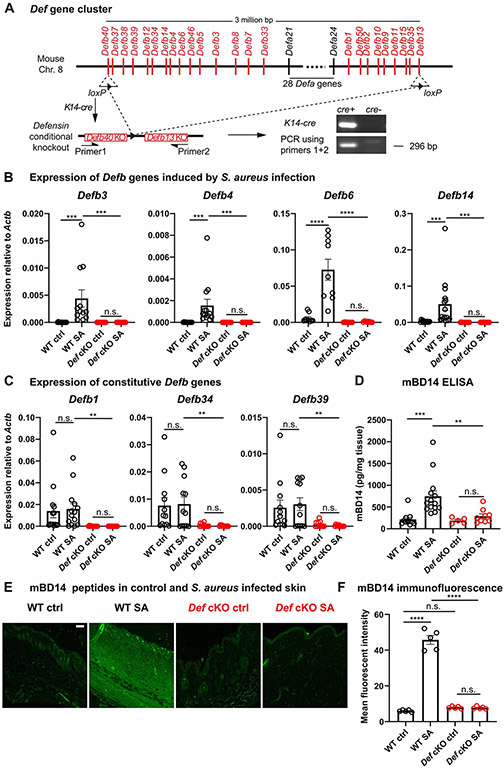
To investigate the roles of defensin-Mrgpra2 interaction in skin immunity, we needed both ligand and receptor KO animals. The defensin family of AMPs can be classified into alpha- or beta- subgroups based on distinct topologies of conserved disulfide bonds (Ganz, 2003). Mouse alpha-defensins are produced by Paneth cells in the intestines (Eisenhauer et al., 1992; Salzman et al., 2003; Wilson et al., 1999), while beta-defensins are expressed and secreted primarily by keratinocytes in the skin (Ali et al., 2001; Harder et al., 2001; Midorikawa et al., 2003). More than 50 defensin-encoding genes form a 3 million base pair cluster on chromosome 8 in mice (Amid et al., 2009) (Figure 2A). Individual defensins harbor redundant functions, making it uniquely challenging to study the functions of these important AMPs in single KO mice (Morrison et al., 2002; Zhou et al., 2013). Therefore, we sought to delete the entire Def cluster from the skin (Figure 2A). Using CRISPR/cas9, two loxP sites were inserted into the Defb40 and Defb13 loci, located at the two ends of the gene cluster (Figure 2A). This Def flox mouse (Defflox) was then crossed with Keratin14-cre (K14-cre) mice to conditionally knockout (cKO) all Def genes from keratinocytes. PCR and Sanger sequencing of K14-cre+; Defflox/flox skin genomic DNA confirmed successful recombination of the loxP sites and removal of the Def gene cluster (Figure 2A, Figure S2A).
Figure 2. Generation of Defensin cluster conditional knockout mice.
(A) Schematic illustration of the Def gene cluster on mouse Chromosome 8. Two loxP sites were inserted into exons of Defb40 and Defb13 on the two ends of the gene cluster. K14-cre-mediated recombination successfully removed the entire 3 million base pair genomic region from keratinocytes. Genotyping PCR using primers flanking the cluster detected a band corresponding to partial sequence in Defb40, one recombined loxP site, and partial sequence in Defb13.
(B-C) mRNA quantification of infection-induced Defb3 (n=12), Defb4 (n=13), Defb6 (n=9) and Defb14 (n=15) (B) and constitutively expressed Defb1 (n=14), Defb34 (n=12) and Defb39 (n=12) (C) in WT (black) and Def cKO (red) control (Ctrl) skin and 24 hours post S. aureus infection (SA) by qPCR. The amounts of mRNA for each gene were normalized to housekeeping gene Actb.
(D) Quantification of mBD14 peptide in control and S. aureus infected skin of WT and Def cKO animals by ELISA. n=5-14
(E) Representative images of mBD14 immunofluorescence of WT control skin, WT skin infected with S. aureus, Def cKO control skin and Def cKO skin infected with S. aureus. mBD14 was low in uninfected WT skin but was robustly induced 24 hours after infection by S. aureus. In Def cKO animals, infection failed to induce mBD14 production. Scale bar=100μm
(F) Quantification of mean fluorescent intensity of anti-mBD14 immunofluorescence across the thickness of the skin tissue as shown in (E). n=5
Results are presented as mean ± SEM from at least three independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001 n.s. not significant by one-way ANOVA.
See also Figure S2.
The expressions of several Def genes had been shown to be induced by tissue damage or infection (Ahrens et al., 2011; Ali et al., 2001; Bals et al., 1999; Harder et al., 2001; Hinrichsen et al., 2008; Midorikawa et al., 2003; Röhrl et al., 2008; Sumikawa et al., 2006). To validate the deletion of Def genes on the mRNA level, we extracted RNA from both healthy and S. aureus-infected skin. We found that in WT animals, the expressions of Defb3, 4, 6 and 14 were robustly induced post-infection (Figure 2B), while Defb1, 34 and 39 were constitutively expressed in both control and infected skin (Figure 2C). RNA purified from Def cKO mouse skin, on the other hand, contained little or no Defb transcripts (Figure 2B, C). Consistent with previous reports (Adolph et al., 2013; Eisenhauer et al., 1992), Defa transcripts were not observed in the skin (Figure S2B). Further, we examined the amount of mBD14, which activated Mrgpra2 (Figure 1E, H, I) and was among the highest expressed beta-defensins after infection (Figure 2B) (Davis et al., 2014; Harder et al., 2001; Hinrichsen et al., 2008; Midorikawa et al., 2003; Röhrl et al., 2008). Immunofluorescence staining revealed that 24 hours post-S. aureus infection, mBD14 was observed in both epidermal and dermal areas of WT skin around the infection site, whereas infected skin of Def cKO mice had low amounts of mBD14 that could not be distinguished from uninfected controls (Figure 2E, F), as confirmed by ELISA (Figure 2D). These results showed that defensins were efficiently removed in the Def cKO mice on DNA, mRNA and peptide levels.
Disruption of the defensin-Mrgpra2 axis caused dysbiosis and enrichment of Staphylococcus in the skin
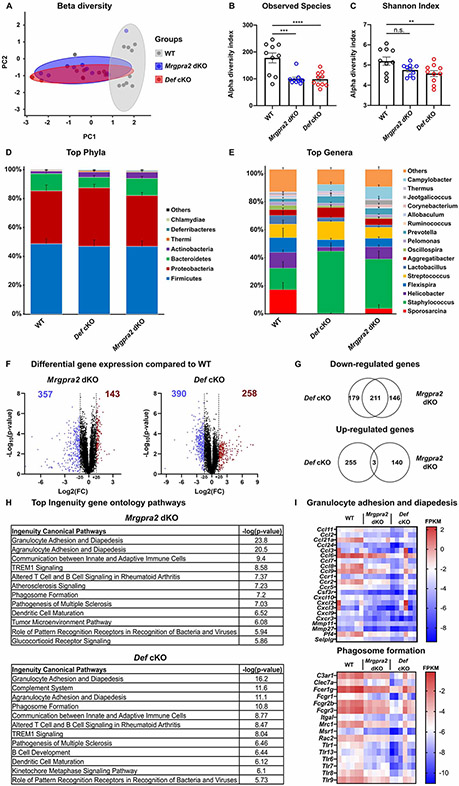
Given the importance of AMPs in innate immunity, we hypothesized that defensin signaling contributed to the maintenance of skin microbiome. To directly test this hypothesis, we analyzed the skin microbiomes of WT, Mrgpra2 dKO and Def cKO animals. Littermates were weaned and genotyped at three weeks old then housed with other mice of the same genotype for six weeks under conventional housing conditions. Skin swabs were taken from the backs and analyzed using 16S rRNA sequencing (Bolyen et al., 2019; Kozich et al., 2013). Beta diversity analyses revealed that the microbiomes of Def cKO and Mrgpra2 dKO mice were similar to each other, but distinct from WT mice (Figure 3A). Alpha diversity, measured by the total number of observed species in each sample and by Shannon indices, was significantly lower in both the Def cKO and Mrgpra2 dKO samples, suggesting a loss of community diversity in these mutants (Figure 3B, C). Phylum and genus level analyses showed that the skin of Def and Mrgpra2 mutant mice were colonized by higher percentages of Staphylococcus (Figure 3D, E, Figure S3B), which accounted for about 12% of WT reads but 31% and 36% of Mrgpra2 dKO and Def cKO reads. In parallel to 16S sequencing which provided information about the relative abundance of each genus, we also used qPCR to quantify the eubacterial 16S gene as a measurement of total bacterial colonization. Compared to WT animals, the skin of both Mrgpra2 and Def mutant animals contained more 16S rRNA (Figure S3A). Given the neutrophil-specific expression of Mrgpra2, we also evaluated the effect of systemic neutrophil depletion on the skin microbiome. WT animals were treated with an anti-Ly6g monoclonal antibody for two weeks, resulting in roughly 70-80% reductions of neutrophils both in the blood and in the skin (Figure S3C). Compared to the animals that received a control antibody, anti-Ly6g treated animals displayed dysbiosis in their skin microbiome similar to the Mrgpra2 and Def mutant animals (Figure S3D), with reduced community alpha diversity (Figure S3E, F) and increased Staphylococcus counts (Figure S3G). These results were consistent with our hypothesis that defensins and their neutrophil receptor Mrgpra2 regulated skin microbiome composition. Disruption in this signaling axis resulted in dysbiosis and increased colonization by Staphylococcus.
Figure 3. Def and Mrgpra2 mutant animals had altered skin microbiomes.
(A-E) 16S sequencing analysis of microbial communities on the skin of WT, Mrgpra2 dKO and Def cKO mice. n=10
(A) Principal coordinates analysis showed that the clustering of Mrgpra2 dKO (blue) and Def cKO (red) microbial communities overlapped with each other but were distinct from WT (grey).
(B) Total numbers of bacterial species observed in WT (black), Mrgpra2 dKO (blue), and Def cKO (red) skin swab samples.
(C) Shannon indices of WT (black), Mrgpra2 dKO (blue), and Def cKO (red) skin swab samples.
(D-E) Stacked bar charts depicting the relative abundance of top phyla (D) and top genera (E) in WT, Mrgpra2 dKO and Def cKO skin microbiota.
(F-I) RNA-seq analysis of gene expression in naïve skin of WT, Mrgpra2 dKO, and Def cKO mice. n=6
(F) Volcano plots illustrating genes with significantly down- (blue) or up-regulated (red) expression in the Mrgpra2 dKO (left) or Def cKO (right) naïve skin compared to WT.
(G) Top: Venn diagram showing overlap of down-regulated gene sets between Mrgpra2 dKO and Def cKO. Bottom: Very few overlap in the up-regulated gene sets.
(H) Top altered Ingenuity gene ontology pathways in Mrgpra2 dKO (top) and Def cKO (bottom) naïve skin compared to WT.
(I) Heat maps depicting fragments per kilobase per million mapped reads (FPKM) counts of genes related to granulocyte adhesion and diapedesis (top) and phagosome formation (bottom). n=6
Results are presented as mean ± SEM from one experiment. *p < 0.05, **p < 0.01, ***p < 0.001, n.s. not significant by one-way ANOVA.
To examine the cause for the difference in skin microbiome between WT and Mrgpra2 and Def mutants, we used RNA sequencing (RNA-seq) to profile the entire stromal and immune landscape of the naïve skin. Whole transcriptome analysis revealed down-regulation of a set of immune-related genes in both Mrgpra2 dKO and Def cKO skin (Figure 3F-I, Figure S3H, I). Ingenuity gene ontology analysis showed that the top altered pathways in both mutants include granulocyte adhesion and diapedesis and pattern recognition receptor signaling (Figure 3H, I, Figure S3H, I). The overlap in down-regulated genes between the Mrgpra2 and Def knockouts (Figure 3G) was consistent with their ligand-receptor relationship. These observations indicated that defensins-Mrgpra2 signaling was critical to maintaining steady baseline immunity required for a balanced and diverse skin microbiota.
In the last decade, with the advancements in microbiome analysis technologies, it has become clear that the gram-positive pathogenic bacterium S. aureus is a major driver of skin dysbiosis (Kennedy et al., 2017; Meylan et al., 2017; Nakatsuji et al., 2017; Williams and Gallo, 2017; Williams et al., 2017b). We mimicked a dysbiotic scenario by topically applying S. aureus (SF8300 strain, 1×109 colony-forming units [CFU]) on the surface of back skin and examined the animals’ ability to restrict and eventually clear the bacteria (Malhotra et al., 2016). Eighteen hours after epicutaneous S. aureus application, a significant number of Ly6G+ neutrophils could be observed in WT skin close to the epidermis and hair follicles near bacterial colonies (Figure S4A, B) (Gallo, 2017; Lone et al., 2015; Matsumoto et al., 2021), affirming the involvement of neutrophils in clearing epicutaneous infection. Def and Mrgpra2 KOs, however, had compromised responses (Figure S4A, B). In vivo imaging of bacterial luminescence revealed that the bacteria were consistently cleared by the WT skin within 4-5 days (Figure S4C, D). In contrast, S. aureus persisted until day 10 in Def and Mrgpra2 mutants and led to skin lesions (Figure S4C-F). Collectively, changes in the mutants’ steady-state microbiome composition and gene expression, as well as their inability to effectively restrict S. aureus on the skin surface all pointed to the importance of this epithelial-neutrophil pathway in regulating the microbial habitat.
Defensins and Mrgpra2 were critical for immunity against bacterial infection and abscess formation
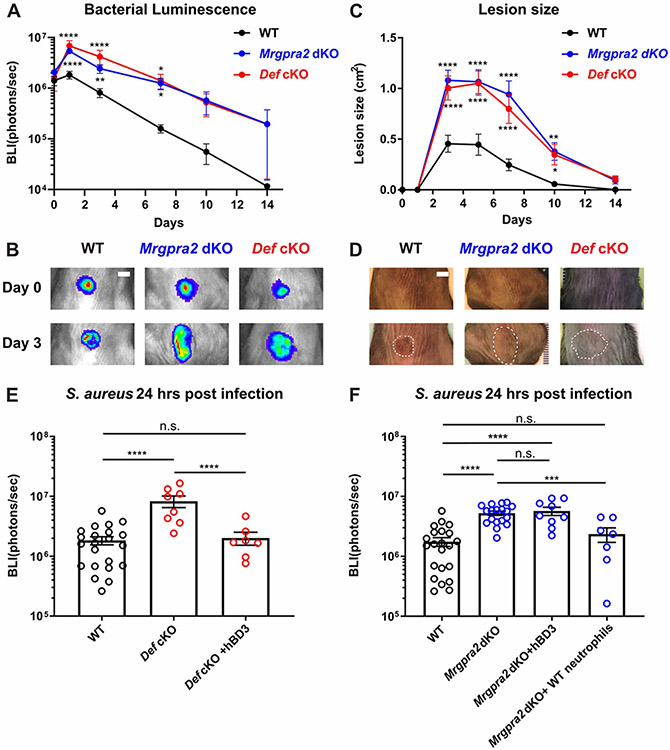
We next analyzed the immunological deficits of the Def and Mrgpra2 mutants in further detail using a well-established intradermal infection model (Miller et al., 2006; Ridder et al., 2022). In this model, a bioluminescent S. aureus strain (USA3000 LAC::lux, 3×107 CFU) was intradermally injected into the back skin, causing the mouse to develop a dermonecrotic lesion (Miller et al., 2007; Ridder et al., 2022; Tkaczyk et al., 2013). Co-housed animals with similar microbiomes were used in all infection studies to control for commensal variations between genotypes (Figure S5A, B) (Archer et al., 2019; Wang et al., 2021; Williams et al., 2017a). We monitored in vivo bacterial bioluminescence for two weeks. Throughout the course of infection, Mrgpra2 dKO and Def cKO animals carried significantly higher bacterial load (Figure 4A-B) that spread to much larger areas (Figure 4C-D). Intradermal injection of human defensin hBD3 at the infection site 6 hours after S. aureus infection was sufficient to restore the antibacterial defect of the Def cKO animals (Figure 4E) but did not rescue the phenotype of the Mrgpra2 dKO animals (Figure 4F), further supporting the role of Mrgpra2 as defensin receptors. On the other hand, adoptive transfer of purified WT neutrophils into Mrgpra2 dKO mice two hours before infection fully rescued the Mrgpra2 dKO phenotype, affirming the neutrophil-intrinsic functions of these GPCRs (Figure 4F).
Figure 4. Mrgpra2 and defensins were critical for anti-S. aureus immunity.
(A) Bacterial luminescence (mean total flux [photons per second]) of WT (black, n=22), Mrgpra2 dKO (blue, n=13) and Def cKO (red, n=8) back skin intradermally infected with S. aureus.
(B) Representative images of in vivo bioluminescence. Scale bar=0.5cm
(C) Mean total infected area (cm2) of WT (black), Mrgpra2 dKO (blue) and Def cKO (red) back skin infected with S. aureus.
(D) Representative photographs of S. aureus-infected skin. Dotted lines indicate the dermonecrotic area caused by the infection. Scale bar=0.5cm
(E) hBD3 injection 6 hours post S. aureus infection was sufficient to rescue the anti-bacterial defect of Def cKO mice. n=7-22
(F) Anti-bacterial defect of Mrgpra2 dKO animals could not be rescued by hBD3 injection but was fully rescued by adoptive transfer of purified WT neutrophils. n=7-22
Results are presented as mean ± SEM from at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. not significant by two-way ANOVA (A and C) and one-way ANOVA (E and F). In (A) and (C), comparisons were made between each mutant and WT.
See also Figure S5.
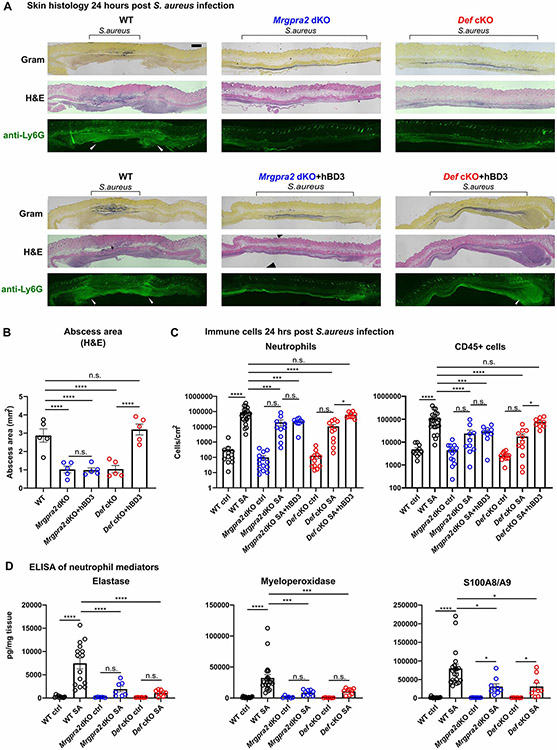
The formation of a neutrophil abscess is a hallmark in anti-S. aureus immunity (Cho et al., 2012; Miller et al., 2006; Miller et al., 2007). In WT animals 24 hours post-intradermal infection, staining with hematoxylin and eosin (H&E) and a neutrophil-specific anti-Ly6G antibody revealed large clusters of neutrophils on the edges of the infection sites, surrounding the bacteria (as shown by Gram staining) (Figure 5A). In both Mrgpra2 dKO and Def cKO animals, however, fewer neutrophils were observed and the bacteria spread to much larger areas, consistent with the in vivo bacterial luminescence imaging result (Figure 5A, Figure 4A, B). The total areas of abscess formed in response to S aureus infections were significantly reduced in the Mrgpra2 and Def mutant animals (Figure 5B). Using flow cytometry (gating strategy in Figure S5C), we quantified the number of immune cells at the infection site 24-hours post-infection. Consistent with previous reports (Cho et al., 2012), the majority of CD45+ leukocytes in WT skin at this time point were neutrophils (Figure 5C). In Mrgpra2 and Def mutant mice, the numbers of neutrophils in the infected skin were reduced by roughly 10 fold (Figure 5C). Consistent with the bacterial clearance phenotype (Figure 4E, F), injection of human defensin hBD3 was sufficient to rescue the neutrophil defects of Def cKO but not Mrgpra2 dKO animals (Figure 5A-C). The reduction in neutrophil infiltration in the infected skin of mutant animals was also evident by lower amounts of neutrophil-specific mediators (Figure 5D) including neutrophil elastase, MPO and calprotectin (S100A8/A9) (Cho et al., 2012; Corbin et al., 2008).
Figure 5. Mrgpra2 dKO and Def cKO animals showed severe defects in neutrophil abscess formation following S. aureus infection.
(A) Top row: Histology staining of WT, Def cKO, and Mrgpra2 dKO mouse skin 24 hours post S. aureus infection using Gram stain (top), H&E (middle), and immunofluorescent staining using a monoclonal antibody against Ly6G (bottom) to show the locations of neutrophils. Black brackets indicate the bacteria band. White arrowheads point to neutrophil abscesses. Scale bar=1mm
Bottom row: hBD3 injection rescued abscess formation in Def cKO but not Mrgpra2 dKO mutant animals.
(B) Quantification of abscess areas based on H&E staining shown in (A). n=5
(C) Flow cytometry analyses of total CD45+ cells and neutrophils in the skin 24 hours post S. aureus infection. Both Def cKO and Mrgpra2 dKO animals showed reduced recruitment of neutrophils. hBD3 injection rescued neutrophil recruitment of Def but not Mrgpra2 mutants. n=9-23
(D) ELISA quantifications of elastase, myeloperoxidase (MPO), and calprotectin (S100A8/A9 dimer) in the skin of WT, Def cKO and Mrgpra2 dKO mice 24 hours post S. aureus infection. n=6-21
Results are presented as mean ± SEM from at least three independent experiments. *p < 0.05, ***p < 0.001, ****p < 0.0001, n.s. not significant by one-way ANOVA.
See also Figure S5.
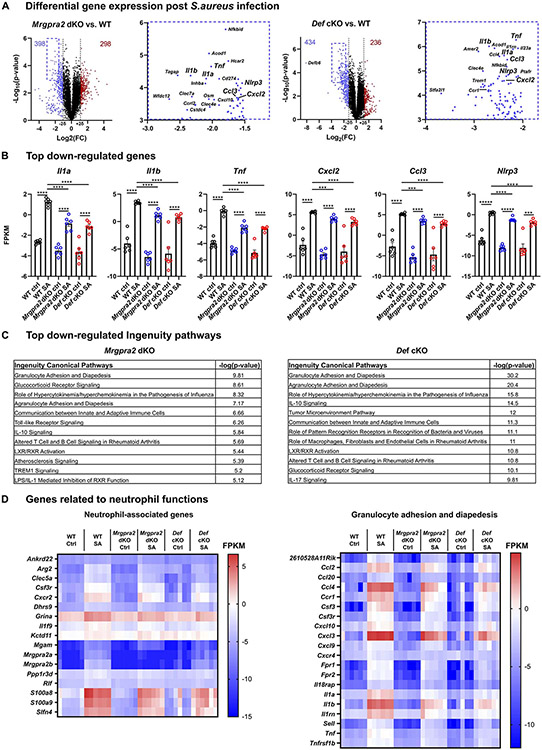
To acquire a comprehensive understanding of the antibacterial immune defects of the Def and Mrgpra2 mutants, we performed RNA-seq of S. aureus-infected skin. Consistent with the neutrophil deficiencies observed by histology and flow cytometry, infected Mrgpra2 and Def mutant animals exhibited altered transcriptomes characterized by a reduction in neutrophil-associated genes and pathways compared to WT (Figure 6A-D, S6A-C) (Ericson et al., 2014). Among the most significantly down-regulated genes in both Mrgpra2 and Def mutants were pro-inflammatory cytokines Il1a, Il1b and Tnf, chemokines Cxcl2 and Ccl3, as well as inflammasome component Nlrp3 (Figure 6A, B). The expressions of several key genes were validated using qPCR (Figure S6C). Ingenuity gene ontology analysis showed that the top pathways affected in both Mrgpra2 dKO and Def cKO include granulocyte adhesion and diapedesis (Figure 6C, D), glucocorticoid receptor signaling, Toll-like receptor signaling and IL-10 signaling, all of which are linked to inflammatory and antimicrobial responses (Figure 6C). Ingenuity also identified a network of inter-related pathways down-regulated in both mutants centering on a few key genes including the pro-inflammatory cytokine IL-1β (Figure S6B). We confirmed the reduced IL-1β in Mrgpra2 dKO and Def cKO infected skin using both qPCR (Figure S6C) and ELISA (Figure S7A). This finding was particularly interesting since we know that mutant animals lacking neutrophil-derived IL-1β showed very similar phenotypes to Def and Mrgpra2 mutants when infected by S. aureus, with significantly higher bacterial burden, larger necrotic lesions, and a significant loss of neutrophil abscess (Cho et al., 2012; Miller et al., 2007). Given the neutrophil-specificity of Mrgpra2 and the central roles of neutrophil-derived cytokines and chemokines in anti-bacterial immunity (Cho et al., 2012; Miller and Cho, 2011), we next sought to understand the effects of defensin-Mrgpra2 interaction on neutrophil cell biology and to identify key effectors downstream of ligand-receptor signaling system.
Figure 6. RNA-seq analysis of WT, Mrgpra2 dKO and Def cKO skin 24 hours post-S. aureus infection.
(A) Volcano plots illustrating genes with significantly down- (blue) or up-regulated (red) expression in the Mrgpra2 dKO (left) or Def cKO (right) infected skin compared to WT. Boxed areas in the upper-left corners were magnified to show the most significantly down-regulated genes in detail.
(B) FPKMs of key pro-inflammatory genes in WT, Mrgpra2 dKO and Def cKO naïve and S. aureus-infected skin. n=6
(C) Top altered Ingenuity gene ontology pathways in Mrgpra2 dKO (top) and Def cKO (bottom) S. aureus infected skin.
(D) Heat maps depicting FPKM of neutrophil-associated genes and genes related to granulocyte adhesion and diapedesis.
Results are presented as mean ± SEM from one experiment. ***p < 0.001, ****p < 0.0001, *****p < 0.00001, n.s. not significant by one-way ANOVA.
See also Figure S6.
Defensin-Mrgpra2 interaction triggered neutrophils to release IL-1β and Cxcl2
Neutrophils are the most numerous leukocytes in circulation and can be rapidly recruited to the site of infection to kill infiltrating pathogens (Mayadas et al., 2014). Uncontrolled neutrophil activity can cause severe tissue damage, thus the activation of these cells generally follows a tightly regulated multi-step process: Upon entering the inflamed tissue, neutrophils can first become “primed” by a variety of microbial-derived PAMPs or cytokines (“signal 1”) (Figure 7B) (Miralda et al., 2017; Wright et al., 2013). Priming leads to the expression of a set of cytokine and chemokines including IL-1β, Cxcl2, Ccl3, and TNFα (Wright et al., 2013) (Figure 7A. C). The primed neutrophils are now poised to respond rapidly to other activating stimuli (“signal 2”) (Figure 7B), release these pro-inflammatory molecules and perform their effector functions. Importantly, several neutrophil-derived factors were shown to primarily act on other neutrophils, thus forming an “autocrine” signal amplification loop that result in a highly coordinated immune response targeted precisely to the site of infection (Ballesteros et al., 2020; Cho et al., 2012; Chou et al., 2010; Lämmermann et al., 2013; Lentini et al., 2020; Sadik et al., 2011).
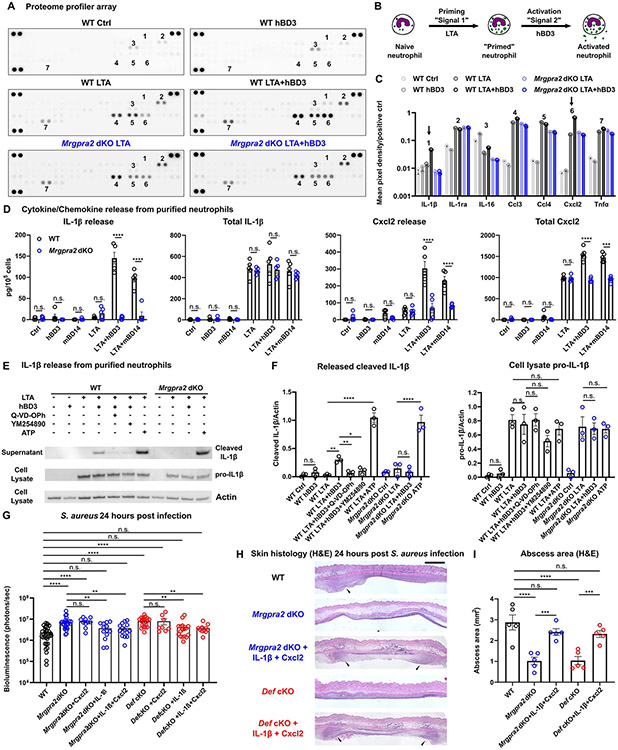
Figure 7. Defensin-Mrgpra2 interaction triggered neutrophil release of IL-1β and Cxcl2.
(A) Proteome profiler array analysis of culture medium of bone marrow neutrophils purified from WT (black) or Mrgpra2 dKO (blue) animals stimulated by control medium, hBD3 only, S. aureus lipoteichoic acid (LTA), or LTA+hBD3. Numbers: 1=IL-1β, 2=IL-1ra, 3=IL-16, 4=Ccl3, 5=Ccl4, 6=Cxcl2, 7=Tnfα. Quantifications are shown in (C).
(B) Schematic illustration of experimental design. Neutrophils are first primed with LTA (“signal 1”), then activated by hBD3 (“signal 2”).
(C) Quantification of mean pixel intensity of proteome profiler arrays shown in (A). Arrows point to IL-1β (1 in A) and Cxcl2 (6 in A), whose releases were enhanced by hBD3. Pixel intensities were normalized to positive controls on the same array.
(D) ELISA quantification of IL-1β and Cxcl2 release from neutrophils. WT neutrophils pretreated with LTA released IL-1β and Cxcl2 in response to hBD3 or mBD14 stimuli (black). Neutrophils purified from Mrgpra2 dKO animals failed to respond to hBD3 (blue). Total IL-1β and Cxcl2 was calculated as the sum of secreted and intracellular amounts. hBD3 or mBD14 alone did not induce the synthesis of IL-1β or Cxcl2. n=5
(E) Western blot showing the release of mature IL-1β (17kDa) released into the supernatant by WT neutrophils when stimulated with hBD3 (5μM). IL-1β release was inhibited by pan-caspase inhibitor Q-VD-OPh and Gq inhibitor YM-254890 and was abolished in the Mrgpra2 mutant neutrophils. ATP (5mM) was used as positive control.
(F) Quantification of western blots of released IL-1β (17kDa) and cellular pro-IL-1β (31kDa) normalized to Actin. n=3
(G) Bacteria bioluminescence 24 hour post S. aureus infection. Cxcl2 alone did not rescue Mrgpra2 or Def KO phenotypes, whereas IL-1β or IL-1β+Cxcl2 together rescued the anti-S. aureus defect of Mrgpra2 dKO and Def cKO mice. n=8-36
(H) H&E staining showing that injecting IL-1β and Cxcl2 restored abscess formation in Mrgpra2 dKO (blue) and Def cKO (red) animals 24 hours post S. aureus infection. Arrow heads point to neutrophil abscesses. Scale bar=1mm
(I) Quantification of abscess areas based on H&E staining shown in (H). n=5
Results are presented as mean ± SEM from at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. not significant by one-way ANOVA.
See also Figure S7.
We utilized a proteome profiler array (R&D systems) to identify key cytokines and chemokines released from neutrophils in response to defensin stimuli. Human hBD3 or its mouse homologue mBD14 alone did not trigger the production or release of any cytokine or chemokine included in the array and thus did not serve as priming signals (Figure 7A, C, D, E). We then used S. aureus-derived lipoteichoic acid (LTA, a Toll-like receptor 2 ligand) as signal 1 (Lotz et al., 2004). LTA stimulus alone triggered the release of more IL-1ra, Ccl3, Ccl4, Cxcl2 and TNFα compared to the control (Figure 7A, C, D). It also induced the production of IL-1β, but the cytokine was not released from the cells (Figure 7A, D). When hBD3 or mBD14 were added following LTA, we saw a significant increase in the amount of IL-1β and Cxcl2 in the extracellular supernatant (Figure 7A, C, D). In contrast, neutrophils purified from Mrgpra2 dKO did not respond to hBD3 or mBD14 (Figure 7A, D), though the priming effects of LTA did not differ between WT or Mrgpra2 dKO neutrophils (Figure 7A, C, D). IL-1β is synthesized and stored as a 31kDa pro form in primed neutrophils and is cleaved and released as a 17kDa mature form in an inflammasome-dependent manner (Martinon et al., 2002; Miller et al., 2007; Shi et al., 2015). We used Western blot to distinguish the 31kDa pro-IL-1β from the 17kDa cleaved IL-1β (Figure 7E). Consistent with the proteome profiler and ELISA results (Figure 7A-D), LTA induced the production of pro-IL-1β in both WT and Mrgpra2 dKO neutrophils, but only WT neutrophils released cleaved IL-1β in response to hBD3 (Figure 7E, F), affirming the notion that defensin activated neutrophils via the Mrgpra2 receptor. We further examined hBD3-mediated IL-1β release using pharmacological inhibitors. Q-VD-OPh, a pan-caspase inhibitor, effectively reduced IL-1β release from WT neutrophils in response to hBD3, indicating that the process was caspases-dependent (Figure 7E, F). The defensin-Mrgpra2 ligand-receptor interaction was also inhibited by a Gq inhibitor YM-254890 (Figure 7E, F), consistent with informatic predictions that Mrgpra2a/b were Gq-coupled GPCRs (Singh et al., 2019).
Last, we tested whether injecting IL-1β and Cxcl2 could alter the immune responses of Mrgpra2 and Def mutant animals. We reasoned that, if these cytokine and chemokine were downstream of defensin-Mrgpra2 signaling, supplying exogenous IL-1β and Cxcl2 would rescue the anti-S. aureus phenotype of the mutant animals. Injection of recombinant mouse Cxcl2 alone did not alter the mutant phenotypes (Figure 7G); whereas IL-1β (17kDa form) alone or co-injecting IL-1β and Cxcl2 significantly reduced the bacterial load in both Mrgpra2 dKO and Def cKO animals 24 hours post-infection (Figure 7G). Histological analyses of post-infection tissues also showed that IL-1β and Cxcl2 restored the neutrophil abscess in the Mrgpra2 and Def mutants (Figure 7H, I). These results showed that IL-1β and Cxcl2 acted downstream of the defensin-Mrgpra2 signaling axis. Given the importance of IL-1β in anti-S. aureus immunity (Cho et al., 2012; Miller et al., 2007) and as a downstream factor of defensin-Mrgpra2 signaling, we were curious whether ll1b mutant animals also harbor altered skin microbiota. Interestingly, 16S sequencing revealed strong skin dysbiosis in the Il1b KO animals (Figure S7B), with drastically reduced community diversity (Figure S7C, D). However, unlike animals lacking defensins, Mrgpra2 (Figure 3E) or neutrophils (Figure S3G), the skin microbiota of Il1b mutants were dominated by Sporosarcina (Figure S7E), which is a common component of the murine gut microbiome (Guo et al., 2022; Zhao et al., 2019). This was likely due to the important roles of IL-1β in intestinal immunity which could potentially interfere with the skin microbiome (Franchi et al., 2012; Molloy et al., 2013).
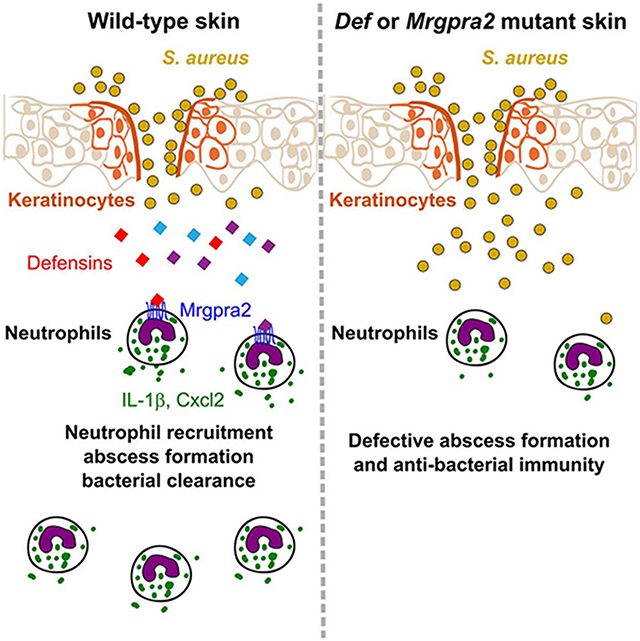
In summary, our results demonstrated that keratinocytes released defensins in response to S. aureus skin infections. These AMPs activated Mrgpra2 receptors on neutrophils and triggered the release of IL-1β and Cxcl2. Additional neutrophils responded to this inflammatory signal, infiltrated into the tissue, released more pro-inflammatory cytokine and chemokines, and effectively propagated the signal. Abolishing the defensin-Mrgpra2 axis by either removing all defensin ligands from the keratinocytes or the Mrgpra2 receptors from the neutrophils disrupted this signaling cascade and caused the severe deficiencies in abscess formation and antibacterial immunity observed in the mutants.
DISCUSSION
Our study revealed an important epithelial to neutrophil signaling pathway that was vital for skin immunity and microbiome homeostasis. The idea that defensins both directly kill bacteria and signal to immune cells via their cell surface receptors has long been appreciated (Ganz, 2003; Röhrl et al., 2010; Subramanian et al., 2013a; Yang et al., 1999; Yang et al., 2017). However, genetic studies of these important AMPs have been hindered by the massive redundancy of this gene family (Morrison et al., 2002; Navid et al., 2012; Zhou et al., 2013). The Def cKO mice allowed us to directly demonstrate the host-intrinsic immune-modulatory functions of defensins beyond direct bacterial killing. Our results clearly showed that removal of all defensins from keratinocytes resulted in severely compromised neutrophil response to infection. The importance of neutrophils in defensin-mediated immunity was further highlighted by the identification of Mrgpra2a and Mrgpra2b as neutrophil-specific defensin receptors. Mrgpra2 dKO mice phenocopied the Def cKO mice in every aspect we analyzed, with similar microbiome dysbiosis, comparable defects in antibacterial immunity and a lack of neutrophil abscess formation post-infection. These results pointed to neutrophils as key effector cells of defensin signaling.
Neutrophils are the most abundant granulocytes in circulation and have been shown to be the key players in a wide range of immune functions (Cassatella et al., 2019; Mayadas et al., 2014; Miller and Cho, 2011; Sadik et al., 2011). Here, we identified a pair of receptors important for neutrophil functions. The Mrgpr family of GPCRs have attracted much attention as key regulators of innate immune functions (Arifuzzaman et al., 2019; Dwyer et al., 2016; McNeil et al., 2015; Pundir et al., 2019). Mrgpra2a and a2b showed highly specific expression in mouse neutrophils (Dwyer et al., 2016; Ericson et al., 2014) and were vital to antibacterial functions. Several key questions remain to be answered. First, the precise intracellular signaling pathways employed by Mrgpra2 remain to be defined. Both bioinformatics prediction (Singh et al., 2019) and our pharmacological inhibition result suggested that Mrgpra2a/b were Gq-coupled GPCRs, and likely engaged Ca2+ as part of the signaling cascade. Importantly, we found that in the context of S. aureus infections, defensins did not serve as priming “signal 1”, but rather acted on primed neutrophils as “signal 2” and that IL-1β and Cxcl2 were key downstream effectors of defensin-Mrgpra2 signaling. Still, the full picture of functional consequences of Mrgpra2 activation in neutrophils remains to be elucidated. Second, RNAscope showed that Mrgpra2+ neutrophils accounted for about half of Ly6g+ cells in the bone marrow. Recent single-cell analyses revealed heterogeneity and tissue specificities in neutrophil populations (Ballesteros et al., 2020; Xie et al., 2020). Detailed analyses using reporter mouse lines will be necessary for understanding important properties of the Mrgpra2+ subpopulation, their development, trafficking, localization and gene expression regulation in response to infections.
Our current study focused on the contribution of neutrophils to defensin-mediated innate immunity upon bacterial infection and skin microbiome homeostasis. However, it is likely that immune cells other than neutrophils contribute to defensin-evoked immunity and that other defensin receptors besides Mrgpra2 function in these immune cells (Röhrl et al., 2010; Subramanian et al., 2013b; Tseng and Hoon, 2022; Yang et al., 1999). Neutrophils do not exist in large numbers in the naïve skin but are rapidly recruited from the blood stream following infections and inflammations (Abdul Hamid et al., 2021; Kolaczkowska and Kubes, 2013). Tissue resident immune cells including mast cells and macrophages are important for the initial phase of neutrophil recruitment before the neutrophil self-amplification loop reaches a critical mass (Arifuzzaman et al., 2019; De Filippo et al., 2013; De Filippo et al., 2008; Kim and Luster, 2015; Nestle et al., 2009; Pundir et al., 2019; Subramanian et al., 2013b). Intra-vital imaging analyses can potentially be used to precisely quantify the roles of defensins and defensin receptors in regulating cellular dynamics following infections (Kolaczkowska and Kubes, 2013). The defensin-neutrophil axis presented in this study added an important piece to the complex and intricate signaling networks at host-microbe interfaces (Kabashima et al., 2019).
The Def gene family is one of the largest gene families in the mouse genome. The exact reason for the organism to require these redundant peptides is unclear. Genetic analysis in Drosophila has revealed convincing functional diversities of AMPs (Hanson et al., 2019; Lazzaro et al., 2020). In the context of anti-S. aureus skin immunity, two mouse peptides mBD3 and mBD14 (and its human homologue hBD3) appeared to be of central importance. Among mouse defensins, mBD3 and mBD14 bear the largest number of positive charges (+10.6 and +11.6, respectively) and are the most potent in both their abilities to directly kill pathogens and activate immune receptors (Midorikawa et al., 2003; Röhrl et al., 2008; Röhrl et al., 2010; Subramanian et al., 2013b). The transcripts of Defb3 and Defb14 were among the most highly up-regulated after S. aureus infections. More intriguingly, in the absence of the entire Def gene cluster, supplying the human peptide hBD3 (homologue of mBD14) alone could restore the S. aureus infection phenotype to one similar to the WT animals. This suggested that despite the apparent genetic redundancy, a single potent peptide could be sufficient to restrict S. aureus infection. In the future, it would be interesting to dissect the roles of various AMPs, their relative importance in combatting different classes of pathogens (including Gram positive and negative bacteria, viruses and fungi), the types of host immune cells they activate and the receptors they employ. Another key question about AMPs that remains to be answered is the relative importance of direct killing versus immune-modulation. The strong loss-of-function phenotypes of the neutrophil receptors Mrgpra2 supported the importance of innate immune cells in response to defensin signaling. However, given that defensins have been clearly shown to be capable of directly killing S. aureus (Harder et al., 2001; Hinrichsen et al., 2008; Midorikawa et al., 2003; Röhrl et al., 2008), both functions of these peptides are probably important in the host’s ability to defeat the bacteria during infections. To definitively distinguish the relative contributions of direct killing versus immune modulation, stringent structure-function analyses will need to be performed to generate peptides lacking one aspect of their function while retaining the other. Ongoing efforts using cryogenic electron microscopy (cryo-EM) to illuminate the structure and ligand binding mechanisms of the MRGPR family receptors might provide an entry point to this difficult and important problem (Cao et al., 2021; Yang et al., 2021).
Last but not least, our finding has important implications in the pathologies of skin diseases. Studies of the skin microbiota have yielded important knowledge about the instructive roles that commensal microbiota play during the development of the host immune system and causal links between dysbiosis and inflammatory skin diseases (Grice et al., 2009; Liu et al., 2017; Naik et al., 2015; Naik et al., 2012; Nakatsuji et al., 2017; Wang et al., 2021; Williams and Gallo, 2017; Williams et al., 2017b). However, host factors defining the microenvironment for the microbiota remain largely unclear. Physical and chemical properties of the skin surface such as moisture and acidity have been known to profoundly influence microbiome composition (Grice et al., 2009). However, influence from the host immune system, whose primary function is to interact with the microbial world, remains largely unknown. Defects in defensin production and neutrophil recruitment have been implicated in skin disease conditions involving severe microbiome dysbiosis (Chiang et al., 2019; Harder et al., 1997; Hollox et al., 2008). Here we established a direct causal link between genetic deletion of the defensin-Mrgpra2 signaling pathway and skin dysbiosis. These findings suggest that normal skin maintains a baseline of immune activity and profoundly influences the microbiome composition on the surface (Matsumoto et al., 2021). Crosstalk between the epithelium and neutrophils through AMPs and their GPCRs are an important component of this baseline immune activity. Exactly how this system maintains a balanced microbiome, permitting colonization by commensal bacteria while restricting the expansion of pathogenic bacteria deserves further investigation. Overall, this study opens avenues to study innate immunity and could lead to improved therapies to treat infections and skin diseases related to AMP deregulation.
Limitations of the study
Due to the large number of genes in the Def cluster and technical difficulty in peptide synthesis, we were unable to test every single defensin for its ability to activate Mrgpra2 or regulate skin immunity. Although the Def cKO mouse is a powerful genetic tool to examine the collective loss-of-function consequences of defensins, it does not explain the diversity and specificity of this important AMP family. This study is limited to skin immunity, and it remains unknown whether other epithelial surfaces including the airway, intestines and urogenital tracts employ similar epithelial-neutrophil axes to maintain healthy microbiota and fight infections.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xinzhong Dong (xdong2@jhmi.edu).
Materials availability
Reagent and materials generated in this study are available from the lead contact without restriction.
Data and code availability
RNA-seq data has been deposited at GEO and are publicly available as of the date of publication. Accession number is listed in the Key Resources Table.
All data supporting the findings of this study are available in the manuscript or the supplementary materials and are available upon request to the lead contact author.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-biotin PE clone 1D4-C5 | Biolegend | Cat# 409004; RRID: AB_10641847 |
| Anti-biotin | Abcam | Cat# ab53494; RRID: AB_867860 |
| Sepharose anti-GFP antibody | Abcam | Cat# ab69314; RRID: AB_1640178 |
| Anti-GFP | Thermo Fisher | Cat# A-11122; RRID: AB_221569 |
| Anti-rabbit IgG HRP-conjugated | R&D systems | Cat# HAF008; RRID: AB_357235 |
| Rat monoclonal anti-mouse Ly6G clone 1A8 | BioXCell | Cat# BE0075-1; RRID: AB_1107721 |
| Rat monoclonal IgG2a isotype control clone 2A3 | BioXCell | Cat# BE0089; RRID: AB_1107769 |
| Anti-mouse CD45 APC-Cy7 clone 30-F11 | Biolegend | Cat# 103116; RRID: AB_312981 |
| Anti-mouse CD11b PE/Dazzle594 Clone M1/70 | Biolegend | Cat# 101256; RRID: AB_2563648 |
| Anti-mouse CD11c PE/Cy7 Clone N418 | Biolegend | Cat# 117318; RRID: AB_493568 |
| Anti-mouse Ly6C APC Clone HK1.4 | Biolegend | Cat# 128016; RRID: AB_1732076 |
| Anti-mouse Ly6G BV421 Clone 1A8 | Biolegend | Cat# 127628; RRID: AB_2562567 |
| Anti-mouse F4/80 BV605 Clone BM8 | Biolegend | Cat# 123133; RRID: AB_2562305 |
| Anti-mouse Siglec F BB515 Clone E50-2440 | BD Bioscience | Cat# 564514; RRID: AB_2738833 |
| Anti-mouse c-Kit BV421 Clone ACK2 | Biolegend | Cat# 135124; RRID: AB_2562237 |
| Anti-mouse ICAM FITC Clone YN/1.7.4 | Biolegend | Cat# 116105; RRID: AB_313696 |
| Anti-mouse F4/80 PE Clone BM8 | Biolegend | Cat# 123110; RRID: AB_893486 |
| Anti-mouse FcεRI PE/Cy7 Clone MAR-1 | Biolegend | Cat# 134318; RRID: AB_10640122 |
| Anti-mouse CD11b BV785 Clone M1/70 | Biolegend | Cat# 101243; RRID: AB_2561373 |
| Anti-mouse CD11c BV605 Clone N418 | Biolegend | Cat# 117334; RRID: AB_2562415 |
| Anti-mouse I-A/I-E BV711 Clone M5/114.15.2 | Biolegend | Cat# 107643; RRID: AB_2565976 |
| Anti-mouse Ly6G PE/Cy7 Clone 1A8 | Biolegend | Cat# 127617; RRID: AB_1877262 |
| Anti-mouse CD11b PE Clone M1/70 | Biolegend | Cat# 101207; RRID: AB_312790 |
| Anti-mouse B220 APC Clone RA3-6B2 | eBioscience | Cat# 17-0452-82; RRID: AB_469395 |
| Anti-mouse Ly6G AF700 Clone 1A8 | Biolegend | Cat# 127622; RRID: AB_10643269 |
| Anti-mouse CD49b PE Clone DX5 | Biolegend | Cat# 108908; RRID: AB_313415 |
| Rabbit anti-mouse BD14 | MyBioSource | Cat# MBS1490249; RRID: AB_2732866 |
| Rat anti-mouse Ly6G | Biolegend | Cat# 127602; RRID: AB_1089180 |
| Rabbit anti-S. aureus | Abcam | Cat# 20920;RRID: AB_445913 |
| Alexa Fluor ® 488 goat anti-rabbit IgG | Thermo Fisher | Cat# A-11008; RRID: AB_143165 |
| Alexa Fluor ® 488 goat anti-rat IgG | Thermo Fisher | Cat# A-11006; RRID: AB_2534074 |
| Alexa Fluor ® 594 goat anti-rabbit IgG | Thermo Fisher | Cat# A-11012; RRID: AB_2534079 |
| Goat anti-mouse IL-1β | R&D systems | Cat# AF-401-NA; RRID: AB_416684 |
| Rabbit anti-cleaved-IL-1β | Cell Signaling Technology | Cat# 63124s |
| Rabbit anti-goat IgG HRP-conjugated | R&D systems | Cat# HAF017; RRID: AB_562588 |
| HRP anti-beta Actin | Abcam | Cat# Ab49900; RRID: AB_867494 |
| Bacterial and virus strains | ||
| Staphylococcus aureus SF8300 | Binh Diep (UCSF) | N/A |
| USA300 | Tammy Kielian (University of Nebraska) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| hBD3 | Anaspec | Cat# AS-60741 |
| Biotin-hBD3 | Anaspec | N/A |
| mBD14 | Wuyuan Lu (University of Maryland Baltimore) | N/A |
| mBD3 | Wuyuan Lu (University of Maryland Baltimore) | N/A |
| S. aureus derived LTA | Invivogen | Cat# Tlrl-pslta |
| Tryptic soy broth | Millipore | Cat# 22092 |
| TRIzol reagent | Thermo Fisher | Cat# 15596018 |
| Liberase TL | Roche | Cat# 05401054001 |
| DNase I | Sigma-Aldrich | Cat# 4536282001 |
| Recombinant IL-1β | R&D systems | Cat# 401-ML |
| NuPAGE™ LDS Sample Buffer | Thermo Fisher | Cat# NP0007 |
| Q-VD-Oph | R&D systems | Cat# OPH001-01M |
| YM254890 | Tocris | Cat# 7352 |
| SYTOX blue | Thermo Fisher | Cat# S34857 |
| SYTOX red | Thermo Fisher | Cat# S34859 |
| SYTOX orange | Thermo Fisher | Cat# S11368 |
| Critical commercial assays | ||
| Mouse Elastase DuoSet ELISA Kit | R&D systems | Cat# DY4517-05 |
| Mouse MPO DuoSet ELISA Kit | R&D systems | Cat# DY3667 |
| Mouse S100A8/A9 DuoSet ELISA Kit | R&D systems | Cat# DY8596-05 |
| Mouse IL-1β DuoSet ELISA Kit | R&D systems | Cat# DY401-05 |
| Mouse Cxcl2 DuoSet ELISA Kit | R&D systems | Cat# DY452-05 |
| Mouse BD14 ELISA Kit | MyBioSource | Cat# MBS2515829 |
| MACS neutrophil isolation kit | Miltenyi Biotec | Cat# 130-097-658 |
| RNAscope® Multiplex Fluorescent Detection Kit v2 | ACDBio | Cat# 323110 |
| RNAscope® Ly6g probe | ACDBio | Cat# 455701 |
| RNAscope® Mrgpra2 probe | ACDBio | Custom-made |
| RNeasy Plus Mini kit | Qiagen | Cat# 74034 |
| Chemotaxis assay kit | Abcam | Cat# Ab235692 |
| Cellular reactive oxygen species detection assay kit | Abcam | Cat# Ab113851 |
| Deposited data | ||
| RNA-seq data | NCBI GEO | GSE178507 |
| Experimental models: Cell lines | ||
| Gα15 HEK 293 cells | This study | N/A |
| Mrgpra2b-GFP; Gα15 HEK 293 cells | This study | N/A |
| MRGPRX3-FLAG; Gα15 HEK 293 cells | This study | N/A |
| MRGPRX4-GFP; Gα15 HEK 293 cells | This study | N/A |
| Experimental models: Mice | ||
| C57BL/6J | Jackson laboratory | Cat# 000664 |
| Mrgpra2 dKO | This study | N/A |
| K14-cre; Defflox/flox | This study | N/A |
| K14-cre | Jackson laboratory | Cat# 018964 |
| Oligonucleotides | ||
| gRNAs targeting Mrgpra2a/b CAAGTTAGTTTTGGTCAATC, GATCTGATACCCTCTTCTGG, TAAACTGGCTATCTACTTGA, TTATCCAATGCTGGGATAAT | This study | N/A |
| Genotyping primers for Mrgpra2 dKO GTCAGCATGATGGTCAC (WT-F), CATAGGGAAGGCCTCATGTCTC (WT-R), CTATGGAGGGATGCTCATTACTG (Mrgpra2 dKO-F), CAAGATCTGACTCCCTCTTCTGG (Mrgpra2 dKO-R) | This study | N/A |
| gRNA targeting Defb13 CCAAGCCAATTTCCTGTGCTGCA | This study | N/A |
| gRNA targeting Defb40 CCATGGGCATTTCTGAATGTGAC | This study | N/A |
| Primers for genotyping Def cluster deletion GATGCAGGTGCCCTGAAATAT (F) CACTGAATAGACCTGACTTCC (R) | This study | N/A |
| RT-PCR primer for Mrgpra2 CCTCCTACACAAGCCAGCAA (F) AAGCACAAGTGAAAGATGATGCT | This study | N/A |
| RT-PCR primer for Gapdh TGTTCCTACCCCCAATGTGT (F) TGTGAGGGAGATGCTCAGTG (R) | Zambrano et al., 2019 | N/A |
| Software and algorithms | ||
| FlowJo | BD | https://www.flowjo.com/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://fiji.sc/ |
| R | R | https://www.r-project.org/ |
| QIIME2 | QIIME2 development team | https://qiime2.org/ |
| STAR - 2.7.3a | Alexander Dobin | https://github.com/alexdobin/STAR/releases/tag/2.7.9a |
| Partek Flow | Partek | https://www.partek.com/ |
| Partek GS v7.0 | Partek | https://www.partek.com/ |
| Spotfire | TIBCO | https://www.tibco.com/products/tibco-spotfire |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All experiments were performed in accordance with protocols approved by the Animal Care and Use Committee at the Johns Hopkins University School of Medicine. 8-10 weeks old, age- and gender-matched male and female mice on the C57BL/6J background were used for experiments. Detailed procedures for the generation of Mrgpra2 dKO and Def cKO mouse lines are described in the METHOD DETAILS section. For the 16S sequencing experiment, WT and mutant littermates were weaned by genotypes at three weeks old then housed with animals of the same genotype for six weeks under conventional housing conditions in a non-helicobacter free facility. Co-housed animals were used in all infection studies.
Bacterial strains
Bioluminescent S. aureus SF8300 strain was used for epicutaneous S. aureus exposure (Figure S4). Bioluminescent USA300 LAC::lux strain was used in the intradermal injection model (Figure 4-7). Bacteria were streaked onto blood tryptic soy agar (TSA) plates (Thermo Fisher Scientific) and grown overnight at 37°C. Single colonies were picked and cultured in tryptic soy broth (TSB, Sigma) at 37°C in a shaking incubator (240 rpm) overnight, followed by a 1:50 subculture at 37°C for 2 hours to obtain mid-logarithmic phase bacteria. The bacteria were pelleted by centrifugation at 5000g, washed 3 times, and resuspended in sterile PBS to the required concentration. OD600 was taken to measure bacterial density. SF8300 was resuspended at 109 CFU/50μL per mouse. USA300 LAC::lux was resuspended at 3×107/100μL per mouse.
METHOD DETAILS
Generation of Mrgpra2 dKO and Def cKO mice
Sequences for all guide RNAs (gRNAs) and genotyping primers are listed in the key resources table. Mrgpra2 double knockout (dKO) animals were generated by the Johns Hopkins University School of Medicine Transgenic Core Laboratory using standard CRISPR/cas9 methods. gRNAs were designed to target the 5’ untranslated regions (UTR) of Mrgpra2a and 3’ UTR of Mrgpra2b and resulted in deletion of the entire genomic region including all exons of both Mrgpra2a and Mrgpra2b. Defflox mice were generated by the Howard Hughes Medical Institute Janelia Research Campus Gene Targeting and Transgenics team using standard CRISPR/cas9 technique. The first loxP cassette was inserted in exon2 of Defb13 by co-injecting Cas9 mRNA, gRNA and a repair template with loxP site flanked by 30 base pairs homology arms on each side. Animals bred homozygous for this Defb13 loxP allele were then mutated with another CRISPR construct targeting exon2 of Defb40 and a second loxP repair template. The resulting Defflox mice were then crossed with K14-cre mice to generate K14-cre; Defflox/flox conditional knock out (cKO) mice. PCR genotyping and Sanger sequencing were performed to confirm successful deletion of the entire cluster.
Fluorescence-activated cell sorting (FACS) of innate immune cells
Dendritic cells were isolated from spleen; mast cells and macrophages were isolated from peritoneal lavage; neutrophils and basophils were isolated from bone marrow; and monocytes and eosinophils were isolated from peripheral blood. Single-cell suspensions from each tissue were subjected to red blood cell lysis using a hypotonic ammonium chloride-potassium (ACK) buffer (Quality Biological). Then, samples were incubated with CD16/CD32 Fc Blocker (Biolegend) for 10 min in FACS buffer (PBS supplemented with 2% fetal bovine serum). Surface staining was performed for 30 min at 4°C with the antibodies listed in Table S1. Dead cells were stained with either SYTOX Blue or SYTOX Red (Thermo Fisher). Cell sorting was performed on the SONY SH800 cell sorter (Sony Biotechnology) with a 100 μm nozzle. Neutrophils were gated as SiglecF−Ly6G+CD11b+CD49b−SSCInt; mast cells were gated as c-Kit+FcεRI+F4/80−ICAM−; basophils were gated as SiglecF−CD49b+FcεRI+c-Kit−SSClow; eosinophils were gated as SiglecF+Ly6G−CD11b− SSChigh; monocytes were gated as TCRβ−B220−CD11b+Ly6G−SSCmed; macrophages were gated as Ly6G−CD11bhighF4/80highICAM+; dendritic cells were gated as B220−CD11chighMHC-II+Ly6G−.
RNAscope® in situ hybridization
Bone marrow was collected from the femur and tibia bones of WT and Mrgpra2 dKO mice. Red blood cells were lysed in ACK lysing buffer (Quality Biological). Remaining cells were cytospun onto poly-L-lysine-coated glass slides (Fisher Scientific) and fixed in 10% neutral buffered formalin. Slides were washed in PBS for 5 min and treated with protease for 15 min at 40°C. Target probes for Ly6g and Mrgpra2b were combined and hybridized for 2 hours at 4°C using the HybEZ Hybridization System. Following amplification and label application according to manufacturer instructions, slides were imaged with a Zeiss LSM700 confocal microscope.
Calcium imaging
HEK293 cells stably expressing a Gα15 subunit were transfected with constructs containing the cDNA of Mrgpra2b, MRGPRX3 or MRGPRX4. Mock-transfected cells were used as controls. Cells were plated onto 96 well plates and grown to 80-90% confluence, then incubated at 37°C with the FLIPR Calcium 5 calcium indicator (Molecular Devices) in Hank’s Balanced Salt Solution (HBSS, Millipore Sigma) with 20 mM HEPES (ThermoFisher) for 1 hour, equilibrated at room temperature for 15 min and imaged on Flex Station 3 (Molecular Devices). Baseline was recorded for 20 sec, peptides were added, and intracellular Ca2+ response was recorded for 150 sec. 2-9 duplicate experiments were averaged and EC50 values were determined by normalizing to the peak response.
Co-IP
HEK cells expressing Mrgpra2b-GFP or MRGPRX4-GFP (negative control) were cultured to confluence in T75 flasks and lysed in co-IP lysis buffer (50mM Tris HCl, pH=7.4, 150 mM NaCl, 1% Triton X-100, 5% Glycerol) at 107 cells/mL. 1mL of cell lysates were incubated with sepharose anti-GFP beads (Abcam) and 10μM biotin-hBD3 or control buffer at 4°C overnight. The beads were spun down and washed with lysis buffer 10 times and protein bound to the beads was extracted using NuPAGE LDS Sample Buffer and analyzed by western blot.
Western blot
Supernatants and cell lysates were loaded on Bolt Bis-Tris Plus gels (Life Technologies), and transferred to PVDF membranes using the iBlot system (Thermo Fisher). Membranes were blocked and incubated with primary antibodies at 4°C overnight.
For co-IP analysis in Figure 1F, rabbit anti-biotin (Abcam, 1:1000) was used to detect biotin-hBD3, rabbit anti-GFP (Thermo Fisher, 1:1000) was used for Mrgpra2b-GFP or MRGPRX4-GFP. For Figure 7E, goat anti mouse IL-1β (R&D systems, 1:1000) was used to detect pro-IL-1β (31kDa), rabbit anti-mouse cleaved-IL-1β (Cell Signaling Technology, 1:500) was used to detect mature IL-1β (17kDa), goat anti-Actin beta (Abcam, 1:1000) was used as loading control. On the next day, secondary antibodies (R&D HRP conjugated anti-goad or anti-rabbit, 1:1000) were applied, followed by SuperSignal™ West Pico Substrate solution (Thermo Scientific) and the blots were imaged on a chemiluminescent blot imager (Cytiva).
KD determination by flow cytometry
Control HEK cells or HEK cells stably expressing Mrgpra2b-GFP were incubated with control medium (complete DMEM) or various concentrations of biotin-hBD3 peptides for three hours at 37°C. Cells were washed in PBS five times, stained using PE-anti-biotin antibody (Biolegend) in FACS buffer for 30 minutes. Mean fluorescent intensity of PE for each sample was measured by flow cytometry.
Protein quantification by ELISA
Snap-frozen skin tissues were weighed and homogenized in PBS using a Pro200 Series homogenizer (Pro Scientific). Elastase and Myeloperoxidase (MPO), S100A8/A9, CXCL1, CXCL2 and IL-1β were measured using ELISA DuoSets (R&D Systems) according to manufacturer’s instructions. mBD14 was measured using an ELISA kit from Abbexa. The same samples were measured for multiple factors.
16S sequencing
Microbiome samples were collected from a 2 cm × 4 cm area on mouse back skin using sterile cotton swabs (Epicentre) pre-moistened with lysis buffer (20 mmol/L tris [pH 8.0], 2 mmol/L EDTA and 1.2% Triton X-100) and immediately frozen in 250 uL of lysis buffer in −80°C. All samples were submitted on a 96 well plate to the University of Michigan Microbial Systems Molecular Biology Laboratory for Illumina 16S rDNA gene sequencing. Briefly, the V4 region of 16S RNA were amplified by PCR, barcoded and sequenced on an Illumina MiSeq platform. Initial informatics analyses were performed by the Johns Hopkins SKCCC Experimental and Computational Genomics Core using the Qiime2 platform (Bolyen et al., 2019). The GreenGenes database was used as reference for taxonomic classification and alpha and beta diversity metrics were computed in Qiime2. Principal coordinate analysis (PCoA) of beta diversity was performed and visualized in R. For neutrophil depletion (Figure S3C-G), mice were intraperitoneally treated with 250μg of anti-Ly6g antibody or isotype control diluted in 100μL PBS every other day for 2 weeks. Skin swabs were collected as described above.
Histology
Skin samples were fixed in 4% paraformaldehyde (PFA) in PBS overnight and submitted to the Johns Hopkins Oncology Tissue Services where they were embedded in paraffin, sliced into 4 μm sections, mounted and stained with hematoxylin and eosin (H&E). Gram stain was performed using the Gram Staining Kit (Sigma Aldrich) according to manufacturer’s instructions. For immunofluorescence labeling, paraffin sections were deparaffinized, underwent heat-mediated antigen retrieval in either Trilogy buffer (Cell Marque) or Tris-EDTA buffer (10mM Tris Base, 1 mM EDTA Solution, 0.05% Tween 20, pH 9.0), blocked in blocking buffer (PBS with 10% goat serum) for 1 hour at room temperature, and then incubated at 4° C overnight with 1 μg/mL rabbit anti-mBD14, rat anti-mouse Ly6G or rabbit anti-S.aureus primary antibodies in blocking buffer. The next day, sections were incubated for 1 hour at room temperature with 1 μg/mL of either AlexaFluor-488 goat anti-rabbit IgG (ThermoFisher, for mBD14 staining), AlexaFluor-488 goat anti-rat IgG (ThermoFisher, for Ly6G staining) or AlexaFluor-594 goat anti-rabbit IgG (ThermoFisher, for S. aureus staining). All slides were washed and mounted with Fluoromount-G with DAPI (ThermoFisher). All microscopy were performed on a Keyence BZ-X710 microscope (Keyence) and analyzed with Image J (National Institutes of Health Research Services Branch).
RNA extraction and quantification
Snap-frozen skin tissue or purified cells were homogenized in Trizol and RNA was extracted using a Direct-zol RNA Miniprep kit (Zymo Research) following manufacturer’s instructions. mRNA was reverse-transcribed into cDNA using a Superscript III kit (Invitrogen) followed by RNase digestion. Quantitative PCR (qPCR) was performed using an EagleTaq mastermix (Roche) and Taqman gene expression probes (Thermo Fisher) corresponding to each gene of interest on a StepOnePlus RT-PCR system (Applied Biosystems). Gene expression was normalized to Actb using the ΔCt method. The same samples were measured for multiple mRNAs. RT-PCR of Mrgpra2 and Gapdh was done using primers listed in the Key Reagent Table.
RNA sequencing
Total RNA was extracted from naïve or S. aureus-infected skin (24 hours post intradermal injection of 3 x 107 CFU USA300 LAC::lux) using Trizol-chloroform and purified using the Qiagen RNeasy Plus Mini kit. 6 control samples and 6 SA (S. aureus-infected) samples of each genotype (WT, Def cKO and Mrgpra2 dKO) were submitted to the Johns Hopkins Transcriptomics and Deep Sequencing Core. Libraries were constructed using the Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit with standard protocol and sequenced on an Illumina NovaSeq 6000 system. Reads were mapped to the mm10 reference mouse genome with STAR using default parameters. Counts were quantified and normalized to fragments per kilobase per million mapped reads (FPKM) using Partek Flow, and one-way ANOVA tests were run between experimental groups. Data were visualized using GraphPad Prism 9.
Mouse models of S. aureus skin infection and quantification by IVIS
For intradermal S. aureus infections, mice were shaved on the back and inoculated with an intradermal injection of mid-logarithmic growth phase bioluminescent S. aureus CA-MRSA strain USA300 LAC::lux as previously described (3 × 107 CFUs/100 μL PBS) using 29-gauge insulin syringes (Dillen et al., 2018). For the epicutaneous swab infection model, 109 CFUs/50 μL PBS of CA-MRSA strain SF8300 was dropped on the shaved back skin and spread into a 2 cm × 2 cm area using a pre-moistened cotton swab (Malhotra et al., 2016). Digital photographs (Nikon Coolpix 5400) of infected skin were taken and lesion sizes were quantified using Image J (National Institutes of Health Research Services Branch) with a millimeter ruler as a reference. For in vivo bioluminescence imaging (BLI) quantification, mice were anesthetized via inhalation of isoflurane (2%), and imaged on a Lumina III IVIS (PerkinElmer); total flux (photons/s) was measured within a 1 × 103-pixel circular region of interest using Living Image software (PerkinElmer). For rescue experiments, hBD3 (Anaspec) was dissolved in PBS to 100 μM and 50 μL of either hBD3 or PBS was intradermally injected at the infection sites 6 hours post-S. aureus injection. For experiments in Figure 7G-I, IL-1β and Cxcl2 (100ng each, R&D) were co-injected with S. aureus (Miller et al., 2007).
Adoptive transfer of neutrophils
Bone marrows were taken from WT animals under sterile conditions and neutrophils were selected using the MACS Neutrophil Isolation Kit (Miltenyi Biotec). Purified neutrophils were resuspended in PBS and 5×106 cells/100uL were retro-orbitally injected into Mrgpra2 dKO animals 2 hour before S. aureus infection.
Flow Cytometry
10-mm skin punch biopsies were minced and digested in 3 mL RPMI containing 100 μg/mL DNase I (Sigma-Aldrich) and 1.67 Wunsch units/mL Liberase TL (Roche) for one hour at 37 °C on a rotor wheel. Single cell suspensions were generated by passing digested skin samples through 100-μm cell strainers and washed in RPMI and PBS. Live versus Dead cells were stained using Live/Dead Fixable Aqua Dead Cell Stain Kit (Thermo Fisher). Cells were treated with CD16/CD32 Fc Block (S17011E, Biolegend) for 10 min before incubation with the antibody cocktail: CD45-APC/Cy7 (30-F11, Biolegend), CD11b-PE/Dazzle594 (M1/70, Biolegend), CD11c-PE/Cy7 (N418, Biolegend), Ly6C-APC (HK1.4, Biolegend), Ly6G-BV421 (1A8, Biolegend), F4/80-BV605 (BM8, Biolegend) and SiglecF-BB515 (E50-2440, BD Bioscience). Data were collected using a CytoFLEX LX (Beckman Coulter) and analyzed using FlowJo (BD). Neutrophils were gated as live CD45+CD11b+Ly6G+.
Isolation and stimulation of bone marrow neutrophils
Neutrophils were isolated from bone marrow by negative selection using the Neutrophil Isolation Kit with magnetic-activated cell sorting (MACS) columns (Miltenyi Biotec) following manufacturer’s instructions. Purity of neutrophils after selection was validated by flow cytometry (>90%). 105-106 cells were used for each condition. To measure elastase and MPO release, 2×105 cells were resuspended in RPMI medium on a round-bottom 96 well plate and incubated with 0, 1, 5 or 10 μM hBD3 peptide (Anaspec, dissolved in RMPI) or 10 μM mBD14 (Dr. Wuyuan Lu) for one hour and the supernatant was analyzed for elastase and MPO concentrations using DuoSet ELISA kits (R&D Systems). Chemotaxis was measured using Boyden chambers with 8 micron filters. 1×105 cells were seeded in the upper chamber and the lower chambers contained control medium, migration inducer (Abcam) or 10 μM hBD3. The cells were incubated at 37°C for 4 hours and the numbers of cells in the lower chamber were counted. For the measurement of NETosis, 105 cells were seeded in a flat-bottom 96 well plate and incubated with control medium, 10 nM PMA, 10 μM Nigericin, 1mM ATP or 10 μM hBD3 for 4 hours. SYTOX Orange dye (ThermoFisher, 0.25 μM) was added to each well, incubated for 5 min and fluorescence was measured at excitation and emission wavelengths of 540 nm and 580 nm on Flex Station 3 (Molecular Devices) (Jiang et al., 2017). Reactive oxygen species (ROS) generation was measured using a Neutrophil/Monocyte Respiratory Burst Assay Kit (Cayman) following manufacturer’s instructions.
To assay IL-1β and Cxcl2 release, cells were pre-incubated with 1 μg/mL S. aureus lipoteichoic acid (LTA, Invivogen) for 2 hours before hBD3 or mBD14 were added at final concentrations of 10 μM. Another 2 hours later, supernatants were taken, and the remaining cells were lysed in RIPA buffer (Sigma). IL-1β and Cxcl2 concentrations in the supernatants and cell lysates were assayed using DuoSet ELISA (R&D Systems). ATP (5 mM) was used as a positive control. For pharmacological inhibitions (Figure 7E, F), Q-VD-Oph (R&D Systems) or YM254890 (Tocris) were added together with LTA to final concentrations of 10μM. Proteome profiler array (R&D Systems) was performed following manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image quantifications were performed using ImageJ (NIH). All statistical analyses were performed using Prism 9 (GraphPad). Single comparisons were made using two-tailed unpaired Student’s t test. Data for multiple comparisons were analyzed using one-way or two-way ANOVA as specifically indicated in the figure legends. All data are presented as mean ± standard error of the mean (SEM) and values of *p<0.05 were considered statistically significant.
Supplementary Material
HIGHLIGHTS.
Orphan GPCRs Mrgpra2a/b are defensin receptors in neutrophils
Defensin-Mrgpra2 signaling preserves skin microbiome diversity
Deletion of defensins or Mrgpra2 impairs neutrophil-mediated bacterial clearance
Activation of Mrgpra2 by defensins triggers neutrophils to release IL-1β and Cxcl2
ACKNOWLEDGEMENTS
The authors thank Dr. Caiying Guo and the Janelia Farm Gene Targeting and Transgenics Team for generating the Defflox mice, Chip Hawkins at the Johns Hopkins Transgenic Core for generating the Mrgpra2 dKO mice, Christopher Blair at the University of Michigan Microbiome Core for 16S sequencing, and Anuj Gupta at the Johns Hopkins SKCCC Experimental and Computational Genomics Core for 16S data analyses, Dongeun Heo and the Johns Hopkins Transcriptomics and Deep Sequencing Core for RNA-seq. The work was supported by the Howard Hughes Medical Institute (X.Z.D.), a Damon Runyon Cancer Research Foundation Fellowship Award to X.T.D. (DRG-2295-17) and NIH grants 1T32AR074920 and R01AR074846 to L.A.G., R01AI146177, R01AR073665, and R01AR069502 to N.K.A., R01AR070116, R01AR077007 and R21AI167047 to B.S.K., T32GM136577 to R.K. and C.V. and 2R37NS054791 to X.Z.D.
Footnotes
DECLARATION OF INTERESTS
X.Z.D. is a co-founder and a scientific advisory board member of Escient Pharmaceuticals, a company focused on developing small molecules targeting MRGPRs. N.K.A. has received previous grant support from Pfizer and Boehringer Ingelheim, and was a paid consultant for Janssen Pharmaceuticals. L.S.M. is currently a full-time paid employee of Janssen Research & Development, the pharmaceutical companies of Johnson & Johnson, and owns Johnson & Johnson stock. L.S.M. performed all work at his prior affiliation at Johns Hopkins University School of Medicine and he has received prior grant support from AstraZeneca, Pfizer, Boehringer Ingelheim, Regeneron Pharmaceuticals, and Moderna Therapeutics, was a paid consultant for Armirall and Janssen Research and Development, was on the scientific advisory board of Integrated Biotherapeutics and is a shareholder of Noveome Biotherapeutics, which are all developing therapeutics against infections (including S. aureus and other pathogens) and/or inflammatory conditions. B.S.K. has served as a consultant for AbbVie, ABRAX Japan, Almirall, Cara Therapeutics, Maruho, Menlo Therapeutics, Pfizer, and Third Rock Ventures. He has also participated on the advisory board for Almirall, Boehringer Ingelheim, Cara Therapeutics, Kiniksa Pharmaceuticals, Menlo Therapeutics, Regeneron Pharmaceuticals, Sanofi Genzyme, and Trevi Therapeutics. He is also Founder, Chief Scientific Officer, and stockholder of Nuogen Pharma, Inc. He is stockholder of Locus Biosciences. C.V. is a paid consultant and stockholder of AstraZeneca. The other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdul Hamid AI, Cara A, Diot A, Laurent F, Josse J, and Gueirard P (2021). Differential Early in vivo Dynamics and Functionality of Recruited Polymorphonuclear Neutrophils After Infection by Planktonic or Biofilm Staphylococcus aureus. Frontiers in Microbiology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph TE, Tomczak MF, Niederreiter L, Ko H-J, Böck J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, et al. (2013). Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens K, Schunck M, Podda GF, Meingassner J, Stuetz A, Schröder JM, Harder J, and Proksch E (2011). Mechanical and metabolic injury to the skin barrier leads to increased expression of murine β-defensin-1, −3, and −14. The Journal of investigative dermatology 131, 443–452. [DOI] [PubMed] [Google Scholar]
- Ali RS, Falconer A, Ikram M, Bissett CE, Cerio R, and Quinn AG (2001). Expression of the peptide antibiotics human beta defensin-1 and human beta defensin-2 in normal human skin. The Journal of investigative dermatology 117, 106–111. [DOI] [PubMed] [Google Scholar]
- Amid C, Rehaume LM, Brown KL, Gilbert JGR, Dougan G, Hancock REW, and Harrow JL (2009). Manual annotation and analysis of the defensin gene cluster in the C57BL/6J mouse reference genome. BMC Genomics 10, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer NK, Jo J-H, Lee SK, Kim D, Smith B, Ortines RV, Wang Y, Marchitto MC, Ravipati A, Cai SS, et al. (2019). Injury, dysbiosis, and filaggrin deficiency drive skin inflammation through keratinocyte IL-1α release. Journal of Allergy and Clinical Immunology 143, 1426–1443.e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arifuzzaman M, Mobley YR, Choi HW, Bist P, Salinas CA, Brown ZD, Chen SL, Staats HF, and Abraham SN (2019). MRGPR-mediated activation of local mast cells clears cutaneous bacterial infection and protects against reinfection. Science Advances 5, eaav0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros I, Rubio-Ponce A, Genua M, Lusito E, Kwok I, Fernández-Calvo G, Khoyratty TE, van Grinsven E, González-Hernández S, Nicolás-Ávila J, et al. (2020). Co-option of Neutrophil Fates by Tissue Environments. Cell 183, 1282–1297.e1218. [DOI] [PubMed] [Google Scholar]
- Bals R, Wang X, Meegalla RL, Wattler S, Weiner DJ, Nehls MC, and Wilson JM (1999). Mouse β-Defensin 3 Is an Inducible Antimicrobial Peptide Expressed in the Epithelia of Multiple Organs. Infection and Immunity 67, 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N, Sørensen OE, and Theilgaard-Mönch K (2007). Neutrophil granules: a library of innate immunity proteins. Trends in Immunology 28, 340–345. [DOI] [PubMed] [Google Scholar]
- Byrd AL, Belkaid Y, and Segre JA (2018). The human skin microbiome. Nature Reviews Microbiology 16, 143–155. [DOI] [PubMed] [Google Scholar]
- Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, et al. (2021). Structure, function and pharmacology of human itch GPCRs. Nature 600, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassatella MA, Östberg NK, Tamassia N, and Soehnlein O (2019). Biological Roles of Neutrophil-Derived Granule Proteins and Cytokines. Trends in Immunology 40, 648–664. [DOI] [PubMed] [Google Scholar]
- Chen YE, Fischbach MA, and Belkaid Y (2018). Skin microbiota-host interactions. Nature 553, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessa C, Bodet C, Jousselin C, Wehbe M, Lévêque N, and Garcia M (2020). Antiviral and Immunomodulatory Properties of Antimicrobial Peptides Produced by Human Keratinocytes. Frontiers in Microbiology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-C, Cheng W-J, Korinek M, Lin C-Y, and Hwang T-L (2019). Neutrophils in Psoriasis. Frontiers in immunology 10, 2376–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Guo Y, Ramos RI, Hebroni F, Plaisier SB, Xuan C, Granick JL, Matsushima H, Takashima A, Iwakura Y, et al. (2012). Neutrophil-derived IL-1β Is Sufficient for Abscess Formation in Immunity against Staphylococcus aureus in Mice. PLOS Pathogens 8, e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou RC, Kim ND, Sadik CD, Seung E, Lan Y, Byrne MH, Haribabu B, Iwakura Y, and Luster AD (2010). Lipid-cytokine-chemokine cascade drives neutrophil recruitment in a murine model of inflammatory arthritis. Immunity 33, 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. [DOI] [PubMed] [Google Scholar]
- Davis SE, Hopke A, Minkin SC Jr., Montedonico AE, Wheeler RT, and Reynolds TB (2014). Masking of β(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infection and immunity 82, 4405–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo K, Dudeck A, Hasenberg M, Nye E, van Rooijen N, Hartmann K, Gunzer M, Roers A, and Hogg N (2013). Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood 121, 4930–4937. [DOI] [PubMed] [Google Scholar]
- De Filippo K, Henderson RB, Laschinger M, and Hogg N (2008). Neutrophil Chemokines KC and Macrophage-Inflammatory Protein-2 Are Newly Synthesized by Tissue Macrophages Using Distinct TLR Signaling Pathways. The Journal of Immunology 180, 4308. [DOI] [PubMed] [Google Scholar]
- Dillen CA, Pinsker BL, Marusina AI, Merleev AA, Farber ON, Liu H, Archer NK, Lee DB, Wang Y, Ortines RV, et al. (2018). Clonally expanded γδ T cells protect against Staphylococcus aureus skin reinfection. The Journal of Clinical Investigation 128, 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DF, Barrett NA, Austen KF, and Immunological Genome Project, C. (2016). Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol 17, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer PB, Harwig SS, and Lehrer RI (1992). Cryptdins: antimicrobial defensins of the murine small intestine. Infection and immunity 60, 3556–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson JA, Duffau P, Yasuda K, Ortiz-Lopez A, Rothamel K, Rifkin IR, Monach PA, and ImmGen C (2014). Gene expression during the generation and activation of mouse neutrophils: implication of novel functional and regulatory pathways. PLoS One 9, e108553–e108553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, and Núñez G (2012). NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol 13, 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL (2017). Human Skin Is the Largest Epithelial Surface for Interaction with Microbes. The Journal of investigative dermatology 137, 1213–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T (2003). Defensins: antimicrobial peptides of innate immunity. Nature Reviews Immunology 3, 710–720. [DOI] [PubMed] [Google Scholar]
- Gerdol M, Schmitt P, Venier P, Rocha G, Rosa RD, and Destoumieux-Garzón D (2020). Functional Insights From the Evolutionary Diversification of Big Defensins. Frontiers in Immunology 11, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, and Segre JA (2011). The skin microbiome. Nat Rev Microbiol 9, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Cao X, and Zhang K (2022). 16S rRNA Gene Sequencing Revealed Changes in Gut Microbiota Composition during Pregnancy and Lactation in Mice Model. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, and Lemaitre B (2019). Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, and Schroder JM (2001). Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. The Journal of biological chemistry 276, 5707–5713. [DOI] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, and Schröder JM (1997). A peptide antibiotic from human skin. Nature 387, 861–861. [DOI] [PubMed] [Google Scholar]
- Hinrichsen K, Podschun R, Schubert S, Schröder JM, Harder J, and Proksch E (2008). Mouse Beta-Defensin-14, an Antimicrobial Ortholog of Human Beta-Defensin-3. Antimicrobial Agents and Chemotherapy 52, 1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PLJM, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PCM, Traupe H, de Jongh G, den Heijer M, et al. (2008). Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet 40, 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Saffarzadeh M, and Scharffetter-Kochanek K (2017). In vitro Demonstration and Quantification of Neutrophil Extracellular Trap Formation. Bioprotocol 7, e2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Honda T, Ginhoux F, and Egawa G (2019). The immunological anatomy of the skin. Nature Reviews Immunology 19, 19–30. [DOI] [PubMed] [Google Scholar]
- Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WHI, Murray D, Jo JH, Segre JA, Kong HH, and Irvine AD (2017). Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 139, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ND, and Luster AD (2015). The role of tissue resident cells in neutrophil recruitment. Trends in immunology 36, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Malachowa N, and DeLeo FR (2018). Neutrophils and Bacterial Immune Evasion. Journal of Innate Immunity 10, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, and Nagao K (2015). Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity 42, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E, and Kubes P (2013). Neutrophil recruitment and function in health and inflammation. Nature Reviews Immunology 13, 159–175. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Applied and Environmental Microbiology 79, 5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, and Miller LS (2012). Host–pathogen interactions between the skin and Staphylococcus aureus. Current Opinion in Microbiology 15, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T, Afonso PV, Angermann BR, Wang JM, Kastenmüller W, Parent CA, and Germain RN (2013). Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Zasloff M, and Rolff J (2020). Antimicrobial peptides: Application informed by evolution. Science 368, eaau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PHA, Ohtake T, Zaiou M, Murakami M, Rudisill JA, Lin KH, and Gallo RL (2005). Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proceedings of the National Academy of Sciences of the United States of America 102, 3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini G, Famà A, Biondo C, Mohammadi N, Galbo R, Mancuso G, Iannello D, Zummo S, Giardina M, De Gaetano GV, et al. (2020). Neutrophils Enhance Their Own Influx to Sites of Bacterial Infection via Endosomal TLR-Dependent Cxcl2 Production. The Journal of Immunology 204, 660. [DOI] [PubMed] [Google Scholar]
- Ley K, Laudanna C, Cybulsky MI, and Nourshargh S (2007). Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nature Reviews Immunology 7, 678–689. [DOI] [PubMed] [Google Scholar]
- Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, Ortines RV, Kao T, Lee SK, Cai SS, Miller RJ, et al. (2017). Staphylococcus aureus Epicutaneous Exposure Drives Skin Inflammation via IL-36-Mediated T Cell Responses. Cell host & microbe 22, 653–666.e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone AG, Atci E, Renslow R, Beyenal H, Noh S, Fransson B, Abu-Lail N, Park J-J, Gang DR, and Call DR (2015). Colonization of Epidermal Tissue by <span class=" named-content genus-species" id="named-content-1">Staphylococcus aureus</span> Produces Localized Hypoxia and Stimulates Secretion of Antioxidant and Caspase-14 Proteins. Infection and Immunity 83, 3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz S, Aga E, Wilde I, van Zandbergen G, Hartung T, Solbach W, and Laskay T (2004). Highly purified lipoteichoic acid activates neutrophil granulocytes and delays their spontaneous apoptosis via CD14 and TLR2. Journal of leukocyte biology 75, 467–477. [DOI] [PubMed] [Google Scholar]
- Malhotra N, Yoon J, Leyva-Castillo JM, Galand C, Archer N, Miller LS, and Geha RS (2016). IL-22 derived from γδ T cells restricts Staphylococcus aureus infection of mechanically injured skin. J Allergy Clin Immunol 138, 1098–1107.e1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, and Tschopp J (2002). The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of proIL-β. Molecular Cell 10, 417–426. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nakagawa S, Zhang L, Nakamura Y, Villaruz AE, Otto M, Wolz C, Inohara N, and Núñez G (2021). Interaction between Staphylococcus Agr virulence and neutrophils regulates pathogen expansion in the skin. Cell Host & Microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayadas TN, Cullere X, and Lowell CA (2014). The multifaceted functions of neutrophils. Annu Rev Pathol 9, 181–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, and Dong X (2015). Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 519, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, Vial Y, Prod'hom G, Greub G, Kypriotou M, et al. (2017). Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. The Journal of investigative dermatology 137, 2497–2504. [DOI] [PubMed] [Google Scholar]
- Midorikawa K, Ouhara K, Komatsuzawa H, Kawai T, Yamada S, Fujiwara T, Yamazaki K, Sayama K, Taubman MA, Kurihara H, et al. (2003). Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infection and immunity 71, 3730–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LS, and Cho JS (2011). Immunity against Staphylococcus aureus cutaneous infections. Nature Reviews Immunology 11, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LS, O'Connell RM, Gutierrez MA, Pietras EM, Shahangian A, Gross CE, Thirumala A, Cheung AL, Cheng G, and Modlin RL (2006). MyD88 Mediates Neutrophil Recruitment Initiated by IL-1R but Not TLR2 Activation in Immunity against Staphylococcus aureus. Immunity 24, 79–91. [DOI] [PubMed] [Google Scholar]
- Miller LS, Pietras EM, Uricchio LH, Hirano K, Rao S, Lin H, O'Connell RM, Iwakura Y, Cheung AL, Cheng G, et al. (2007). Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. Journal of immunology (Baltimore, Md : 1950) 179, 6933–6942. [DOI] [PubMed] [Google Scholar]
- Miralda I, Uriarte SM, and McLeish KR (2017). Multiple Phenotypic Changes Define Neutrophil Priming. Front Cell Infect Microbiol 7, 217–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, Quinones M, Dzutsev AK, Gao JL, Trinchieri G, et al. (2013). Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G, Kilanowski F, Davidson D, and Dorin J (2002). Characterization of the Mouse Beta Defensin 1, <em>Defb1</em>, Mutant Mouse Model. Infection and Immunity 70, 3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. (2015). Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Science translational medicine 9, eaah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, and Gallo RL (2012). Antimicrobial peptides: old molecules with new ideas. The Journal of investigative dermatology 132, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navid F, Boniotto M, Walker C, Ahrens K, Proksch E, Sparwasser T, Müller W, Schwarz T, and Schwarz A (2012). Induction of regulatory T cells by a murine β-defensin. Journal of immunology (Baltimore, Md : 1950) 188, 735–743. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Di Meglio P, Qin J-Z, and Nickoloff BJ (2009). Skin immune sentinels in health and disease. Nature Reviews Immunology 9, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Haase I, and Nestle FO (2014). Mechanisms regulating skin immunity and inflammation. Nature Reviews Immunology 14, 289–301. [DOI] [PubMed] [Google Scholar]
- Pundir P, Liu R, Vasavda C, Serhan N, Limjunyawong N, Yee R, Zhan Y, Dong X, Wu X, Zhang Y, et al. (2019). A Connective Tissue Mast-Cell-Specific Receptor Detects Bacterial Quorum-Sensing Molecules and Mediates Antibacterial Immunity. Cell Host & Microbe 26, 114–122.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder MJ, McReynolds AKG, Dai H, Pritchard MT, Markiewicz MA, and Bose JL (2022). Kinetic Characterization of the Immune Response to Methicillin-Resistant Staphylococcus aureus Subcutaneous Skin Infection. e0006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrl J, Yang D, Oppenheim JJ, and Hehlgans T (2008). Identification and Biological Characterization of Mouse beta-defensin 14, the orthologue of human beta-defensin 3. The Journal of biological chemistry 283, 5414–5419. [DOI] [PubMed] [Google Scholar]
- Röhrl J, Yang D, Oppenheim JJ, and Hehlgans T (2010). Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. Journal of immunology (Baltimore, Md : 1950) 184, 6688–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik CD, Kim ND, and Luster AD (2011). Neutrophils cascading their way to inflammation. Trends Immunol 32, 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KM, Paterson Y, and Bevins CL (2003). Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422, 522–526. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Inoue A, Gutkind JS, Russell RB, and Raimondi F (2019). PRECOG: PREdicting Coupling probabilities of G-protein coupled receptors. Nucleic Acids Research 47, W395–W401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian H, Gupta K, Lee D, Bayir AK, Ahn H, and Ali H (2013a). β-Defensins activate human mast cells via Mas-related gene X2. Journal of immunology (Baltimore, Md : 1950) 191, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian H, Gupta K, Lee D, Bayir AK, Ahn H, and Ali H (2013b). β-Defensins activate human mast cells via Mas-related gene X2. Journal of immunology (Baltimore, Md : 1950) 191, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumikawa Y, Asada H, Hoshino K, Azukizawa H, Katayama I, Akira S, and Itami S (2006). Induction of beta-defensin 3 in keratinocytes stimulated by bacterial lipopeptides through toll-like receptor 2. Microbes and infection 8, 1513–1521. [DOI] [PubMed] [Google Scholar]
- Tkaczyk C, Hamilton MM, Datta V, Yang XP, Hilliard JJ, Stephens GL, Sadowska A, Hua L, O’Day T, Suzich J, et al. (2013). Staphylococcus aureus Alpha Toxin Suppresses Effective Innate and Adaptive Immune Responses in a Murine Dermonecrosis Model. PLoS One 8, e75103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng PY, and Hoon MA (2022). Specific β-Defensins Stimulate Pruritus through Activation of Sensory Neurons. The Journal of investigative dermatology 142, 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, Lakshmikanth T, Forsstrom B, Edfors F, Odeberg J, et al. (2019). A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366, eaax9198. [DOI] [PubMed] [Google Scholar]
- Wang G, Sweren E, Liu H, Wier E, Alphonse MP, Chen R, Islam N, Li A, Xue Y, Chen J, et al. (2021). Bacteria induce skin regeneration via IL-1β signaling. Cell Host Microbe 29, 777–791 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams H, Crompton RA, Thomason HA, Campbell L, Singh G, McBain AJ, Cruickshank SM, and Hardman MJ (2017a). Cutaneous Nod2 Expression Regulates the Skin Microbiome and Wound Healing in a Murine Model. The Journal of investigative dermatology 137, 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, and Gallo RL (2017). Evidence that Human Skin Microbiome Dysbiosis Promotes Atopic Dermatitis. The Journal of investigative dermatology 137, 2460–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Nakatsuji T, and Gallo RL (2017b). Staphylococcus aureus : Master Manipulator of the Skin. Cell Host & Microbe 22, 579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, and Parks WC (1999). Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117. [DOI] [PubMed] [Google Scholar]
- Wright HL, Thomas HB, Moots RJ, and Edwards SW (2013). RNA-seq reveals activation of both common and cytokine-specific pathways following neutrophil priming. PLoS One 8, e58598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, Yu H, Park S-Y, Guo R, Ren Q, et al. (2020). Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol 21, 1119–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schröder JM, Wang JM, Howard OM, et al. (1999). Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286, 525–528. [DOI] [PubMed] [Google Scholar]
- Yang D, Han Z, and Oppenheim JJ (2017). Alarmins and immunity. Immunol Rev 280, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang G-X, Chen X, Liu L, Yan X, et al. (2021). Structure, function and pharmacology of human itch receptor complexes. Nature 600, 164–169. [DOI] [PubMed] [Google Scholar]
- Zambrano S, Möller-Hackbarth K, Li X, Rodriguez PQ, Charrin E, Schwarz A, Nyström J, Wernerson A, Lal M, and Patrakka J (2019). GPRC5b Modulates Inflammatory Response in Glomerular Diseases via NF-κB Pathway. Journal of the American Society of Nephrology : JASN 30, 1573–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang S, Li B, Li W, Wang J, Chen Z, Yang J, Tan H, and Li J (2019). Alterations of the Mice Gut Microbiome via Schistosoma japonicum Ova-Induced Granuloma. Frontiers in Microbiology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YS, Webb S, Lettice L, Tardif S, Kilanowski F, Tyrrell C, MacPherson H, Semple F, Tennant P, Baker T, et al. (2013). Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice. PLOS Genetics 9, e1003826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data has been deposited at GEO and are publicly available as of the date of publication. Accession number is listed in the Key Resources Table.
All data supporting the findings of this study are available in the manuscript or the supplementary materials and are available upon request to the lead contact author.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-biotin PE clone 1D4-C5 | Biolegend | Cat# 409004; RRID: AB_10641847 |
| Anti-biotin | Abcam | Cat# ab53494; RRID: AB_867860 |
| Sepharose anti-GFP antibody | Abcam | Cat# ab69314; RRID: AB_1640178 |
| Anti-GFP | Thermo Fisher | Cat# A-11122; RRID: AB_221569 |
| Anti-rabbit IgG HRP-conjugated | R&D systems | Cat# HAF008; RRID: AB_357235 |
| Rat monoclonal anti-mouse Ly6G clone 1A8 | BioXCell | Cat# BE0075-1; RRID: AB_1107721 |
| Rat monoclonal IgG2a isotype control clone 2A3 | BioXCell | Cat# BE0089; RRID: AB_1107769 |
| Anti-mouse CD45 APC-Cy7 clone 30-F11 | Biolegend | Cat# 103116; RRID: AB_312981 |
| Anti-mouse CD11b PE/Dazzle594 Clone M1/70 | Biolegend | Cat# 101256; RRID: AB_2563648 |
| Anti-mouse CD11c PE/Cy7 Clone N418 | Biolegend | Cat# 117318; RRID: AB_493568 |
| Anti-mouse Ly6C APC Clone HK1.4 | Biolegend | Cat# 128016; RRID: AB_1732076 |
| Anti-mouse Ly6G BV421 Clone 1A8 | Biolegend | Cat# 127628; RRID: AB_2562567 |
| Anti-mouse F4/80 BV605 Clone BM8 | Biolegend | Cat# 123133; RRID: AB_2562305 |
| Anti-mouse Siglec F BB515 Clone E50-2440 | BD Bioscience | Cat# 564514; RRID: AB_2738833 |
| Anti-mouse c-Kit BV421 Clone ACK2 | Biolegend | Cat# 135124; RRID: AB_2562237 |
| Anti-mouse ICAM FITC Clone YN/1.7.4 | Biolegend | Cat# 116105; RRID: AB_313696 |
| Anti-mouse F4/80 PE Clone BM8 | Biolegend | Cat# 123110; RRID: AB_893486 |
| Anti-mouse FcεRI PE/Cy7 Clone MAR-1 | Biolegend | Cat# 134318; RRID: AB_10640122 |
| Anti-mouse CD11b BV785 Clone M1/70 | Biolegend | Cat# 101243; RRID: AB_2561373 |
| Anti-mouse CD11c BV605 Clone N418 | Biolegend | Cat# 117334; RRID: AB_2562415 |
| Anti-mouse I-A/I-E BV711 Clone M5/114.15.2 | Biolegend | Cat# 107643; RRID: AB_2565976 |
| Anti-mouse Ly6G PE/Cy7 Clone 1A8 | Biolegend | Cat# 127617; RRID: AB_1877262 |
| Anti-mouse CD11b PE Clone M1/70 | Biolegend | Cat# 101207; RRID: AB_312790 |
| Anti-mouse B220 APC Clone RA3-6B2 | eBioscience | Cat# 17-0452-82; RRID: AB_469395 |
| Anti-mouse Ly6G AF700 Clone 1A8 | Biolegend | Cat# 127622; RRID: AB_10643269 |
| Anti-mouse CD49b PE Clone DX5 | Biolegend | Cat# 108908; RRID: AB_313415 |
| Rabbit anti-mouse BD14 | MyBioSource | Cat# MBS1490249; RRID: AB_2732866 |
| Rat anti-mouse Ly6G | Biolegend | Cat# 127602; RRID: AB_1089180 |
| Rabbit anti-S. aureus | Abcam | Cat# 20920;RRID: AB_445913 |
| Alexa Fluor ® 488 goat anti-rabbit IgG | Thermo Fisher | Cat# A-11008; RRID: AB_143165 |
| Alexa Fluor ® 488 goat anti-rat IgG | Thermo Fisher | Cat# A-11006; RRID: AB_2534074 |
| Alexa Fluor ® 594 goat anti-rabbit IgG | Thermo Fisher | Cat# A-11012; RRID: AB_2534079 |
| Goat anti-mouse IL-1β | R&D systems | Cat# AF-401-NA; RRID: AB_416684 |
| Rabbit anti-cleaved-IL-1β | Cell Signaling Technology | Cat# 63124s |
| Rabbit anti-goat IgG HRP-conjugated | R&D systems | Cat# HAF017; RRID: AB_562588 |
| HRP anti-beta Actin | Abcam | Cat# Ab49900; RRID: AB_867494 |
| Bacterial and virus strains | ||
| Staphylococcus aureus SF8300 | Binh Diep (UCSF) | N/A |
| USA300 | Tammy Kielian (University of Nebraska) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| hBD3 | Anaspec | Cat# AS-60741 |
| Biotin-hBD3 | Anaspec | N/A |
| mBD14 | Wuyuan Lu (University of Maryland Baltimore) | N/A |
| mBD3 | Wuyuan Lu (University of Maryland Baltimore) | N/A |
| S. aureus derived LTA | Invivogen | Cat# Tlrl-pslta |
| Tryptic soy broth | Millipore | Cat# 22092 |
| TRIzol reagent | Thermo Fisher | Cat# 15596018 |
| Liberase TL | Roche | Cat# 05401054001 |
| DNase I | Sigma-Aldrich | Cat# 4536282001 |
| Recombinant IL-1β | R&D systems | Cat# 401-ML |
| NuPAGE™ LDS Sample Buffer | Thermo Fisher | Cat# NP0007 |
| Q-VD-Oph | R&D systems | Cat# OPH001-01M |
| YM254890 | Tocris | Cat# 7352 |
| SYTOX blue | Thermo Fisher | Cat# S34857 |
| SYTOX red | Thermo Fisher | Cat# S34859 |
| SYTOX orange | Thermo Fisher | Cat# S11368 |
| Critical commercial assays | ||
| Mouse Elastase DuoSet ELISA Kit | R&D systems | Cat# DY4517-05 |
| Mouse MPO DuoSet ELISA Kit | R&D systems | Cat# DY3667 |
| Mouse S100A8/A9 DuoSet ELISA Kit | R&D systems | Cat# DY8596-05 |
| Mouse IL-1β DuoSet ELISA Kit | R&D systems | Cat# DY401-05 |
| Mouse Cxcl2 DuoSet ELISA Kit | R&D systems | Cat# DY452-05 |
| Mouse BD14 ELISA Kit | MyBioSource | Cat# MBS2515829 |
| MACS neutrophil isolation kit | Miltenyi Biotec | Cat# 130-097-658 |
| RNAscope® Multiplex Fluorescent Detection Kit v2 | ACDBio | Cat# 323110 |
| RNAscope® Ly6g probe | ACDBio | Cat# 455701 |
| RNAscope® Mrgpra2 probe | ACDBio | Custom-made |
| RNeasy Plus Mini kit | Qiagen | Cat# 74034 |
| Chemotaxis assay kit | Abcam | Cat# Ab235692 |
| Cellular reactive oxygen species detection assay kit | Abcam | Cat# Ab113851 |
| Deposited data | ||
| RNA-seq data | NCBI GEO | GSE178507 |
| Experimental models: Cell lines | ||
| Gα15 HEK 293 cells | This study | N/A |
| Mrgpra2b-GFP; Gα15 HEK 293 cells | This study | N/A |
| MRGPRX3-FLAG; Gα15 HEK 293 cells | This study | N/A |
| MRGPRX4-GFP; Gα15 HEK 293 cells | This study | N/A |
| Experimental models: Mice | ||
| C57BL/6J | Jackson laboratory | Cat# 000664 |
| Mrgpra2 dKO | This study | N/A |
| K14-cre; Defflox/flox | This study | N/A |
| K14-cre | Jackson laboratory | Cat# 018964 |
| Oligonucleotides | ||
| gRNAs targeting Mrgpra2a/b CAAGTTAGTTTTGGTCAATC, GATCTGATACCCTCTTCTGG, TAAACTGGCTATCTACTTGA, TTATCCAATGCTGGGATAAT | This study | N/A |
| Genotyping primers for Mrgpra2 dKO GTCAGCATGATGGTCAC (WT-F), CATAGGGAAGGCCTCATGTCTC (WT-R), CTATGGAGGGATGCTCATTACTG (Mrgpra2 dKO-F), CAAGATCTGACTCCCTCTTCTGG (Mrgpra2 dKO-R) | This study | N/A |
| gRNA targeting Defb13 CCAAGCCAATTTCCTGTGCTGCA | This study | N/A |
| gRNA targeting Defb40 CCATGGGCATTTCTGAATGTGAC | This study | N/A |
| Primers for genotyping Def cluster deletion GATGCAGGTGCCCTGAAATAT (F) CACTGAATAGACCTGACTTCC (R) | This study | N/A |
| RT-PCR primer for Mrgpra2 CCTCCTACACAAGCCAGCAA (F) AAGCACAAGTGAAAGATGATGCT | This study | N/A |
| RT-PCR primer for Gapdh TGTTCCTACCCCCAATGTGT (F) TGTGAGGGAGATGCTCAGTG (R) | Zambrano et al., 2019 | N/A |
| Software and algorithms | ||
| FlowJo | BD | https://www.flowjo.com/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://fiji.sc/ |
| R | R | https://www.r-project.org/ |
| QIIME2 | QIIME2 development team | https://qiime2.org/ |
| STAR - 2.7.3a | Alexander Dobin | https://github.com/alexdobin/STAR/releases/tag/2.7.9a |
| Partek Flow | Partek | https://www.partek.com/ |
| Partek GS v7.0 | Partek | https://www.partek.com/ |
| Spotfire | TIBCO | https://www.tibco.com/products/tibco-spotfire |