Abstract
The nucleolus is a multi‐compartment, non‐membrane‐bound organelle within the nucleus. Nucleolar assembly is influenced by proteins capable of phase separation. Xenopus laevis oocytes contain hundreds of large nucleoli that provide experimental access for nucleoli that is unavailable in other systems. Here we detail methods to streamline the in vivo analysis of the compartmentalization of nucleolar proteins that are suspected of phase separation. The nucleolus is the main hub of ribosome biogenesis and here we present data supporting the division of proteins into nucleolar domains based on their function in ribosome biogenesis. We also describe the use of vital dyes such as Hoechst 33342 and Thioflavin T in nucleolar staining. Additionally, we quantify nucleolar morphology changes induced by heat shock and actinomycin D treatments. We suggest these approaches will be valuable in a variety of studies that seek to better understand the nucleolus, particularly those regarding phase separation. These approaches may also be instructive for other studies on phase separation, especially in the nucleus.
Keywords: biomolecular condensates, cell nucleolus, fluorescence microscopy, oocytes, Xenopus laevis
1. INTRODUCTION
Distinct compartments within eukaryotic cells allow the efficient organization of many cellular processes. Some of this separation comes from membrane‐bound organelles, such as mitochondrion or nuclei. However, a recent paradigm shift in understanding compartmentalization relates to the formation of non‐membrane‐bound structures mediated by protein condensates and aggregates (Fassler et al., 2021; Hyman et al., 2014; Maji et al., 2009). Some of the critical, early studies on protein condensates were carried out in vitro and established rosters of proteins that would aggregate either by themselves or with aid from a co‐aggregate like RNA (Han et al., 2012; Kato et al., 2012). Proteins that can form condensates typically have low complexity and intrinsically disordered regions (IDRs), which are thought to be necessary for protein condensate formation (Alberti et al., 2019; Boeynaems et al., 2019; Feric et al., 2016; Kato et al., 2012; Kato & McKnight, 2017). Condensates may take the form of liquid–liquid phase‐separated droplets, gels, and amyloids, all of which have been reported in vitro and in vivo. In vivo studies on protein aggregates have used yeast, nematodes, fruit flies, tissue culture cells, and Xenopus laevis oocytes (Banani et al., 2016; Berry et al., 2018; Boke et al., 2016; Brangwynne et al., 2011; Decker & Parker, 2012; Fassler et al., 2021; Feric et al., 2016; Hayes & Weeks, 2016; McSwiggen et al., 2019). Studies that described the phase separation of two nucleolar proteins, nucleoplasmin (Npm1) and fibrillarin (Fbl), provided a visually memorable example of protein phase separation within a nucleolus from X. laevis oocytes (Feric et al., 2016; Hayes et al., 2018). Here we will discuss our use of X. laevis oocytes in studying nucleolar condensates and the advantages of the system.
Historical studies outlined three distinct nucleolar domains using electron microcopy which correspond to sequential steps in ribosome biogenesis. The domains are the fibrillar center where rRNA transcription occurs, the dense fibrillar component where rRNA is modified and processing begins, and the granular component where processing continues and ribosome assembly occurs (Fawcett, 1981; Mais & Scheer, 2001). The nucleolar domains track with the phase separation of sentinel proteins such as Fbl in the dense fibrillar component and Npm1 in the granular component (Feric et al., 2016). However, as important as Npm1 and Fbl are, there are more than 100 proteins in the nucleolus that play a role in ribosome biogenesis and other cellular activities, many of which are suspected of phase separation (Banski et al., 2010; Boisvert et al., 2007; Chen & Huang, 2001; Hayes et al., 2018; Hayes & Weeks, 2016).
We carried out a proteomic study to identify aggregating proteins in the nucleus based on isolation conditions that allow soluble proteins to diffuse away, selectively retaining proteins either in aggregates or stably associated with aggregates (Carroll & Lehman, 1991; Hayes et al., 2018; Paine et al., 1983). Many of these proteins were also identified in studies aimed at characterizing protein aggregates or in studies on the nucleolus (Andersen et al., 2005; Chen & Huang, 2001; Feric et al., 2016; Kar et al., 2011; Kato et al., 2012; Lam & Trinkle‐Mulcahy, 2015; Wang et al., 2019).
The approach and data we present here examine the compartmentalization of proteins in the nucleolus. mRNA encoding a protein of interest in frame with a monomeric fluorescent protein was injected into Xenopus oocytes. The ability to direct protein synthesis controlled by injection of mRNA into Xenopus oocytes has been used for over 50 years (Gurdon et al., 1971). The method has been refined over time by dozens of laboratories to produce protein that is synthesized in a living cell, where posttranslational modification, processing, distribution, and interaction with other proteins are all possible. Our particular interest is in the distribution and behavior of phase‐separating nucleolar proteins.
2. MATERIALS AND METHODS
2.1. Oocyte removal and preparation
Wild‐type X. laevis frogs were obtained from Xenopus‐1 (Dexter Michigan, USA), Nasco, or from the National Xenopus Resource (at the Marine Biological Laboratory RRID:SCR_013731, Woods Hole, MA). We note that the Nasco Colony is now managed by Xenopus‐1. Frogs were anesthetized by immersion in 0.08%–0.1% tricaine (FDA approved, Syndel, USA) in water buffered with NaHCO3 (pH 7.2) for 15 min. Ovary was aseptically removed and placed in Oocyte Ringers Solution 2 (OR2, 82.5 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 5 mM HEPES, and NaOH to pH 7.8). Follicle cells were removed either manually or with collagenase.
2.2. Collagenase treatment
Ovary was sectioned into lobes and placed in 0.2% collagenase (Worthington, type I) in Ca2+‐free OR2 and rocked at room temperature for 60–90 min. Oocytes were then washed in Ca2+‐free OR2 supplemented with 0.1% BSA three times followed by five washes with OR2 at room temperature (5 min each). Residual follicle cells were manually removed with watchmaker forceps. Prior to mRNA injection or other treatment, oocytes were left in OR2 at 13°C overnight to recover from collagenase treatment. Oocytes that have follicle cells removed manually can be injected without overnight recovery (Smith et al., 1991). All protocols were approved by the University of Iowa Office of Animal Resources and the Institutional Animal Care and Use Committee.
2.3. Oocyte mRNA injection
mRNA encoding each desired fluorescent fusion protein (10 nl containing 100 ng/μl) was injected directly into each defolliculated oocyte (stage V–VI) using a glass needle paired with a Singer MK‐1 (Somerset, England) and an Inject+Matic injector (Geneva, Switzerland). After injection, oocytes were incubated in Oocyte Culture Medium (OCM; 60% Leibovitz's L‐15 medium [containing L‐glutamine], 100 μg/ml Pen‐Strep, 0.4 mg/ml [all components purchased from Sigma, USA]) at 13°C for at least 24 h prior to microscopy or treatment.
2.4. Fluorescent fusion protein constructs and in vitro synthesis of mRNA
Fbl and Npm1 fluorescent fusion‐encoding plasmids (inserted into pCS2+) were gifts from Cliff Brangwynne (Princeton University, Princeton, New Jersey) and were described in Feric et al. (2016). Other fluorescent fusion constructs were made by amplifying a gene of interest (purchased from Transomic Technologies) using NEB's Q5 PCR and Gibson Assembly Cloning mix (New England Biolabs, E5510S) to insert it into pRN3P (Zernicka‐Goetz et al., 1996). The RN3P plasmid was a gift from John Gurdon (Cambridge University, UK). Plasmids were then transformed into Escherichia coli DH5α. Some clones with large IDRs required growth in NEB Stable Competent E. coli (High Efficiency) (Gar1 and Ncl). See Table S1 for a comprehensive list of proteins, their product numbers, and the primers used to make fluorescent fusions.
The final plasmids have the following features (listed in 5′➔3′ order): either an Sp6 or T3 RNA polymerase promoter, a β‐globin 5′ ribosome binding site, our cDNA gene of interest, an in‐frame fusion of a fluorescent protein (mGFP5, or RFPs either mKate, mRed, or mCherry [see supplemental table 1]), the 3′ β‐globin untranslated region, and a span of 30 A's and 30 G's. We note that for studies on protein condensates, monomeric fluorescent proteins help avoid the appearance of aggregation due to the fluorescent protein itself forming multimers (Costantini et al., 2012). We used fluorescent proteins reported by others to be monomeric. A large database of fluorescent proteins including known aggregation tendencies can be found at https://www.fpbase.org/ (Lambert, 2019). A sample plasmid map can be seen in Figure S1. We tested several of the proteins with the fluorescent protein fusions placed on the N‐terminal end and saw similar results. DNA templates for in vitro synthesis of mRNA were made using Q5 polymerase (NEB) directed PCR with either m13F and m13R primers for pRN3P‐based clones or M13R and custom primer CATTCTGCCTGGGGACGTC for pCS2+ plasmids. mRNA encoding the fluorescent fusion protein was made using either the SP6 (for Fbl and Npm1) or T3 (for all others) mMessage mMachine in vitro transcription kit following the recommended protocol with lithium chloride RNA extraction (Invitrogen). The concentration of the mRNA was determined using a Nanodrop spectrophotometer (Thermo Scientific) and full‐length product was confirmed using gel electrophoresis. mRNA aliquots were stored at −80°C until use.
2.5. Microscopy
Nuclei were manually isolated from oocytes under a dissection microscope in OR2 using watchmaker's forceps. Representative samples were often previewed using a Zeiss Discovery V8 dissecting microscope with fluorescence. Samples that were analyzed at higher magnification were immediately mounted on a glass microscope slide (in a well with petrolatum sides) and covered with a cover slip. The Zeiss filter sets we used were 1 (Hoechst), 37 (GFP and RNA stains), 43 (RFP), and 47 (ThioT). Images were acquired with an AxioPlan or ApoTome fluorescence microscope running Axio Vision software (RRID:SCR_002677) and an AxioCam Mrm or AxioCam MrC5 camera (Zeiss). The images were acquired using Zeiss's ApoTome with a 0.25 μm slice size unless otherwise stated. ApoTome images are comparable to confocal microscopy images. Images were processed with Zen 3.3 (blue edition) for further analysis. Images acquired with a 63× objective have 2752 × 2208 pixels for the image size of 198.32 μm × 159.12 μm.
2.6. Image analysis
For each experimental condition, 15 or more nucleoli from each of three nuclei were imaged. Each experiment was repeated at least three times using oocytes from three different frogs. We note that the level of autofluorescence of samples varies from frog to frog. To ensure any fluorescence measured was from fluorescent fusions and not autofluorescence, images were taken of un‐injected nuclei at the beginning of each experiment. We typically used fluorescent fusions of either Fbl and Npm1 across all frogs to account for normal variation in size, shape, and number of dense fibrillar components per nucleolus. Differences in size, shape, component proportion, etc., in Figure 1 are all within normal nucleoli variation. We concentrated our studies on nucleoli with multiple dense fibrillar components in each nucleolus.
FIGURE 1.
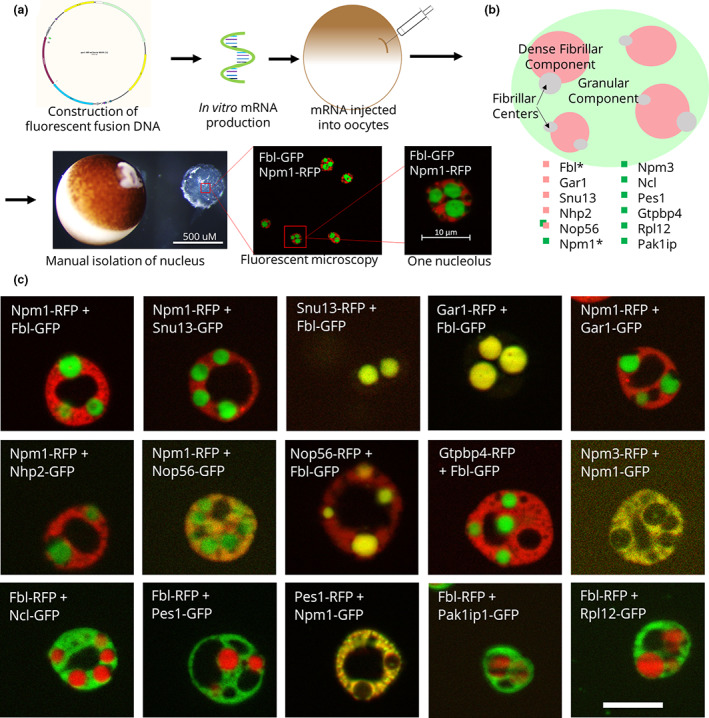
Fluorescent fusion protein nucleolar localization. (a) Schematic of fluorescent fusion protein expression within Xenopus laevis oocyte nucleoli, showing the expression of the canonical domain markers Npm1 (fused with RFP) and Fbl (fused with GFP). (b) Model of the localization of each of these proteins. *Canonical domain markers. (c) Images of representative nucleoli with the labeled fluorescent fusion expressed with either Npm1 or Fbl, the canonical domain markers for the granular component and the dense fibrillar component, respectively. Scale bar = 10 μm
2.7. Nucleolar staining
Nucleolar DNA was stained with Hoechst 33342 (ThermoFisher). We stained nucleolar RNA using either Sybr Green II (ThermoFisher, a gel stain) or Syto Green (ThermoFisher, a cell stain). We used Thioflavin T (Sigma Aldrich) as an indicator of possible protein amyloid structure. Hoechst (20 μM), Sybr Green II (to 1×), Syto Green (500 nM), or Thioflavin T (50 μM) was added to the OR2, and nuclei were soaked for 1–2 min, rinsed in OR2, and then imaged immediately as described above.
2.8. Heat shock treatment
Oocytes were placed in OCM preheated at 37°C for 1 h (Bienz & Gurdon, 1982; Bouche et al., 1981). Control oocytes were incubated at 13°C (incubation temperature) for 1 h (note: room temperature controls produced the same results). For heat shock recovery experiments, oocytes were placed at 13°C for 1 h after heat shock. Immediately following treatment, nuclei were imaged as described above. For each dataset, color channels in images were split and then the punctate spots, marked by a dense fibrillar component protein, were manually counted in each nucleolus to determine if there was a change in the ratio of punctate spots per nucleolus before and after treatment. Nucleoli that had too many punctate spots to reasonably count were given a value of 20 for statistical purposes. An unpaired t‐test of this ratio using GraphPad Prism (RRID:SCR_002798) was used to determine if the change was significant (p < .05).
2.9. Actinomycin D treatment
Oocytes were incubated in 20 μg/ml actinomycin D (Sigma Aldrich) in OR2 for 6 h at room temperature (Mais & Scheer, 2001; Scheer et al., 1975). Afterwards, nuclei were isolated in OR2 and immediately imaged as described above. Images were blinded, and the area of the granular component and dense fibrillar component was determined using ImageJ (RRID:SCR_003070) by first splitting the color channels and then using the “Analyze Particles” function after setting a threshold that was consistent for every image in the dataset. Additionally, the number of dense fibrillar components per nucleolus was counted. RStudio (RRID:SCR_000432) was used to determine if there was a significant difference between the areas and the number of dense fibrillar components per nucleolus before and after treatment using a t‐test (p < .05).
2.10. Identification of published proteome data and protein analysis
We found it useful to examine published proteomic data as we planned experiments. For example, we used the supplemental tables in Hayes et al. (2018) as a starting point to identify nucleolar proteins with a nuclear pool resistant to rapid diffusion out of the nucleus during isolation in OR2 (Hayes et al., 2018). We also found the whole egg proteome and the nuclear proteomes in the supplemental data published by Wühr et al. (2014) very helpful as well as supplemental protein data in Kato et al. (2012) and Han et al. (2012). There is a growing resource of proteomic data for phase‐separating proteins within the entire cell, including the nucleolus. These proteomes are almost always found in supplemental information published with papers of interest. Another resource for proteomic information specific to Xenopus can be found on Xenbase (https://www.xenbase.org, RRID:SCR_003280) under the protein expression tab. There, data accumulated by Peshkin et al. (2019) can be viewed for a variety of Xenopus proteins. We also predicted the IDRs, nuclear localization sequences (NLSs), and nucleolar localization sequences (NoLSs) of each protein of interest using the predictors PONDR‐fit, seqNLS, and NoD, respectively (Lin & Hu, 2013; Scott et al., 2010; Xue et al., 2010; RRID:SCR_000997). Visualization of monomer structures used the artificial intelligence‐based protein structure predictions in AlphaFold (Jumper et al., 2021). As of May 11, 2022, over 5000 predicted structures of X. laevis proteins have been predicted at https://alphafold.com/search/text/Xenopus%20laevis?suggested=true.
3. RESULTS
We chose proteins for this study from Hayes et al.'s (2018) proteomic study as described above and made a monomeric fluorescent protein fusion for each. A comprehensive list of these proteins, their functions, and information about the presence of IDRs, NLS, and NoLSs can be found in Table 1. mRNA of each fluorescent fusion, along with mRNA encoding a domain marker (either Npm1 or Fbl), was made in vitro and then injected into stage V–VI oocytes (Figure 1a). Oocytes were incubated in OCM at 13°C for at least 24 h (up to 96 h) prior to imaging to allow the fluorescent fusions to be expressed, modified, and localized with their endogenous counterparts. Oocytes can also be incubated at 18°C for accelerated protein expression, with a shorter survival period.
TABLE 1.
Comprehensive list of the proteins shown in this study (column 1) with the function of each protein (column 2) and information about the presence of a predicted intrinsically disordered region (IDR), nuclear localization sequence (NLS), or nucleolar localization sequence (NoLS) as predicted by Pondr‐Fit, seqNLS, and NoD, respectively
| Protein | Function | Citation | Size (kDa) | Presence of IDR/NLS/NoLS (Y = yes, N = no, M = minor [<25 aa disordered]) | ||
|---|---|---|---|---|---|---|
| IDR | NLS | NoLS | ||||
| Fbl a | Member of the rRNA methylation box C/D complex | Iyer‐Bierhoff et al. (2018) | 35 | Y | N | N |
| Npm1 a | Ribosome nuclear export | Lindstrom (2011) | 40 | Y | Y | N |
| Gar1 | Member of the rRNA pseudouridination H/ACA complex | Hamma and Ferre‐D'Amare (2010) | 22 | Y | N | N |
| Nhp2 | Member of the rRNA pseudouridination H/ACA complex | Hamma and Ferre‐D'Amare (2010) | 16 | M | N | Y |
| Snu13 | Member of the rRNA methylation box C/D complex | Watkins and Bohnsack (2012) | 14 | M | N | N |
| Nop56 | Member of the rRNA methylation box C/D complex | van Nues et al. (2011) | 59 | Y | Y | Y |
| Pes1 | Member of the PeBoW complex, involved with cleavage of rRNA | Rohrmoser et al. (2007) | 67 | Y | Y | Y |
| Gtpbp4 | Late maturation of the 60X ribosomal subunit | Fuentes et al. (2007) | 46 | Y | Y | Y |
| Npm3 | Negative regulator of Npm1 | Huang et al. (2005) | 19 | Y | Y | N |
| Ncl | Multi‐functional protein, ribosome assembly and pre‐rRNA transcription | Bouvet et al. (1998); Ginisty et al. (1998) | 73 | Y | Y | Y |
| Rpl12 | Large ribosomal subunit | Imami et al. (2018) | 18 | M | N | Y |
| Pak1ip1 | Large ribosomal subunit assembly | Sloan et al. (2013) | 41 | M | Y | Y |
Canonical domain markers.
Nuclei were isolated from the oocytes in OR2. Unfixed nuclei were then imaged using ApoTome fluorescence microscopy. The contents and activities within each domain have previously been proposed to represent the sequential steps of ribosome biogenesis, where proteins involved in early, intermediate, and late stages localize to the fibrillar center, dense fibrillar component, and granular component, respectively (Figure 1b). Figure 1c shows representative nucleoli containing fluorescent fusion proteins, including examples of dense fibrillar component‐ and granular component‐localizing proteins. The proteins that co‐localized with Fbl in the dense fibrillar component include those involved in rRNA modification, such as Snu13, Gar1, Nhp2, and Nop56. Proteins involved in ribosome assembly, maturation, and export, such as Gtpbp4, Npm3, Ncl, and Pes1, co‐localize with Npm1 in the granular component. We note that the crisp phase separation seen when comparing Fbl and Npm1 as markers of the dense fibrillar component and the granular component, respectively, extends to some but not all the nucleolar proteins. For example, while Nop56 primarily localizes to the dense fibrillar component, it also partially partitions into the granular component (Figure 1c). Dense fibrillar component localization was anticipated, as Nop56 is part of the same rRNA methylation complex as the canonical dense fibrillar component domain marker Fbl. New models of the nucleolus will need to account for proteins that drift between compartments. While the proteins that join the dense fibrillar component seem homogeneously distributed at the resolution obtained, the granular component protein localization was not as evenly overlapping (see Npm1 and Pes1 image from Figure 1c). These findings indicate that a more nuanced model may need to include additional sub‐compartments.
In addition to fluorescent protein fusions studies, DNA or RNA within the nucleolus can be visualized using fluorescent stains that diffuse into the nucleus. Nuclei were isolated in OR2, and then stains were added to the solution. Hoechst 33342 stains DNA, including the extra‐chromosomal rDNA in the fibrillar center of nucleoli. This makes it an easily accessible marker for the fibrillar center (Figure 2). We have shown previously that Sybr Green II stains nucleoli (Hayes & Weeks, 2016), and here we show that Syto Green can also be used to stain RNA within the nucleolus. Both primarily stain the granular component (Figure 2). Syto Green was slightly brighter than Sybr Green II at the recommended concentrations.
FIGURE 2.
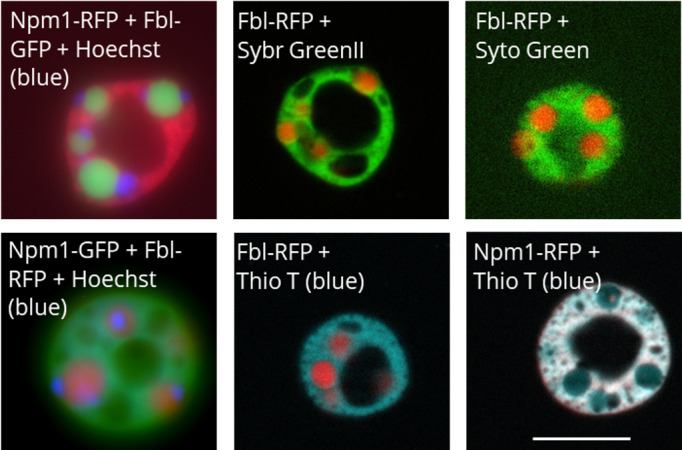
Representative images of Hoechst 33342, Sybr Green II, Syto Green, and Thioflavin T nucleolar staining as indicated by the label on each image. The Hoechst images are whole field fluorescence images, while all other images are apotome section fluorescence images. Scale bar = 10 μm
Thioflavin T is a probe that was developed to identify amyloid structures in neurodegenerative diseases such as Alzheimer's disease and we have detailed its use in oocytes previously (Hayes & Weeks, 2016; Vassar & Culling, 1959). Thioflavin T is a small molecule that fluoresces when bound to amyloid structures (Vassar & Culling, 1959). In solution, Thioflavin T molecules have free rotation around a central carbon bond that is restricted when bound to amyloid structures, which releases energy as fluorescence. While Thioflavin T is commonly used to detect amyloid structures, some ordered RNA structures can also be stained by it (Xu et al., 2016). In vitro phase separation studies have shown that RNA has the ability to increase the order of protein condensates (Feric et al., 2016; Kato et al., 2012). Whether Thioflavin T is indicating amyloid structures or ordered RNA, it does stain the nucleolus, primarily the granular component.
The nucleolus is dynamic and must be able to respond to various stresses. Here we show how morphological changes after heat shock and actinomycin D treatment can be assessed (Figures 3, 4, 5). Heat shock invokes an overall stress response. As part of this response, ribosomal RNA processing is compromised and total rRNA accumulation is reduced (Labhart & Reeder, 1987). Actinomycin D inhibits RNA polymerase I, which stops the production of new rRNA in stage V–VI oocytes (Roger et al., 2002). We compared nucleolar morphology changes after heat shock and actinomycin D treatment, as both compromise ribosome biogenesis. While we expected the morphological changes induced by the two treatments to be similar, we found that they cause almost opposite phenotypes.
FIGURE 3.
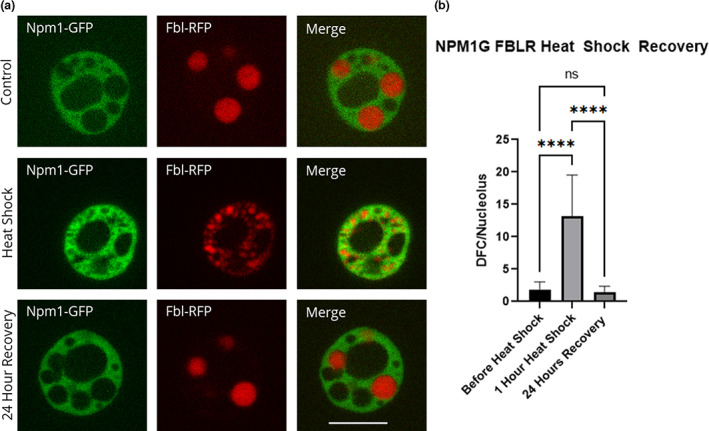
Recovery of fibrillarin after heat shock. Oocytes were heat shocked at 37°C for 1 h (controls were left at 13°C) in OCM and then recovered at 13°C for 24 h in OCM. (a) Representative images of nucleoli expressing fluorescently labeled Npm1 and Fbl after heat shock treatment and recovery. All images are of unfixed samples. (b) Graphical representation of a heat shock and recovery treatment showing the number of punctate spots in the control vs. heat shock vs. recovery as represented by the dense fibrillar component protein fibrillarin (p < .05). Scale bar = 10 μm
FIGURE 4.
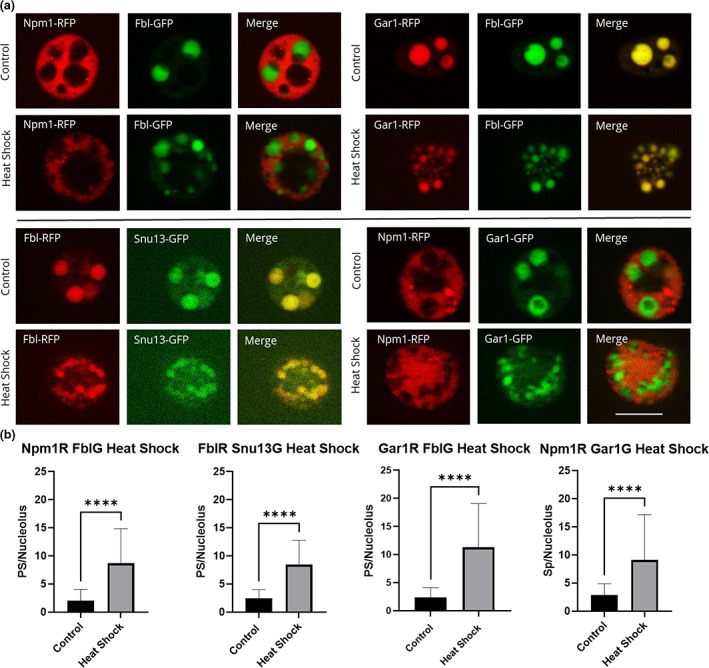
Nucleolar proteins after heat shock. Oocytes were heat shocked at 37°C for 1 h (controls were left at 13°C) in OCM for 1 h. (a) Representative images of nucleoli expressing fluorescently labeled proteins after heat shock treatment. RFP‐fused proteins are shown in the first column of, GFP‐fused proteins are shown in the second column, the merged images are shown in the third column. All images are of unfixed samples. Scale bar = 10 μm. (b) Graphical representation of a dataset of heat shock treatments showing the number of punctate spots per nucleolus from oocytes treated at 13°C (control) or 37°C (heat shock) for 1 h as represented by the dense fibrillar component localization protein indicated in either red or green. PS = punctate spots, which are partial dense fibrillar components. Each experiment was repeated three times, all producing significant changes between untreated and heat shocked nucleoli (p < .05). Scale bar = 10 μm
FIGURE 5.
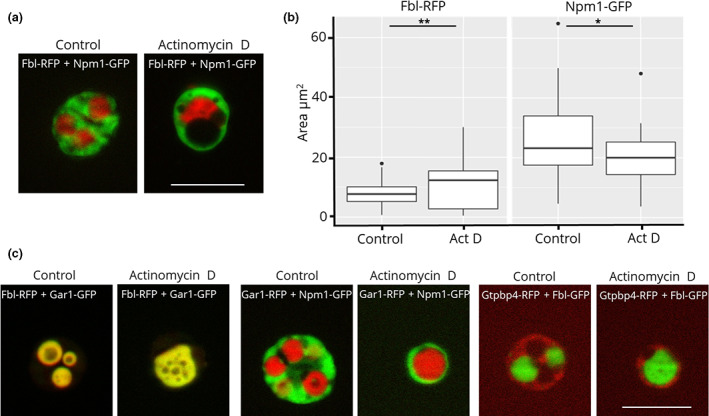
(a) Fluorescence microscopy images of actinomycin D treatment of nucleoli from oocytes injected with Fbl‐RFP and Npm1‐GFP. (b) Quantification of the area occupied by Fbl and Npm1 before and after actinomycin D treatment. (c) Representative images of actinomycin D‐treated Fbl‐RFP/Gar1‐GFP, Gar1‐mCherry/Npm1‐GFP, and Gtpbp4‐mCherry/Fbl‐GFP. Scale bar = 10 μm
Oocytes were heat shocked at 37°C for 1 h. To determine optimal heat shock treatment, we tested heat shock durations from 20 min to 2 h. We found that the greatest morphological changes occur after 1 h and the difference between 1 and 2 h was negligible. Optimization of temperature was done previously (Bienz & Gurdon, 1982; Labhart & Reeder, 1987). We also found that oocytes ruptured when heat shocked at temperatures higher than 37°C. After heat shock, nuclei were isolated and nucleoli were imaged (Figures 3a and 4a). We manually counted the number of punctate spots in the widest apotome slice of nucleoli comparing heat shocked versus control oocytes (Figures 3b and 4b). We counted the number of punctate spots per nucleolus and found that the dense fibrillar components breaks apart and begin to diffuse into the granular component after heat shock (Figures 3 and 4). Heat shock morphology can be reversed by incubation at 13°C. As shown in Figure 3, Fbl and Npm1 are restored to their positions in the dense fibrillar component and granular component, respectively, after recovery. Other proteins that localize to the dense fibrillar component were tested and mimic the pattern shown for Fbl in response to heat shock (Figure 4).
In contrast to heat shock, incubating oocytes in 20 μg/ml actinomycin D for 6 h increases the area of and decreases the number of dense fibrillar components per nucleolus (Figure 5a,b). Additionally, the area occupied by the granular component, as marked by Npm1, decreases upon actinomycin D treatment. Other proteins that localize to the dense fibrillar component or the granular component follow the same trend after treatment, as they co‐localize with Fbl and Npm1, respectively (Figure 5c). These results mirrored earlier studies in HeLa cells that were observed via electron microscopy (Reynolds et al., 1964); however, X. laevis oocyte nuclei provide a system where unfixed samples are observed and isolation and treatment conditions can be easily modified. Previous studies have suggested that dense fibrillar component condensing is due to the collapse of the inactive chromosomal rDNA pulling the tethered proteins of the dense fibrillar components together (Tchelidze et al., 2017). Xenopus laevis oocytes have hundreds of nucleoli that surround circular, extra‐chromosomal rDNA, yet the collapse of the dense fibrillar component after actinomycin D treatment still occurs. It is important to note that actinomycin D treatment 72 h or more post‐mRNA injection did not always produce statistically significant differences in the ratio of dense fibrillar components per nucleolus. This was because as the oocytes age, the control groups in some experiments had a lower number of dense fibrillar components per nucleolus. We recommend actinomycin D treatment experiments be carried out 24–48 h post‐mRNA injection. We saw the reversal of the morphology changes caused by heat shock treatment after a period of recovery; however, we were unable to reverse the changes induced by actinomycin D treatment.
4. DISCUSSION
4.1. Advantages and limitations of this method
There are many advantages of using X. laevis oocytes to study multi‐protein nucleolar phase separation, a major advantage being size. Stage VI Xenopus oocytes and their nuclei have diameters of ~1.3 mm and 400 μm, respectively (Brangwynne et al., 2011; Dumont, 1972; Weber & Brangwynne, 2015). Their nucleoli are similar in size to a whole HeLa cell, providing spatial resolution of nucleolar domains that would require electron microscopy for most other cell types. While the increased nucleolar size lends increased resolution to nucleolar compartments, fluorescence microscopy is still limited to light resolution. Samples do not need to be fixed prior to imaging, which is particularly beneficial when studying phase separation, as phase separation properties may be altered by fixation. Because imaging is done immediately following treatments, data collection is limited to the amount of time a fluorescence microscope is immediately available. Additionally, access to an ApoTome, confocal, or similar fluorescence microscope is helpful, as fluorescence generated from different focal planes may make domain separation unclear.
Another advantage to using late‐stage Xenopus oocytes in these studies is the accessibility of large numbers of oocytes. Portions of the ovary can be removed at different intervals in order to assess samples from the same frog multiple times (Pearl et al., 2012). Additionally, each oocyte has hundreds of nucleoli, increasing the sample size of nucleoli available for analysis (Brown & Dawid, 1968; Hausen & Riebesell, 1991). Nucleolar condensates from various stages of oogenesis can be analyzed in this system; however, nucleoli cannot be assessed at various stages of the cell cycle as oocytes are non‐dividing cells.
Examining nucleolar phase separation in OR2 isolated nuclei also provides a system of biologically formed, mixed component condensates, difficult or impossible to reproduce in in vitro analysis of phase separation. Proteins that have been assembled into biologically relevant compartments can be observed for alterations as one changes their environment. For example, we feature studies using OR2, which has about one half of the physiologic salt concentration and likely stabilizes electrostatic interactions. Nuclei incubated in OR2 also start to polymerize nuclear actin and loosen the molecular weight restriction for free diffusion through nuclear pores, from an estimated 58 to over 150 kDa. Within an hour, 90% of nuclear protein is lost to diffusion (Hayes et al., 2018; Hayes & Weeks, 2016; Paine et al., 1983). Proteins in non‐membrane‐bound particles, like nucleoli, coil bodies, pearls, and speckles, are retained (Hayes & Weeks, 2016). We note that many of the domain‐specific localization we show in Figure 1 can be disrupted by isolation in 300 mM salt. Studies analyzing electrostatic effects on phase‐separating proteins are ongoing. Small molecules such as ATP are also depleted when nuclei are isolated in OR2. In previous studies, we took advantage of this in an analysis of the effect of ATP concentration on nucleolar protein compartmentalization of Npm1 and Fbl (Hayes et al., 2018). A similar study could be done with other small molecules depleted by OR2 isolation. Nuclear content can be retained when nuclei are isolated from oocytes in oil or in buffers specifically formulated to retain nuclear content (Carroll & Lehman, 1991; Gall et al., 2004; Scalenghe et al., 1978).
4.2. Expertise required to implement the method and helpful Xenopus tools
To carry out these studies, animal training specific to X. laevis is required. Expertise in fluorescence microscopy and proficiency in standard molecular biology techniques (for synthesis of fluorescent fusion clones, synthesis of mRNA, and troubleshooting) are also needed to implement these methods.
Xenbase (https://www.xenbase.org, RRID:SCR_003280) is an invaluable tool for any Xenopus research. On Xenbase information is available about the expression, phenotype, isoforms, interactomes, antibodies for, and literature related to a specific protein of interest. Additionally, Xenbase has a wealth of protocols and information on reagents specifically for Xenopus research. We utilized numerous Xenopus‐specific protocols from Cold Spring Harbor (http://cshprotocols.cshlp.org/cgi/collection/xenopus). The books The Early Development of Xenopus Laevis: An Atlas of the Histology and Xenopus laevis: Practical uses in cell and molecular biology were also instrumental for reagents, protocols, and understanding the development of X. laevis (Hausen & Riebesell, 1991; Kay & Peng, 1991).
4.3. Importance of new findings and applications of the method
The nucleolus is the main hub for ribosome biogenesis, which is a vital process within the cell. Our understanding of the role protein aggregation plays in ribosome biogenesis has primarily been based on canonical domain markers, yet nucleolar condensates contain many proteins. Here we show the localization of numerous nucleolar proteins that are suspected of phase separation, including Gar1, Nhp2, Snu13, Nop56, Pes1, Ncl, Npm3, Gtpbp4, Rpl12, and Pak1ip1. From these studies we created a more nuanced model of the nucleolus.
The nucleolus is dynamic and must be able to respond to various types of stress. It is affected by various cell states, including cell division, aging, and various diseases. Additionally, outside of ribosome biogenesis, the nucleolus is a site where both useful and toxic aggregating proteins are regulated (Frottin et al., 2019). Xenopus laevis oocytes are a useful tool to gain a clearer picture of the nucleolus in various stress and disease states. Here we show the drastic difference in response elicited by two conditions known to decrease ribosome biogenesis, heat shock and actinomycin D treatment. Furthermore, we were able to examine these changes using various nucleolar proteins.
This system can also be used to assess how biologically relevant mutations in nucleolar proteins affect localization to or the structure of the nucleolus under various conditions. Many of the proteins examined here are members of multi‐protein complexes and we are currently examining the relationship between complex formation and phase separation. These studies are instructive for other multi‐compartment, non‐membrane‐bound organelles. Many of these techniques could be directly applied to other nuclear non‐membrane‐bound organelles.
AUTHOR CONTRIBUTIONS
EDL and DLW conceived and designed the project. EDL and INP acquired and analyzed the data. DLW did all microinjection. EDL and DLW wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supporting information
Figure S1
Table S1
ACKNOWLEDGMENTS
Fbl and Npm1 fluorescent fusion proteins were a gift from Cliff Brangwynne's lab (Princeton University). RN3P plasmid used as backbone for fluorescent fusion proteins was a gift from John Gurdon (Cambridge University). Ashley Goll and Francesca Spencer provided technical assistance. We thank our University of Iowa collaborators in the Metabolism of Neurodegenerative Disease (MIND) consortium and the Molecular Aggregation Network (MAggNET) consortium for helpful discussions. Funding: EL recognizes stipend support from a predoctoral Pharmacological Training Grant (NIH T32). The project has been supported by grant R01 GM124063 NIGMS and an Iowa Center for Aging Pilot Grant to DLW.
Lavering, E. D. , Petros, I. N. , & Weeks, D. L. (2022). Component analysis of nucleolar protein compartments using Xenopus laevis oocytes. Development, Growth & Differentiation, 64(6), 306–317. 10.1111/dgd.12794
Communicating Editor: Aaron Zorn
This article is part of the special issue “Versatile utilities of amphibians.”
REFERENCES
- Alberti, S. , Gladfelter, A. , & Mittag, T. (2019). Considerations and challenges in studying liquid‐liquid phase separation and biomolecular condensates. Cell, 176(3), 419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J. S. , Lam, Y. W. , Leung, A. K. , Ong, S. E. , Lyon, C. E. , Lamond, A. I. , & Mann, M. (2005). Nucleolar proteome dynamics. Nature, 433(7021), 77–83. 10.1038/nature03207 [DOI] [PubMed] [Google Scholar]
- Banani, S. F. , Rice, A. M. , Peeples, W. B. , Lin, Y. , Jain, S. , Parker, R. , & Rosen, M. K. (2016). Compositional control of phase‐separated cellular bodies. Cell, 166, 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banski, P. , Kodiha, M. , & Stochaj, U. (2010). Chaperones and multitasking proteins in the nucleolus: Networking together for survival? Trends in Biochemical Sciences, 35(7), 361–367. 10.1016/j.tibs.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Berry, J. , Brangwynne, C. P. , & Haataja, M. (2018). Physical principles of intracellular organization via active and passive phase transitions. Reports on Progress in Physics, 81(4), 046601. 10.1088/1361-6633/aaa61e [DOI] [PubMed] [Google Scholar]
- Bienz, M. , & Gurdon, J. B. (1982). The heat‐shock response in xenopus oocytes is controlled at the translational level. Cell, 29(3), 811–819. 10.1016/0092-8674(82)90443-3 [DOI] [PubMed] [Google Scholar]
- Boeynaems, S. , Holehouse, A. S. , Weinhardt, V. , Kovacs, D. , van Lindt, J. , Larabell, C. , van den Bosch, L. , Das, R. , Tompa, P. S. , Pappu, R. V. , & Gitler, A. D. (2019). Spontaneous driving forces give rise to protein‐RNA condensates with coexisting phases and complex material properties. Proceedings of the National Academy of Sciences of the United States of America, 116(16), 7889–7898. 10.1073/pnas.1821038116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert, F. M. , van Koningsbruggen, S. , Navascues, J. , & Lamond, A. I. (2007). The multifunctional nucleolus. Nature Reviews. Molecular Cell Biology, 8(7), 574–585. 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- Boke, E. , Ruer, M. , Wühr, M. , Coughlin, M. , Lemaitre, R. , Gygi, S. P. , Alberti, S. , Drechsel, D. , Hyman, A. A. , & Mitchison, T. J. (2016). Amyloid‐like self‐assembly of a cellular compartment. Cell, 166(3), 637–650. 10.1016/j.cell.2016.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche, G. , Raynal, F. , Amalric, F. , & Zalta, J. P. (1981). Unusual processing of nucleolar rna synthesized during a heat‐shock in cho cells. Molecular Biology Reports, 7(4), 253–258. 10.1007/bf00805762 [DOI] [PubMed] [Google Scholar]
- Bouvet, P. , Diaz, J. J. , Kindbeiter, K. , Madjar, J. J. , & Amalric, F. (1998). Nucleolin interacts with several ribosomal proteins through its RGG domain. Journal of Biological Chemistry, 273(30), 19025–19029. 10.1074/jbc.273.30.19025 [DOI] [PubMed] [Google Scholar]
- Brangwynne, C. P. , Mitchison, T. J. , & Hyman, A. A. (2011). Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proceedings of the National Academy of Sciences, 108, 4334–4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. D. , & Dawid, I. B. (1968). Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science, 160(3825), 272–280. 10.1126/science.160.3825.272 [DOI] [PubMed] [Google Scholar]
- Carroll, D. , & Lehman, C. W. (1991). DNA recombination and repair in oocytes, eggs, and extracts. Methods in Cell Biology, 36, 467–486. 10.1016/s0091-679x(08)60292-7 [DOI] [PubMed] [Google Scholar]
- Chen, D. , & Huang, S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. The Journal of Cell Biology, 153(1), 169–176. 10.1083/jcb.153.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini, L. M. , Fossati, M. , Francolini, M. , & Snapp, E. L. (2012). Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic, 13(5), 643–649. 10.1111/j.1600-0854.2012.01336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C. J. , & Parker, R. (2012). P‐bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspectives in Biology, 4(9), a012286. 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont, J. N. (1972). Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. Journal of Morphology, 136(2), 153–179. 10.1002/jmor.1051360203 [DOI] [PubMed] [Google Scholar]
- Fassler, J. S. , Skuodas, S. , Weeks, D. L. , & Phillips, B. T. (2021). Protein aggregation and disaggregation in cells and development. Journal of Molecular Biology, 433(21), 167215. 10.1016/j.jmb.2021.167215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett, D. (1981). The cell (2nd ed.). WB Saunders. ISBN:0721635849 [Google Scholar]
- Feric, M. , Vaidya, N. , Harmon, T. S. , Mitrea, D. M. , Zhu, L. , Richardson, T. M. , Kriwacki, R. W. , Pappu, R. V. , & Brangwynne, C. P. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell, 165(7), 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin, F. , Schueder, F. , Tiwary, S. , Gupta, R. , Körner, R. , Schlichthaerle, T. , Cox, J. , Jungmann, R. , Hartl, F. U. , & Hipp, M. S. (2019). The nucleolus functions as a phase‐separated protein quality control compartment. Science, 365(6451), 342‐+. 10.1126/science.aaw9157 [DOI] [PubMed] [Google Scholar]
- Fuentes, J. L. , Datta, K. , Sullivan, S. M. , Walker, A. , & Maddock, J. R. (2007). In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Molecular Genetics and Genomics, 278(1), 105–123. 10.1007/s00438-007-0233-1 [DOI] [PubMed] [Google Scholar]
- Gall, J. G. , Wu, Z. , Murphy, C. , & Gao, H. (2004). Structure in the amphibian germinal vesicle. Experimental Cell Research, 296, 28–34. 10.1016/j.yexcr.2004.03.017 [DOI] [PubMed] [Google Scholar]
- Ginisty, H. , Amalric, F. , & Bouvet, P. (1998). Nucleolin functions in the first step of ribosomal RNA processing. EMBO Journal, 17(5), 1476–1486. 10.1093/emboj/17.5.1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon, J. B. , Lane, C. D. , Woodland, H. R. , & Marbaix, G. (1971). Use of frog eggs and oocytes for the study of messenger RNA and its translation in living cells. Nature, 233(5316), 177–182. 10.1038/233177a0 [DOI] [PubMed] [Google Scholar]
- Hamma, T. , & Ferre‐D'Amare, A. R. (2010). The box H/ACA ribonucleoprotein complex: Interplay of RNA and protein structures in post‐transcriptional RNA modification. Journal of Biological Chemistry, 285(2), 805–809. 10.1074/jbc.R109.076893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, T. W. , Kato, M. , Xie, S. , Wu, L. C. , Mirzaei, H. , Pei, J. , Chen, M. , Xie, Y. , Allen, J. , Xiao, G. , & McKnight, S. (2012). Cell‐free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell, 149(4), 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hausen, P. , & Riebesell, M. (1991). The early development of Xenopus laevis: An atlas of the histology. Springer‐Verlag. ISBN:0‐387‐53740‐6 [Google Scholar]
- Hayes, M. H. , Peuchen, E. H. , Dovichi, N. J. , & Weeks, D. L. (2018). Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. eLife, 7, e35224. 10.7554/eLife.35224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, M. H. , & Weeks, D. L. (2016). Amyloids assemble as part of recognizable structures during oogenesis in Xenopus. Biol Open, 5(6), 801–806. 10.1242/bio.017384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N. , Negi, S. , Szebeni, A. , & Olson, M. O. J. (2005). Protein NPM3 interacts with the multifunctional nucleolar protein B23/nucleophosmin and inhibits ribosome biogenesis. Journal of Biological Chemistry, 280(7), 5496–5502. 10.1074/jbc.M407856200 [DOI] [PubMed] [Google Scholar]
- Hyman, A. A. , Weber, C. A. , & Ulicher, F. (2014). Liquid‐liquid phase separation in biology. Annual Review of Cell and Developmental Biology, 30, 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Imami, K. , Milek, M. , Bogdanow, B. , Yasuda, T. , Kastelic, N. , Zauber, H. , Ishihama, Y. , Landthaler, M. , & Selbach, M. (2018). Phosphorylation of the ribosomal protein RPL12/uL11 affects translation during mitosis. Molecular Cell, 72(1), 84. 10.1016/j.molcel.2018.08.019 [DOI] [PubMed] [Google Scholar]
- Iyer‐Bierhoff, A. , Krogh, N. , Tessarz, P. , Ruppert, T. , Nielsen, H. , & Grummt, I. (2018). SIRT7‐dependent Deacetylation of Fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Reports, 25(11), 2946–2954.e5. 10.1016/j.celrep.2018.11.051 [DOI] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. , Tunyasuvunakool, K. , Bates, R. , Žídek, A. , Potapenko, A. , Bridgland, A. , Meyer, C. , Kohl, S. A. A. , Ballard, A. J. , Cowie, A. , Romera‐Paredes, B. , Nikolov, S. , Jain, R. , Adler, J. , … Hassabis, D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583‐589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, B. , Liu, B. , Zhou, Z. , & Lam, Y. W. (2011). Quantitative nucleolar proteomics reveals nuclear re‐organization during stress‐ induced senescence in mouse fibroblast. BMC Cell Biology, 12, 33. 10.1186/1471-2121-12-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Han, T. W. , Xie, S. , Shi, K. , du, X. , Wu, L. C. , Mirzaei, H. , Goldsmith, E. J. , Longgood, J. , Pei, J. , Grishin, N. V. , Frantz, D. E. , Schneider, J. W. , Chen, S. , Li, L. , Sawaya, M. R. , Eisenberg, D. , Tycko, R. , & McKnight, S. (2012). Cell‐free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell, 149(4), 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , & McKnight, S. L. (2017). Cross‐beta polymerization of low complexity sequence domains. Cold Spring Harbor Perspectives in Biology, 9(3), a023598. 10.1101/cshperspect.a023598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, B. K. , & Peng, H. B. (1991). Xenopus laevis: Practical uses in cell and molecular biology. Methods in Cell Biology, 36, 663–669. ISBN: 0‐12‐564136‐2 [PubMed] [Google Scholar]
- Labhart, P. , & Reeder, R. H. (1987). Heat shock stabilizes highly unstable transcripts of the Xenopus ribosomal gene spacer. Proceedings of the National Academy of Sciences of the United States of America, 84(1), 56–60. 10.1073/pnas.84.1.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, Y. W. , & Trinkle‐Mulcahy, L. (2015). New insights into nucleolar structure and function. F1000Prime Rep, 7, 48. 10.12703/P7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, T. J. (2019). FPbase: A community‐editable fluorescent protein database. Nature Methods, 16(4), 277–278. 10.1038/s41592-019-0352-8 [DOI] [PubMed] [Google Scholar]
- Lin, J. R. , & Hu, J. J. (2013). SeqNLS: Nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One, 8(10), e76864. 10.1371/journal.pone.0076864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom, M. S. (2011). NPM1/B23: A Multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochemistry Research International, 2011, 1–16. 10.1155/2011/195209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais, C. , & Scheer, U. (2001). Molecular architecture of the amplified nucleoli of Xenopus oocytes. Journal of Cell Science, 114(4), 709–718. 10.1242/jcs.114.4.709 [DOI] [PubMed] [Google Scholar]
- Maji, S. K. , Perrin, M. H. , Sawaya, M. R. , Jessberger, S. , Vadodaria, K. , Rissman, R. A. , Singru, P. S. , Nilsson, K. P. , Simon, R. , Schubert, D. , Eisenberg, D. , Rivier, J. , Sawchenko, P. , Vale, W. , & Riek, R. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science, 325(5938), 328–332. 10.1126/science.1173155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen, D. T. , Mir, M. , Darzacq, X. , & Tjian, R. (2019). Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes & Development, 33(23–24), 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine, P. L. , Austerberry, C. F. , Desjarlais, L. J. , & Horowitz, S. B. (1983). Protein loss during nuclear isolation. The Journal of Cell Biology, 97(4), 1240–1242. 10.1083/jcb.97.4.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, E. J. , Grainger, R. M. , Guille, M. , & Horb, M. E. (2012). Development of xenopus resource centers: The national xenopus resource and the european xenopus resource center. Genesis, 50(3), 155–163. 10.1002/dvg.22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin, L. , Lukyanov, A. , Kalocsay, M. , Gage, R. M. , Wang, D. , Pells, T. J. , Karimi, K. , Vize, P. D. , Wühr, M. , & Kirschner, M. W. (2019). The protein repertoire in early vertebrate embryogenesis. bioRxiv, 571174. 10.1101/571174 [DOI] [Google Scholar]
- Reynolds, R. C. , Hughes, B. , & Montgomery, P. O. (1964). Nucleolar caps produced by actinomycin D. Cancer Research, 24(7), 1269‐1277. PMID: 14216161. [PubMed] [Google Scholar]
- Roger, B. , Moisand, A. , Amalric, F. , & Bouvet, P. (2002). rDNA transcription during Xenopus laevis oogenesis. Biochemical and Biophysical Research Communications, 290(4), 1151–1160. 10.1006/bbrc.2001.6304 [DOI] [PubMed] [Google Scholar]
- Rohrmoser, M. , Hölzel, M. , Grimm, T. , Malamoussi, A. , Harasim, T. , Orban, M. , Pfisterer, I. , Gruber‐Eber, A. , Kremmer, E. , & Eick, D. (2007). Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Molecular and Cellular Biology, 27(10), 3682–3694. 10.1128/mcb.00172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalenghe, F. , Buscaglia, M. , Steinheil, C. , & Crippa, M. (1978). Large scale isolation of nuclei and nucleoli from Vitellogenic oocytes of Xenopus laevis. Chromosoma, 66, 299–308. https://doi.org.10.1007/BF00328531 [DOI] [PubMed] [Google Scholar]
- Scheer, U. , Trendelenburg, M. F. , & Franke, W. W. (1975). Effects of actinomycin‐d on association of newly formed ribonucleoproteins with cistrons of ribosomal‐rna in triturus oocytes. Journal of Cell Biology, 65(1), 163–179. 10.1083/jcb.65.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, M. S. , Boisvert, F. M. , McDowall, M. D. , Lamond, A. I. , & Barton, G. J. (2010). Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Research, 38(21), 7388–7399. 10.1093/nar/gkq653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, K. E. , Bohnsack, M. T. , & Watkins, N. J. (2013). The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Reports, 5(1), 237–247. 10.1016/j.celrep.2013.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L. D. , Xu, W. L. , & Varnold, R. L. (1991). Oogenesis and oocyte isolation. Methods in Cell Biology, 36, 45–60. 10.1016/s0091-679x(08)60272-1 [DOI] [PubMed] [Google Scholar]
- Tchelidze, P. , Benassarou, A. , Kaplan, H. , O'Donohue, M. F. , Lucas, L. , Terryn, C. , Rusishvili, L. , Mosidze, G. , Lalun, N. , & Ploton, D. (2017). Nucleolar sub‐compartments in motion during rRNA synthesis inhibition: Contraction of nucleolar condensed chromatin and gathering of fibrillar centers are concomitant. PLoS One, 12(11), e0187977. 10.1371/journal.pone.0187977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues, R. W. , Granneman, S. , Kudla, G. , Sloan, K. E. , Chicken, M. , Tollervey, D. , & Watkins, N. J. (2011). Box C/D snoRNP catalysed methylation is aided by additional pre‐rRNA base‐pairing. EMBO Journal, 30(12), 2420–2430. 10.1038/emboj.2011.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar, P. S. , & Culling, C. F. A. (1959). Fluorescent stains, with special reference to amyloid and connective tissues. Archives of Pathology, 68(5), 487–498. PMID:13841452 [PubMed] [Google Scholar]
- Wang, M. , Bokros, M. , Theodoridis, P. R. , & Lee, S. (2019). Nucleolar sequestration: Remodeling nucleoli into amyloid bodies. Frontiers in Genetics, 10, 1179. 10.3389/fgene.2019.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N. J. , & Bohnsack, M. T. (2012). The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdisciplinary Reviews‐Rna, 3(3), 397–414. 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- Weber, S. C. , & Brangwynne, C. P. (2015). Inverse size scaling of the nucleolus by a concentration‐dependent phase transition. Current Biology, 25(5), 641–646. 10.1016/j.cub.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr, M. , Freeman, R. M. , Presler, M. , Horb, M. E. , Peshkin, L. , Gygi, S. P. , & Kirschner, M. W. (2014). Deep proteomics of the Xenopus laevis egg using an mRNA‐derived reference database. Current Biology, 24, 1467–1475. 10.1016/j.cub.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S. J. , Li, Q. , Xiang, J. F. , Yang, Q. F. , Sun, H. X. , Guan, A. J. , Wang, L. , Liu, Y. , Yu, L. , Shi, Y. , Chen, H. , & Tang, Y. (2016). Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G‐quadruplexes. Scientific Reports, 6, 24793. 10.1038/srep24793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, B. , Dunbrack, R. L. , Williams, R. W. , Dunker, A. K. , & Uversky, V. N. (2010). PONDR‐FIT: A meta‐predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta, 1804(4), 996–1010. 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka‐Goetz, M. , Pines, J. , Ryan, K. , Siemering, K. R. , Haseloff, J. , Evans, M. J. , & Gurdon, J. B. (1996). An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development, 122(12), 3719–3724. 10.1242/dev.122.12.3719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1


